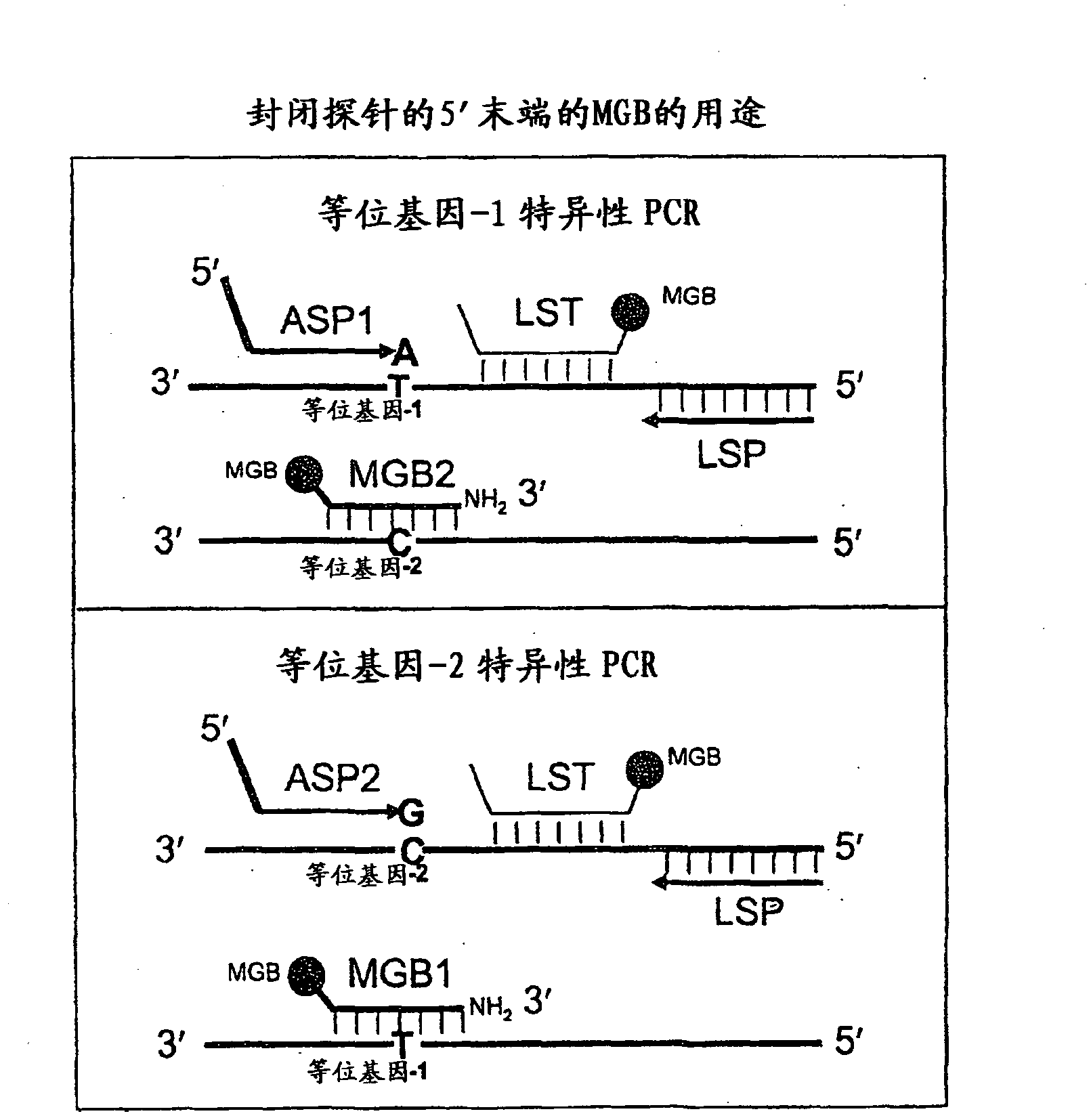
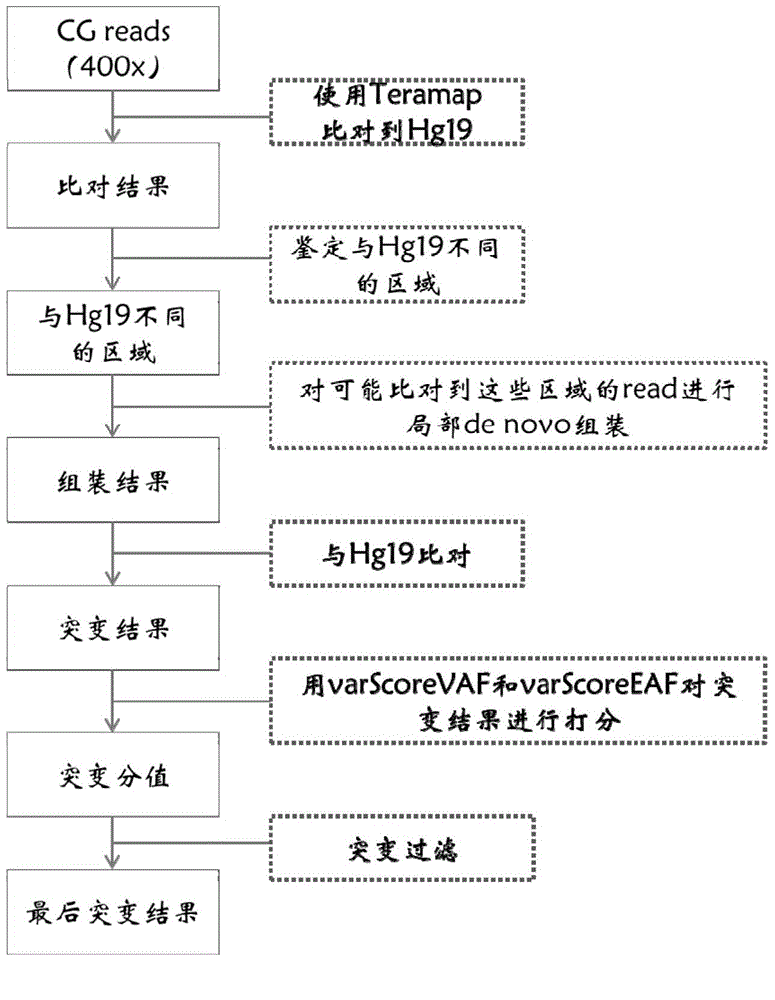
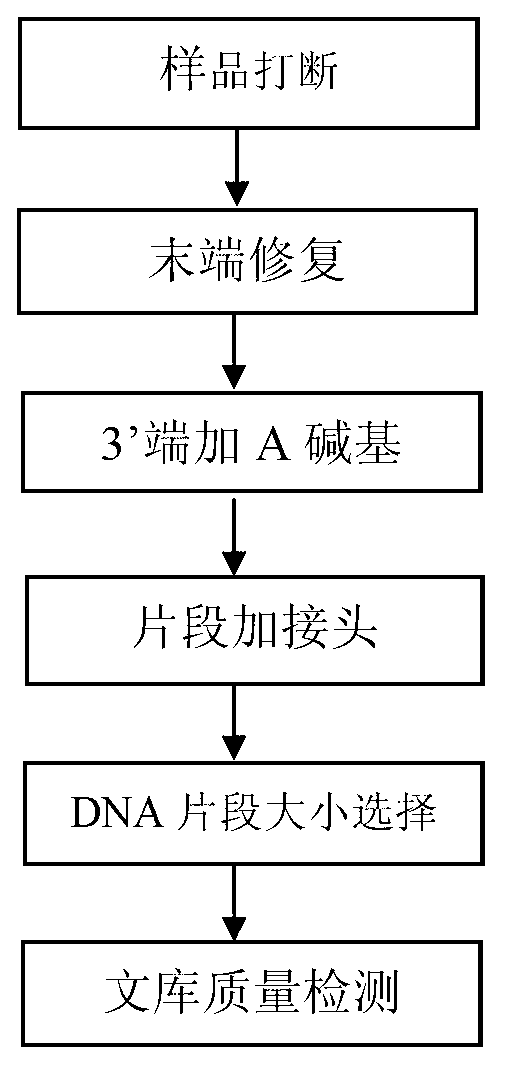
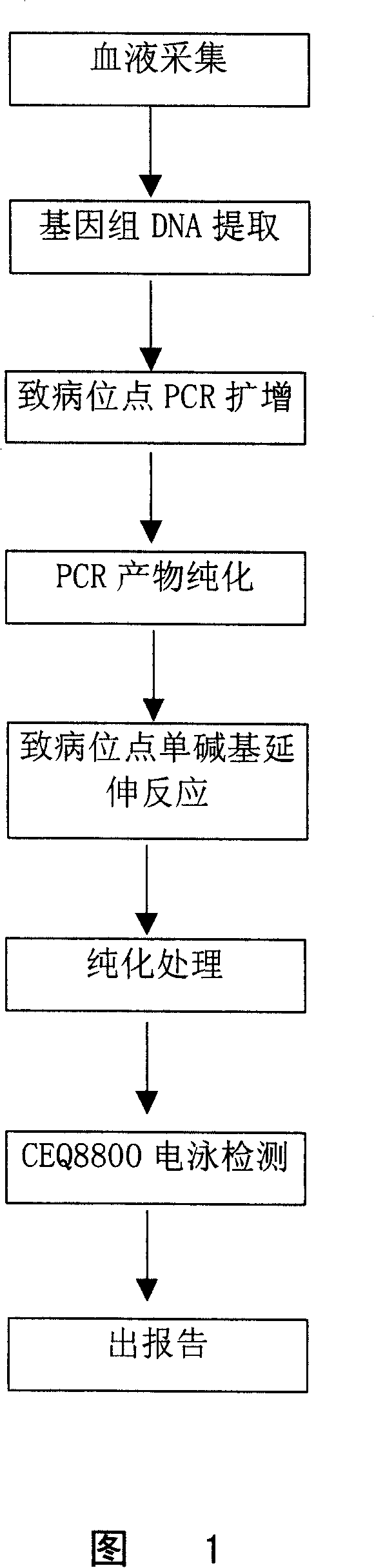
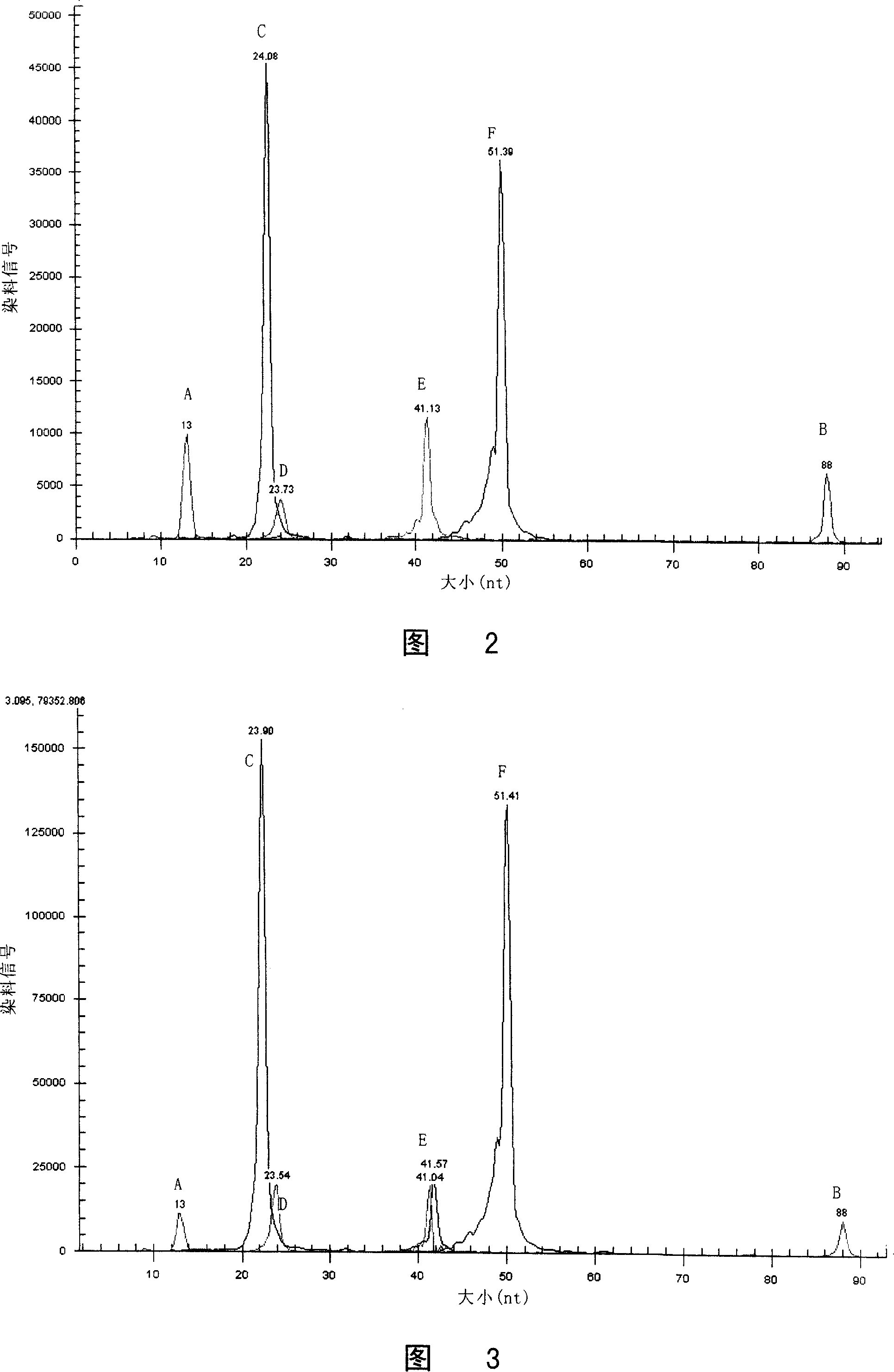
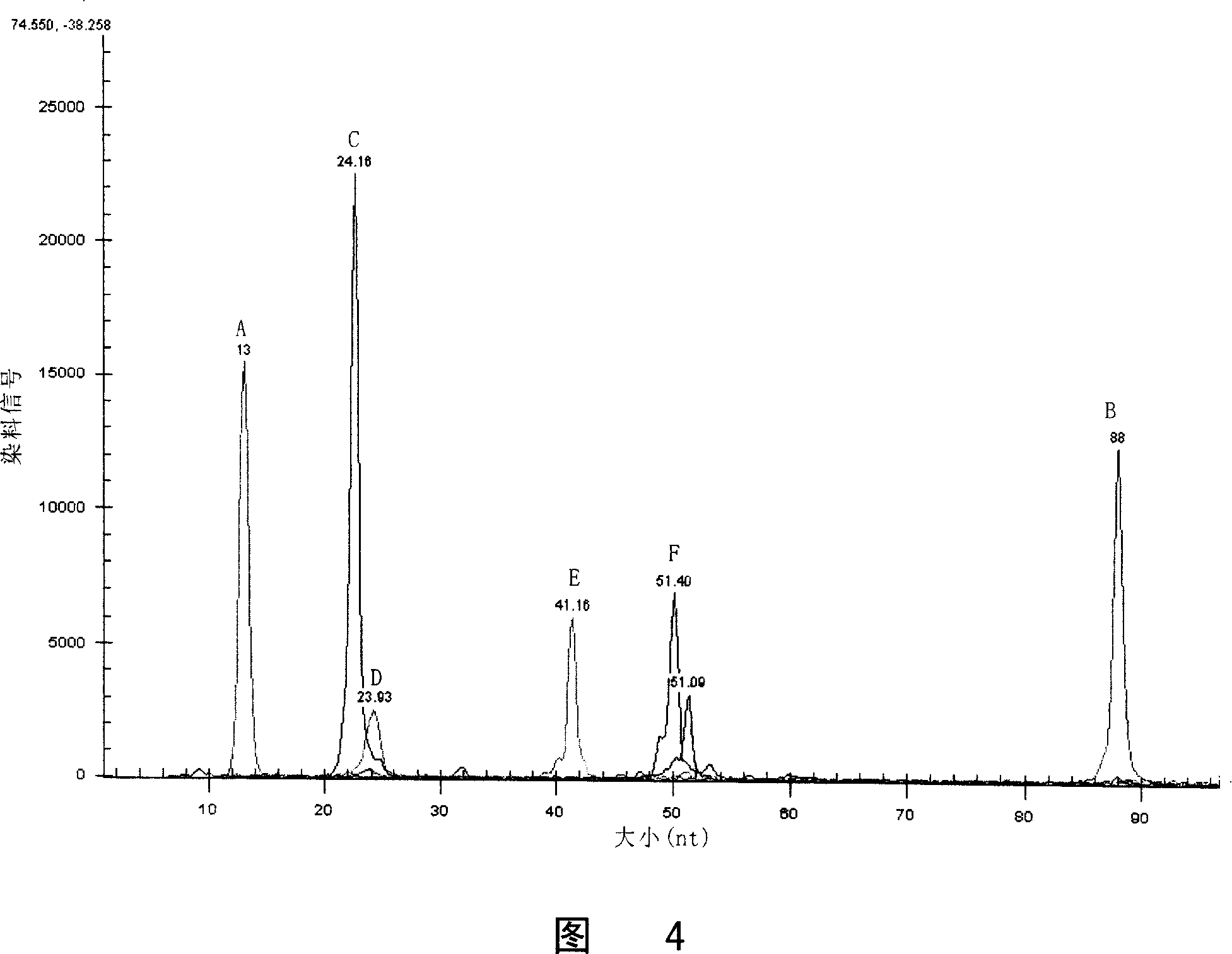
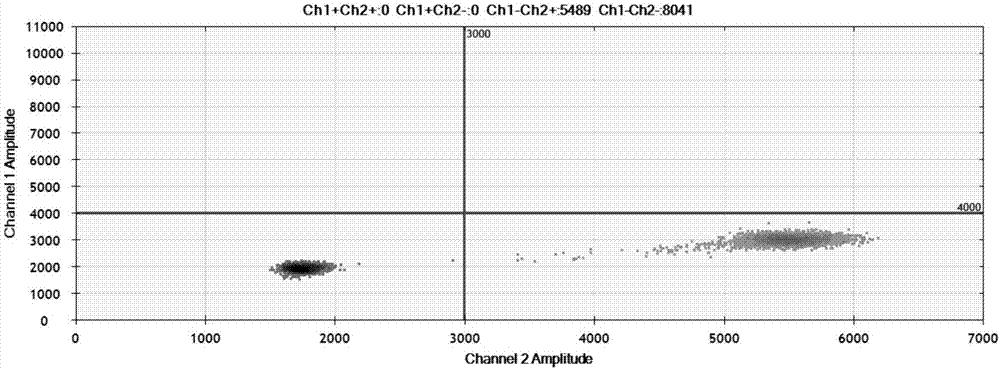
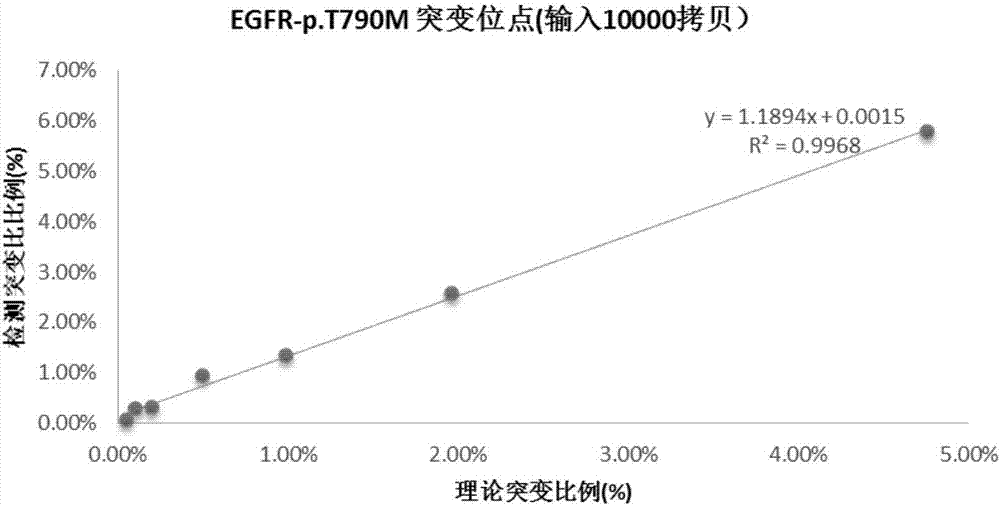
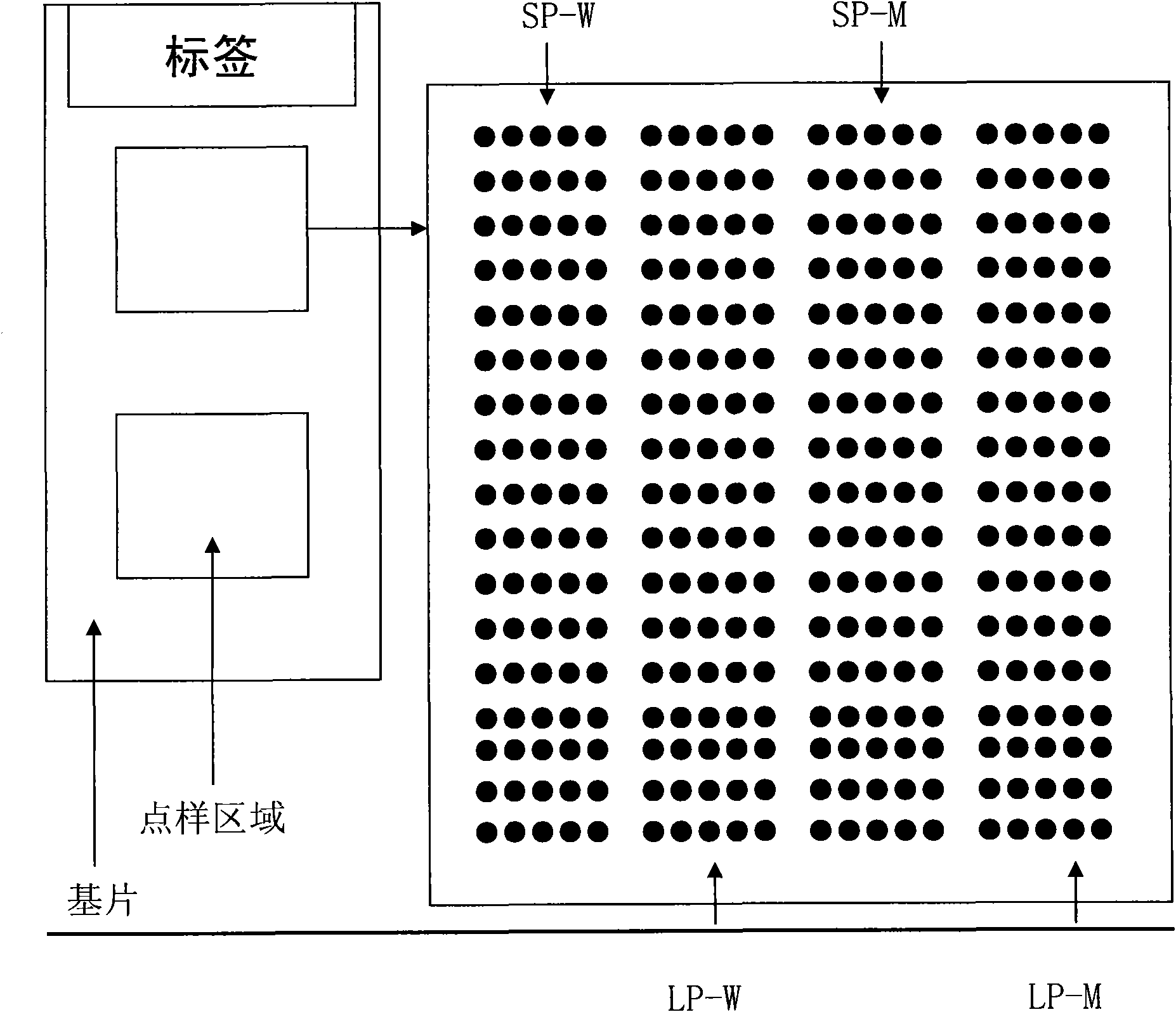
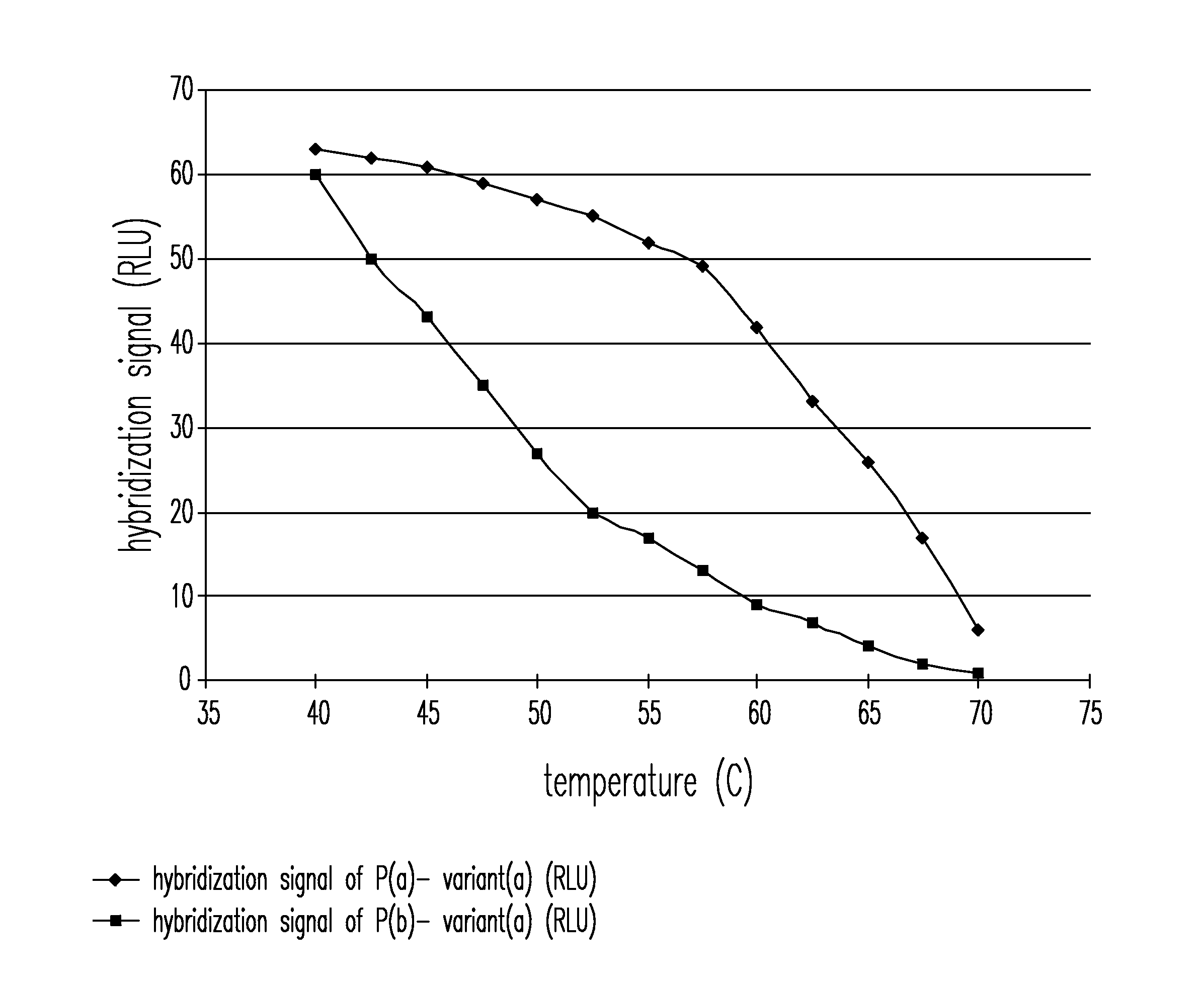
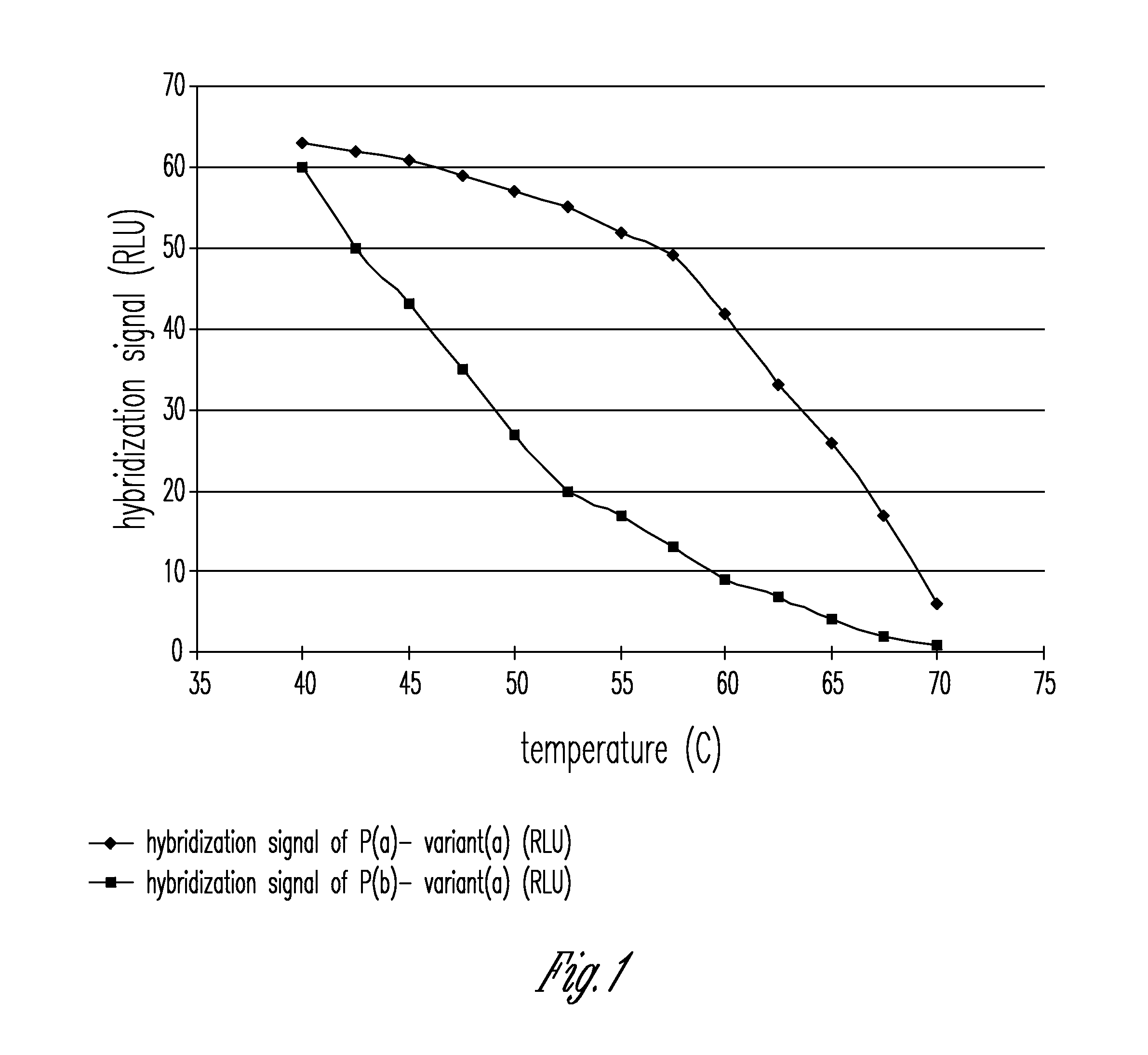
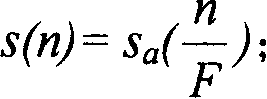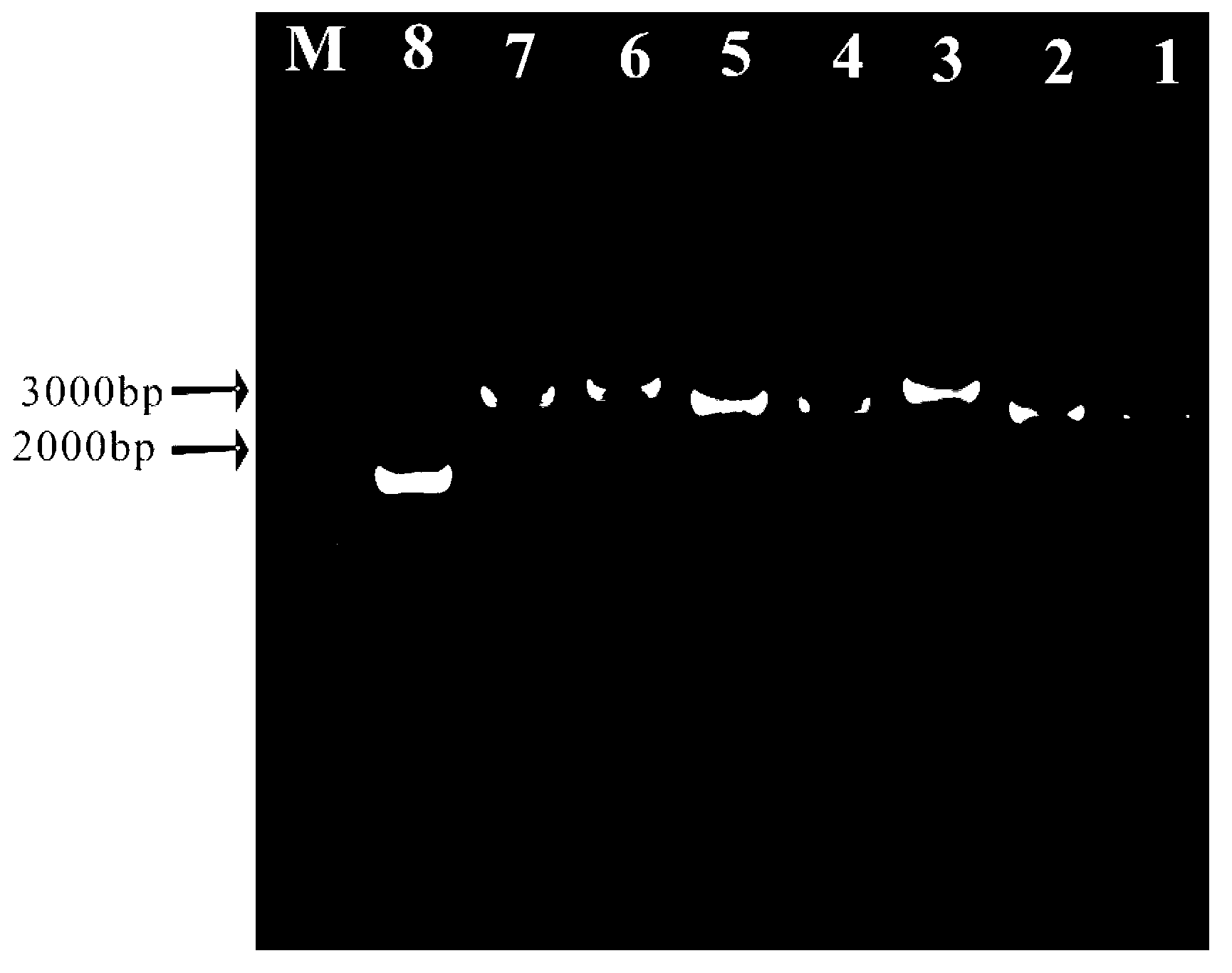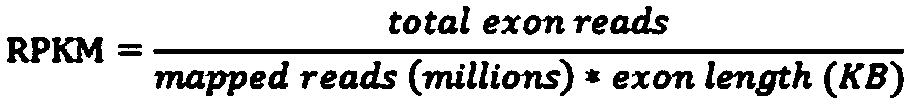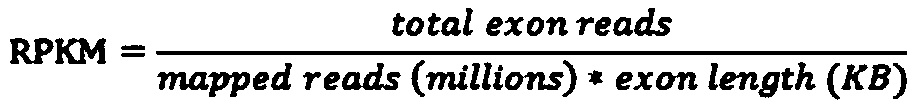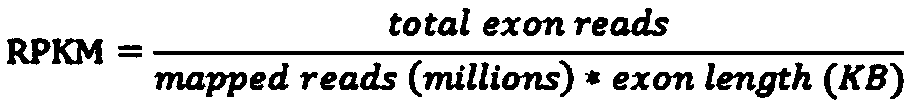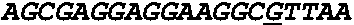Patents
Literature
