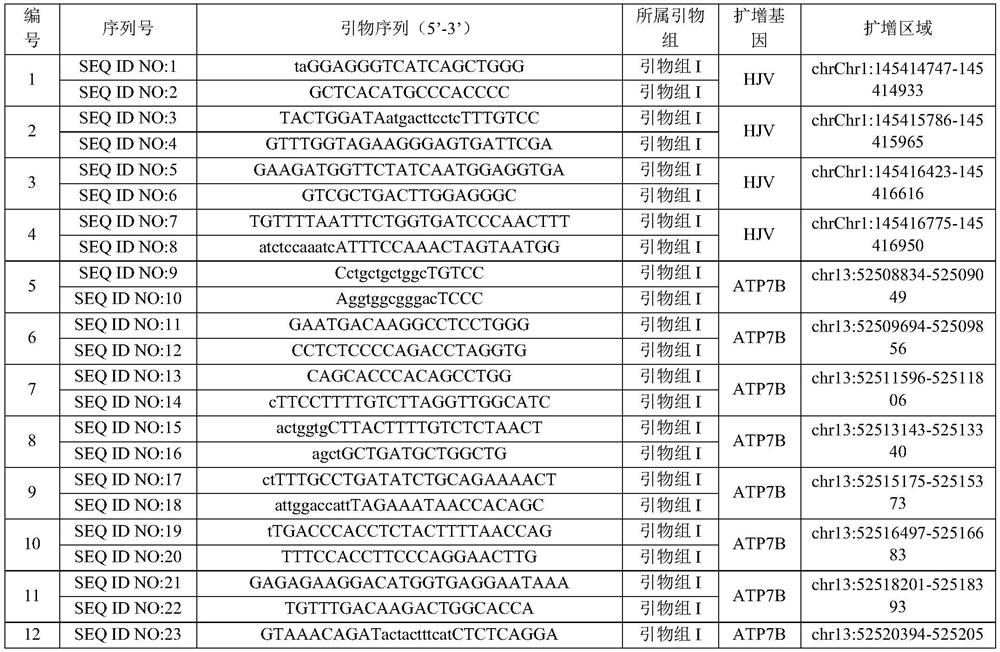
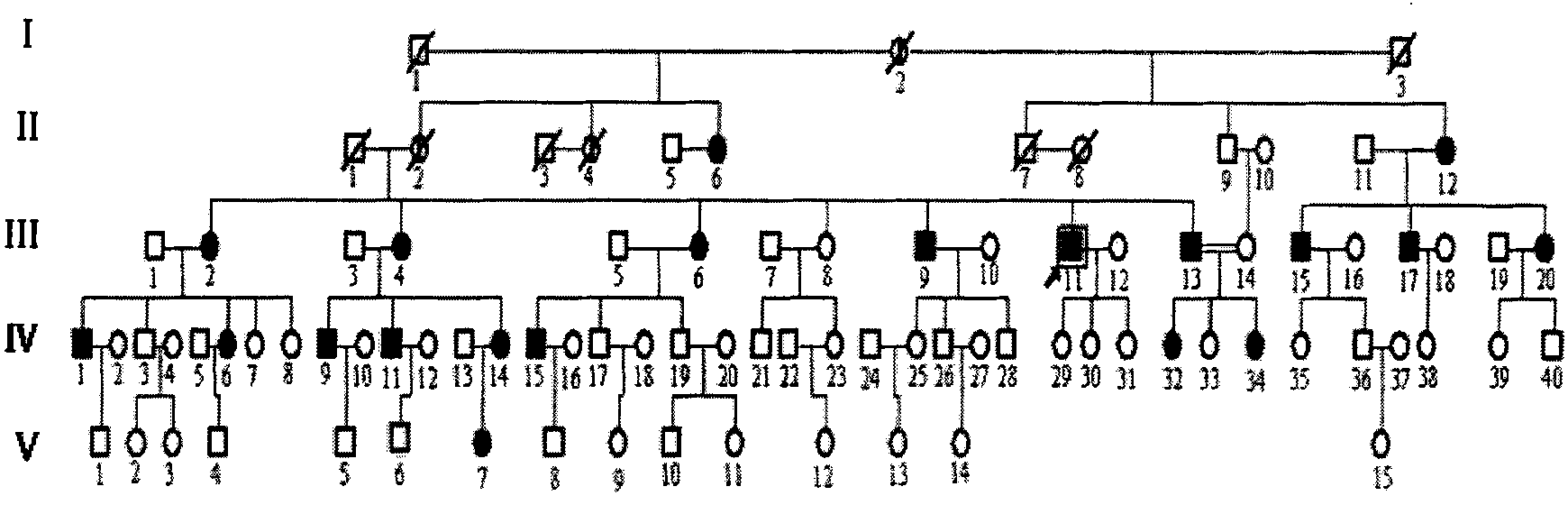
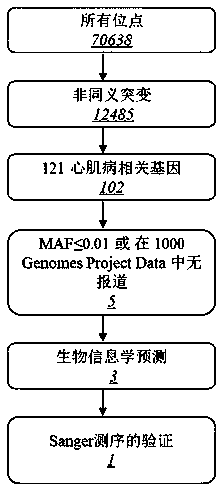
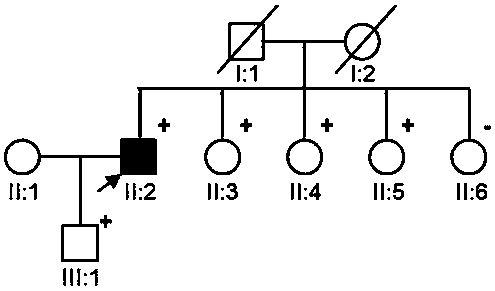
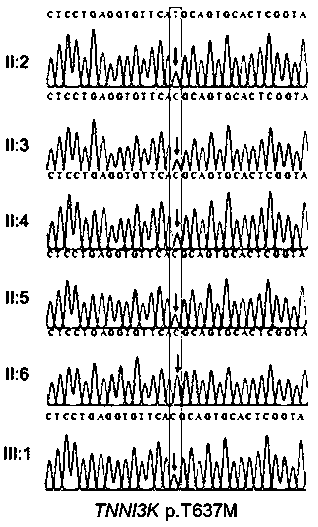
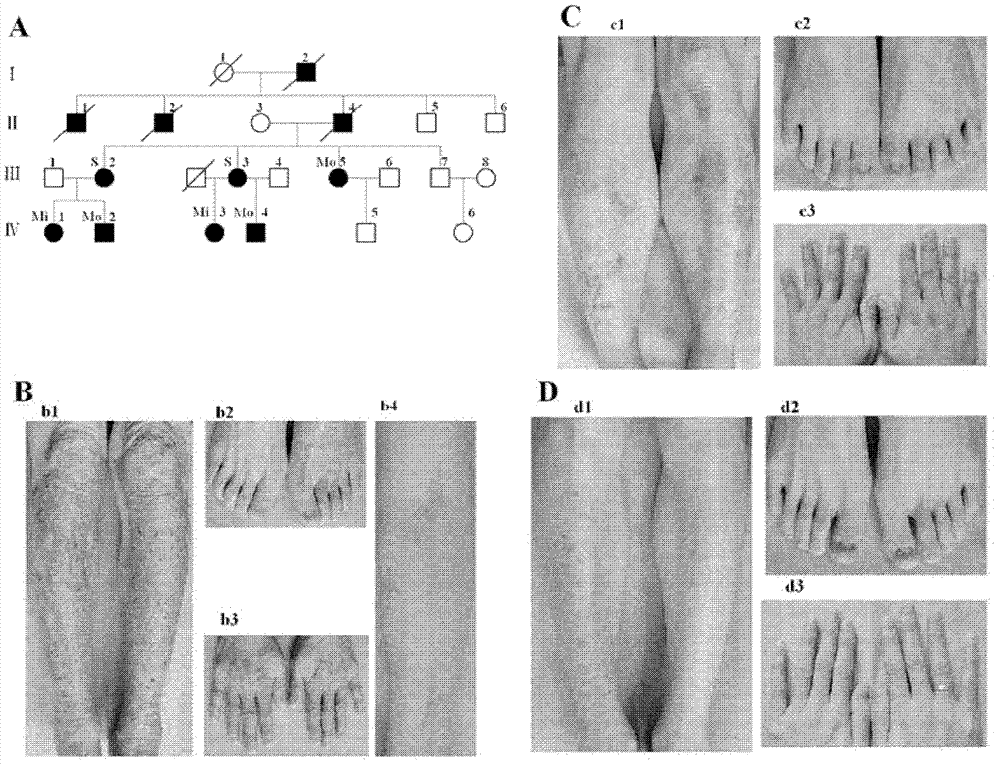
Patents
Literature
138 results about "New mutation" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
New mutation. new mu·ta·tion. redundant term for a heritable trait present in the offspring but in neither parent, that is, not a preexisting mutant form inherited.
Method and device for detecting single sample body cell mutation sites in abnormal tissue and storage medium
ActiveCN107491666AHigh sensitivityStrong specificityProteomicsGenomicsSingle sampleMutation detection
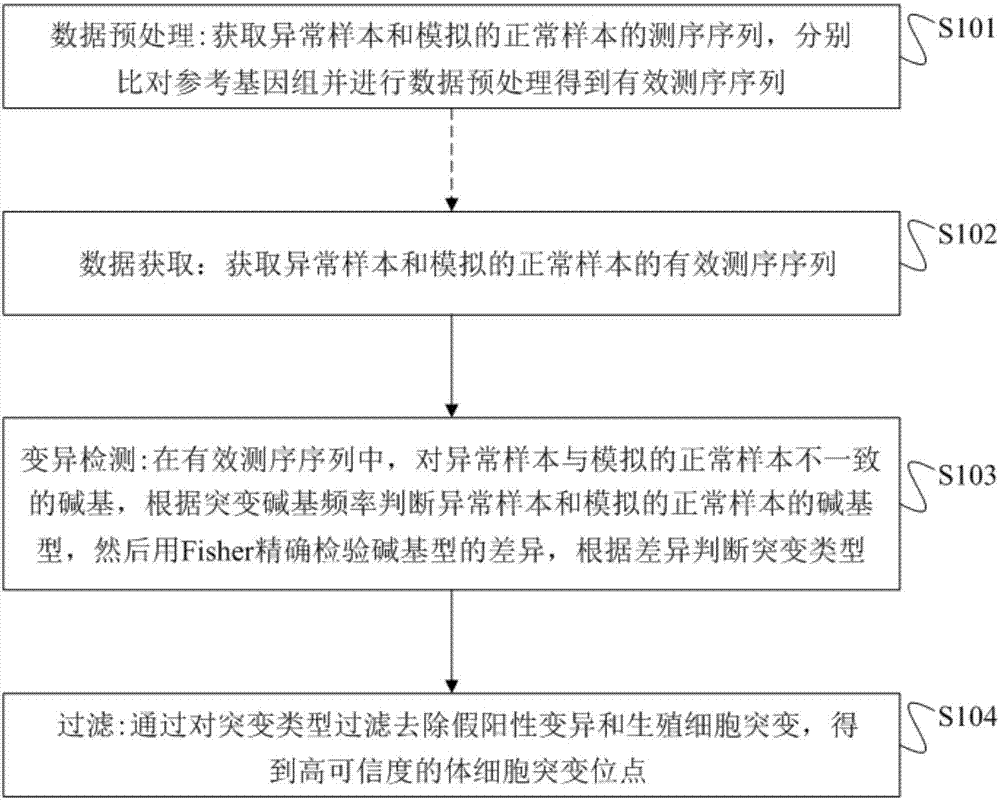
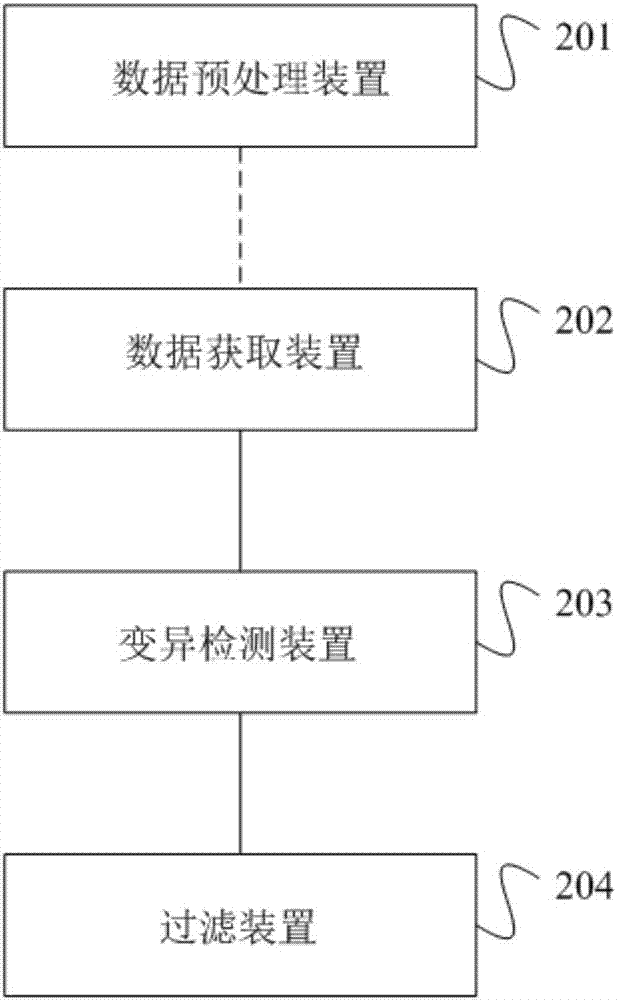
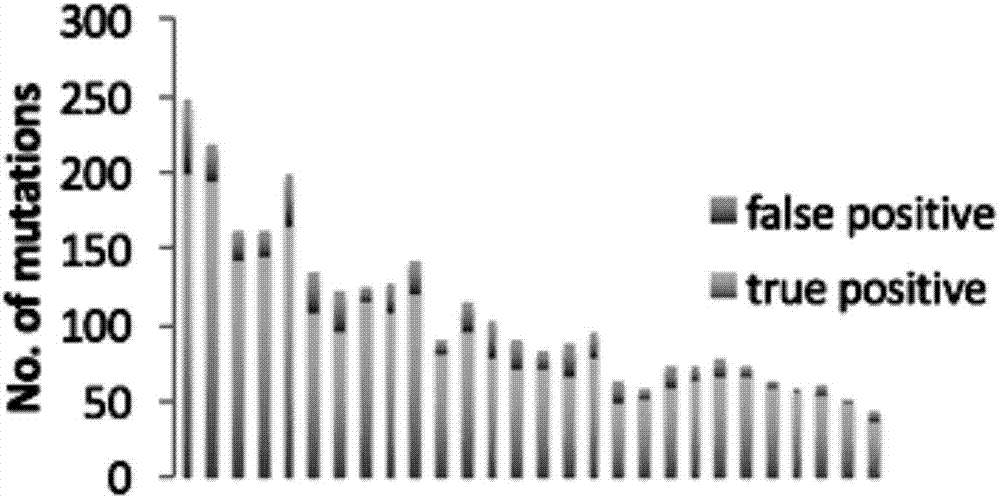
The invention provides a method and device for detecting single sample body cell mutation sites in abnormal tissue and a storage medium. The method includes the following steps that the effective sequencing sequence of an abnormal sample and the effective sequencing sequence of a simulated normal sample are obtained; in the effective sequencing sequences, basic groups, different from basic groups of the simulated normal sample, of the abnormal sample are obtained, according to the mutation basic group frequency, the types of the basic groups of the abnormal sample and the simulated normal sample are judged, the FISHER is then used for accurately detecting the difference of the types of the basic groups, and according to the difference, the mutation type is judged; by filtering the mutation types, false positive mutation and germ cell mutation are removed, and high-reliability somatic cell mutation sites are obtained. The method has the advantages of being high in sensitivity and specificity, has high sensitivity in mutation detection of known mutation genes, and new mutation genes can be found.
