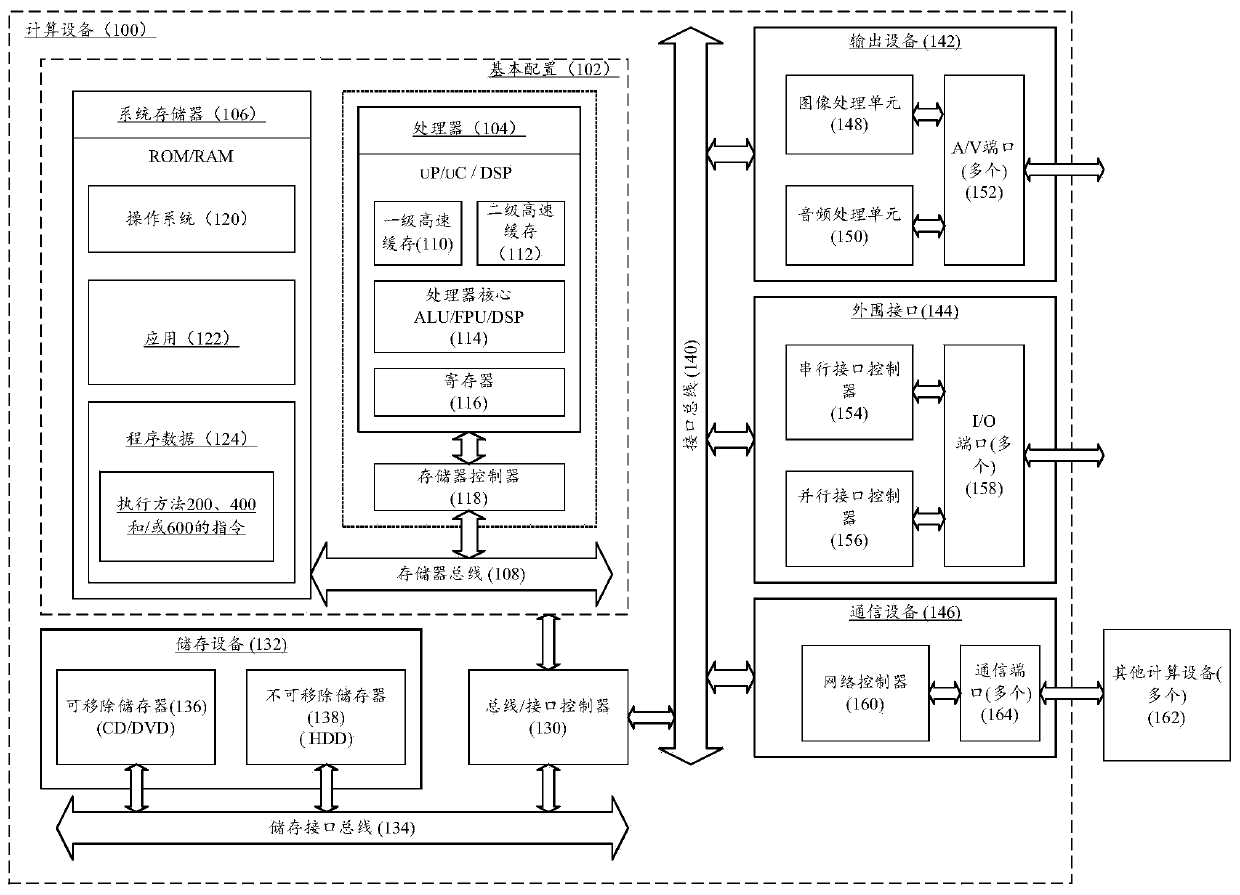
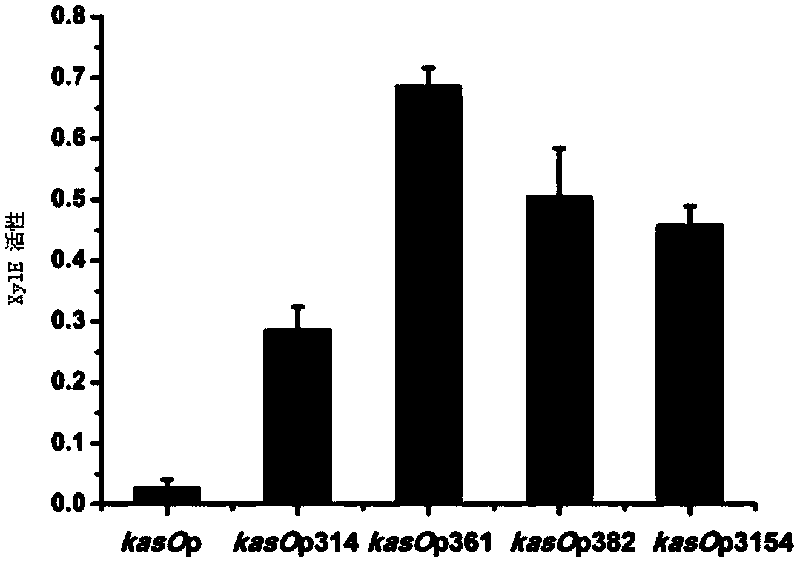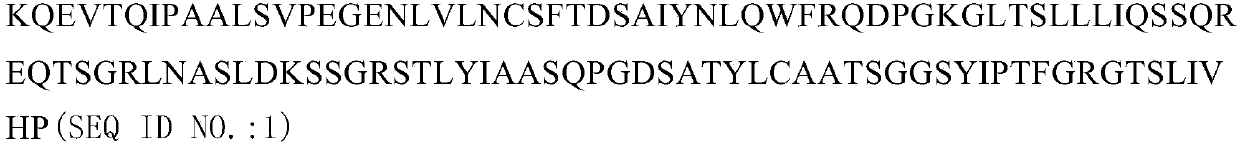Patents
Literature
100 results about "Molecular sequence" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Molecular Sequence Data. Descriptions of specific amino acid, carbohydrate, or nucleotide sequences which have appeared in the published literature and/or are deposited in and maintained by databanks such as GENBANK, European Molecular Biology Laboratory (EMBL), National Biomedical Research Foundation (NBRF), or other sequence repositories.
Methods and systems for annotating biomolecular sequences
A method of annotating biomolecular sequences. The method comprises (a) computationally clustering the biomolecular sequences according to a progressive homology range, to thereby generate a plurality of clusters each being of a predetermined homology of the homology range; and (b) assigning at least one ontology to each cluster of the plurality of clusters, the at least one ontology being: (i) derived from an annotation preassociated with at least one biomolecular sequence of each cluster; and / or (ii) generated from analysis of the at least one biomolecular sequence of each cluster thereby annotating biomolecular sequences.
Owner:COMPUGEN
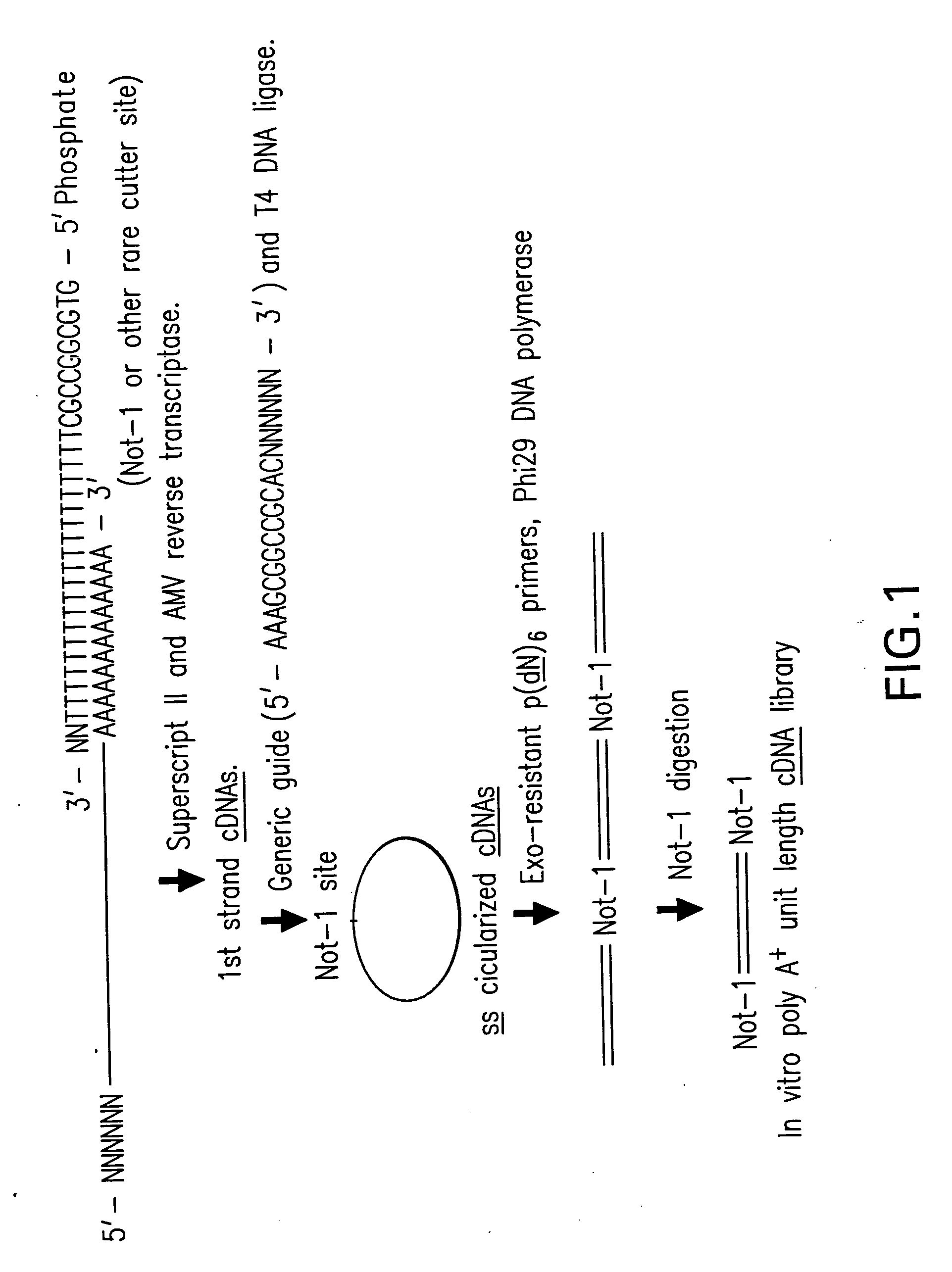
Rolling circle amplification of RNA
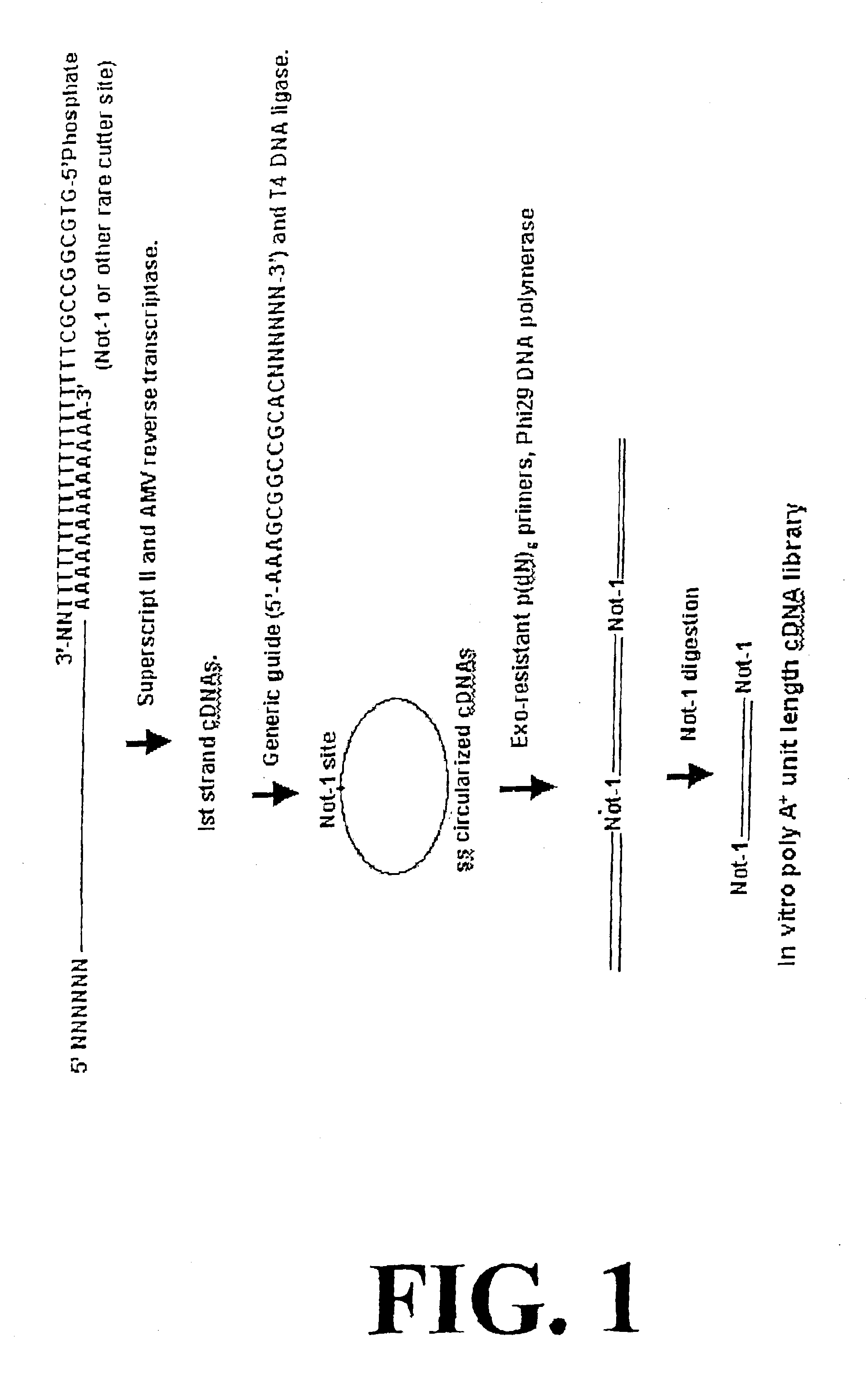
InactiveUS6977153B2Increase productionOptimization orderMicrobiological testing/measurementFermentationTandem repeatDouble strand
Disclosed are compositions and methods for amplification of RNA molecules. The disclosed method involves synthesizing first strand cDNA molecules from RNA molecules, circularizing the first strand cDNA molecules and replicating the circularized first strand cDNA molecules using rolling circle replication. The method can be aided by the use of specialized primers for cDNA synthesis and specialized probes for circularizing the first strand cDNA molecules. The method can be used to replicate and amplify multiple RNA molecules, such as all RNA molecules in a sample or all mRNA molecules in a sample, or be used to replicate and amplify specific RNA molecules. Rolling circle replication of the circularized first strand cDNA molecules results in long DNA strands containing tandem repeats of the cDNA sequence. The tandem sequence DNA can be used directly (for detection of sequences, for example), further amplified, or used any other purpose. Double-stranded tandem sequence DNA can be used to produce unit lengths of the cDNA sequence. Tandem sequence DNA can also be transcribed to produce transcripts having sequence complementary to or matching the sequence of RNA molecules.