920 results about "Mutation detection" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
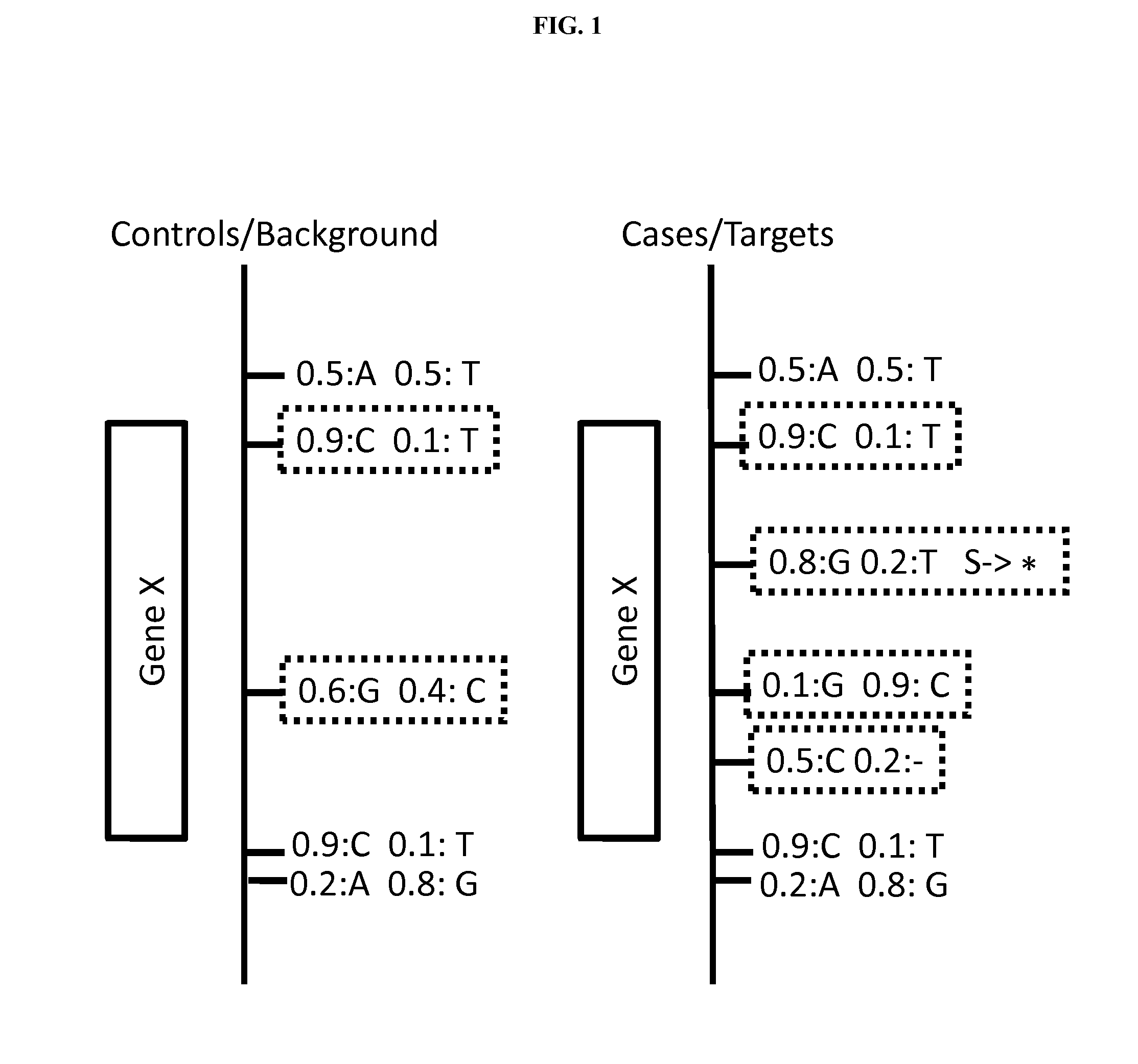
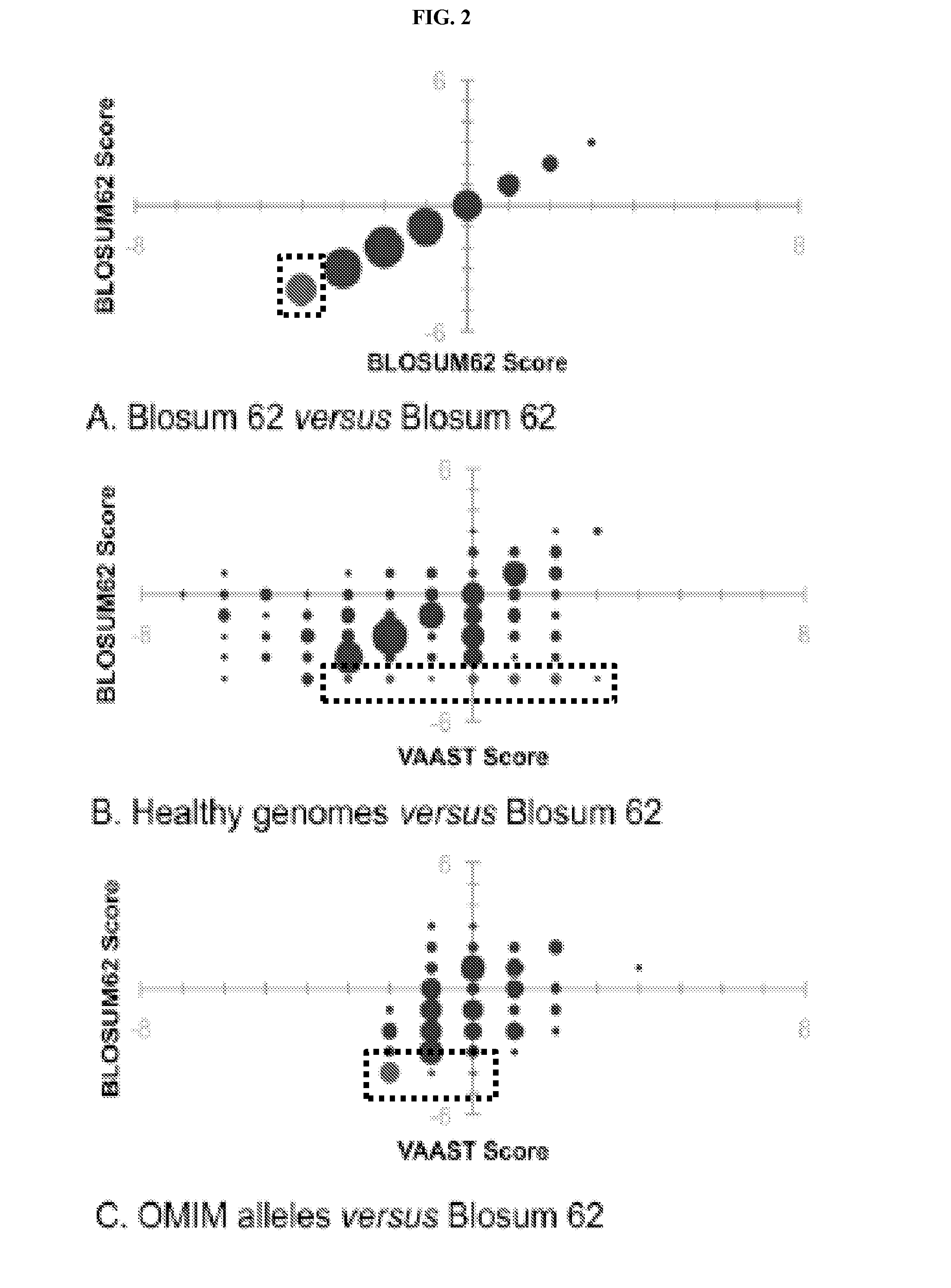
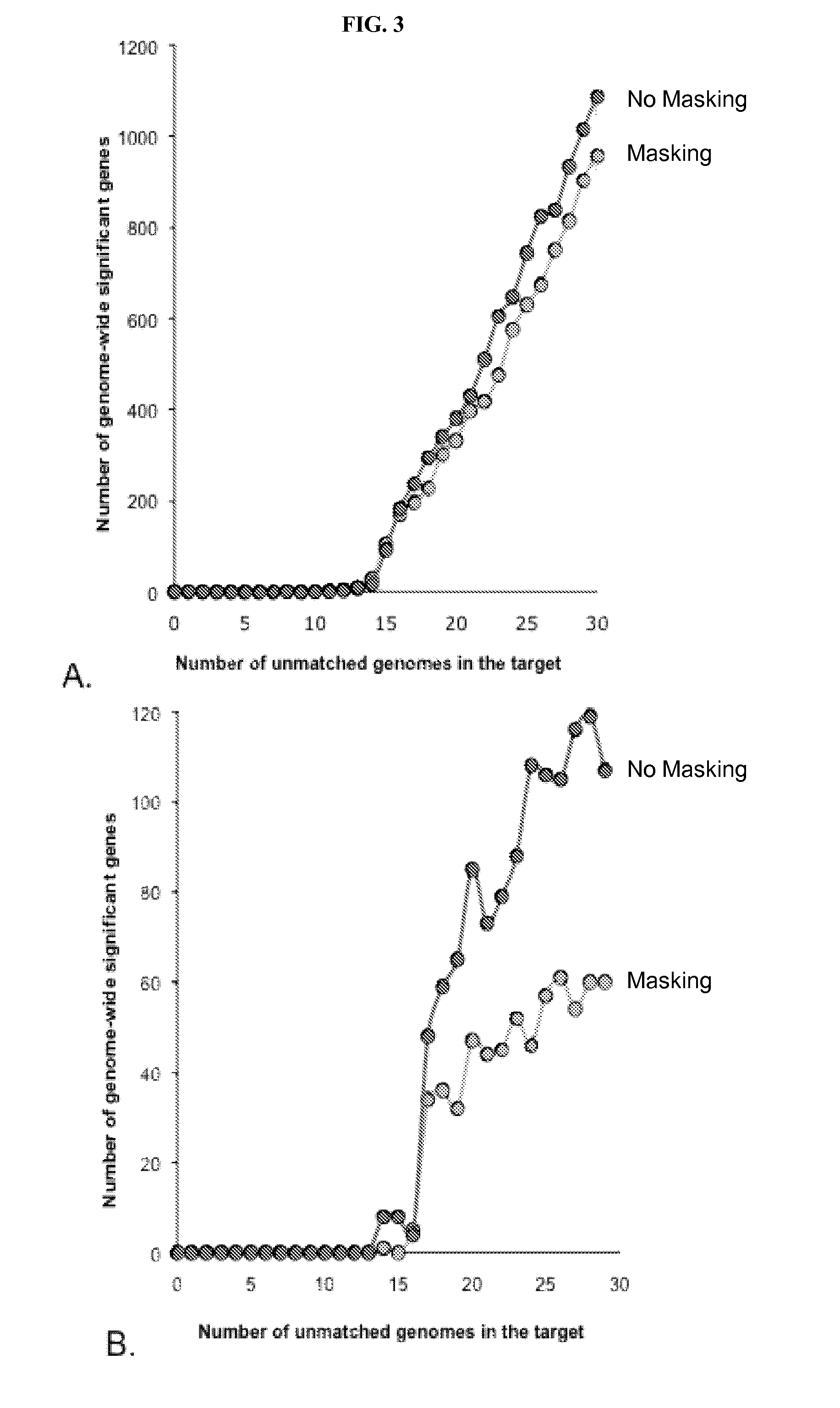
Variant annotation, analysis and selection tool
Disclosed are methods for detecting and / or prioritizing phenotype-causing genomic variants and related software tools. The methods include genomic feature based analysis and can combine variant frequency information with sequence characteristics such as amino acid substation. The methods disclosed are useful in any genomics study; for example, rare and common disease gene discovery, tumor growth mutation detection, personalized medicine, agricultural analysis, and centennial analysis.