Owner:深圳裕康医学检验实验室
Probes, method and chip for detecting alpha and/or beta-thalassemia mutation based on whole-gene capture sequencing and application of such probes, such method and such chip
ActiveCN106591441AEnables detection of deletions in large regionsMicrobiological testing/measurementDNA/RNA fragmentationBeta thalassemiaNew mutation
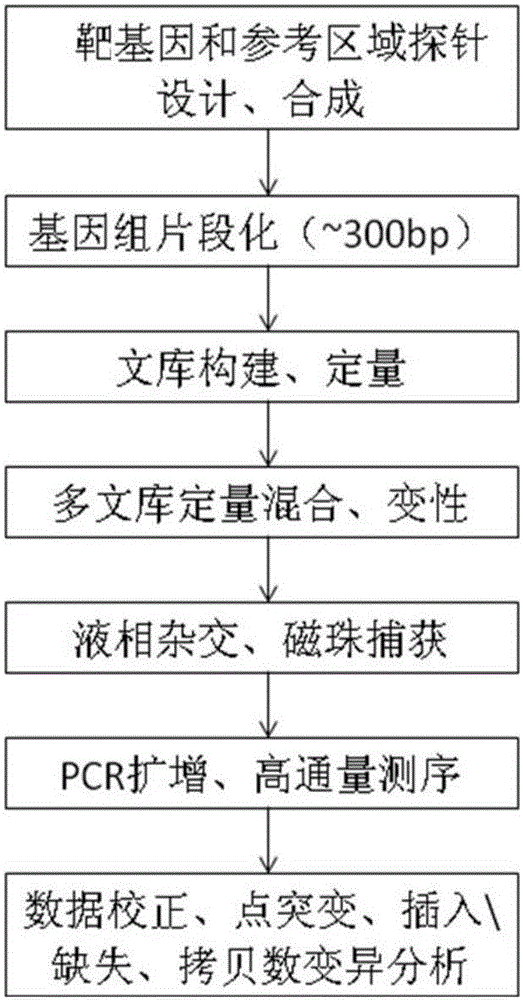
The invention provides primers, a method and a chip for detecting alpha and / or beta-thalassemia point mutation and deletion mutation based on whole-gene capture sequencing and application of such primers, such method and such chip. The primers, the method, the chip and application thereof have the advantages that through designing of capture probes, relevant genes involved in alpha-thalassemia and beta-thalassemia are enriched and all mutation information including SNP and indel in full-length sequences of genes is detected; through addition of autosome, X-chromosome and Y-chromosome regions as well as upstream and downstream regions of coded genes as references, structure variations such as SNV and CNV are detected; compared with existing various hotspot mutation site detection technologies, the method is capable of detecting hotspot mutation information as well as some rare mutations and undiscovered new mutation types to detect and analyze full-length sequence specificity of target genes, fully covers the mutation types and makes up the defect that a conventional detection method easily causes missing detection of low-frequency mutations and rare mutations greatly.
Owner:SHENZHEN E GENE TECH
Targeting-based new generation sequencing deafness gene detection set and kit, and detection method
InactiveCN106399504AWide coverageImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationData informationTest sample
The invention discloses a targeting-based new generation sequencing deafness gene detection set and kit, and a detection method. The deafness gene detection set comprises 258 genes, 81 non CDS regions and a whole mitochondrial group. The detection method comprises the steps: designing primers for a part of or all of deafness disease genes and loci; with a to-be-tested sample DNA as a template, carrying out PCR amplification with the primers, and constructing a deafness detection gene library based on the amplified product; and according to the deafness detection gene library, establishing a sequencing template, carrying out high-throughput sequencing, and analyzing sequencing data information. The invention also discloses the related kit. Not only can conventional known mutations be detected out, but also new mutation types also can be detected out; in addition, sequencing and analysis of a large number of objective regions also can be completed within a short period of time, and positions, possible to generate pathogenic mutation, of all exons and regulatory regions of the hundreds of genes associated with deafness are subjected to related sequencing and analyzing.
Owner:SUZHOU BASECARE MEDICAL DEVICE CO LTD
PKD1 gene mutation detection kit and detection method
ActiveCN104531883AComprehensive detection rangeLow mismatch rateMicrobiological testing/measurementSequence analysisNephrosisPKD1
The invention provides a primer set, a kit and a detection reaction system for detecting PKD1 gene mutation through the long fragment PCR and high-throughput sequencing technology, a non-diagnosis-purpose method for external PKD1 gene mutation detection, a non-diagnosis-purpose method for external PKD1 gene analysis and a method for detecting new mutation sites on the PKD1 gene. According to the method, the primer set is used for carrying out long fragment PCR amplification on the PKD1 gene of a sample, and detecting or analyzing is carried out through high-throughput sequencing. The autosomal dominant genetic polycystic nephrosis (adult polycystic nephrosis) can be diagnosed through the assistance of a detection result, previous unknown new mutation on a plurality of PKD1 real genes can be obtained and supplied to doctors or researchers so that the relevance between the mutation and the adult polycystic nephrosis can be studied.
Owner:北京圣谷智汇医学检验所有限公司
Biological information analysis method for human single-gene genetic disease detection
InactiveCN110931081AAnalyze content clearlyEasy to analyzeProteomicsGenomicsCandidate Gene Association StudyGenotype
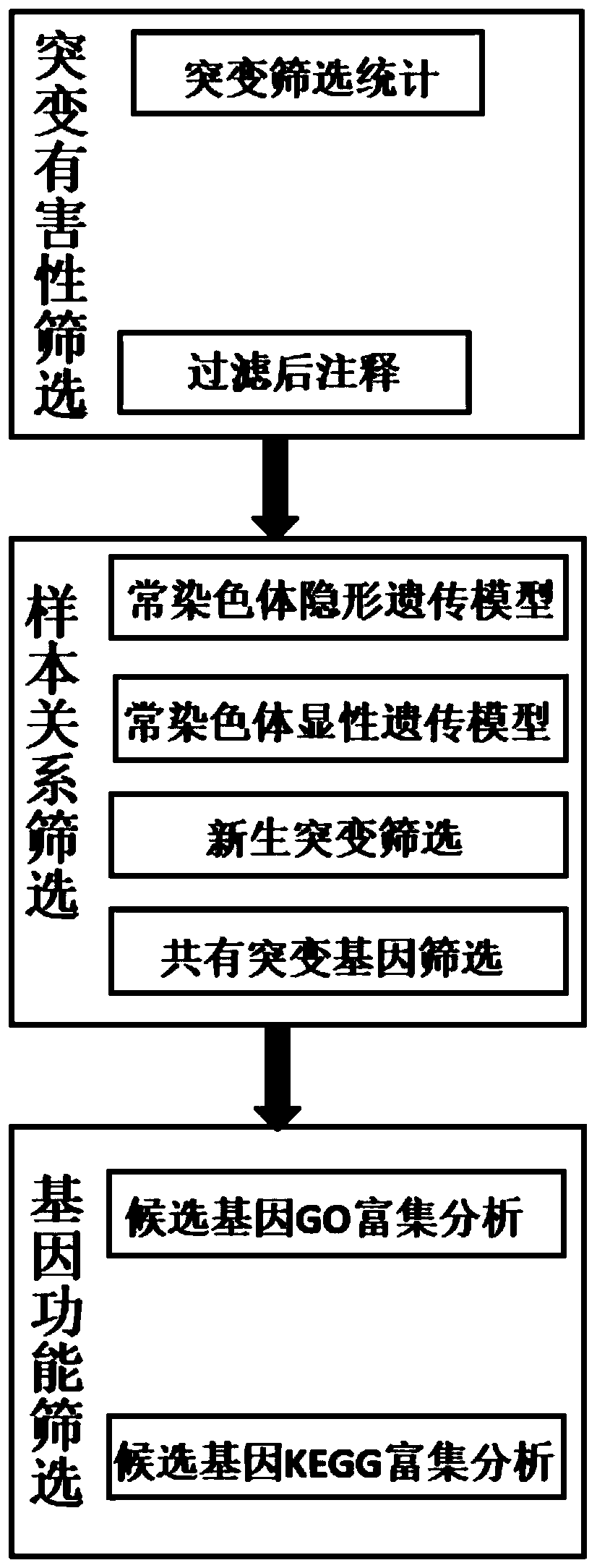
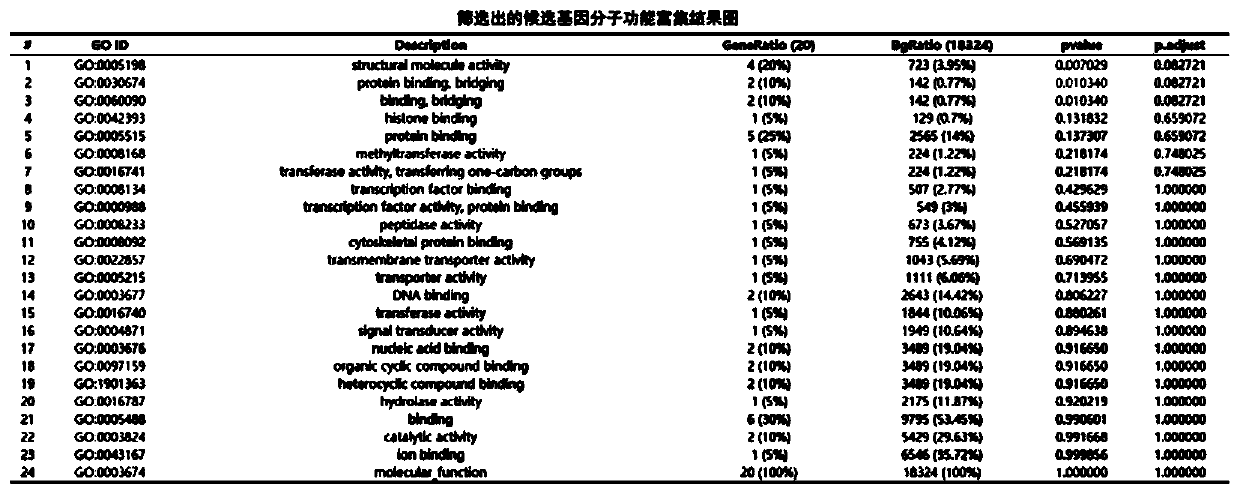
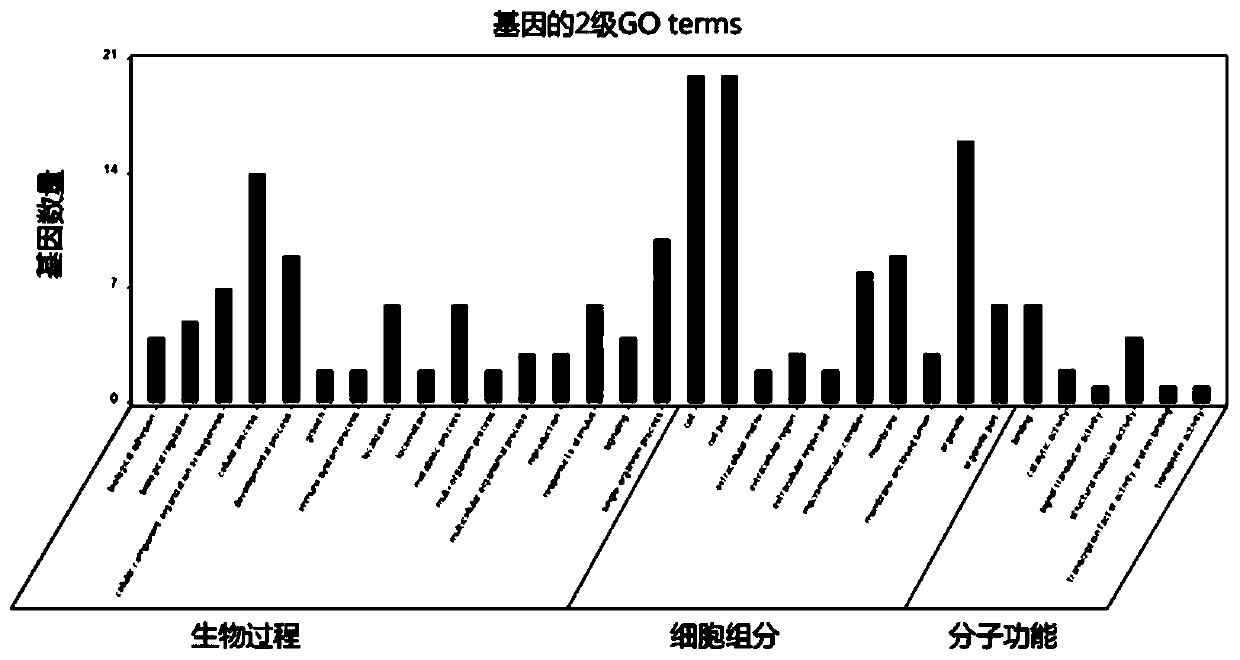
The invention provides a biological information analysis method for human single-gene genetic disease detection. The method comprises the following steps: S1, screening mutation harmful sites; S2, screening the sample relationship of human single-gene genetic disease detection; S3, performing gene function screening. According to the method, harmful mutation sites can be quickly and clearly screened from human exon sequencing data; the annotation analysis on the detected SNP, InDel and other genomic variations and an external database is carried out to determine the genomic position, variationfrequency, protein harmfulness, genotype heterozygosity, functional pathways and other information of the variations; the candidate genes are screened by using an autosomal invisible genetic model, an autosomal dominant genetic model, new mutation screening and common mutation gene screening, so the candidate genes are determined, GO and KEGG enrichment analysis is performed on the candidate genes, and powerful evidences are provided for determining the candidate genes related to diseases.
