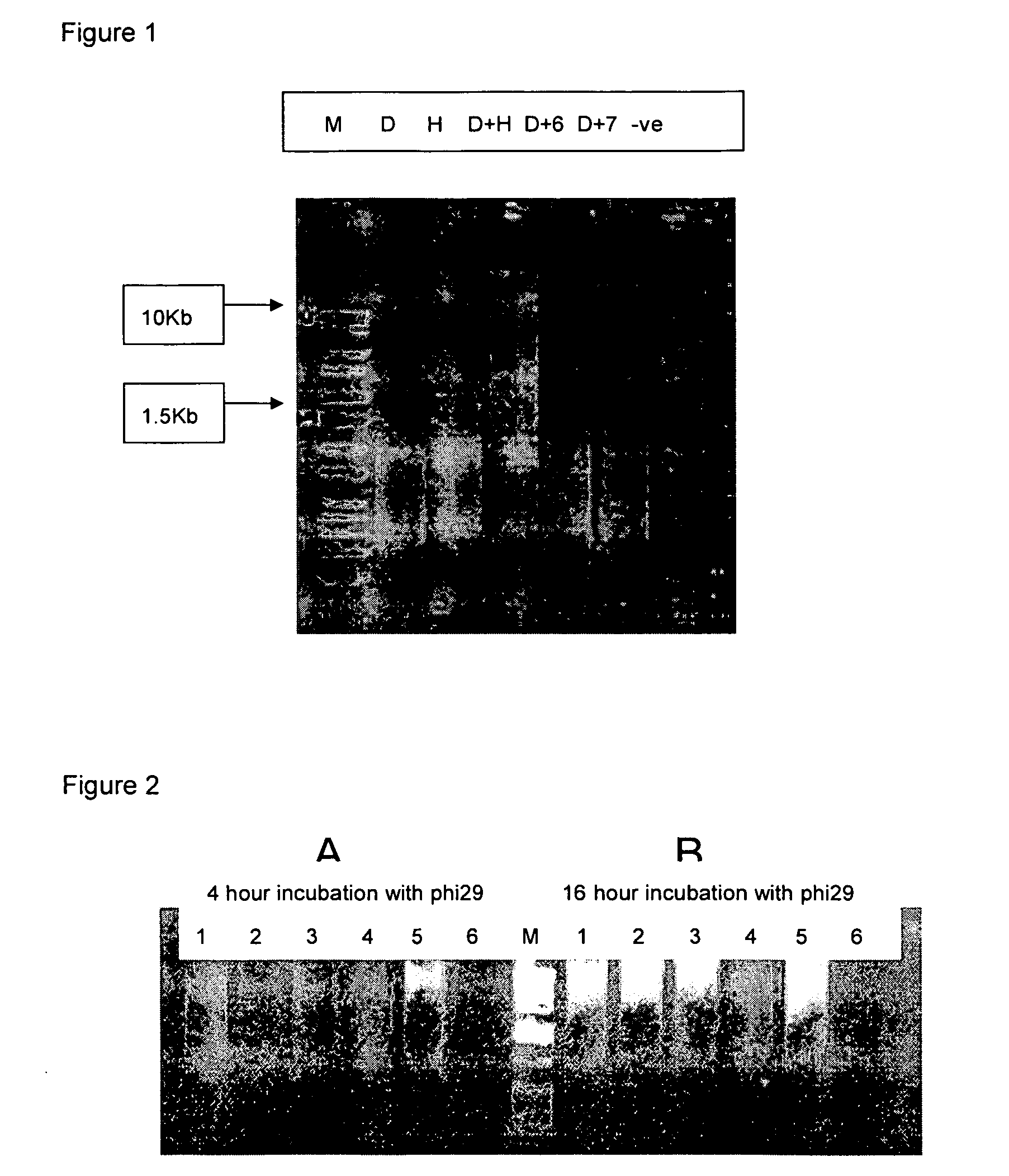
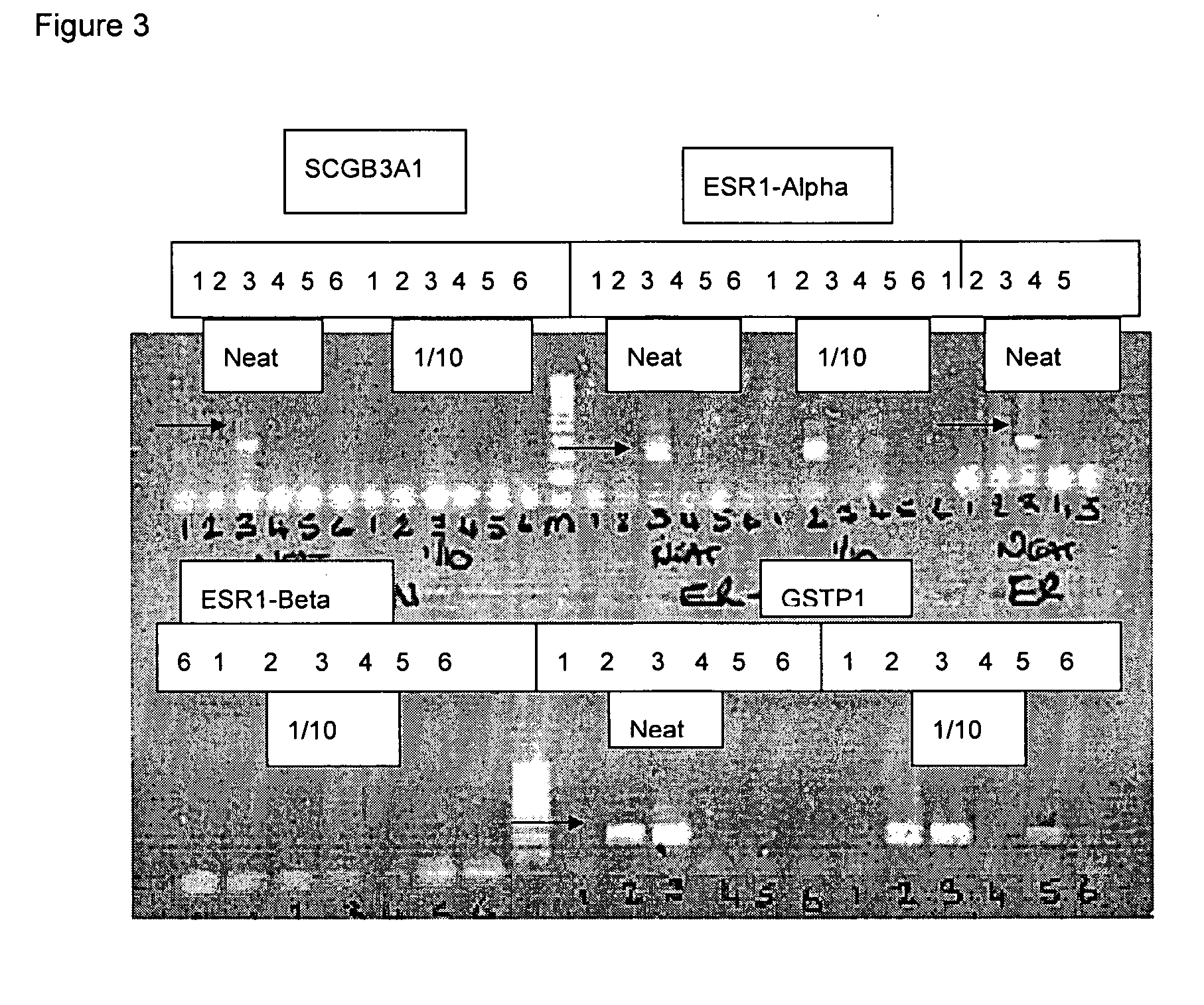
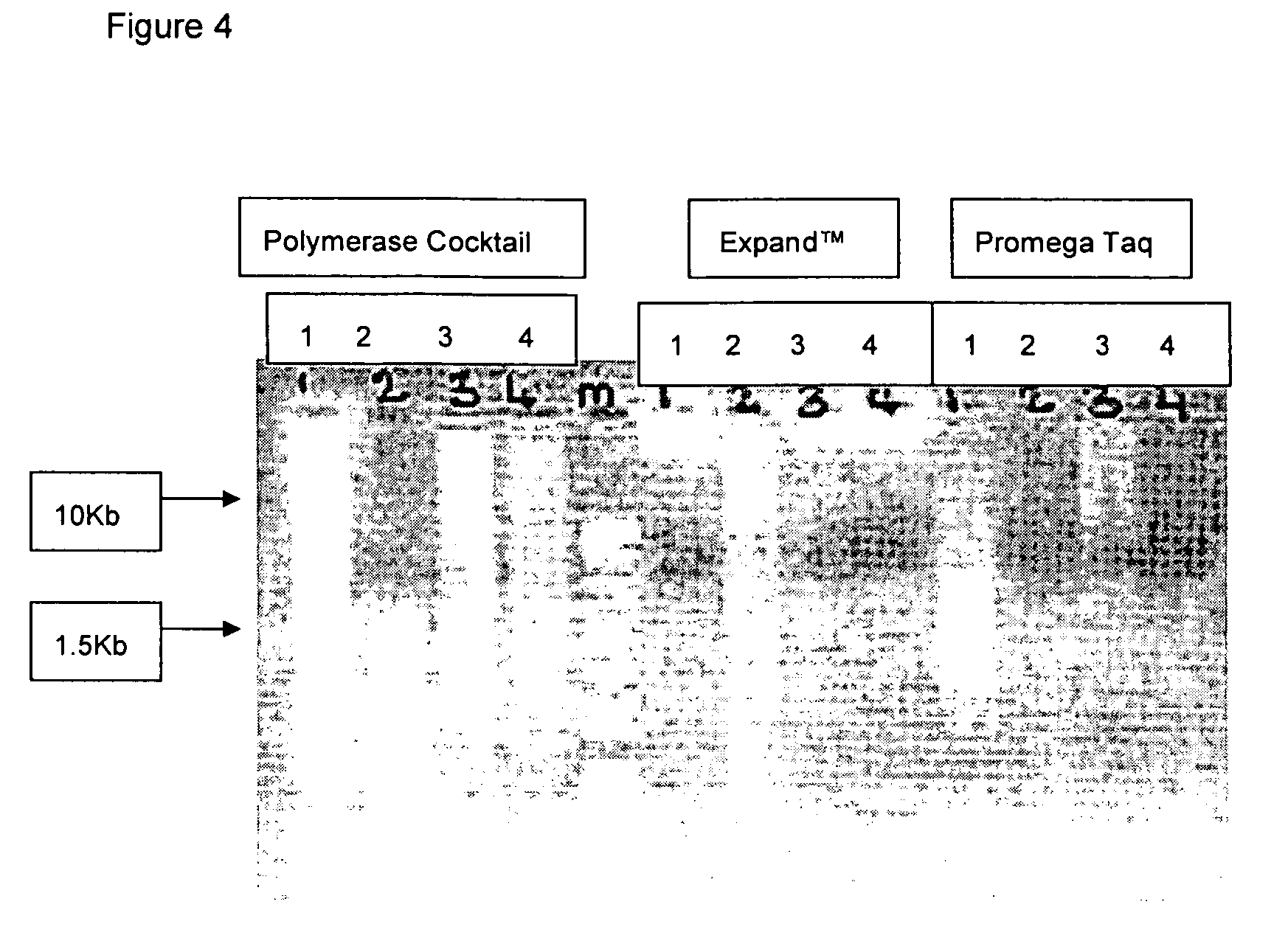
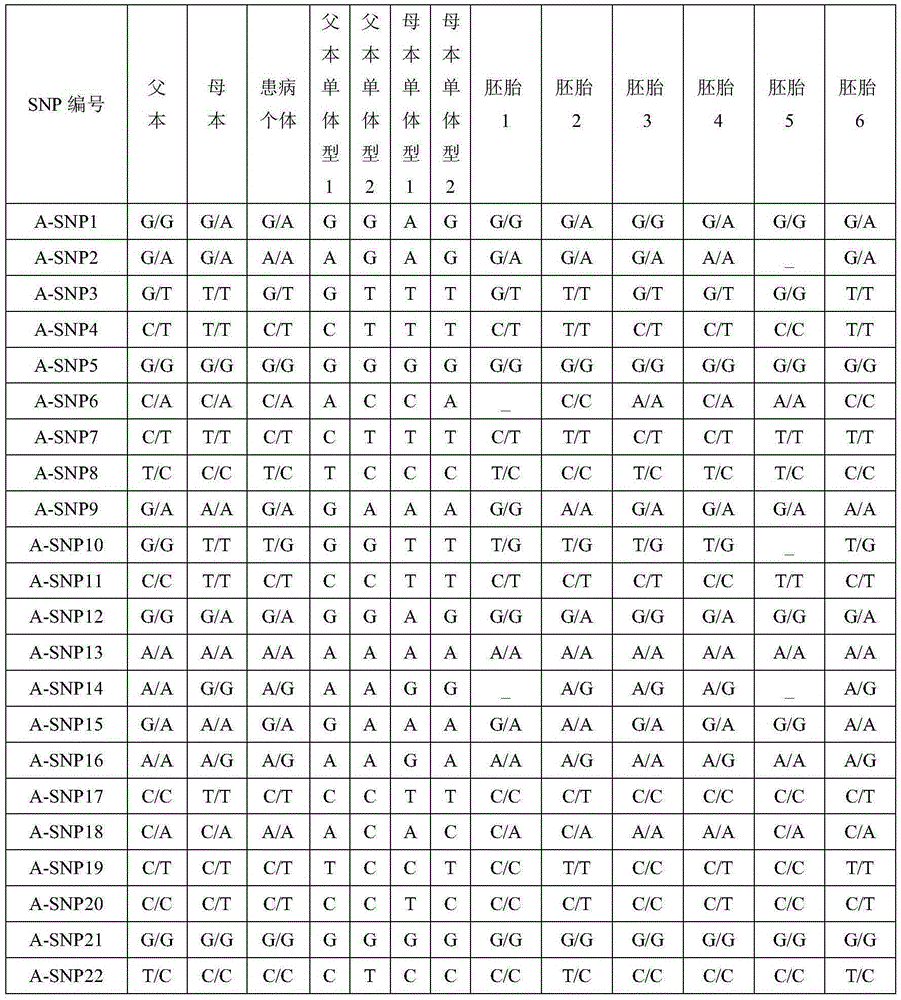
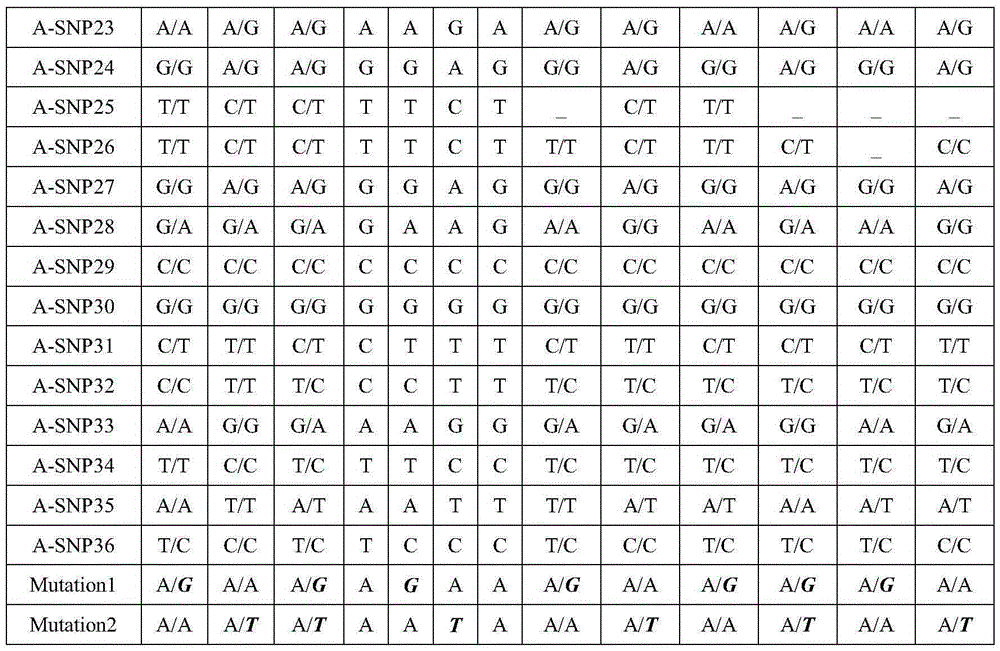
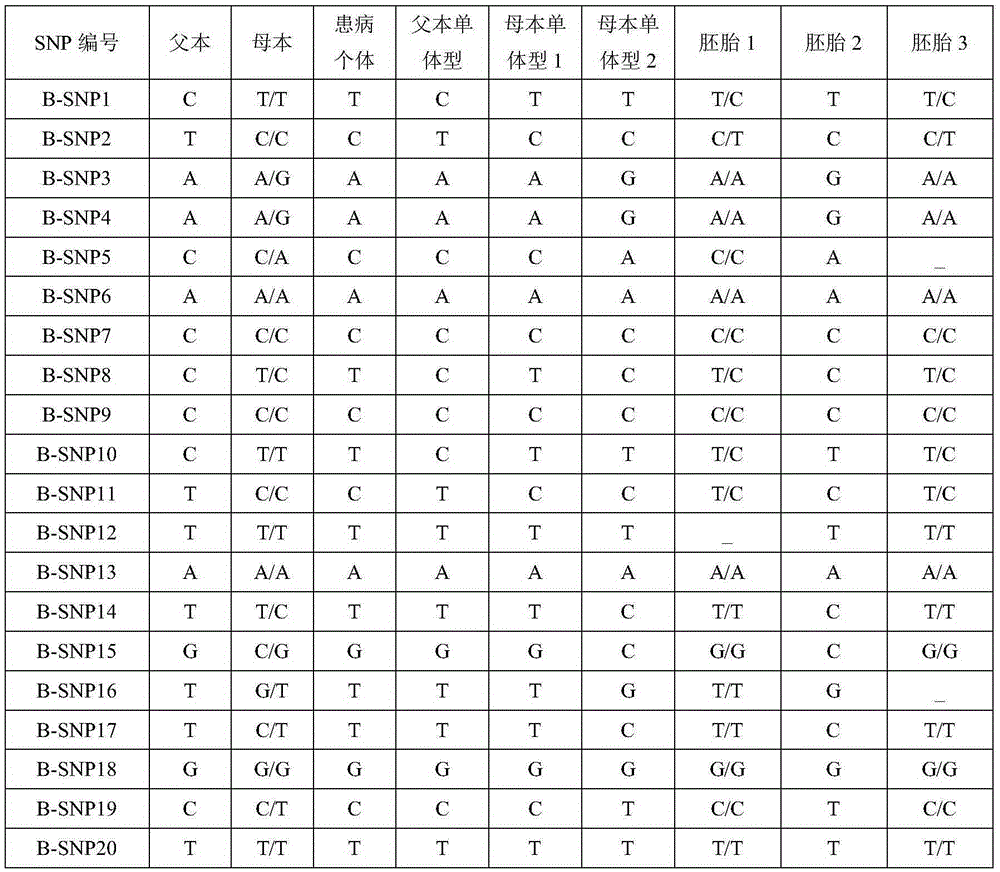
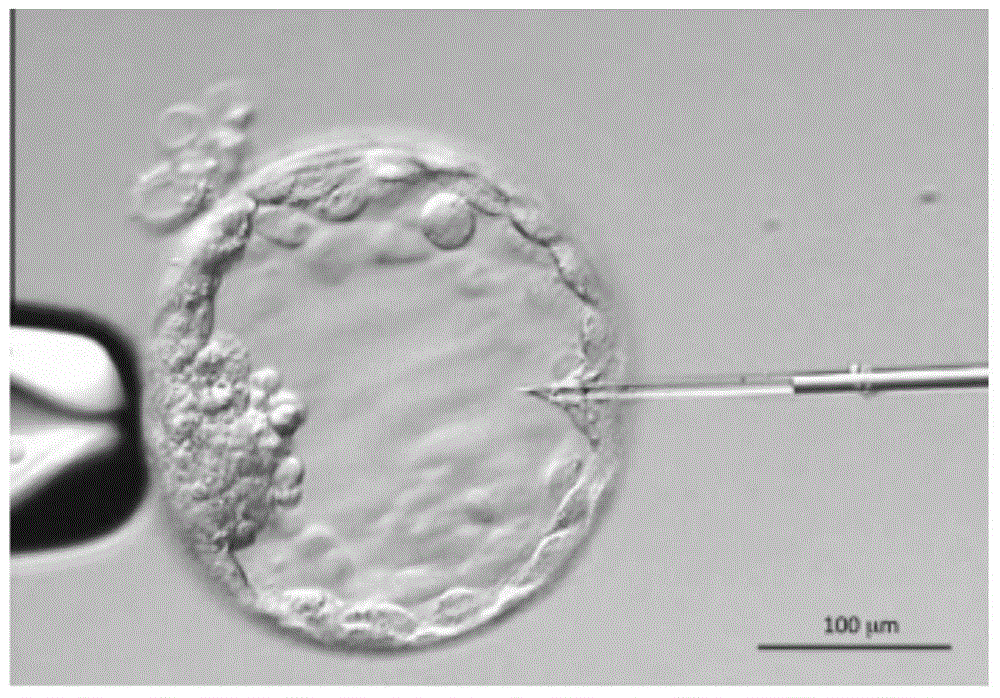
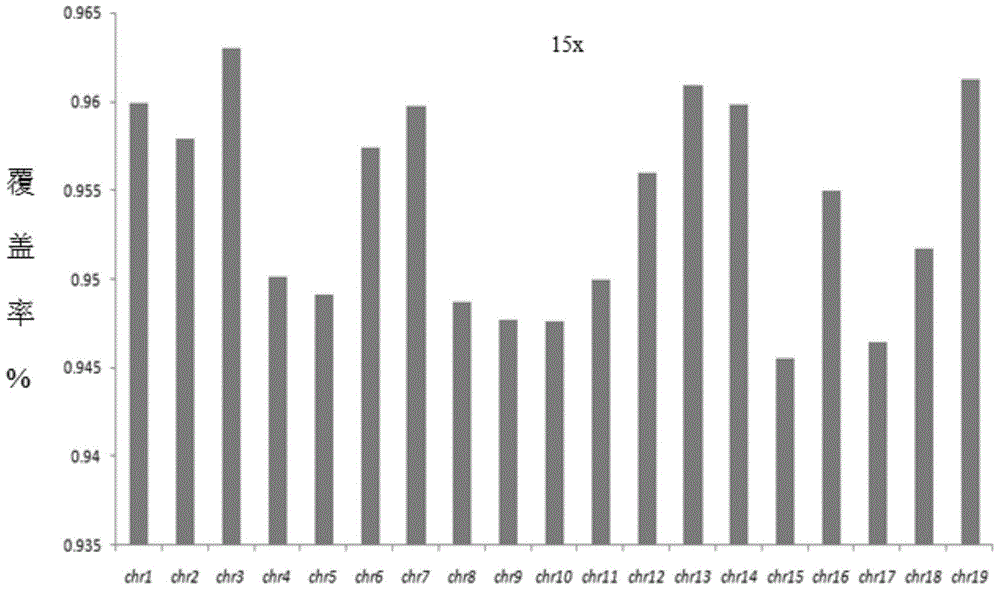
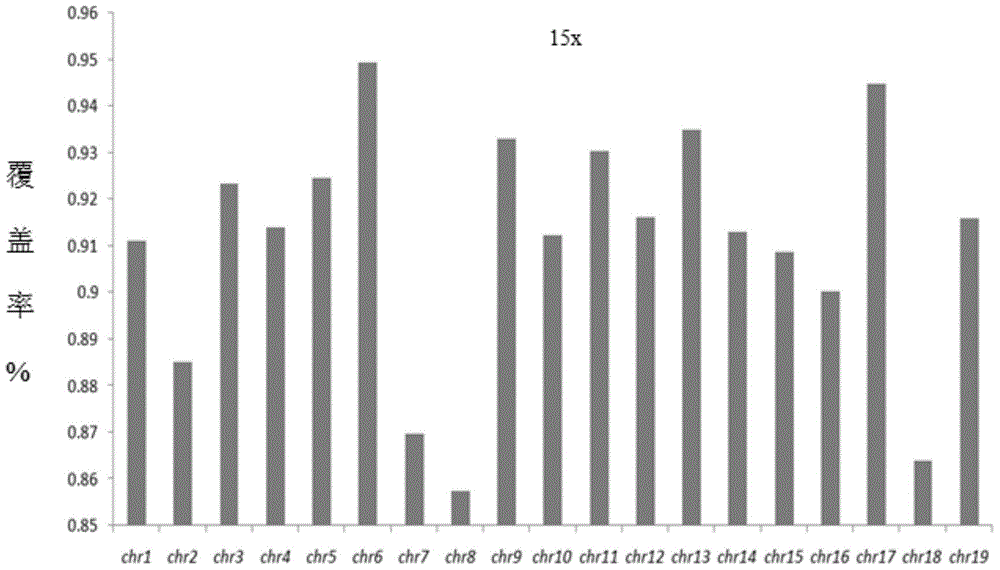
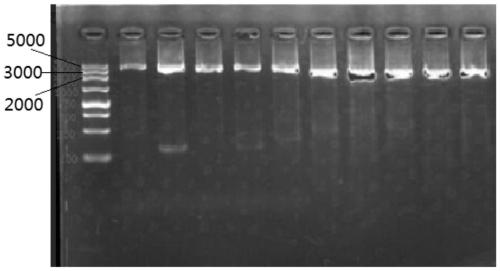
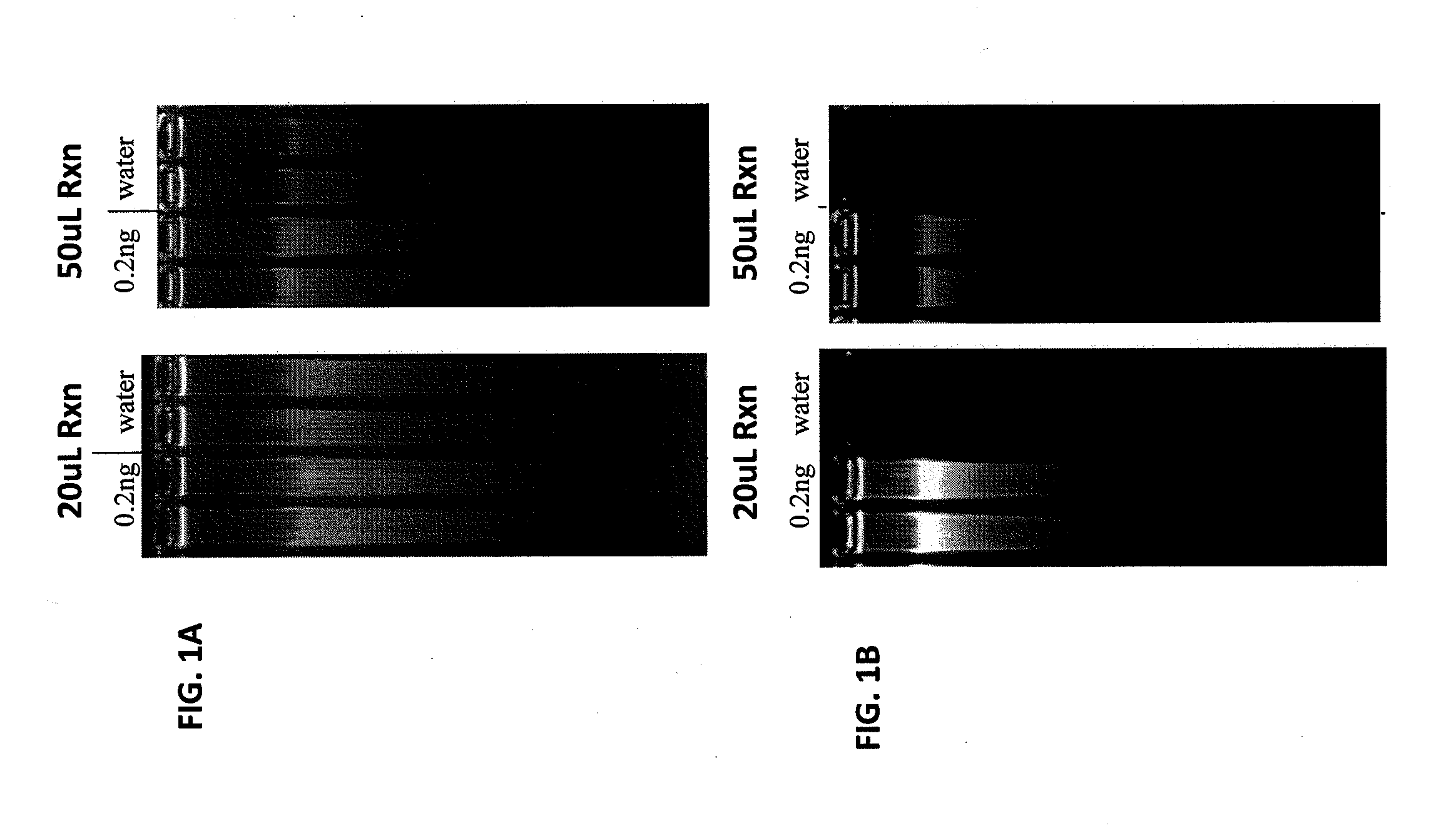
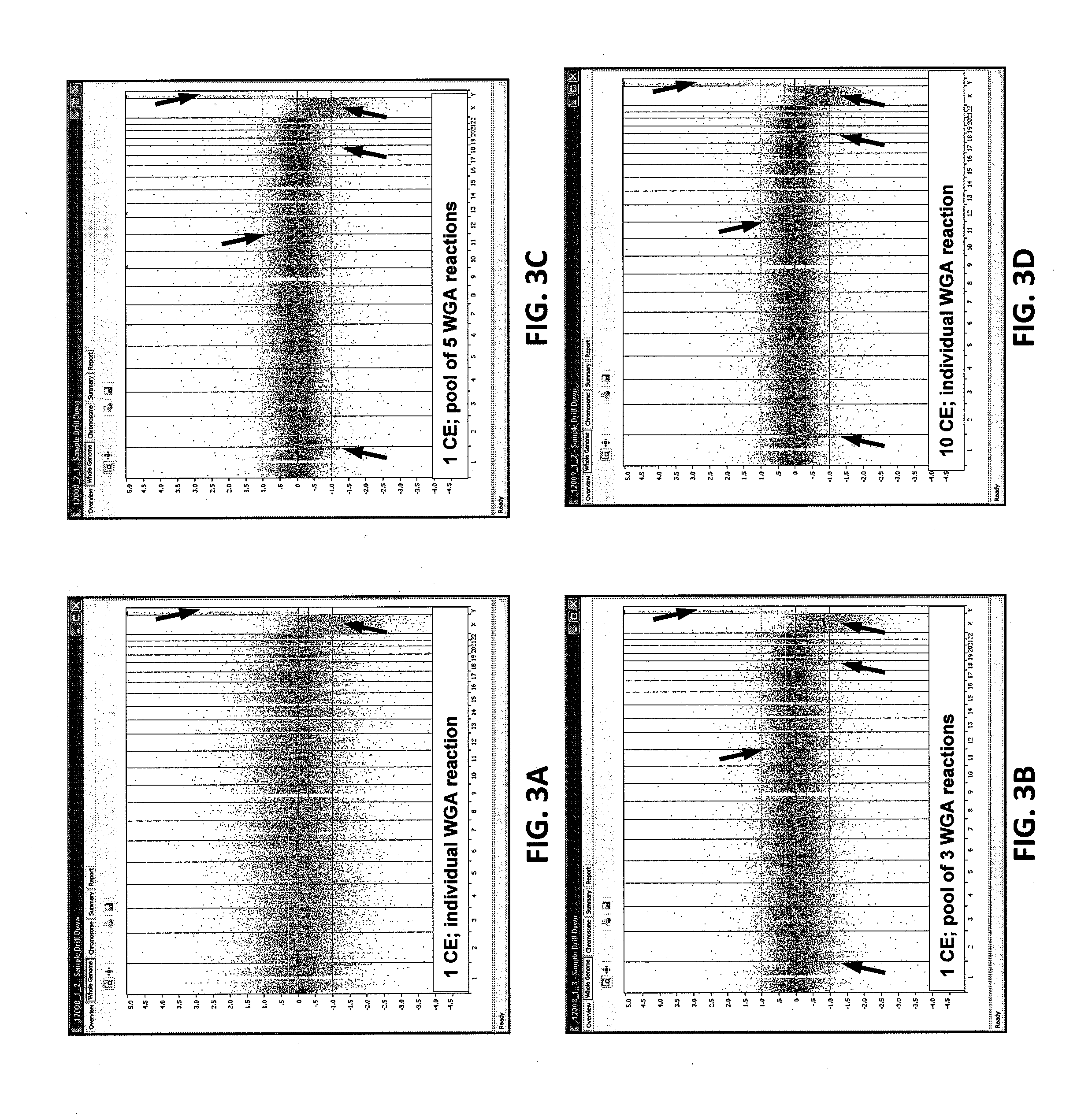
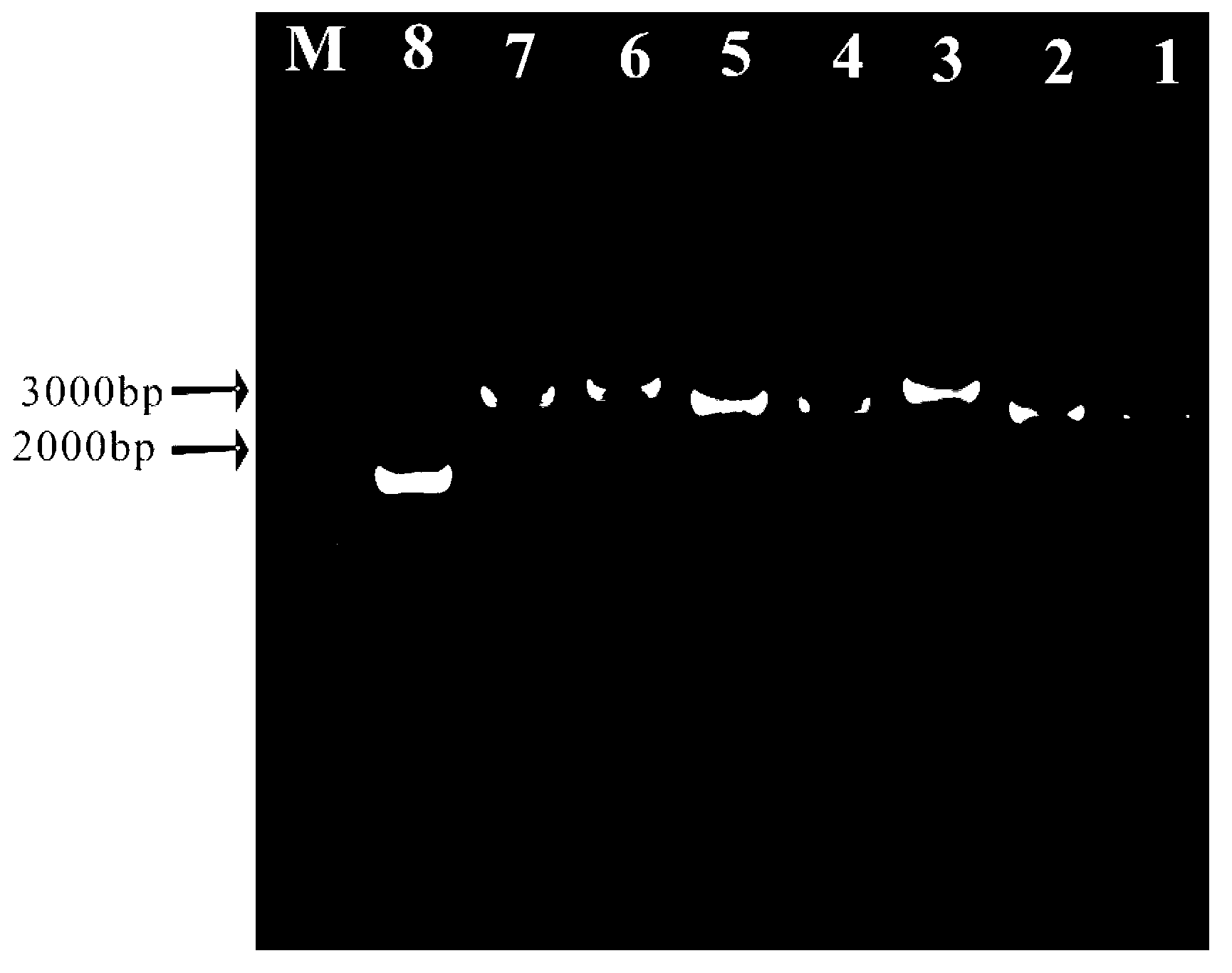Patents
Literature
198 results about "Genome amplification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
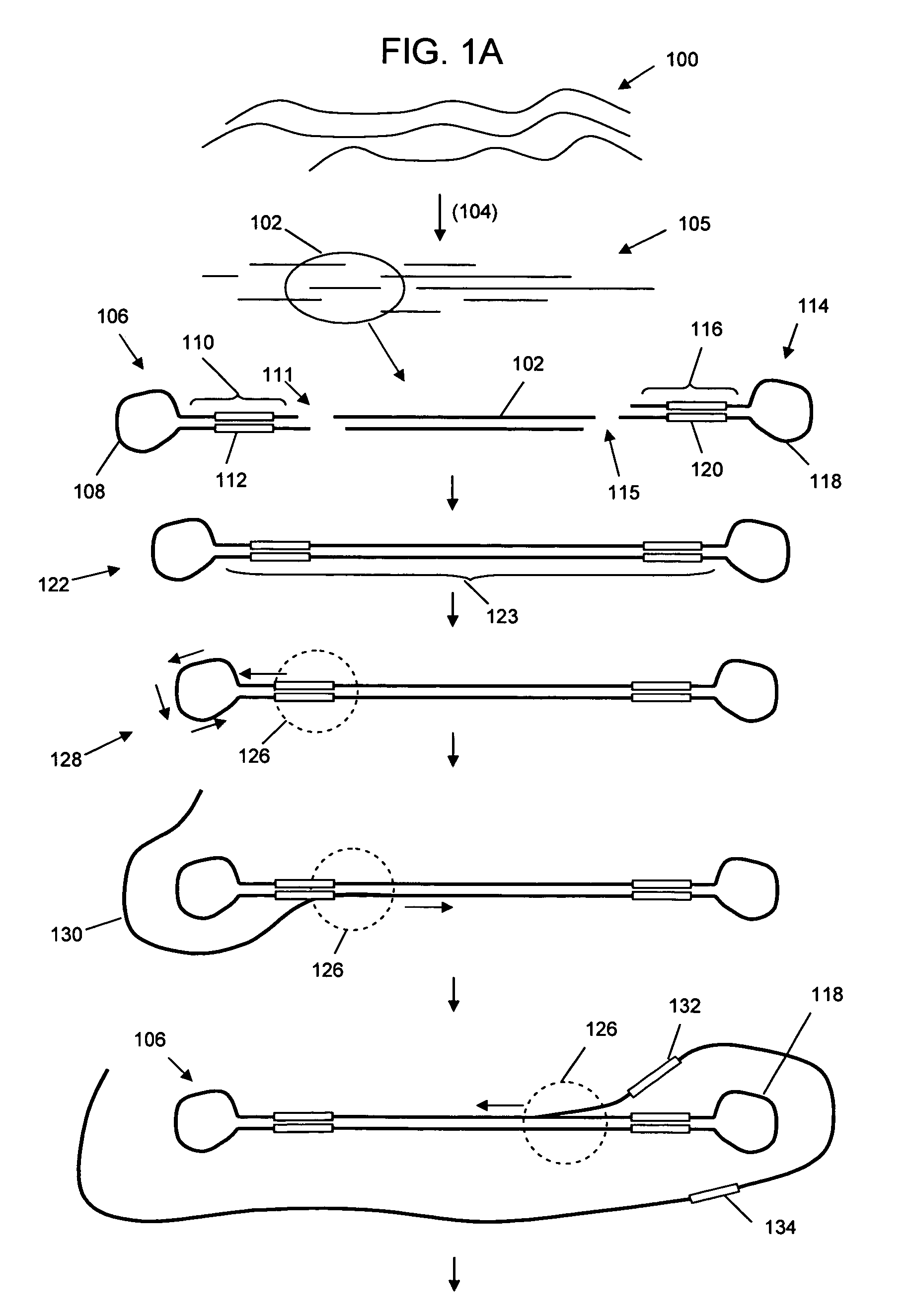