Owner:QIAGEN GMBH
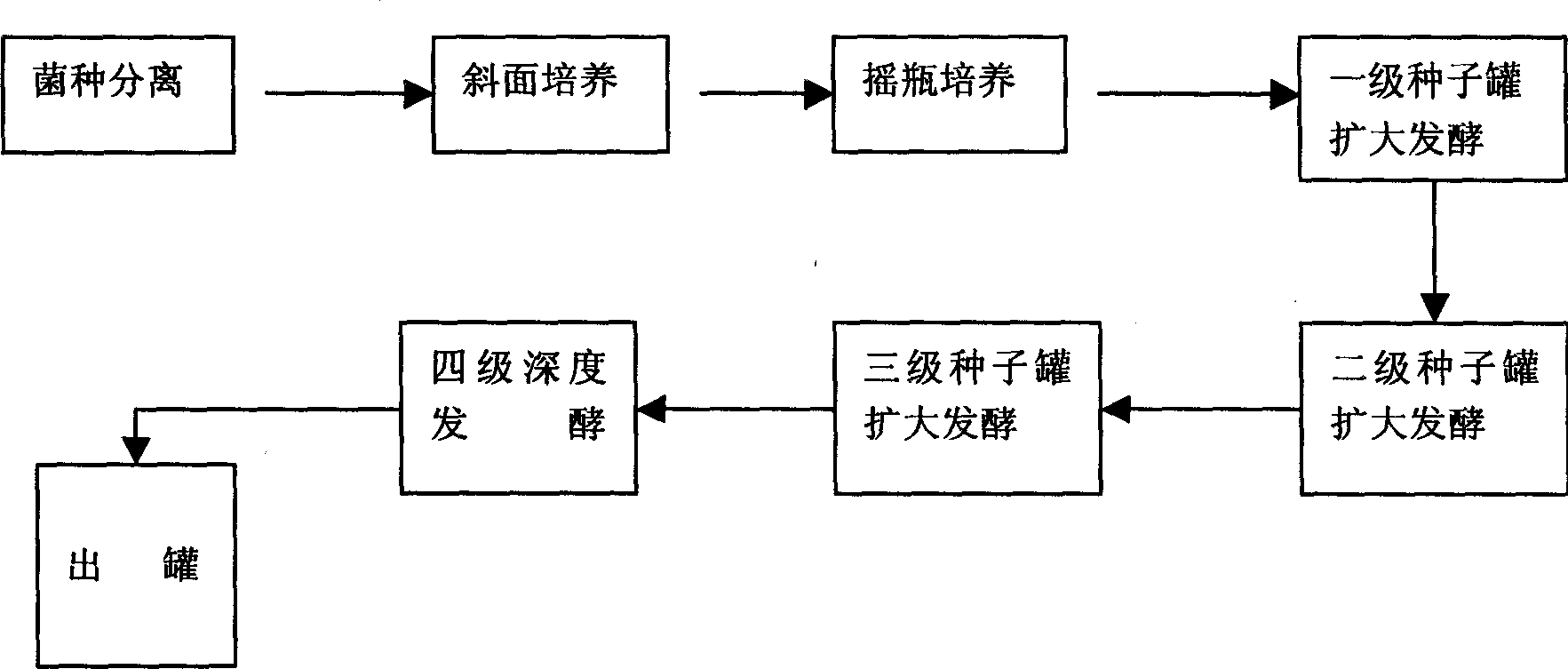
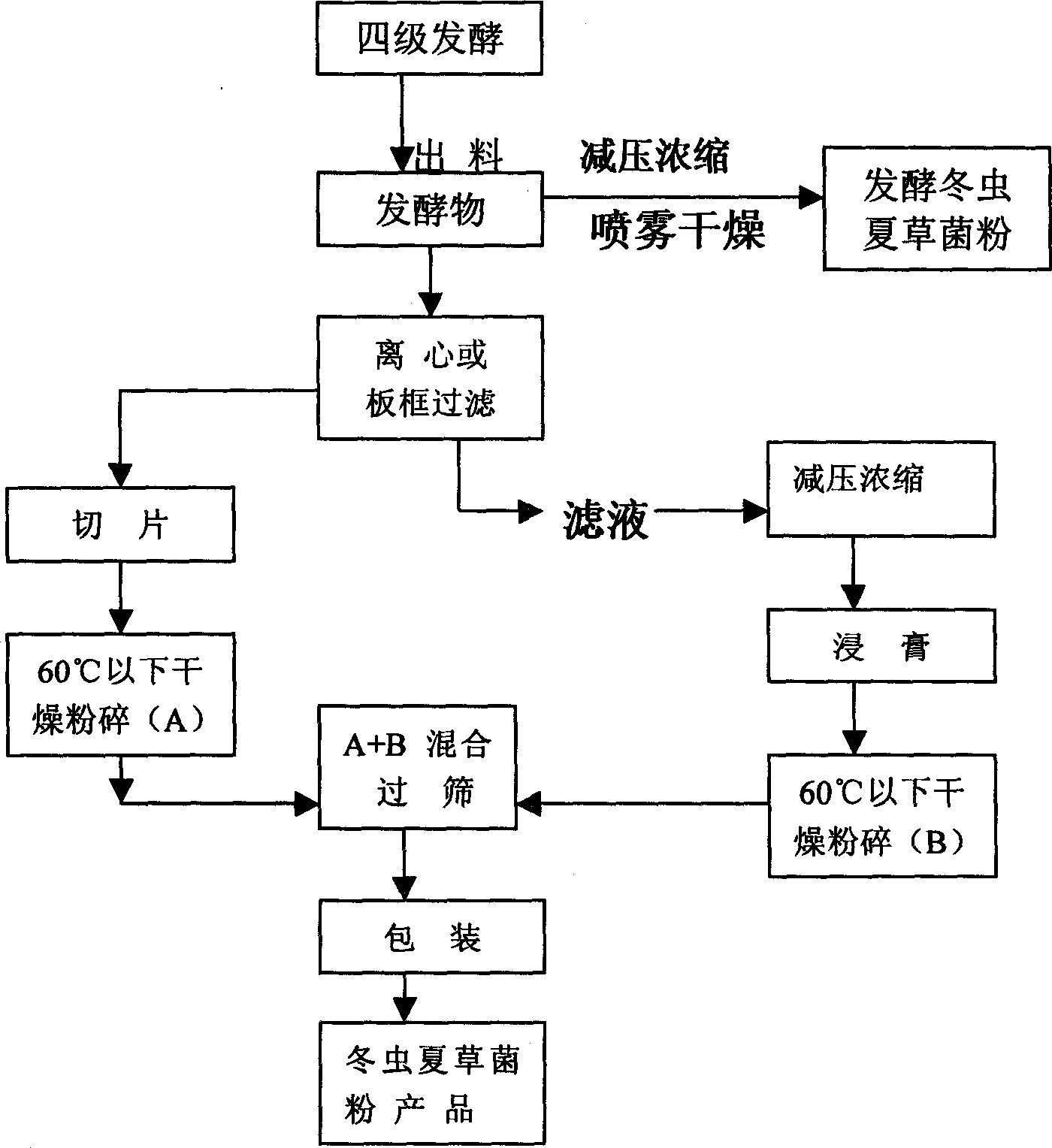
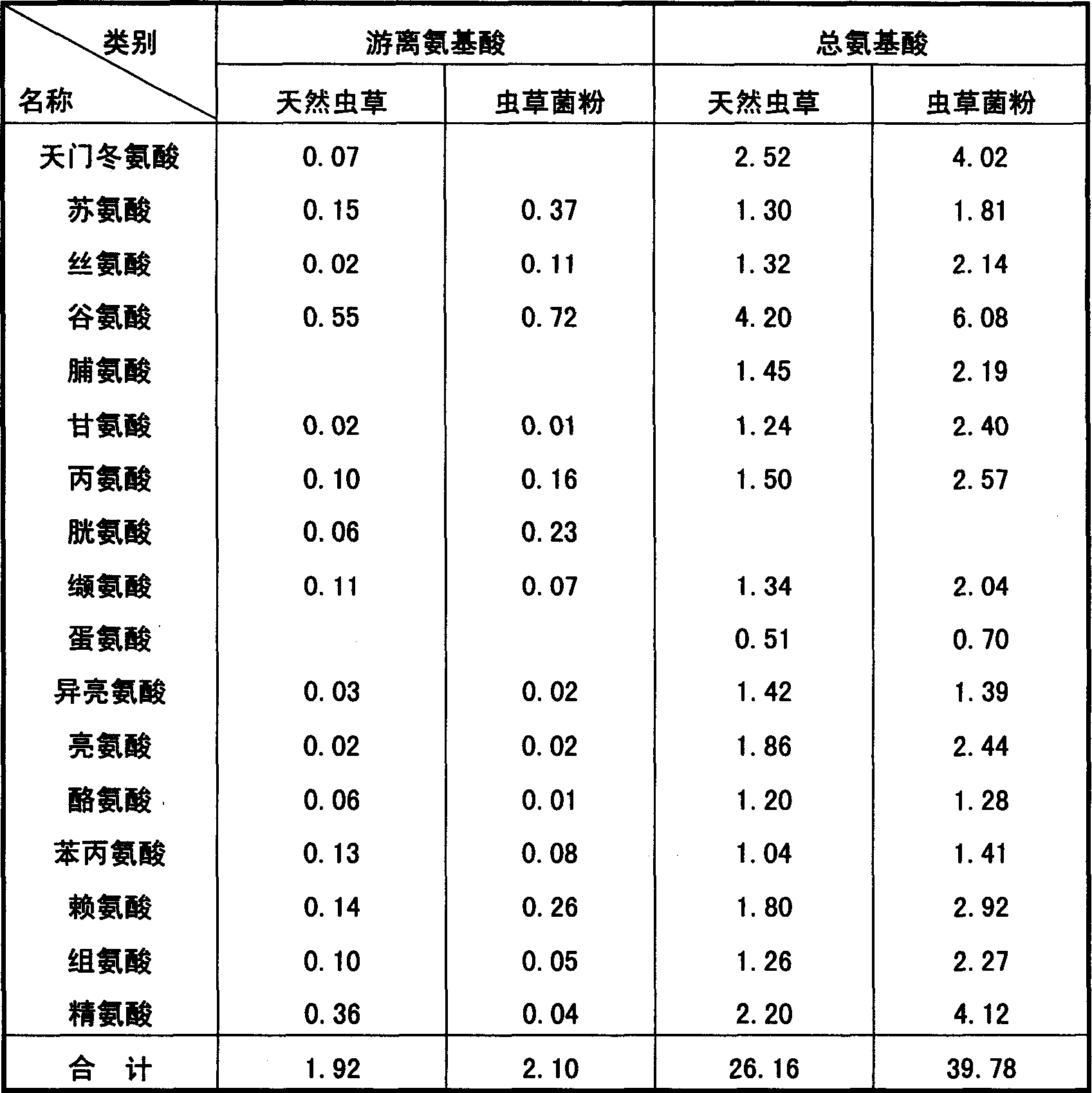
Ferment for producing aweto in large scale and technique for processing power of fungus
This invention describes the large-scale fermentation and processing of Chinese caterpillar fungus. In this invention, hirsutella sinensis strains are used as the fermentation strain and cultivated in the inclined plate and shaking-bottle. 3-5% of potato, 0.5-2% of soybean powders, 0.01-0.02% of soifnum muicfun, 0.01-0.02% of Chinese yam, 0.5-2% of protein, 0.5-5% of glucose, a small amount of dipotassium hydrogen phosphorus and magnesium sulfate, and water culture medium are successively directed into all levels of fermenter for fermentation and processing. The yield of the fermented product is 20% higher than that in the present processes, the fermentation periodic time is 12 days shorter, the cost is 20% lower and the pollution rate is 2% lower. Through the DNA molecular sequence examination by the institute of microbes, Chinese Academy of Sciences, the constitution of the fermented product is the same with that of natural Chinese caterpillar fungus, and the content of cordycepin, adenosine, Chinese caterpillar fungus polyoses, sterol, biological base, amino acids and trace elements is much higher than that in natural Chinese caterpillar fungus. The pharmacological action of the fermented product is better than that of natural Chinese caterpillar fungus.
Owner:YUEWANGQINGZANG PHARMA QINGHAI
Rolling circle amplification of RNA
InactiveUS20060188893A1Optimization orderIncrease productionMicrobiological testing/measurementFermentationDouble strandTandem repeat
Owner:KUMAR GYANENDRA +1
Method and apparatus for chemical and biochemical reactions using photo-generated reagents
This invention provides method and apparatus for performing chemical and biochemical reactions in solution using in situ generated photo-products as reagent or co-reagent. Specifically, the method and apparatus of the present invention have applications in parallel synthesis of molecular sequence arrays on solid surfaces.