Owner:GUANGZHOU GENE DENOVO BIOTECH
CYP4V2 gene mutant and application thereof
InactiveCN103374575AEfficient screeningScreening treatment is effectiveBioreactor/fermenter combinationsBiological substance pretreatmentsBIETTI CRYSTALLINE DYSTROPHYCYP4V2 gene
Owner:THE FIRST AFFILIATED HOSPITAL OF THIRD MILITARY MEDICAL UNIVERSITY OF PLA
Novel pathogenetic gene mutation of hypertrophic cardiomyopathy and use thereof
InactiveCN1865440AMicrobiological testing/measurementGenetic engineeringHypertrophic cardiomyopathyNew mutation
The invention relates to several new mutations of pathogenic genes for hypertrophic cardiomyopathy, and their use in the treatment and diagnosis of hypertrophic cardiomyopathy.
Owner:FUWAI HOSPITAL OF CARDIOVASCULAR DESEASE CHINESE ACAD OF MEDICAL SCI
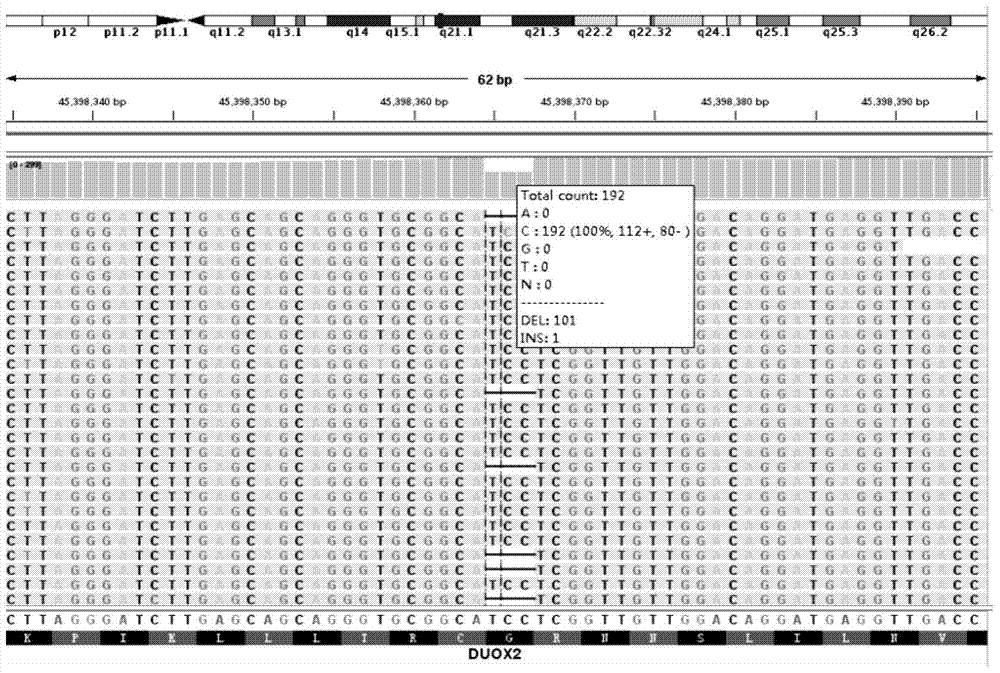
Detection probe for DUOX2 gene mutation and detection method thereof
InactiveCN104120187AQuick checkComprehensive detection effectMicrobiological testing/measurementGenetic material ingredientsDUOX2 geneNew mutation
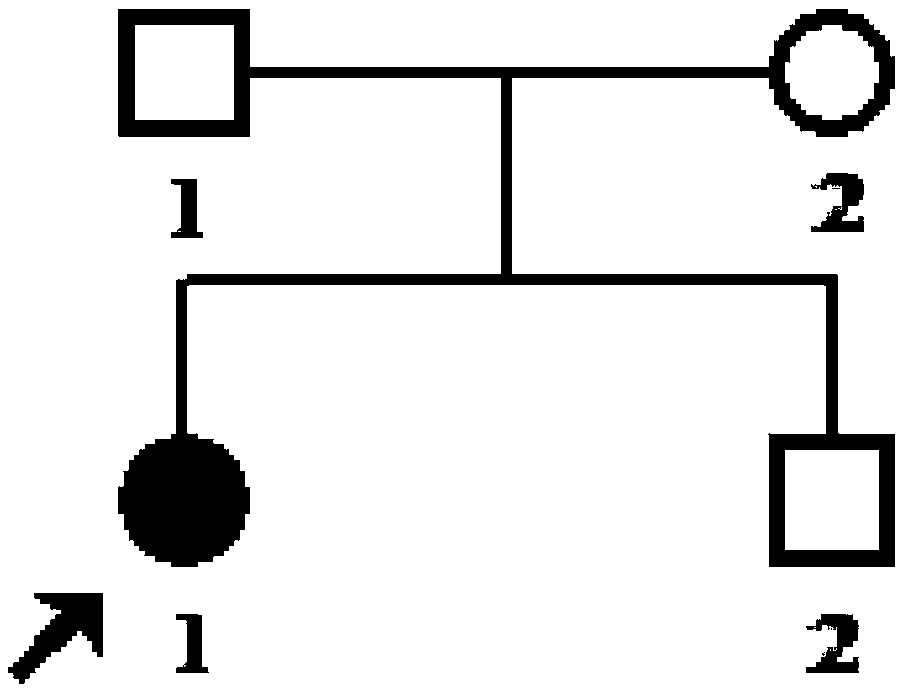
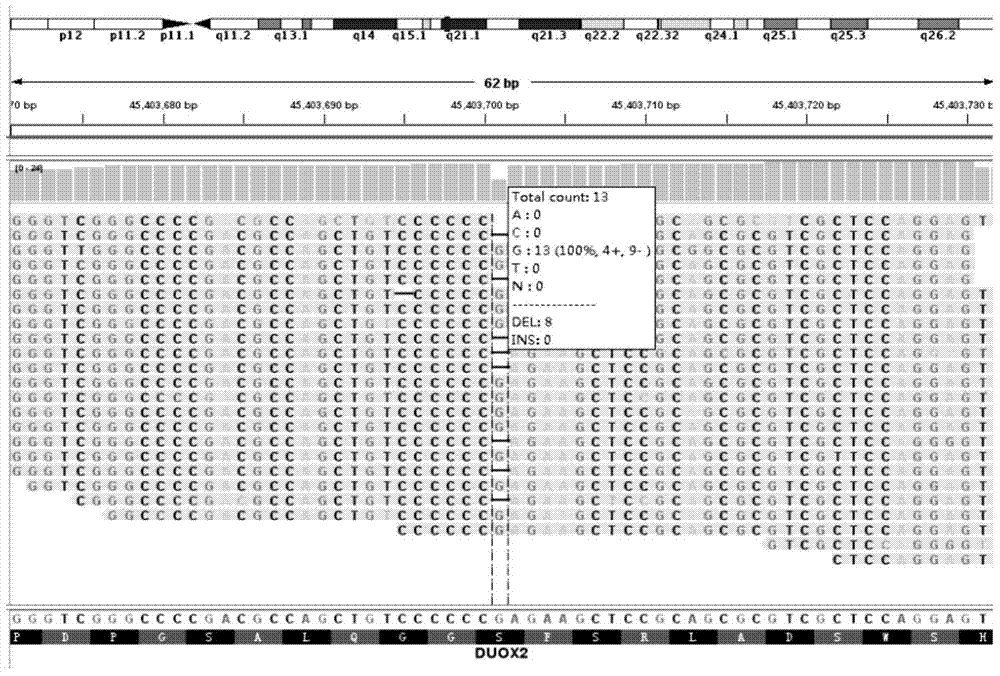
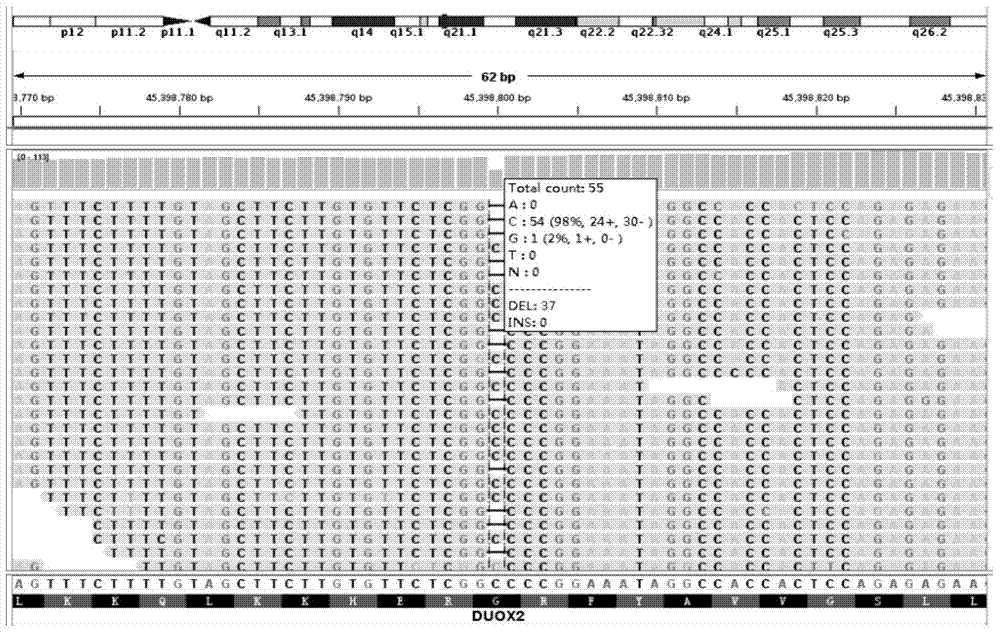
The invention discloses a detection probe for DUOX2 gene mutation and a detection method thereof, particularly relates to a method for detecting DUOX2 gene mutation related to a congenital hypothyroidism (CH) pathogenic gene, and belongs to the technical field of molecular biology. A PCR primer sequence of a DUOX2 gene for detecting the CH is a nucleotide sequence as shown in SEQ ID No.1-SEQ ID No.18 in the sequence table. The detection probe has the advantages that the DUOX2 gene causing the CH is detected by utilizing new generation sequencing, and 24 new mutation sites which are not yet reported home or abroad are discovered. The detection probe applied to clinical gene mutation detection is beneficial to etiological analysis and diagnosis of CH, further recognition of mutation spectrum of the CH gene and further research on pathogenesis of CH.
Owner:MATERNAL & CHILD HEALTH HOSPITAL OF GUANGXI ZHUANG AUTONOMOUS REGION GUANGXI ZHUANG AUTONOMOUS REGION
CYP4V2 gene mutant and application thereof
InactiveCN103374574AEfficient screeningScreening treatment is effectiveBioreactor/fermenter combinationsBiological substance pretreatmentsDrugPathogenic genes
The invention provides a CYP4V2 gene mutant and an application thereof, belonging to the field of medical molecular biology. The invention applies to protect the separated new CYP4V2 mutant pathogenic gene of Bietti crystalline dystrophy (BCD) and a mutant polypeptide thereof on the one hand. The gene is characterized in that compared with genes coding wild type CYP4V2 proteins, such as nucleotide sequences shown in SEQ ID NO:1, a nucleic acid sample of the mutant pathogenic gene has the characteristic that the (1027) basic group suffers from T>G mutation. Based on the separated pathogenic mutant gene, the invention further provides a method, system and kit for screening biological samples susceptible to BCD as well as a method for screening medicines for treating or preventing BCD.
Owner:THE FIRST AFFILIATED HOSPITAL OF THIRD MILITARY MEDICAL UNIVERSITY OF PLA
New mutant protein related to dilated cardiomyopathy, new mutant gene and application thereof
ActiveCN112028983AReduce birthPromote research and developmentMicrobiological testing/measurementGenetic engineeringMutated proteinNew mutation
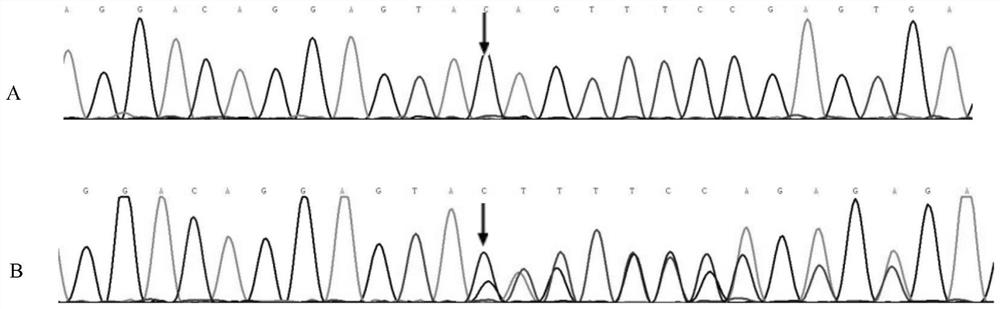
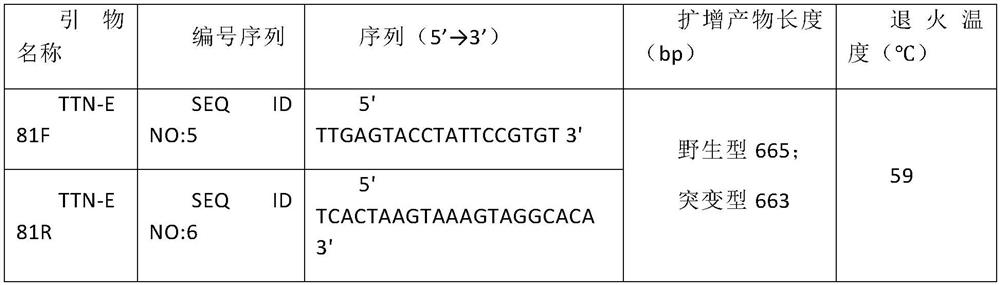
The invention relates to a new mutant protein related to dilated cardiomyopathy, a new mutant gene and application thereof, discloses a mutant protein and a mutant TTN gene, has c.20292-20293 heterozygosis deletion variation compared with a human TTN gene, verifies that the mutation is related to dilated cardiomyopathy through research data, and protects the mutant protein and the gene based on arequest. And a kit for detecting dilated cardiomyopathy is disclosed. The mutant TTN gene provided by the invention can distinguish patients with dilated cardiomyopathy from normal people, and can beused as a biomarker for clinical auxiliary diagnosis of dilated cardiomyopathy. Carriers of the variation can be screened in the early stage, and prenatal and postnatal guidance and genetic counselingare provided for subjects. And a possible drug treatment target is provided for human to overcome the dilated cardiomyopathy.