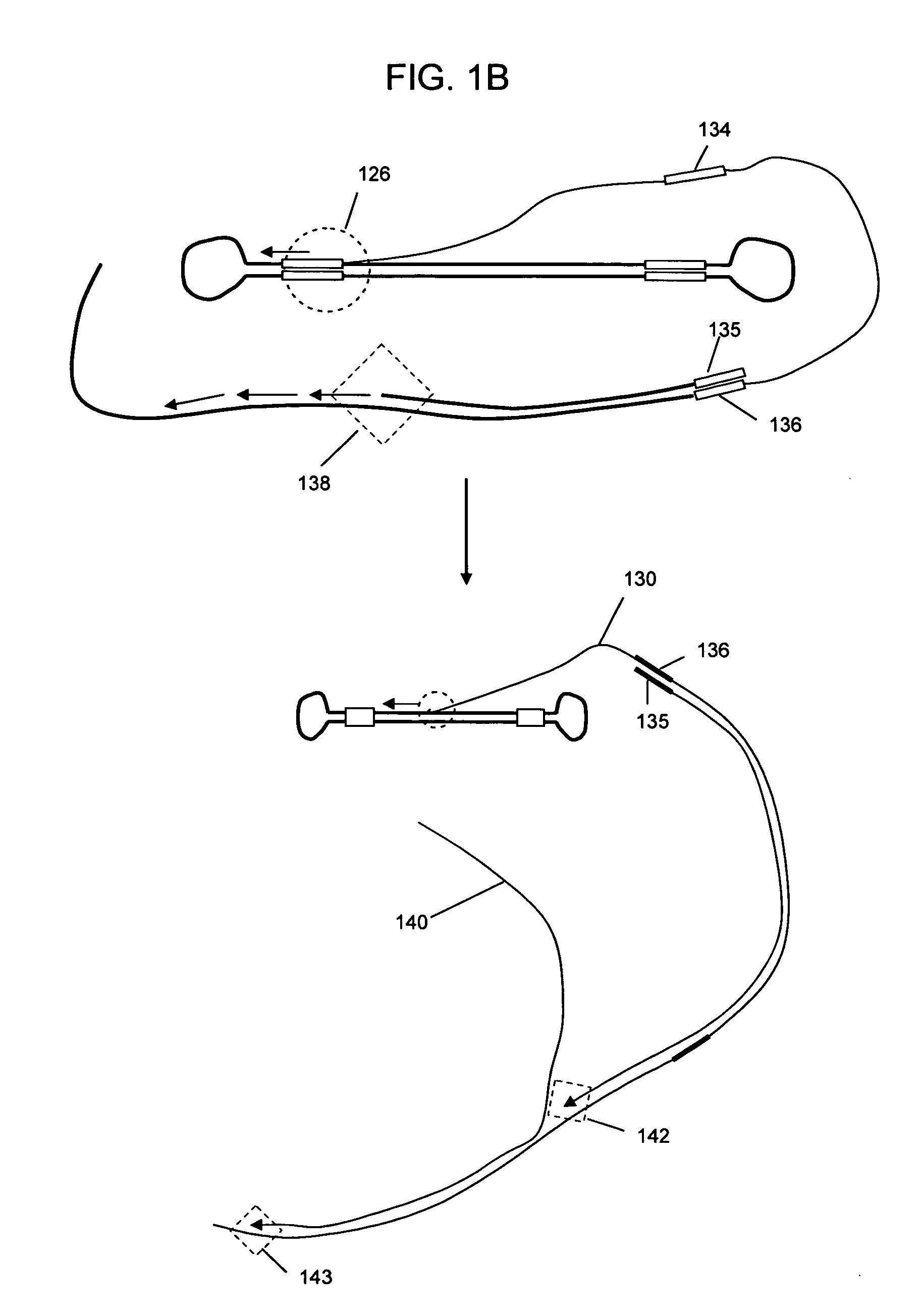
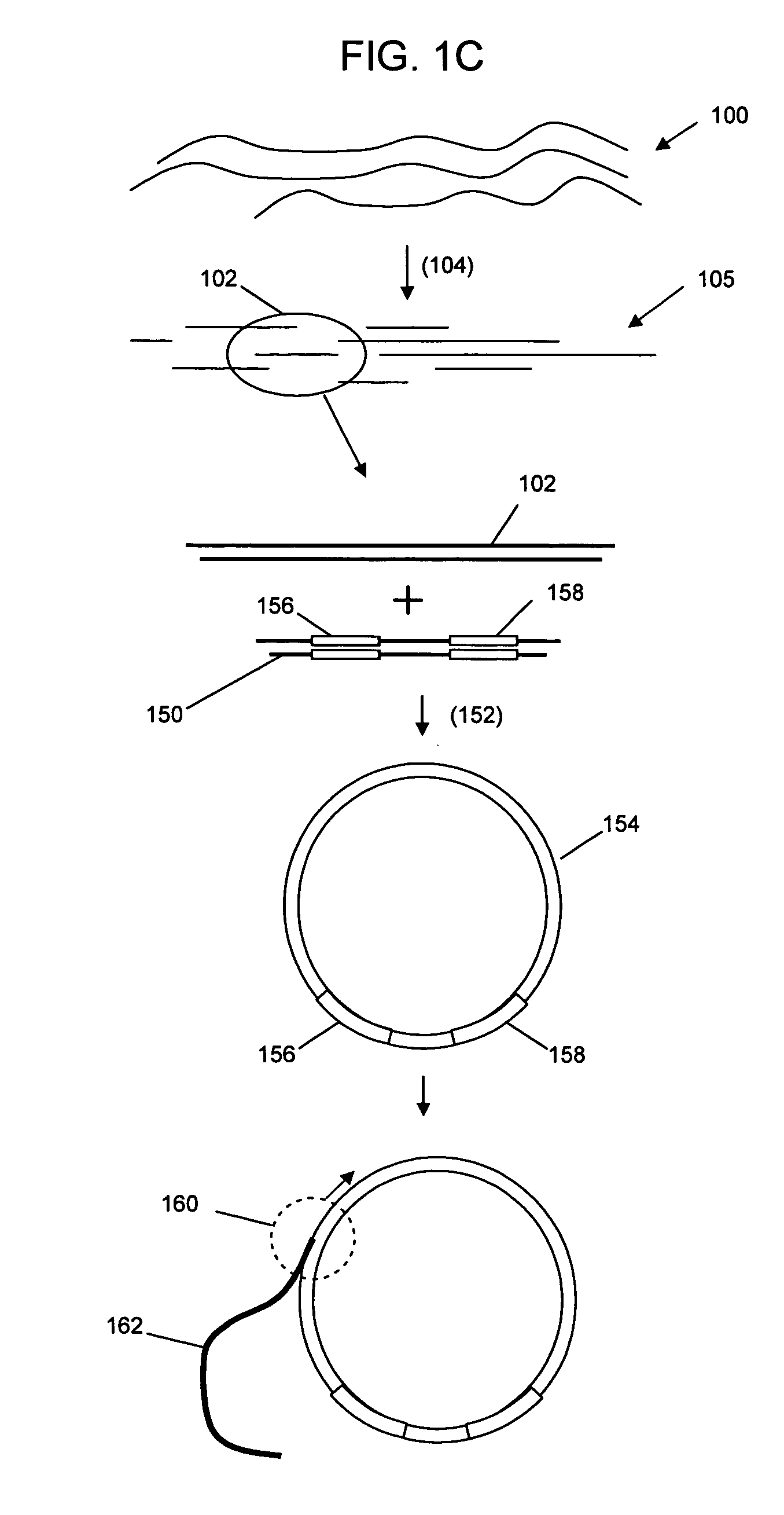
Universal selective genome amplification and universal genotyping system
InactiveUS20050019776A1Enriches SNPsImprove reuseMicrobiological testing/measurementFermentationSmall fragmentGenomic DNA
The invention relates to methods for isolating and amplifying small fragments of genomic DNA for genotyping polymorphisms in human populations.
Owner:COMPLETE GENOMICS INC
Selective genome amplification
The invention provides methods and compositions for amplifying selected polynucleotides, especially selected subsets of restriction fragments. Generally, methods of the invention are implemented by ligating adaptors containing at least