Owner:RGT UNIV OF MICHIGAN
Molecule substitution label sequencing parallel detection method-oligomictic nucleic acid coding label molecule library micro-sphere array analysis
InactiveCN101100764AImprove reliabilityGreat advantageMicrobiological testing/measurementLibrary member identificationBiotechnologyMolecular replacement
Molecular displacement label sequence parallel inspection - oligonucleo coded label molecular pool micro-ball array analysis includes: using artificial sequential oligo nucleo-labeled DNA coded molecular pool, coating micro-balls with ligands in correspondent pools to form catching micro-balls, caught ligands combined with labeled ciding molecular pools to form labeled micro-balls array pools. Sample molecules competitively combined with micro-balls catching ligands when sample contains molecules same as those in labeled micro-balls array pools, calculating amount of displaced coded molecular sequence computing kinds and mounts of molecules to be tested. It verifies high flux of inspection with good repeatability, high reliability and sensitivity, so that it is suitable to precisely parallel quantitative analysis concerned with cancer-inhibiting genes and cancer genes pools, cell factor pools, etc.
Owner:北京万达因生物医学技术有限责任公司
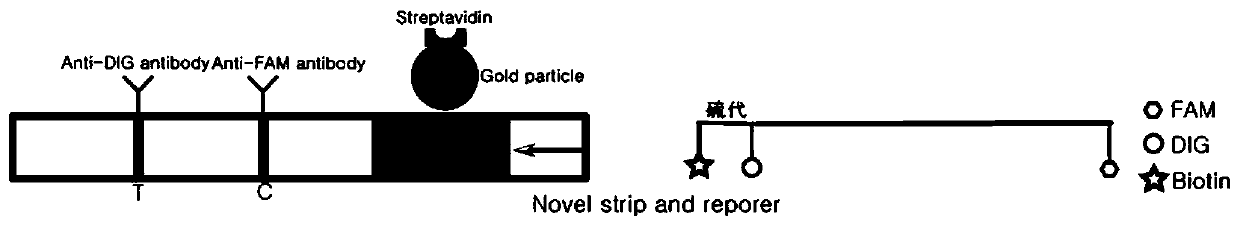
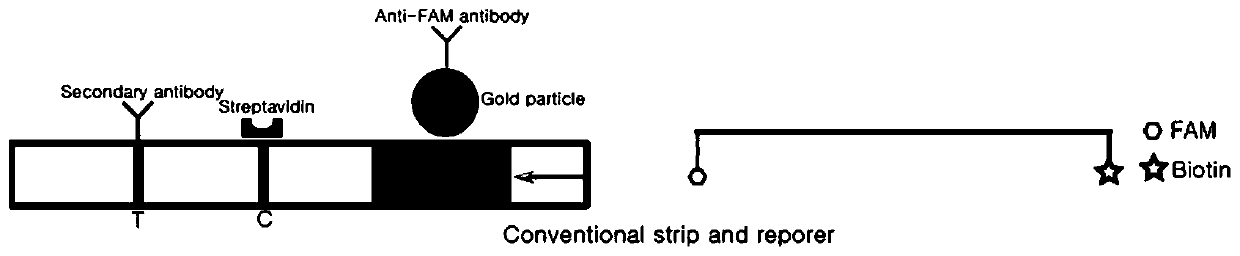
Detection method based on CRISPR/Cas and nucleic acid test paper and human papilloma virus detection kit
PendingCN111560482AHigh sensitivityStrong specificityMicrobiological testing/measurementMicroorganism based processesNucleic acid testSingle strand
The invention relates to the technical field of gene detection, in particular to a nucleic acid detection method based on CRISPR / Cas and nucleic acid test paper and a human papilloma virus rapid detection kit. The human papilloma virus detection kit comprises crRNA, Cas12, RPA upstream and downstream primers, a buffer system and a single-stranded DNA reporter molecule, the sequence of the reportermolecule is 5 '-NNN......-3', the length of the reporter molecule is 3-30 basic groups, FAM, DIG and Biotin labeling is carried out, and meanwhile modification for preventing DNA enzyme or cas12 degradation after activation is carried out on nucleic acid molecules. The CRISPR / Cas and nucleic acid test paper-based nucleic acid detection method and the human papilloma virus rapid detection kit havethe advantages that innovative reporter molecule design and modification are adopted, and corresponding test paper strips are combined, so that non-specific color development is completely eradicated, and the kit has remarkable significance in clinical practical application.
Owner:亚能生物技术(深圳)有限公司
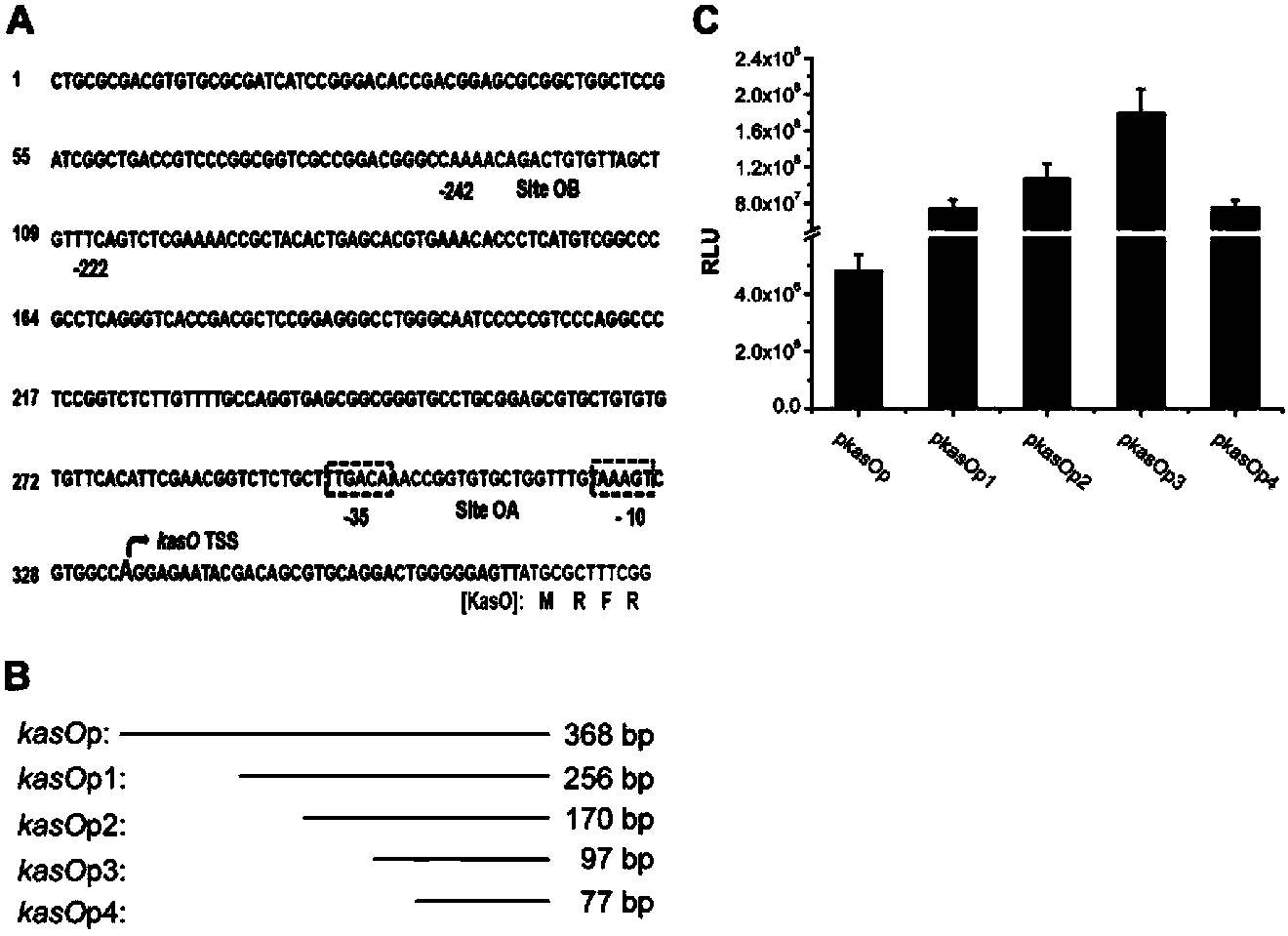
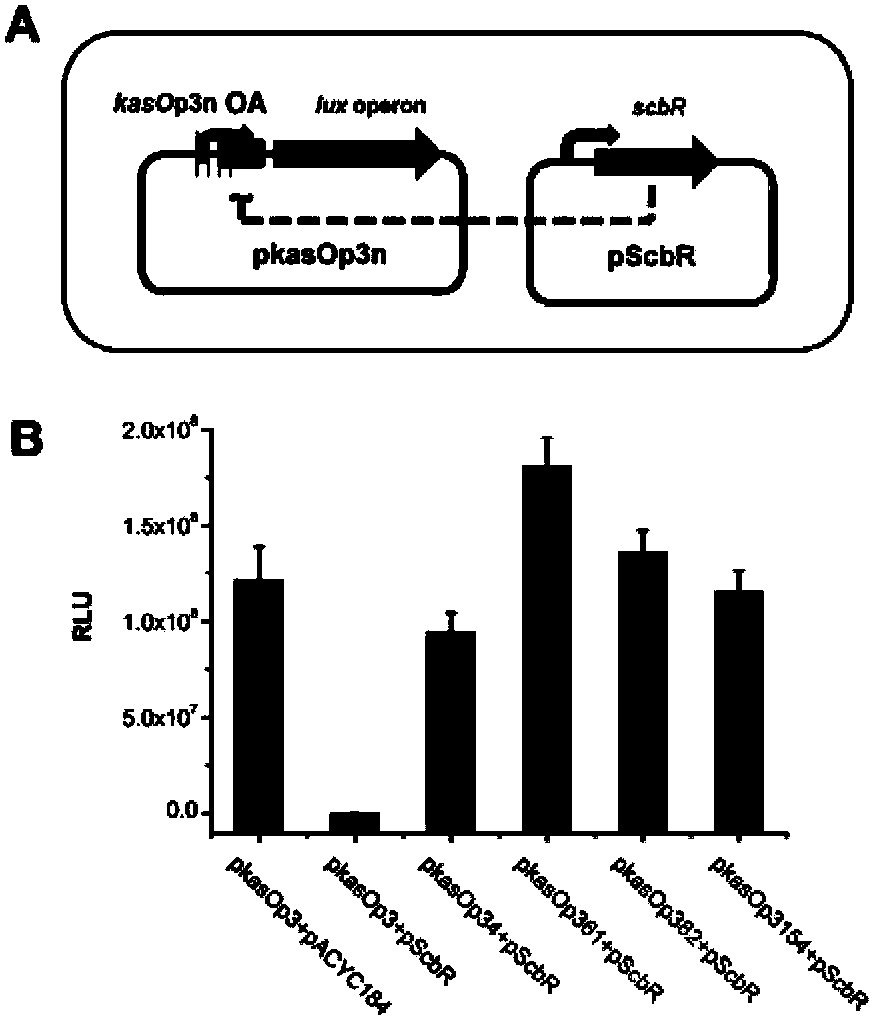
Streptomycete constitutive promoter and applications thereof
InactiveCN103421778AIncrease productionIncreased transcript levelsBacteriaMicroorganism based processesAgricultural scienceStreptomyces venezuelae
The invention discloses a streptomycete constitutive promoter and applications thereof, and provides an DNA fragment. The DNA fragment is any one selected from the following DNA molecules in 1)-3): 1) an DNA molecule represented by the sequence 1 in a sequence table; 2) an DNA molecule with promoter functions and hybrid with the DNA molecule shown in 1) under strict conditions; 3) an DNA molecule with more than 90% of homology with the DNA molecule shown in 1) and with promoter functions. The experiments shows that a strong constitutive promoter kasO*p is screened out, and the promoter and the erythromycin promoter ermE*p are compared in representation of transcriptional level and expression level in streptomyces coelicolor, streptomyces avermitilis and streptomyces venezuelae, and the results show that the modified promoter kasO*p is better than the the erythromycin promoter ermE*p.
Owner:INST OF MICROBIOLOGY - CHINESE ACAD OF SCI
Rice glume development gene promoter p-TRI1 and application thereof
The invention discloses a rice glume development gene promoter p-TRI1 and application thereof. The promoter provided by the invention comes from ordinary cultivated rice O. sativa L. 93-11 which belongs to Oryza species, and the promoter is a DNA molecule of the following 1), 2) or 3): 1) a DNA molecule shown in Sequence 1 in a sequence table; 2) a DNA molecule which crosses with a DNA sequence limited by 1) or 2) under strict condition and has the promoter function; and 3) a DNA molecule which has more than 90 percent of homology with the DNA sequence limited by 1) or 2) and has the promoter function. The discovery of the promoter and the elucidation of the promoter function have important theoretical and practical significance for studying flower organ development mechanism of rice, especially the glume development mechanism and for breeding rice varieties of specific grain types. The promoter has important theoretical and practical significance for studying the molecular mechanism of rice glume development and for rice grain type molecular breeding. The promoter has wide application and market prospect in the field of agricultural.
