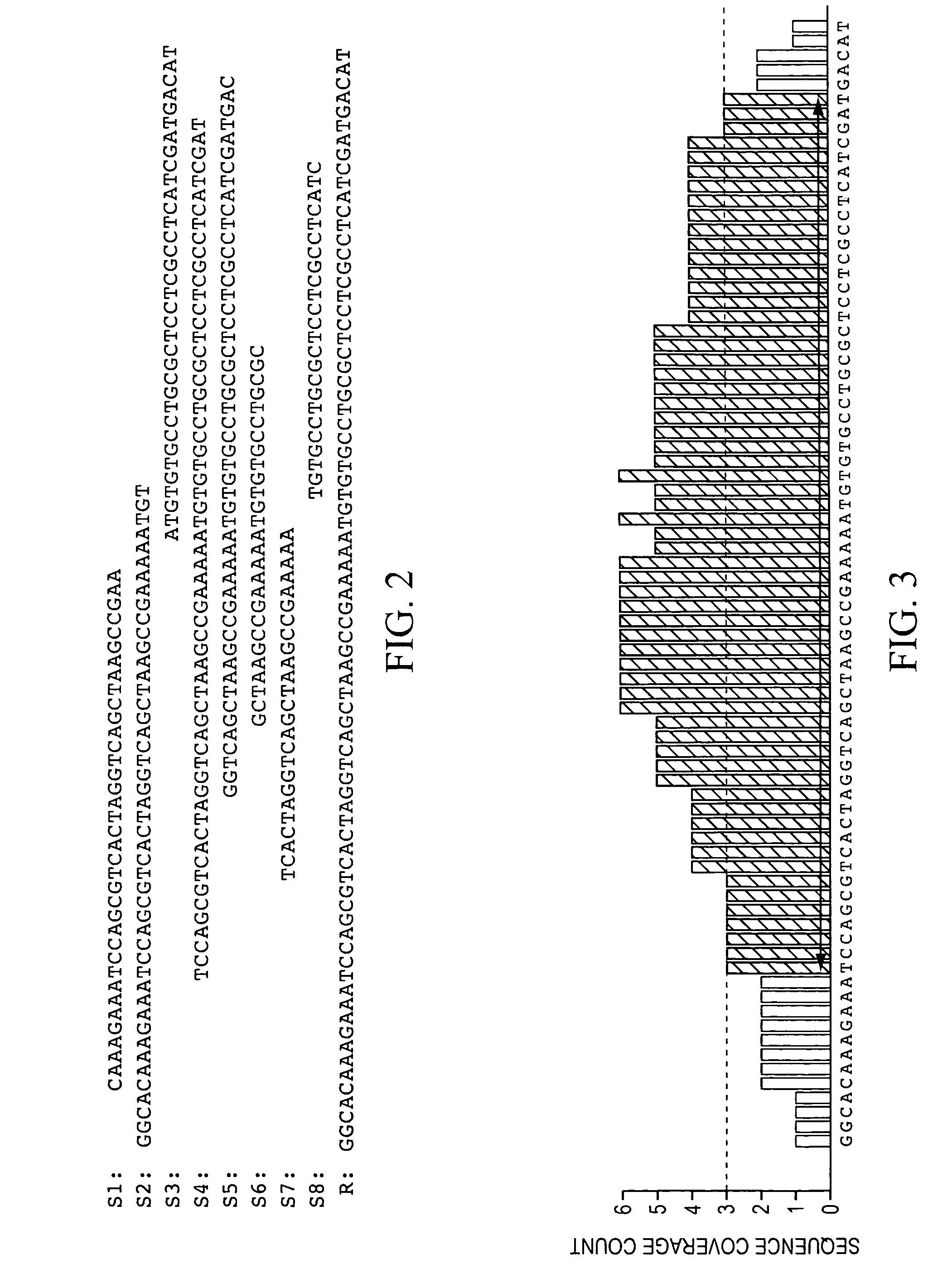
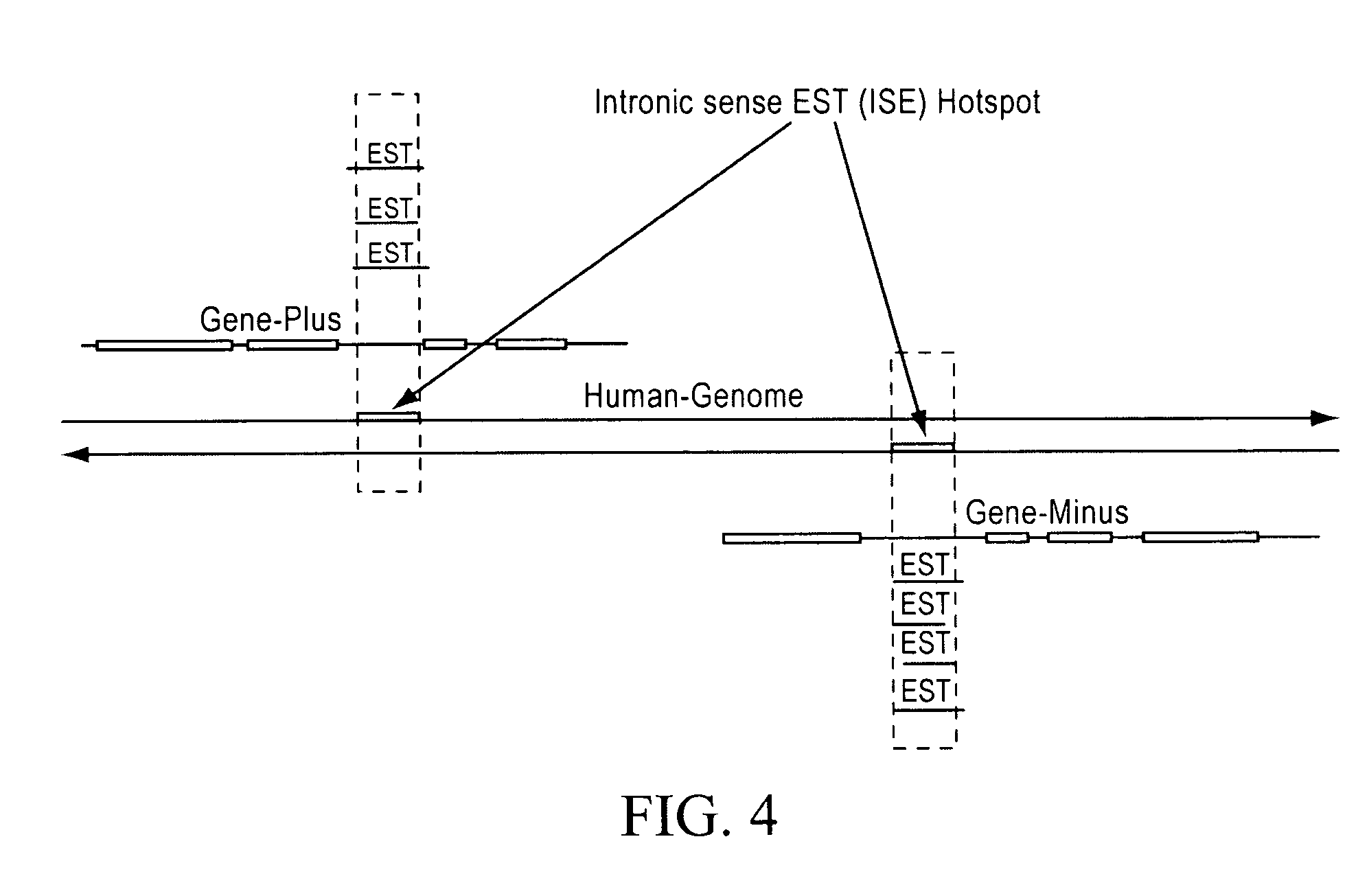
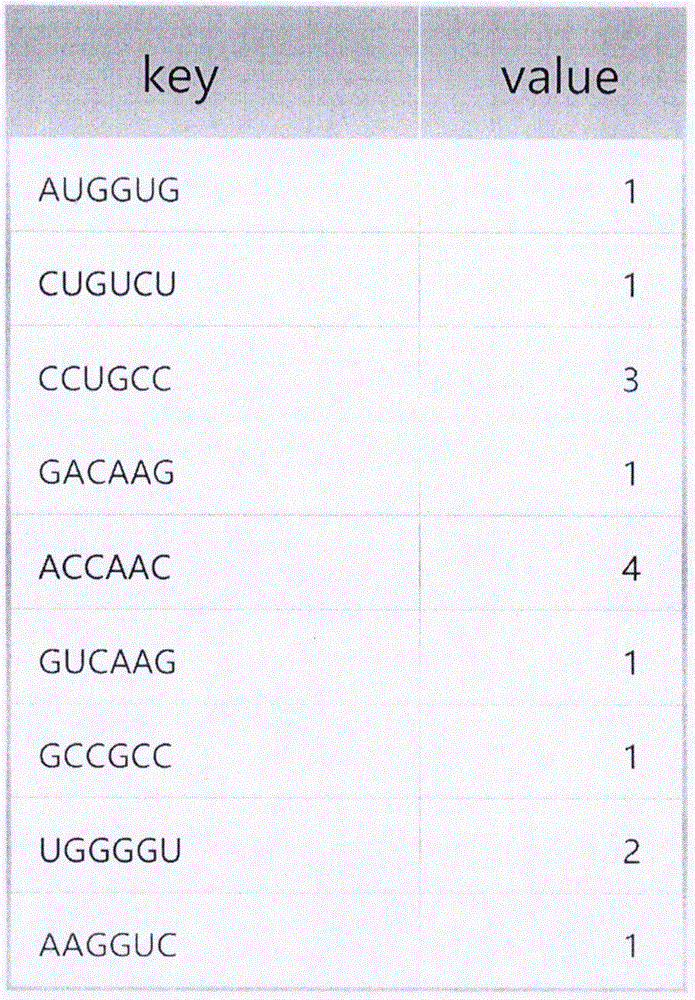
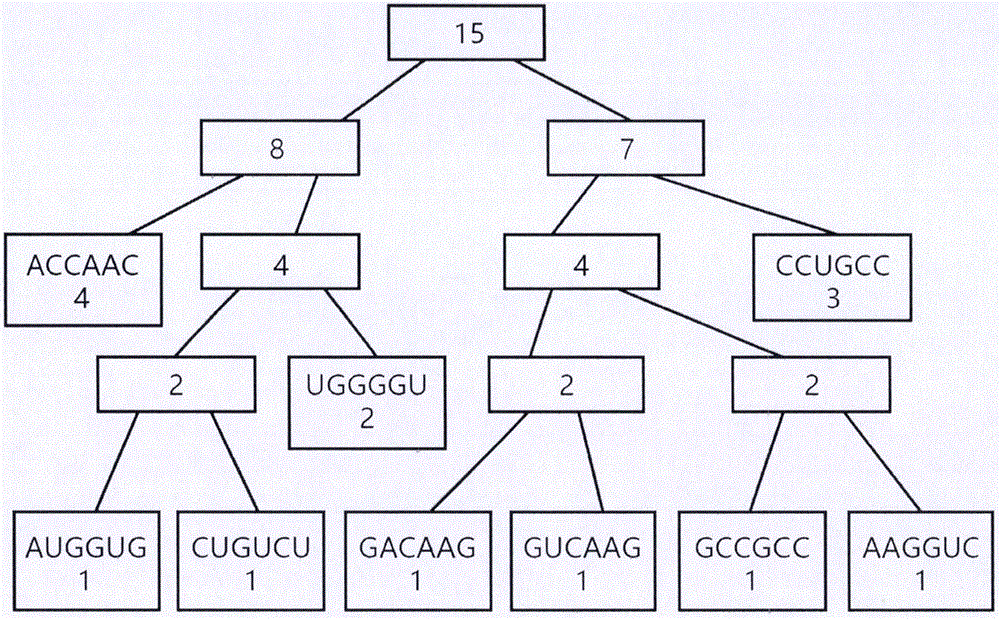
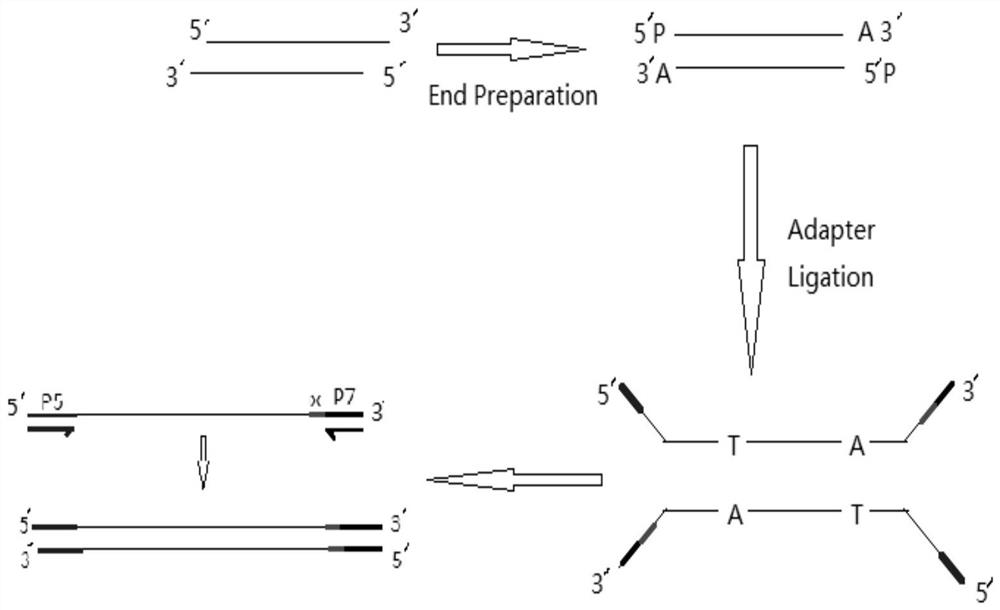
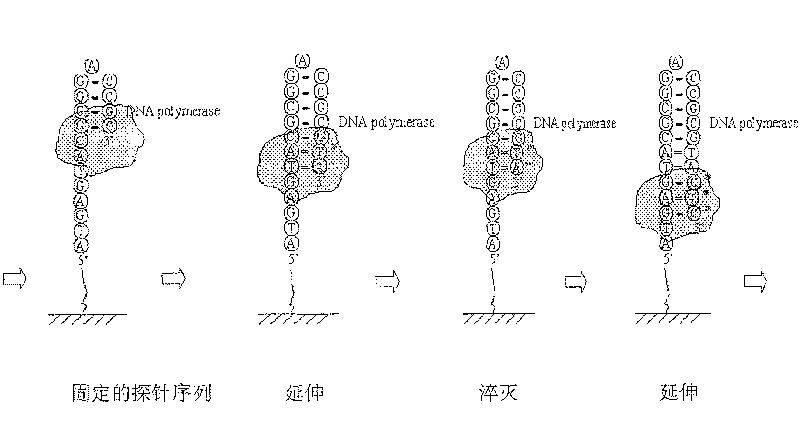
Patents
Literature
38 results about "Large-Scale Sequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
The process of determining of the precise sequence of nucleotides over a very large piece of DNA, possibly the entire genome of an organism.