Owner:UNIV OF UTAH RES FOUND +1
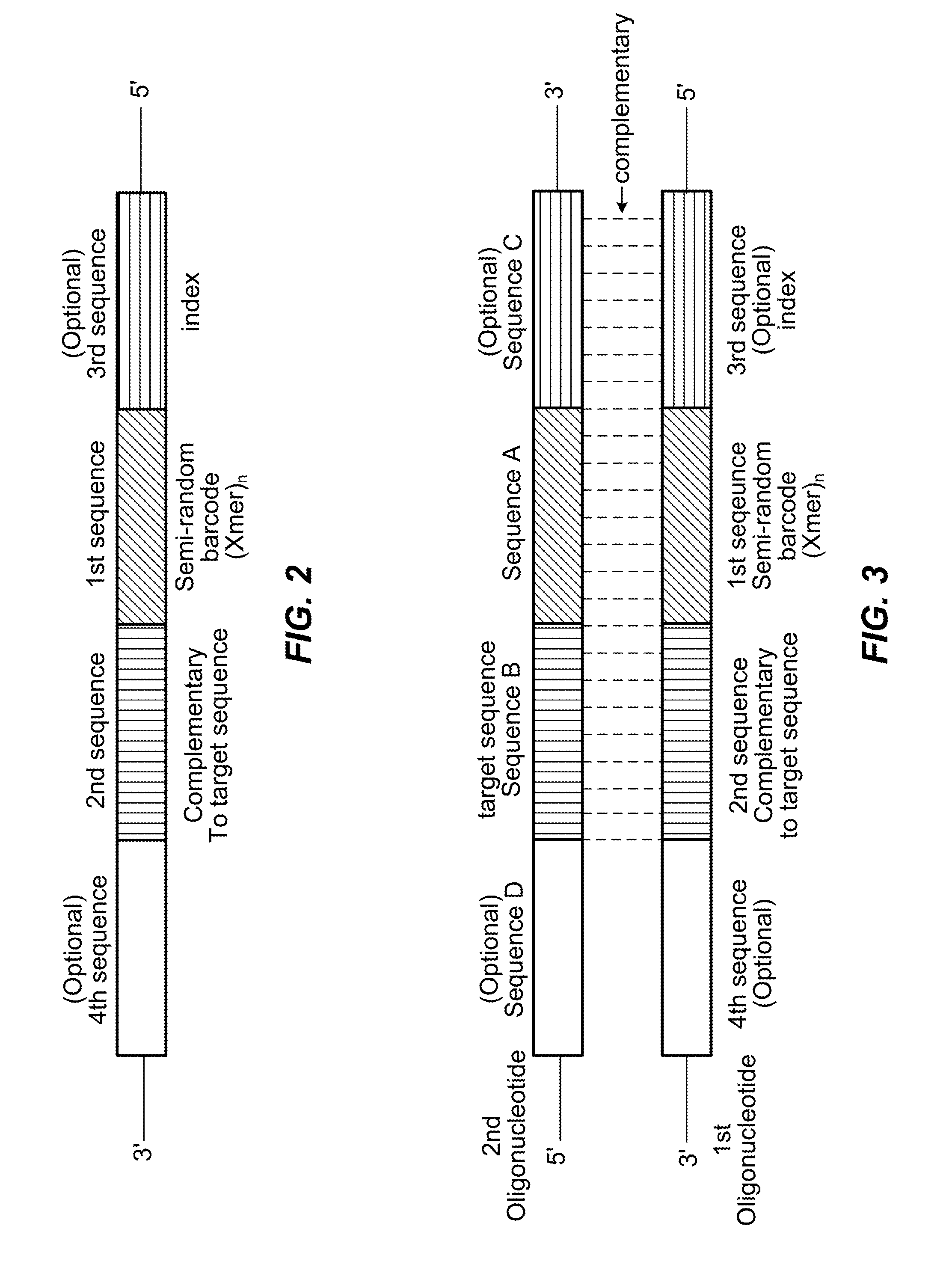
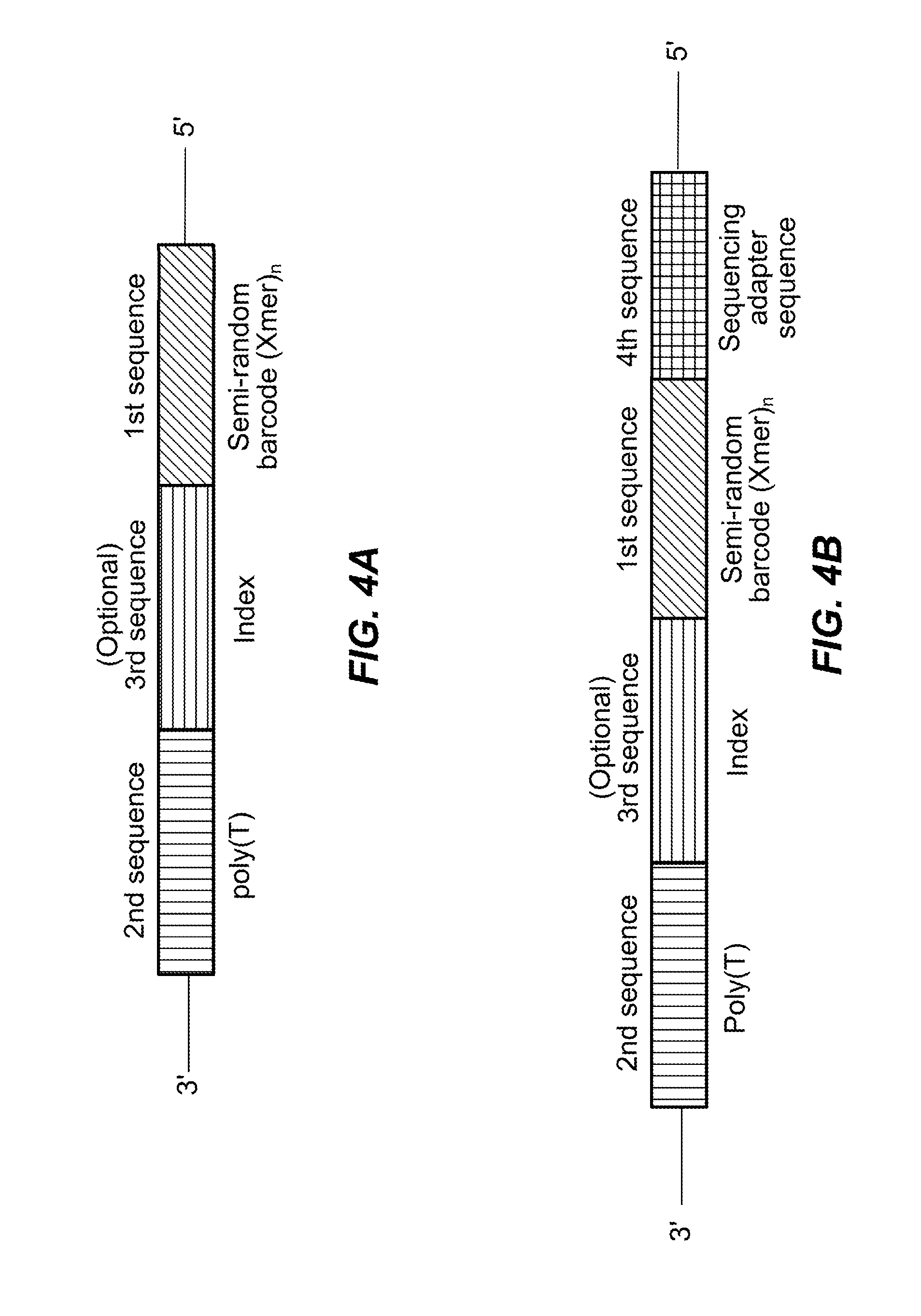
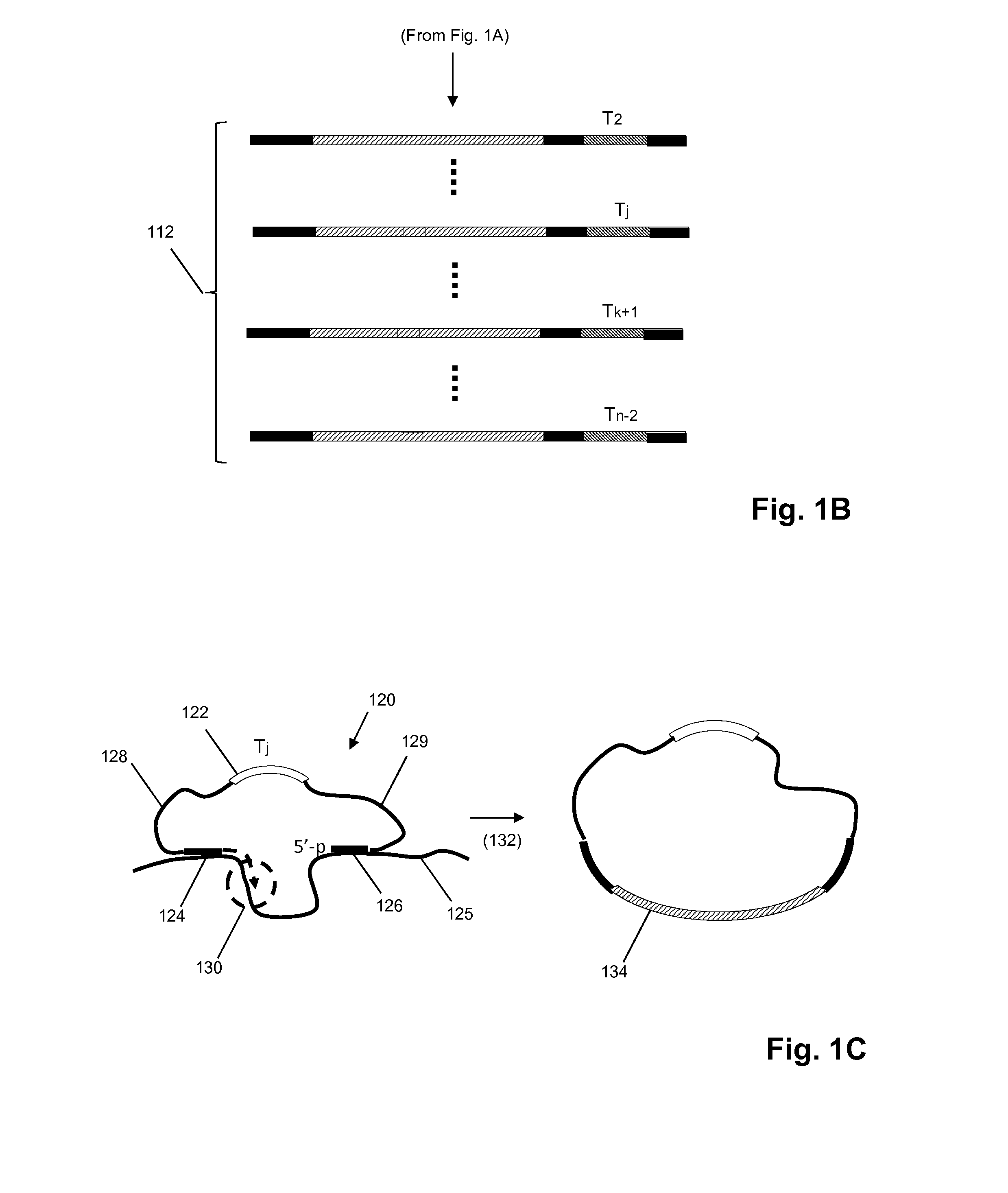
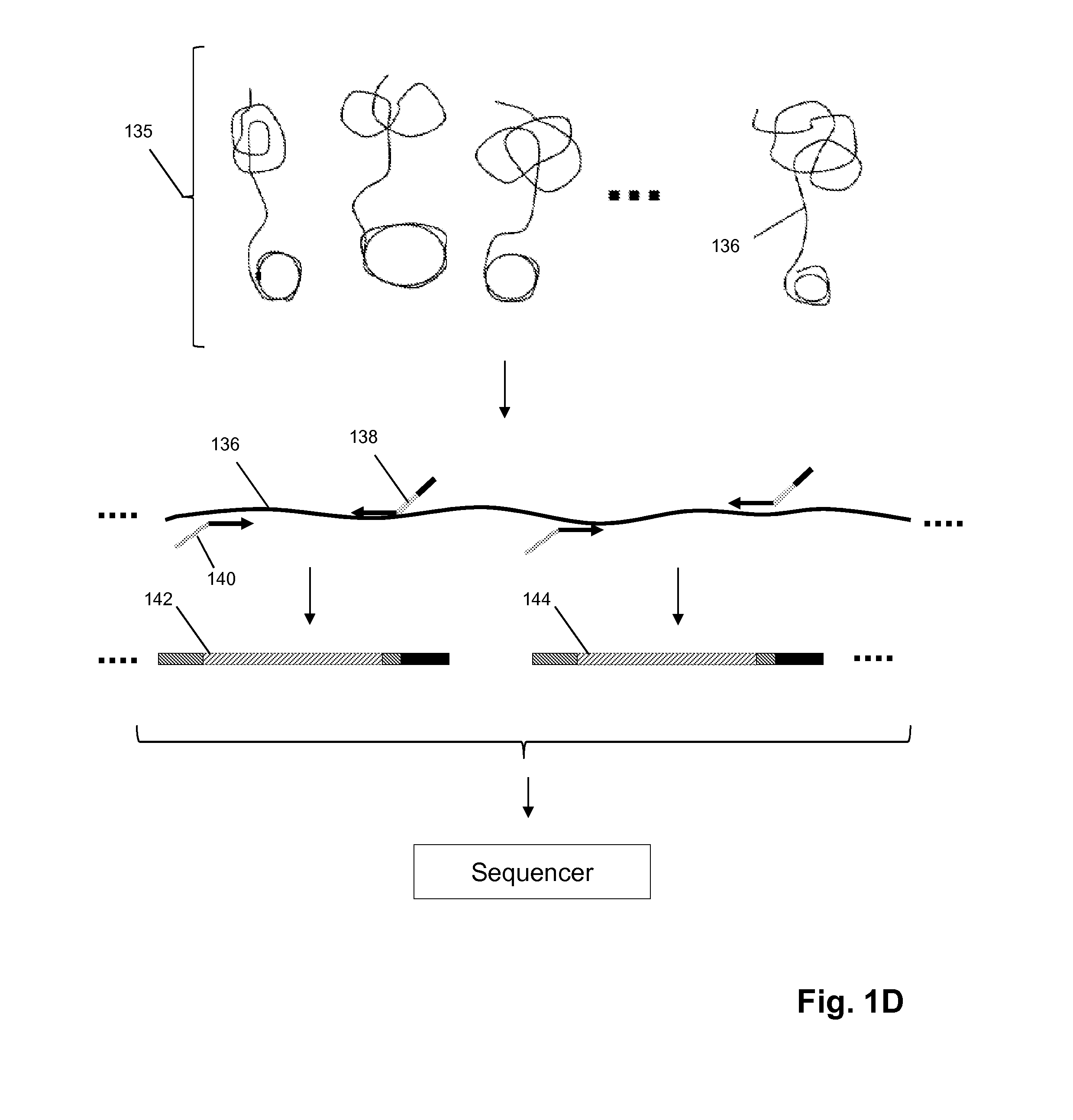
Semi-random barcodes for nucleic acid analysis
PendingUS20160017320A1Nucleotide librariesMicrobiological testing/measurementMutation detectionBarcode
The present disclosure provides oligonucleotides that comprise semi-random barcode sequences. Such oligonucleotides may be incorporated into reverse transcription primers, PCR primers, or portions of sequencing adapters in preparing sequencing libraries. The resulting sequencing libraries can be used for accurate sequencing, including DNA or RNA counting and mutation detection. Methods and kits for preparing sequencing adapters and sequencing libraries are also provided.
Owner:QIAGEN SCIENCES LLC
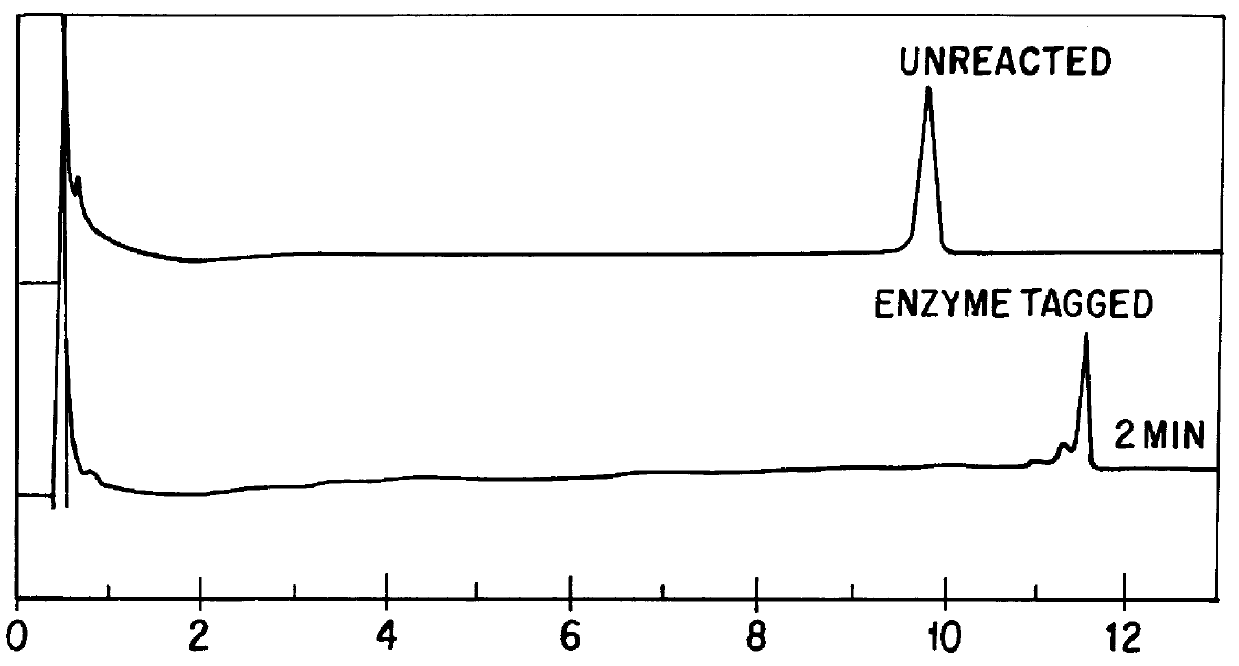
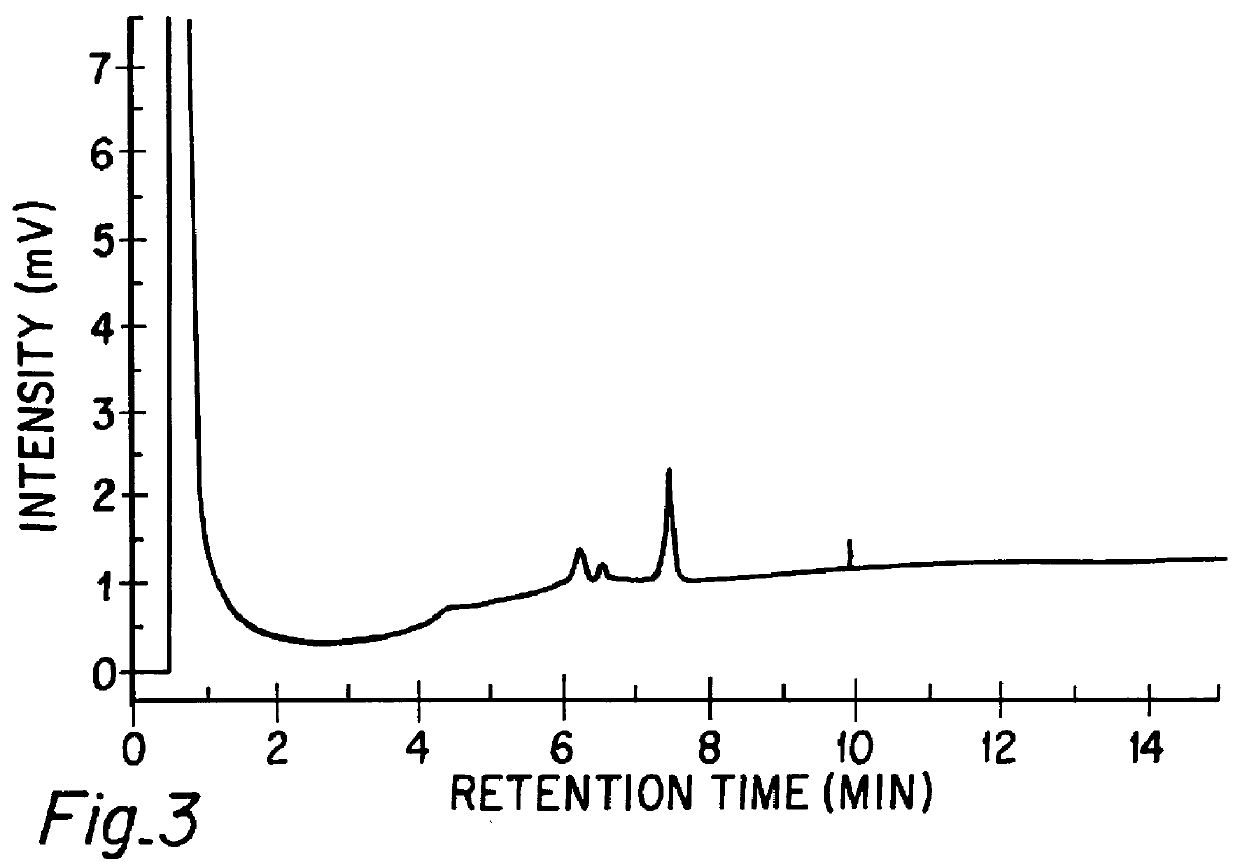
Chromatographic method for mutation detection using mutation site specifically acting enzymes and chemicals
InactiveUS6027898AHighly reproducibleEasy to implementSugar derivativesMicrobiological testing/measurementChromatographic separationRetention time
A method for analyzing a sample of double stranded DNA to determine the presence of a mutation therein comprises contacting the sample with a mutation site binding reagent, and chromatographically separating and detecting the product. The chromatographic separation can be performed using Matched Ion Polynucleotide Chromatography, size exclusion chromatography, ion exchange chromatography, or reverse phase chromatography. The mutation site binding reagent can be an enzyme or a non-proteinaceous chemical reagent. In one embodiment, a mutation site binding reagent binds to the site of mutation and alters the chromatographic retention time. In another embodiment, a mutation site binding reagent cleaves at the site of mutation, resulting in an increase in the number of fragments.