Owner:百世诺(北京)医学检验实验室有限公司
Attenuated streptococcus suis vaccines and methods of making and use thereof
The present invention provides attenuated S. suis strains that elicit an immune response in animal S. suis, compositions comprising said strains, methods of vaccination against S. suis, and kits for use with such methods and compositions. The invention further provides novel, mutagenically-induced mutations in S. suis genes, which are useful in the production of novel attenuated S. suis bacterial strains.
Owner:BOEHRINGER INGELHEIM ANIMAL HEALTH USA INC
Thalassemia mutant gene detection primer pair and kit, and application and library construction methods thereof
InactiveCN108796061AFacilitate popularization of applicationsReduce testing costsMicrobiological testing/measurementLibrary creationAgricultural scienceThalassemia
Disclosed are a thalassemia mutant gene detection primer pair and kit, and application and library construction methods thereof. The thalassemia mutant gene detection primer pair comprises primer sequences shown as SEQ ID NO: 1-8. The thalassemia mutant gene detection primer pair can be applied to simultaneously detecting a number of mutation types and help discover new mutation types, and when combined with second-generation sequencing, can greatly reduce the detecting cost and improve throughput, thereby achieving significant cost advantages and being beneficial to popularized application ofthalassemia mutant gene detection.
Owner:BGI CLINICAL LAB (SHENZHEN) CO LTD
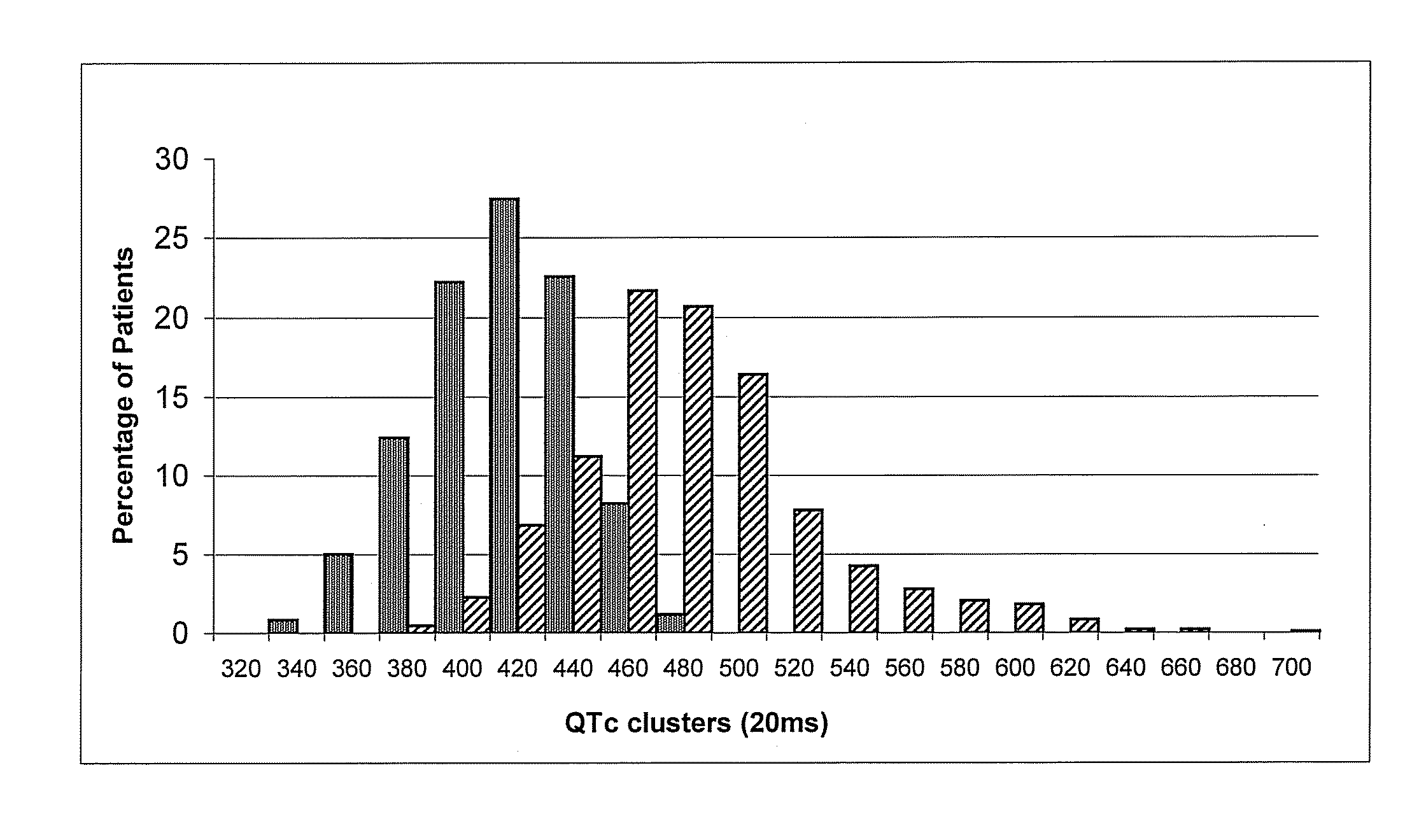
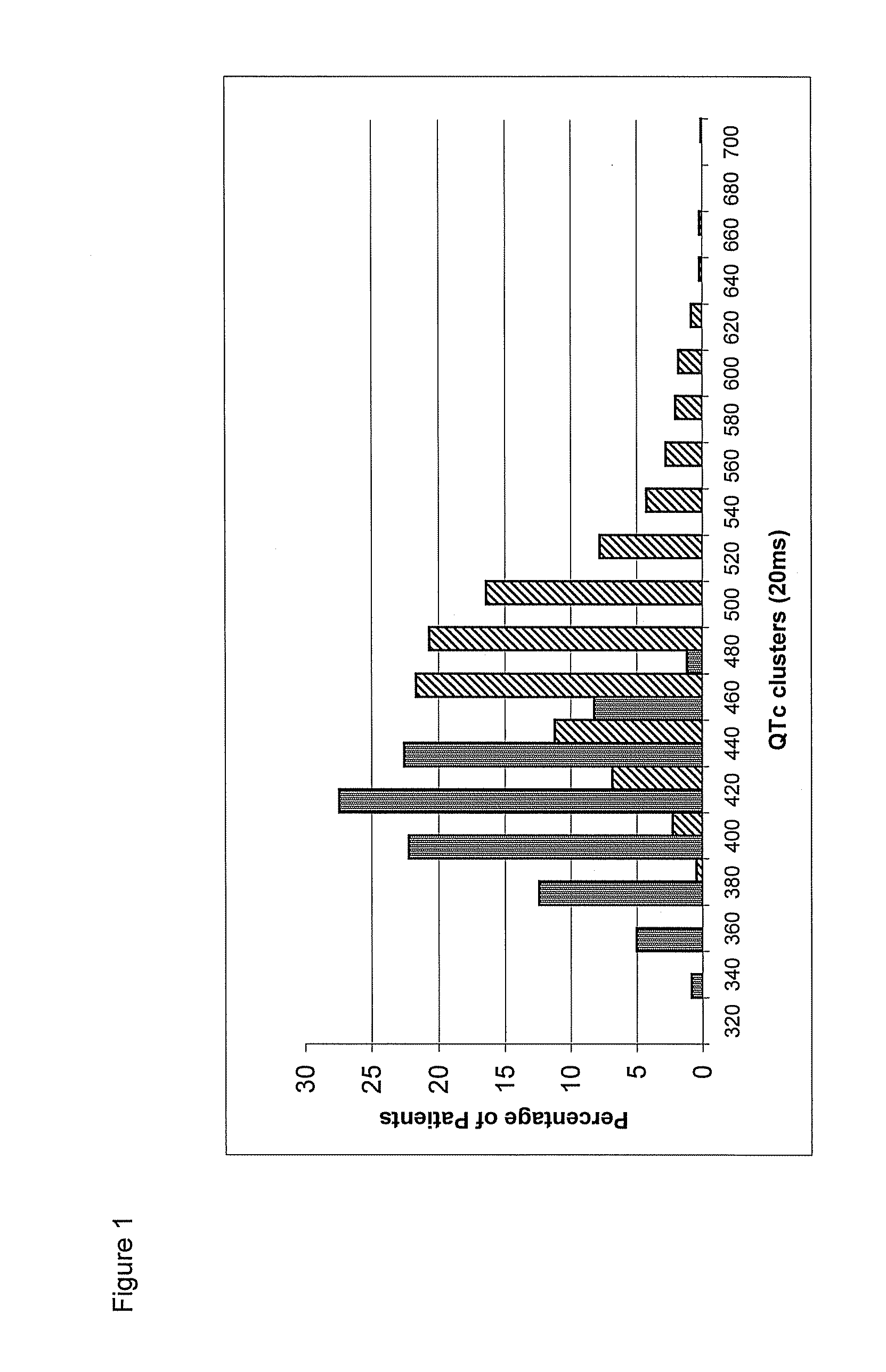
Mutations Associated with the Long QT Syndrome and Diagnostic Use Thereof
The present invention is based on the identification of new mutations in KCNQ1 (also termed KvLQTI), KCNH2 (also termed HERG), SCN5A, KCNE1 (also termed minK), KCNE2 (also termed MiRP) genes that encode ionic channels involved in cardiac electrical activity and are potentially responsible for the Long QT Syndrome. According to a main aspect, the invention relates to nucleic acids, oligonucleotides and polynucleotides and mRNA, containing sequences of KCNQ1, KCNH2 SCN5A, KCNE1, KCNE2 genes and cDNAs in a mutated form and to respective variant proteins thereof. A preferred embodiment of the present invention is represented by a diagnostic method based on the identification of a group of about 70 non-private mutations in the KCNQ1, KCNH2 and SCN5A genes, detected at high frequency. The method, which is able to identify about 40% of the probands, is non exclusively based on identification of mutations that are described and characterized in this invention where said identification has both prognostic and diagnostic value for the Long QT Syndrome.
Owner:FOND SALVATORE MAUGERI CLINICA DEL LAVORO E DELLA RIABILITAZIONE +1
Primer composition, kit composed of primer composition and application
ActiveCN107058538ASimple methodLow costMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceHypertrophic cardiomyopathy
The invention belongs to the technical field of biology, relates to medical molecular diagnosis and biotechnology and relates to a primer composition, a kit composed of the primer composition and application. The primer composition comprises a real-time fluorescence PCR (Polymerase Chain Reaction) primer for detecting a new mutation site c.622 greater than G of a TAZ gene and a Taqman-MGB (Minor Groove Binder) probe sequence. The primer composition can be used for detecting a plurality of case samples and has the characteristics of simplicity, rapidness, accuracy and economical efficiency and the like; a new method is established for clinical early-stage molecular diagnosis and prevention of hypertrophic cardiomyopathy.