Owner:CHINA AGRI UNIV
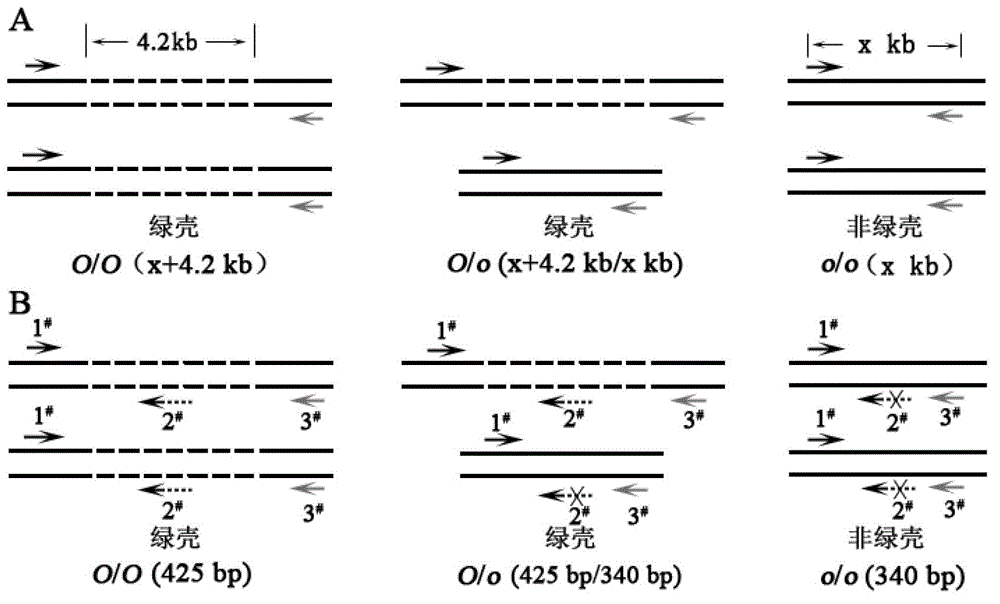
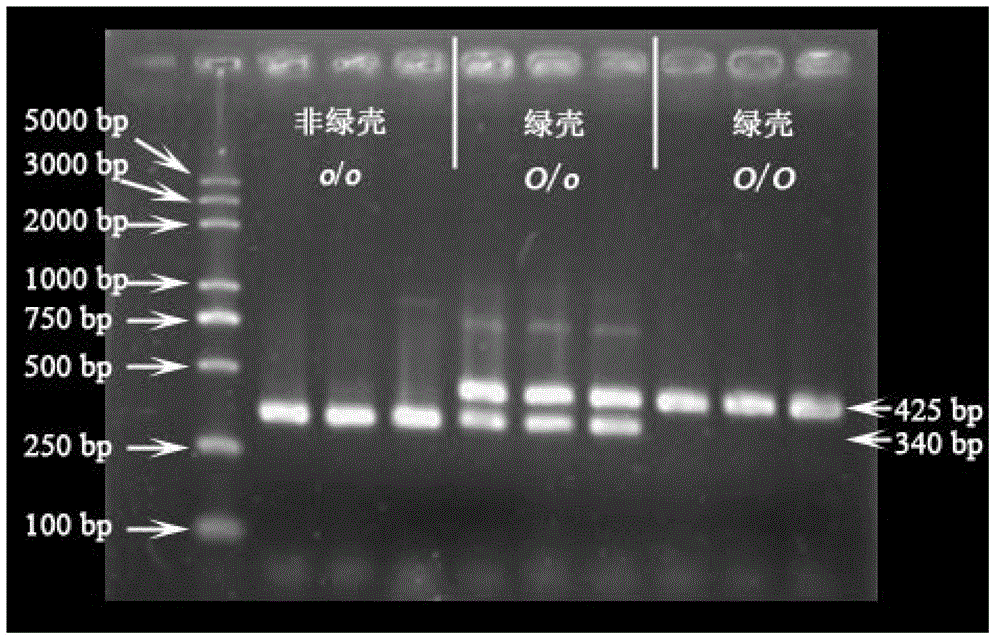
Method for distinguishing green shell egg laying chicken and chicken green shell egg genotype as well as special fragment and primers
ActiveCN102876796AShort identification timeLow costMicrobiological testing/measurementDNA/RNA fragmentationMultiplex pcrsPolymerase chain reaction
The invention discloses a method for distinguishing green shell egg laying chicken and chicken green shell egg genotype as well as a special fragment and primers. The invention provides a method for identification or auxiliary identification of whether chicken to be detected is green shell egg chicken or not, in order to detect the genome DNA (deoxyribonucleic acid), if objective DNA molecules (sequence 4) are contained in the genome DNA, the chicken to be detected belongs to or is candidated as the green shell egg chicken, and when the objective DNA molecules are not contained in the genome DNA, the chicken to be detected does not belong to or is not candidated as the green shell egg chicken. Experiments prove that a 425bp short fragment is adopted, three primers are designed and synthesized according to the short fragment and are used for multiple PCR (polymerase chain reaction) amplification, whether the chicken to be detected lays green shell eggs or not can be distinguished through detecting whether amplification products contain the short fragment or not, and meanwhile, the chicken green shell egg genotype can be distinguished.
Owner:CHINA AGRI UNIV
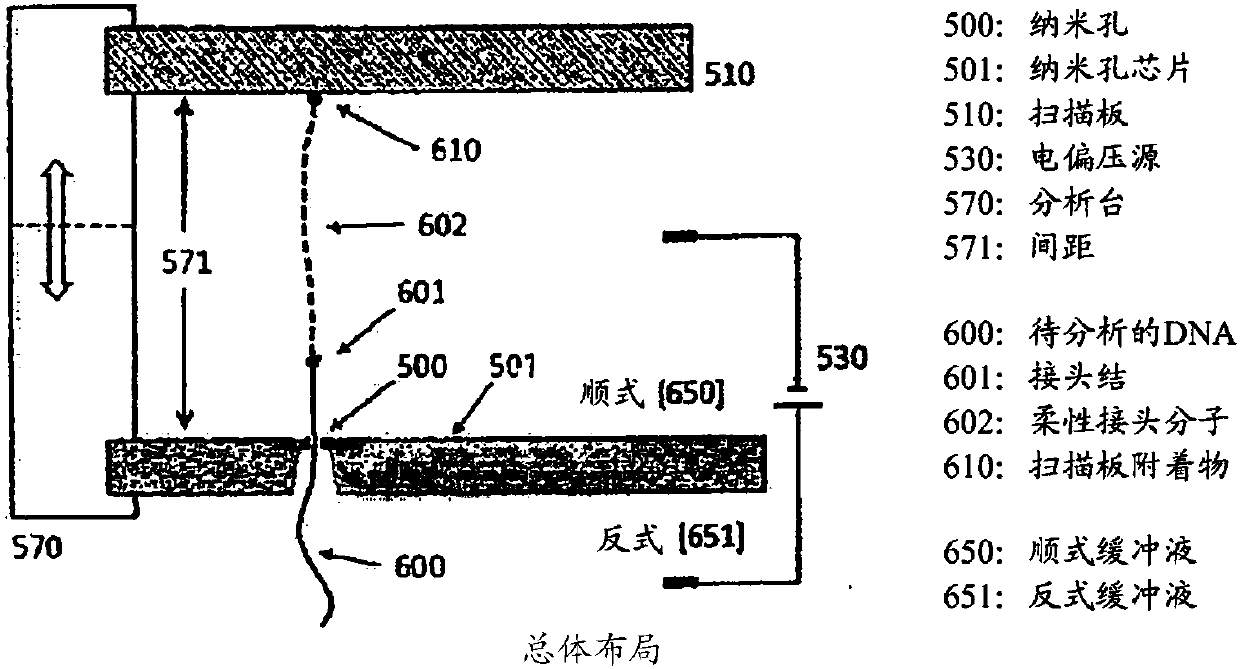
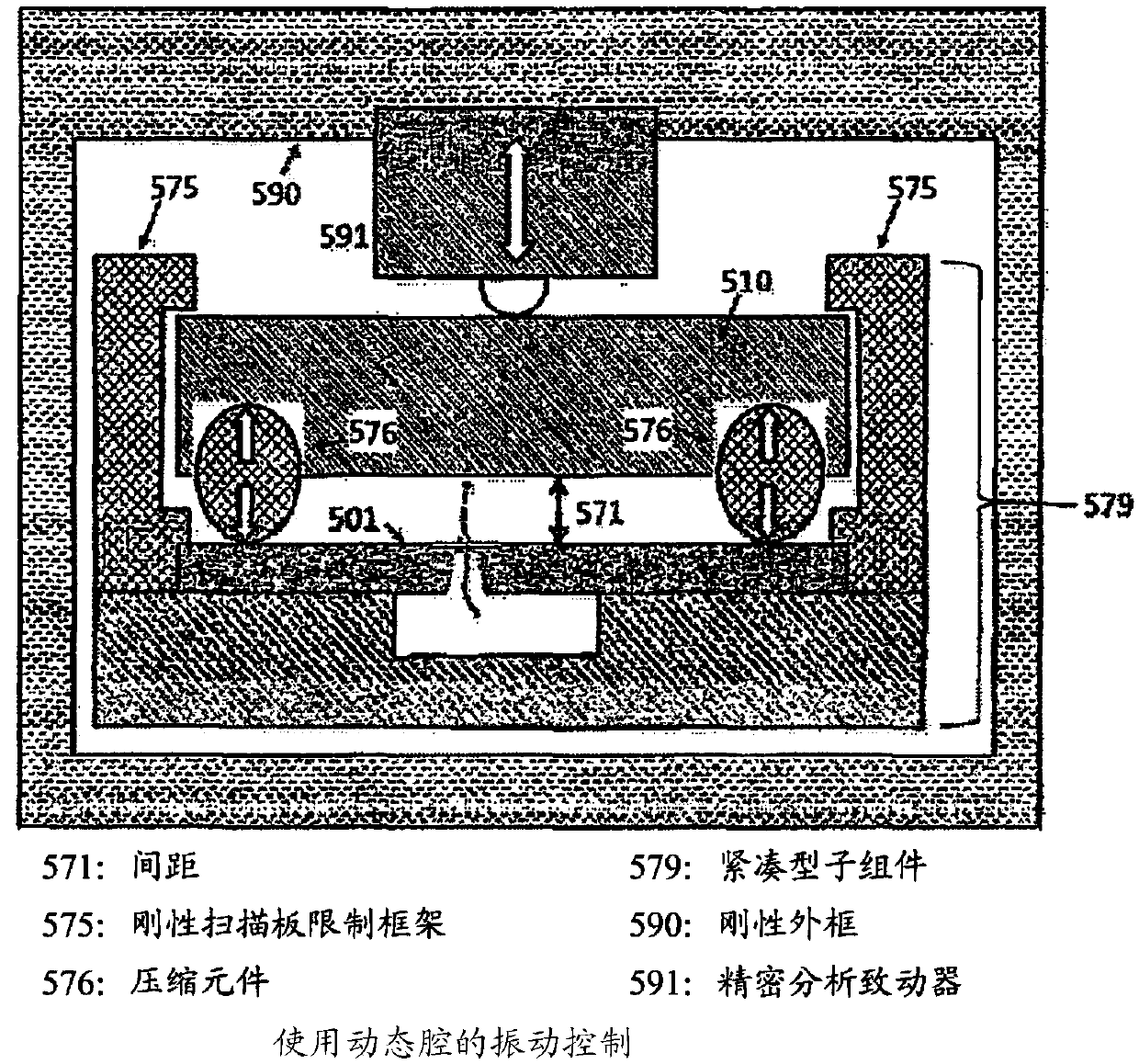
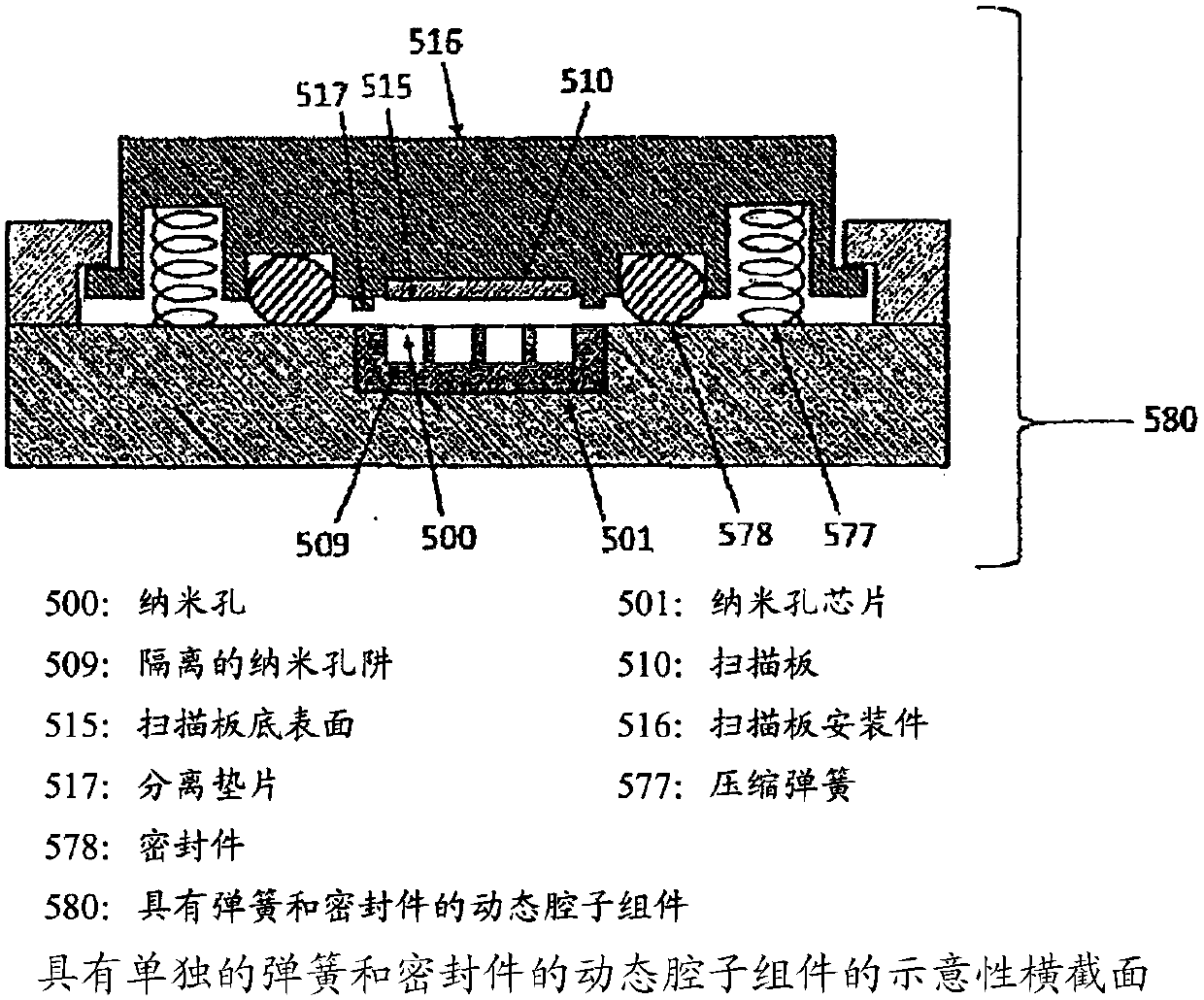
Methods and systems for controlling dna, RNA and other biological molecules passing through nanopores
The present disclosure provides, in one aspect, a device and a method for unit sequencing and / or analysis of a molecular sequence comprising attaching the molecular sequence to a plate and controllingthe progression of the molecular sequence through a pore of a nanopore chip, wherein the separation distance between the nanopore chip and the scan plate is controlled by a precision mechanical drive, and the molecular sequence is sensed as it progresses through the nanopore.