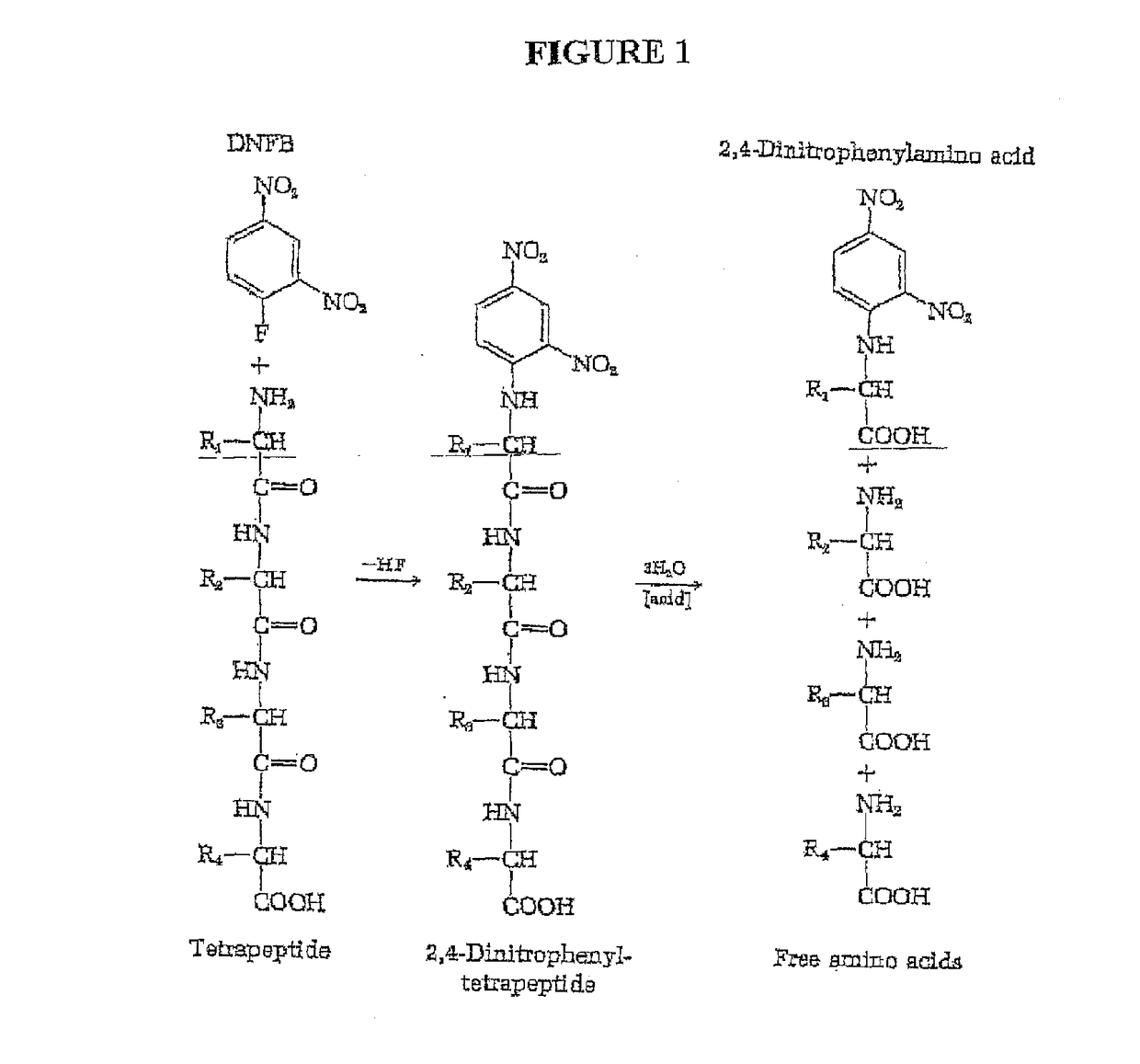
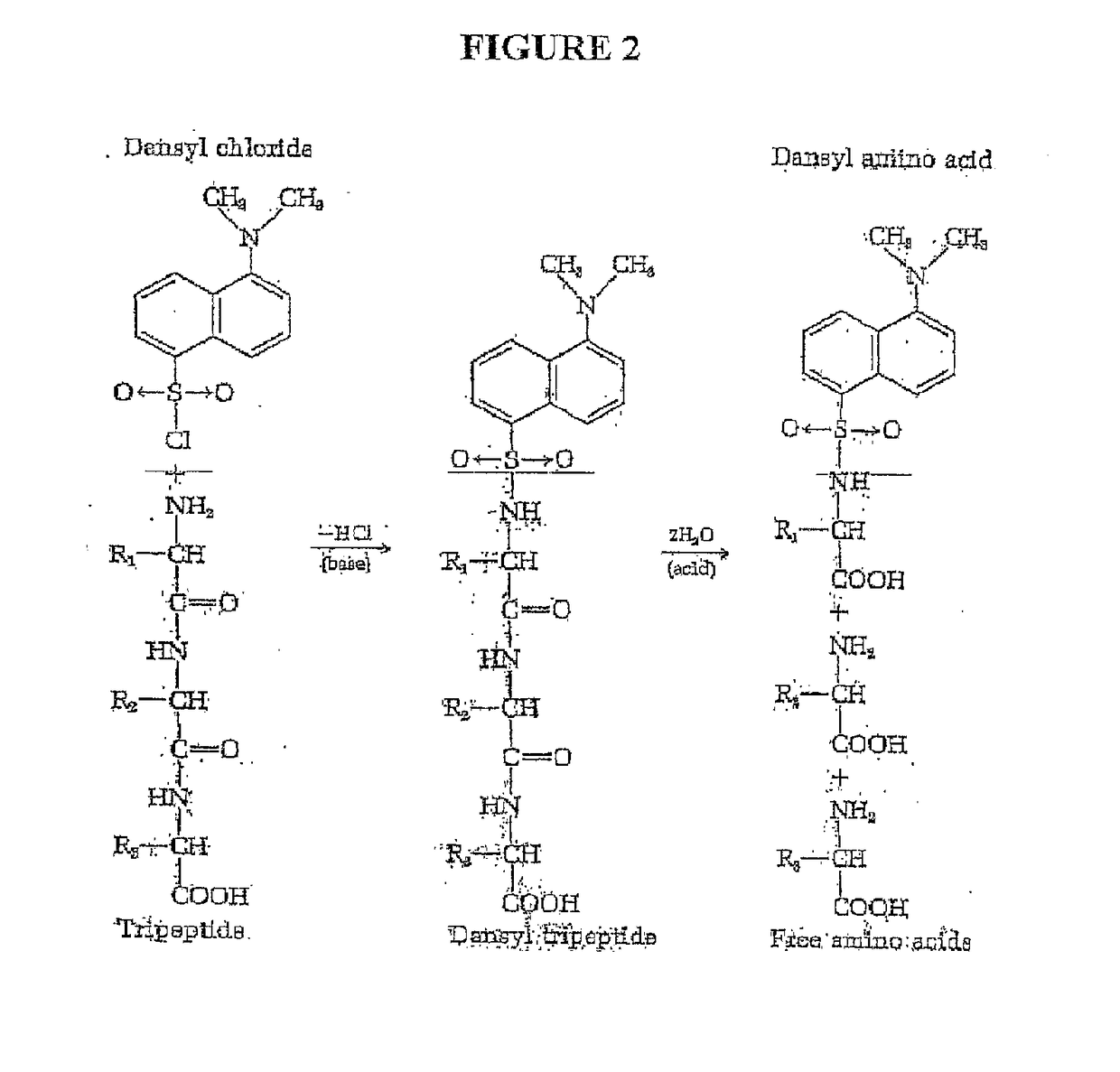
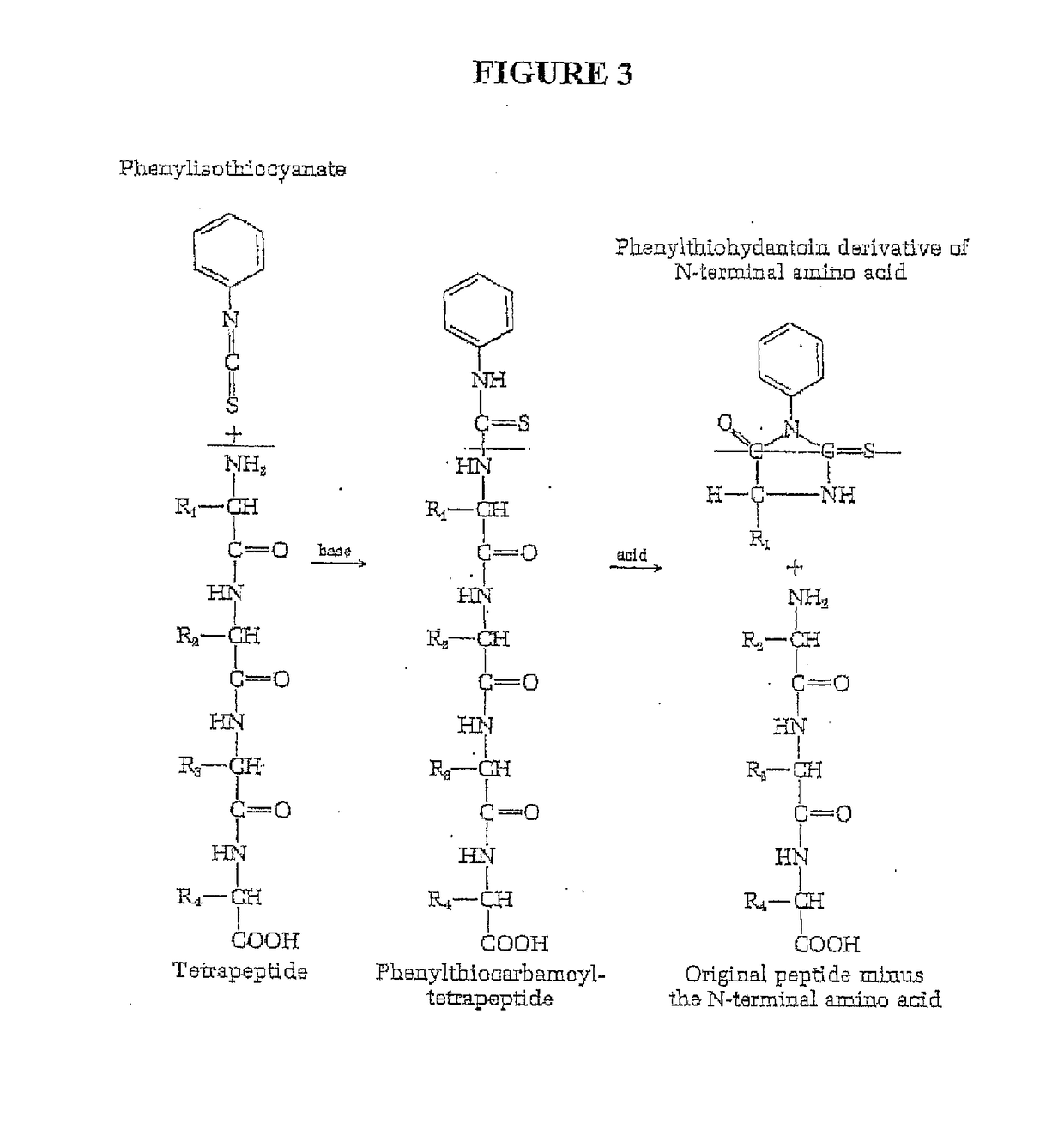
Single molecule peptide sequencing
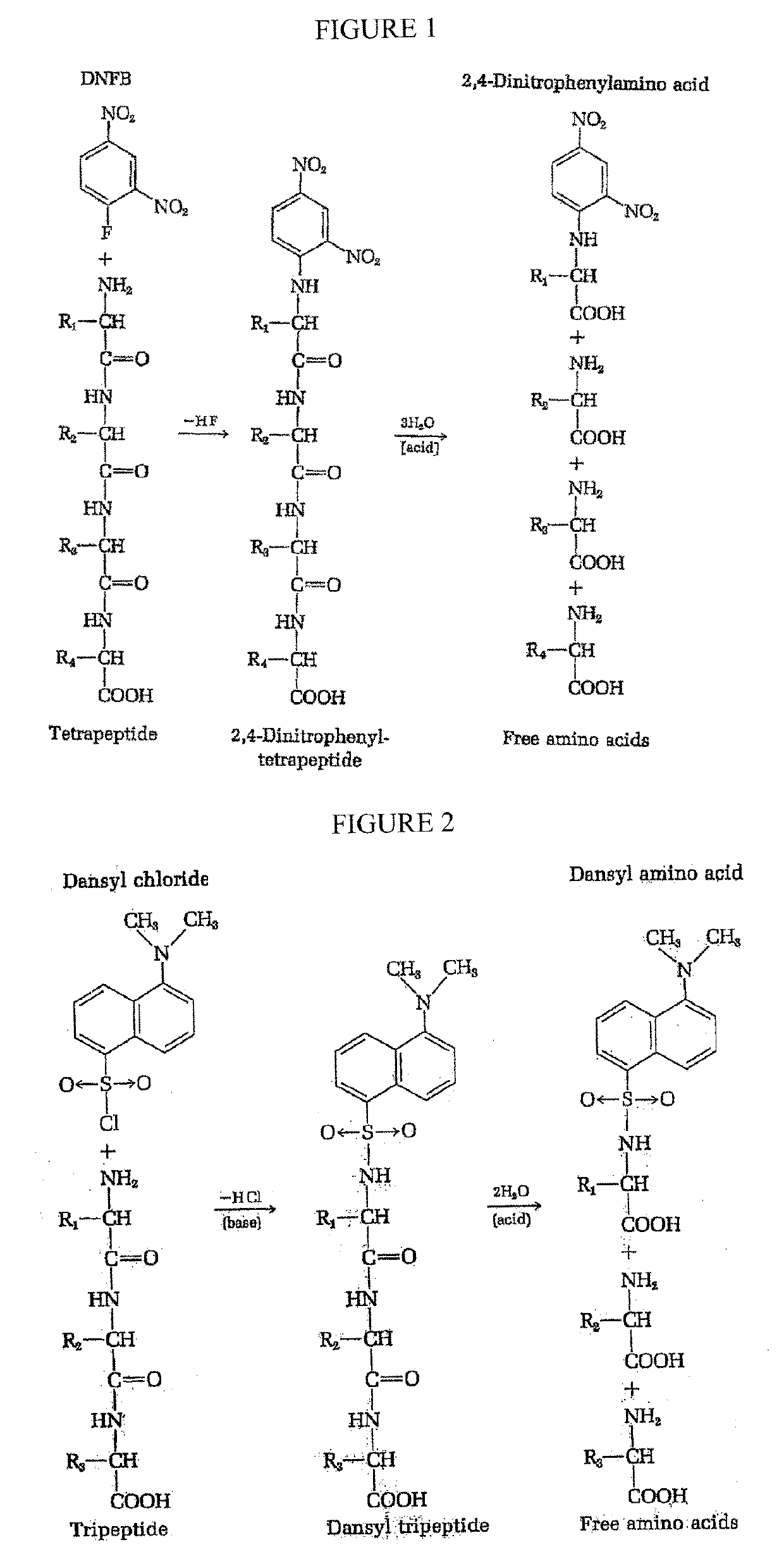
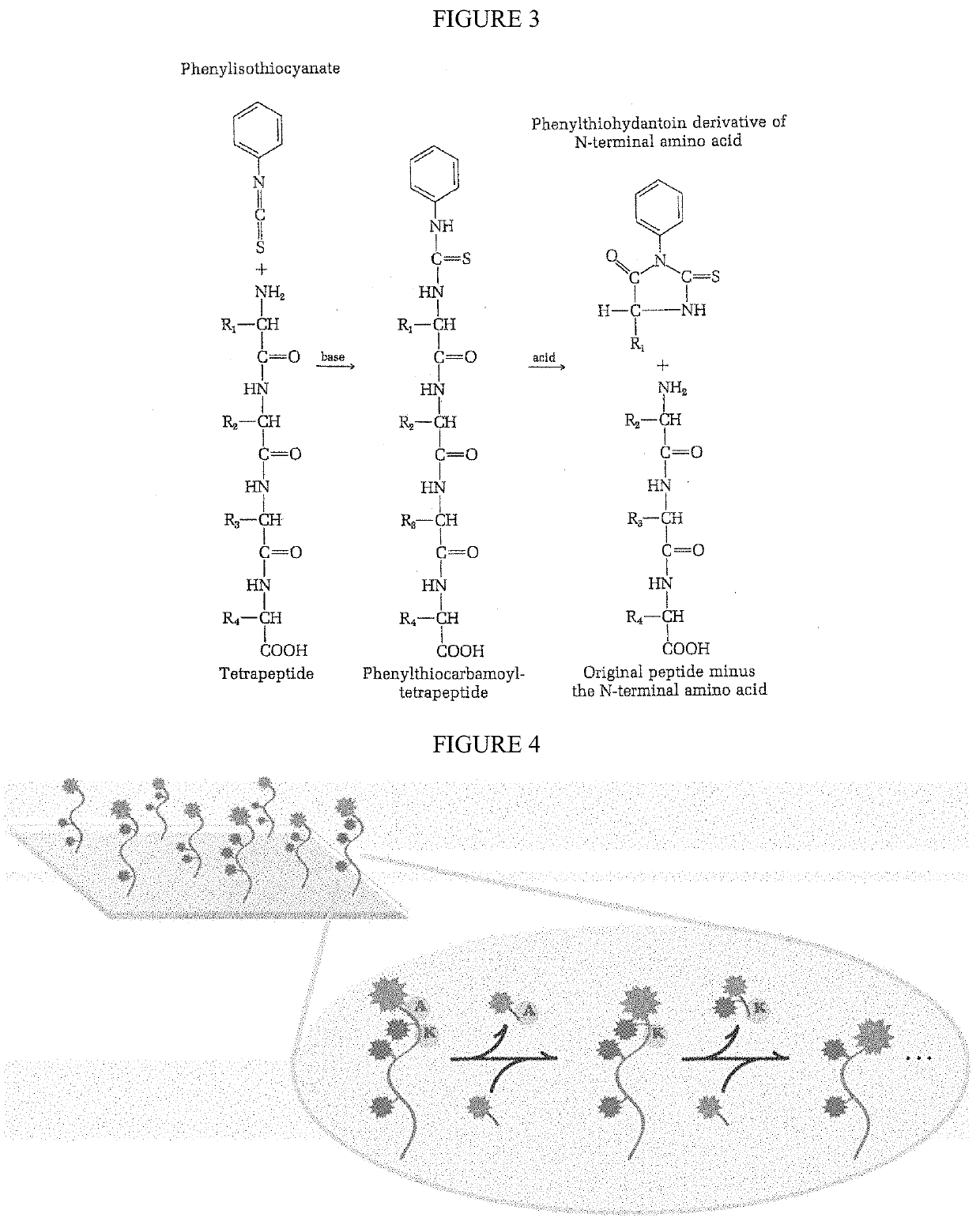
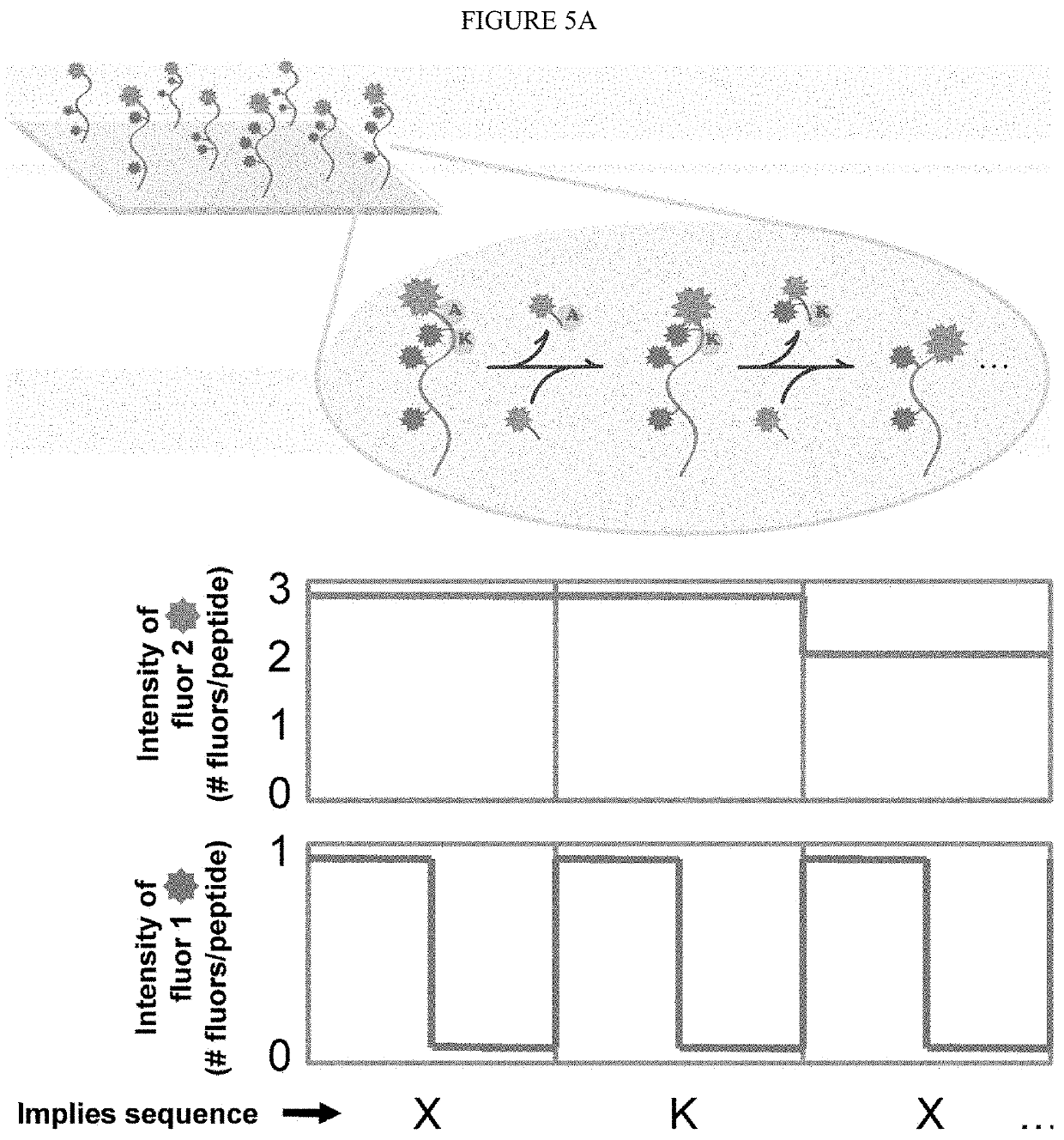
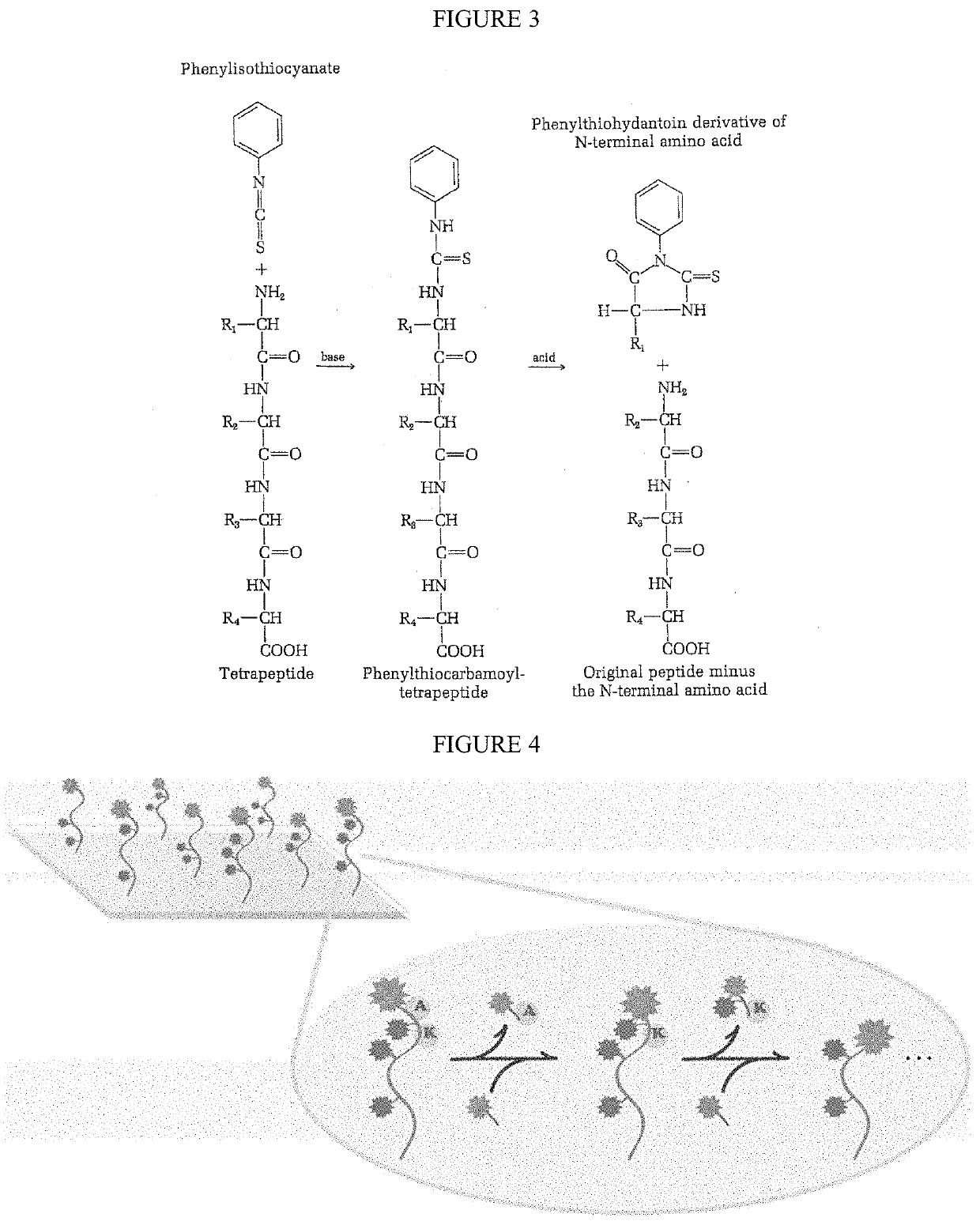
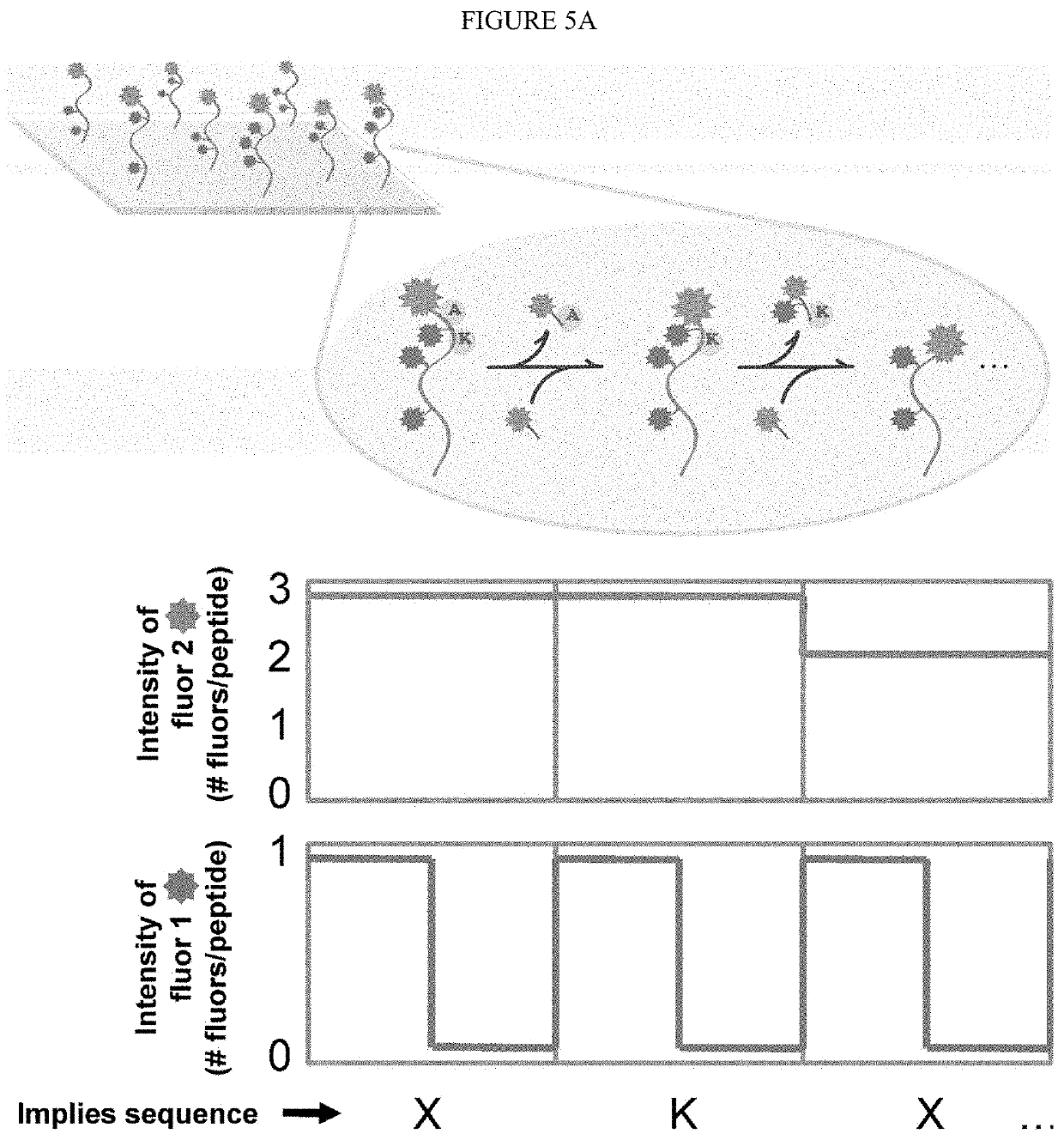
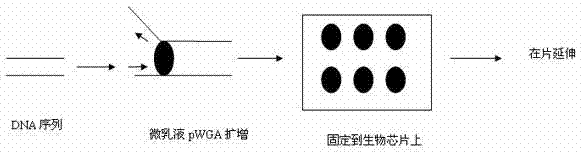
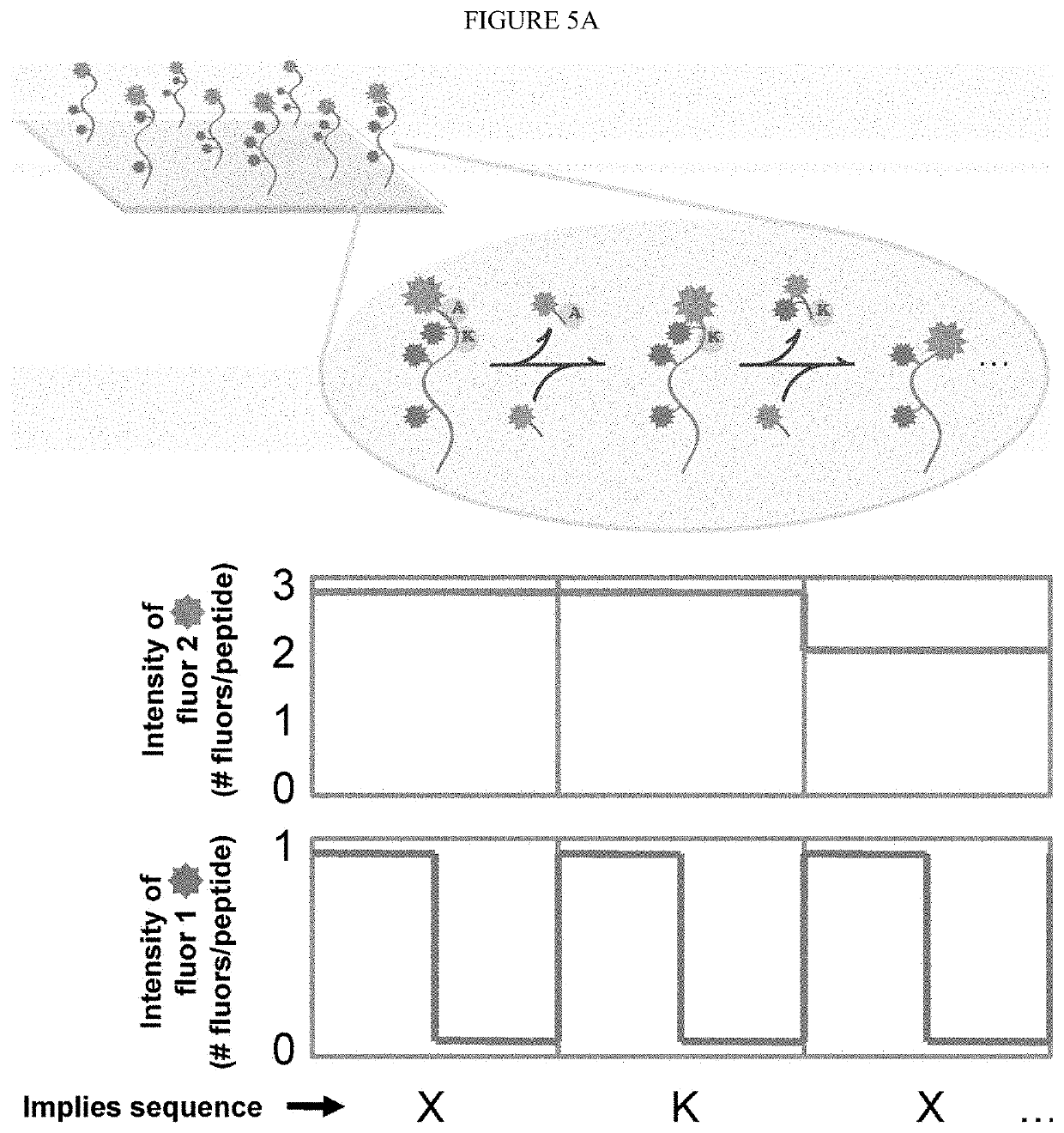
The present invention relates to the field of identifying proteins and peptides, and more specifically large-scale sequencing of single peptides in a mixture of diverse peptides at the single molecule level. The present invention also relates to methods for identifying amino acids in peptides, including peptides comprising unnatural amino acids. In one embodiment, the present invention contemplates labeling the N-terminal amino acid with a first label and labeling an internal amino acid with a second label. In some embodiments, the labels are fluorescent labels. In other embodiments, the internal amino acid is Lysine. In other embodiments, amino acids in peptides are identified based on the fluorescent signature for each peptide at the single molecule level.