Owner:KUNMING UNIV OF SCI & TECH
New mutant disease-causing gene of Alport syndrome, encoded protein and application thereof
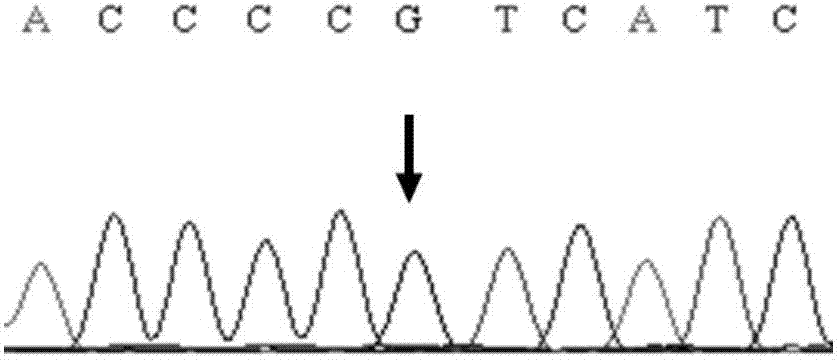
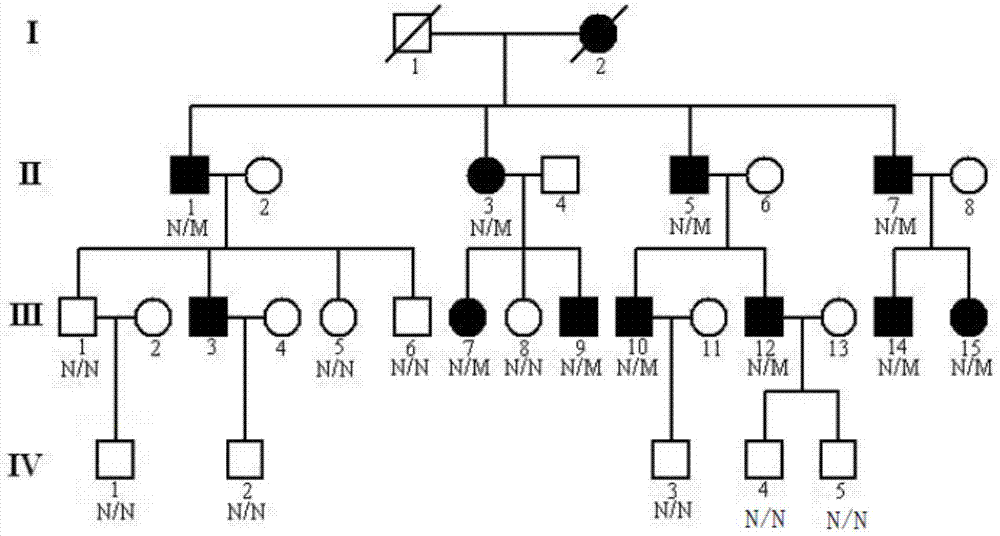
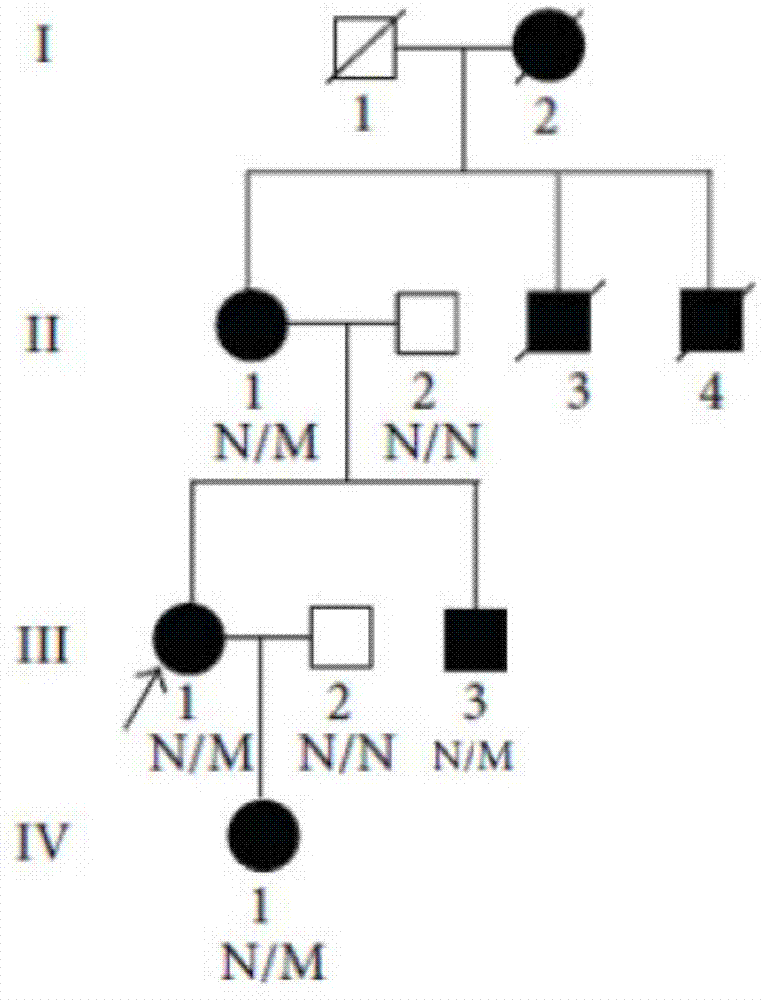
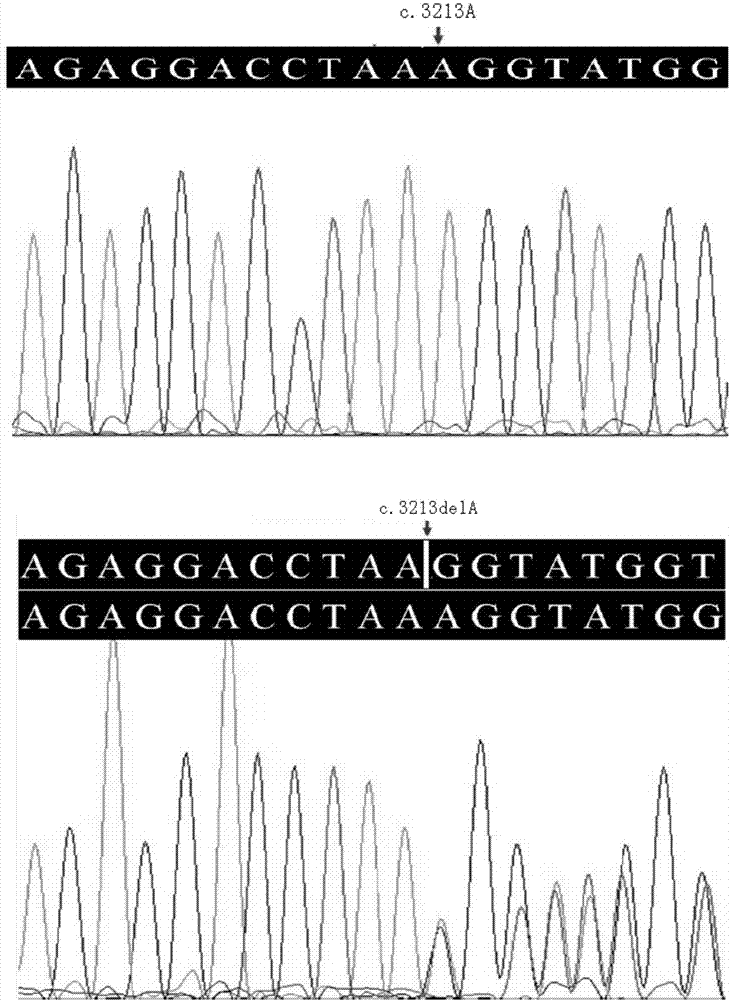
ActiveCN104212806AQuick forecastRapid diagnosisSenses disorderConnective tissue peptidesDisease riskResearch Object
In the invention, two new mutations of two disease-causing genes relative to Alport syndrome are identified through an exon sequencing method. The inventor, by employing two ATS families as research objects, particularly performs exon sequencing and comparison respectively to diseased individuals and non-diseased individuals in the two ATS family and finds a frame shift deletion mutation (c.3213delA,p.Lys1071fs*5) on a COL4A4 gene of one family and a frame shift deletion mutation (c499delC,p.Pro167Glnfs*36) on a COL4A5 gene of the other family, wherein the two frame shift deletion locus are actual disease-causing locus of the two families. On the basis of above, the invention provides a mutant COL4A4 gene and a mutant COL4A5 gene, encoded protein and application thereof and includes carriers, host cells and kits of the mutant COL4A4 gene and / or the mutant COL4A5 gene. By means of the mutant COL4A4 gene and / or the mutant COL4A5 gene, molecular diagnosis and diseasing risk evaluation of the Alport syndrome can be carried out. The two mutant genes and the encoded proteins thereof also can be used as medicine targets for treating the Alport syndrome.
Owner:石家庄华大医学检验实验室有限公司 +1
Primer composition, kit and method for detecting susceptibility gene mutation of hemochromatosis and hepatolenticular degeneration
ActiveCN111647654AInterpretation is clear and objectiveImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationMedicineExon
The invention discloses a primer composition, a kit and a method for detecting susceptibility gene mutation of hemochromatosis and hepatolenticular degeneration. The invention firstly discloses the primer composition for detecting the susceptibility gene mutation of hemochromatosis and hepatolenticular degeneration. The invention further discloses the kit containing the primer composition and themethod for detecting the susceptibility gene mutation of hemochromatosis and hepatolenticular degeneration. The method for detecting the susceptibility gene mutation of hemochromatosis and hepatolenticular degeneration integrates gene mutation and copy number mutation information of all exon and intron cleavage regions of susceptibility genes of hemochromatosis and hepatolenticular degeneration inChinese population, and has the advantages of high detection flux, high specificity and sensitivity, 100% of sequencing coverage, more than 30x of depth, clear and objective gene detection result interpretation, good accuracy and repeatability, low cost, simple operation and easy popularization; and the method can not only detect known high-incidence mutation, but also discover new mutation sites.
Owner:BEIJING FRIENDSHIP HOSPITAL CAPITAL MEDICAL UNIV
Gene relevant mutational site of mitochondrial deaf and application
InactiveCN101875938AHelps to rule outAids in diagnosisMicrobiological testing/measurementFermentationGenomicsNew mutation
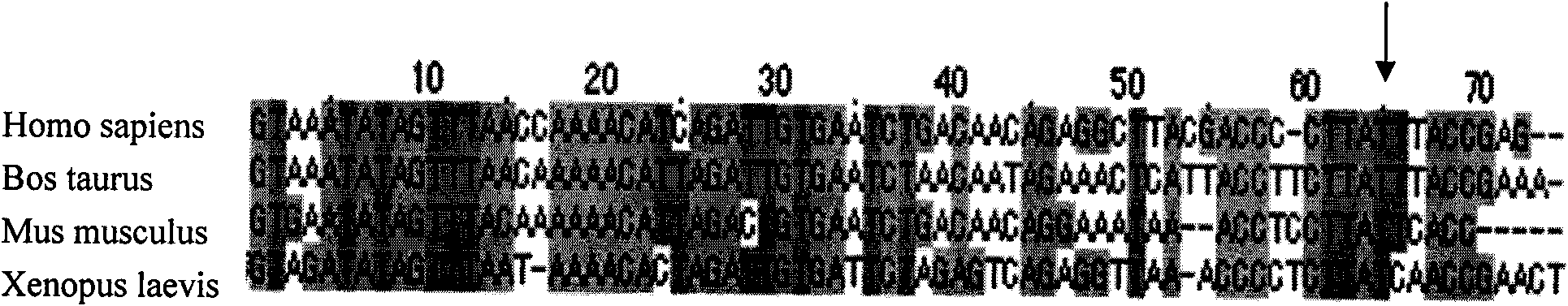
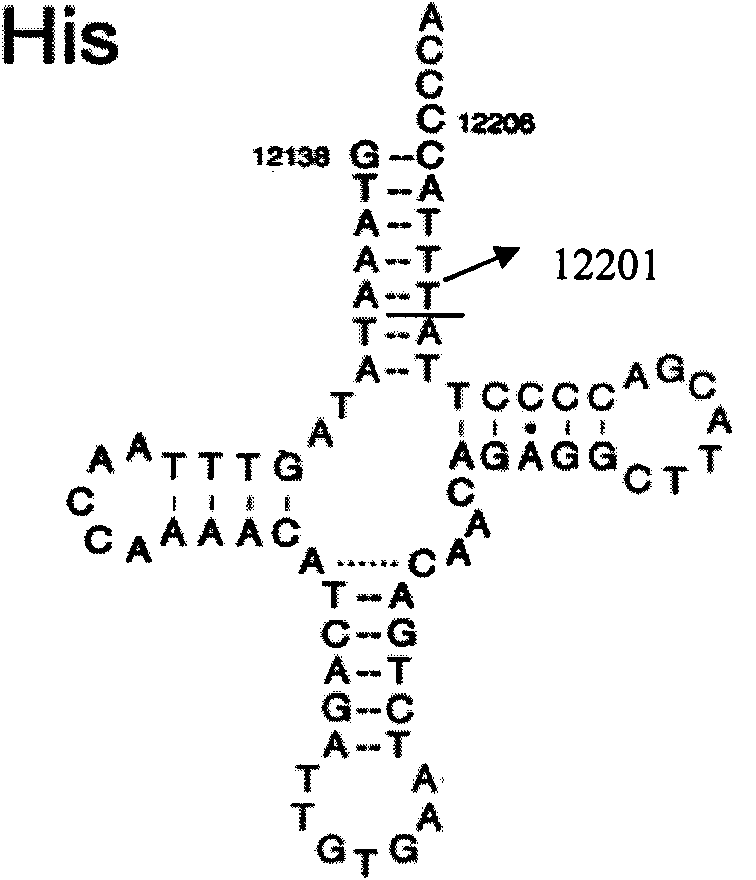
The invention belongs to the field of genomics, particularly relating to the deaf relevant mutation 12201 T>C of mitochondrial gene and application. The sequence in which the deaf relevant mutation 12201 T>C of mitochondrial gene of the invention is positioned has the structure of sequence 1. The invention can adopt the deaf relevant mutation of mitochondrial gene to preferentially detect the mitochondria mutational site of deaf patients, which is favourable for eliminating or diagnosing deaf caused by mitochondrial gene mutation; a diagnostic probe prepared by deaf relevant new mutation of the mitochondrial gene, a primer for introducing mutation to cause enzyme cutting site and a kit are adopted to conveniently and quickly screen deaf relevant to mitochondria. The invention provides the reference base for function change caused by gene mutation and provides a favorable chance for disclosing the molecular mechanism of deaf caused by the mitochondria DNA mutation.
Owner:EYE & ENT HOSPITAL SHANGHAI MEDICAL SCHOOL FUDAN UNIV
Nucleic acid sequence for detecting deaf genes and applications thereof
ActiveCN105936940AAccurate detectionImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationNucleotideNucleic acid sequencing
The invention discloses a nucleic acid sequence for detecting the deaf genes and applications thereof, and belongs to the fields of gene sequencing and biological examination. A set of specific primers for detecting the deaf genes comprises nucleic acid sequences represented by SEQ ID No.1 to SEQ ID No.17. A novel specific multiple PCR primer, which comprises whole exon with a total length of 9497 bp and is capable of detecting four deaf genes at the same time, is designed. Furthermore, a method using the specific multiple PCR primer to carry out detection is also provided. Compared with the prior art, a high-throughput sequencing technology is adopted to detect the deaf genes, and all mutation sites on four deaf genes can be rapidly and accurately detected, including the hot spot mutations and new mutations. The provided detection method has the characteristics of high throughput, low cost, and high detection rate.
Owner:成都凡迪医疗器械有限公司
Novel Complex Mutations in the Epidermal Growth Factor Receptor Kinase Domain
ActiveUS20130121996A1BiocideOrganic active ingredientsEpidermal Growth Factor Receptor KinaseAbnormal tissue growth
Six new mutations were found in exon 19 of the EGFR gene, the exon that is often mutated in tumors. The invention comprises methods of detecting the mutations, methods of prognosis and methods of predicting response to treatment based on the presence of absence of the mutations.
