Method for searching for new mutation/SNP locus based on double-nucleotide pyrosequeneing-by-synthesis
A two-nucleotide, pyrosequencing technology, applied in the biological field, can solve problems such as not easy to find, and achieve the effect of increasing the length of measurement and broadening the scope of analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
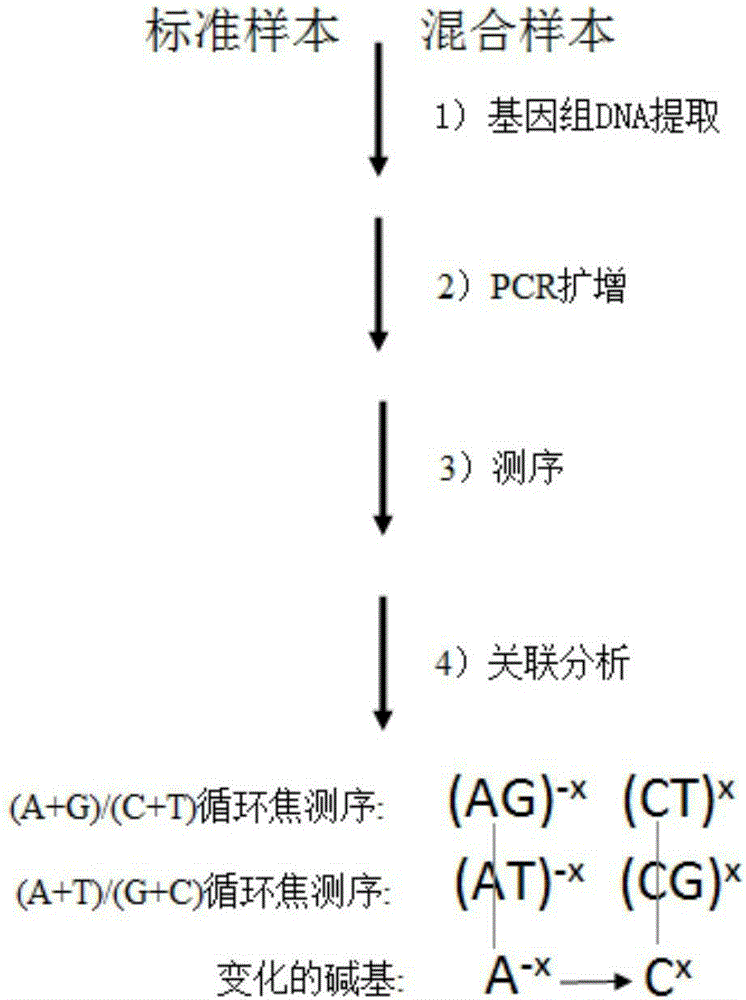
[0032] 1) Genomic DNA extraction: According to the source of the sample, select mature experimental steps to extract genomic DNA from standard samples and mixed samples.
[0033] 2) PCR: Design a pair of PCR primers, one of which is modified with biotin at 5', and amplify the fragment to be sequenced.
[0034] 3) Sequencing: The PCR amplification products of the standard sample and the mixed sample were divided into two parts, and reacted with streptavidin-modified magnetic beads to prepare a single-stranded DNA template; the sequencing primers were hybridized with the single-stranded DNA template, Specific two-nucleotide synthetic pyrosequencing was performed.
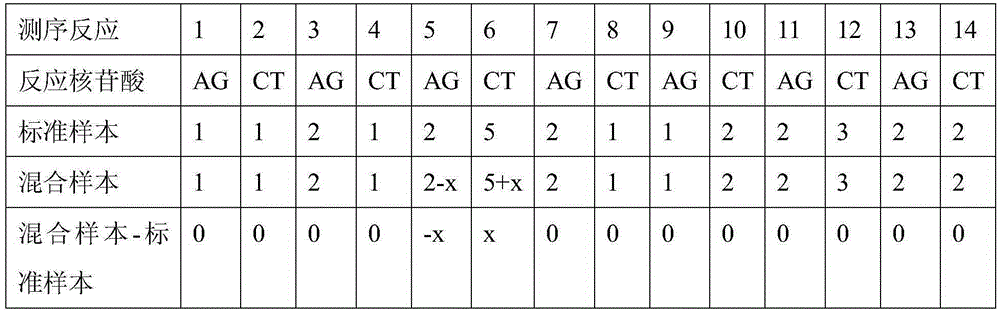
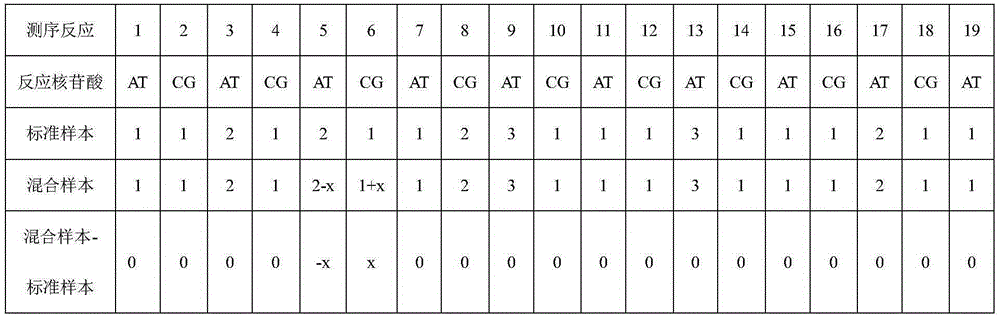
[0035] 4) Correlation analysis: Correct the pyrosequencing information of the two nucleotide synthesis, and perform division and subtraction operations on the two corrected mixed sample sequencing information and the known sequence samples to find out the differences in the information and derive The specific base ch...
Embodiment 2
[0054] A method for finding new mutations / SNPs from a sequence of the human sample UGT1A1 gene, the specific methods include:
[0055] (1) Select a single human blood sample confirmed by the sequence information as the standard sample, and 100 mixed blood samples from different people as the analysis sample;
[0056] (2) Genomic DNA in peripheral blood was extracted by traditional protein kinase K and phenol / chloroform extraction;
[0057] (3) PCR amplification: PCR primer 1: 5'-biotin-CCCTGCTACCTTTGTGGACT-3' (SEQ ID NO: 3), PCR primer 25'-CATTATGCCCGAGACTAACAAA-3' (SEQ ID NO: 4) polymerized with 200ng genomic DNA, 0.2mMdNTP, 1UTaqDNA Enzyme, 1× amplification buffer, 1.8mM MgCl 2 50 μL PCR amplification system was used for amplification. The amplification conditions were: initial denaturation at 95°C for 5 min; 40 thermal cycles: denaturation at 94°C for 30 s, annealing at 57°C for 45 s, extension at 72°C for 45 s, and final extension at 72°C for 7 min. Each sample was ampli...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com