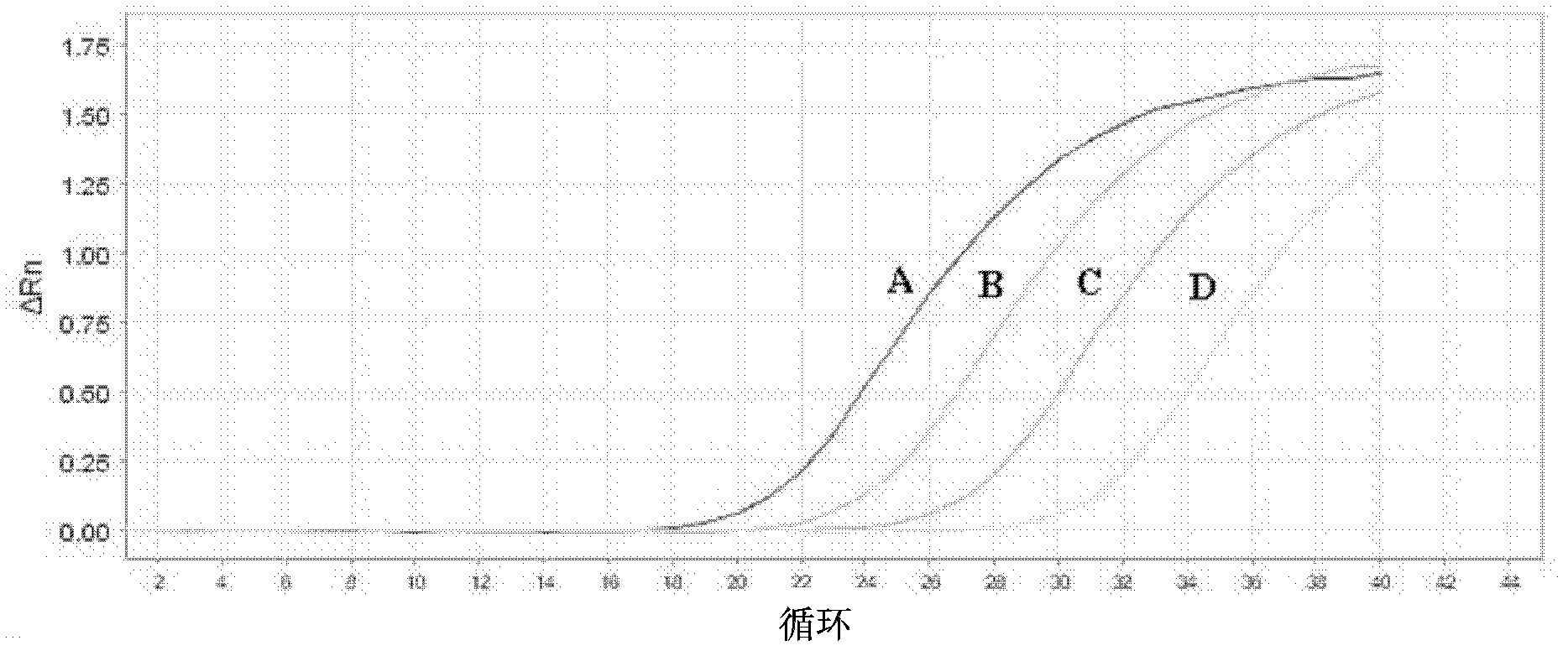Method of detecting gene mutation based on Blocker primers and ARMS primers, and kit
A kit and mutant technology, applied in the field of molecular biology, can solve the problems of inability to effectively distinguish mutant templates from wild-type templates, reduce wild-type blocking efficiency, and mutant detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
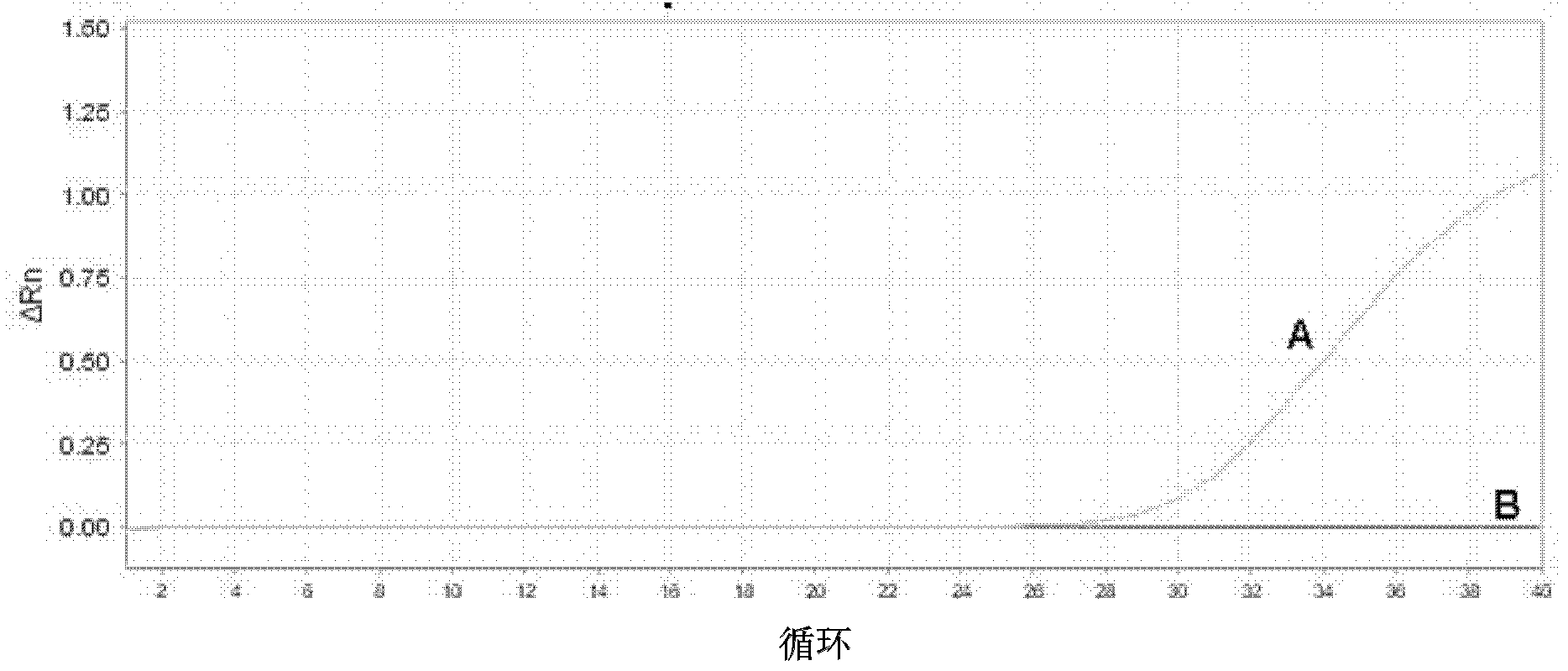
[0072] Example 1 One-step detection method of ARMS fluorescence quantitative PCR for mutation blocking enrichment of EGFR gene point mutation T790M
[0073] 1. Primer and Probe Design
[0074] 1) Blocker primer design
[0075] Primers were designed near the mutation site (including the mutation site), and the 3' end was modified with dideoxy bases to prevent its extension and increase its annealing temperature. The relevant parameters are: Tm value 63.0°C-80.0°C, GC value 40.0%-65.0%, primer size 25±10bp.
[0076] The designed Blocker primer sequences are as follows:
[0077] Blocker primer: 5'-TGCAGCTCATCACGCAGCTCATG-3'ddC (SEQ ID NO: 1)
[0078] 2) ARMS primer design
[0079] Design primers that can amplify a 60-150bp fragment, the relevant parameters are: Tm value 55.0°C-60.0°C, GC value 40.0%-60.0%, primer size 20±3bp. The 3' end of the upstream ARMS primer is located at the mutation site and is consistent with the mutant gene. In order to improve the specificity, a m...