Owner:ROCHE MOLECULAR SYST INC
New mutation site related to isoniazide resistance of mycobacterium tuberculosis and application thereof
InactiveCN103820440AImproving Molecular DetectionShorten diagnostic timeMicrobiological testing/measurementMicroorganism based processesNew mutationDrug resistance
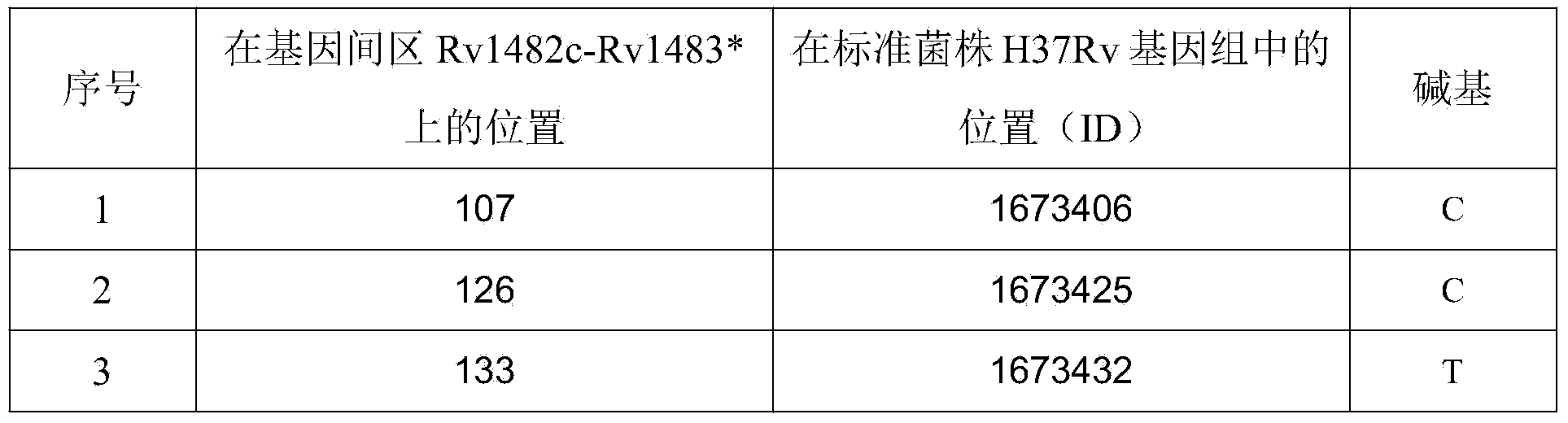
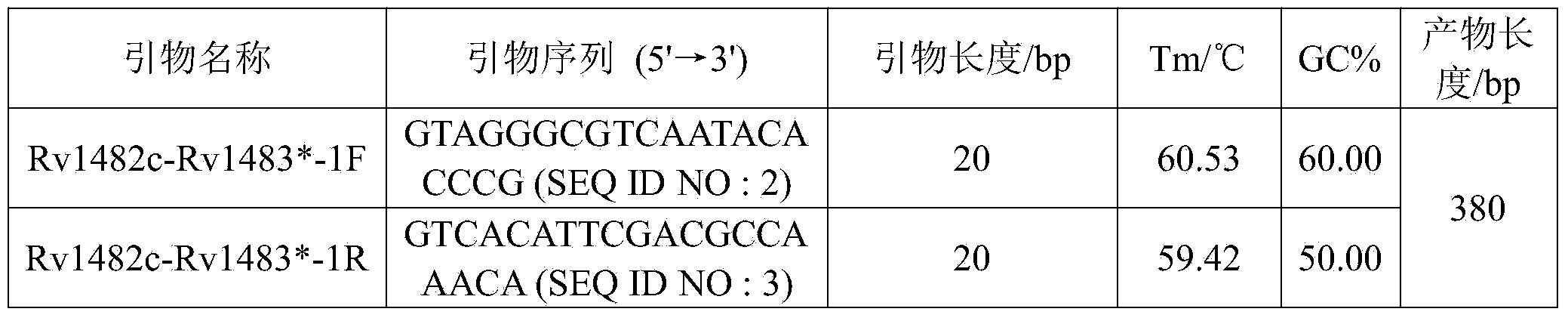
The invention discloses a new mutation site related to isoniazide resistance of mycobacterium tuberculosis and an application thereof. The new mutation site is positioned at a 1673425th site of a standard strain H37Rv genome, that is a 126th base in an intergenic region of Rv1482c to Rv1483*, and the generated point mutation comprises that a base C is mutated into a base T or a base A. The found new mutation site can be used as a detection target applied in detection of the isoniazide resistance of mycobacterium tuberculosis, improves the molecular detection level of the isoniazide resistance of mycobacterium tuberculosis, shortens the diagnosis time of patients, and saves the treating time and cost for the patients. The mycobacterium tuberculosis drug-resistant mutation site is explained to possibly occur in a non-coding region, the base mutation of the intergenic region is prompted to be possibly related to the drug resistance, and a drug-resistant biomarker also exists. The invention provides a new idea for finding out a new and more effective mycobacterium tuberculosis drug-resistant molecular marker.
Owner:广东省结核病控制中心
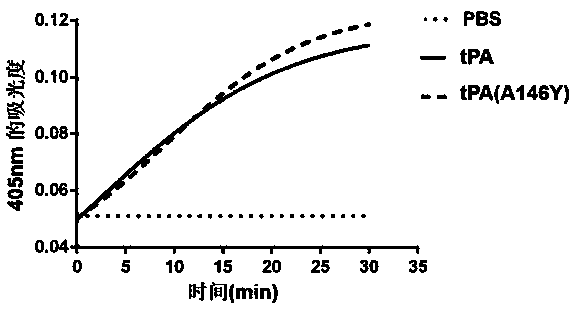
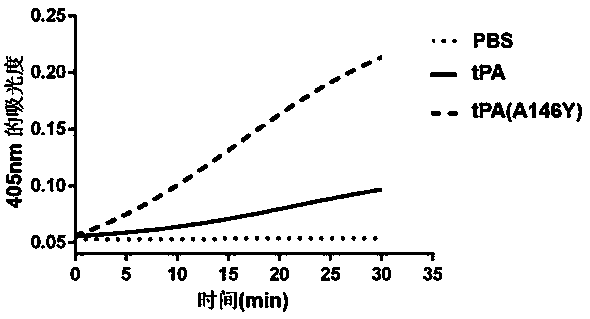
Tissue-type plasminogen activator mutant and application thereof
ActiveCN107760660AImprove the immunityPeptide/protein ingredientsRespiratory disorderDiseaseThrombus
The invention provides a tissue-type plasminogen activator mutant and application thereof. The mutant comprises an A146 locus which is a brand new mutation site, referring to substitution, deletion oraddition of at least one or more amino acid residues comprising the A146. The mutant disclosed by the invention has the effects of obviously resisting ability inhibited by an endogenous inhibitor (PAI-1) (Plasminogen Activator Inhibitor 1) and enhanced ability of activating plasminogen. The mutant can be applied to preparing medicines for treating thrombotic diseases comprising acute myocardial infarction, acute pulmonary embolism, cerebral apoplexy, phlebothrombosis and the like.
Owner:FUZHOU UNIV
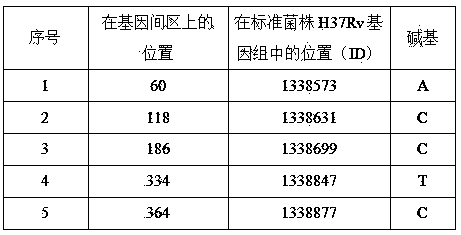
New mutation site related to streptomycin resistance of mycobacterium tuberculosis and application thereof
InactiveCN103820439AImproving Molecular DetectionShorten diagnostic timeMicrobiological testing/measurementDNA/RNA fragmentationNew mutationStreptomycin
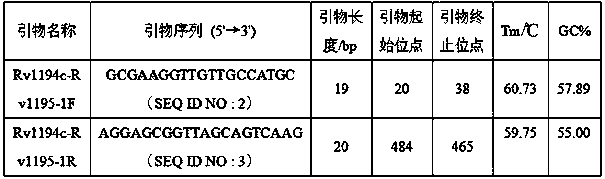
The invention discloses a new mutation site related to streptomycin resistance of mycobacterium tuberculosis and an application thereof. The new mutation site is positioned at a 1338631st site of a standard strain H37Rv genome, that is a 47th base in an intergenic region of Rv1194c to Rv1195, and the generated point mutation comprises that a base C is mutated into a base A. The found new mutation site can be used as a detection target applied in detection of the streptomycin resistance of mycobacterium tuberculosis, improves the molecular detection level of the streptomycin resistance of mycobacterium tuberculosis, shortens the diagnosis time of patients, and saves the treating time and cost for the patients. The mycobacterium tuberculosis drug-resistant mutation site is explained to possibly occur in a non-coding region, the base mutation of the intergenic region is prompted to be possibly related to the drug resistance, and a drug-resistant biomarker also exists. The invention provides a new idea for finding out a new and more effective mycobacterium tuberculosis drug-resistant molecular marker.
Owner:广东省结核病控制中心
Kit comprising primer pair for detecting genetic marker related to male infertility
InactiveCN104928385AEnables diagnostic testingMicrobiological testing/measurementMale infertilityLarge-Scale Sequencing
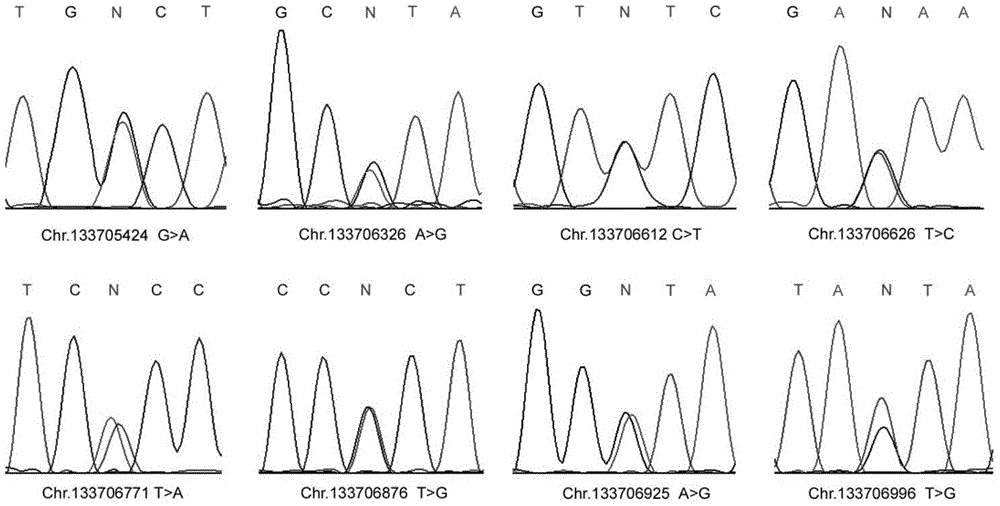
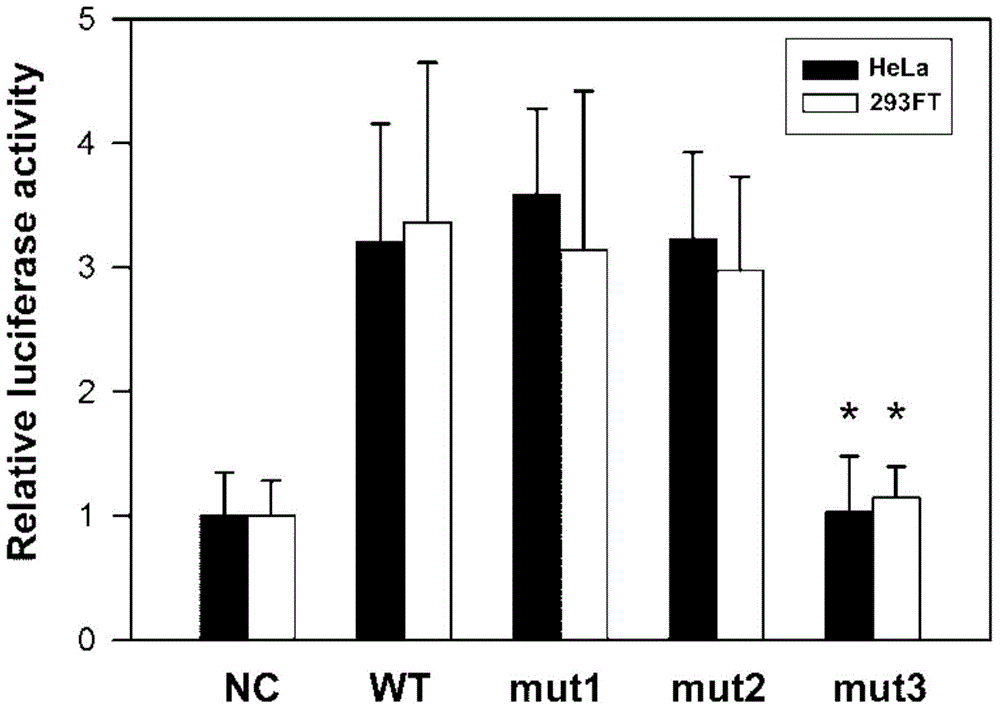
The invention discloses a male infertility related genetic marker, the sequence of which is that of the UBE2B gene. The binding site of the UBE2B gene's promoter region and the transcription factor SP1 undergoes mutation, and the mutation site is one or more of Chr.133706771T>A, Chr.133706876T>G and Chr.133706925A>G. By large-scale sequencing, four new mutation sites of the UBE2B gene's promoter region are selected from male infertility patients, a transcription factor plasmid is cotransfected into cells together with a wild type plasmid and a mutant plasmid respectively, and experiments find significant function differences and show that mutation of the UBE2B gene's promoter region is relevant to male infertility (especially azoospermia). Therefore, diagnosis detection of male infertility can be realized through the mutation.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
Primer combination for detecting TNNI3K gene mutation and application thereof
PendingCN110846409AConvenient methodLow costMicrobiological testing/measurementDNA/RNA fragmentationMedicineExon
The invention discloses a primer combination for detecting TNNI3K gene mutation. The primer combination is characterized by comprising a primer group and a probe set which are used for detecting c.1910C>T mutation in TNNI3K gene. The primer combination has the advantages that the all-exon sequencing technology is used to acquire dilated cardiomyopathy pathogenic gene new mutation, and the fluorescent quantitative PCR technology is used to simply, fast and accurately detect the TNNI3K-c.1910C>T new mutation; multiple case samples can be detected in one step, and a new method is built for the clinical early molecular diagnosis and prevention of dilated cardiomyopathy.