Owner:UNIVERSAL SEQUENCING TECH CORP
T cell receptor (TCR) capable of recognizing short peptide of PRAME antigen
The invention provides a T cell receptor (TCR) capable of specifically recognizing short peptide EVLVDLFLK derived from a PRAME antigen, particularly provides the TCR which can bond with an EVLVDLFLK-HLA-A*1101 complex presented to cell surfaces. The invention also provides a nucleic acid molecular sequence coding the TCR and a vector containing the nucleic acid molecular sequence.
Owner:GUANGZHOU INST OF BIOMEDICINE & HEALTH CHINESE ACAD OF SCI
Plant tissue specific expression promoter and application of plant tissue specific expression promoter
The invention discloses a plant tissue specific expression promoter and an application of the plant tissue specific expression promoter. A DNA fragment provided by the invention is any one of the following DNA molecules from (1)-(3): (1) a DNA molecule shown in a sequence 1 of a sequence table, (2) a DNA molecule which is hybridized with the DNA molecule in (1) under the strict condition and has the function of a promoter, and (3) a DNA molecule which has more than 90% of homology with the DNA molecule in (1) and has the function of the promoter. Experiments show that the invention provides the DNA fragment which is the promoter, can be highly expressed at roots, hypocotyl (especially the joints of leaves and the hypocotyl), and the joints of the leaves and stems of arabidopsis, and cannot be expressed in the leaves, flowers or pods. Experiments indicate that the promoter is a specific promoter which can be expressed in specific plant tissue, and can play an important role in the artificial control genetic breeding, the expression of related genes of stress resistance and stress tolerance, and the transgenic research on the cultivation of plants with the stress resistance and the stress tolerance.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Drug molecule recommendation system for regulating and controlling disease targets based on a deep learning, computer equipment and storage medium
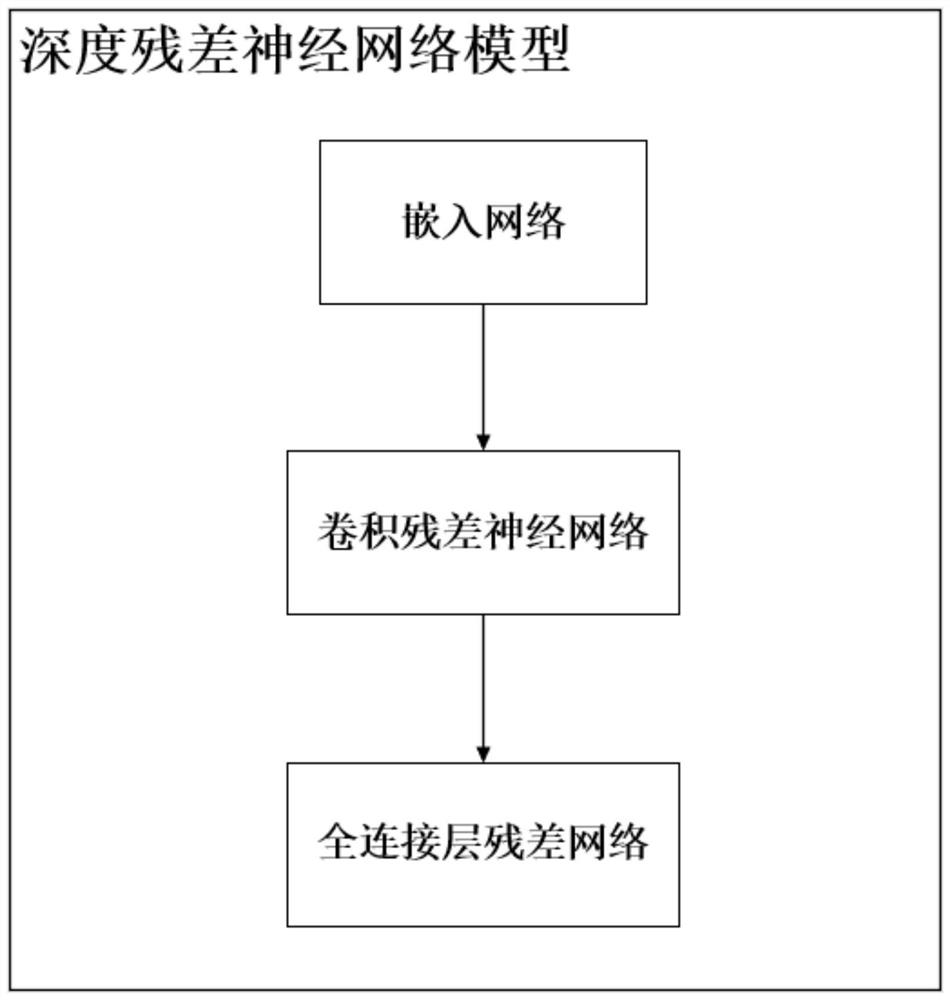
InactiveCN112562790AShorten the R&D processProcessing speedBiostatisticsNeural architecturesDiseaseResidual neural network
The invention discloses a drug molecule recommendation system for regulating and controlling disease targets based on a deep learning method, and belongs to the technical field of drug relocation, convolutional neural networks and residual networks. The system comprises a deep residual error network model, and the deep residual error network model comprises an embedded network, a convolution residual error neural network and a full connection layer residual error network. The embedded network converts a drug molecule SMILES sequence or a protein amino acid sequence into a binary matrix. The convolution residual neural network comprises three convolutional layers, an addition layer and a maximum pooling layer, and is a network represented by a 'learning' drug molecule SMILES sequence or protein amino acid sequence feature, the input of the network is a binary matrix representing a drug molecule or protein, and the output of the network is a feature representation vector of the drug molecule or protein. The full connection layer residual network comprises three full-connection layers, two dropout layers and an addition layer, the input is a splicing vector expressed by drug moleculeand protein characteristics and an actual binding affinity value of the drug molecule and the protein characteristics, and the output is a binding affinity prediction value of the drug molecule and the protein.
Owner:CHINA UNIV OF PETROLEUM (EAST CHINA)
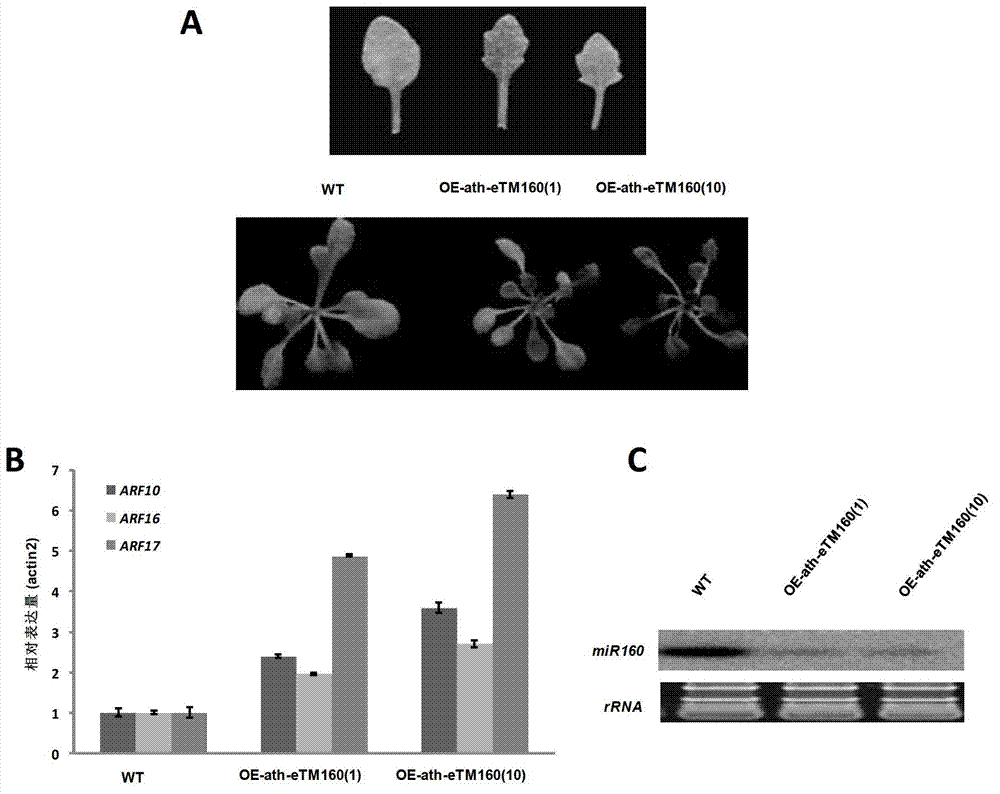
Application of ath-eTM160 in inhibiting functions of microRNA160
ActiveCN102864146AInhibit expressionHigh expressionBacteriaFermentationNucleotideArabidopsis thaliana
The invention discloses application of ath-eTM160 in inhibiting functions of microRNA160. The DNA (deoxyribonucleic acid) disclosed by the invention is any one of the following (1)-(3): (1) DNA molecules disclosed as 8th-187th nucleotides from the 5' tail end in Sequence 1 in the sequence table; (2) DNA molecules with identical functions, which can be hybridized with the DNA sequences defined in (1) under strict conditions; and (3) DNA molecules with identical functions, which has at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98% or at least 99% homology with the DNA sequences defined in (1). The experiment proves that the gene segment ath-eTM160, which is obtained from Arabidopsis thaliana, can inhibit the expression of microRNA160 in the Arabidopsis thaliana after being transferred into Arabidopsis thaliana for overexpression.
Owner:INST OF GENETICS & DEVELOPMENTAL BIOLOGY CHINESE ACAD OF SCI
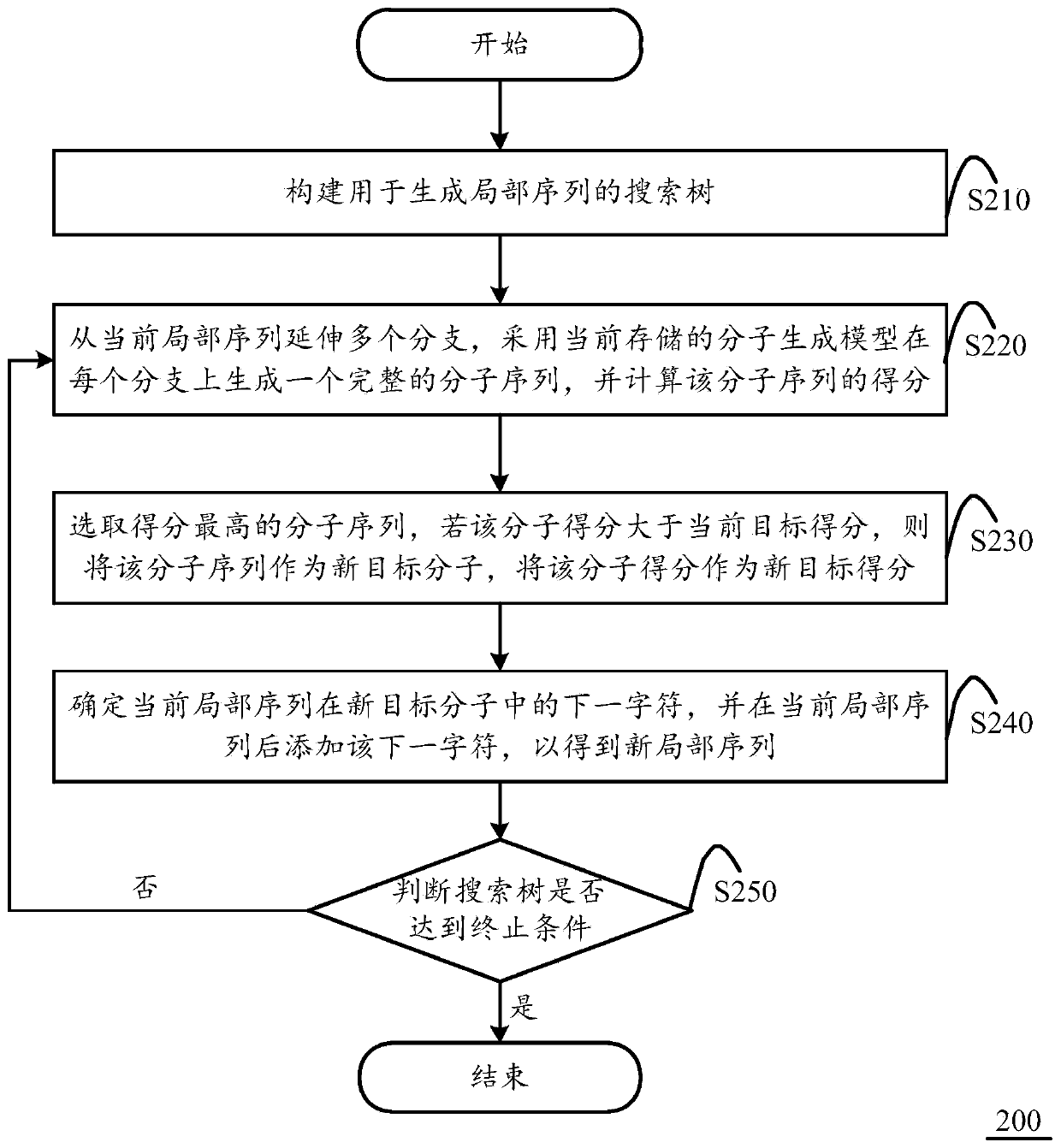
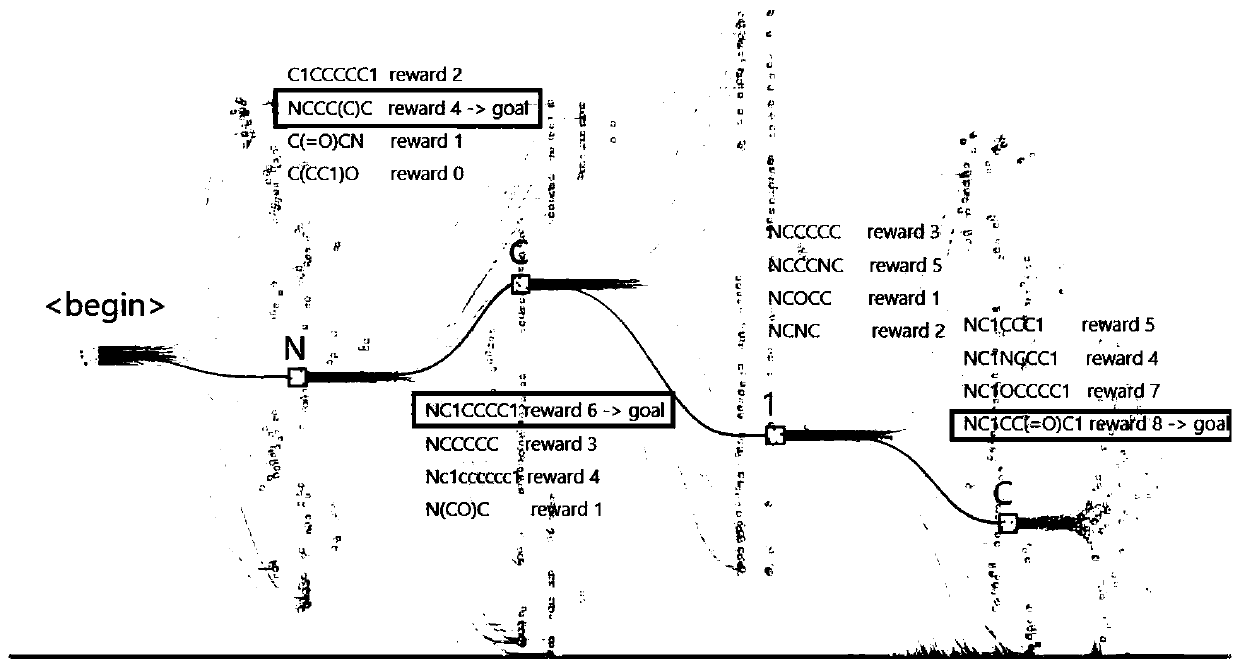
Molecular sequence generation method and apparatus, and computing equipment
ActiveCN111370074AImprove scoreImprove performanceMolecular designNeural architecturesAlgorithmTheoretical computer science
The invention discloses a molecular sequence generation method. The molecular sequence generation method is executed in computing equipment and comprises the following steps of: constructing a searchtree for generating a local sequence, wherein an initial local sequence is a null character positioned on a root node; allowing a plurality of branches to extend from the current local sequence, generating a complete molecule on each branch by adopting a current molecule generation model, and calculating a score of each molecule; selecting a molecular sequence with the highest score, and if the molecular score of the molecular sequence is greater than the current target score, taking the molecule with the molecular sequence as a new target molecule and the molecular score as a new target score; determining a next character of the current local sequence in the new target molecule to add the next character so as to obtain a new local sequence; and setting the new local sequence as a currentlocal sequence, and circularly executing the above steps until no new target molecule is generated and the local sequence of the search tree is the same as a final target molecule. The invention further discloses a corresponding molecular sequence generation device and the computing equipment.