one promoter sequence to such fragments under conditions that promote the formation of closed single stranded or double stranded structures, which are capable of serving as cyclical templates for transcription.
Owner:AGENCY FOR SCI TECH & RES
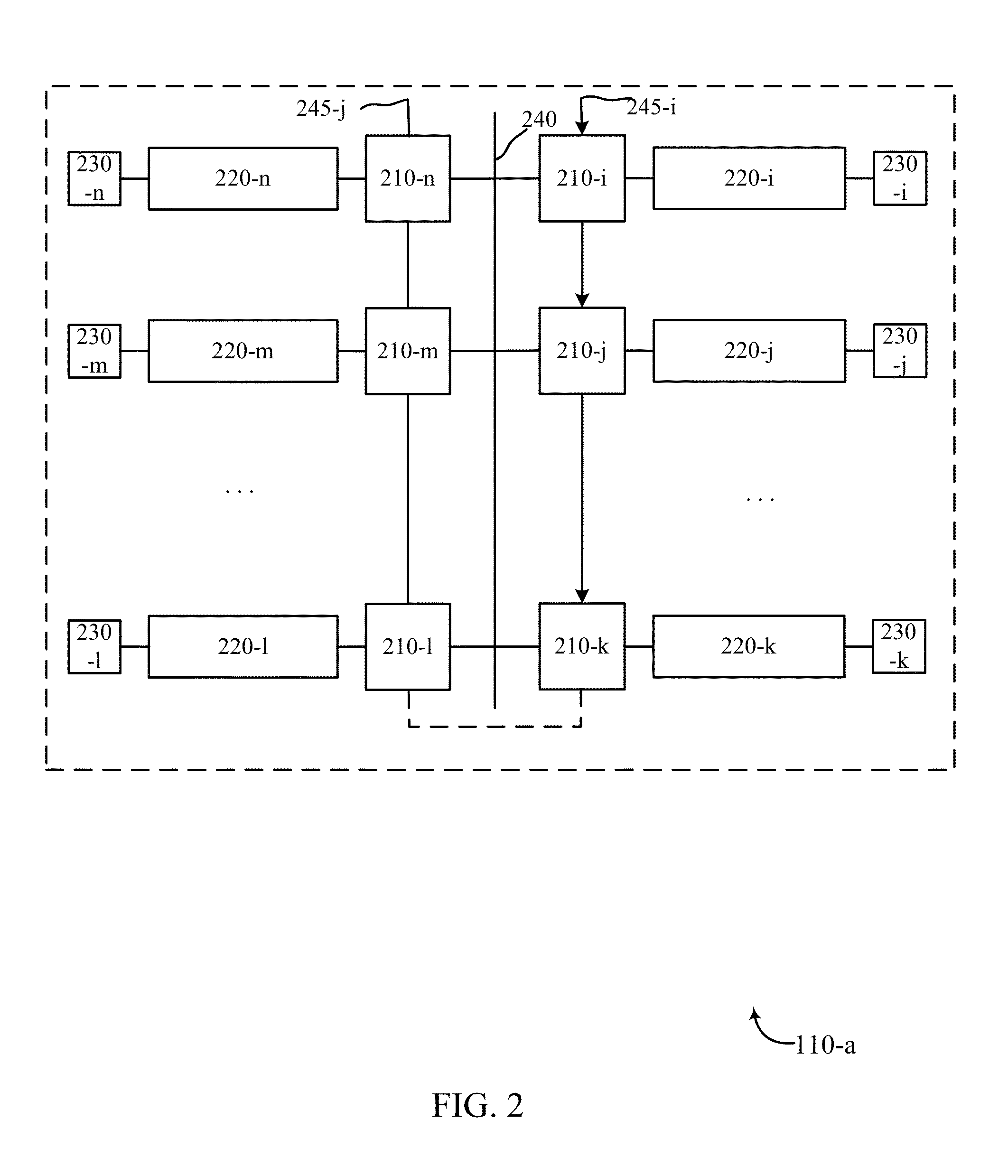
Micro fluidic chip for sorting and whole genome amplification of single cell
PendingCN106065391AEnables in situ fluorescence identificationAchieve crackingBioreactor/fermenter combinationsBiological substance pretreatmentsControl layerEngineering
The invention provides a micro fluidic chip for sorting and whole genome amplification of a single cell. The micro fluidic chip comprises a four-layer structure, wherein the four layers are stacked together in order and mutually sealed, and the structure comprises the following layers from top to bottom: a top cover, a micro valve control layer, a valve plate layer and a channel layer; and the bottom of the micro fluidic chip is externally provided with a magnet. The invention also provides a system for screening and identifying cells as well as amplification and analysis of a single cell, and the system is matched with the chip. The micro fluidic chip and the corresponding detection system can be used for sorting target cells from a large amount of cells quickly, simply and cheaply, and amplifying and analyzing whole genome DNA of a single cell therein.
