Mitochondria genome amplified universal primer mixture as well as design and amplification method thereof
A mitochondrial genome and universal primer technology, applied in the field of genetic engineering, can solve the problems of high cost, high sequencing throughput, and easy mixing of mitochondrial pseudogenes, etc., and achieve the effect of reduced cost and low computational complexity of splicing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] The design of embodiment 1 universal primer
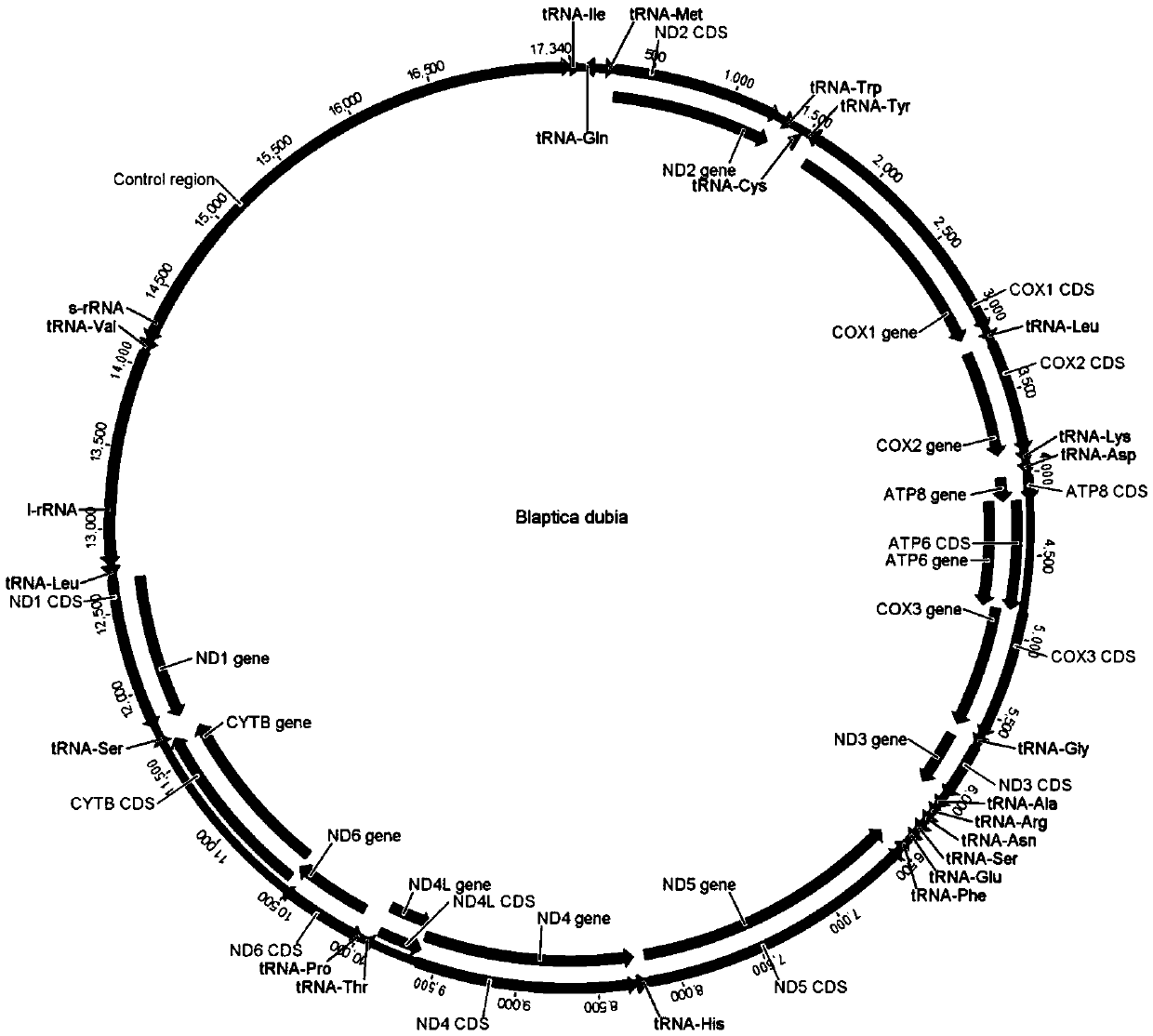
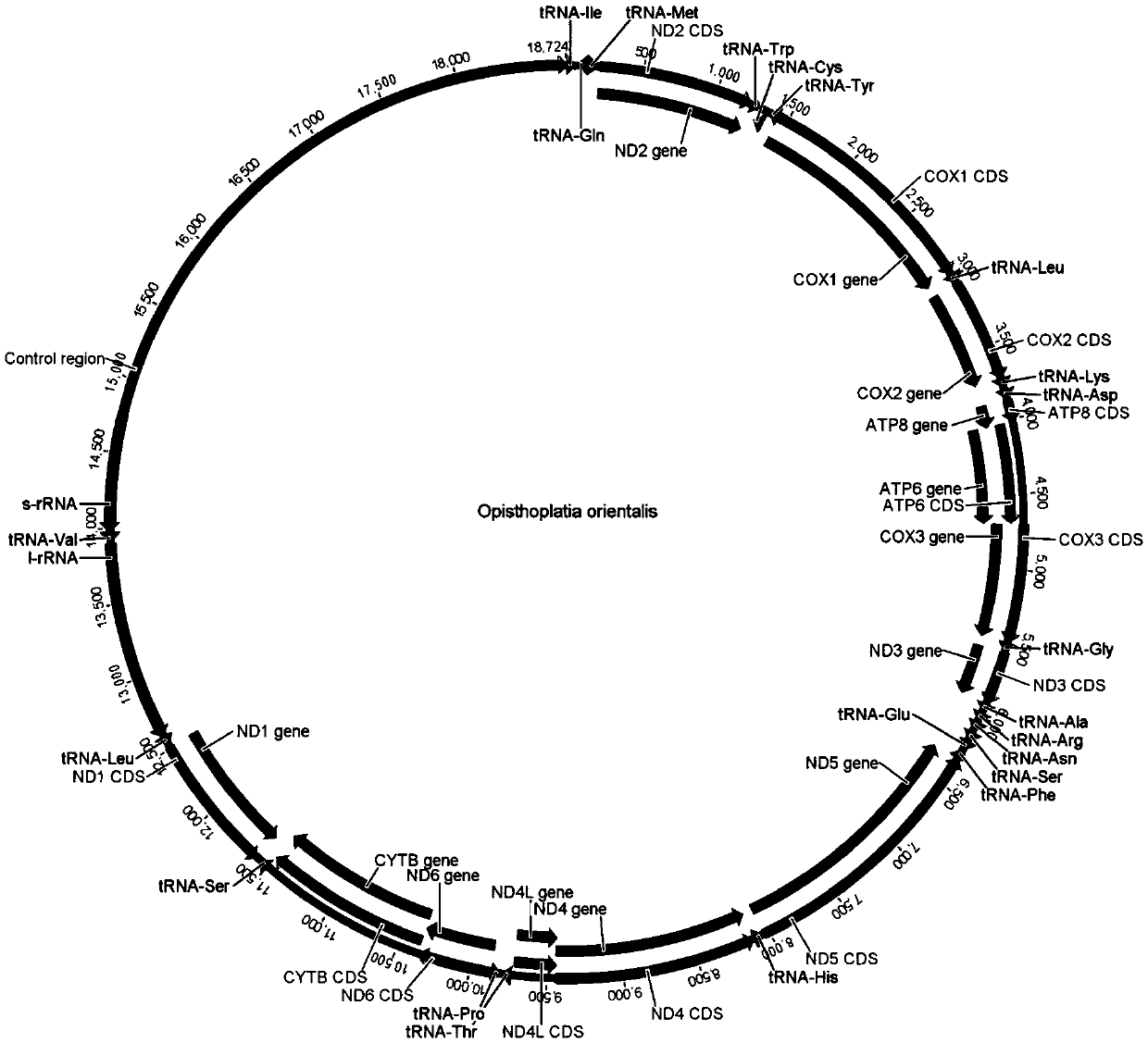
[0082]Log in to the GenBank (http: / / www.ncbi.nlm.nih.gov) database on the NCBI website to search for ND2, COI, COII, COIII, ND5, ND4, cytB, 16S and 12S of some invertebrate mitochondrial genomes that have been determined Gene sequence, load the sequence into BioEdit software, use the ClustalW algorithm to perform multiple sequence alignment analysis, find the conserved sequence, load the searched conserved sequence into the Oligo6 software, manually debug and design the invertebrate mitochondrial genome amplification Primers (see Table 2). After obtaining the designed primers, PremierPrimer5.0 software can be used to investigate the possibility of forming homologous or heterologous dimers among the primers and exclude those with a high possibility of forming homologous or heterologous dimers. The two bases at the 3' end of the primer are thio-modified to prevent the 3'-5' exonuclease activity of the 3' end of the primer by ...