Embodiment 2
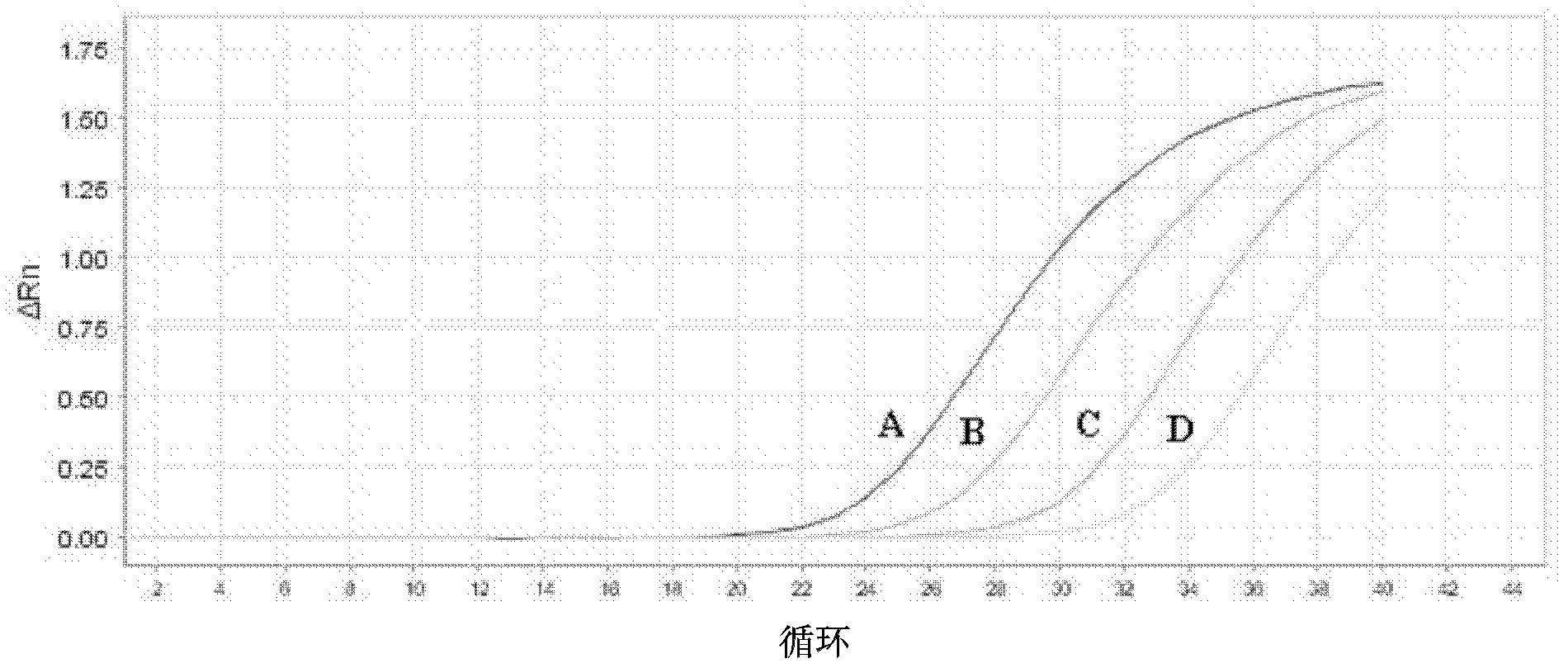
[0098] Example 2 EGFR gene exon 19 deletion E746_A750 deletion mutation mutation blocking enrichment ARMS fluorescent quantitative PCR one-step detection method
[0099] 1. Primer and Probe Design
[0100] 1) Blocker primer design
[0101] Primers were designed near the deletion mutation region (including the deletion sequence), and the 3' end was modified with dideoxy bases to prevent its extension and increase its annealing temperature. The relevant parameters are: Tm value 65.0°C-80.0°C, GC value 40.0%-65.0%, primer size 25±10bp.
[0102] The designed Blocker primer sequences are as follows:
[0103] Blocker primer: 5'-CAAGGAATTAAGAGAAGCAACATC-3'ddC (SEQ ID NO: 9)
[0104] 2) ARMS primer design
[0105] Design primers that can amplify a 60-150bp fragment, the relevant parameters are: Tm value 55.0°C-60.0°C, GC value 40.0%-60.0%, primer size 20±3bp. The 3' end of the upstream ARMS primer is located at the mutation site and is consistent with the mutant gene. In order to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com