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
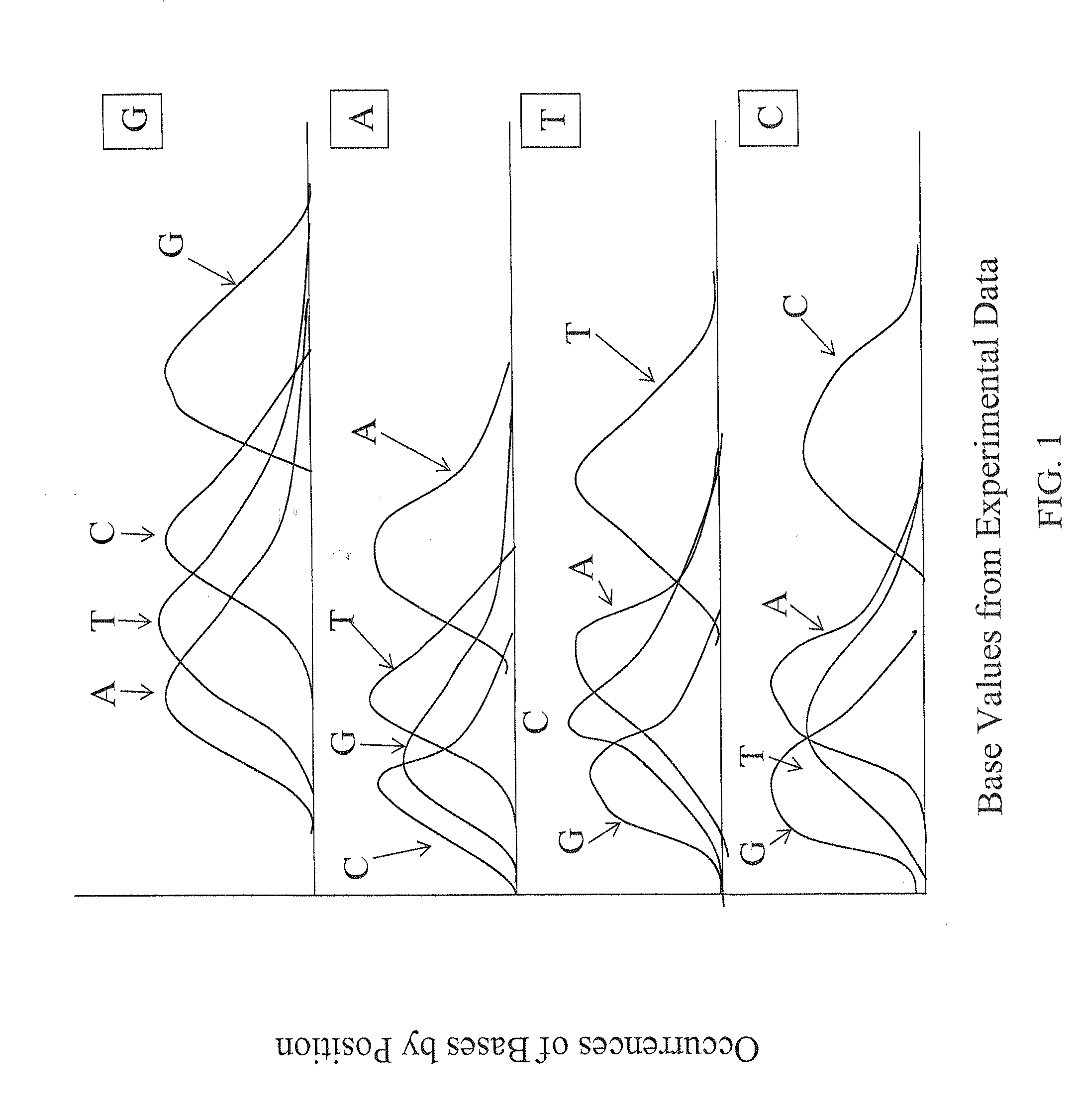
Methods of nucleic acid identification in large-scale sequencing
InactiveUS20090105961A1Improve accuracyAccurate measurementMicrobiological testing/measurementBiological testingData setLarge-Scale Sequencing
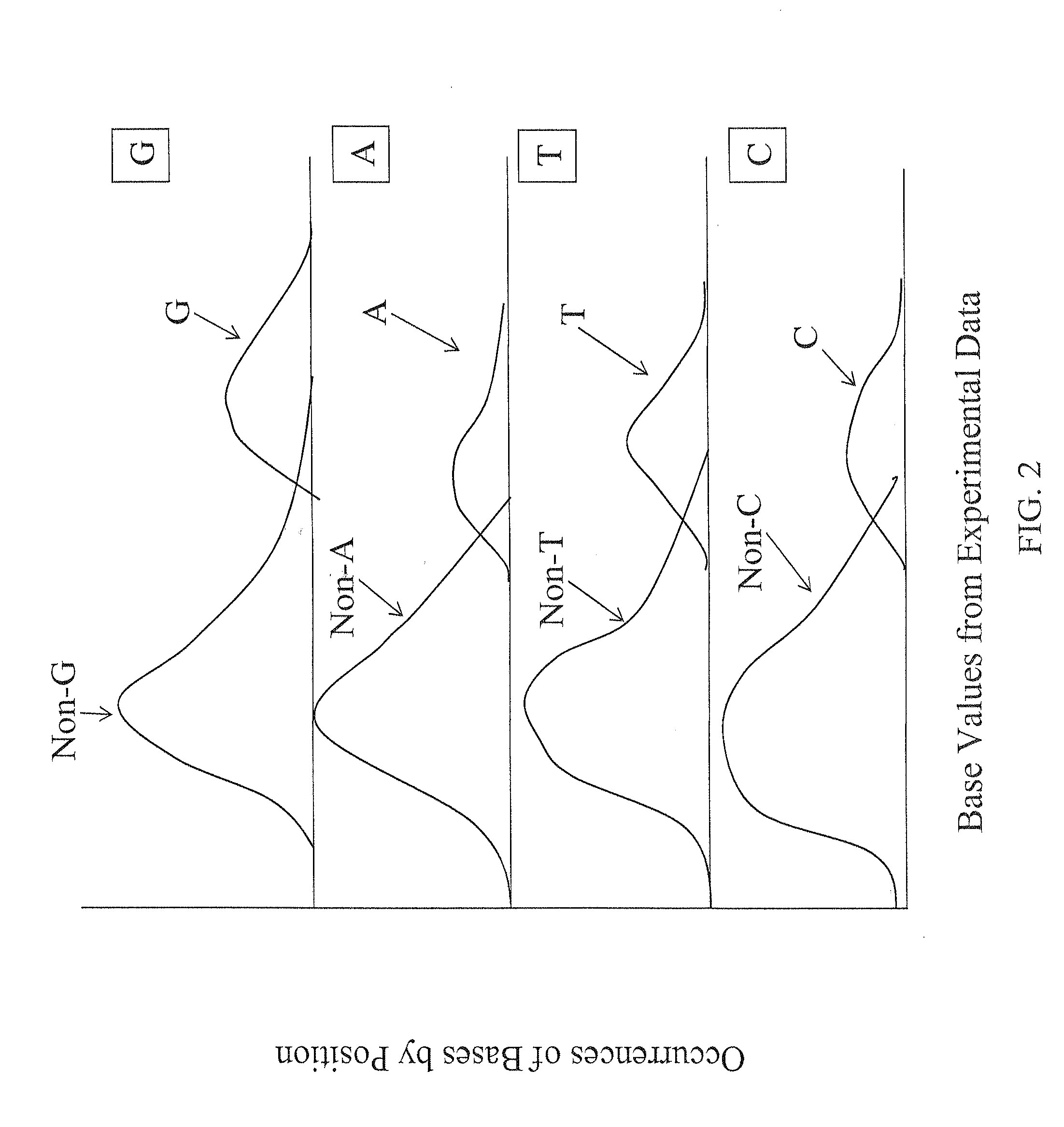
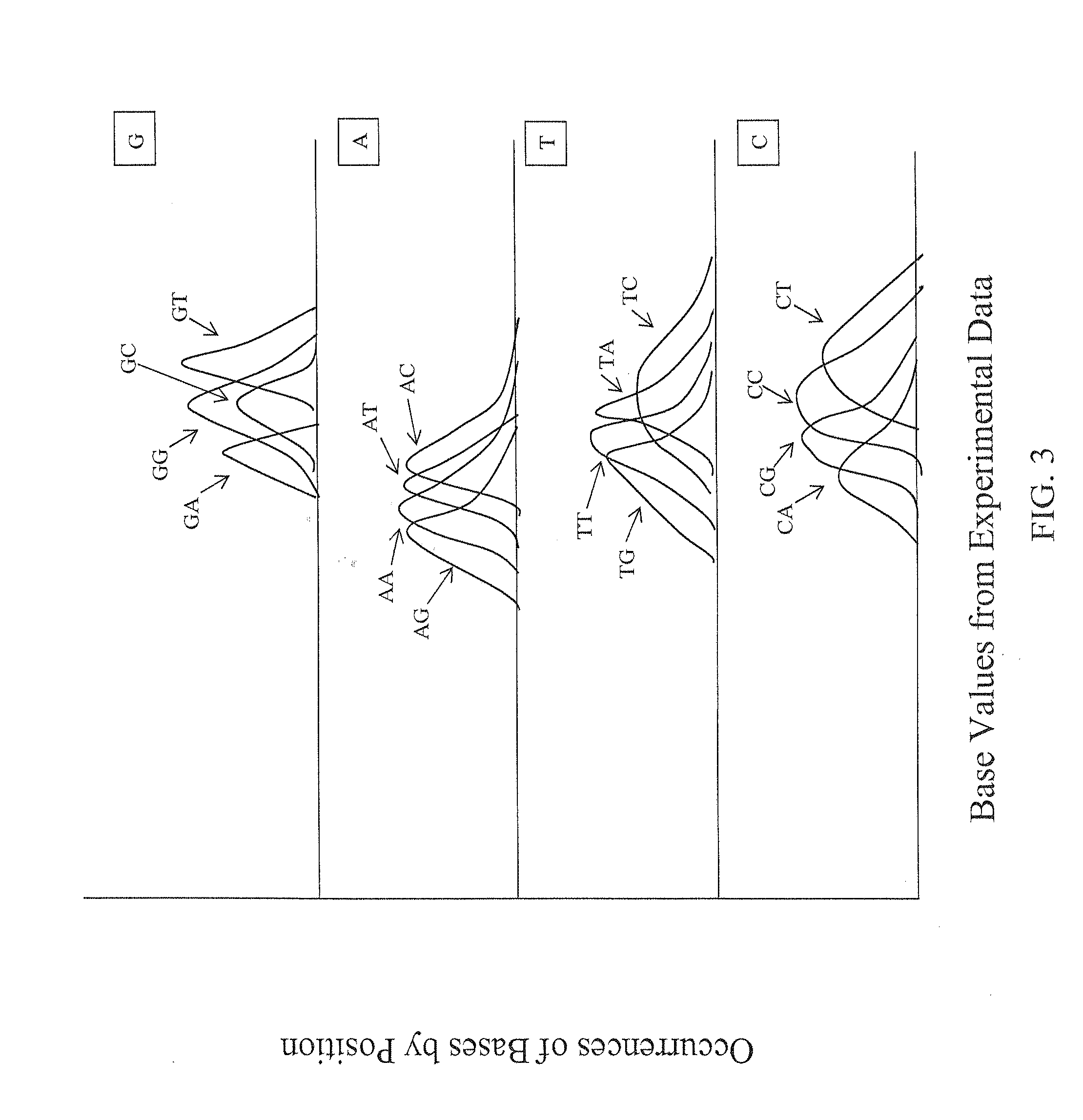
The present invention provides methods for determining a base probability in a target nucleic acid within an experimental data set. The methods of the invention provide specific methods of improving accuracy of base calling for experimental sequencing data compared to conventional methods. The experimental base values used in the methods of the present invention provide relative base probabilities within an experimental data set that are robust and uniformly optimal regardless of the experimental conditions.