Owner:THE NAT CENT FOR NANOSCI & TECH NCNST OF CHINA
Mitochondria genome amplified universal primer mixture as well as design and amplification method thereof
ActiveCN105506103AEfficient and specific enrichmentAchieve deep coverageMicrobiological testing/measurementDNA/RNA fragmentationGenomic informationMitophagy
The embodiment of the invention discloses a universal primer mixture for animal mitochondria genome amplification as well as a design and amplification method thereof, and a kit comprising the universal primer mixture. By utilizing the primer mixture or the kit and the animal mitochondria genome amplified method and combining the novel high throughput sequencing technique, the animal mitochondria genome information can be efficiently obtained in low cost.
Owner:TIANJIN UNIV OF TRADITIONAL CHINESE MEDICINE
Amplification method, primer, sequencing method and mutation detection method of mitochondria whole genome DNA (Deoxyribonucleic Acid)
InactiveCN103173441ALow costMicrobiological testing/measurementDNA preparationDiseaseMutation detection
The invention discloses a primer pair and a kit for amplification of a mitochondria whole genome. The invention further discloses an amplification method, a sequencing method and a mutation detection method of the mitochondria whole genome DNA (Deoxyribonucleic Acid). According to the invention, the direct amplification of the given primer is combined with the direct sequencing method, so that a mitochondria whole genome sequence can be obtained; all mutations of the mitochondria genes can be detected by one step for being used for detecting the diseases caused by most of the mitochondria gene mutations.
Owner:BGI GENOMICS CO LTD
Selection of embryo of test tube baby through sequencing by single cell genome of polar body or embryo
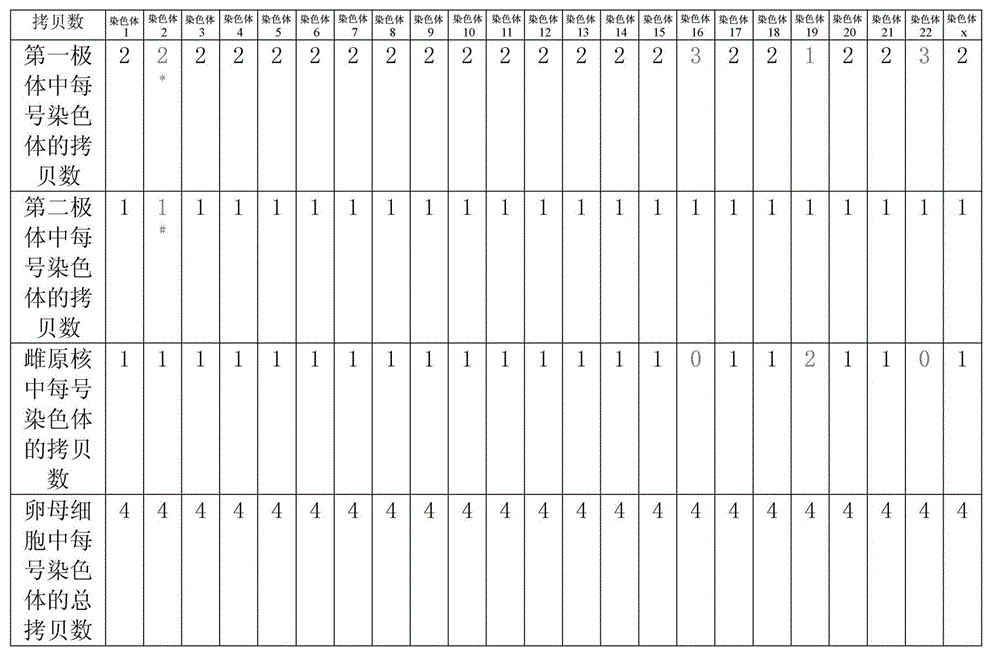
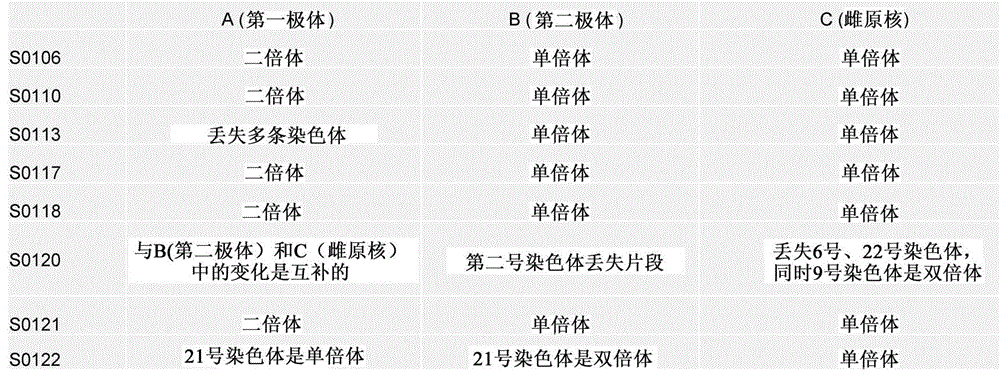
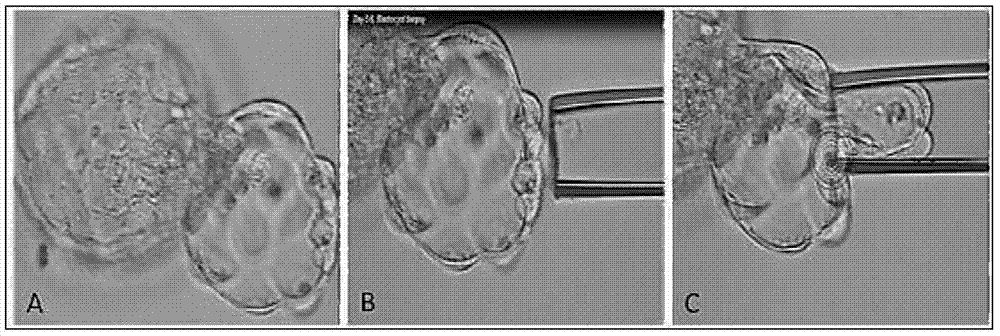
Disclosed is a method for using a first polar body and a second polar body as well as a single-cell embryo to carry out whole-genome non-exponential amplification and high-throughput genome sequencing, so as to perform preimplantation genetic diagnosis for genetic disease and testing for pathogenic genes causing repeated miscarriages. The method of the present invention comprises the following steps: (1) obtaining oocytes and embryos, and carrying out separation and genome amplification of first and second polar bodies and single-cell embryos; (2) establishing a genome sequencing library and sequencing, and carrying out bioinformatic analysis of the genome to obtain a gene spectrum and information concerning the number of copies of chromosomes and fragments thereof; (3) determining the chromosome ploidy of the polar bodies and embryos as well as information concerning defects, replication and point mutation in the chromosome fragments; (4) selecting normal or suitable embryos for implantation.
Owner:HARVARD UNIV +2
Method for detecting embryo chromosome abnormalities by using blastula-stage embryo cells
InactiveCN104711362AAvoid harmA large amountMicrobiological testing/measurementHigh cellCell separation
The invention discloses a method for performing genome amplification by using embryo blastula-stage cells, performing chromosome detection on the preimplantational embryo by combining a high-flux sequencing technique and screening out the chromosome normal embryo. The method can comprehensively and completely analyze the genetic variation information of the embryo genome, thereby instructing the preimplantational embryo selection, reducing the hereditary diseases and enhancing the success rate of test tube babies. The method comprises the following steps: blastula-stage trophocyte separation; genome amplification; DNA (deoxyribonucleic acid) segmentation; and Proton library establishment, mounting sequencing and sequencing data analysis. By using the blastula-stage embryo to perform trophocyte separation detection, the method avoids the injuries of cleavage-stage cell separation to the embryo, obtains higher cell quantity than the cleavage stage, and enhances the success rate and amplification effect of genome amplification. After the blastula-stage embryos are subjected to the natural elimination process, the high-quality blastula-stage embryo is selected for detection, thereby saving the cost.
Owner:SUZHOU BASECARE MEDICAL DEVICE CO LTD
Real-time quantitative PCR method for detecting components of meat product
ActiveCN108624659AImprove accuracyImprove reliabilityMicrobiological testing/measurementBiotechnologyGenomic DNA
The invention discloses a real-time quantitative PCR method for detecting components of meat product. The real-time quantitative PCR method comprises the following steps: extracting genomic DNA of a meat product sample and a standard sample; designing PCR amplification primers required by the genomic DNA of the meat product sample and the standard sample according to the polymorphism difference ofmitochondrial NADH oxidoreductase 1, correspondingly preparing a PCR reaction system for PCR amplification, and reading the Ct value during the amplification process; judging the reliability of PCR amplification according to the contrast between the value Ct of the NADH oxidoreductase 1 in PCR amplification products and the Ct value of 18S rRNA; and judging whether the meat product sample contains any component to be detected according to the difference delta Ct of the Ct value of the NADH oxidoreductase 1 of the genomic DNA amplification products of the meat product sample and the standard sample. The real-time quantitative PCR method can be used for simultaneously detecting one or more of the components with different species sources in the meat product, and has accurate and reliable detection results.
Owner:WUHAN POLYTECHNIC UNIVERSITY
Methods for genome amplification
A method for whole genome amplification comprising (a) treating genomic DNA with a modifying agent which modifies cytosine bases but does not modify 5′-methyl-cytosine bases under conditions to form single stranded modified DNA; (b) providing a population of random X-mers of exonuclease-resistant primers capable of binding to at least one strand of the modified DNA, wherein X is an integer 3 or greater; (c) providing polymerase capable of amplifying double stranded DNA, together with nucleotides and optionally any suitable buffers or diluents to the modified DNA; and (d) allowing the polymerase to amplify the modified DNA.
Owner:HUMAN GENETIC SIGNATURES PTY LTD
Method for conducting SNP-haplotype analysis by means of multiplex PCR technology
PendingCN105385755AEfficient determinationAccurately determineMicrobiological testing/measurementWork cycleEmbryo
The invention discloses a method for conducting SNP-haplotype analysis by means of a multiplex PCR technology. Designed primers are combined into one tube for single primer amplification when synthesis is conducted, and meanwhile whole genome amplification is completed through an MALBAC technology; when multiplex PCR amplification is conducted, a touch down PCR amplification program is adopted, high-throughput sequencing and reasonable data analysis are combined, only one set of SNP detection is needed, different schemes do not need to be designed according to different objects, the working cycle is shortened, and the cost is reduced. In addition, according to the method for conducting the SNP-haplotype analysis by means of the multiplex PCR technology, family and embryonic genome SNP information is captured, embryo SNP information can be effectively and accurately determined, SNP-haplotype analysis is conducted to determine whether an embryo carries mutation sites of a genetic gene or not, and the defects that in the prior art, the technological cost is high, the cycle is long, and the application range is small are effectively overcome.