Owner:ADS BIOTEC INC
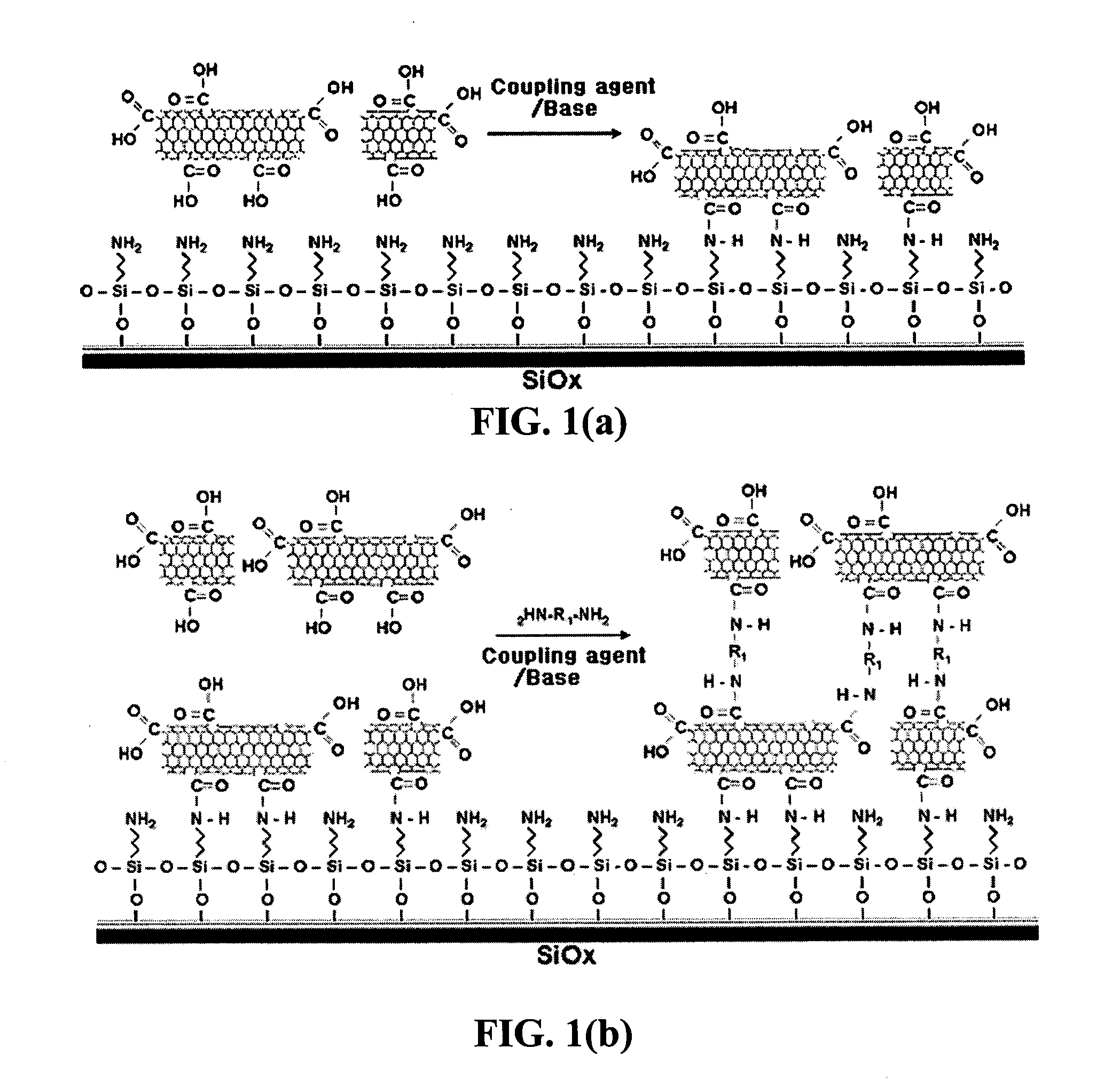
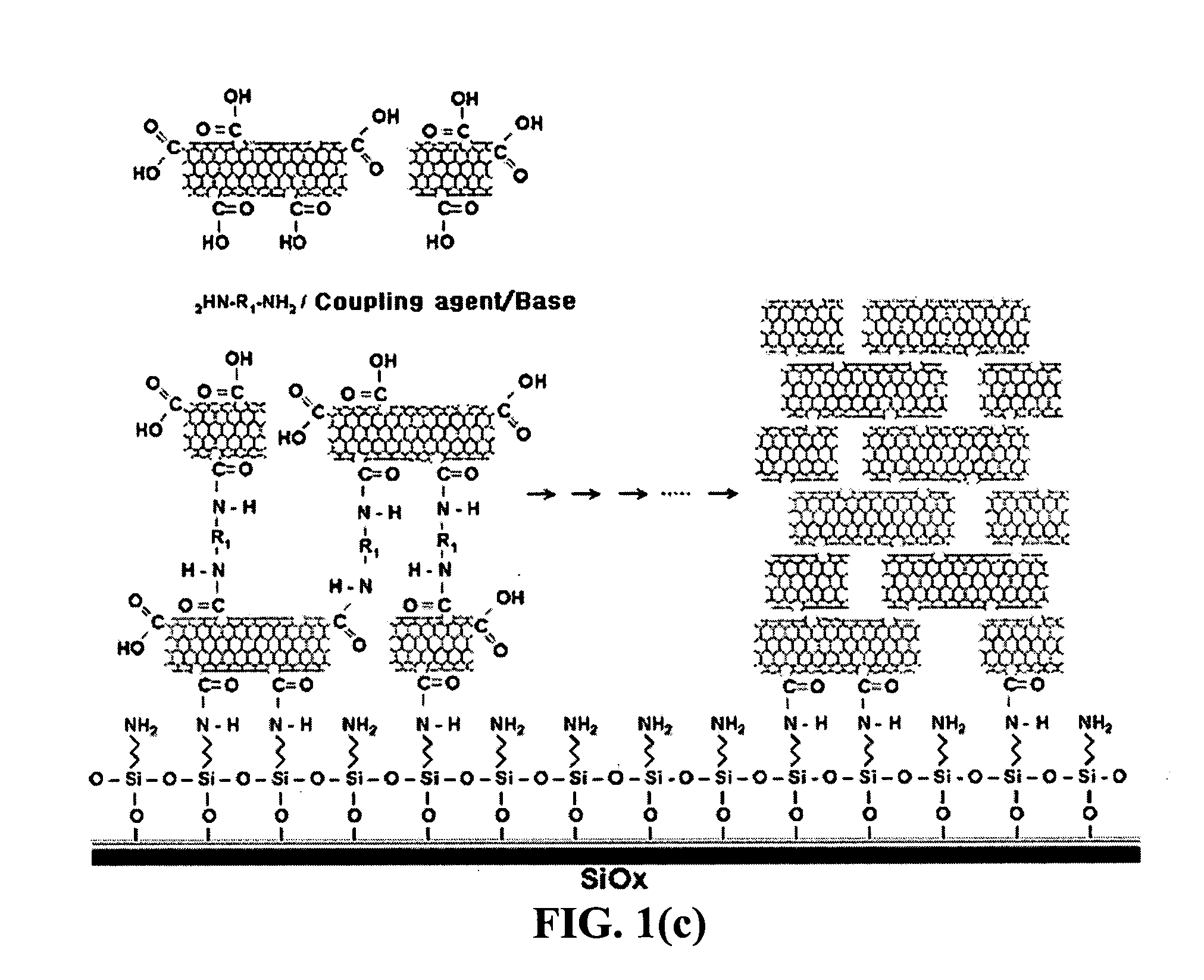
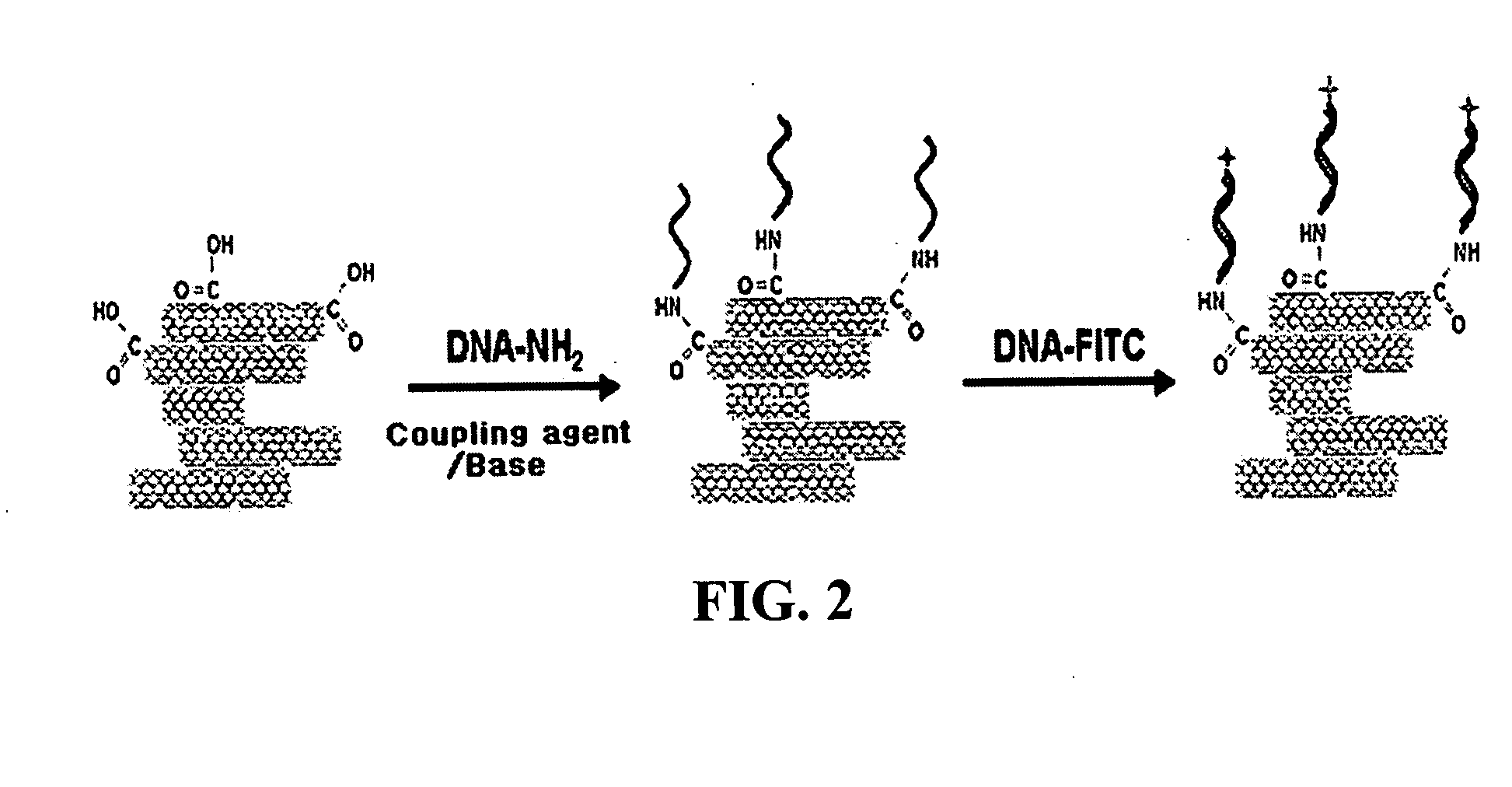
Method for fabricating a biochip using the high density carbon nanotube film or pattern
InactiveUS20050019791A1Improve defectsUtility and advantageBioreactor/fermenter combinationsMaterial nanotechnologyElectrical conductorFluorescence
The present invention relates to a CNT-biochip comprising a bio-receptor which is attached by means of an exposed chemical functional group on a surface of a high density CNT film or pattern which is produced by laminating repeatedly carbon nanotubes (CNT) by chemical bond on the substrate modified with amine groups, and a method for fabricating the same. According to the present invention, it is possible to fabricate various types of CNT-biochips by chemical or physicochemical bonding of various bio-receptors to a CNT pattern (or film) containing exposed carboxyl groups or a CNT pattern (or film) modified by various chemical functional groups. Also, it is possible to fabricate a CNT-biochip comprising bio-receptors attached evenly with high density on a surface of a CNT film where chemical functional groups are abundant and present evenly. Further, the CNT-biochip is applicable to next generation biochips which measure an electrical or electrochemical signal using both conductor and semiconductor properties of the CNT, thereby not needing labeling. Particularly, upon fluorescent measurement of DNA hybridization using the CNT-DNA chip according to the present invention, it is possible to show more distinct signals, thereby producing excellent results. The CNT-DNA chip is useful for genotyping, mutation detection, pathogen identification and the like.
Owner:KOREA ADVANCED INST OF SCI & TECH
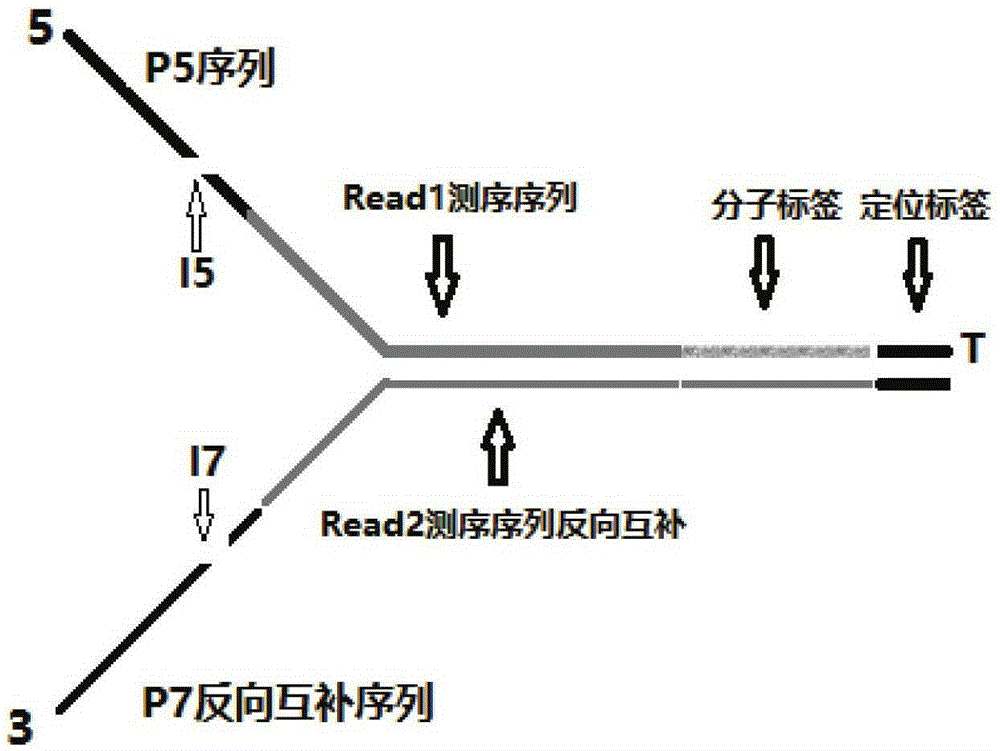
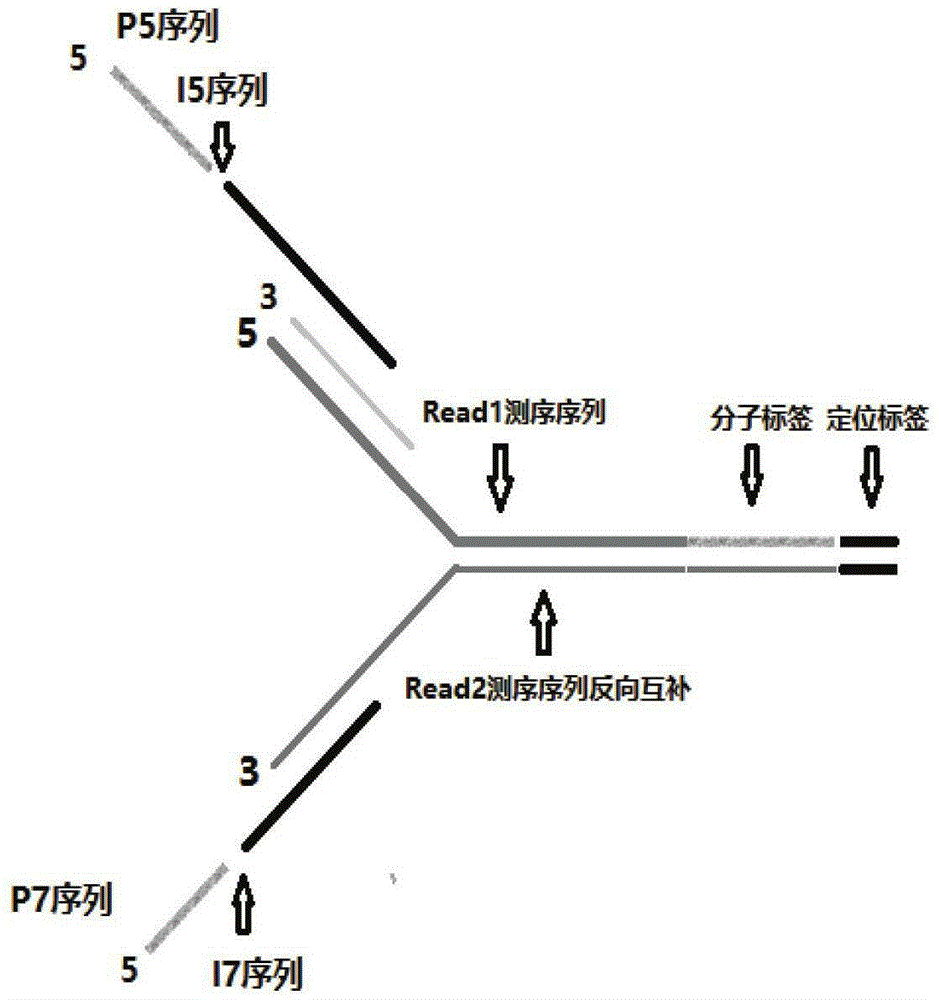
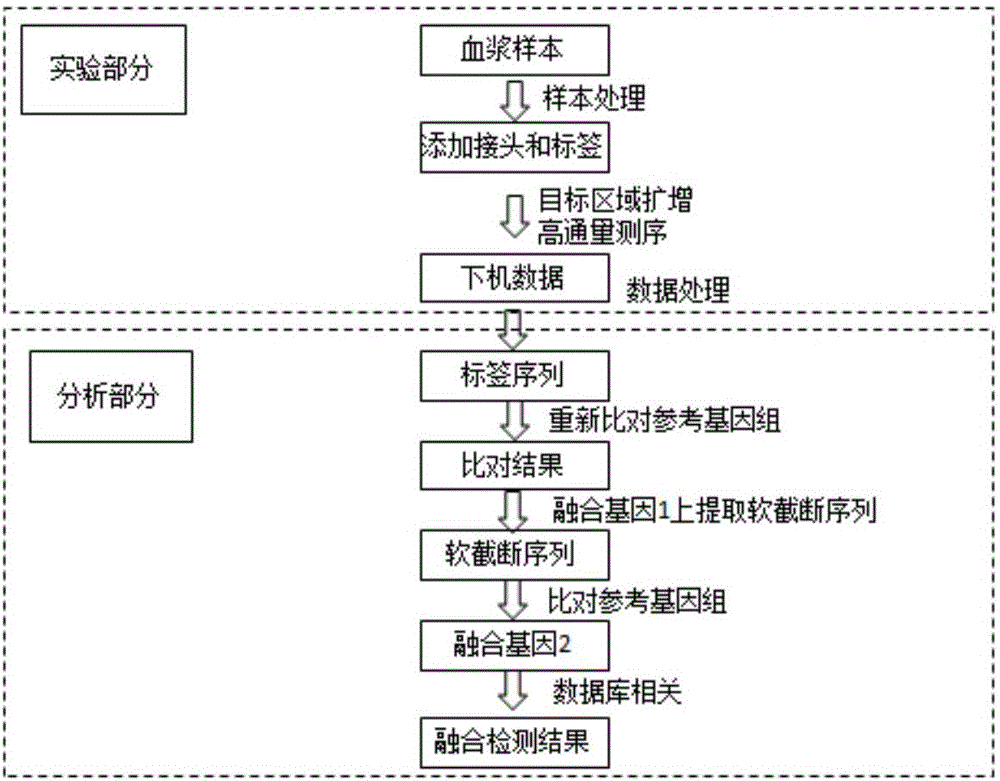
Double-label joint sequence for detection of tumor mutation and detection method
ActiveCN106086162AEasy to distinguishIncrease sample sizeMicrobiological testing/measurementLibrary creationMutation detectionBioinformatics
The invention discloses a double-label joint sequence for detection of tumor mutation, and is characterized in that the joint sequence is synthesized by a joint primer P5 and a joint primer P7, wherein the joint primer P5 has the sequence shown in SEQ ID NO:1, and the joint primer P7 has the sequence shown in SEQ ID NO:2. The invention also provides a database building method and a sequencing method. With use of a double-label joint, the tumor mutation rate of 1*10<-5> can be accurately detected, the tumor mutation detection sensitivity is effectively improved, and with combination of the flux of high-flux sequencing, one-time sequencing can detect multiple mutation loci of multiple genes.
Owner:AMOY DIAGNOSTICS CO LTD
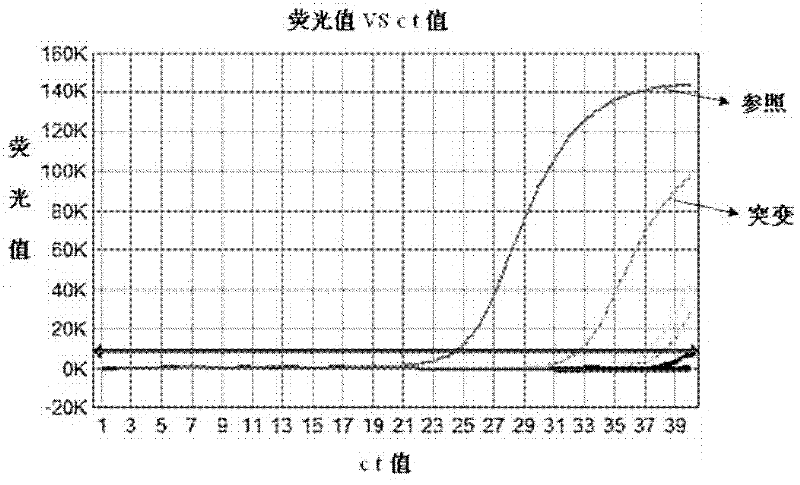
ARMS-qPCR (Allele Refractory Mutation System-quantitative Polymerase Chain Reaction) detection kit for KRAS (Kirsten Rat Sarcoma Viral Oncogene Homolog) gene mutation subtype and detection method
InactiveCN102367478AIncreased sensitivityQuick checkMicrobiological testing/measurementViral OncogenePositive control
The invention relates to the field of molecular biology and aims to provide an ARMS-qPCR (Allele Refractory Mutation System-quantitative Polymerase Chain Reaction) detection kit for KRAS (Kirsten Rat Sarcoma Viral Oncogene Homolog) gene mutation subtype and a detection method. The kit comprises a qPCR hybrid reaction solution, a locked nucleic acid retardant probe, a reference primer, an ARMS primer and a positive control sample, wherein the qPCR hybrid reaction solution comprises a PCR buffer solution, dNTPs (Deoxynucleotide Triphosphates), MgCl2, GoldStarbest Taq enzyme, a universal PCR reverse primer and a universal TaqMan probe. The kit provided by the invention can be used for rapidly and accurately detecting specific locus mutation of KRAS genes in various cancer tissues with high sensitivity, has high sensitivity, and can be used for detecting genome DNA with various tissue origins, specially free DNA segments adopting cell-free systems, such as blood serum and blood plasma, orother body fluid origins, wherein the genome DNA is derived from cell systems. Compared with direct sequencing and other mutation detection technologies, the kit and the detection method thereof havethe advantages of strong specificity, high sensitivity, simplicity and rapidness in operation, high throughput, safety, definiteness and objectivity in result identification and the like for detecting the KRAS gene mutation.
