Nucleic acid sequencing method based on fluorescence quenching
A technology of fluorescence quenching and nucleic acid sequencing, applied in the field of biomedicine, can solve the problems of large DNA damage, high cost and toxicity, and achieve the effect of simple and effective implementation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
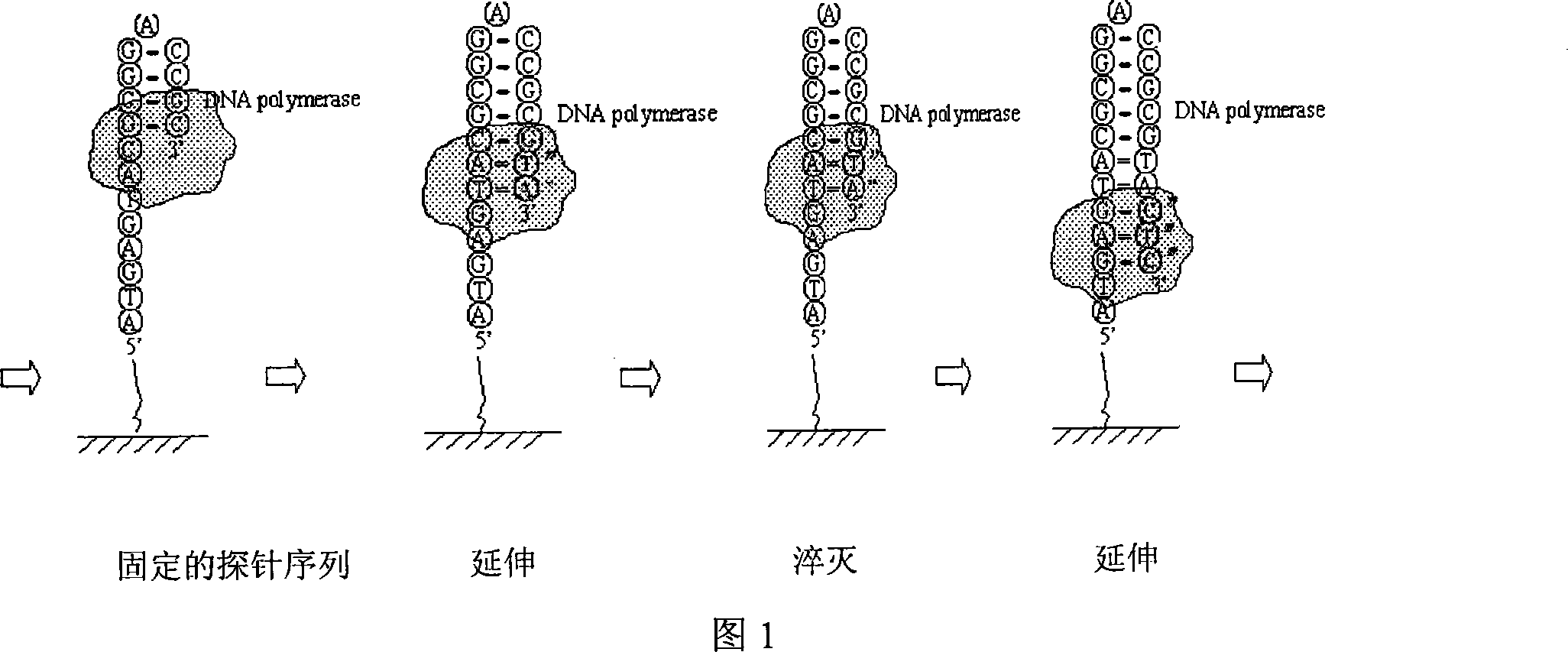
[0022] 1. Template preparation: through rolling circle formation sequence
[0023] 5’-P-TAGCTAGAATCAAAAATGTTGAGTACGACGAATCTGTATGCTAATGCGGCGTGATGTATTAT
[0024] GCGTATAGAAATAATACAGA-3' and
[0025] 5'-ACCTTTATGTCAACATTTTTGATTCTAGCTATCTGTATTATTTTCACCTAGCTT-3'
[0026] The concentration is 10um and 4ul each is mixed and then denatured, and the ring is formed after natural refolding. Ligation is performed by using T4 ligase. Add probe Acry-probe (100um) 0.3ul (E3: 5'-Acry-(T) 10 ATTAGCATACAGATTCGTCGTACT-3'), through denaturation, hybridization after natural renaturation. Then add i00×BSA, dNTP, and Bst enzyme respectively, and roll the circle at 40 degrees for 30 hours.
[0027] The rolling circle products were dissolved with acrylamide gel and fixed on acrylamide gel-modified glass slides.
[0028] 2. Detection: use 1uM Cy5-probe (5'-Cy5-GCGGCGTGATGTA) to hybridize with the immobilized product [3] . After scanning, clear images with high fluorescence intensity can be obta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com