Owner:北京晶泰科技有限公司
Endosperm specific expression promoter
The invention discloses an endosperm specific expression promoter. The endosperm specific expression promoter is any one of (1) a DNA (deoxyribonucleic acid) molecule as shown in a sequence l of a sequence table, (2) a DNA molecule which is hybridized with a definitive DNA sequence under a strict condition and has a promoter function, and (3) a DNA molecule which has more than 90% of homology with the DNA sequence defined in (1) or (2) and has a promoter function. The experiment proves that the endosperm specific expression promoter provided by the invention can be used for driving the specific expression of a target gene in plant endosperm. The invention provides a useful core component for the study on expression, adjustment and control of plant genes in grains and improvement of crops by using a genetic engineering method. The endosperm specific expression promoter has important significance for cultivation of new crop varieties through specific improvement and transformation of crop seeds.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Optimized triterpenoid synthase coding gene and application thereof
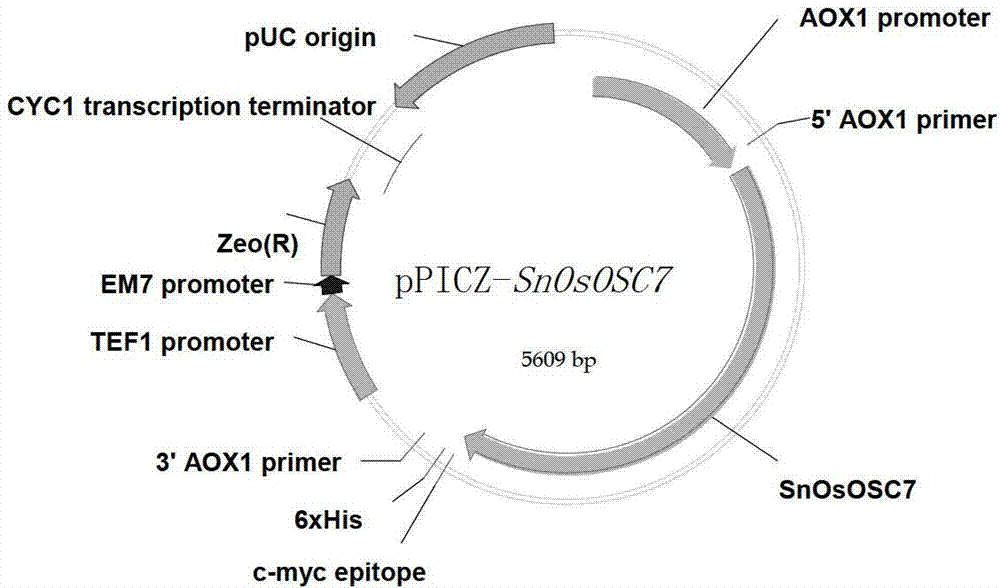
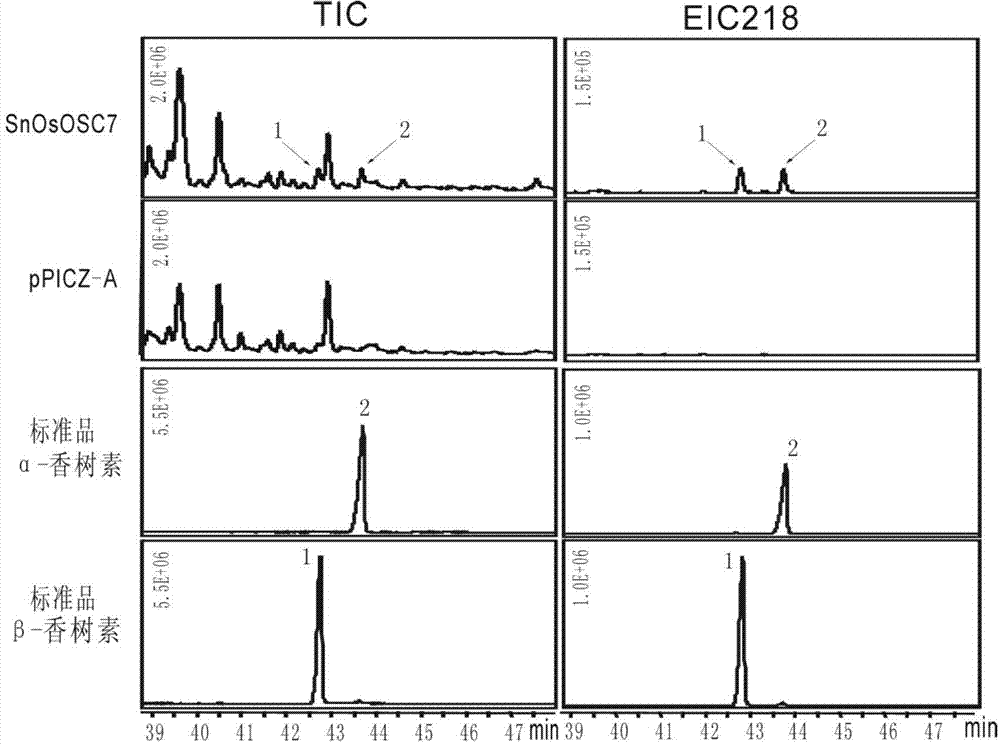
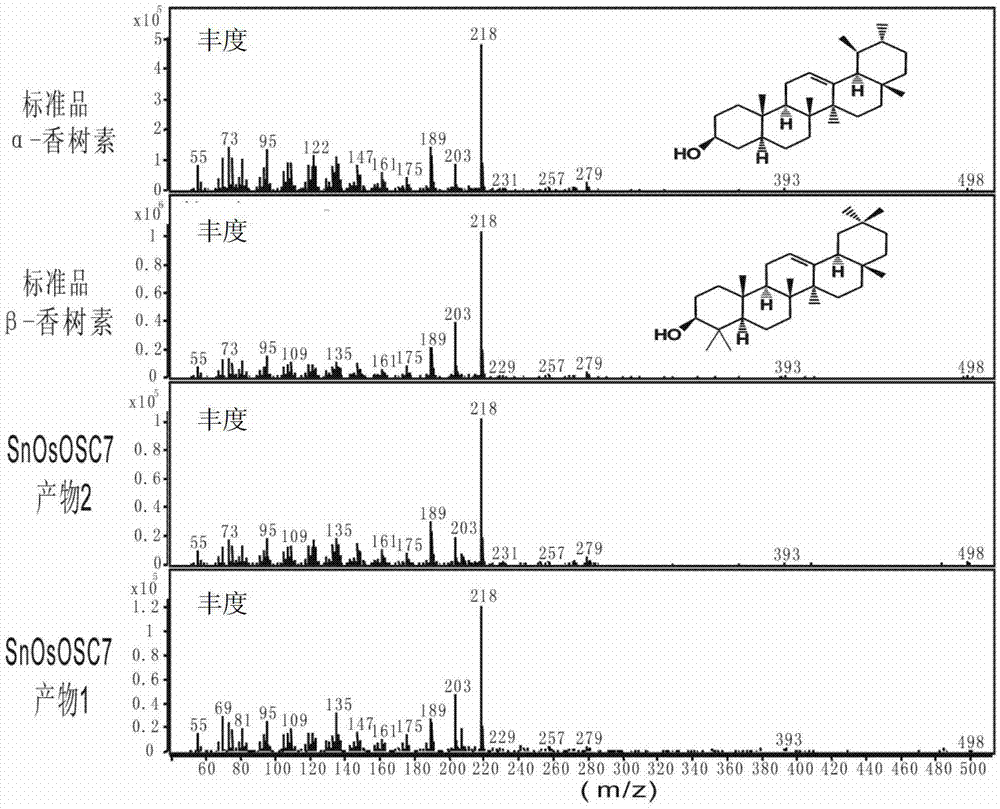
The invention discloses an optimized triterpenoid synthase coding gene and application thereof. A DNA (Deoxyribonucleic Acid) molecule provided by the invention is any one of DNA molecules as follows: (1) the DNA molecule shown in a sequence I of a sequence table; (2) the DNA molecule which is hybridized with the DNA sequence defined in (1) and is coded and synthesized into a gap-associated protein of a triterpene compound under strict conditions; and (3) the DNA molecule which has at least 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98% or 99% homology with the DNA sequence defined in (1) and is used for synthesizing the gap-associated protein of the triterpene compound. Experiments prove that the sequence of rice SnOsOSC7 gene subjected to codon optimization, which is provided by the invention, can be used for preparing the triterpene compound alpha-amyrin and / or beta-amyrin.