Owner:YIKON GENOMICS SHANGHAI CO LTD +1
Method for detecting embryonic chromosome abnormality by virtue of blastochyle free DNA
InactiveCN104450923AProbability of small developmental abnormalitiesSimple and fast operationMicrobiological testing/measurementFragment sizeEmbryo
The invention relates to a method for detecting embryonic chromosome abnormality by virtue of blastochyle free DNA. The method comprises the following steps: acquiring blastochyle free DNA, detecting the blastochyle DNA, carrying out whole genome amplification of the free DNA, analyzing a product of the whole genome amplification, implementing fragmenting treatment on genome DNA, carrying out quantitative analysis and fragment size analysis on fragmented target DNA, constructing a library, sequencing by virtue of a computer and analyzing biological information. By virtue of high-throughput sequencing, the method disclosed by the invention can be used for overcoming shortcomings of a conventional DNA analysis method which is merely used for researching partial region of a single cell genome, and is capable of completely analyzing the genetic information of the single cell genome; the method is simple and convenient to operate, time-saving and efficient; meanwhile, by using the blastochyle free DNA as a detection sample, the method is convenient and safe to sample, so that the probability of later embryonic development abnormality is reduced and embryo is protected from being influenced in later development.
Owner:SUZHOU BASECARE MEDICAL DEVICE CO LTD
Fish mitochondria genome-wide DNA amplification
InactiveCN104911182AAvoid high demandsModerate amplification lengthDNA preparationDNA/RNA fragmentationFisheryDna amplification
The invention provides fish mitochondria genome-wide DNA amplification. The invention firstly provides a primer pair for fish mitochondria genome-wide DNA amplification. The invention further provides a method for fish mitochondria genome-wide DNA amplification based on the primer combination. By the adoption of the amplification method, clone products of various fish mitochondria genomes can be obtained successfully. In addition, a primer pair obtained through designing and screening has the excellent characteristics that the amplification performance is high and the number of by-products generated is small.
Owner:CHINESE ACAD OF FISHERY SCI
Single-cell genome analysis method and kit
ActiveCN102533960AEfficient analysisQuality improvementMicrobiological testing/measurementHousekeeping geneResearch strategies
The invention discloses a single-cell genome analysis method and a kit. The method comprises the following steps of: a, acquiring a complete cell genome DNA (Deoxyribonucleic Acid); b, carrying out single-cell whole genome amplification; c, carrying out quantitative detection and qualitative detection, wherein the qualitative determination comprises that single-cell whole genome amplification product is determinated by adopting a Housekeeping Gene detection method. The kit contains a specific primer of the single cell Housekeeping Gene. By adopting the method disclosed by the invention, genetic variation information of a single cell genome can be comprehensively and completely analyzed, and an effective research strategy can be provided for the single cell genome of a brand-new specie; meanwhile, qualitative and quantitative detection screening steps are introduced, and many unqualified amplification samples can be removed, thus quality of a downstream upper computer sequencing library is controlled and unnecessary waste can be greatly avoided.
Owner:BGI SHENZHEN CO LTD +1
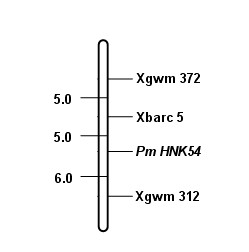
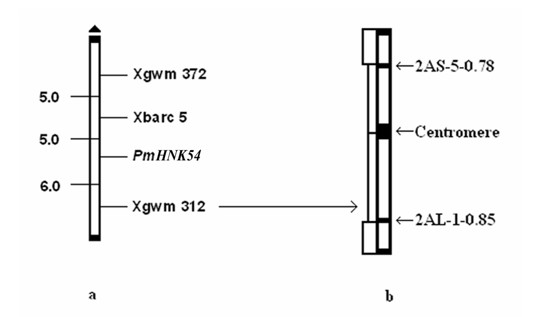
Marker primer interlocked with wheat powdery mildew resistance gene PmHNK54 and application thereof
InactiveCN101988064AReduce dosageStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationStainingGel electrophoresis
The invention relates to a marker primer interlocked with a wheat powdery mildew resistance gene PmHNK54 and application thereof in molecular marker-assisted selection. The sequence of the marker primer on a genetic map is Xbarc5, PnHNK54 and Xgwm312, and the genetic distances between Xbarc5 and PnHNK54 and between Xgwm312 and PnHNK54 are respectively 5.0cM and 6.0cM. 10 mu l of PCR (Polymerase Chain Reaction) reaction system during SSR (Simple Sequence Repeat) marker selection comprises 1 mu l of 10*buffer, 0.2 mu L of 10nM / mu l dNTP, 0.2 mu l of 20u M / mu L primer, 0.1 mu l of 5U / mu lTaq enzyme, 8.1 mu l of deionized water and 0.4 mu l of 20 ng / mu l genome DNA. An amplification procedure comprises the following steps of: pre-denaturing at 94 DEG C for 3 min, denaturing at 94 DEG C for 1 min, renaturing at 55 or 60 DEG C for 1 min, extending at 72 DEG C for 2 min, and repeating for 40 cycles; extending at 72 DEG C for 10 min; and performing 6% denaturing polyacrylamide gel electrophoresis or 8% nondenaturing polyacrylamide gel electrophoresis on amplification products, observing and taking pictures after silver nitrate dyeing. The invention finds the position of the powdery mildew resistance gene PmHNK54 in wheat parent material Zheng 9754, has the advantages of strong marker specificity, high stability, cost saving and high selection efficiency and is applicable to large scale, high throughput and automation.
Owner:HENAN ACAD OF AGRI SCI
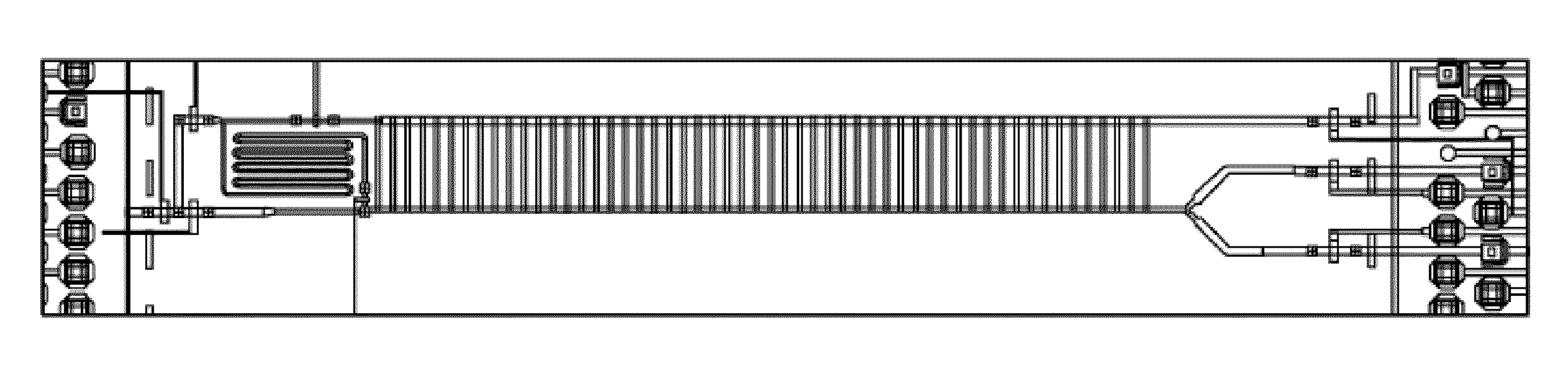
Methods, systems and devices for multiple single-cell capturing and processing using microfluidics
InactiveUS20130302884A1Bioreactor/fermenter combinationsHeating or cooling apparatusReal-Time PCRsMRNA Sequencing
Methods, systems, and devices are described for multiple single-cell capturing and processing utilizing microfluidics. Tools and techniques are provided for capturing, partitioning, and / or manipulating individual cells from a larger population of cells along with generating genetic information and / or reactions related to each individual cell. Different capture configurations may be utilized to capture individual cells and then processing each individual cell in a multi-chamber reaction configuration. Some embodiments may provide for specific target amplification, whole genome amplification, whole transcriptome amplification, real-time PCR preparation, copy number variation, preamplification, mRNA sequencing, and / or haplotyping of the multiple individual cells that have been partitioned from the larger population of cells. Some embodiments may provide for other applications. Some embodiments may be configured for imaging the individual cells or associated reaction products as part of the processing. Reaction products may be harvested and / or further analyzed in some cases.
Owner:FLUIDIGM CORP
High-flux single-cell whole genome amplification method
InactiveCN107400705AFully automatedAchieve scaleMicrobiological testing/measurementDNA preparationHigh fluxBioinformatics
The invention discloses a high-flux single-cell whole genome amplification method. The method comprises the steps as follows: a cell suspension with preset cell density is distributed to each micropore of a nano microporous chip by use of a nano micro liquid distribution platform; a cell lysis buffer is distributed to micropores by use to the micro liquid distribution platform to lyse cells in the micropores; a neutralization solution is distributed to micropores by use to the micro liquid distribution platform for a neutralization reaction; a whole genome amplification reagent is distributed to micropores by use to the micro liquid distribution platform for a whole genome amplification reaction. The method has the characteristics of being high in flux, low in cost, single-cell and integrated, and automatic and large-scale single-cell whole genome amplification can be realized.
Owner:MGI TECH CO LTD
Selective genome amplification
The invention provides methods and compositions for amplifying selected polynucleotides, especially selected subsets of restriction fragments. Generally, methods of the invention are implemented by ligating adaptors containing at least one promoter sequence to such fragments under conditions that promote the formation of closed single stranded or double stranded structures, which are capable of serving as cyclical templates for transcription.
Owner:AGENCY FOR SCI TECH & RES
Methods for genome amplification
A method for whole genome amplification comprising (a) treating genomic DNA with a modifying agent which modifies cytosine bases but does not modify 5′-methyl-cytosine bases under conditions to form single stranded modified DNA; (b) providing a population of random X-mers of exonuclease-resistant primers capable of binding to at least one strand of the modified DNA, wherein X is an integer 3 or greater; (c) providing polymerase capable of amplifying double stranded DNA, together with nucleotides and optionally any suitable buffers or diluents to the modified DNA; and (d) allowing the polymerase to amplify the modified DNA.
Owner:HUMAN GENETIC SIGNATURES PTY LTD
A kind of spider mitochondrial genome complete sequence amplification primer and amplification method
ActiveCN104531688BEfficient and specific amplificationLow costDNA preparationDNA/RNA fragmentationDNA extractionMitophagy
Owner:CHINA JILIANG UNIV
Trace and complex DNA amplification method
The present invention relates to a high sensitivity, high fidelity, high uniformity and high efficiency amplification technology of trace and complex DNA. The technology system is constant temperature DNA amplification optimized by high concentration trehalose and guided by DNA polymerase, wherein balanced and efficient amplification of DNA in different regions of the whole genome or a DNA library sequence can be maintained in the context of effective inhibition on non-specific amplification by the amplification system. The method has the following characteristics that: sensitivity is high; balanced amplification of different region sequences can be maintained, and the amplification product has high fidelity (minimum deviation) and high uniformity; and the total reaction volume has high flexibility, and can be conveniently adjusted according to requirements. In addition, the technology provides broad application prospects in single cell level and trace genome DNA amplification and gene detection, pretreated function genome associated DNA library (spectrum) analysis, and other fields.
Owner:SHANGHAI GREENGLOBE BIOTECH
GII.P12/GII.3 recombinant norovirus genome amplification primer and amplification method
The invention discloses a GII.P12 / GII.3 recombinant norovirus genome amplification primer and an amplification method. The amplification method comprises the following steps: by taking six pairs of amplification primers as forward and reverse primers of amplification primers respectively, and by taking RNA of GII.P12 / GII.3 recombinant norovirus as a template, performing RT-PCR amplification so as to obtain amplification products respectively, performing nucleotide sequencing on the amplification products by using the six amplification primers and two sequencing primers II.12-Seq1R and II.3-Seq6F, and then carrying out jointing and comparison, thereby obtaining full-length sequences of a GII.P12 / GII.3 recombinant norovirus genome. For the GII.P12 / GII.3 recombinant norovirus which occurs frequently in China, a sectional amplification strategy of '4+1+1'is designed according to three opening reading frames included in the genome, corresponding amplification primers are designed according to conserved regions, the amplification primers are applied to practical detection samples, and GII.P12 / GII.3 recombinant norovirus genome sequences can be acquired. The GII.P12 / GII.3 recombinant norovirus genome amplification primer and the amplification method can be widely applied to fields such as medical treatment and public health and inspection and quarantine with norovirus detection requirements, and related scientific fields.