Owner:COMPLETE GENOMICS INC
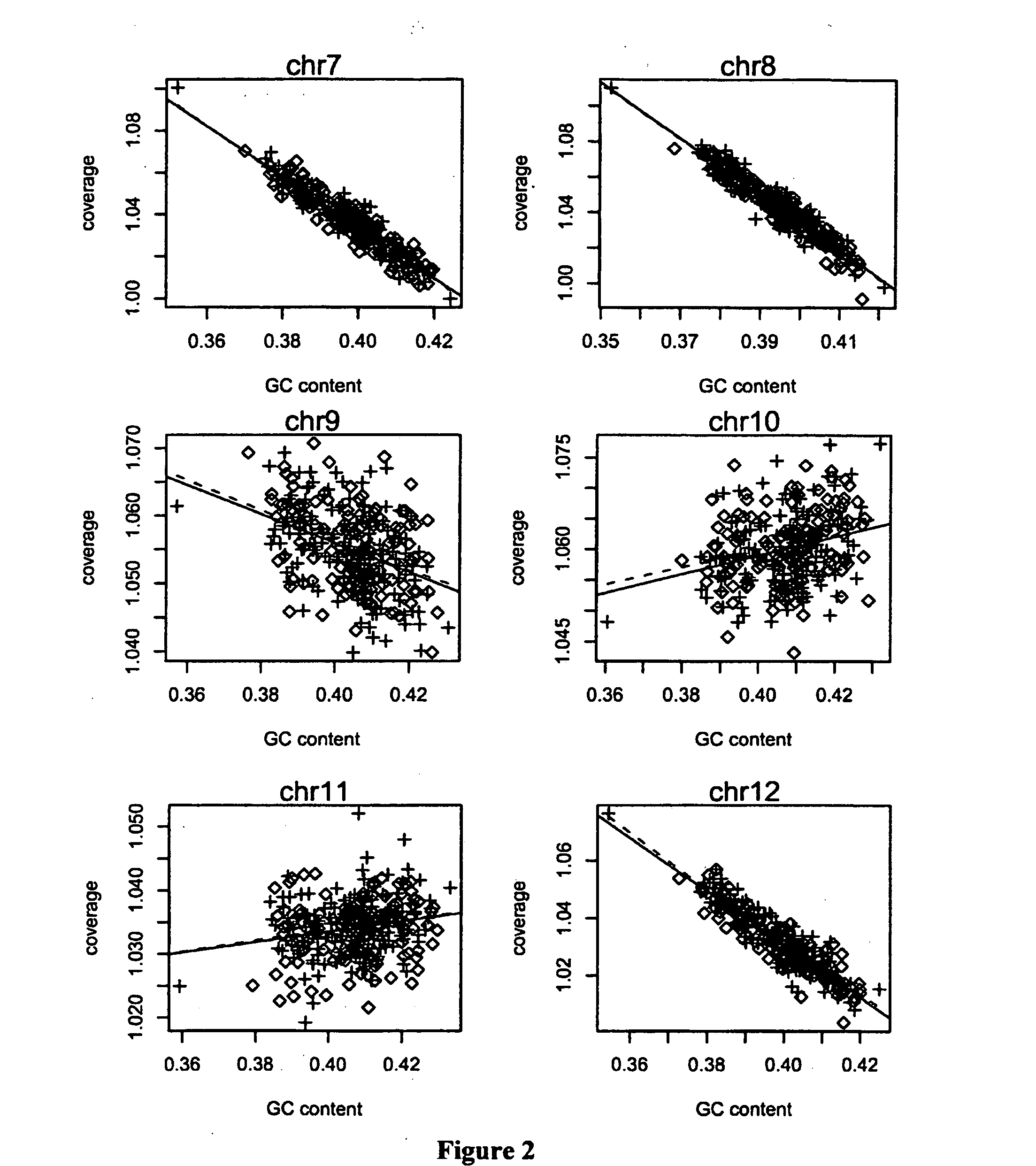
Method and device for carrying out GC correction on chromosome sequencing results
ActiveCN104120181ABioreactor/fermenter combinationsBiological substance pretreatmentsHereditary disordersLarge-Scale Sequencing
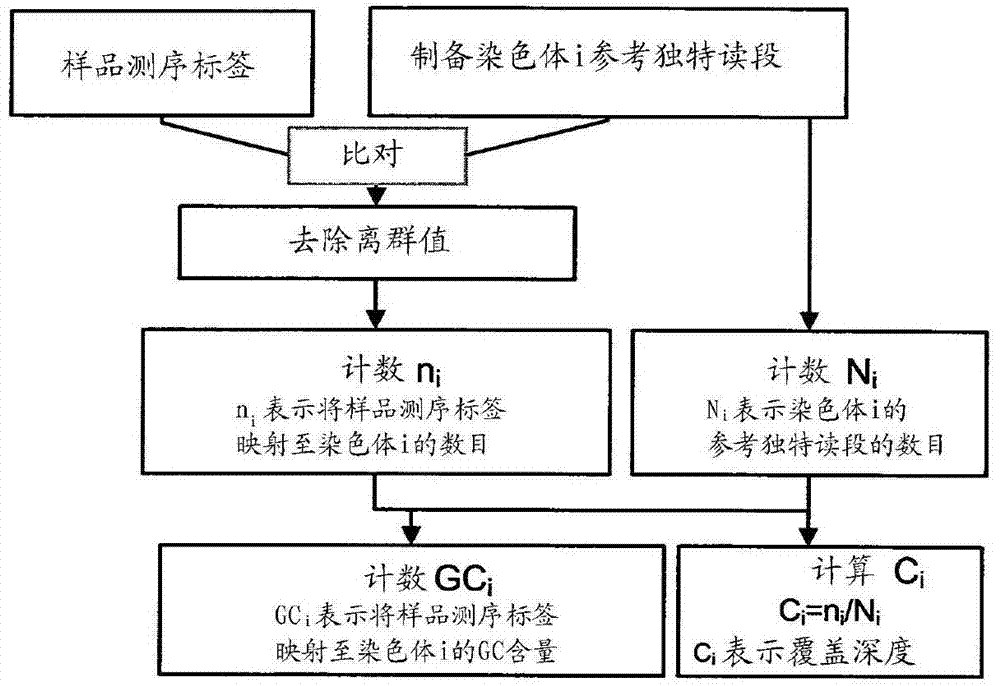
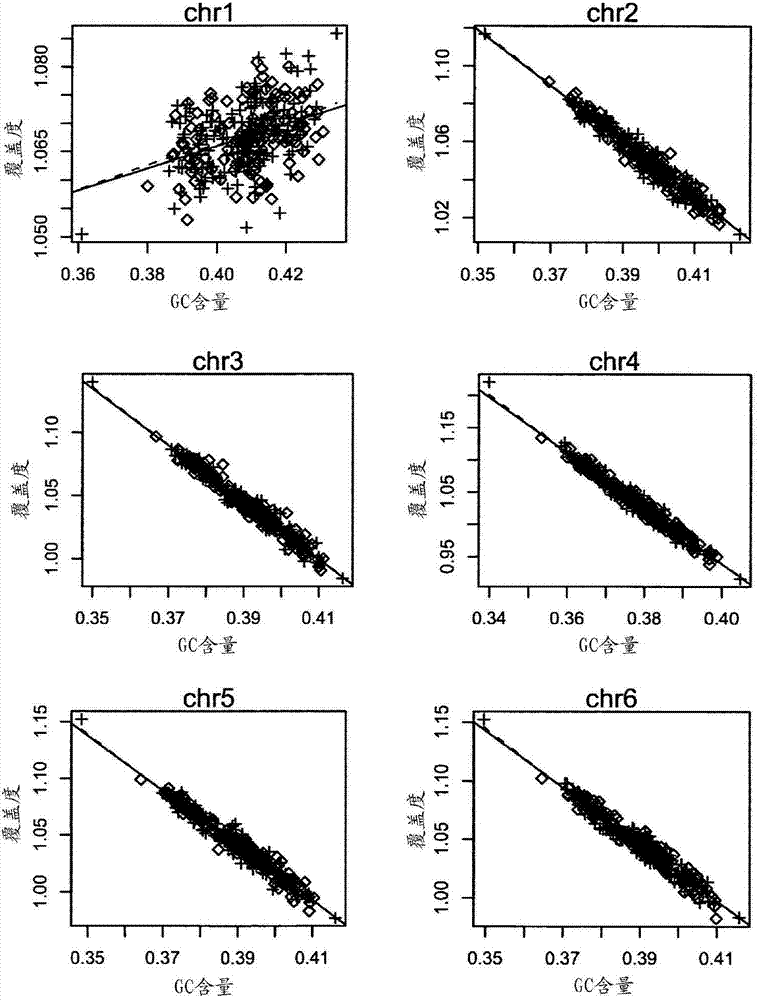
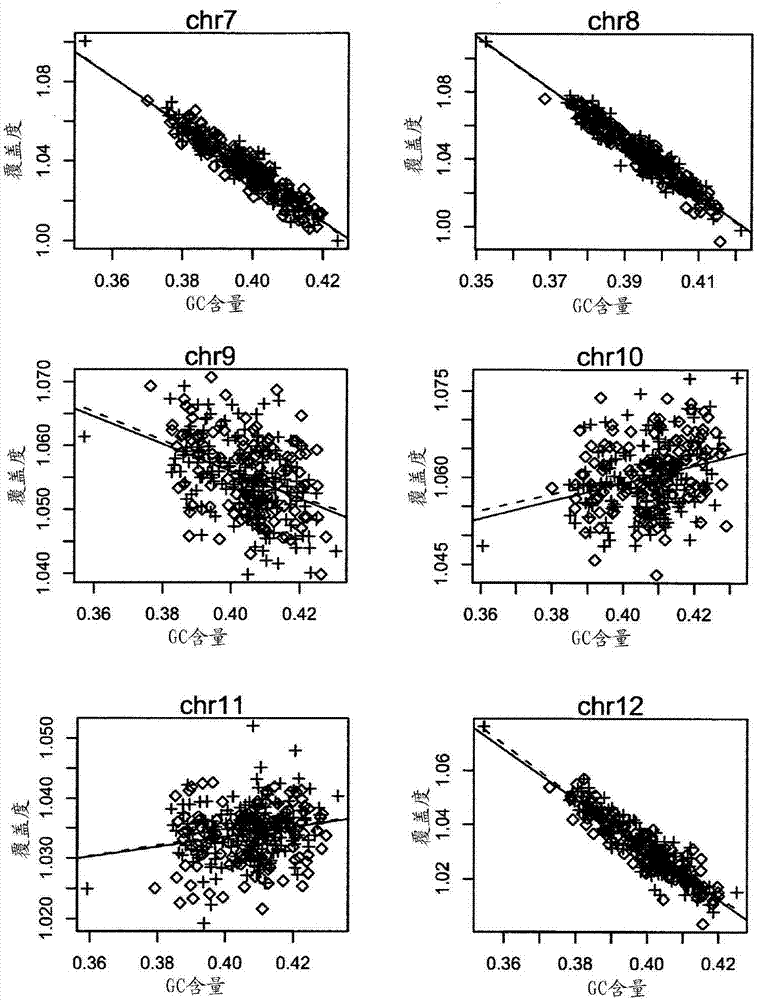
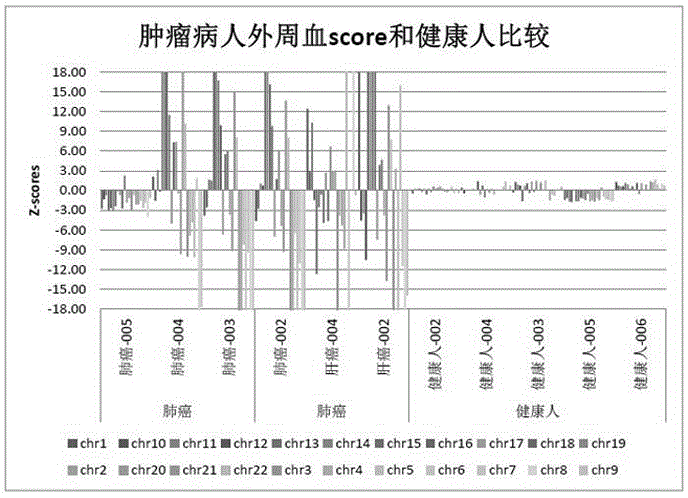
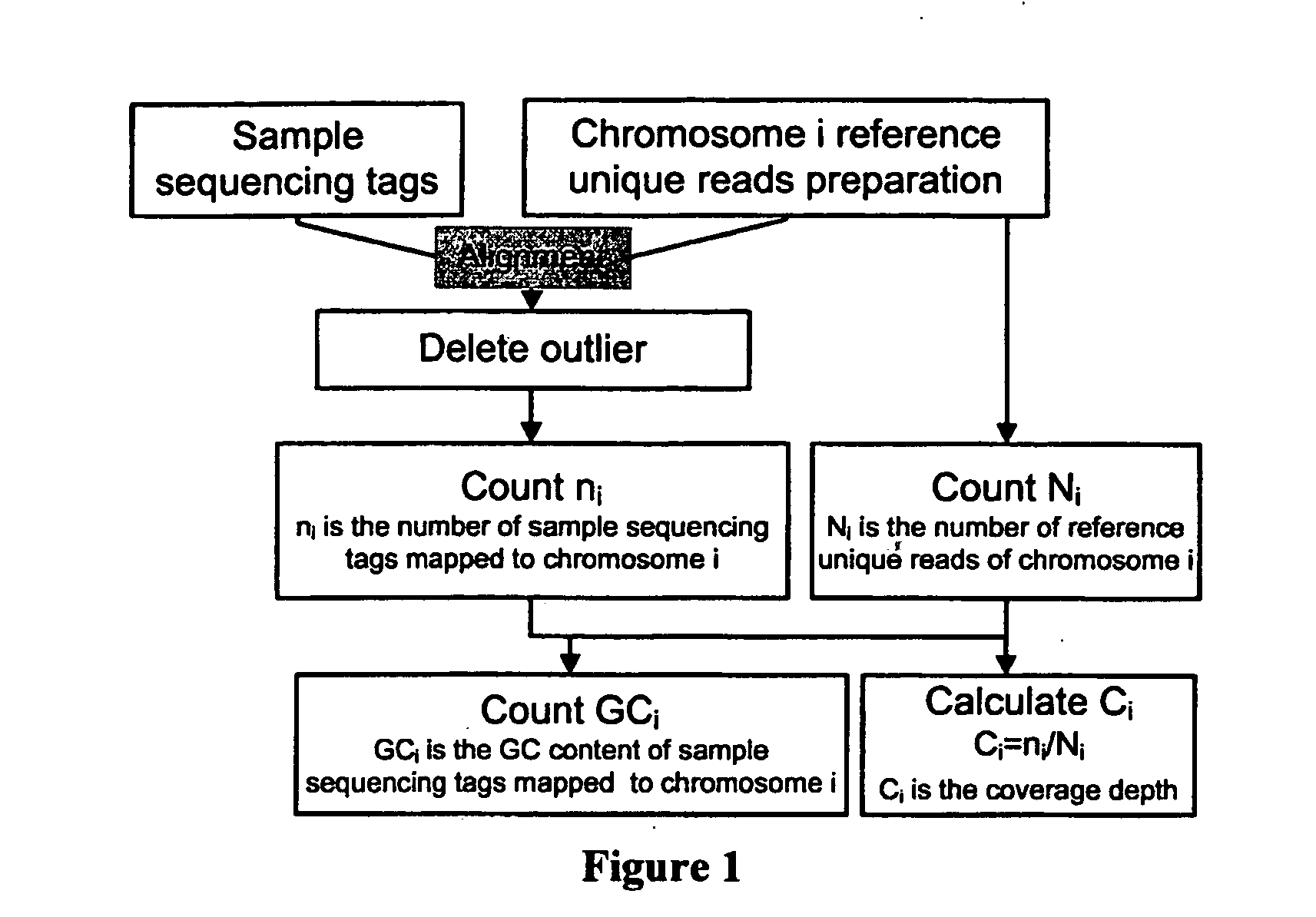
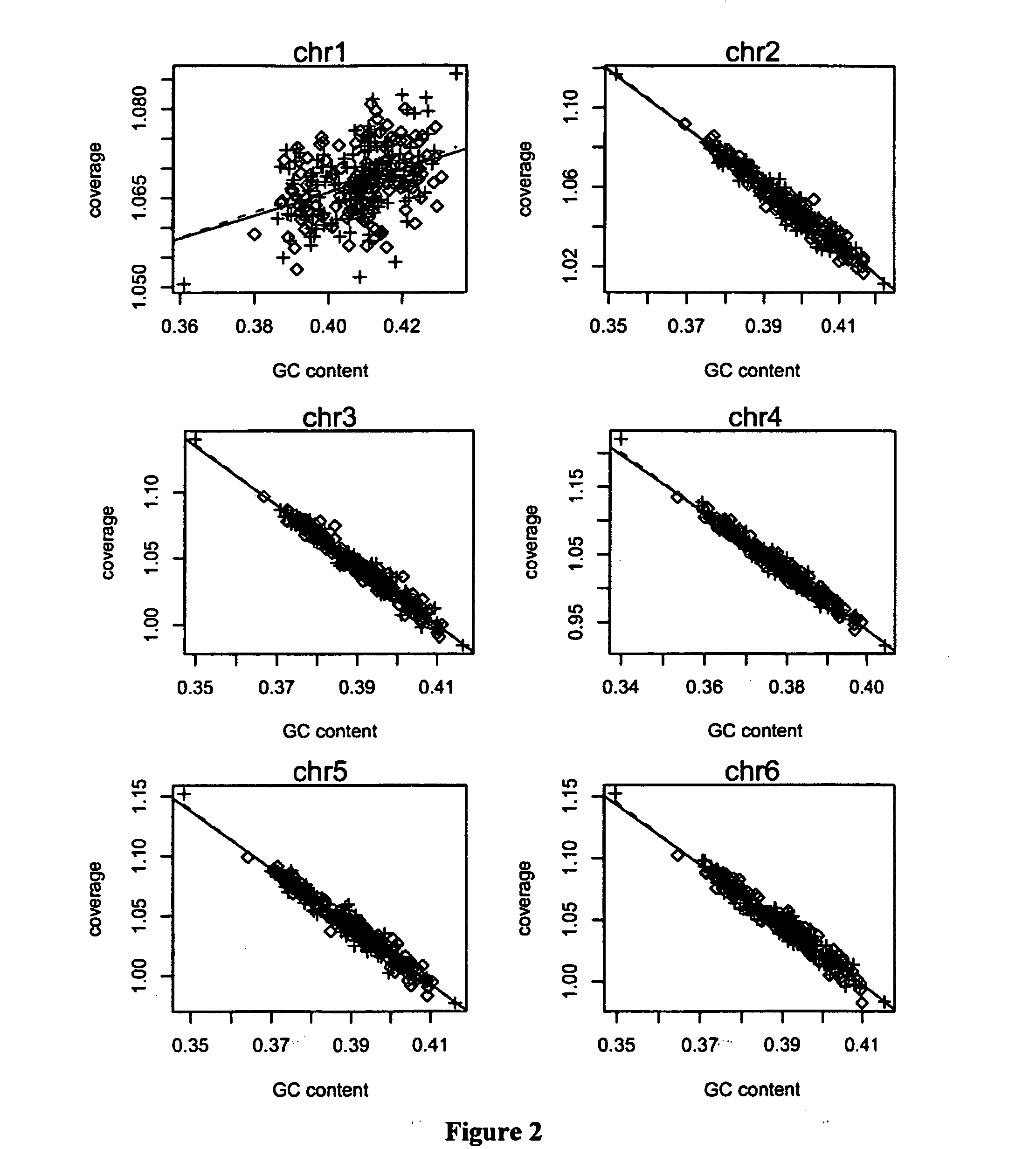
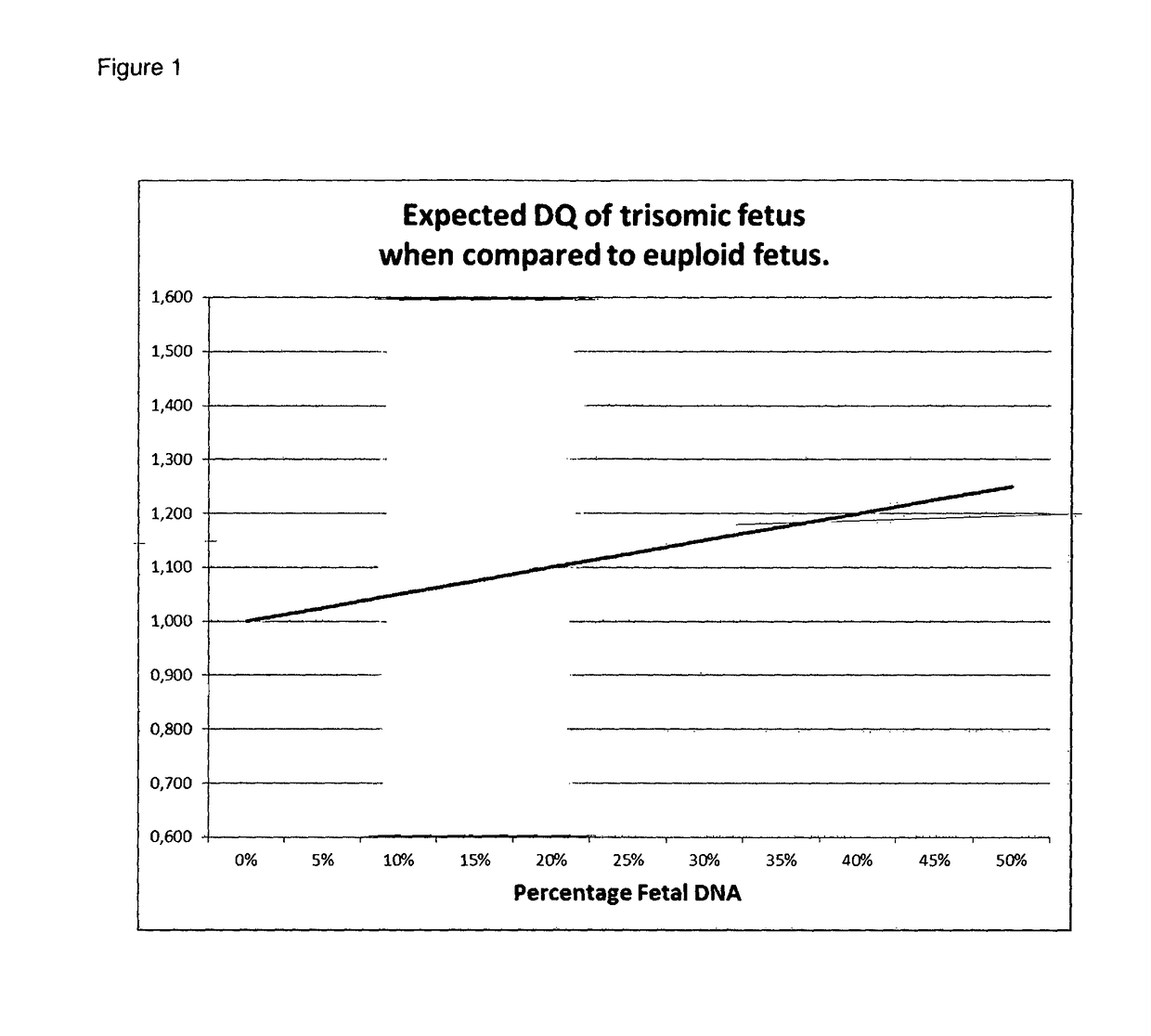
The invention relates to a non-diagnostic method for noninvasively detecting fetal hereditary disorder by sequencing nucleotides from a maternal biological sample on a large scale, and further provides a method for eliminating sequencing result GC bias caused by chromosome GC content difference. The method disclosed by the invention not only enables detection to be more accurate, but also provides a non-diagnostic comprehensive method for detecting fatal aneuploidy including sex chromosomal disease cases, for example, XO, XXX, XXY, XYY, and the like.