Owner:ZHEJIANG UNIV
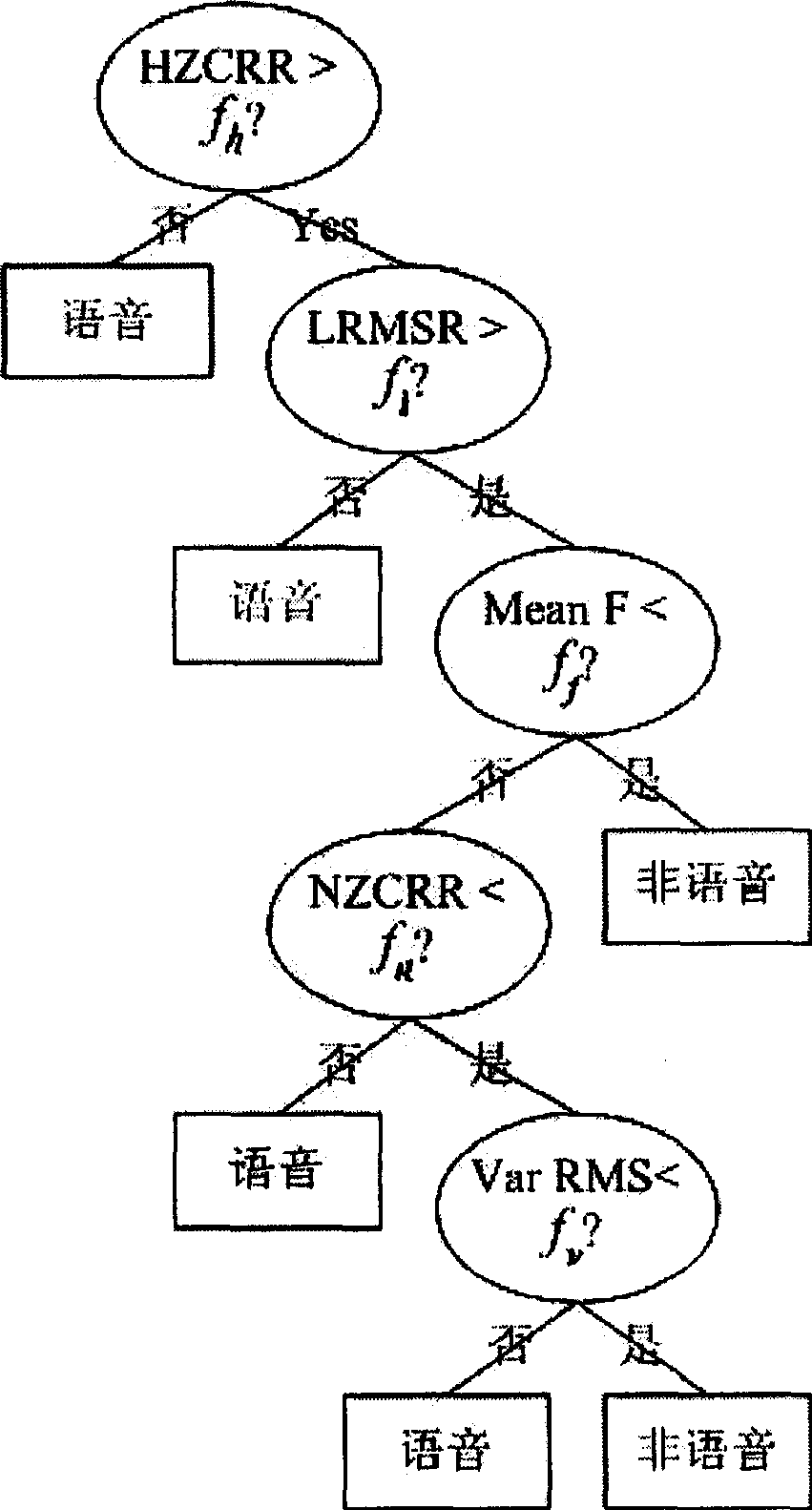
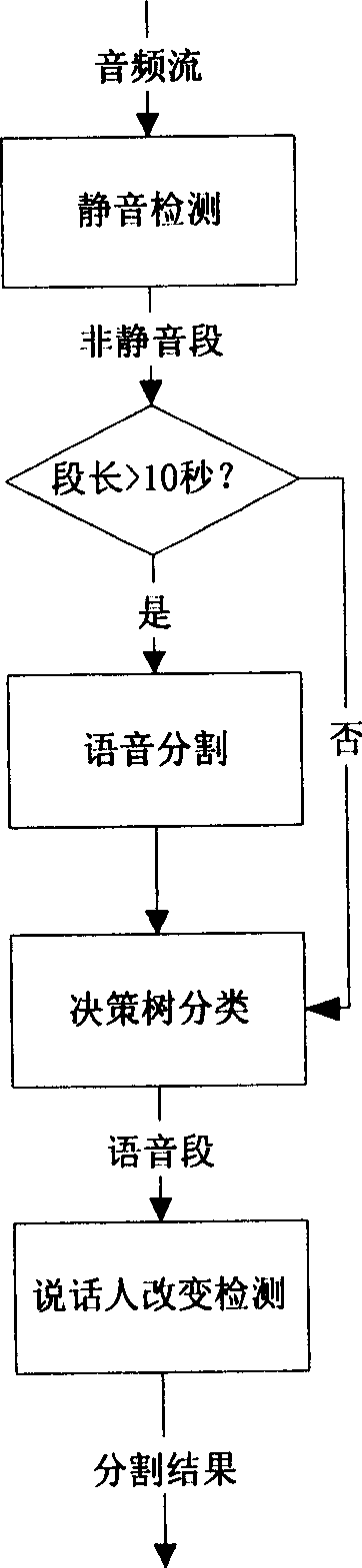
Audio frequency splitting method for changing detection based on decision tree and speaking person
InactiveCN1716380AImprove speech detection accuracyImprove accuracySpeech recognitionMutation detectionAudio frequency
The audio splitting method based on the decision tree and the talker change detection includes the first adaptive silencing detection to find out the mute in audio frequency and the coarse splitting of audio frequency signal with the mute; the subsequent mutation detection for fine splitting of audio frequency signal and classifying the audio segment into phonetic part and non-phonetic part with the decision tree; and the final detecting the talker change point in the phonetic segment to obtain the final splitting result. The present invention performs phonetic detection via combining two methods of mute detection and mutation detection and adopting phonetic / non-phonetic decision tree to raise the accuracy of phonetic detection, and performs the talker change detection in phonetic segment with saving in calculation time.
Owner:ZHEJIANG UNIV
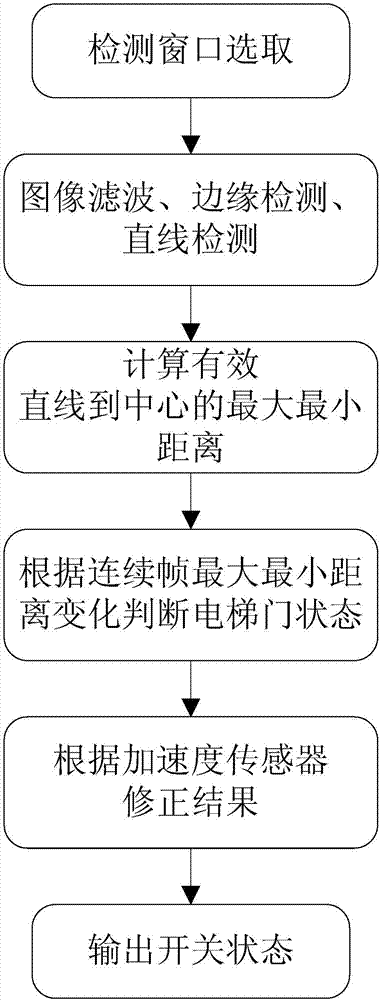
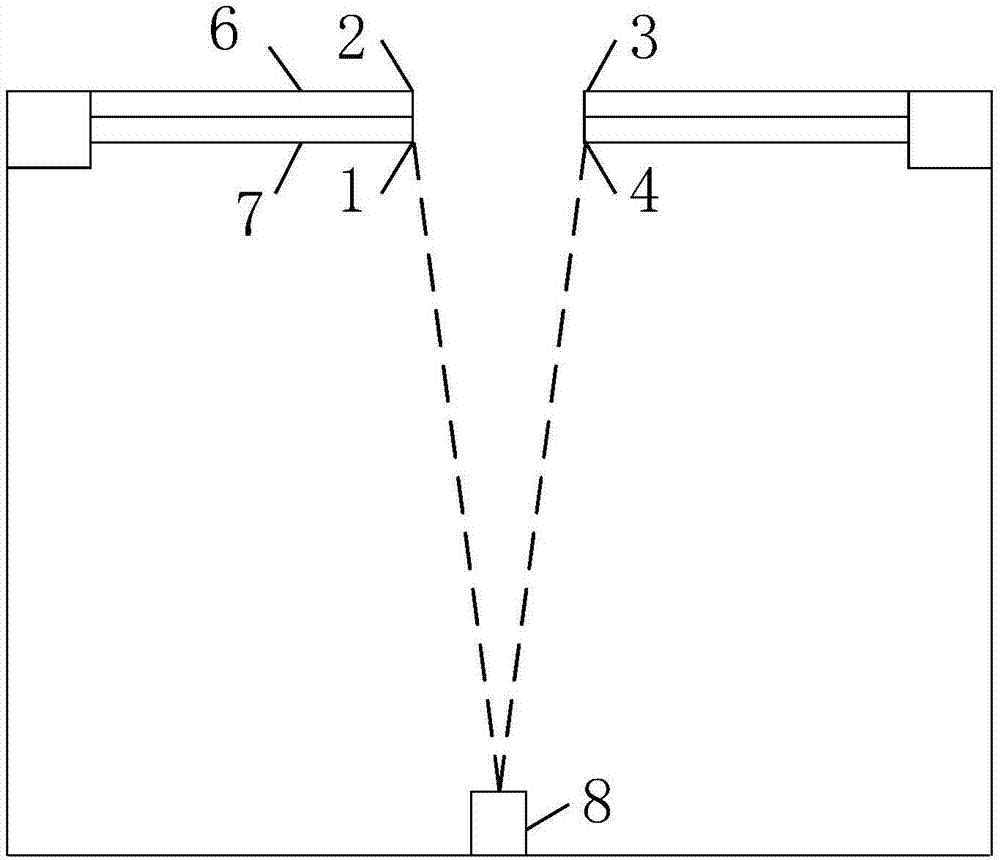
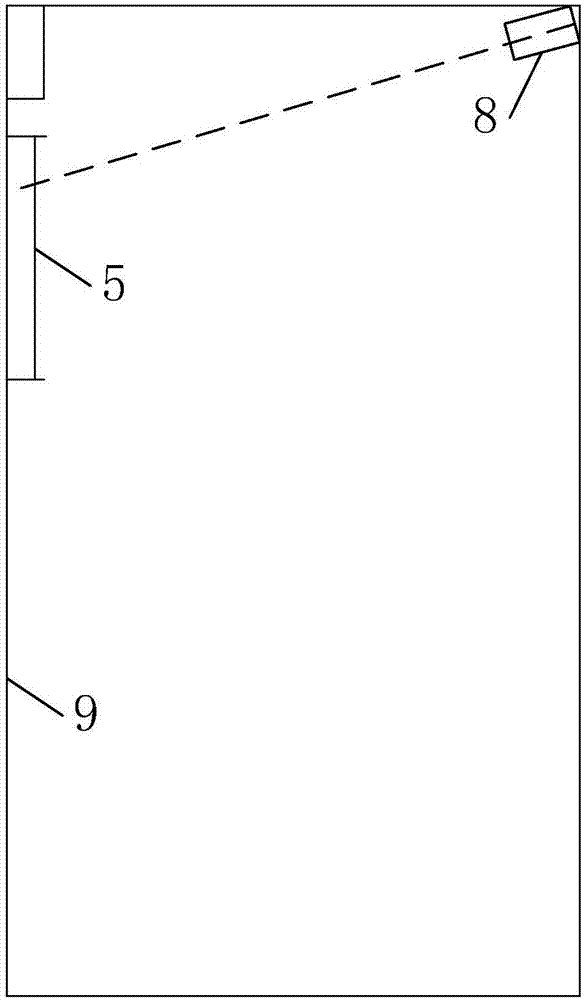
Elevator door opening and closing detection method based on camera images
ActiveCN106986248AIntegrity guaranteedCalculation speedImage enhancementImage analysisCamera imageMutation detection
The invention discloses an elevator door opening and closing detection method based on camera images. According to the detection method, an elevator, an acceleration sensor and an image analyzing and processing system are involved, an elevator door is arranged on the elevator and comprises a landing door and a car door, the image analyzing and processing system comprises an image filtering module, an edge detection module, a binarization processing module, a linear detection module, an effective linear extraction module and a logic analyzing and processing module, and a camera is arranged at the top of the elevator and in electrical communication connection with the image analyzing and processing system. Elevator door opening and closing detection is carried out through the boundary information of the elevator door, the detection method has the advantage of being high in calculation speed and can be applied to an embedded platform, and the instantaneity, accuracy and stability of the detection method are fully considered. According to the detection method, the boundary features of original images and the boundary features of binary images are fused, meanwhile, the interference in multiple aspects is eliminated through light ray detection, distance mutation detection, distance filtering and the like, and the robustness of the detection is improved.
Owner:SICHUAN CHANGHONG ELECTRIC CO LTD
c-KIT gene mutation detection liquid-phase chip
ActiveCN101984070AImprove signal-to-noise ratioImplement parallel detectionMicrobiological testing/measurementSignal-to-noise ratio (imaging)Mutation detection
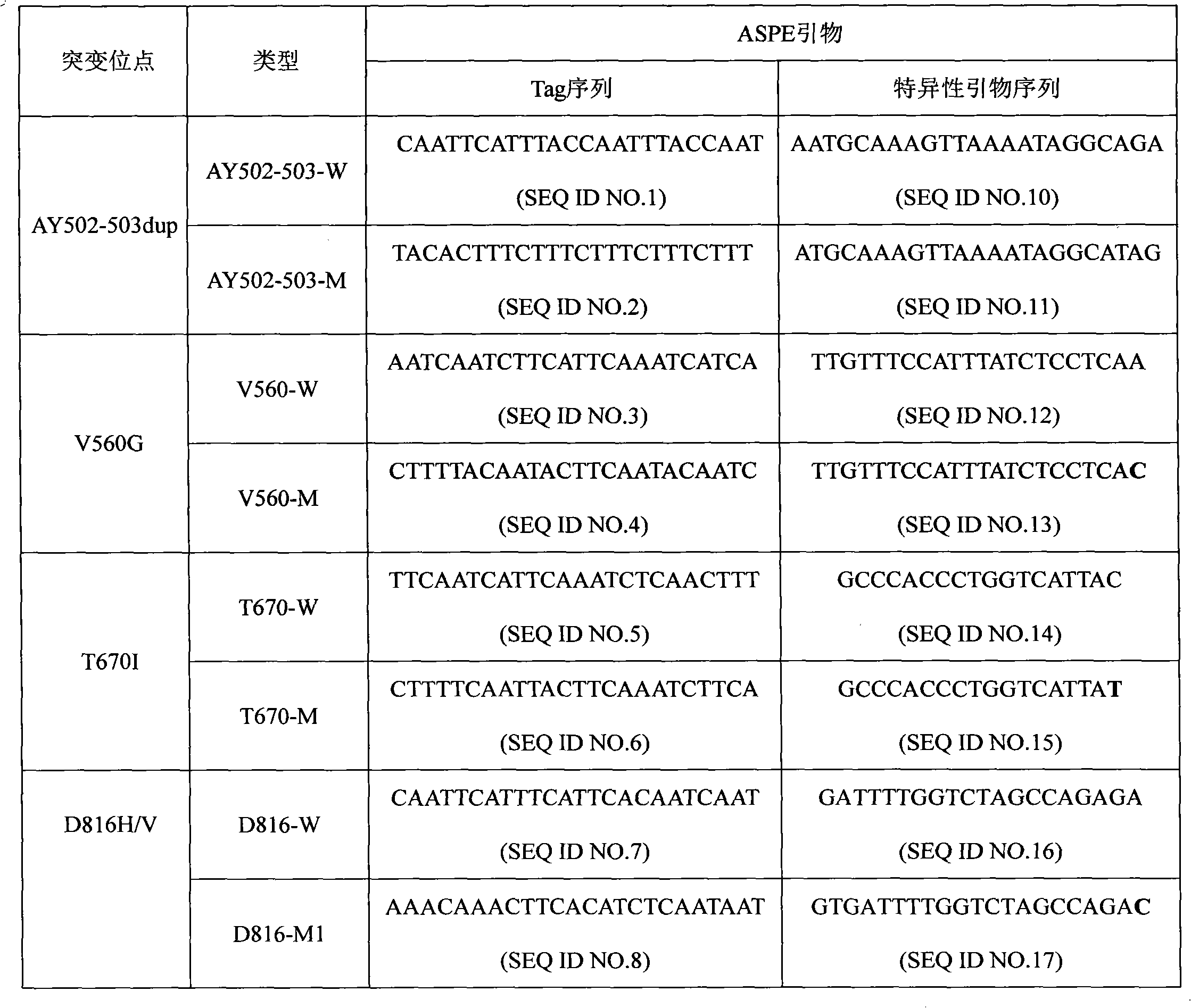
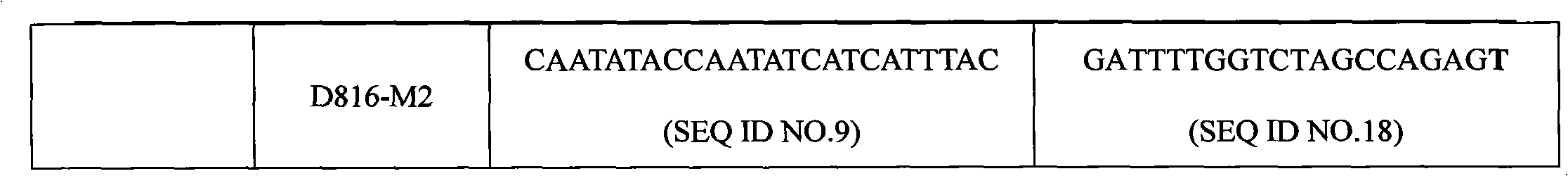
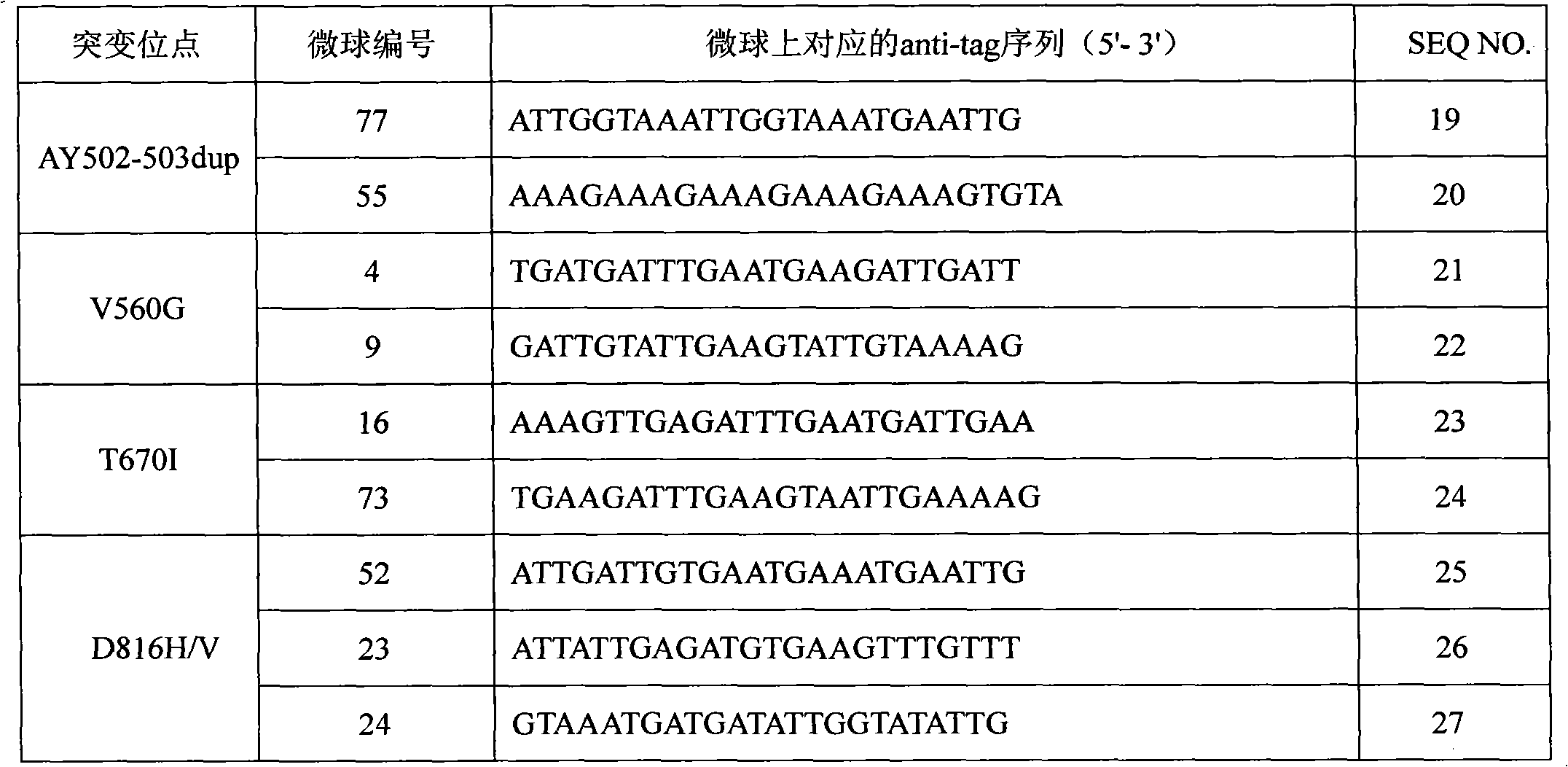
The invention discloses a c-KIT gene mutation detection liquid-phase chip, which comprises tag sequence at 5' terminal and ASPE (Allele Specific Primer Extension) primers consisting of specific primers specific to mutational sites at 3' terminal. The specific primers consist of sequences shown in SEQ ID No.10 and SEQ ID No.11 specific to AY502-503dup mutational sites, sequences shown in SEQ ID No.12 and SEQ ID No.13 specific to V560G mutational site, sequences shown in SEQ ID No.14 and SEQ ID No.15 specific to T670I mutational site and / or sequences shown in SEQ ID No.16-SEQ ID No.18specific to D816H / V mutational site, the tag sequence which is selected from sequences shown in SEQ ID No.1-SEQ ID No.9, microballoons which have different color codings and are respectively enveloped with a specific anti-tag sequence and amplification primers. The coincidence ratio of the result of sequencing method with that of the c-KIT gene mutation detection liquid-phase chip reaches up to 100 percent. The c-KIT gene mutation detection liquid-phase chip can simultaneously detect a plurality of mutational sites with excellent signal to noise ratio.