Owner:GUANGDONG INST OF MICROORGANISM +1
Primer system and method for amplifying avian influenza virus whole genomes
The invention provides a primer system and a method for amplifying avian influenza virus whole genomes. Specifically, the primer system comprises a primer pair, namely UAIV-F and UAIV-R, for amplifying the avian influenza virus whole genomes, as well as eight segmental primer pairs; and the primer pair comprises forward sequence: 5'-AGCRAAAGCAGG, and backward sequence: 3'-AGTAGAAACAAG. According to the first method, virus RNA is taken as a template, and one-step RT-PCR amplification is carried out by virtue of the UAIV-F and the UAIV-R, so as to obtain the avian influenza virus whole genomes; and according to the second method, amplification is carried out by taking virus RNA as a template so as to obtain cDNA and then amplification of eight segments of the avian influenza virus whole genomes is carried out respectively by virtue of the eight segmental primer pairs. The primer system and the method disclosed by the invention are applicable to amplification of all avian influenza virus whole genomes, including the genomes of the various subtype avian influenza viruses as well as quite conserved non-coding regions at two ends of the genomes; therefore, the invention is broad in applicability.
Owner:ZHAOQING DAHUANONG BIOLOGIC PHARMA +1
Genotyping chip for legionella pneumophila, and kit for detection of legionella pneumophila
ActiveCN102424862AMake up for the lack of detection rangeLow costNucleotide librariesMicrobiological testing/measurementNucleotideGenomic DNA
The present invention provides a genotyping chip for legionella pneumophila, and a kit for detection of the legionella pneumophila. The chip and the kit are mainly provided according to the 11 serotypes of the legionella pneumophila, wherein the 11 serotypes comprise O1, O2, O3, O4, O5, O6, O7, O11, O12, O13 and O15. The gene chip comprises a solid phase carrier and oligonucleotide probes fixed on the solid phase carrier, wherein the oligonucleotide probes comprise DNA fragments selected from wzm gene, wzt gene and wecA gene, or complementary DNA fragments of the wzm gene, the wzt gene and the wecA gene, wherein the wzm gene, the wzt gene and the wecA gene have significant biological evolution advantages. According to the present invention, the genomic DNA of the sample requiring detection is amplified by the designed primers; the resulting amplified genomic DNA is labeled; the resulting labeled genomic DNA is subjected to hybridization with the gene chip; according to the resulting hybridization signals, the different serotypes of the legionella pneumophila can be detected. With the gene chip of the present invention, the purpose of the detection of the serotypes of the legionella pneumophila can be achieved, the operation is convenient, the accuracy is high, the repeatability is strong, and the accurate medical medication guidance is provided.
Owner:NANKAI UNIV
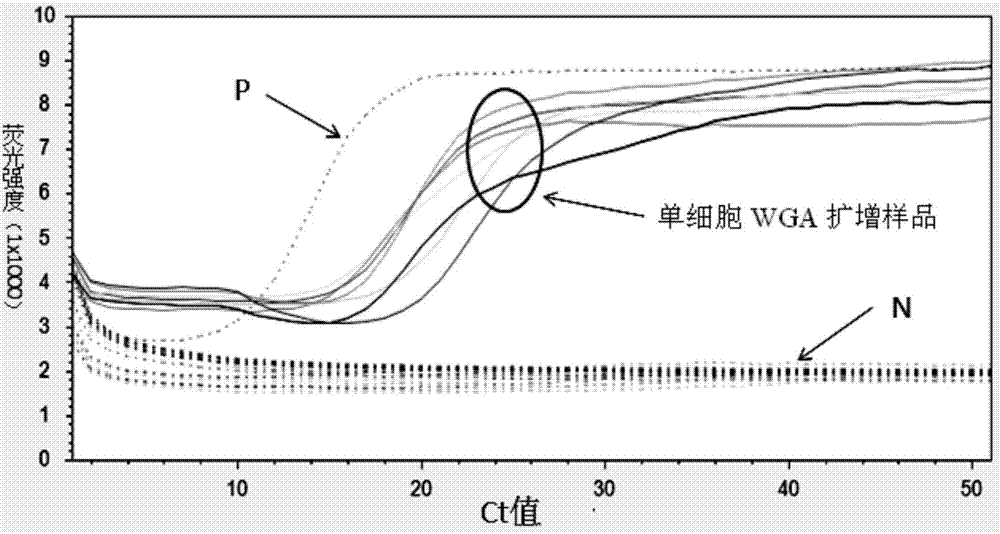
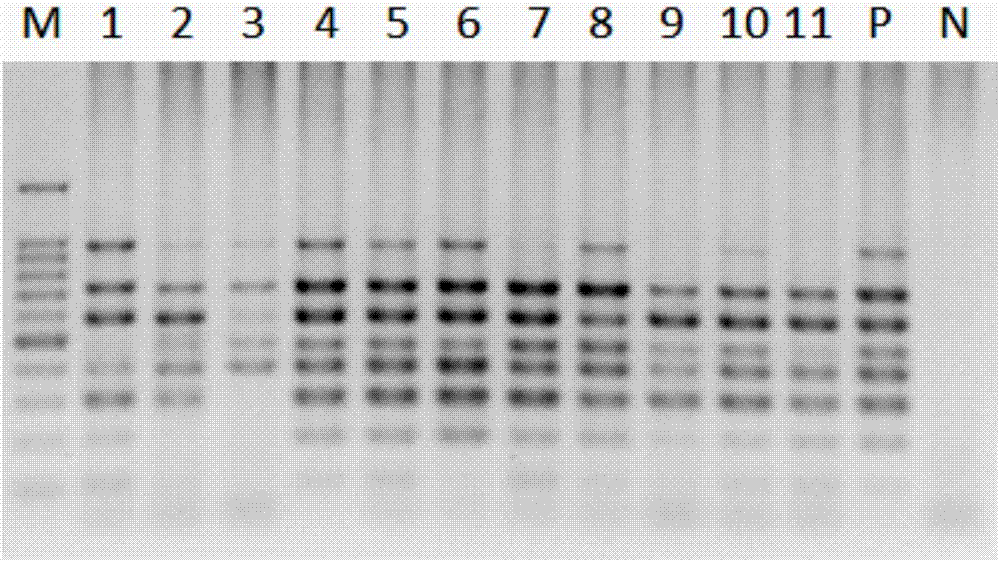
Method and kit for quality appraisal for amplification products after single cell genome amplification
InactiveCN104762405AReasonable designGuaranteed success rateMicrobiological testing/measurementA-DNAQuality appraisal
The invention relates to a method and a kit for quality appraisal for amplification products after single cell genome amplification. The method comprises the following steps: separating and splitting single cells to obtain the whole-genome DNA of the single cells; carrying out single cell whole-genome amplification on the whole-genome DNA to obtain whole-genome amplification products; placing specific primers in conserved segments on 5 pairs of genomes in an amplification system, carrying out PCR amplification by taking the whole-genome amplification products as a template, carrying out electrophoresis detection on 5 amplification products, judging the quality of the whole-genome amplification products according to a detection result, and showing a result that 3-5 amplification products of uniformly distributed strips on an electrophoretogram are amplification product samples meeting sequencing requirements; constructing a DNA sequencing library on the amplification products meeting the sequencing requirements; carrying out high-throughput sequencing on the DNA sequencing library.
Owner:PEKING JABREHOO MED TECH CO LTD +1
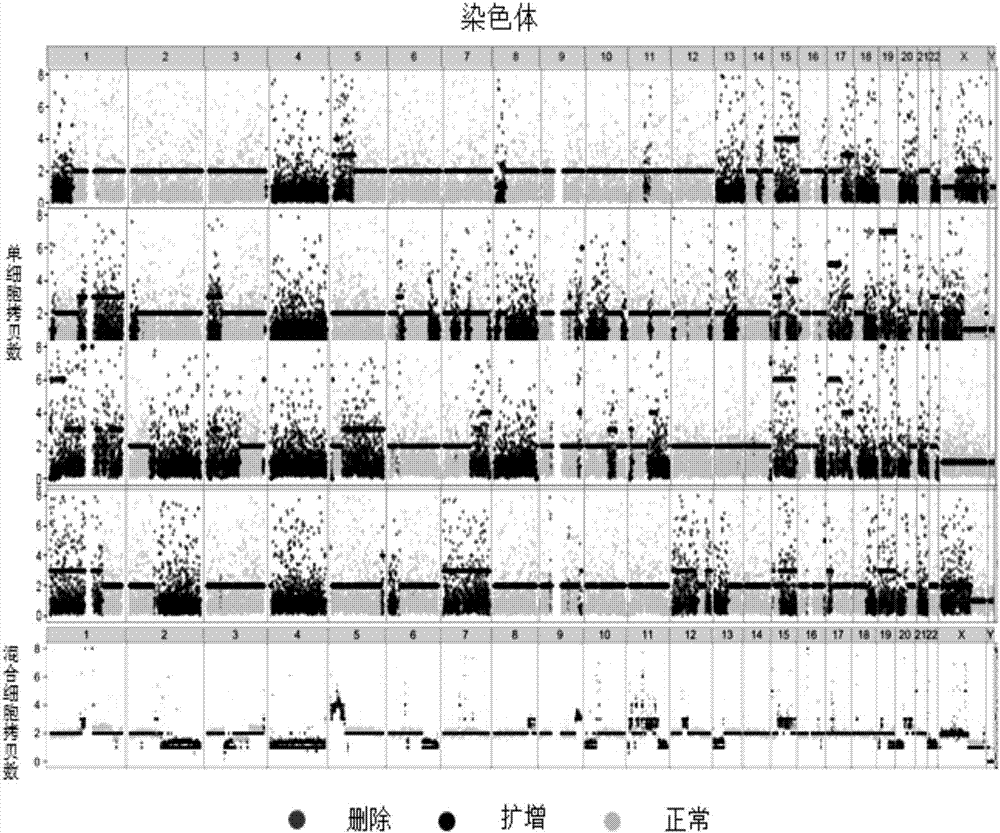
Method for detecting tumor single cell somatic mutation
ActiveCN110093417AAdd dimensionMicrobiological testing/measurementGenome evolutionMutation detection
The invention discloses a method for detecting tumor single cell somatic mutation. The method combines the population polymorphism site information of the tumor single cell and the genomic copy numberchange information for analysis, takes full account of the random base substitutions that may be introduced by a single-cell amplification technology and the systematic biases that may be introducedby corresponding genomic amplification and sequencing technologies, improves the accuracy of somatic mutation detection, and has important significance for the heterogeneity description of tumor genome and the information of tumor genome evolution.
Owner:PEKING UNIV
Method for analysis of multiple regions of DNA in single cells of uncultured microorganisms
InactiveUS20090186778A1Nucleotide librariesMicrobiological testing/measurementMicroorganismCulture cell
Described herein are methods for single cell sorting and DNA analysis which permit metabolic mapping of taxonomically diverse microbial cells. Methods described herein encompass procedures for single-cell separation of individual uncultured cells, such as aquatic microbial cells, by fluorescence-activated cell sorting (FACS), subsequent single cell whole genome amplification (WGA), and downstream analyses of multiple regions of DNA.
Owner:BIGELOW LAB FOR OCEAN SCI
Multiple primers, kit and method for high-throughput sequencing of enterovirus
InactiveCN110387438AComprehensive detection effectPerfect analysis techniquesMicrobiological testing/measurementDNA/RNA fragmentationEnterovirusGenotype
The invention discloses multiple primers and a kit for high-throughput detecting of a whole genome of enterovirus, and a high-throughput sequencing analysis method. Compared with the prior art, specific primers do not need to be used for detecting a certain generic type one by one in a targeted mode any more, but multiple PCR is utilized to conduct genomic amplification on various enterovirus subtypes through an experiment, the virus subtypes are accurately distinguished and identified, and the accuracy rate is higher, and repeatability is better; high-depth sequencing is adopted, the enough detection depth is ensured, and thus false positive is lower; and through the detection method, the gene types of the enterovirus are rapidly identified, genome variable site, diversity and recombination analysis of viruses can be further provided, and the important scientific basis of prevention and control over the enterovirus is provided.