Owner:INST OF BOTANY CHINESE ACAD OF SCI
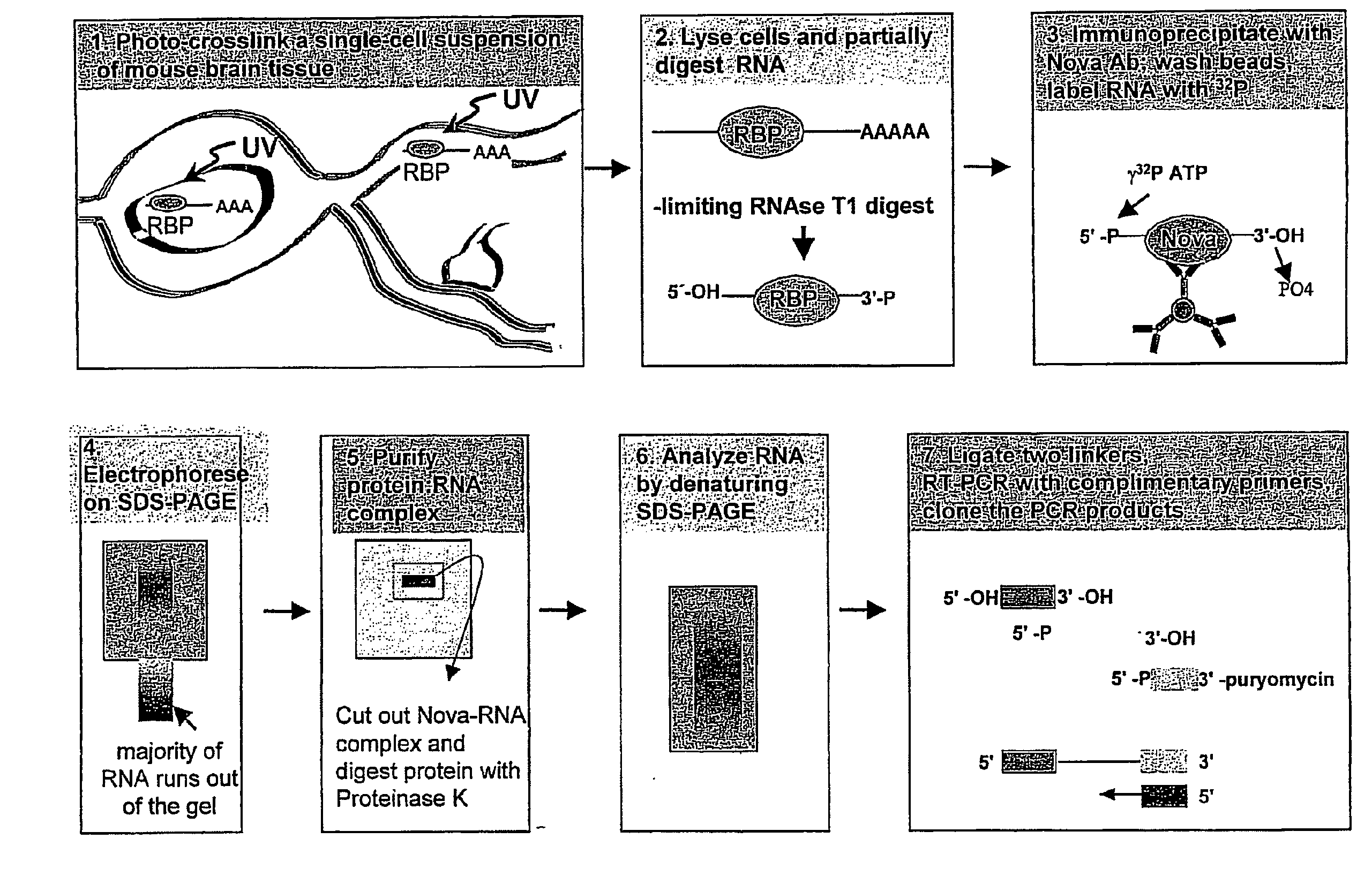
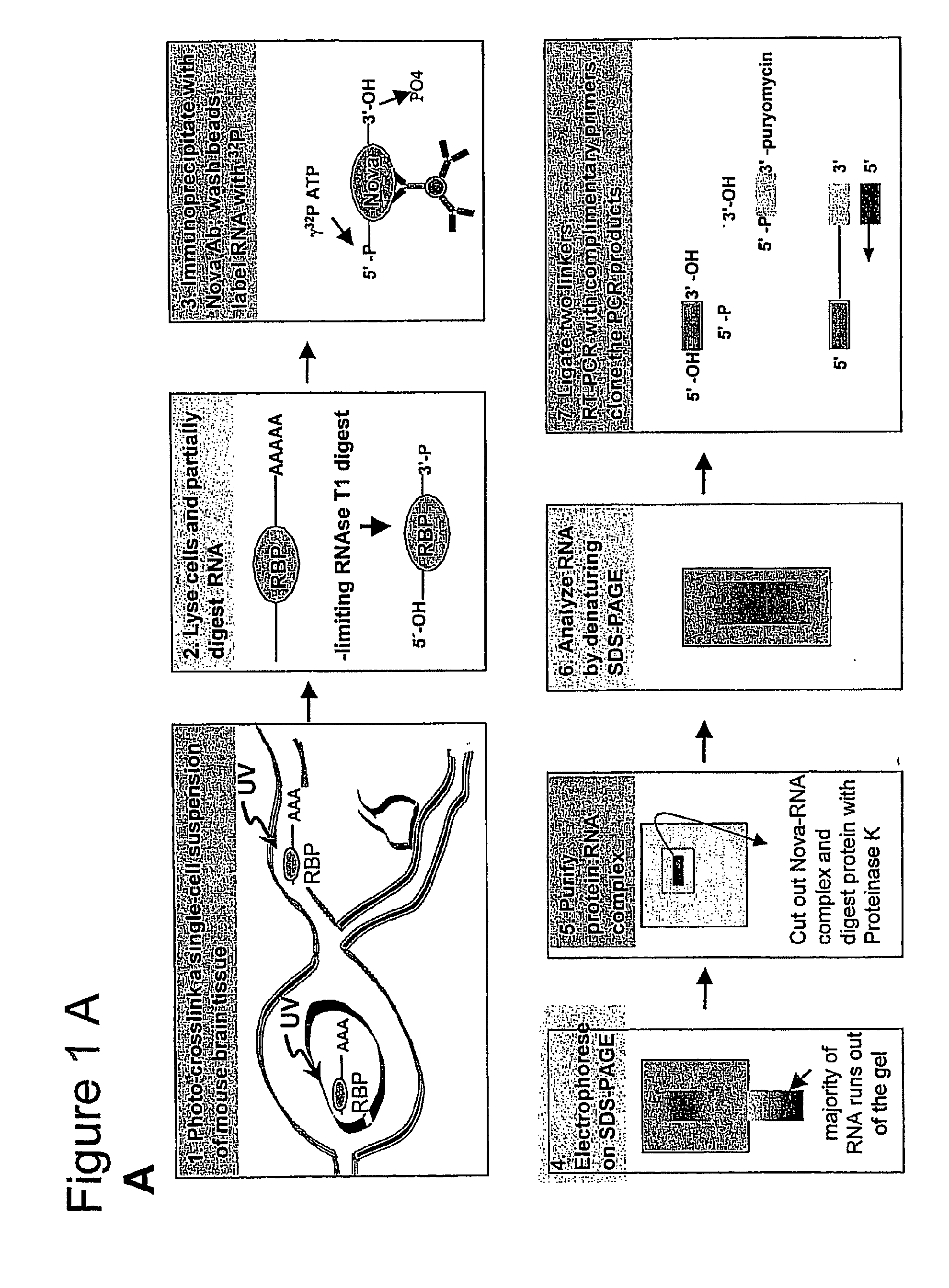
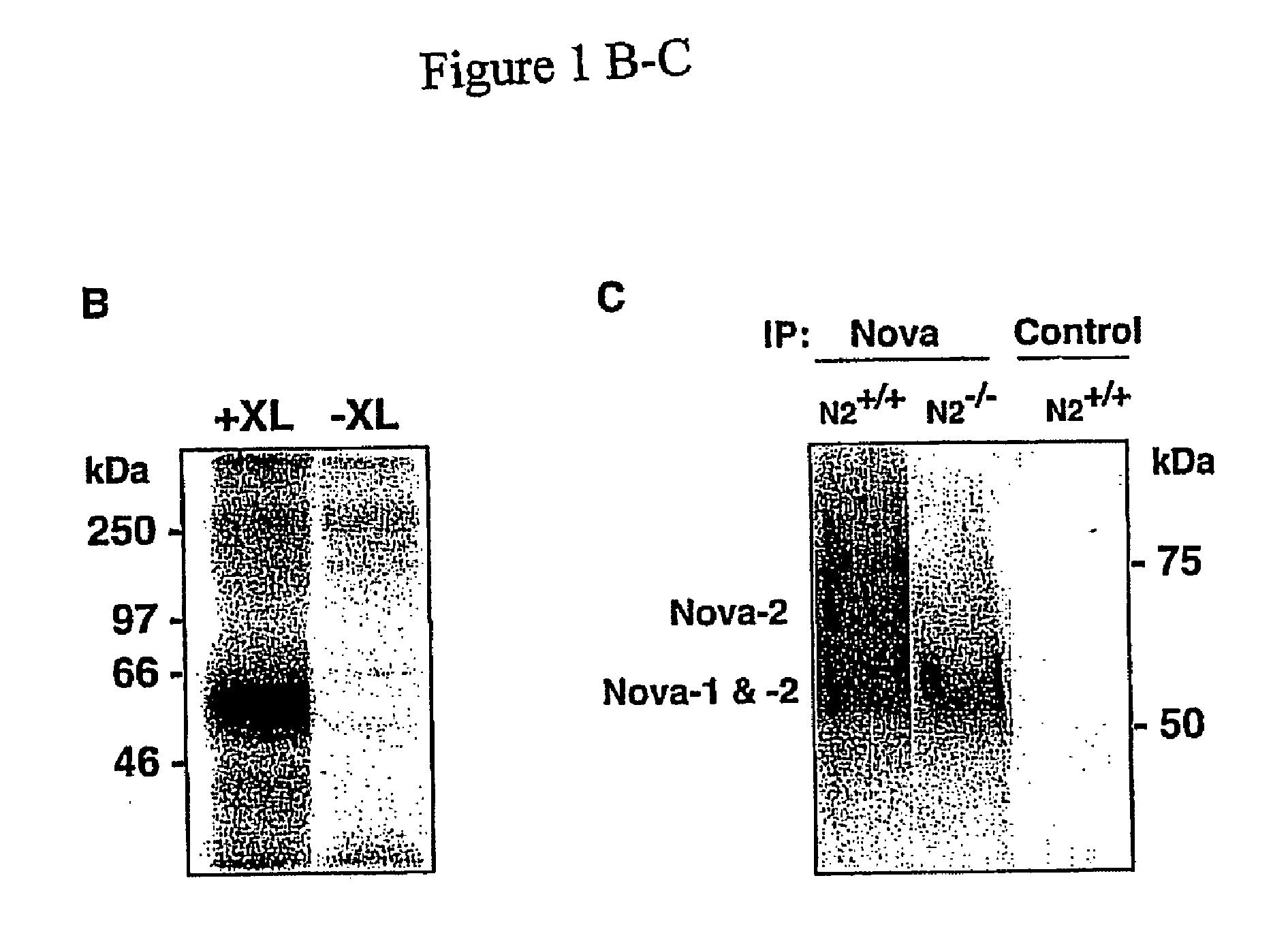
Method of Purifying RNA Binding Protein-RNA Complexes
The present invention provides methods for purifying RNA molecules interacting with an RNA binding protein (RBP), and the use of such methods to analyze a gene expression profile of a cell. The invention also provides sequences of RNA molecules that mediate binding to an RBP, proteins encoded by the sequences, a method of identifying the sequences, and the use of the sequences in a screen to identify bioactive molecules. The invention also provides RNA motifs found among the sequences and compounds that bind the RNA motifs. In addition, the invention provides methods of treating diseases associated with a function of an RNA binding protein.