Owner:SUREXAM BIO TECH
High sensitivity mutation detection using sequence tags
InactiveUS20160115532A1Improve accuracyHigh sensitivityMicrobiological testing/measurementNucleotideMutation detection
The invention is directed to methods for increasing the sensitivity of high throughput sequencing, particularly for distinguishing true rare mutations from amplification, sequencing and other sample processing errors that occur in sequencing techniques. In one aspect, methods of the invention includes steps of (a) preparing templates from nucleic acids in a sample; (b) labeling by sampling the templates to form tag-template conjugates, wherein substantially every template of a tag-template conjugate has a unique sequence tag; (c) linearly amplifying the tag-template conjugates; (d) generating a plurality of sequence reads from the linearly amplified tag-template conjugates; and (e) determining a nucleotide sequence of each of the nucleic acids based on the frequencies, or numbers, of each type of nucleotide at each nucleotide position of each plurality of sequence reads having identical sequence tags.
Owner:ADAPTIVE BIOTECH
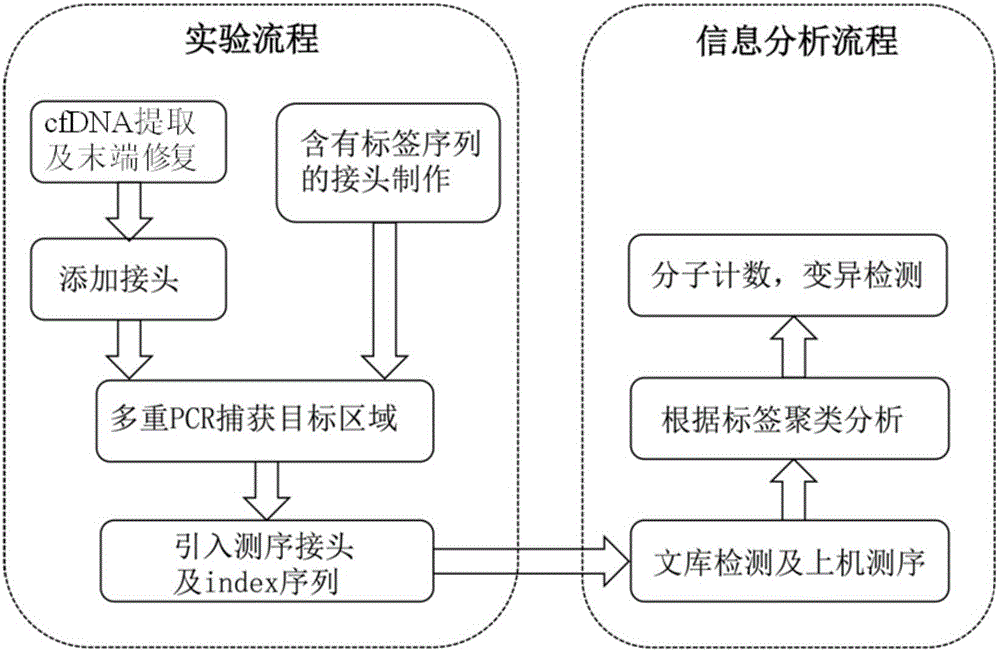
Construction method of ctDNA ultra-low-frequency mutation detection library, kit and analysis method of library detection data
InactiveCN106834275AIncrease diversityEasy to solveMicrobiological testing/measurementBioinformaticsMagnetic beadSmall fragment
The invention discloses a construction method of a ctDNA ultra-low-frequency mutation detection library, a kit and an analysis method of library detection data. The construction method comprises steps as follows: S1, cfDNA is extracted from whole blood; S2, the terminal of cfDNA is restored and an A basic group is added to the 3'terminal; S3, the terminal of the cfDNA obtained in S2 is connected with connectors containing random label sequences; S4, primers for multiplex PCR (polymerase chain reaction) are designed according to the sequences of the connectors and an object region for object region capturing; S5, a PCR product in S4 is subjected to magnetic bead purification, and small-fragment DNA not subjected to non-specific amplification and primer dimer are removed; S6, a product in S5 is subjected to PCR amplification, an index sequence is introduced, and the ctDNA ultra-low-frequency mutation library is obtained. According to the method, PCR mistakes and sequencing mistakes are eliminated, and the detection specificity is improved.
Owner:天津诺禾医学检验所有限公司
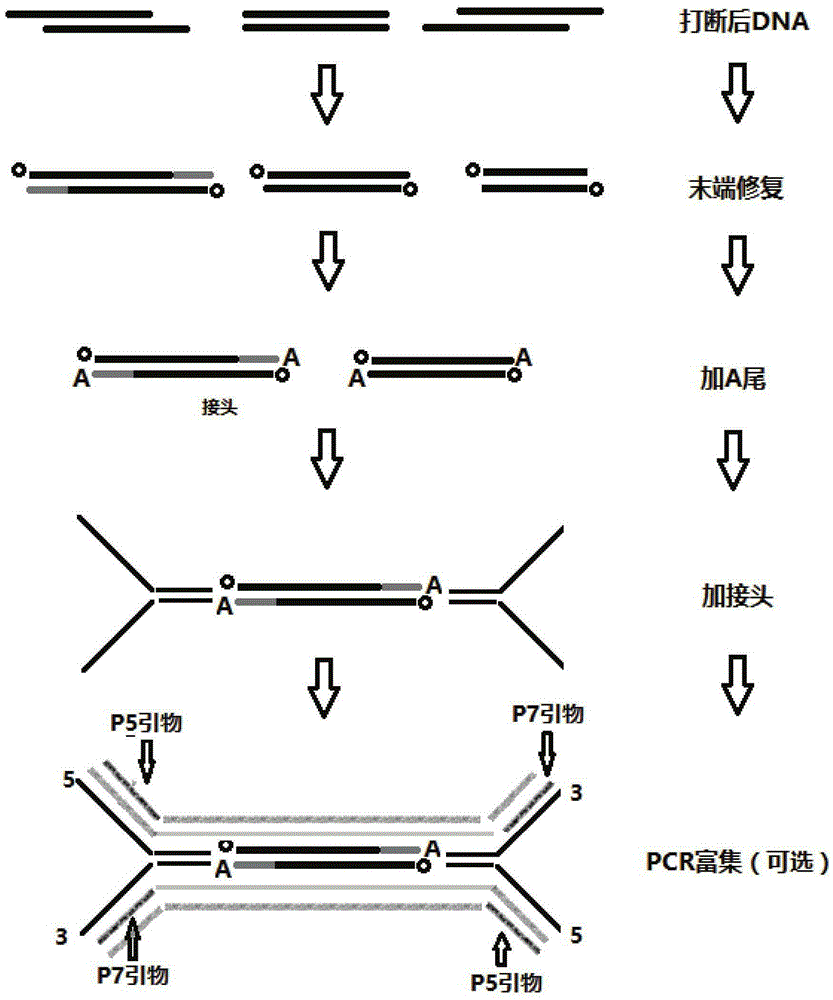
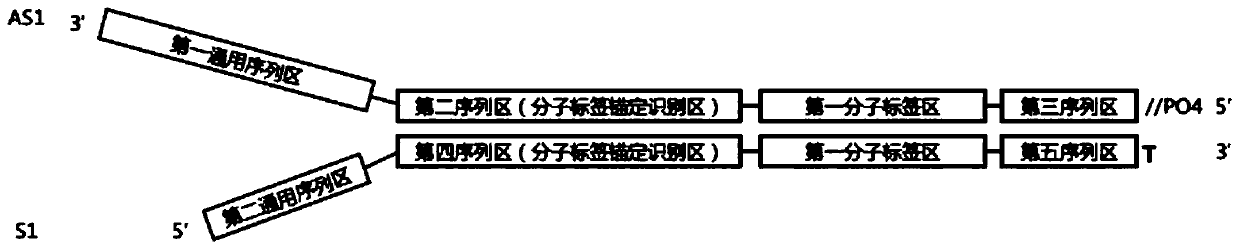
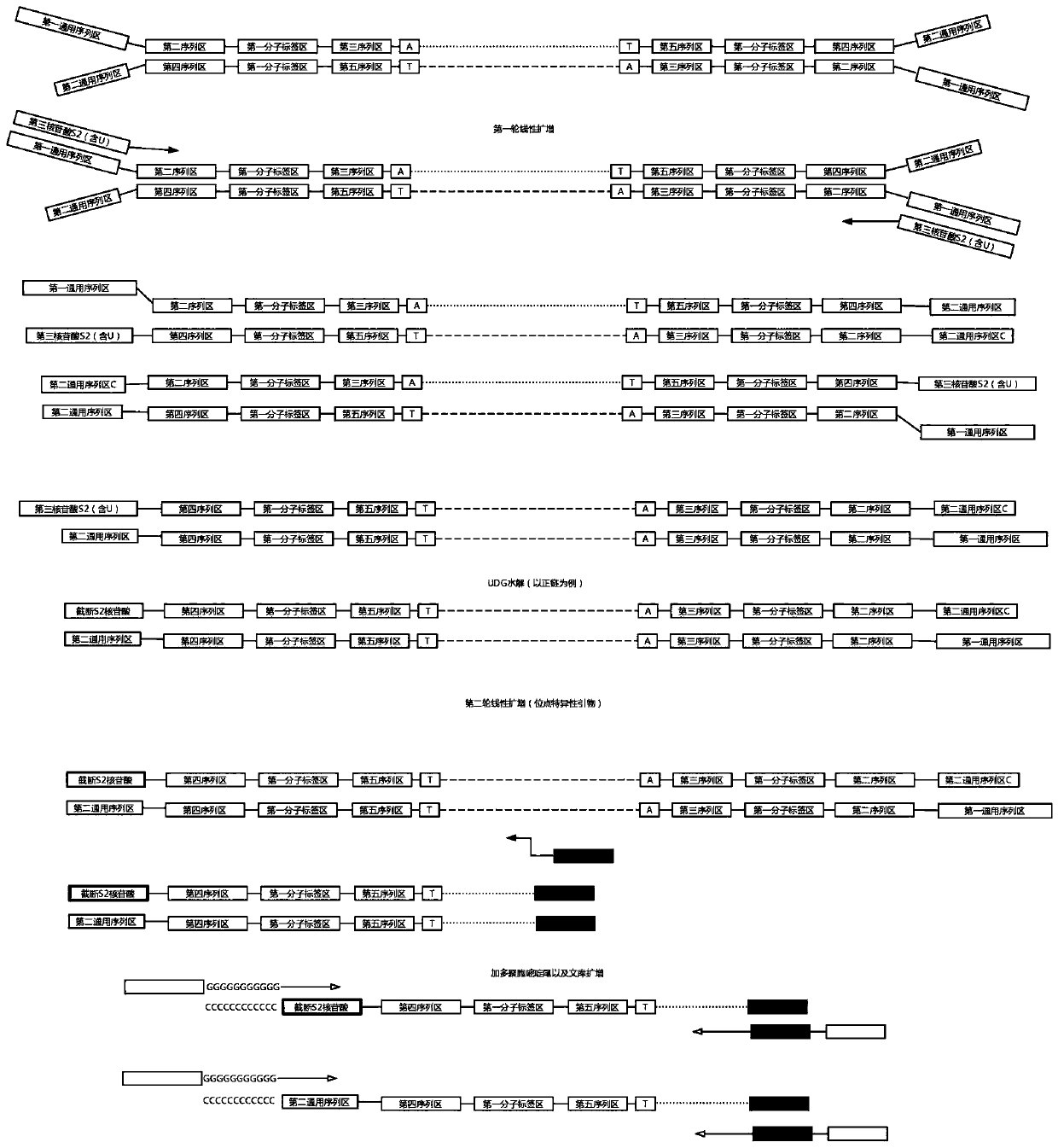
Construction method of high-flux sequencing library and reagent kit for library construction
ActiveCN110734908AAchieve rearrangementAvoid the phenomenon of heterogeneous amplificationNucleotide librariesMicrobiological testing/measurementMultiplexMutation detection
The invention provides a construction method of a high-flux sequencing library and a reagent kit for library construction. The reagent kit comprises one or more of the following components: a high-flux sequencing Y-shaped joint, a universal primer for single-end linear PCR amplification, a biotin labeling specific primer for single-end linear multiplex PCR amplification, forward and backward library amplification primers, a UDG enzyme and the like. The invention relates to a method for constructing a targeting dimolecular identifier (UMI (unique molecular identifier) and chain unique molecularidentifier) high-flux sequencing library based on the reagent kit. According to the method, double error correcting mechanisms of a random UMI and a chain unique molecular identifier having sequencepolymorphism in a targeting sequencing system based on multiplex PCR amplification are realized, and disadvantages of most of conventional multiplex PCR amplification targeting sequencing systems areavoided, so that all false positives and false negatives in mutation detection are avoided, and high-sensitivity high-accuracy high-depth detection can be performed on low-frequency nucleic acid mutation in samples.
Owner:福州福瑞医学检验实验室有限公司
Cancer gene mutation and gene amplification detection
ActiveCN104630375AAmplified equalizationImprove efficiencyMicrobiological testing/measurementDNA/RNA fragmentationMultiplexMedicine
The invention provides a primer set for simultaneously detecting mutation of a plurality of regions of two or more exons of one or more cancer drive genes in a sample on the basis of multiplex PCR (polymerase chain reaction) and high-flux sequencing techniques, a kit comprising the primer set, and a method for amplifying the cancer drive gene or detecting the cancer drive gene by using the primer set and / or kit. The mutation detection result can be used for instructing cancer clinic medication, auxiliary diagnosis or prognosis judgment on certain cancers.
Owner:北京圣谷智汇医学检验所有限公司
Lung cancer somatic mutation detection and analysis method based on high-throughput sequencing technique
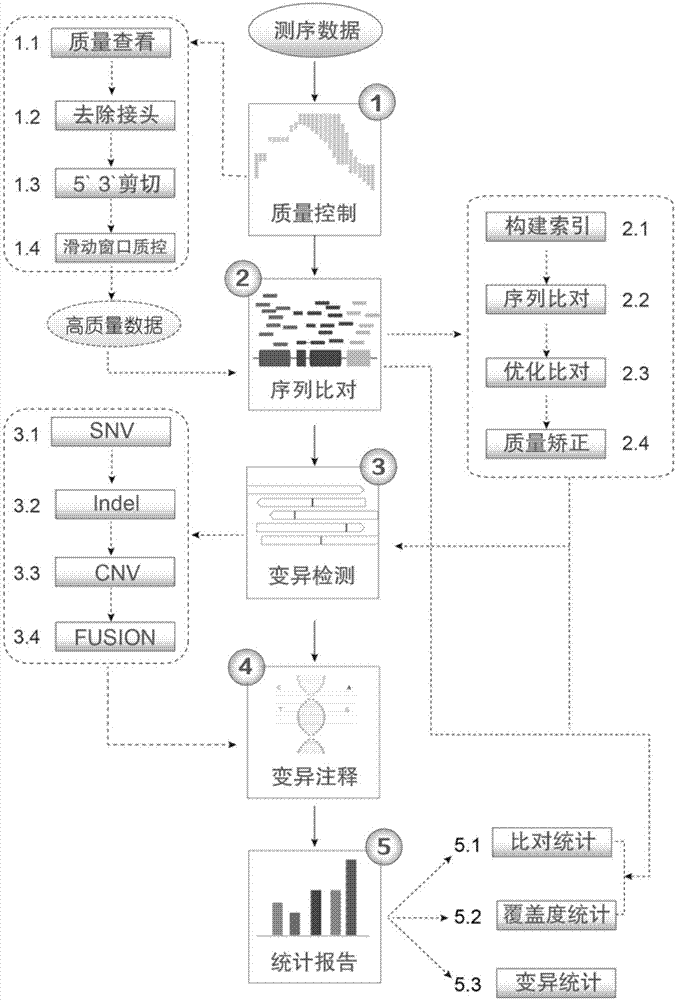
InactiveCN107391965AEfficient detectionAnalytical results are reliableSequence analysisSpecial data processing applicationsMutation detectionLung cancer
The invention discloses a lung cancer somatic mutation detection and analysis method based on a high-throughput sequencing technique. The method comprises the steps of (1) controlling quality; (2) comparing sequences; (3) detecting mutations; (4) annotating the mutations; (5) reporting the results. Compared with the prior art, the method of the invention has the advantages that the detection results are reliable; the detection content is diversified; the detection method is flexible; the data results are visualized.
Owner:SHANGHAI PASSION BIOTECHNOLOGY CO LTD
Methods to detect cross-contamination between samples contacted with a multi-array substrate
InactiveUS20050048505A1Bioreactor/fermenter combinationsBiological substance pretreatmentsGenomicsMutation detection
Methods and compositions for detecting cross-contamination between samples contacted with different arrays of a multi-array substrate are provided. The methods involve contacting sample to arrays of a multi-array substrate that contains cross-contamination probes in each of its arrays, and evaluating the resultant sample contacted arrays for cross-contamination between the samples. In many embodiments, the arrays of the multi-array substrate contain a set of cross-contamination probes for a corresponding set of cross-contamination targets in the sample(s). Kits and systems are provided for performing the invention. The subject methods may be used in a variety of different applications, such as gene expression analysis, DNA sequencing, mutation detection, as well as other genomics and proteomics applications.
Owner:AGILENT TECH INC
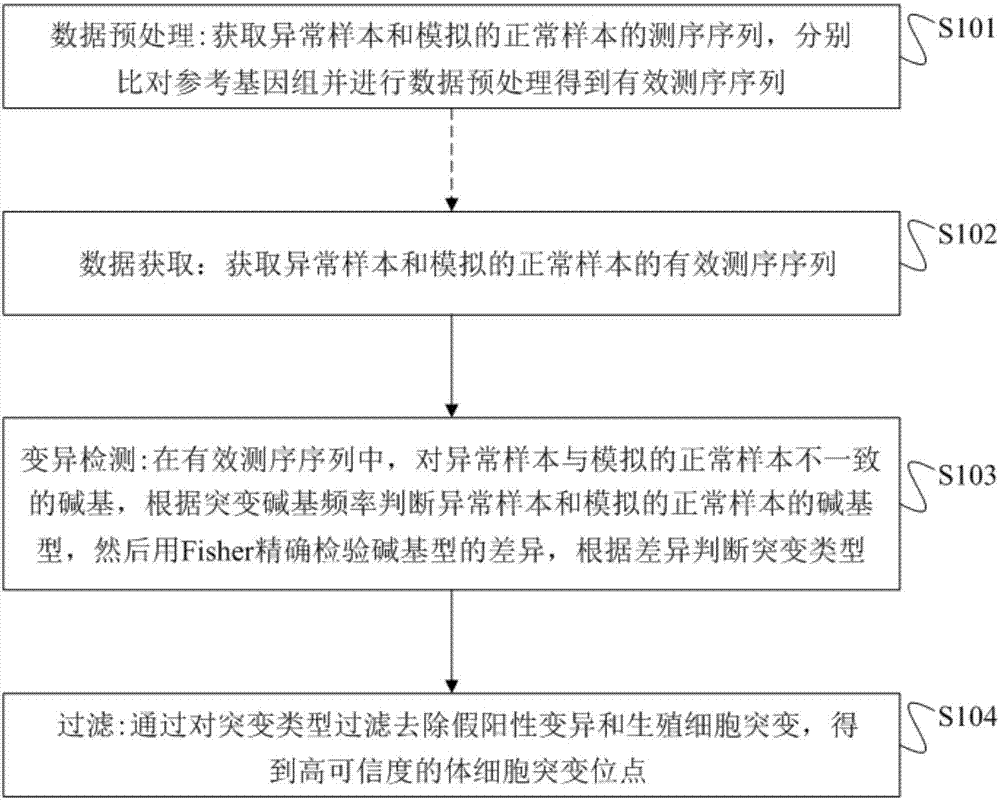
Method and device for detecting single sample body cell mutation sites in abnormal tissue and storage medium
ActiveCN107491666AHigh sensitivityStrong specificityProteomicsGenomicsSingle sampleMutation detection
The invention provides a method and device for detecting single sample body cell mutation sites in abnormal tissue and a storage medium. The method includes the following steps that the effective sequencing sequence of an abnormal sample and the effective sequencing sequence of a simulated normal sample are obtained; in the effective sequencing sequences, basic groups, different from basic groups of the simulated normal sample, of the abnormal sample are obtained, according to the mutation basic group frequency, the types of the basic groups of the abnormal sample and the simulated normal sample are judged, the FISHER is then used for accurately detecting the difference of the types of the basic groups, and according to the difference, the mutation type is judged; by filtering the mutation types, false positive mutation and germ cell mutation are removed, and high-reliability somatic cell mutation sites are obtained. The method has the advantages of being high in sensitivity and specificity, has high sensitivity in mutation detection of known mutation genes, and new mutation genes can be found.
