New mutation site related to streptomycin resistance of mycobacterium tuberculosis and application thereof
A bacillus streptomycin, tuberculosis branch technology, applied in the direction of microbial determination/examination, DNA/RNA fragmentation, recombinant DNA technology, etc., can solve the problems of increasing the chance of drug-resistant strains spreading, taking a long time, affecting the diagnosis and treatment of patients, etc. Achieve the effect of improving the level of molecular detection, shortening the time of diagnosis, and saving the time and cost of treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] 1. Prediction of base mutation sites associated with STR drug resistance
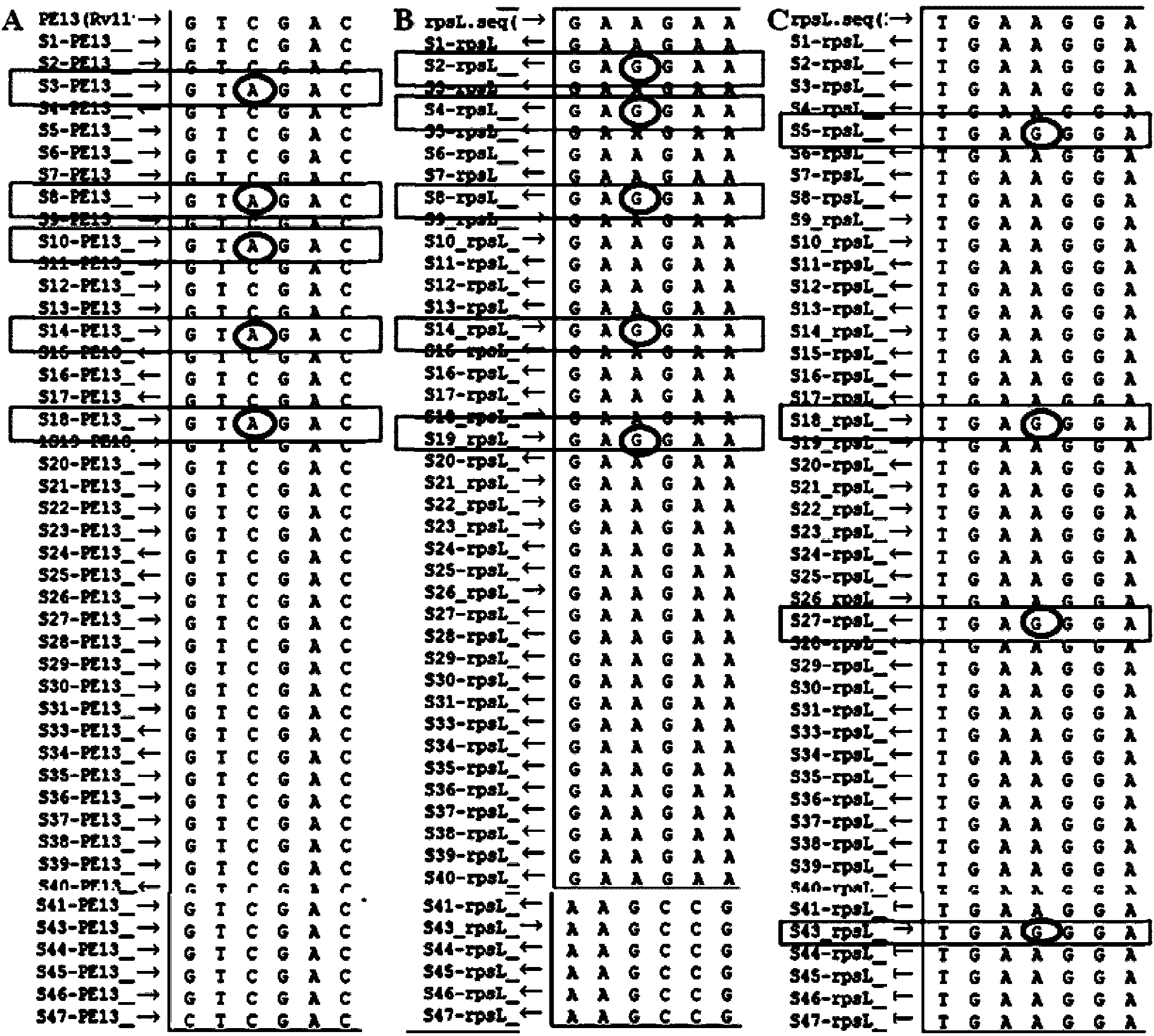
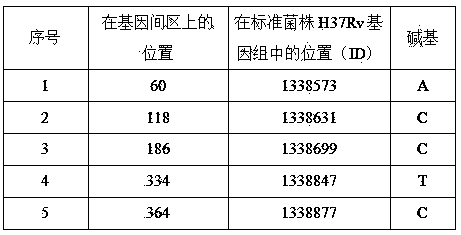
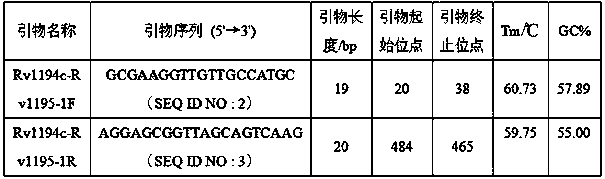
[0026] First, using deep-sequencing technology, the whole genome of 161 strains of Mycobacterium tuberculosis from China was sequenced, including 44 strains of sensitive strains, 94 strains of multidrug-resistant strains (MDR), and 23 strains of extensively drug-resistant strains (XDR). The H37Rv standard Mycobacterium tuberculosis strain genome sequence was used as a control for bioinformatics analysis, and a batch of new genes closely related to drug resistance and single nucleotide polymorphism (SNP) sites in the intergenic region (IGR) were found, among which The single-base mutation site in the intergenic region Rv1194c-Rv1195 has a high correlation with streptomycin (STR) resistance, with a P value of 0.002, as shown in Table 1.
[0027] Table 1 Correlation between the single base mutation site in the intergenic region Rv1194c-Rv1195 and different drug resistance
[0028]
[0029] Tab...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com