Owner:BGI GENOMICS CO LTD
Corn whole genome SNP chip and application thereof
ActiveCN108004344AEffective SNP siteMicrobiological testing/measurementDNA/RNA fragmentationLarge-Scale SequencingGermplasm
The invention relates to the field of molecular biology and genome breeding, and particularly discloses a corn whole genome SNP chip and application thereof. SNP loci covering a genetic expression area and SNP loci which are uniformly distributed in the whole genome are collected, after sifting, a high-throughput Infinium SNP chip of Illumina is prepared, and the chip comprises 500 probes of SNP markers shown in a detection list 1. By means of the selected SNP loci based on a large-scale sequencing result, specific signals which have reasonable frequency distribution, a low miss rate and low false positives are obtained, wherein 2 / 3 SNP loci are distributed in all the corn warm zone and tropical zone materials, and can be effectively used for selecting and utilizing the tropical zone materials; 426 SNP loci are functional markers, and are obviously relevant to kernel substance accumulation gene expression; an Infinium platform of Illumina has the advantages of being high in SNP typingdetection rate, high in repetitive rate, simple in detection and high in accuracy. The corn whole genome SNP chip can be used for corn germplasm resource molecular marker fingerprint analysis, corn linkage map construction, QTL locating, corn breeding material background selection and the like.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Cancer detection kit based on large-scale data mining and detection method
InactiveCN105653898AGroundbreaking improvementSecurity Enhancements and ImprovementsBiostatisticsSequence analysisCancer cellLibrary preparation
The invention discloses a cancer detection kit based on large-scale mining and a detection method, and belongs to the technical field of biomedical detection. The kit comprises a DNA extraction reagent, a high-throughput sequencing library preparation reagent, gene sequence alignment software and chromosome cover degree calculation software. The method comprises the steps that firstly, peripheral blood is collected from a subject and plasma is separated out; DNA polymerase is used for amplification and a sequencing library is prepared; large-scale sequencing is performed on the prepared library; sequence alignment is performed on a sequencing result, statistic is performed on the distribution situation on a genome, and whether a cancer cell genomic sequence from tumor tissue exists or not is judged, so that whether a detection object carries a cancer or not is judged. According to the cancer detection kit and the detection method, the defect that existing cancer screening specificity and sensitivity are poor is overcome, the method causes no irradiation or wounds, and cancer detection can be achieved only through 4-10 mL of peripheral blood.
Owner:江苏格致生命科技有限公司
Nucleic acid sequencing method based on fluorescence quenching
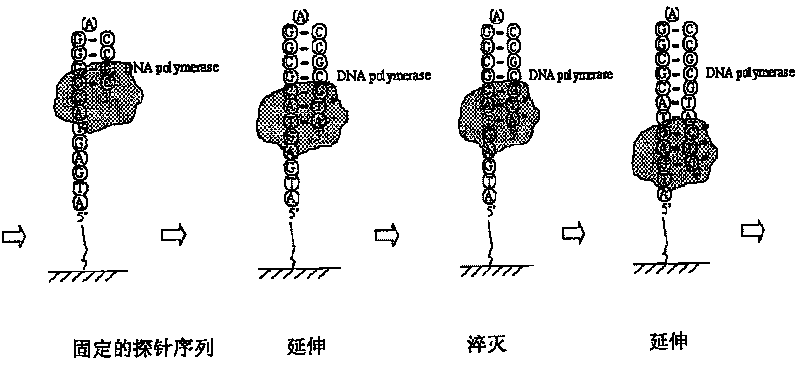
InactiveCN101168773ALow costImprove throughputMicrobiological testing/measurementFluorescencePolymerase L
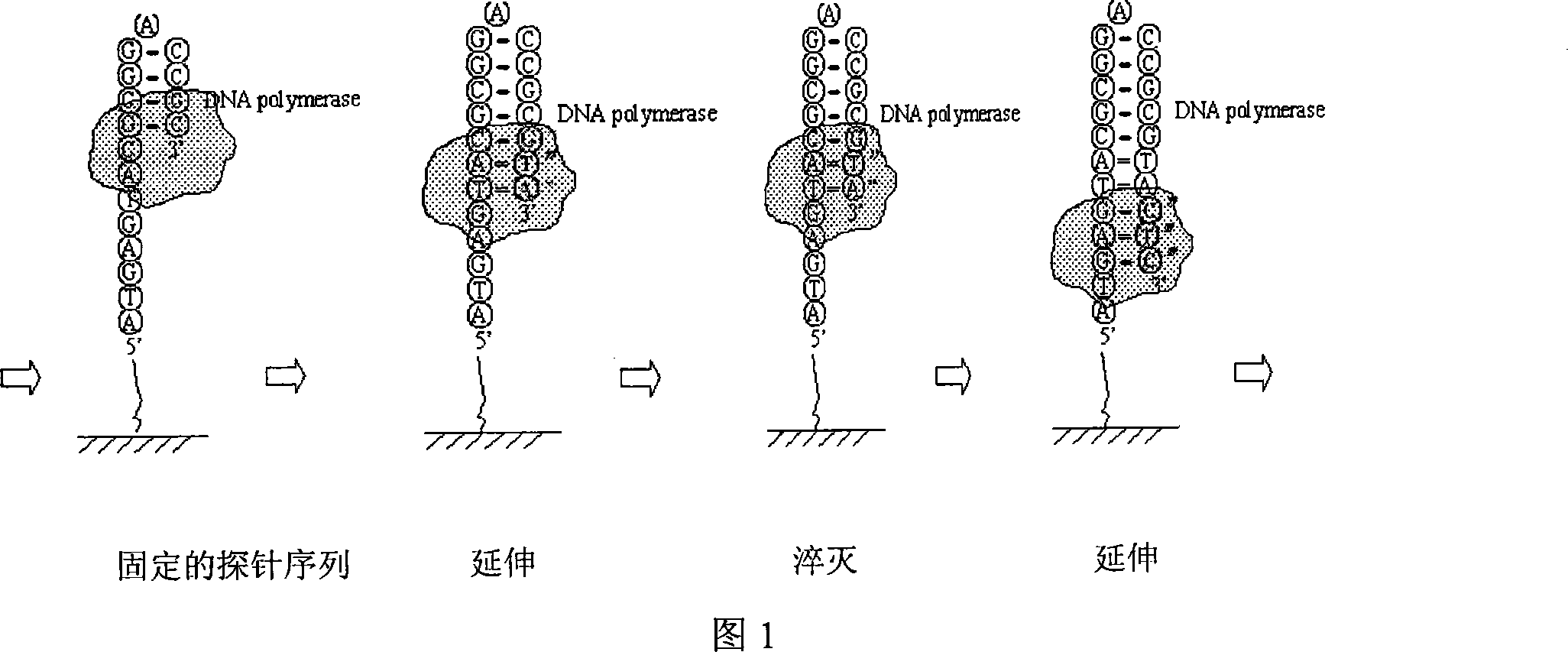
A nucleic acid sequencing method based on fluorescence quenching is: 1) preparing a sequencing template: the nucleic acid fragments to be sequenced, where the nucleic acid fragments include deoxyribonucleic acid, ribonucleic acid or their nucleic acid sequence amplification products, cloned products, and The product is immobilized on the carrier, 2) On-chip extension: Extend with reaction buffer, polymerase and fluorescently labeled dNTP mixture, 3) Fluorescence quenching: Use a fluorescent quencher to quench the fluorescence, 4) On-chip Extension: use reaction buffer, polymerase and fluorescently labeled dNTP mixture to extend, 5) Image recognition: use Matlab to extract feature points from the obtained extended image, remove noise points to achieve chip analysis or other commercial The optimized software directly processes the image. The invention combines nucleic acid microarray chip technology, and can perform large-scale sequencing of nucleic acids with high throughput, rapidity and low cost.
Owner:SOUTHEAST UNIV
Noninvasive detection of fetal genetic abnormality
ActiveUS20140099642A1Bioreactor/fermenter combinationsBiological substance pretreatmentsLarge-Scale SequencingFetal aneuploidy
The current invention is directed to methods for noninvasive detection of fetal genetic abnormalities by large-scale sequencing of nucleotides from maternal biological sample. Further provided are methods to remove GC bias from the sequencing results according to the difference in GC content of a chromosome. The current invention not only makes the detection much more accurate but also represents a comprehensive method for fetal aneuploidy detection including sex chromosome disorders such as XO, XXX, XXY, and XYY, etc.
Owner:BGI GENOMICS CO LTD
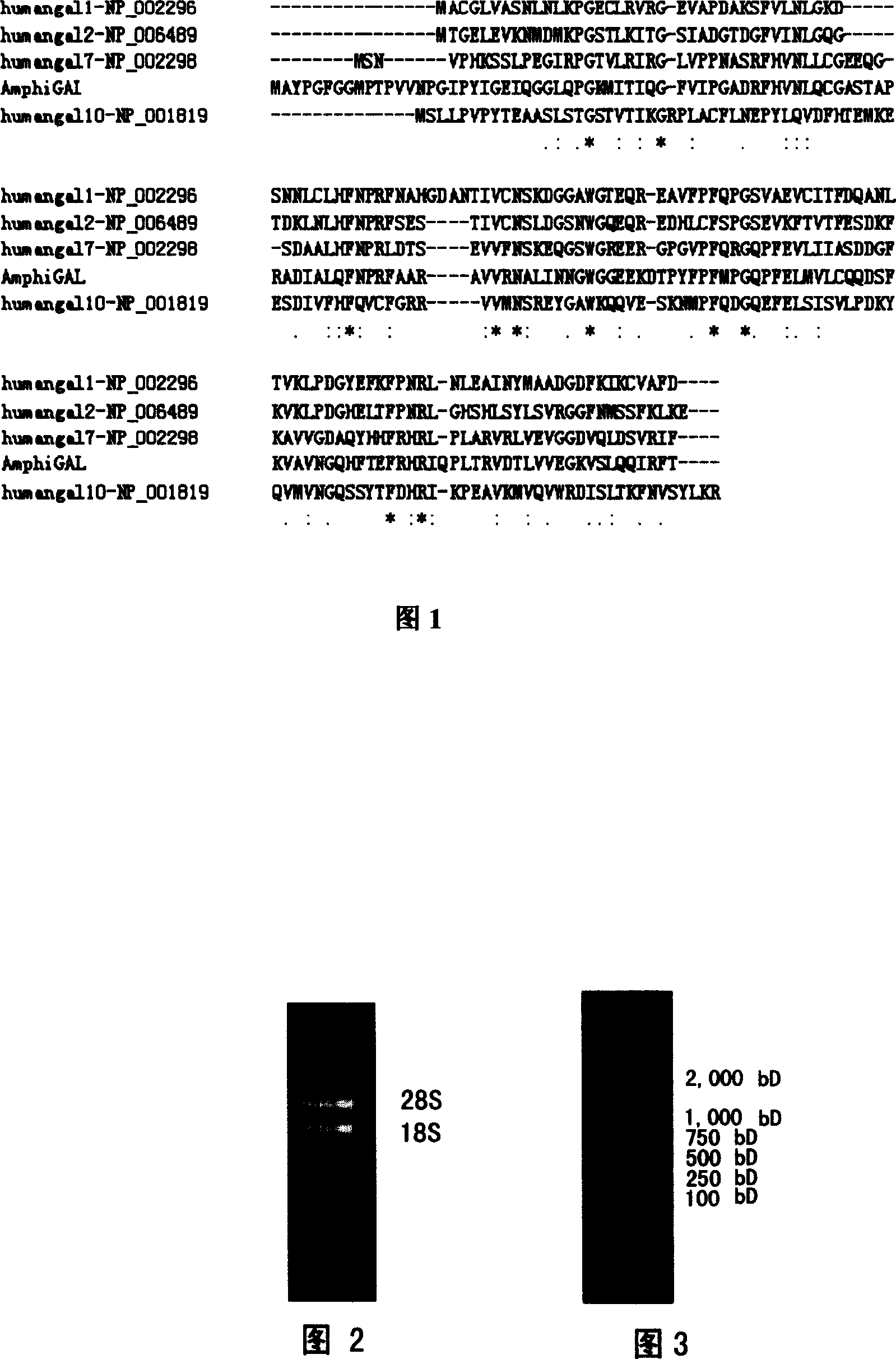
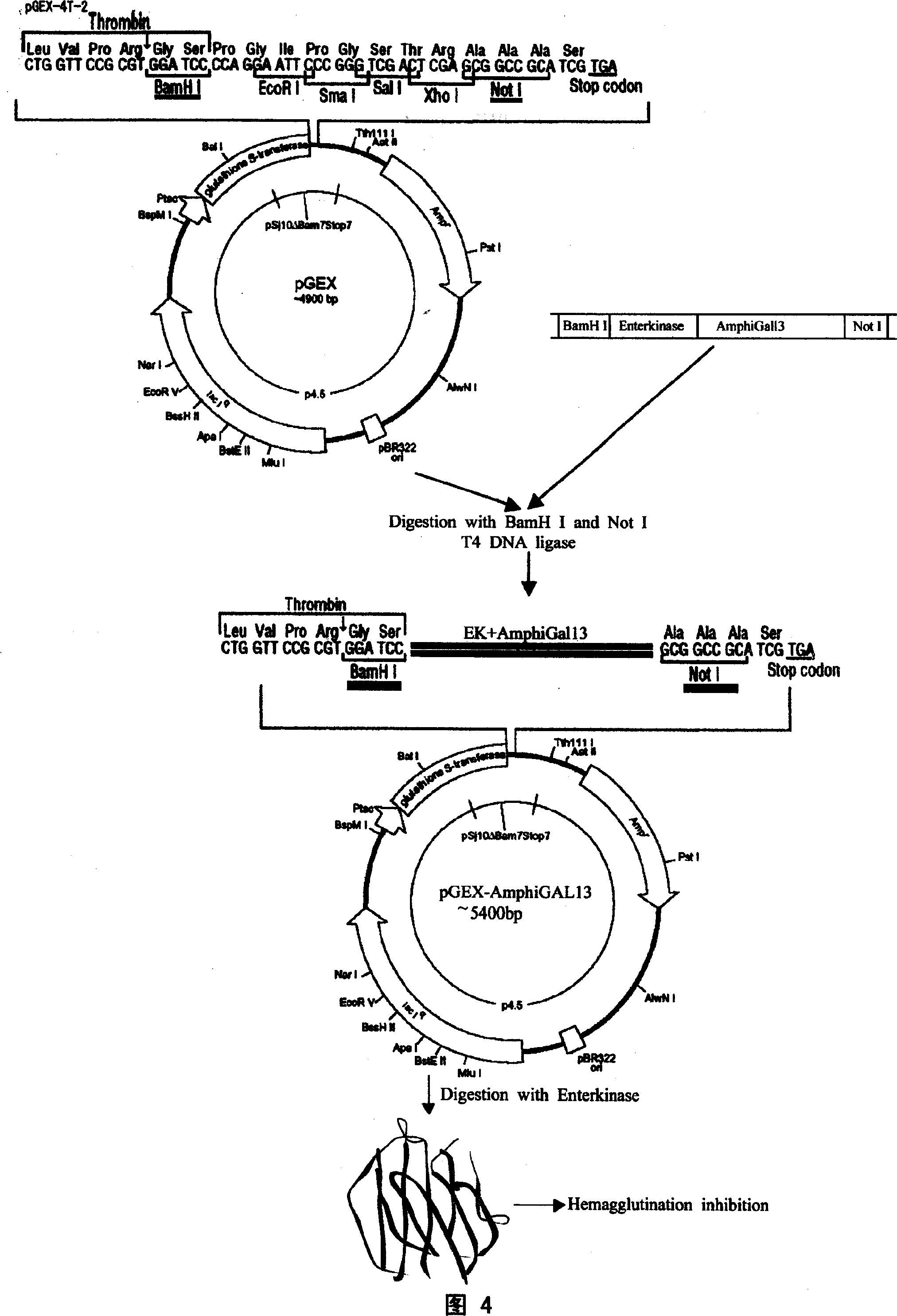
Amphioxus galactose lectin AmphiGAL13-gene and its use
Chinese amphioxus galactose agglutinin AmphiGAL13 gene and its production are disclosed. The process is carried out by constructing cDNA library, large-scaled sequencing, and separating out AmphiGAL13 gene from digestive tube of Chinese amphioxus. It expresses recombinant amphioxus GST-PGAL13 protein by GST fusion and obtains agglutinin by EK enzyme digestion. It can be used to develop and research for natural small molecule biological granular preparation.