Owner:深圳裕康医学检验实验室
Methods, compositions, and kits for detecting allelic variants
ActiveCN102428190AMicrobiological testing/measurementDNA/RNA fragmentationNucleotideMutation detection
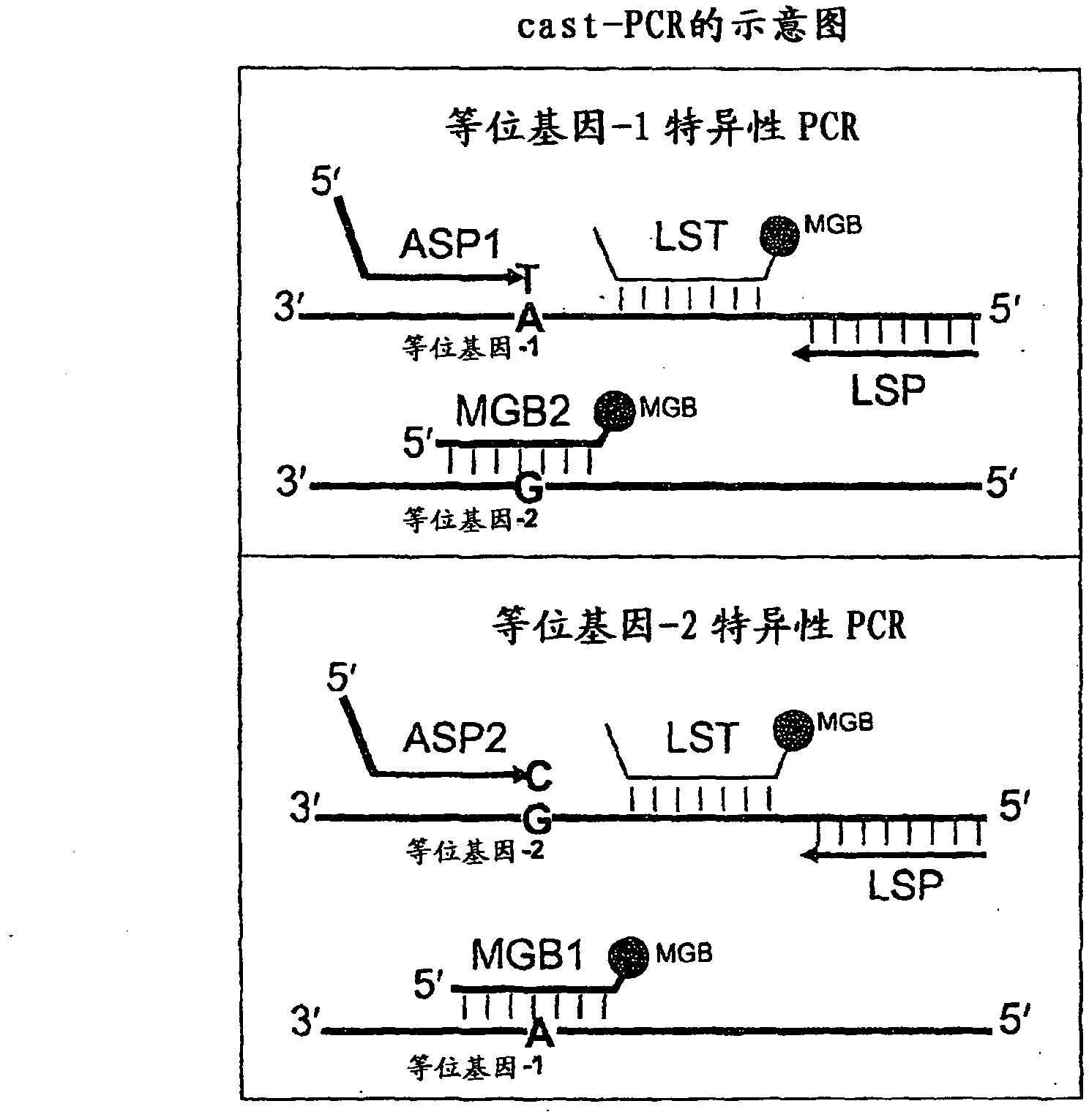
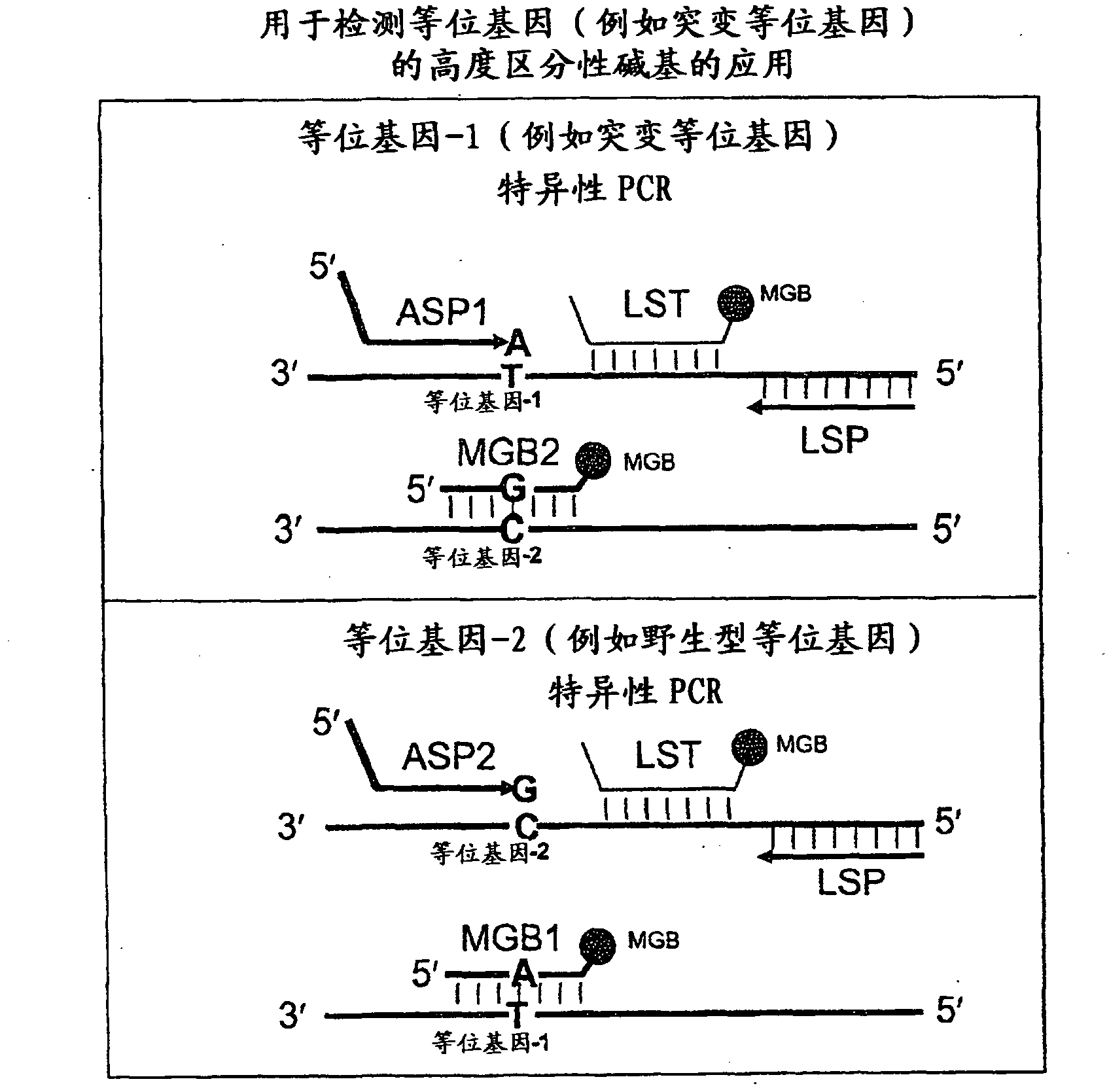
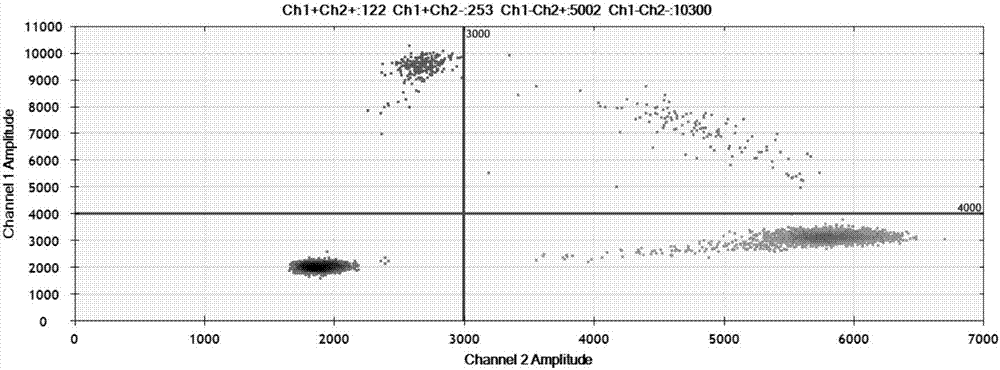
In some embodiments, the present inventions relates generally to compositions, methods and kits for use in discriminating sequence variation between different alleles. More specifically, in some embodiments, the present invention provides for compositions, methods and kits for quantitating rare (e.g., mutant) allelic variants, such as SNPs, or nucleotide (NT) insertions or deletions, in samples comprising abundant (e.g., wild type) allelic variants with high specificity and selectivity. In particular, in some embodiments, the invention relates to a highly selective method for mutation detection referred to as competitive allele-specific TaqMan PCR ('cast-PCR').
Owner:LIFE TECH CORP
Chip and method for capturing target sequences of tumor susceptibility genes, and mutation detection method
ActiveCN105779572ARealize detectionHigh detection throughputMicrobiological testing/measurementAbnormal tissue growthHigh flux
The invention discloses a chip and method for capturing target sequences of tumor susceptibility genes, and a mutation detection method. The chip is a liquid chip which binds with probe groups capable of simultaneously capturing at least 5, preferably at least at least 10, preferably at least at least 20, preferably at least at least 30, preferably at least at least 50, preferably at least at least 80, preferably at least at least 100, preferably at least at least a10, or preferably all of 115 genetic tumor susceptibility genes as shown in a table 1. The mutation detection method comprises a step of capturing the target sequences of the genetic tumor susceptibility genes by using the liquid chip and a step of carrying out sequencing by using G2 high-flux sequencing technology so as to find out mutation sites. The mutation detection method has the advantages of a wide application scope, high efficiency, comprehensiveness and easy operation, can detect replacement of a single base, insertion or deletion of a single base / multiple bases and deletion / amplification of large fragments and is capable of realizing high-efficiency comprehensive detection of common tumor susceptibility gene mutation.
Owner:SHENZHEN HUADA GENE INST
B-raf gene mutation detection kit
ActiveCN104099425AGuaranteed accuracyMonitor for false negativesMicrobiological testing/measurementForward primerB-RAF Gene Mutation
The invention relates to a B-raf gene mutation detection kit. The B-raf gene mutation detection kit comprises quality-control primer probe internal-standard mixed liquor and detection primer probe internal-standard mixed liquor, wherein the quality-control primer probe internal-standard mixed liquor comprises a quality-control primer pair, a B-raf gene specific probe, an internal-standard primer pair, an internal-standard specific probe and an internal-standard template, and the detection primer probe internal-standard mixed liquor comprises a B-raf gene V600E mutation detection specific primer pair, a B-raf gene specific probe, an amplification blocking nucleic acid sequence, an internal-standard primer pair, an internal-standard specific probe and an internal-standard template. The B-raf gene mutation detection kit has the advantages that an amplification refractory mutation system is combined with a wild type amplification blocking nucleic acid sequence with a phosphorylated terminal, deoxyinosine is introduced into detection of a B-raf gene V600E mutation detection specific ARMS (amplification refractory mutation system) forward primer to enable quality-control PCR (polymerase chain reaction) and detection PCR to be performed for detection of samples parallelly, and each reaction system can have an internal-standard system capable of effectively avoiding false negative and intra-assay variation; the B-raf gene mutation detection kit is low in cost, high in sensitivity and more capable of controlling intra-assay and inter-assay variation.
Owner:国九堂山东阿胶有限公司
Amplification method, primer, sequencing method and mutation detection method of mitochondria whole genome DNA (Deoxyribonucleic Acid)
InactiveCN103173441ALow costMicrobiological testing/measurementDNA preparationDiseaseMutation detection
The invention discloses a primer pair and a kit for amplification of a mitochondria whole genome. The invention further discloses an amplification method, a sequencing method and a mutation detection method of the mitochondria whole genome DNA (Deoxyribonucleic Acid). According to the invention, the direct amplification of the given primer is combined with the direct sequencing method, so that a mitochondria whole genome sequence can be obtained; all mutations of the mitochondria genes can be detected by one step for being used for detecting the diseases caused by most of the mitochondria gene mutations.
Owner:BGI GENOMICS CO LTD
Method for detecting multiple endocrine adenoma II gene mutation
InactiveCN101148684AMicrobiological testing/measurementBiological testingRet geneMultiple endocrine adenomas
The present invention belongs to the field of biotechnology, and discloses one method of in vitro determining whether to have RET gene variation in the nucleic acid sample, one kit for determining RET gene variation and one process of obtaining RET gene amplifying product. The method of the present invention is accurate, simple, fast and stable.
Owner:RUIJIN HOSPITAL AFFILIATED TO SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
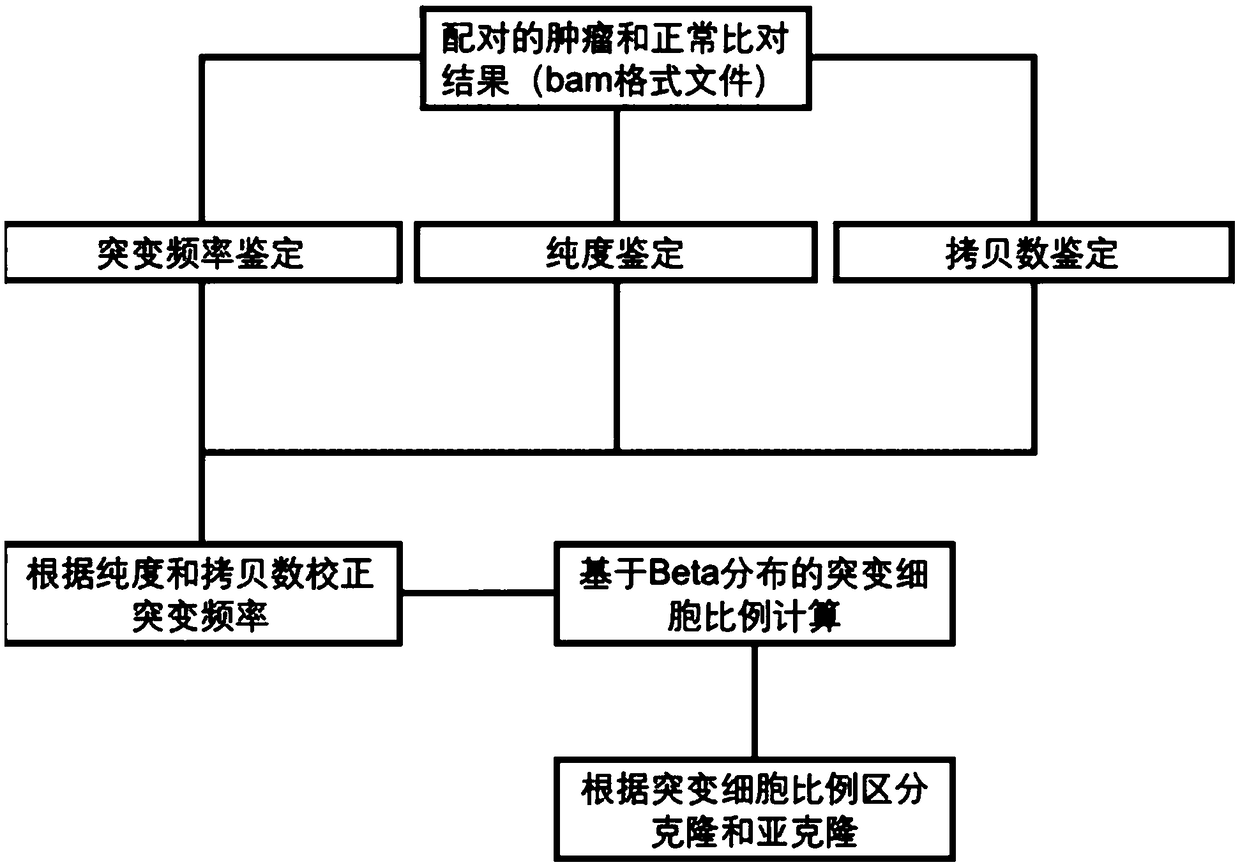
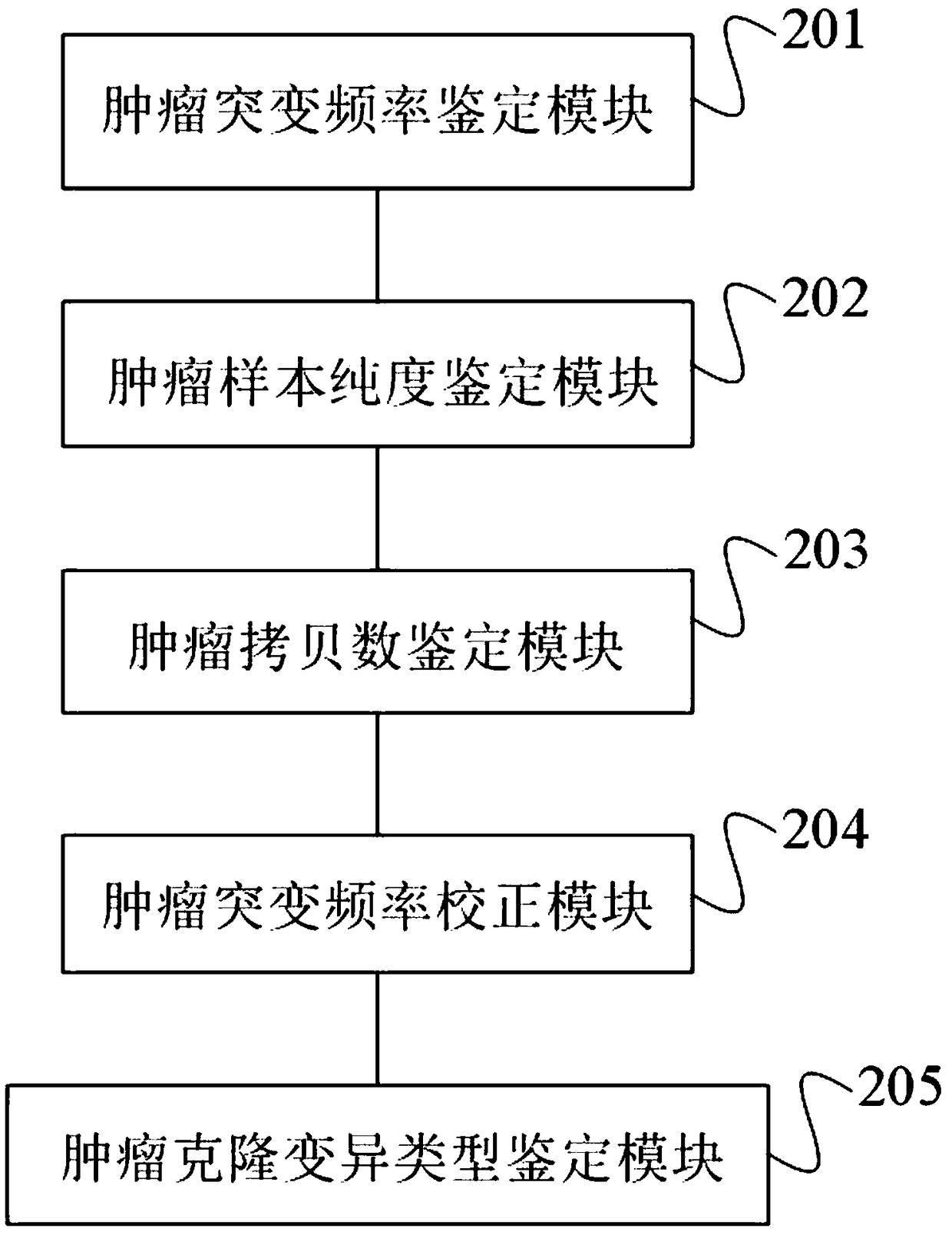
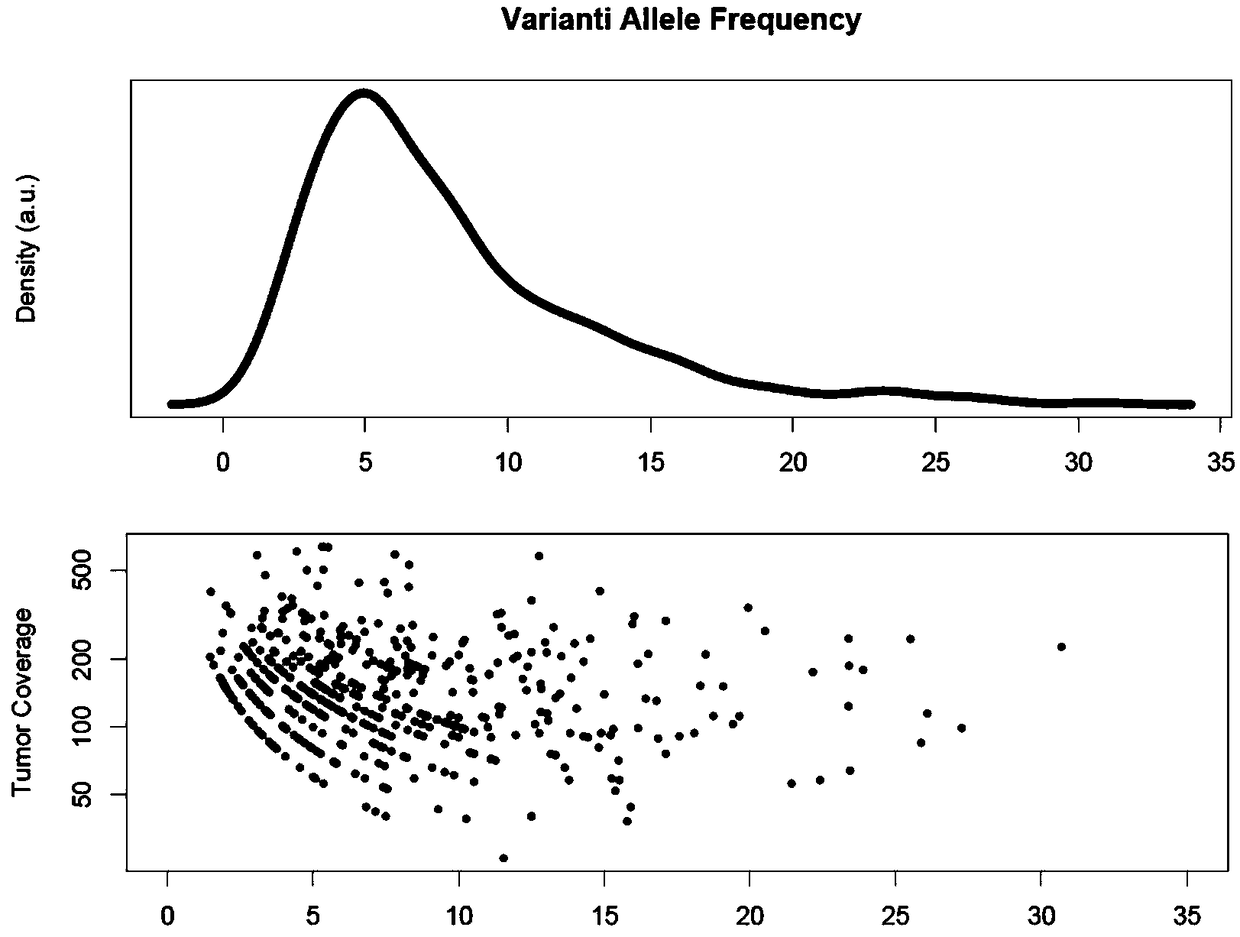
Tumor cloning mutation detection method and device based on next-generation sequencing and memory medium
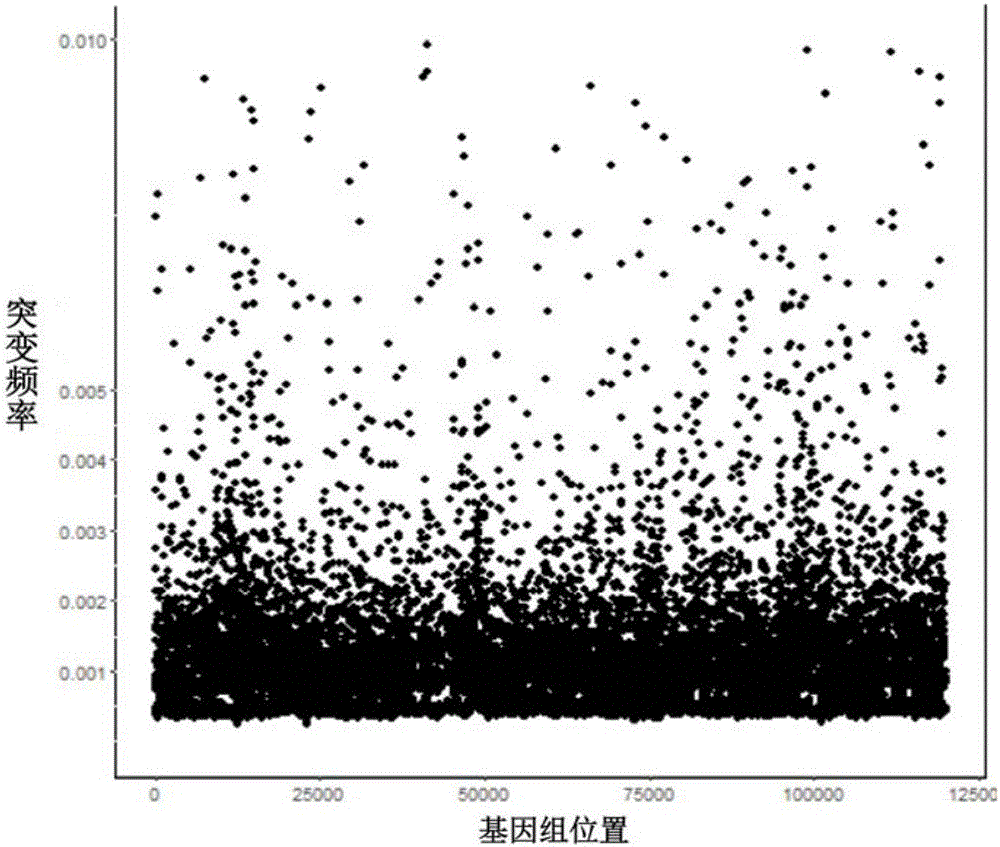
ActiveCN108733975AAvoid influenceAccurate tumor clonal mutation type detection resultsSpecial data processing applicationsMutation frequencyMutation detection
The invention discloses a tumor cloning mutation detection method and device based on next-generation sequencing and a memory medium. The method provided by the invention comprises the steps of carrying out mutation detection on a comparison file of paired tumor and normal samples through utilization of mutation detection software, computing a mutation frequency, and selecting segments with high sequencing quality as a statistics result; carrying out copy number and purity detection on the paired tumor and normal samples through utilization of purity detection software; combining small segments into big segments, and annotating the copy number in a mutation area; and computing a proportion of the mutation in a tested tumor tissue through utilization of a beta distribution model according to tumor sample purity and copy number detection results, thereby judging a tumor cloning mutation type. According to the method provide by the invention, influences of the sample purity and multiploidon the detection are avoided, the mutation type detection is relatively accurate, the subcloning mutation with clinical significance can be effectively identified, and the foundation for accurately and deeply researching a tumor cloning evolution process and searching a tumor therapy molecular mechanism is laid.