Owner:KUNMING UNIV OF SCI & TECH
New mutation of PEB (Phosphatidylethanolamine Binding Protein) virulence gene and application thereof
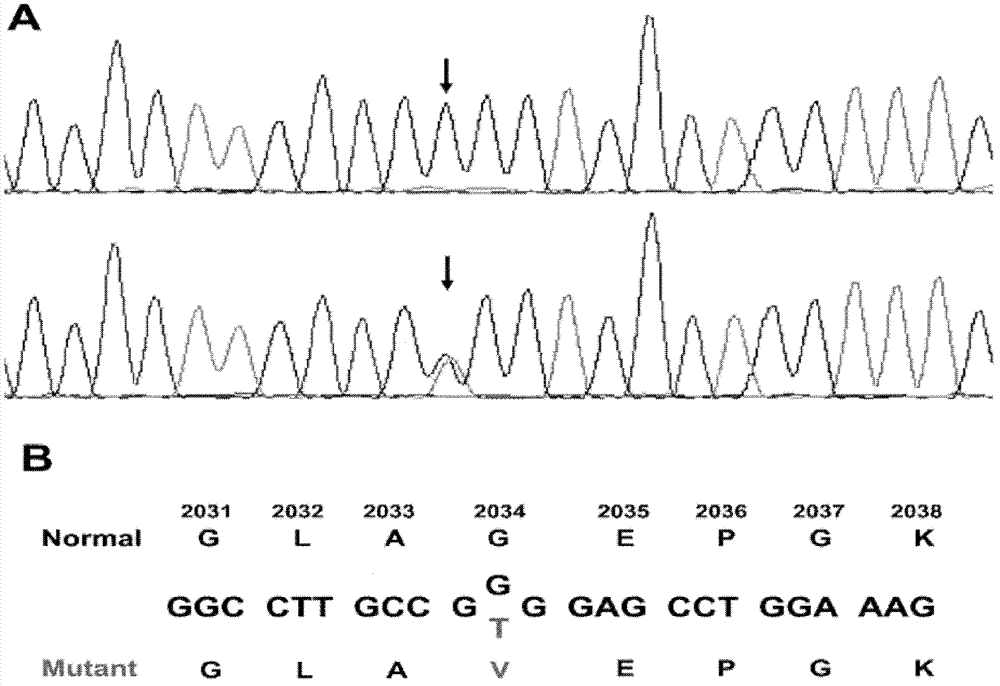
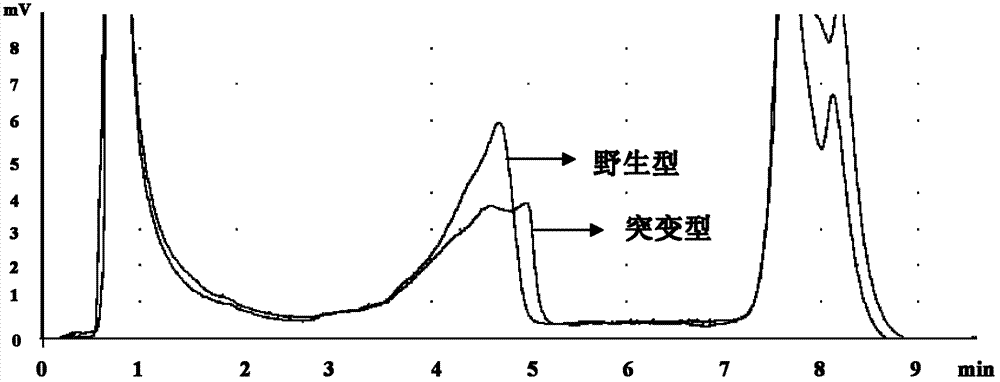
The invention relates to a new mutation of a PEB (Phosphatidylethanolamine Binding Protein) virulence gene and an application thereof. Specifically, a COL7A1 new mutation of a PEB virulence gene is found for the first time by virtue of large-scale screening by the inventor. The inventor takes PEB affected pedigree as a research target, performs exome sequencing and comparison on affected individuals and non-affected individuals in the pedigree and accidently finds a missense mutation (G>T) in the 73rd exon of the COL7A1 gene; due to the mutation, glycocoll Gly of the 2034th of the COLA1 protein is mutated as valine Val; the mutation is highly conservative in the patients; and by applying the mutation, the anterior epidenmoiysis bulbse can be detected.
Owner:BGI GENOMICS CO LTD
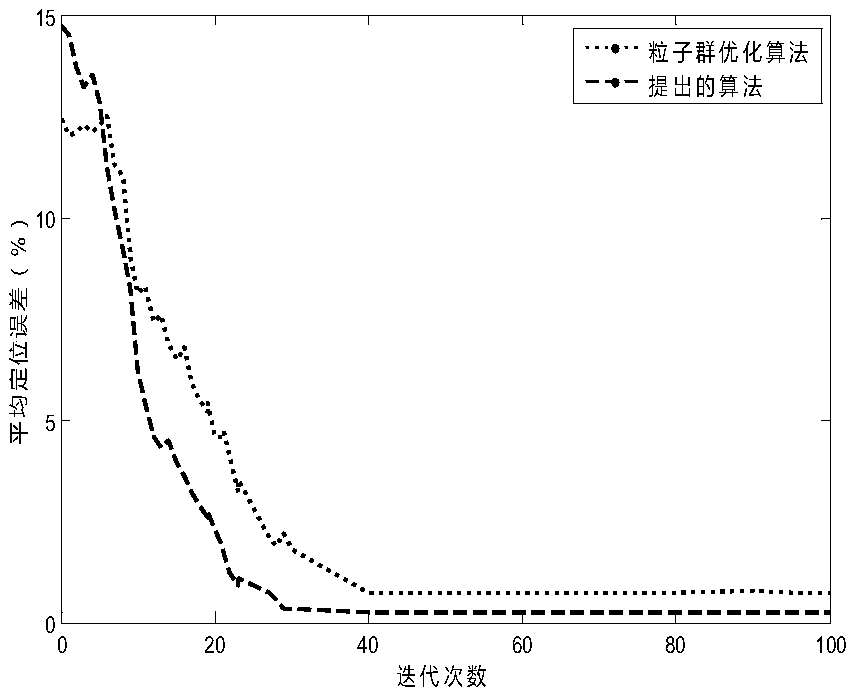
Particle swarm positioning algorithm based on adaptive differential
InactiveCN105517150ADiversity guaranteedImprove exploration abilityNetwork topologiesDifferential algorithmNew mutation
The invention relates to a particle swarm positioning algorithm based on adaptive differential. The particle swarm positioning algorithm mainly comprises relation establishment between signal intensities and distances, least squares algorithm position computation, and a new search method, and includes receiving the distance information of an anchor node by using a unknown node, computing a position by using a least squares algorithm, generating a new swarm by using an improved adaptive differential algorithm, performing local search by using a particle swarm algorithm and a new mutation strategy, comparing with adaptive values and repeatedly performing iteration in order to achieve asymptotic convergence, and finally obtaining the position of the unknown node. The diversity of the swarms generated by the new differential algorithm is guaranteed and it increases rate of convergence and positioning precision to perform the local search by the combination of the particle swarm algorithm and the differential algorithm.
Owner:JIANGNAN UNIV
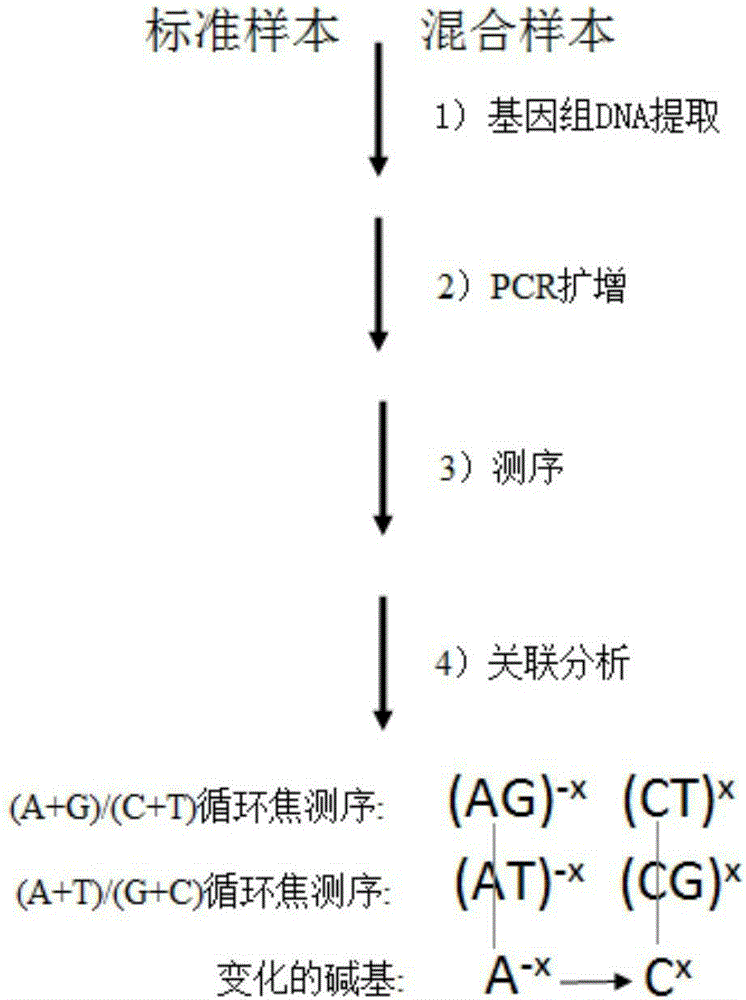
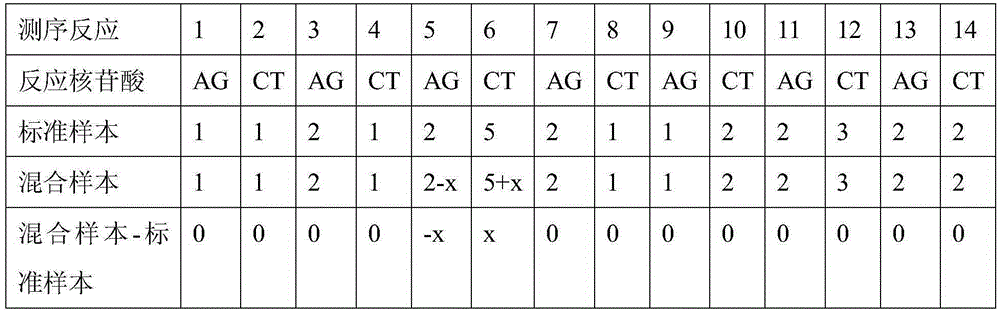
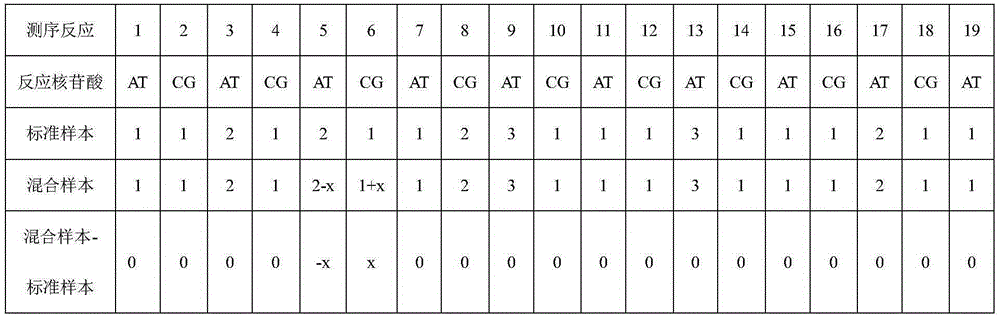
Method for searching for new mutation/SNP locus based on double-nucleotide pyrosequeneing-by-synthesis
ActiveCN105256030AIncrease measurement lengthExpand the scope of analysisMicrobiological testing/measurementGenomic DNASingle strand
The invention discloses a method for searching for a new mutation / SNP locus based on double-nucleotide pyrosequeneing-by-synthesis. According to the method, PCR is conducted on the genomic DNAs of a known sequence sample and a mixed sample, pyrosequeneing is conducted on obtained single-stranded PCR products according to a specific double-nucleotide adding method, each sample acquires two groups of double-nucleotide pyrosequeneing-by-synthesis information, and correction is conducted; the two groups of corrected mixed sample sequencing information are compared with the two groups of corrected known sequence sample sequencing information; if neither the two groups of mixed sample sequencing information nor the two groups of known sequence sample sequencing information are changed, it is indicated that the mixed sample sequencing information is consistent with the known sequence sample sequencing information, and no new mutation / SNP locus exists; if the mixed sample sequencing information is different from at least one of the two groups of known sequence sample sequencing information, it is indicated that the mixed sample sequencing information is not totally consistent with the known sequence sample sequencing information, and it shows that a new mutation / SNP locus exists.
Owner:SOUTHEAST UNIV
Universal primer, probe and test kit for intestinal virus nucleic acid testing
PendingCN111534637AStrong degeneracyImprove the detection rateMicrobiological testing/measurementDNA/RNA fragmentationDiseaseNucleic acid detection
The invention relates to the technical field of biological detection, in particular to a universal primer, a probe and a test kit for intestinal virus nucleic acid testing. The primer and the fluorescent probe are designed according to a new mutation site of an intestinal virus 5'UTR sequence, the primer and the probe for the universal nucleic acid testing of the intestinal virus provided by the invention are strong in degeneracy, and a degeneracy base is introduced at the 8-base position of the 3' end of an upstream prime; and a degenerate base is introduced at the 16-base position of the 5'end of a downstream primer according to T base mutation. RRT-PCR detection is carried out by adopting the primer and the probe disclosed by the invention, so that the primer and the probe have relatively high detection rate and high sensitivity on the novel intestinal virus generated by mutation of the intestinal virus 5'UTR sequence; and a scientific basis is provided for diagnosis of related diseases of intestinal virus infection.