Owner:SUN YAT SEN UNIV
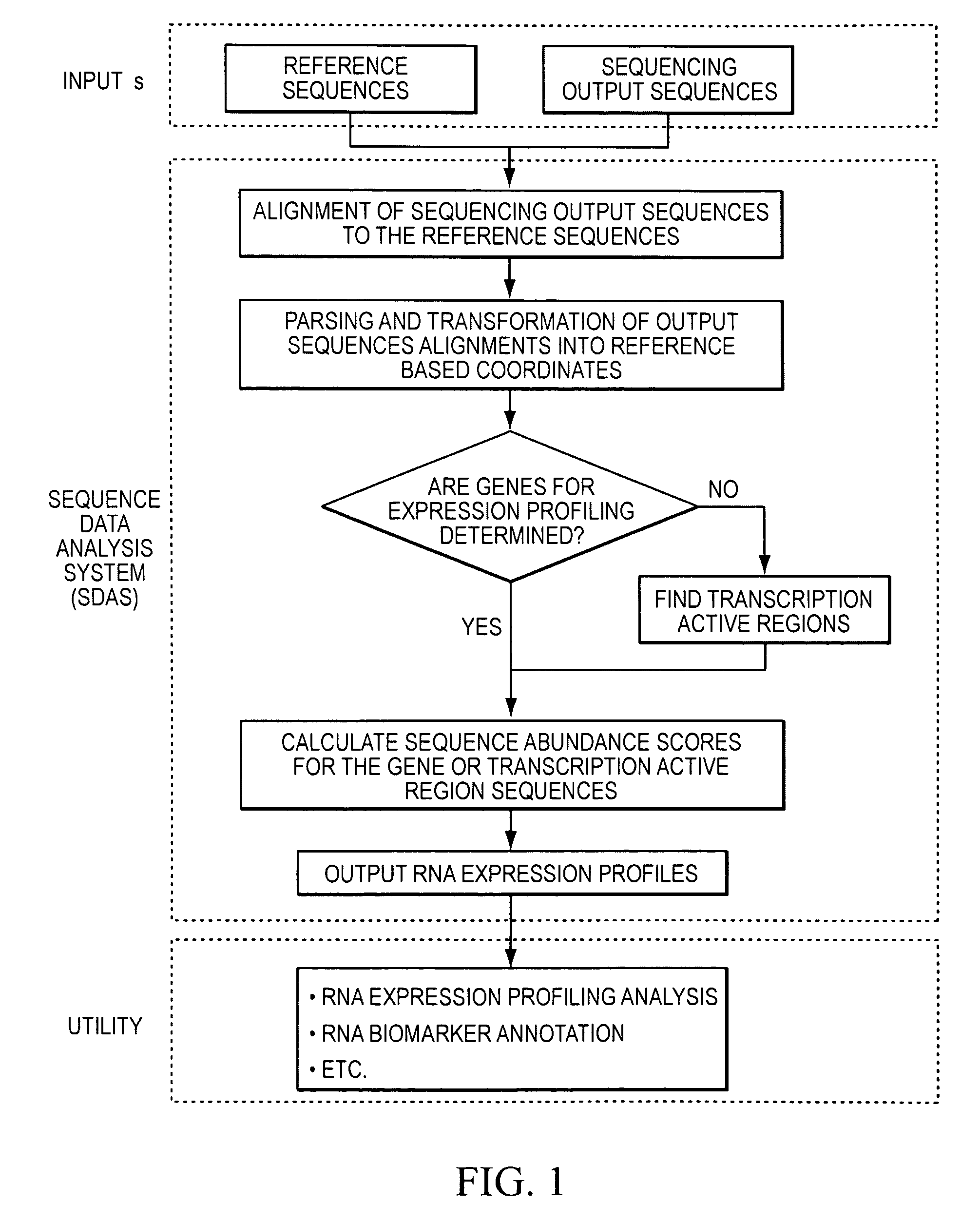
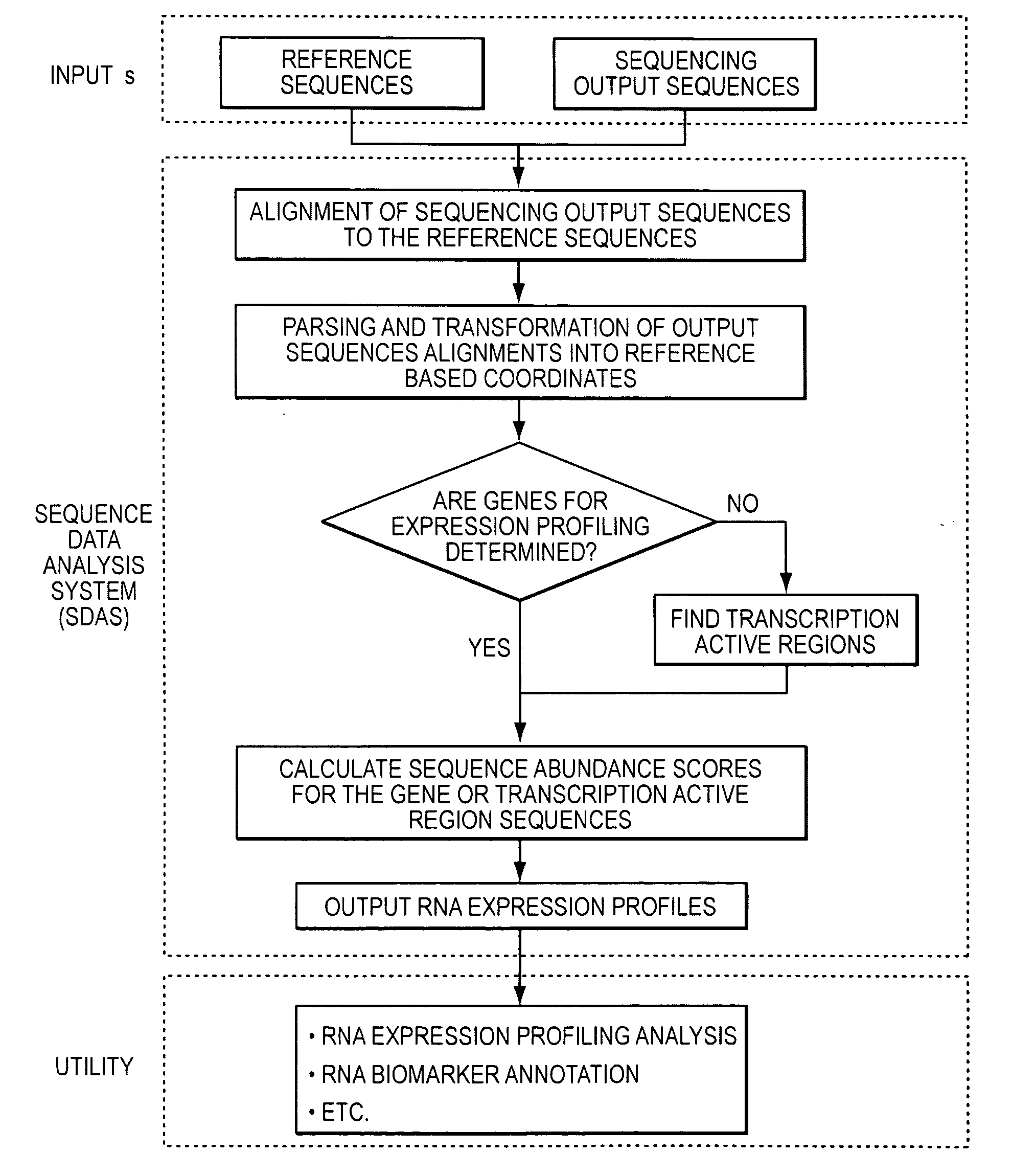
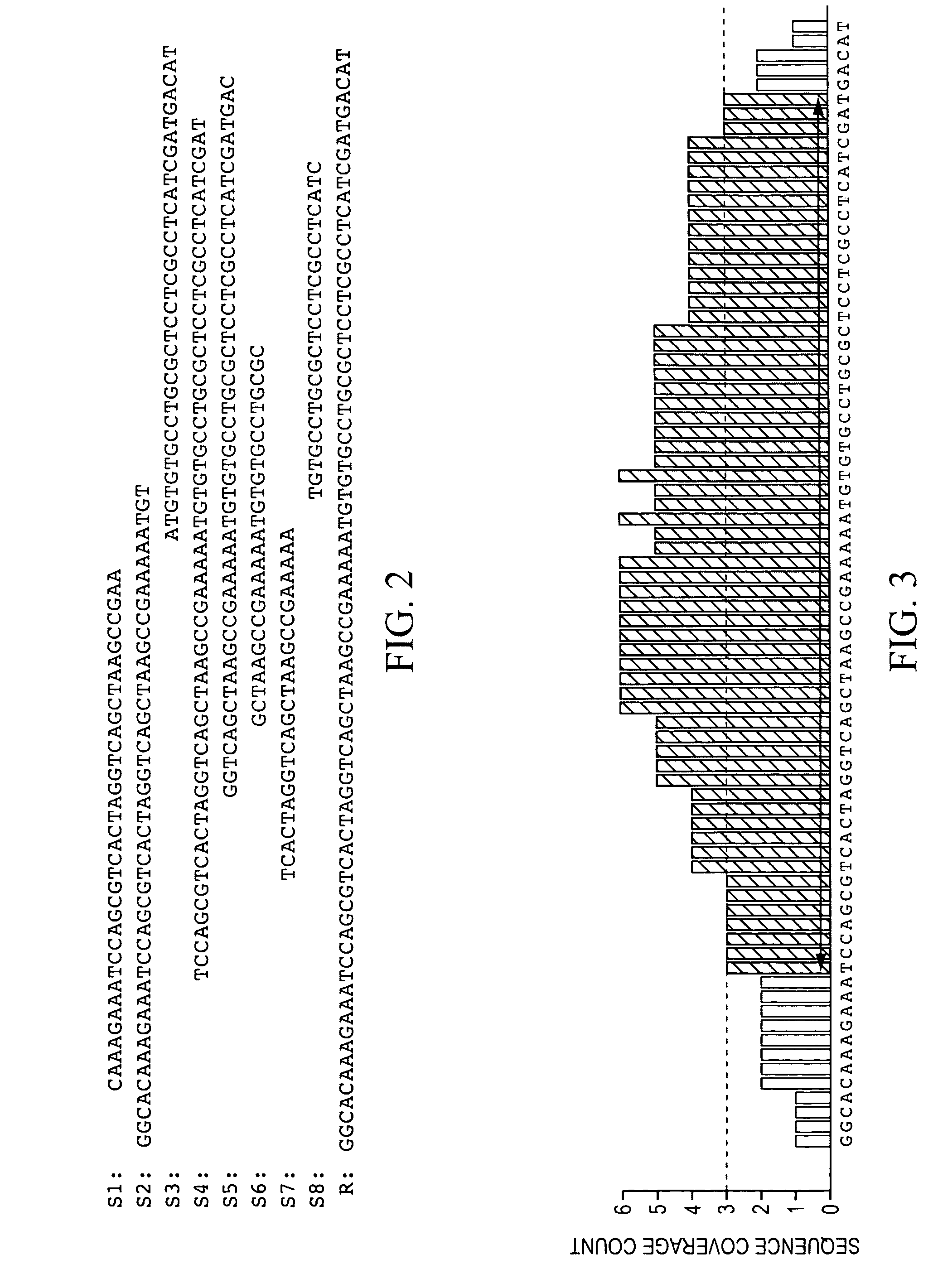
Method and system to characterize transcriptionally active regions and quantify sequence abundance for large scale sequencing data
Owner:GENOMIC HEALTH INC
Sequencing data compression method based on duplex codon
ActiveCN106067824ASmall footprintReduce data transfer timeCode conversionData compressionLarge-Scale Sequencing
The invention discloses a sequencing data compression method based on a duplex codon. The sequencing data compression method comprises the following steps of (1) cutting; (2) establishing of a hash table; (3) ordering; (4) establishing of a Huffman tree; (5) coding; and (6) calculation of the compression ratio, wherein the compression ratio of the algorithm is 8M / L. Space occupation of sequencing data is greatly reduced so that the sequencing data compression method plays a key role in storage and transmission of the large-scale sequencing data, and transmission time of local and cloud-side data can be saved.
Owner:洛阳晶云信息科技有限公司
Kit comprising primer pair for detecting genetic marker related to male infertility
InactiveCN104928385AEnables diagnostic testingMicrobiological testing/measurementMale infertilityLarge-Scale Sequencing
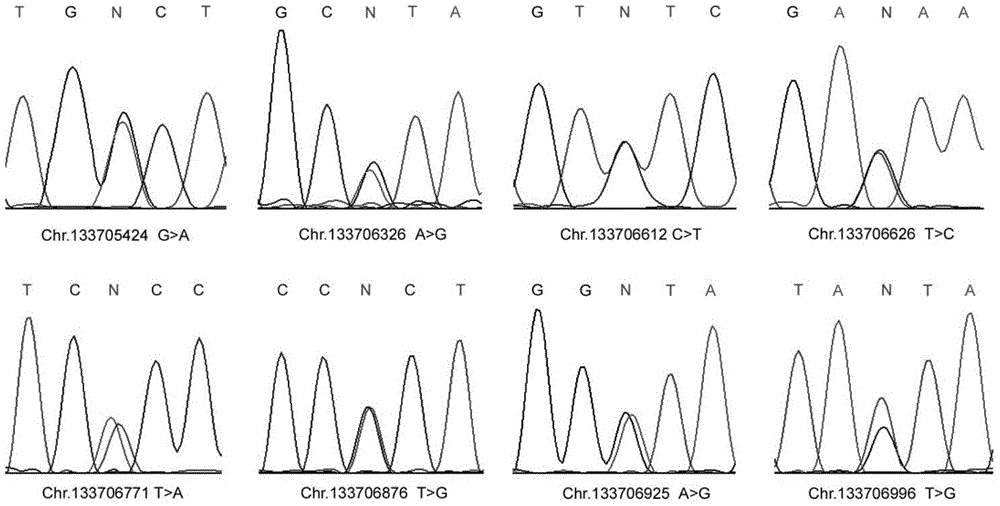
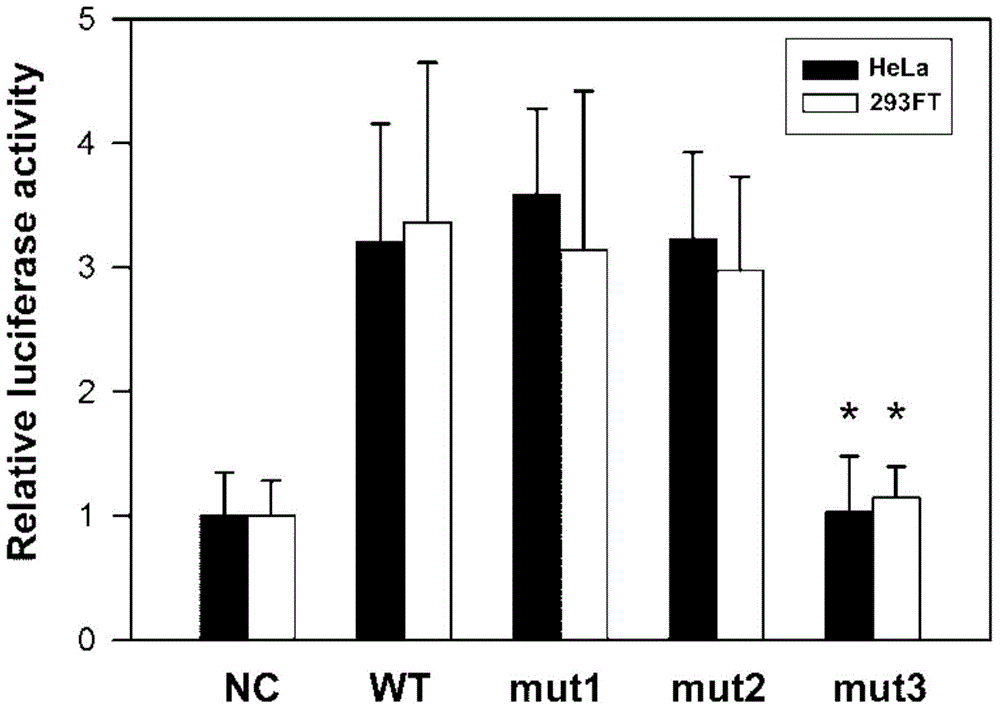
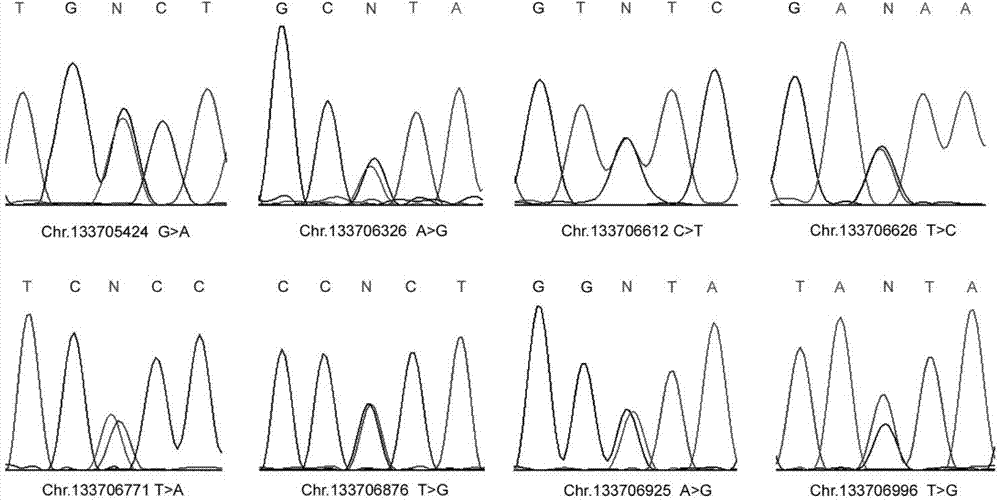
The invention discloses a male infertility related genetic marker, the sequence of which is that of the UBE2B gene. The binding site of the UBE2B gene's promoter region and the transcription factor SP1 undergoes mutation, and the mutation site is one or more of Chr.133706771T>A, Chr.133706876T>G and Chr.133706925A>G. By large-scale sequencing, four new mutation sites of the UBE2B gene's promoter region are selected from male infertility patients, a transcription factor plasmid is cotransfected into cells together with a wild type plasmid and a mutant plasmid respectively, and experiments find significant function differences and show that mutation of the UBE2B gene's promoter region is relevant to male infertility (especially azoospermia). Therefore, diagnosis detection of male infertility can be realized through the mutation.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
Central body located gene and its use in medicinal region
InactiveCN1657537APromote proliferationPromote cancerOrganic active ingredientsSugar derivativesTumour tissueDisease
A centrosome located gene cep is disclosed, which is prepared through sequencing, biological information analysis, and gene technique. It can be used as the candidate gene for treating the diseases is hemopoietic system and the generation and transfer of tumor, or as the target point of genetically engineered medicine.