Owner:深圳裕策生物科技有限公司
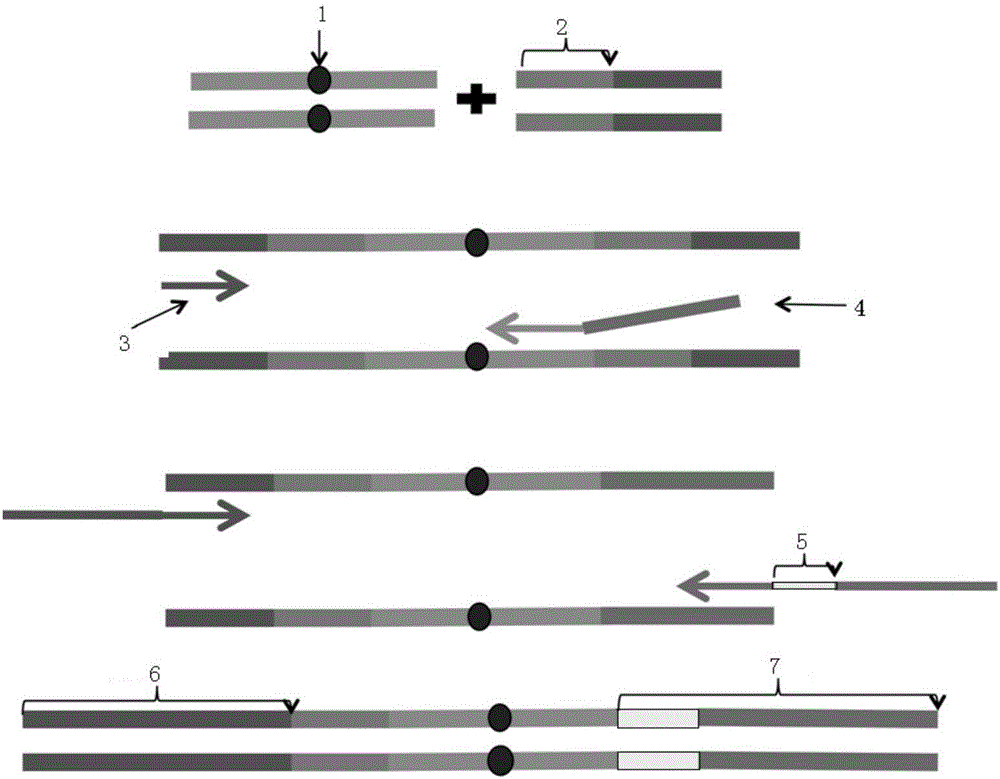
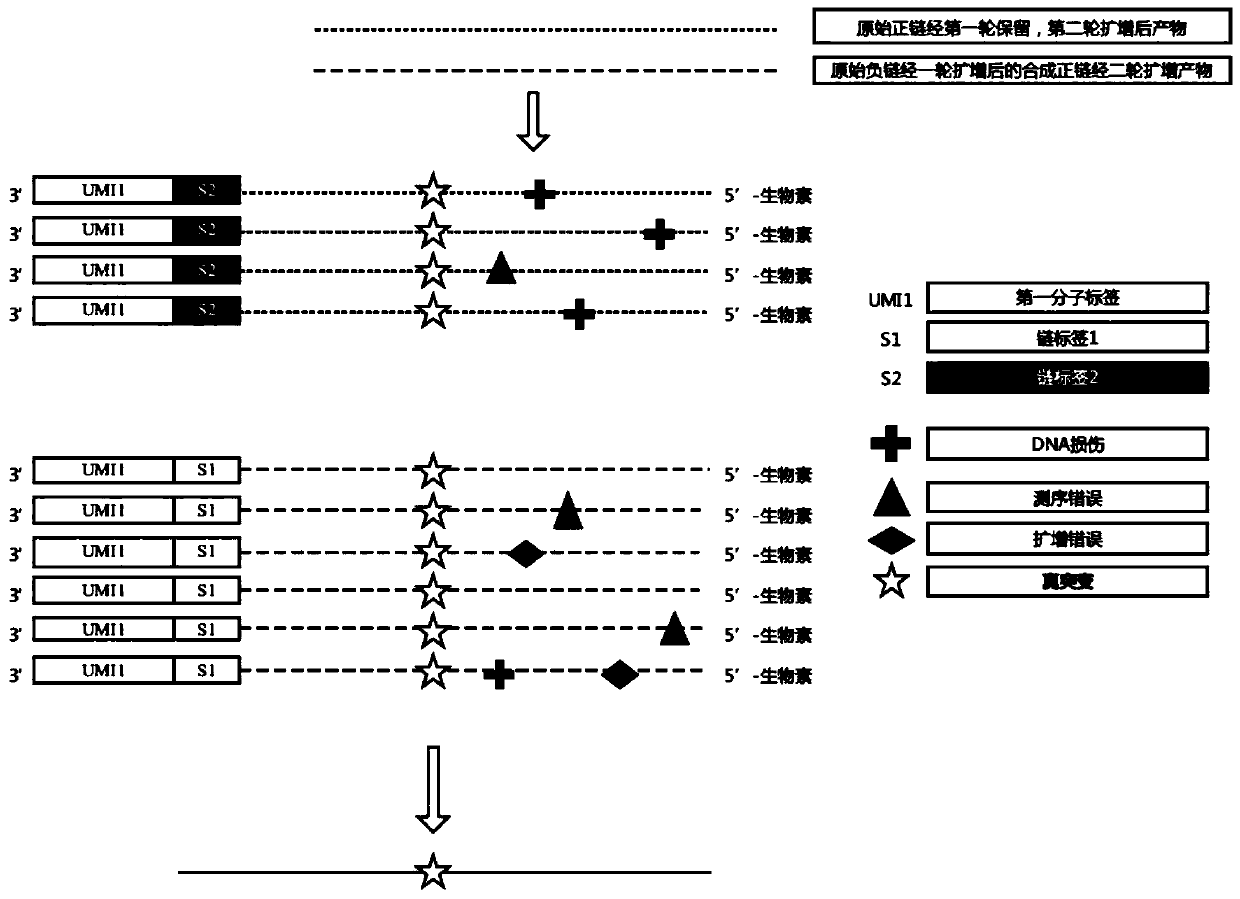
Sequencing joint and preparation method and application thereof in ultra-low frequency mutation detection
ActiveCN105861710AImprove detection accuracyNucleotide librariesMicrobiological testing/measurementMutation detectionDouble strand
The invention provides a sequencing joint and a preparation method and application thereof in ultra-low frequency mutation detection. The sequencing joint comprises a library amplification primer sequence, a target fragment amplification primer sequence and an error prompting sequence which are connected sequentially, the error prompting sequence is close to one side of a target fragment, the library amplification primer sequence is located on one side away from the target fragment, and the error prompting sequence is the sequence of a known base sequence. The error prompting sequence of the known base sequence is added on one side close to the target fragment, so that the error prompting sequence can add a specific foreign marker on each double-strand DNA template. Following sequencing data of the target fragment can be obtained conveniently, mutation introduced in the sequencing or library amplification step is screened or removed according to the fact whether the sequencing sequences have same error prompting sequences, then the sites with variation in the same positions of the two chains are determined to be real mutation, and the site with the mutation on one chain is identified as the amplification or sequencing error, so that the mutation detection efficiency is improved.
Owner:BEIJING KEXUN BIOTECH CO LTD
Transcriptome-based tumor neoantigen identification method
ActiveCN108491689AImprove accuracyComprehensive identificationSequence analysisSpecial data processing applicationsSample sequenceTumor-specific antigen
The invention discloses a transcriptome-based tumor antigen identification method. The method comprises four steps of: obtaining an RNA sample of a patient tumor tissue, and carrying out library construction and amplification on the RNA sample to obtain an RNA sample sequencing result of the tumor tissue; aligning short read segments of the RNA sample sequencing result to a human reference genometo obtain an RNA alignment result; calculating gene expression quantity according to the RNA alignment result, and carrying out mutation detection and prediction of fusion gene events according to theRNA alignment result; and predicting transcriptome HLA typing according to the alignment result, wherein calculation of the gene expression quantity, mutation detection and prediction of the fusion gene events are carried out according to a specified order or simultaneously carried out; and using the gene expression quantity of a transcriptome sample, depth of transcriptome mutation sites in a whole-exon sequencing sample and binding force of neonatal short peptides and the patient HLA typing as an analysis result to submit the same to a downstream analyst. The invention provides the method capable of identifying a tumor-specific antigen of an individual sample from tumor patient transcriptome NGS data.
Owner:HANGZHOU NEOANTIGEN THERAPEUTICS CO LTD
Allele-specific mutation detection assay
InactiveUS20060088872A1Rapid responseDifference in efficiencyHydrolasesMicrobiological testing/measurementSpecific detectionOligomer
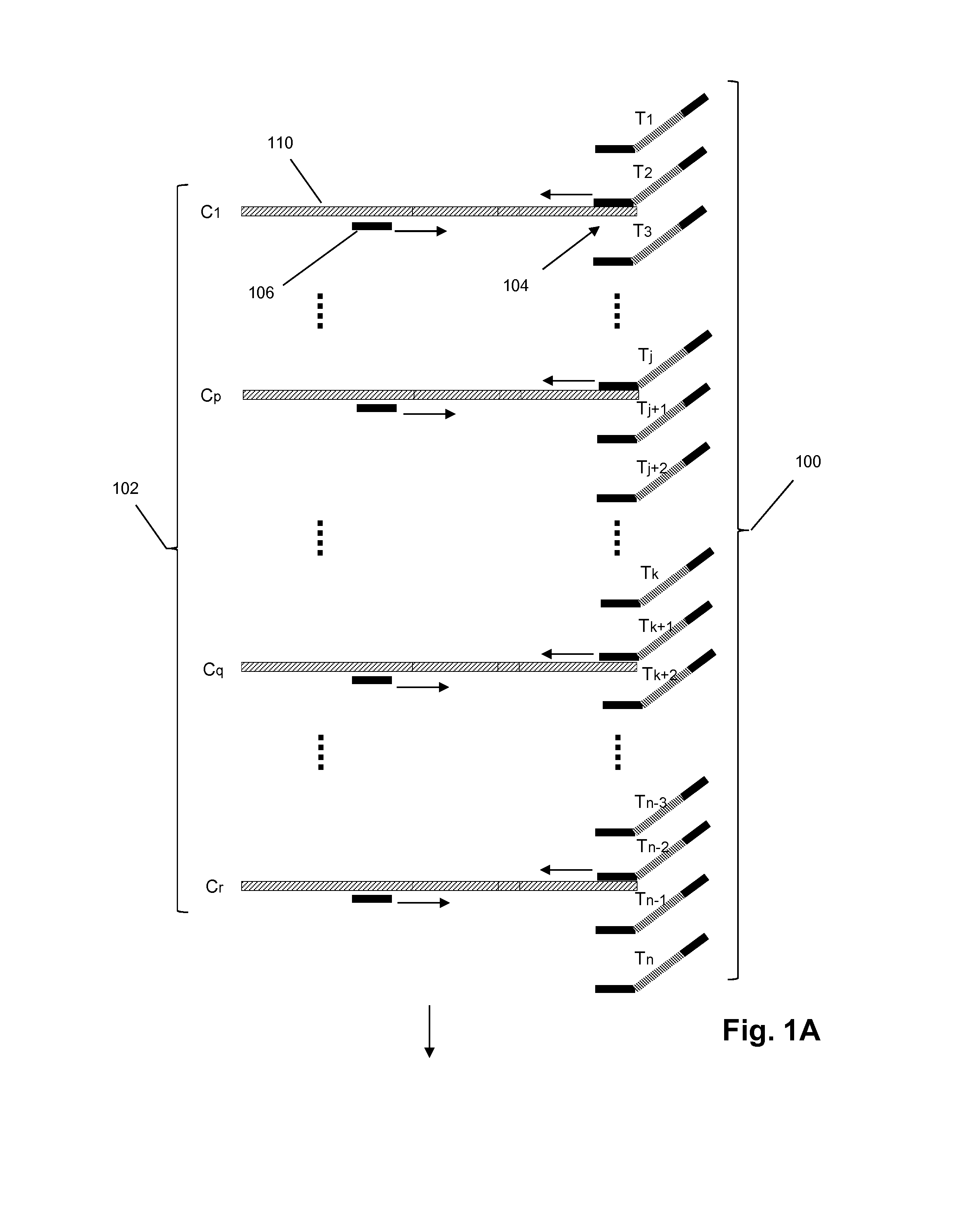
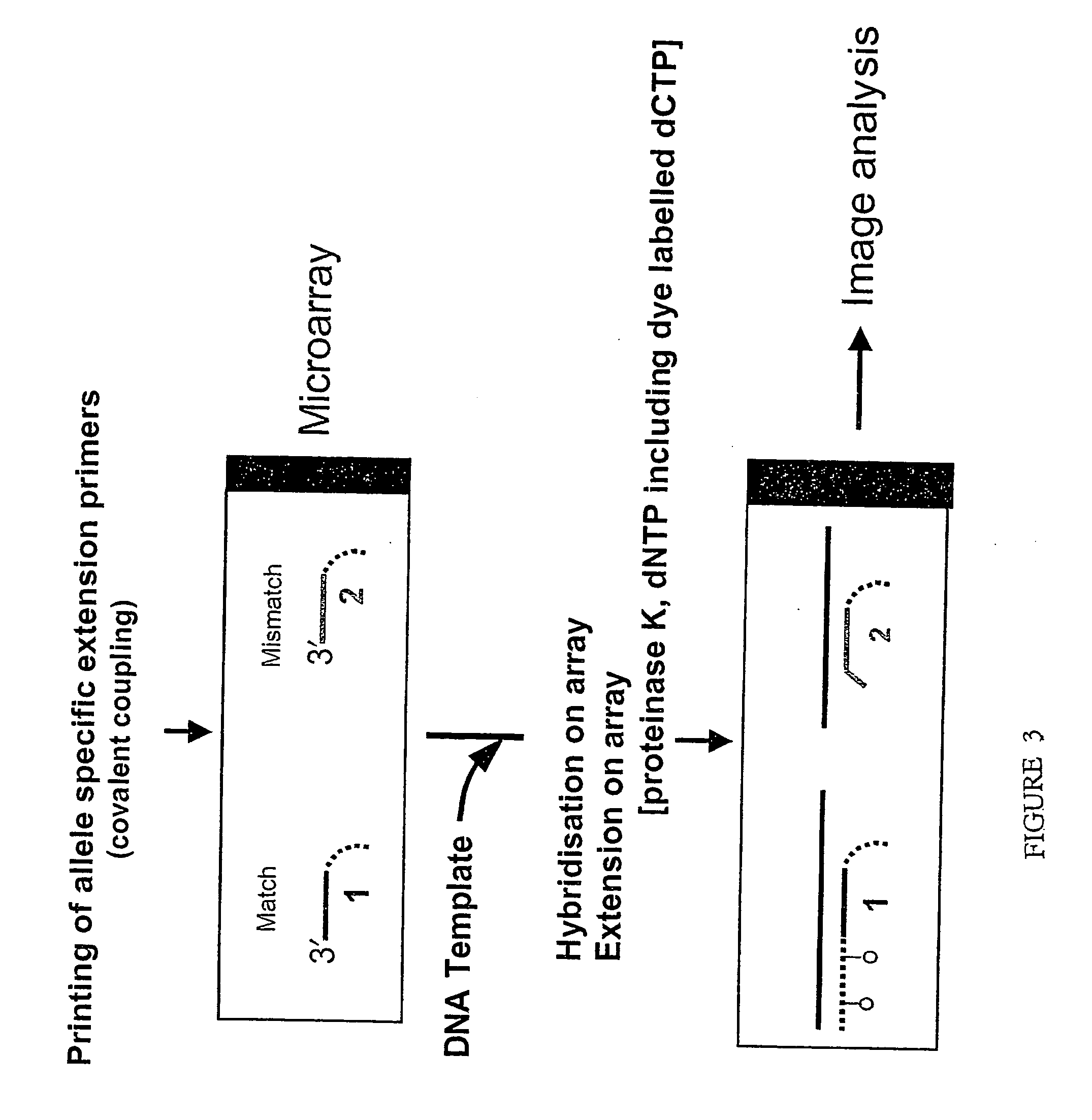
The present invention relates to a method of detecting a base at a pre-determined position in a nucleic acid molecule by performing enzymatic detection reactions using base-specific detection oligomers, where each oligomer is specific for a particular base at the predetermined position, and then comparing the enzymatic detection reactions to determine which base is present at the position, with an enzyme-disabling agent being present during the enzymatic detection reaction. In preferred embodiments the enzymatic detection reaction is an oligomer elongation extension reaction, catalysed by, among others, polymerase or ligase. Also disclosed are methods of performing the assay in a liquid phase and on microarrays.
Owner:AHMADIAN AFSHIN +1
Low-frequency gene fusion detection method and device
ActiveCN106676182ASolving the False Negative/False Positive ProblemBioreactor/fermenter combinationsBiological substance pretreatmentsSmall fragmentMagnetic bead
The invention discloses a low-frequency gene fusion detection method and a low-frequency gene fusion detection device. The detection method comprises the following steps: S1, extracting cfDNA from whole blood; S2, performing tail end repairing on the cfDNA and adding A basic group at the 3' end; S3, connecting a joint containing a random label sequence; S4, designing a multiplex-PCR primer and performing target area capture; S5, performing magnetic bead purification on the PCR product in the S4, removing small fragment DNA not subjected to non-specific amplification and a primer dimer, and introducing an index sequence to obtain a ctDNA ultralow-frequency mutation library; S7, performing computer sequencing; S8, performing clustering analysis on data after library sequencing through the random label sequence; and S9, entering variation detection analysis after the quality control of the remaining data is qualified. By application of the technical scheme of the invention, the low-frequency mutation detection sensitivity is improved.
Owner:BEIJING NOVOGENE TECH CO LTD
EGFR gene 20 exon T790M and C797S mutation detection primers, probes and method
InactiveCN107083438AReduce distractionsIncrease the Tm valueMicrobiological testing/measurementDNA/RNA fragmentationSpecific detectionMutation detection
The invention discloses a primer pair for specific amplification of EGFR gene 20 exon T790M and C797S loci. The sequence of the primer pair is shown in SEQ ID No:1-2. The invention further discloses a probe set for specific detection of the T790M and C797S loci. The probe set comprises a wild type probe and a mutant type probe, and the sequences are shown in SEQ ID No:3-7. The invention further discloses a reagent kit comprising the primer pair and the probe set, and a method for adopting the primer pair and the detection set for detecting EGFR gene 20 exon T790M and C797S mutation. By designing the specific primers and probes, the 3' end of the probes is connected with a minor groove binder and a non-fluorescent quenching group, the interference of background signals is lowered, hybridization of the probes and a template is stabilized, a Tm value of the probes is increased, and therefore the mutation detection sensitivity is improved.
Owner:上海捷易生物科技有限公司
Double-probe gene mutation detecting method based on allele special amplification as well as special chip and kit thereof
ActiveCN101619352AEasy to detectAvoid the hassle of markingMicrobiological testing/measurementGenotypeOligonucleotide
The invention relates to a method for identifying gene mutation types in the field of gene analysis as well as a special chip and a kit thereof. The gene mutation detecting method comprises the following steps: taking a genome to be detected from a human tissue as a template, carrying out multiple allele special PCR amplification by a primer group that is designed aiming at special mutant sites and DNA polymerase without 3'-5' end exonclease activity, then hybridizing the obtained PCR product and an oligonucleotide probe (allele special probe) on the gene chip, and confirming mutation types of all gene sites according to the hybridizing result. The allele special probe is designed aiming at special gene types of gene mutant sites to be detected. The invention can detect gene mutations in comprehensive, systemic and high-flux ways and has light environmental pollution as well as simple and rapid operation compared with PCR-RFLP and a sequencing method.
Owner:CENT SOUTH UNIV
Method for single nucleotide polymorphism and mutation detection using real time polymerase chain reaction microarray
ActiveUS20120088294A1Maximum efficiencyHigh strengthBioreactor/fermenter combinationsHeating or cooling apparatusNucleotideMutation detection
A method and apparatus for real-time, simultaneous, qualitative measurement of one or more single nucleotide polymorphisms in one or more target nucleic acids is provided. This method involves combining a polymerase chain reaction (PCR) technique with an evanescent wave technique.
Owner:HONEYWELL INT INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com