Owner:广东省公共卫生研究院 +1
Methods and compositions for isothermal whole genome amplification
Owner:PROGENITY INC
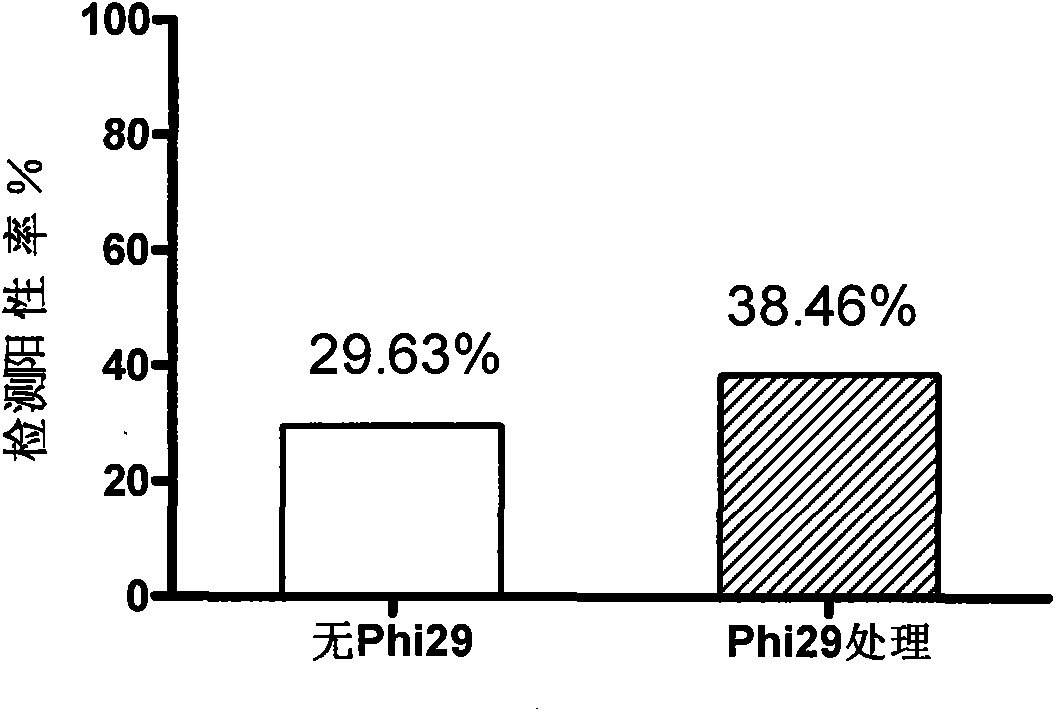
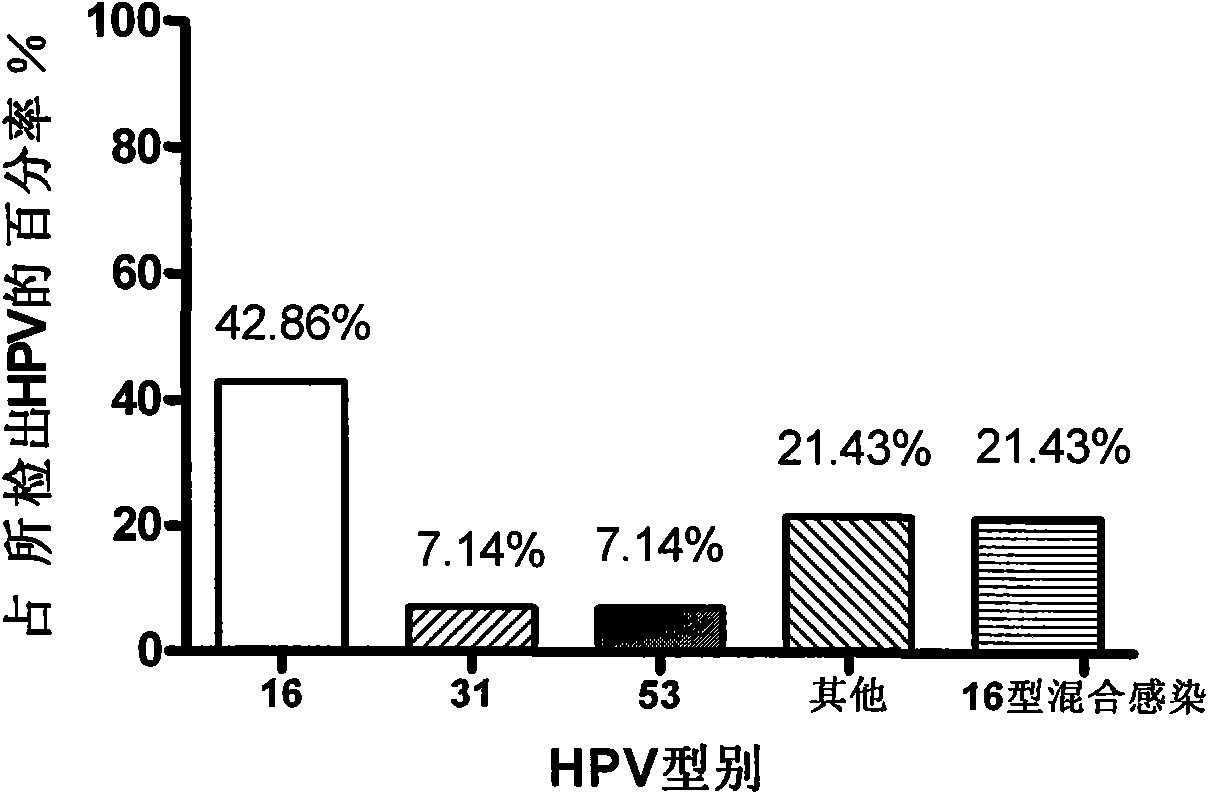
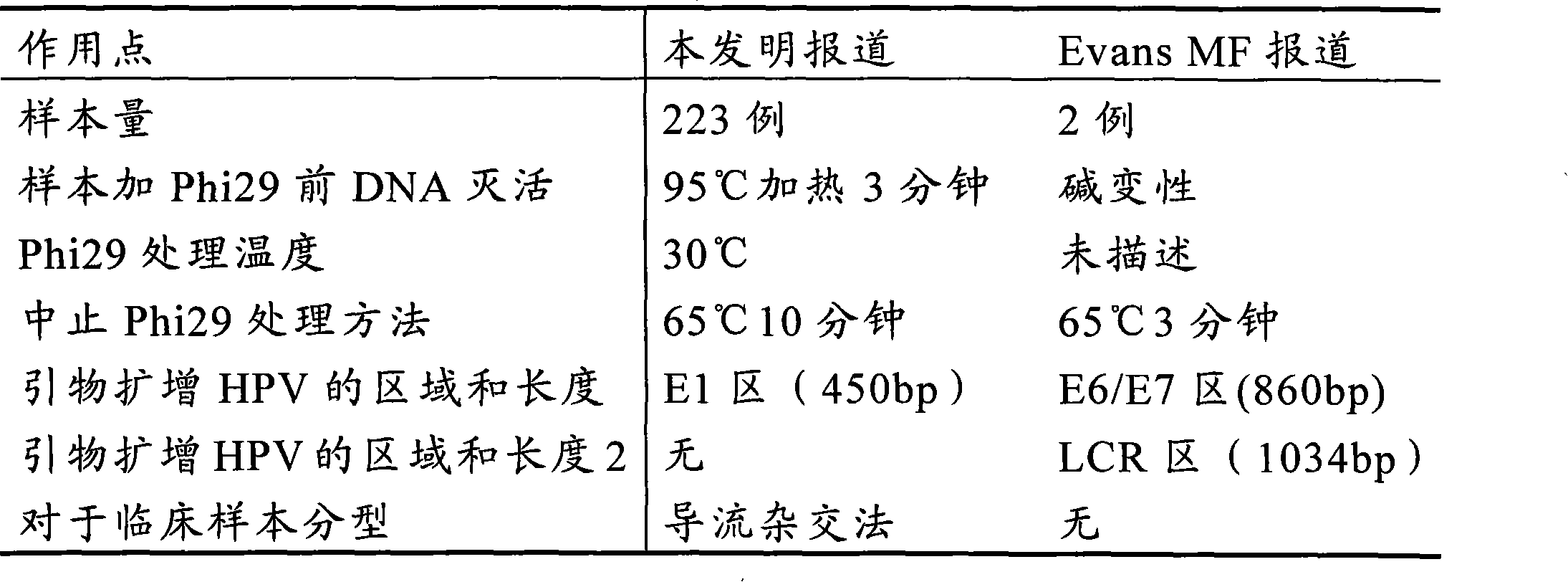
Method for detecting HPV and application of Phi29DNA polymerase in detecting HPV
InactiveCN101619364AImprove the detection rateAmplified equalizationMicrobiological testing/measurementHuman papillomavirusPolymerase L
The invention discloses a method for detecting human papillomavirus (HPV) in a sample, which sequentially comprises the following steps: A) extracting DNA in the sample; B) amplifying the extracted DNA in step A); C) amplifying PCR; and D) detecting viruses. The amplifying of the step B) is to carry out multiple displacement amplification on all the genes of the extracted DNA with a Phi29DNA polymerase. The invention also discloses application of the Phi29DNA polymerase in detecting the human papillomavirus. The detection method can obviously improve the detection rate of the HPV. The sample does not need to be purified when the Phi29DNA polymerase is used to amplify the DNA. A gene group is uniformly amplified; the amplifying yield is stable; the operation is simple; the method is independent of a PCR reaction; and no amplified product having specificity is generated.
Owner:SHANGHAI INST OF IMMUNOLOGY +1
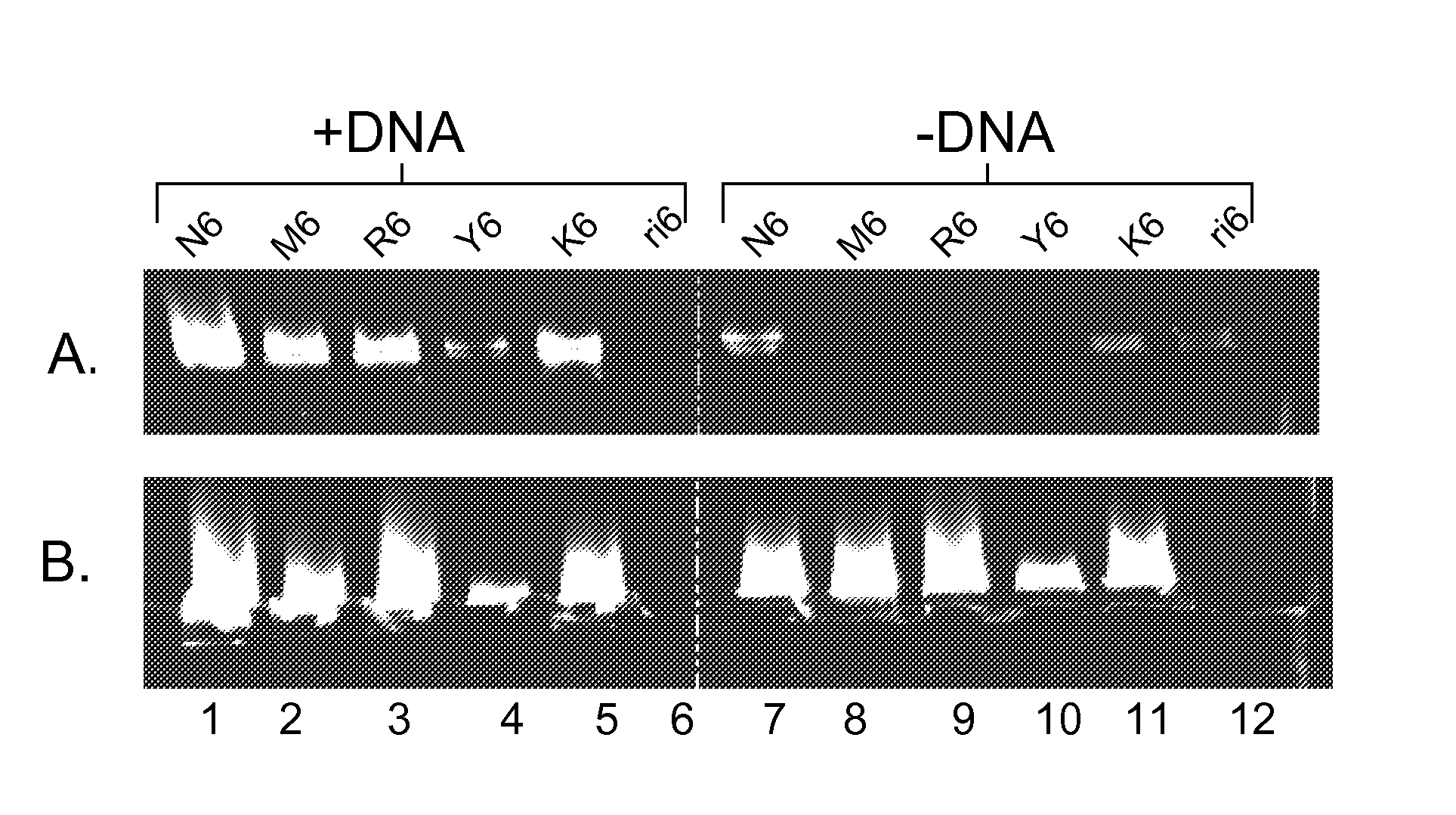
Methods for high specificity whole genome amplification and hybridization
Methods for amplifying genomic DNA using semi-random primers that consist of different combinations of two non-complementary bases are disclosed. In a preferred aspect all of the primers in the collection are composed entirely of the same two non-complementary bases. In preferred aspects the DNA is amplified using R6, Y6, M6 and K6. The amplification is by a strand displacing polymerase and the amplification product may be hybridized to a high complexity array of probes without further complexity reduction. In some aspects, additives are included in the hybridization to reduce non-specific hybridization. The hybridization pattern obtained is preferably analyzed for allele specific hybridization to determine genotype. The primers in the collection are selected so that there is a minimum of self complementarity between any two primers in the collection, minimizing the occurrence of hybrids between primers.
Owner:AFFYMETRIX INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com