Owner:RADIOLOGY INST ACAD OF MILITARY MEDICINE SCI PLA
Single molecule peptide sequencing
ActiveUS20200018768A1OmicsCarrier-bound/immobilised peptidesLarge-Scale SequencingProtein Sequence Determination
Identifying proteins and peptides, and more specifically large-scale sequencing of single peptides in a mixture of diverse peptides at the single molecule level is an unmet challenge in the field of protein sequencing. Herein are methods for identifying amino acids in peptides, including peptides comprising unnatural amino acids. In one embodiment, the N-terminal amino acid is labeled with a first label and an internal amino acid is labeled with a second label. In some embodiments, the labels are fluorescent labels. In other embodiments, the internal amino acid is Lysine. In other embodiments, amino acids in peptides are identified based on the fluorescent signature for each peptide at the single molecule level.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
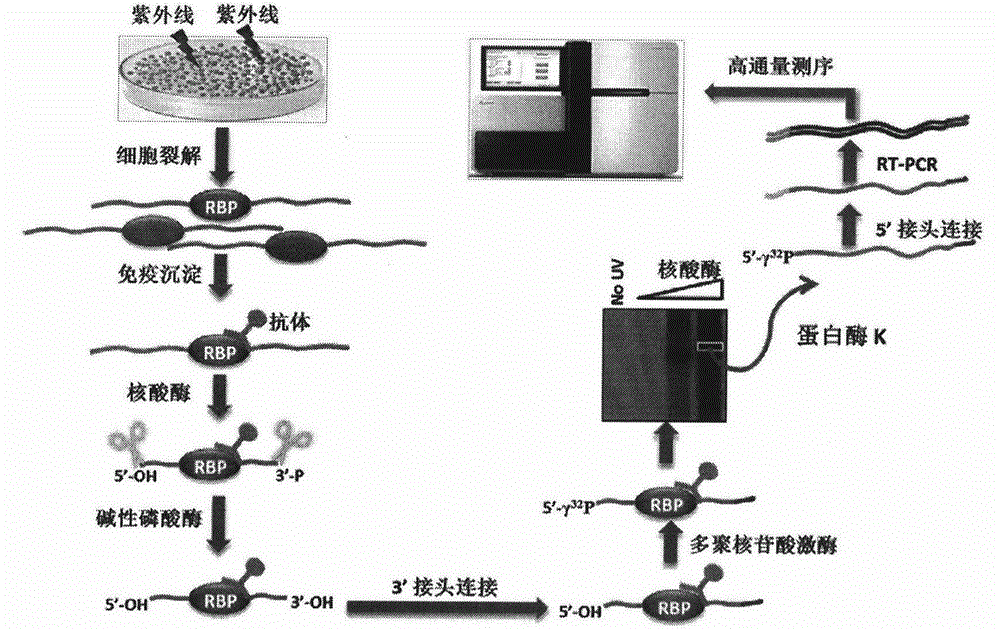
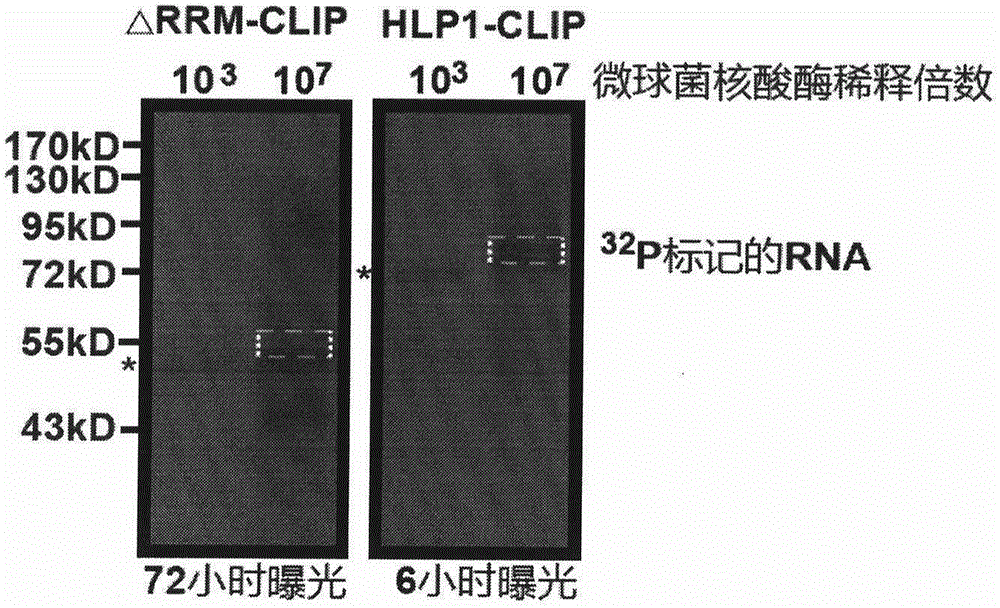
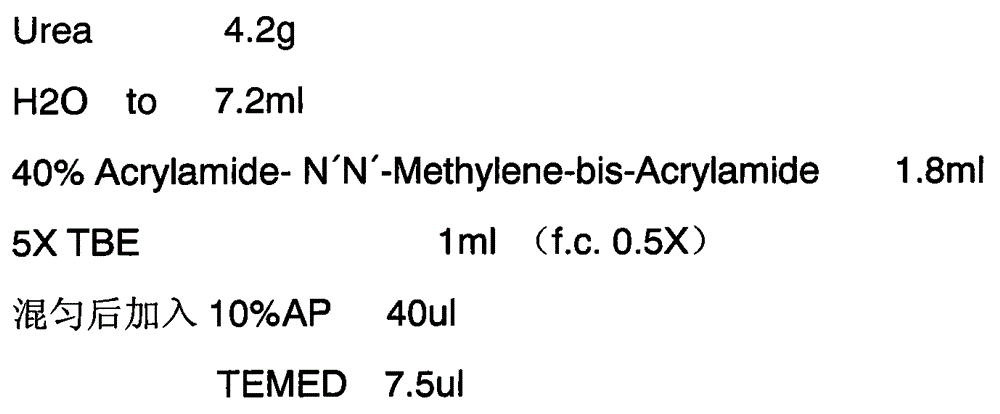
Method for identifying type and site of RNA binding with protein in plant
The invention relates to a method for identifying the type and site of a RNA binding with protein in a plant. The method mainly comprises the following steps: immobilizing the in-vivo binding state of protein and nucleic acid through ultraviolet crosslinking; subjecting immunoprecipitated RNAs to digestion with nuclease into small fragments; removing non-specifically bound RNAs by using SDS-PAGE and transferring methods; and carrying out reverse-transcription PCR amplification and large-scale sequencing by using adaptors connected with two ends of the RNA. The method has the advantages that 1) ultraviolet crosslinking is carried out in vivo, so real in-vivo binding conditions can be reflected; 2) since a covalent bond formed by ultraviolet crosslinking is irreversible, the length of nucleic acid binding with protein can be reduced through digestion by nuclease, and subsequent multi-step purification can greatly improve a ratio of signal to noise; 3) most non-specifically bound RNAs in a precipitation sample are removed through SDS-PAGE separation and subsequent transferring process removes free RNAs; and 4) the two ends of the RNA are connected with the adaptors, which makes subsequent PCR amplification and large-scale sequencing to be possible.
Owner:INST OF GENETICS & DEVELOPMENTAL BIOLOGY CHINESE ACAD OF SCI
Single molecule peptide sequencing
ActiveUS20220091130A1OmicsCarrier-bound/immobilised peptidesLarge-Scale SequencingProtein Sequence Determination
Identifying proteins and peptides, and more specifically large-scale sequencing of single peptides in a mixture of diverse peptides at the single molecule level is an unmet challenge in the field of protein sequencing. Herein are methods for identifying amino acids in peptides, including peptides comprising unnatural amino acids. In one embodiment, the N-terminal amino acid is labeled with a first label and an internal amino acid is labeled with a second label. In some embodiments, the labels are fluorescent labels. In other embodiments, the internal amino acid is Lysine. In other embodiments, amino acids in peptides are identified based on the fluorescent signature for each peptide at the single molecule level.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
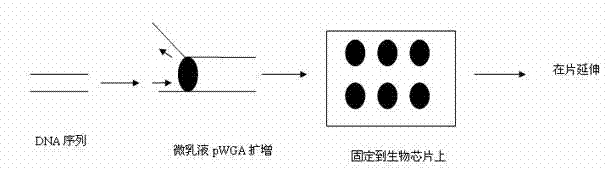
DNA Sequencing Method Based on Microemulsion Isothermal Amplification on Biochip
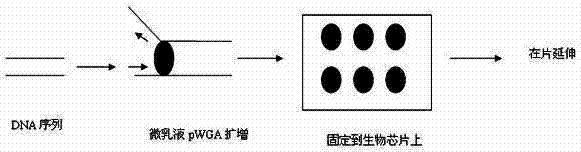
InactiveCN102260747ARapid large-scale sequencingHigh-sensitivity large-scale sequencingMicrobiological testing/measurementBiotin-streptavidin complexModified dna
The invention discloses a DNA (deoxyribonucleic acid) sequencing method based on micro emulsion isothermal amplification on a biochip. The method comprises the following steps of: firstly, modifying the surface of a slide, performing corresponding aldehyde group, amino group or streptavidin modification on a DNA fragment to be detected; then amplifying the mixture of the modified DNA fragment to be detected and T7gp4 primase in a micro emulsion; then fixing the amplified product to the modified slide for providing a template for sequencing; and finally, performing on-chip extension sequencingon the template fixed to a biochip. In the invention, micro emulsion isothermal amplification and biochip technology are combined, so that large-scale sequencing can be performed on DNA quickly with high flux and high sensitivity.
Owner:HANGZHOU DIANZI UNIV
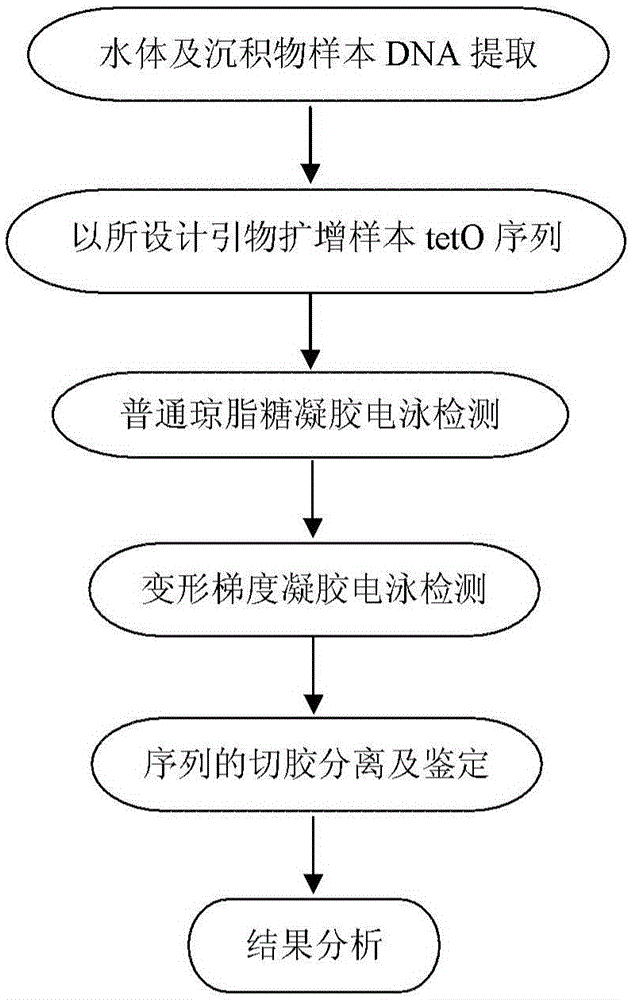
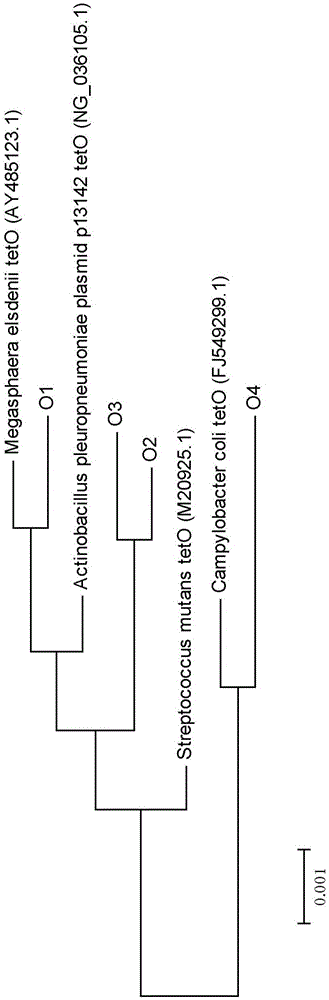
Primer sequence for detecting tetracycline resistant gene tetO in water body and sediments in classified mode and corresponding detecting method
ActiveCN105132576AEasy to operateLow costMicrobiological testing/measurementDNA/RNA fragmentationLarge-Scale SequencingAgricultural science
The invention provides a primer sequence for detecting a tetracycline resistant gene tetO in a water body and sediments in a classified mode and a corresponding detecting method for detecting the tetracycline resistant gene tetO in the water body and the sediments through the primer sequence. The primer sequence comprises a tetO-F, a tetO-R and a tetO-F-GC. The PCR amplification and denaturing gradient gel electrophoresis technology are conducted through the primer sequence, and then the aim of detecting the tetO sequence in the classified mode is achieved. By designing a primer and applying the PCR-DGGE technology to the classified identification of the tetO sequence, the steps, highly consuming labor, time and expenses, of the separation and purification of DNA, the connection and conversion of carriers, the screening of positive clone, the large-scale sequencing of sequences and the like are omitted compared with a traditional method, the comprehensive tetO sequence information in a sample can be obtained, the whole operation becomes easy and rapid, and cost is saved.
Owner:RES CENT FOR ECO ENVIRONMENTAL SCI THE CHINESE ACAD OF SCI
Amphioxus galactose lectin AmphiGAL13-gene and its use
InactiveCN100535115CPeptide/protein ingredientsPeptide preparation methodsDigestive canalEnzyme digestion
Owner:SUN YAT SEN UNIV
Multiple cancer risk susceptibility mutation site, application thereof and produced kit thereof
InactiveCN102311952AIncreased sensitivityImprove stabilityMicrobiological testing/measurementDNA/RNA fragmentationMaterial resourcesGenotype
The invention discloses a multiple cancer risk susceptibility mutation site, an application thereof and a produced kit thereof. The research verifies that a multiple cancer family chromosome 3q24-26 area is tightly linked with a disease site, gene mutation screening verifies that ATG upstream-91bp of human TSC22D2 gene T>C mutation (c.-91T>C) is co-separated from pathotype in a multiple cancer family, According to large scale sequencing verification in local normal population, the mutation is negative. The genotype provides that the mutation site can be used for screening the tumor heredity susceptible population and predicting the prevalence risk of tumor. The invention also provides a kit for screening the relative mutation. The kit has the advantages of high sensitivity, good specificity, high flux and the like. Compared with the traditional sequencing method, a PCR product is not required for purifying, and the method is capable of minimizing the interference of artificial factors like cross-contamination, simultaneously the labor resources, the financial resources and the material resources can be saved.
Owner:CENT SOUTH UNIV
Male infertility related genetic marker
InactiveCN104293781AEnables diagnostic testingMicrobiological testing/measurementDNA/RNA fragmentationMale infertilityLarge-Scale Sequencing
The invention discloses a male infertility related genetic marker, the sequence of which is that of the UBE2B gene. The binding site of the UBE2B gene's promoter region and the transcription factor SP1 undergoes mutation, and the mutation site is one or more of Chr.133706771T>A, Chr.133706876T>G and Chr.133706925A>G. By large-scale sequencing, four new mutation sites of the UBE2B gene's promoter region are selected from male infertility patients, a transcription factor plasmid is cotransfected into cells together with a wild type plasmid and a mutant plasmid respectively, and experiments find significant function differences and show that mutation of the UBE2B gene's promoter region is relevant to male infertility (especially azoospermia). Therefore, diagnosis detection of male infertility can be realized through the mutation.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
Single molecule peptide sequencing
ActiveUS11435358B2OmicsCarrier-bound/immobilised peptidesLarge-Scale SequencingProtein Sequence Determination
Identifying proteins and peptides, and more specifically large-scale sequencing of single peptides in a mixture of diverse peptides at the single molecule level is an unmet challenge in the field of protein sequencing. Herein are methods for identifying amino acids in peptides, including peptides with unnatural amino acids. In one embodiment, the N-terminal amino acid is labeled with a first label and an internal amino acid is labeled with a second label. In some embodiments, the labels are fluorescent labels. In other embodiments, the internal amino acid is Lysine. In other embodiments, amino acids in peptides are identified based on the fluorescent signature for each peptide at the single molecule level.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Fetal chromosomal aneuploidy diagnosis
ActiveUS9598730B2Reduce capacityLow costMicrobiological testing/measurementBiostatisticsLarge-Scale SequencingPrenatal diagnosis
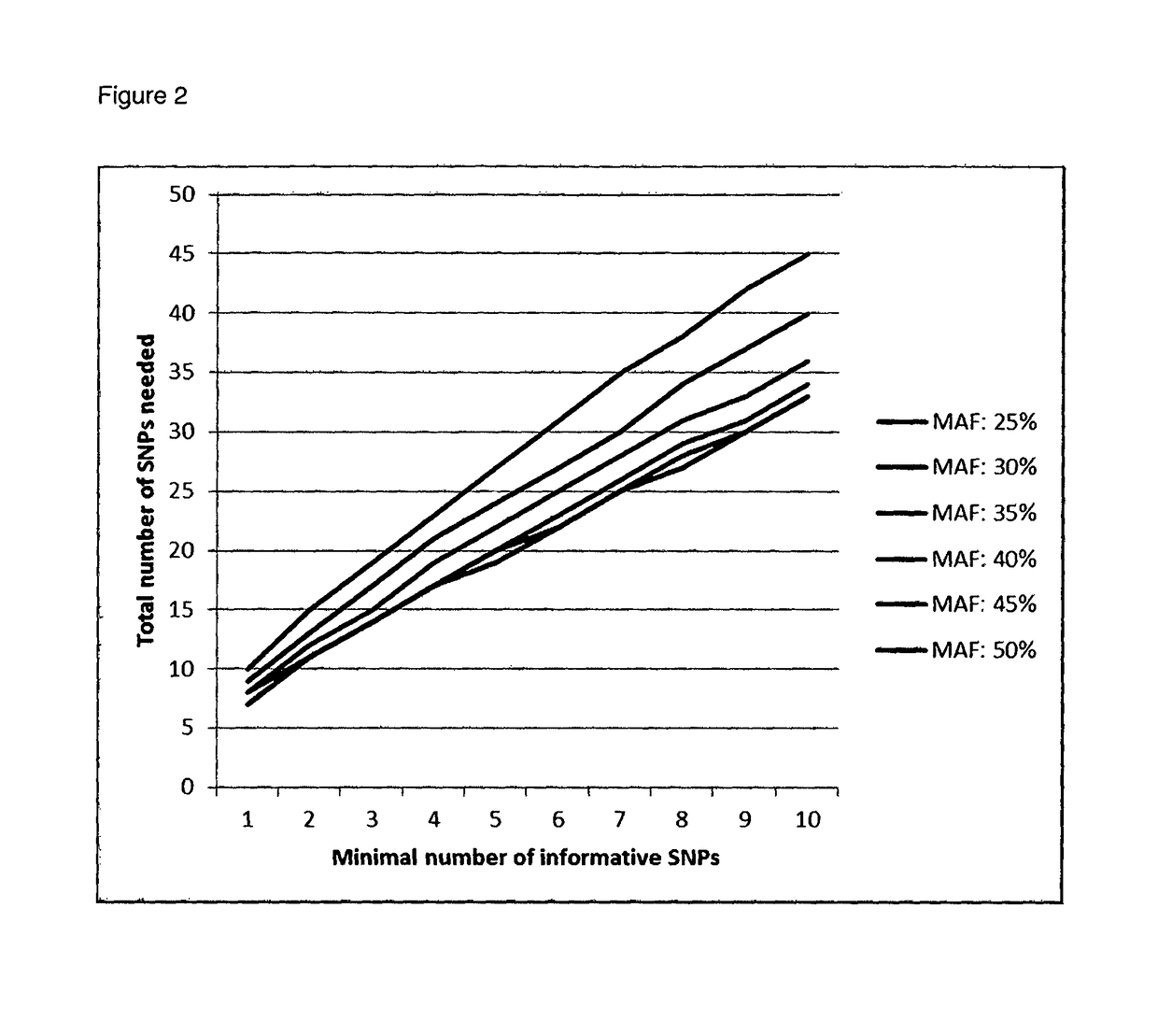
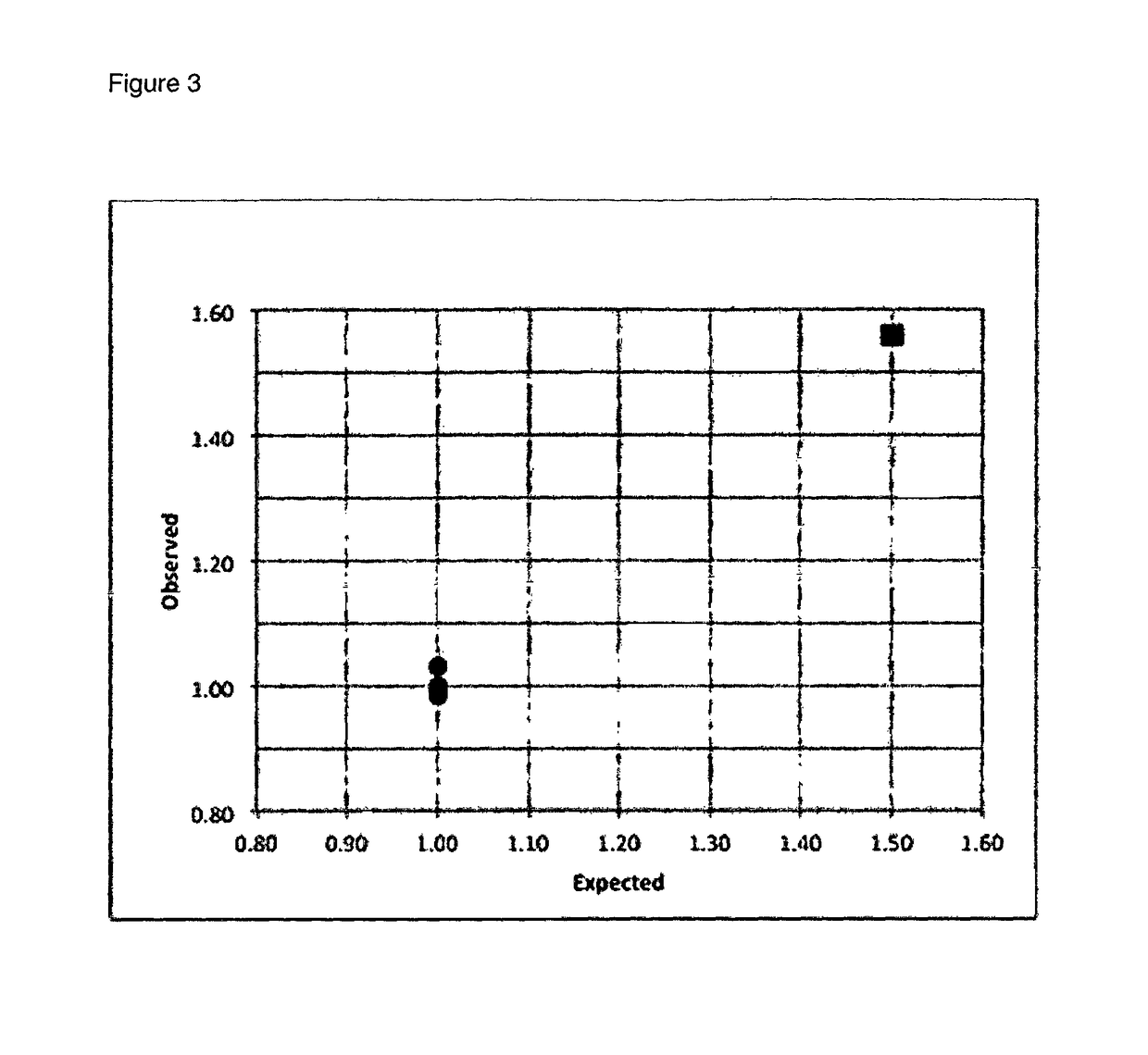
The invention relates to prenatal detection methods using non-invasive techniques. In particular, it relates to prenatal diagnosis of a fetal chromosomal aneuploidy by detecting fetal and maternal nucleic acids in a maternal biological sample. More particularly, the invention applies multiplex PCR to amplify selected fractions of the respective chromosomes of maternal and fetal chromosomes. Respective amounts of suspected aneuploid chromosomal regions and reference chromosomes are determined from massive sequencing analysis followed by a statistical analysis to detect a particular aneuploidy.