Embodiment 2
[0084] Embodiment 2 phenol chloroform method extraction animal total DNA
[0085] Take 100 mg of chopped fresh animal tissue (if it is a tiny animal, keep the necessary residues before using), grind with liquid nitrogen, and resuspend in 100 μL of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH=8.0), After adding 5 μL of 10% SDS, 0.8 μL of 25 mg / ml RnaseA, and 1 μL of 10 mg / ml ProK, incubate at 50° C. for 60 minutes. Then add 55 μL of phenol, 55 μL of chloroform:isoamyl alcohol (24:1), mix well and centrifuge at 12000 g for 6 minutes, and keep the supernatant. Then add 100 μL of chloroform:isoamyl alcohol (24:1), mix well, centrifuge at 12000g for 6 minutes, and keep the supernatant. Add 270 μL of absolute ethanol to the above solution, store at -80°C for 20 minutes, and then centrifuge at 12,000 g for 20 minutes to retain the precipitate. Add 500 μL of pre-cooled 70% ethanol, store at -80°C for 10 minutes, and centrifuge at 12,000 g for 20 minutes to retain the precipitate. Incub...
Embodiment 3
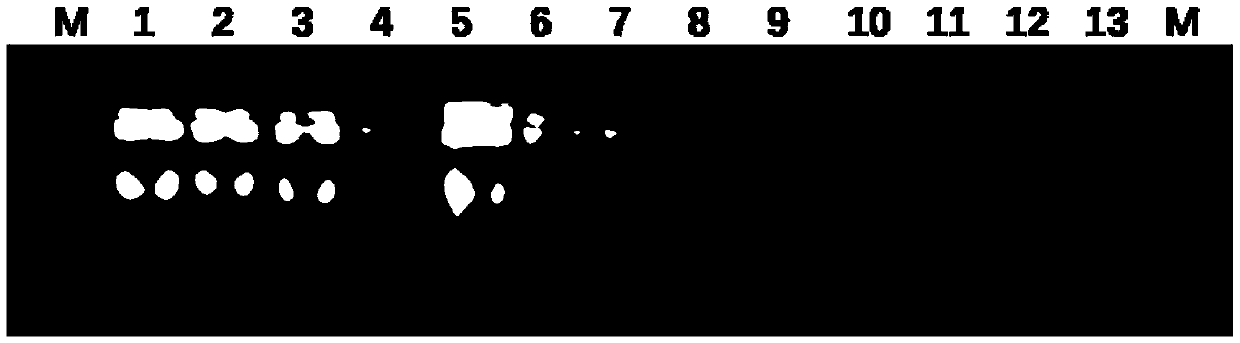
[0087] Example 3 Amplification of the Mitochondrial Genome
[0088] Precisely measure 1 μL of total animal DNA (10-100 ng) obtained by the method of Example 2, add 4 μL of sample buffer, heat at 94° C. for 3 minutes, and then slowly cool to room temperature. Then add 14 μL reaction buffer, 0.5 μLPhi29 DNA polymerase (stock solution concentration: 10 U / μL), 0.5 μL pyrophosphatase (stock solution concentration: 0.04 U / μL), incubate at 30°C for 12 to 18 hours, and then Incubate at 65°C for 10 minutes to terminate the reaction. The enriched mitochondrial genomes of 12 animals were obtained. Among them, the composition of the sample buffer: 50 μL of primer mixture (0.5 μL of 100 μM primers, a total of 18 primers, and 41 μL of ddH 2 O), 125 μL of 2× annealing buffer (80mM Tris-HCl pH7.6, 20mM MgCl 2 ), and 25 μL of ddH 2 O, a total of 200 μL, aliquot 50 μL per centrifuge tube and store at -20°C. The composition of the reaction buffer: 100 μL of Phi2910 × buffer, 40 μL of 10 mM ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com