Owner:江苏派森杰生物科技有限公司 +1
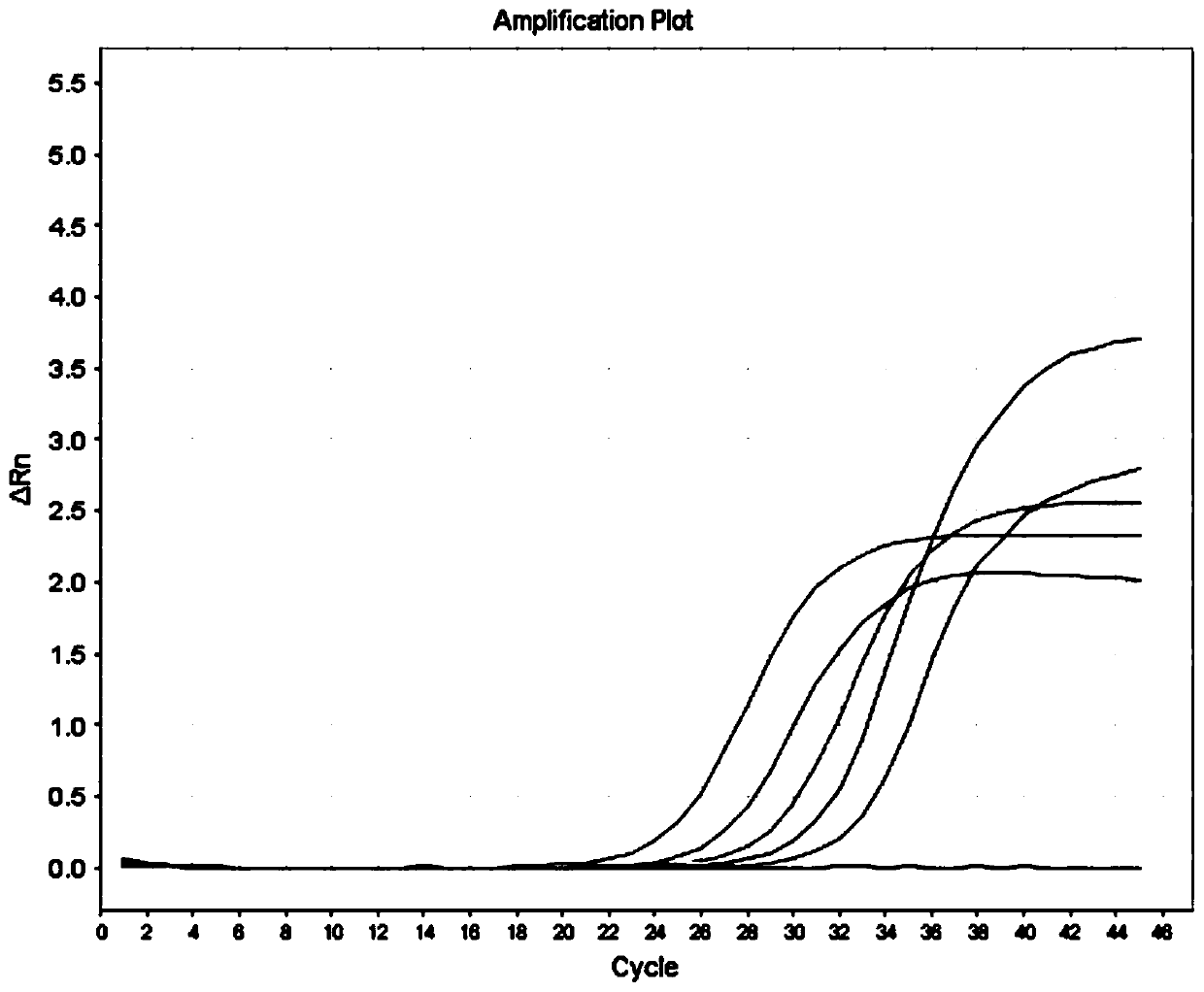
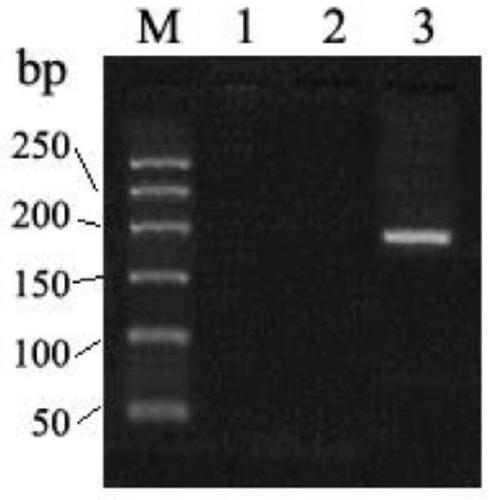
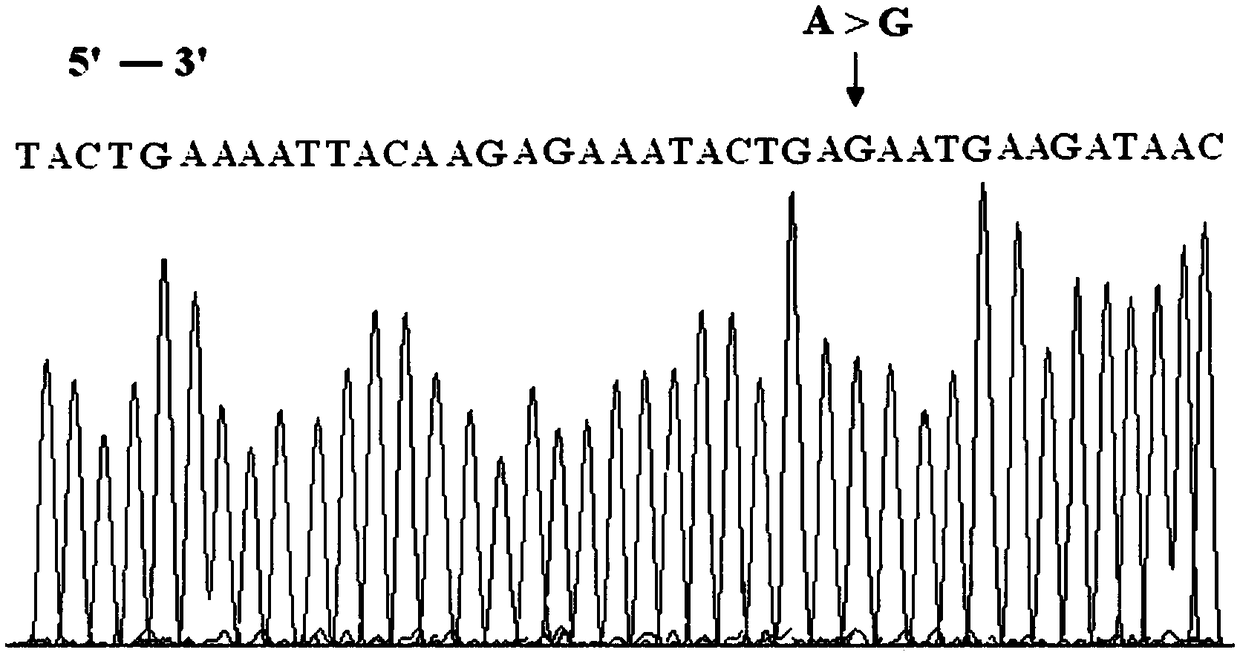
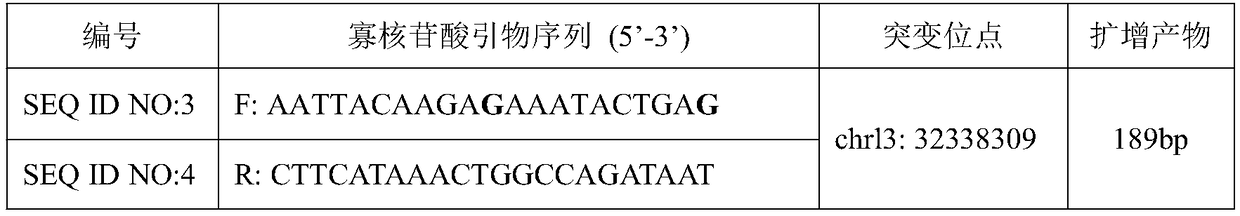
BRCA2 g. 32338309A>G mutant and application thereof in breast cancer assisted diagnosis
ActiveCN109457031AEasy diagnosisIncreased sensitivityMicrobiological testing/measurementDNA/RNA fragmentationMutantBiology
The invention discloses a BRCA2 g. 32338309A>G mutant and application thereof in breast cancer assisted diagnosis. Compared with the sequence of normal BRCA2 genes, the sequence of the BRCA2 g. 32338309A>G mutant has g. 32338309A>G locus mutation. The invention further discloses a specific primer for detecting the BRCA2 g. 32338309A>G mutant, a kit containing the specific primer, and application of the BRCA2 g. 32338309A>G mutant in breast cancer assisted diagnosis. The BRCA2 g. 32338309A>G mutant provides a new mutant sit of breast cancer pathogenesis to assist breast cancer diagnosis.
Owner:WUXI NO 5 PEOPLES HOSPITAL
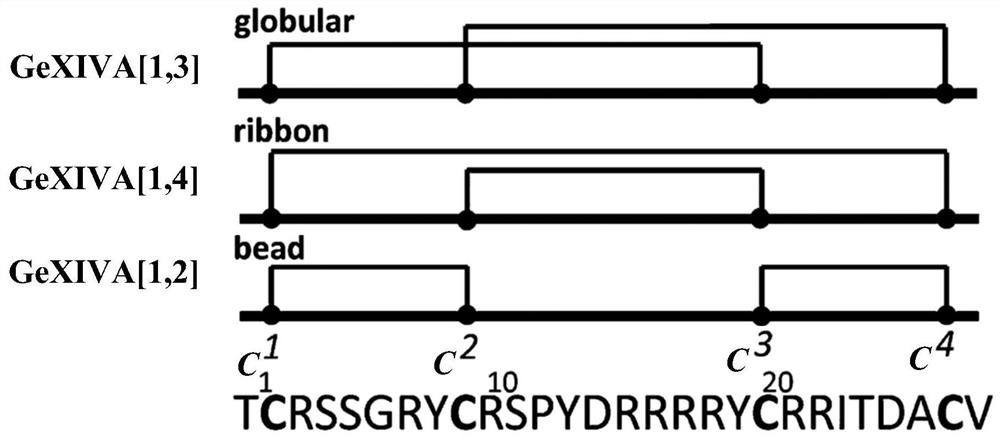
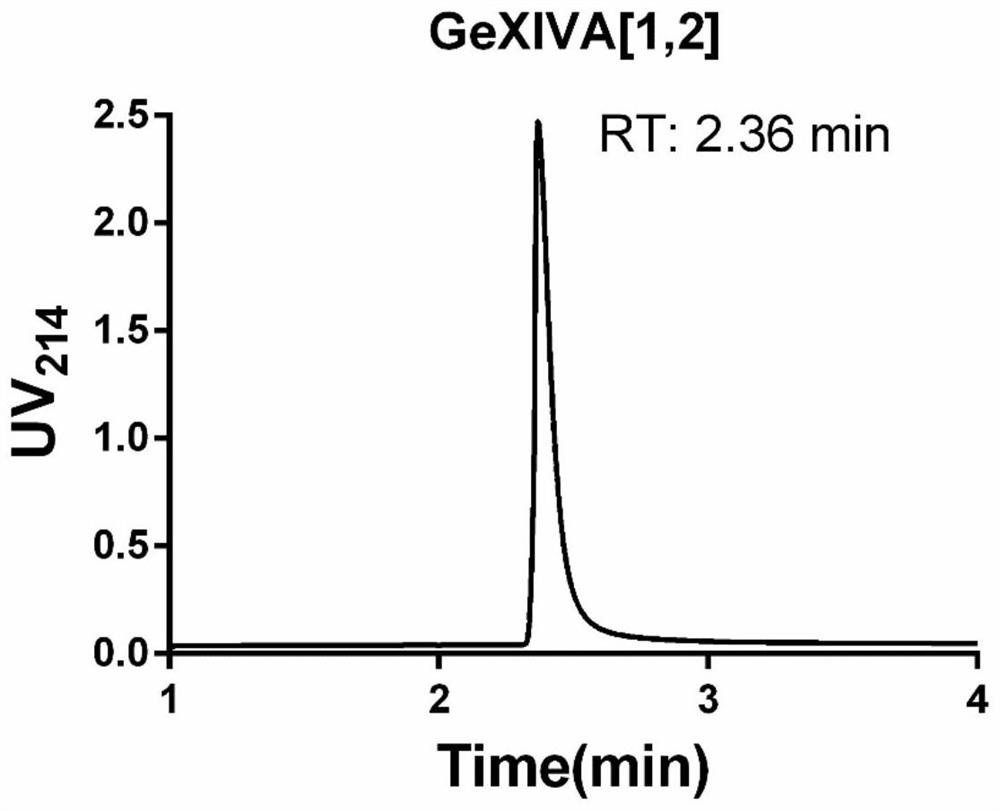
Novel alpha O-conotoxin peptide GeXIVA new mutant as well as pharmaceutical composition and application thereof
ActiveCN112010959AHigh activityHigh selectivityNervous disorderPeptide/protein ingredientsConotoxinPharmaceutical drug
The invention belongs to the field of biochemistry, molecular biology and neurosciences, and relates to a novel alpha O-conotoxin GeXIVA mutant and a pharmaceutical composition and application thereof. Specifically, the invention relates to a separated polypeptide. The amino acid sequence of the polypeptide is obtained by removing one or more amino acids in a mother sequence or replacing one or more amino acids in the mother sequence with the same number of L-type amino acids or D-type amino acids, wherein the mother sequence is SEQ ID NO:1, SEQ ID NO:62 or SEQ ID NO:63; and the polypeptide has an activity of blocking alpha9alpha10nAChR. The polypeptide disclosed by the invention can specifically block the alpha9alpha10nAChR, and has a good application prospect in the aspects of preparinganalgesic drugs, drugs for treating related mental diseases and cancers, neuroscience tool drugs and the like.
Owner:HAINAN UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com