Owner:AGILENT TECH INC
A Sequencing Data Compression Method Based on Double Codons
ActiveCN106067824BSmall footprintReduce data transfer timeCode conversionData compressionLarge-Scale Sequencing
Owner:洛阳晶云信息科技有限公司
DNA (deoxyribonucleic acid) sequencing method based on micro emulsion isothermal amplification on biochip
InactiveCN102260747BRapid large-scale sequencingHigh-sensitivity large-scale sequencingMicrobiological testing/measurementBiotin-streptavidin complexPrimase
The invention discloses a DNA (deoxyribonucleic acid) sequencing method based on micro emulsion isothermal amplification on a biochip. The method comprises the following steps of: firstly, modifying the surface of a slide, performing corresponding aldehyde group, amino group or streptavidin modification on a DNA fragment to be detected; then amplifying the mixture of the modified DNA fragment to be detected and T7gp4 primase in a micro emulsion; then fixing the amplified product to the modified slide for providing a template for sequencing; and finally, performing on-chip extension sequencingon the template fixed to a biochip. In the invention, micro emulsion isothermal amplification and biochip technology are combined, so that large-scale sequencing can be performed on DNA quickly with high flux and high sensitivity.
Owner:HANGZHOU DIANZI UNIV
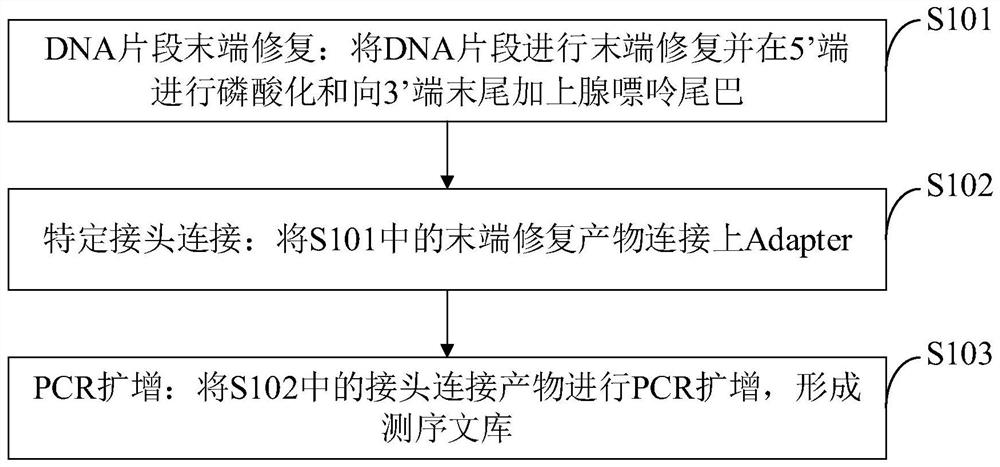
16S rDNA library construction method, 16S rDNA library and application
PendingCN114318552ALow costSuitable for large-scale sequencingNucleotide librariesMicrobiological testing/measurementLarge-Scale SequencingEngineering
The invention belongs to the technical field of genetic engineering high-throughput sequencing, and discloses a 16S rDNA library construction method, a 16S rDNA library and application, and the 16S rDNA library construction method comprises the following steps: repairing the tail end of a DNA fragment; specific joint connection; and carrying out PCR amplification. According to the 16S rDNA library construction method provided by the invention, the cost can be greatly reduced by a next-generation sequencing technology represented by illumina, and the 16S rDNA library construction method is suitable for large-scale sequencing. The essence of 16S rDNA library establishment is to perform PCR enrichment and screening on a specific region by using enzyme and primers, and the library establishment method is lower in cost compared with methods such as a machine interruption method and the like. The 16S rDNA not only can reflect the difference between different bacteria genus, but also can easily obtain the sequence by utilizing a sequencing technology, and is widely applied to detection and identification of pathogenic bacteria at present. According to the 16SrDNA library building method disclosed by the invention, a library building system is obtained after multiple experiments, so that the library building time and cost can be effectively saved.
Owner:上海长为数据技术有限公司
Amphioxus dimethyl arginine hydrolase AmphiDDAH gene and its application
The present invention relates to Amphioxus gene AmphiDDAH, its coded protein, and the application of the protein in preparing medicine for treating cardiac and cerebral vascular diseases and arthritis. By means of cDNA library constitution and large scale sequencing process, the present invention separates Amphioxus neurula to obtain the Amphioxus gene AmphiDDAH with the DNA sequence shown in <400>1 of the sequence list. The gene coded protein, PDDAH, has amino acid sequence shown in <400>2 of the sequence list. The recombinant Amphioxus protein PDDAH has NO creation regulating function and may be used in preparing medicine for treating cardiac and cerebral vascular diseases and arthritis.
Owner:SUN YAT SEN UNIV
Nucleic acid sequencing method based on fluorescence quenching
InactiveCN101168773BLow costImprove throughputMicrobiological testing/measurementLarge-Scale SequencingFluorescence
The invention relates to a nucleic acid sequence detecting method based on fluorescence quenching. The method includes the steps of: firstly, sequence detecting template preparation: the product of anucleic acid fragment to be detected comprises deoxyribonucleic acid, ribonucleic acid or a nucleic acid sequence enlarged product, the cloned product comprises deoxyribonucleic acid and ribonucleic acid are fixed on carrying agent, secondly, extension in the carrying agent: the mixed fluid of reaction buffer fluid, polymerase and fluorescence marked dNTP are used to perform extension, thirdly, fluorescence quenching: fluorescence quenching agent is used to perform fluorescence quenching, fourthly, extension in the carrying agent: the mixed fluid of the reaction buffer fluid, the polymerase and the fluorescence marked dNTP are used to perform extension, fifthly, image recognition: characteristic point extraction is performed to the obtained extended images, the noise point is eliminated torealize analysis to the chip, or other commercialized software is directly used to perform image processing. The invention combines the nucleic acid micro array chip technology, and can perform large-scale nucleic acid sequencing quickly in high flux and at low cost.
Owner:SOUTHEAST UNIV
Method and system to characterize transcriptionally active regions and quantify sequence abundance for large scale sequencing data
This invention provides a quantitative method to determine transcriptionally active regions and quantify sequence abundance from large scale sequencing data. The invention also provides a system based on reference sequences to design and implement the method. The system processes large scale sequence data from high throughput sequencing, generates transcriptionally active region sequences as necessary, and quantifies the sequence abundance of the gene or transcriptionally active region. The method and system are useful for many analyses based on RNA expression profiling.
Owner:GENOMIC HEALTH INC
Amphioxus dimethyl arginine hydrolase AmphiDDAH gene and its application
The present invention relates to Amphioxus gene AmphiDDAH, its coded protein, and the application of the protein in preparing medicine for treating cardiac and cerebral vascular diseases and arthritis. By means of cDNA library constitution and large scale sequencing process, the present invention separates Amphioxus neurula to obtain the Amphioxus gene AmphiDDAH with the DNA sequence shown in <400>1 of the sequence list. The gene coded protein, PDDAH, has amino acid sequence shown in <400>2 of the sequence list. The recombinant Amphioxus protein PDDAH has NO creation regulating function and may be used in preparing medicine for treating cardiac and cerebral vascular diseases and arthritis.
Owner:SUN YAT SEN UNIV
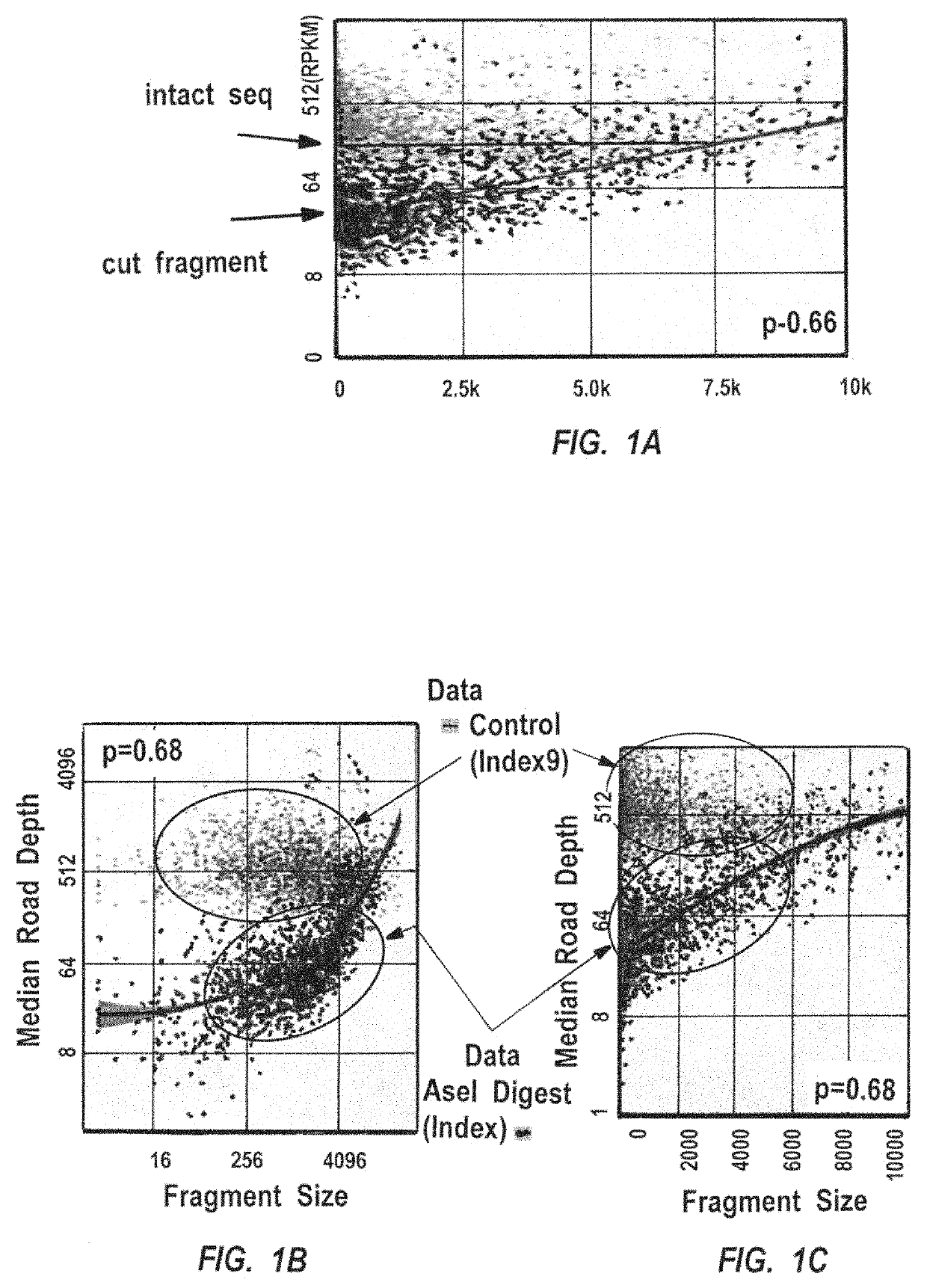
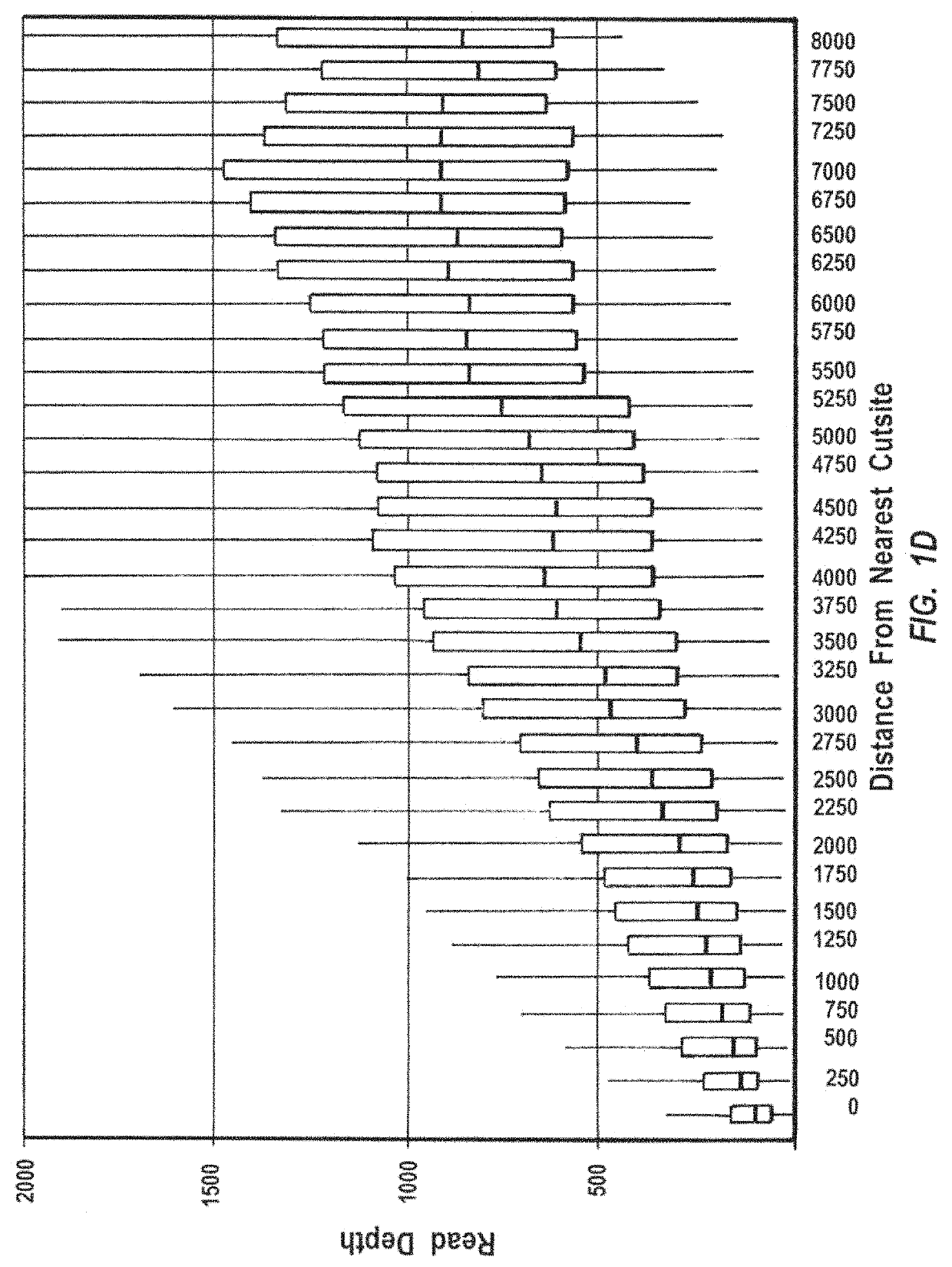
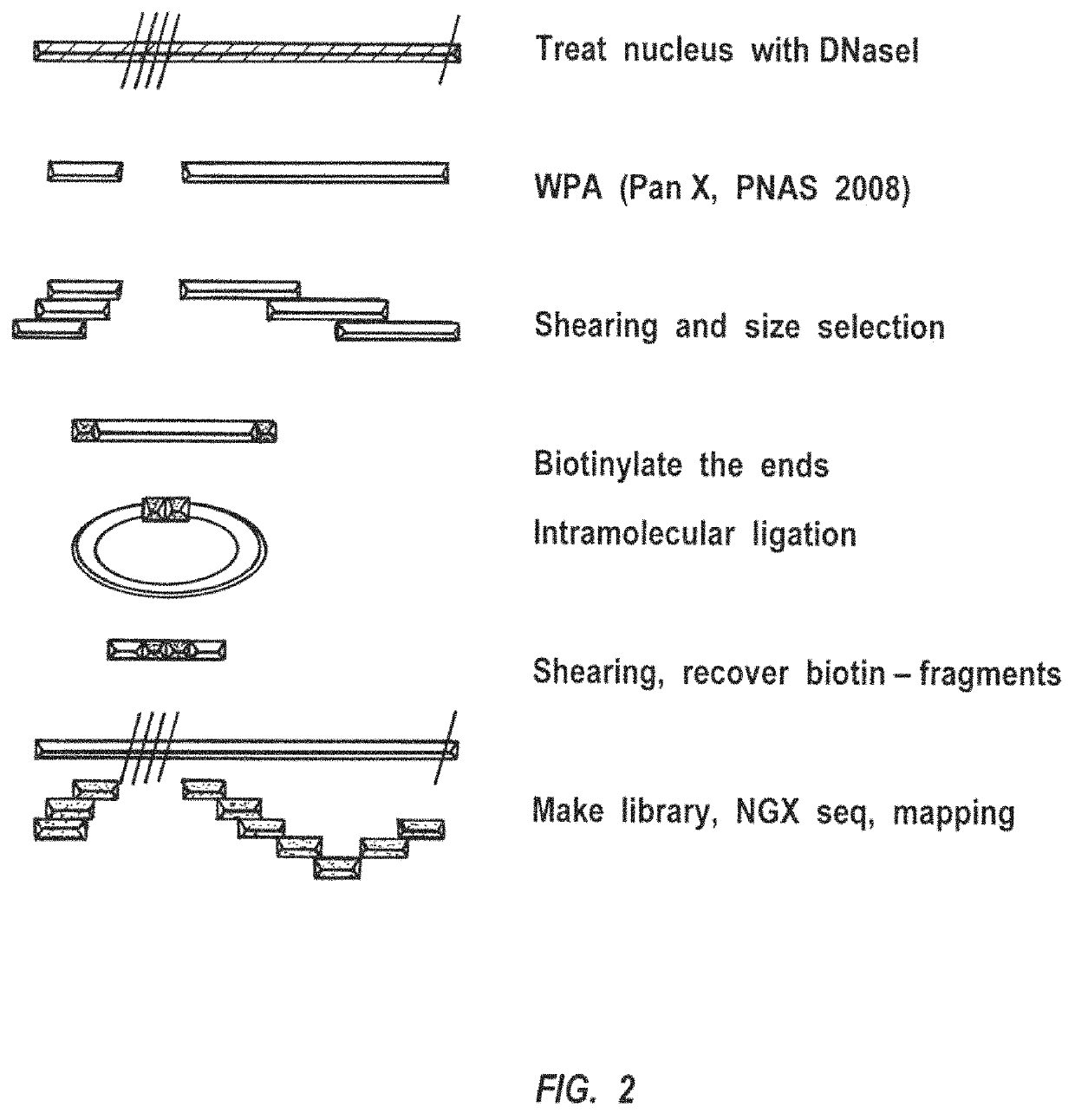
Methods for closed chromatin mapping and DNA methylation analysis for single cells
ActiveUS10480021B2Less efficiently ligatedImprove efficiencyMicrobiological testing/measurementFermentationDNA methylationLarge-Scale Sequencing
Methods of identifying DNase I Hyper-Resistant Sites (DHRS), or in board sense, highly compact chromatin and characterizing the DNA methylation status of DMRs such as CpG islands and CpG island shores are provided. The methods are particularly useful for analysis of genomic DNA from low quantities of cells, for example, less than 1,000 cells, less than 100 cells, less than 10 cells, or even one cell, and can be used to generate chromatin and methylation profiles. The downstream analyses include in parallel massive sequencing, microarray, PCR and Sanger sequencing, hybridization and other platforms. These methods can be used to generate chromatin and DNA methylation profiles in drug development, diagnostics, and therapeutic applications are also provided.
Owner:YALE UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com