Owner:THE ROCKEFELLER UNIV
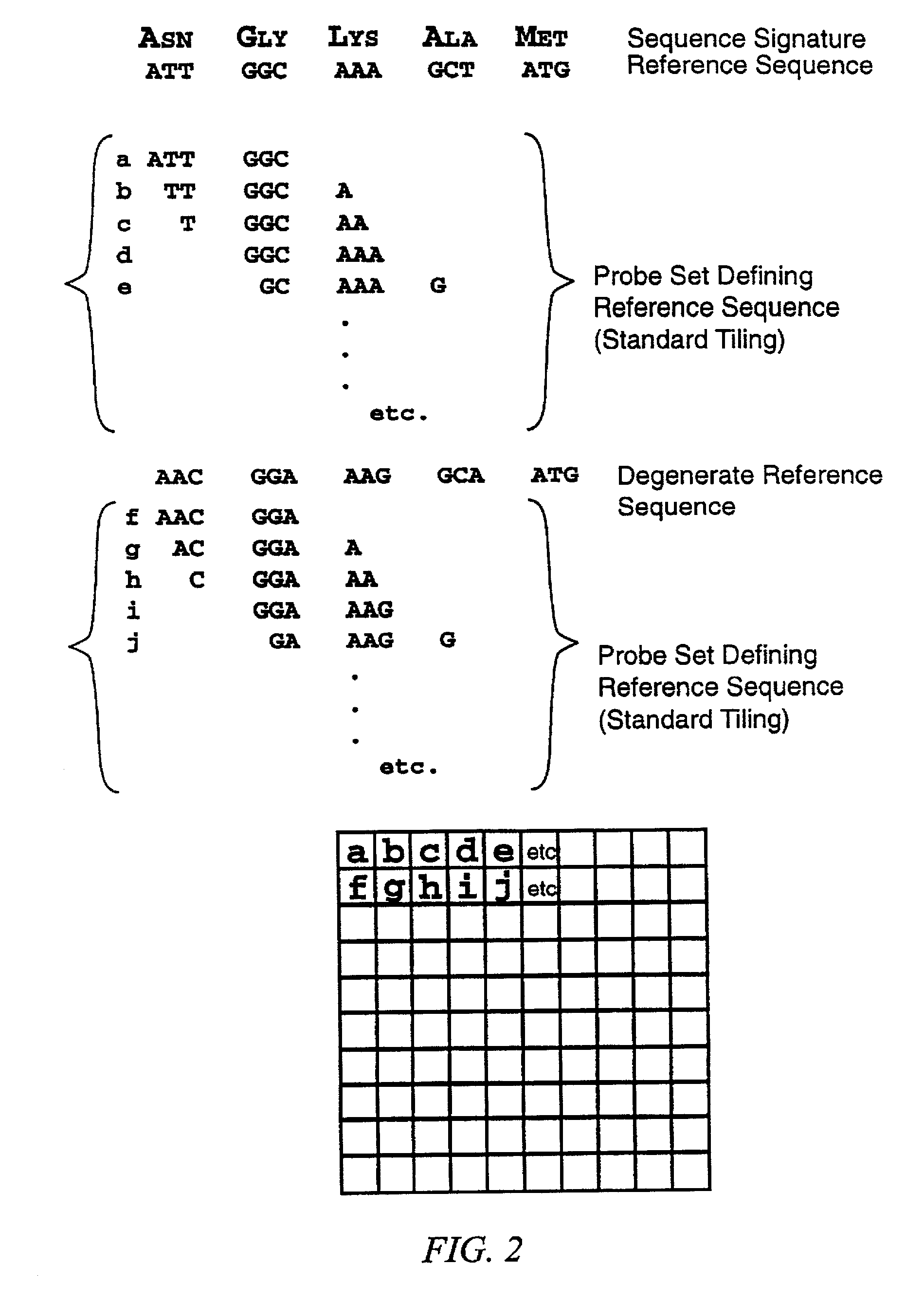
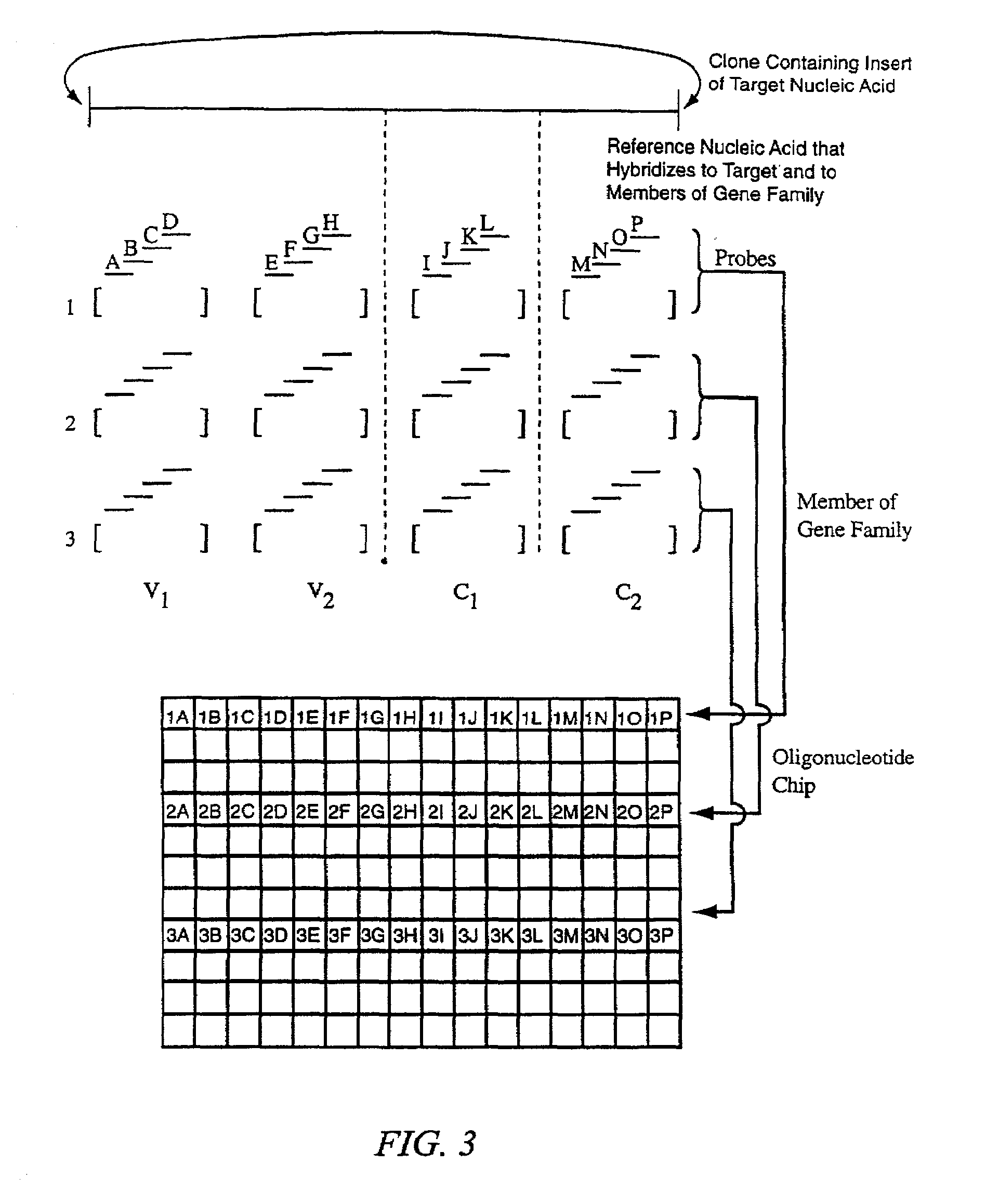
Identification of molecular sequence signatures and methods involving the same
Novel means and methods for analyzing hybridization data derived from hybridization assays between a target nucleic acid and differently sequenced polynucleotide probes involve selecting probe sets that define reference sequences for sequence signatures and deriving useful data about the nature of the target nucleic acid molecule based on its hybridization to the probes. The methods are useful for determining whether the target contains a nucleic acid or polypeptide sequence signature, whether the target encodes a member of a gene family, or whether the target is derived from one of any number of genes.
Owner:AFFYMETRIX INC
Identification method for wild oudemansiella species
ActiveCN103667478APrecise positioningImprove detection accuracyMicrobiological testing/measurementOudemansiellaMicrostructure
An identification method for wild oudemansiella species comprises: (1) field acquisition; (2) external form identification; (3) microstructure identification; (4) molecular sequence identification; and (5) comprehensive identification by integrating the external form characteristics and the species for comparing the microstructure and the molecular sequences and accurately determining the species of wild oudemansiella. Compared with routine morphological identification, the identification method provided by the invention has the advantages of high identification accuracy and accurate positioning and is representative among identification methods for our country oudemansiella species.
Owner:SHANGHAI ACAD OF AGRI SCI
Use of magnetic bead supported matrix and MS for judging mass spectrometry polypeptide spectrum and protein fingerprint
InactiveCN101169416AAccurate resolutionHigh resolutionComponent separationBiological testingProtein profilingHuman body
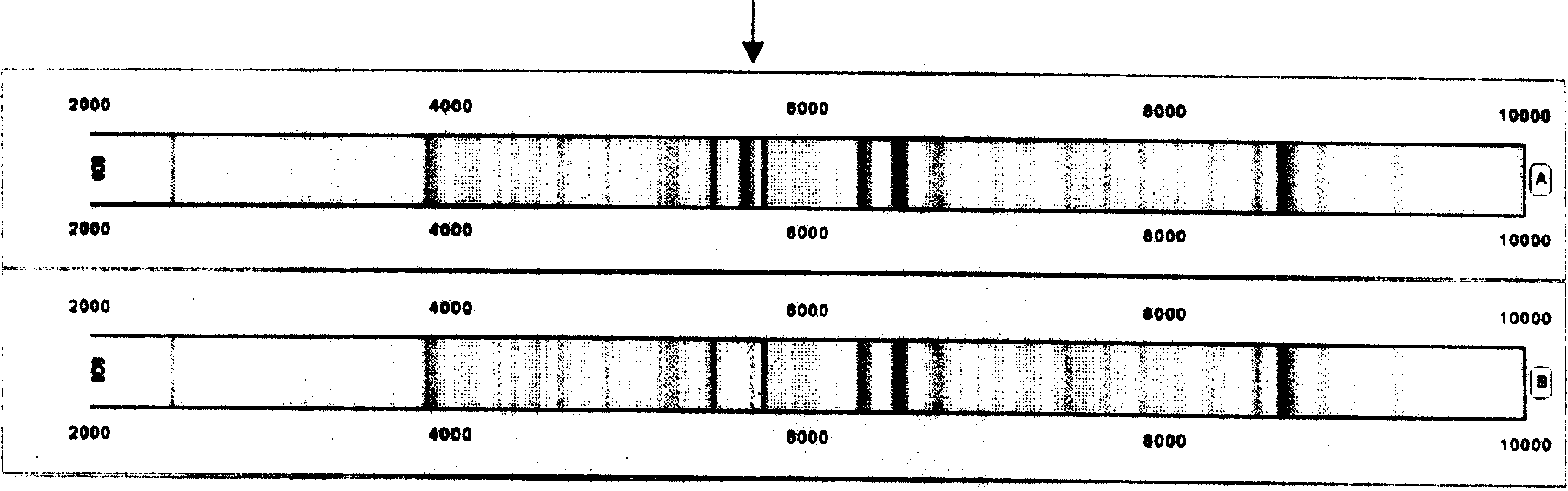
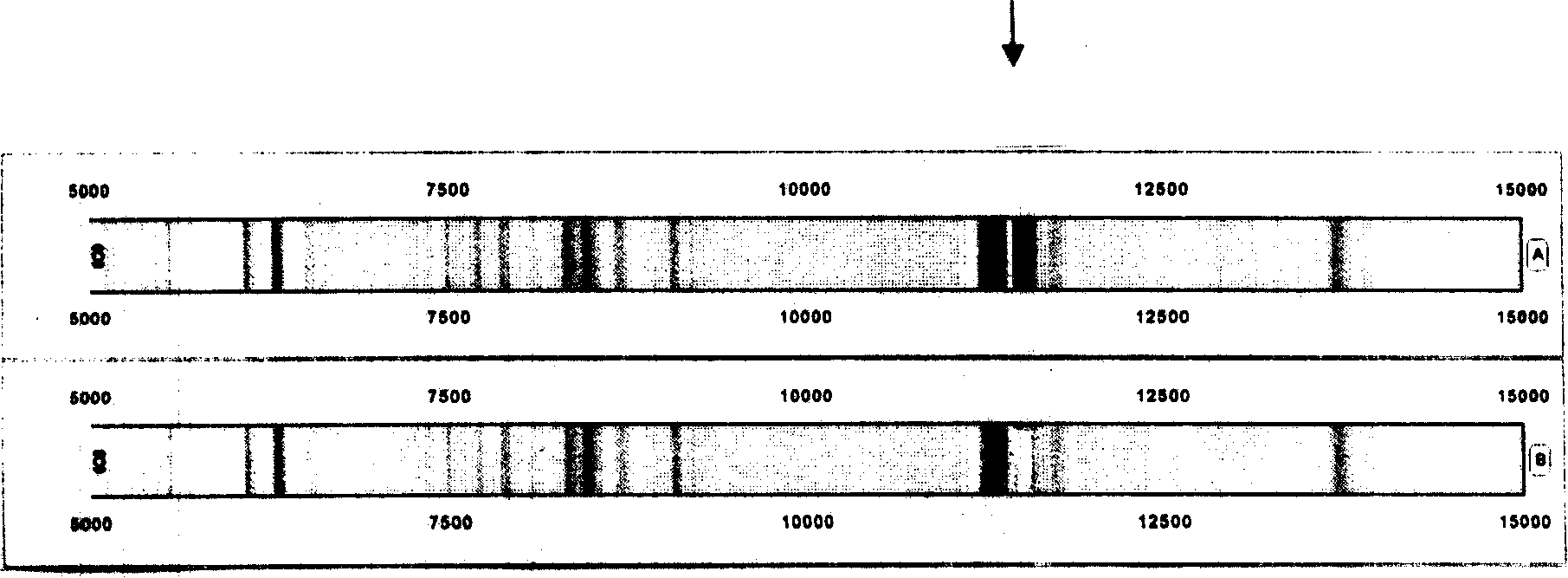
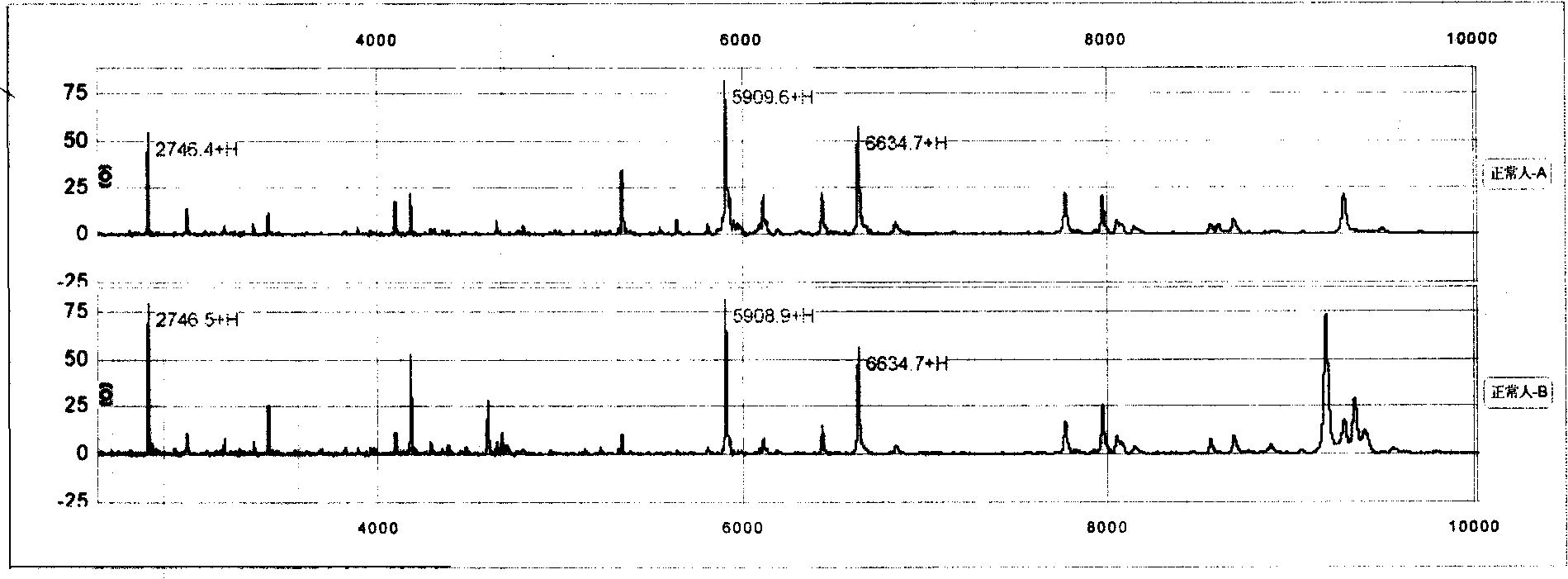
The invention relates to a proteome acquiring method which utilizes the method of bead supporting stroma to capture the protein group in the biology samples, and utilizes the magnetic separator to separate the bead and the sample without eccentric samples, and then utilizes the matter hidden method to analyze. The type of the protein molecular weight obtained by the analysis is a bar code format (protein fingerprint) which can be read by a computer. The invention provides a meaning which confirms the matter hidden bar code format in a certain sample. The confirmations of the bar code format (protein fingerprint) and the meaning of the bar code belt (molecular weight) adopt the addition and subtraction, namely adding or subtracting a certain matter to confirm the meanings of the mass spectrum molecular weight or the biological sign chart. The method of the invention can be used for the identification of the protein fingerprint of the normal people and patients. The significant meaning of the protein fingerprint altas is that the protein fingerprint altas regularly inspects the dynamic changes of mass spectrum polypeptide chart, protein fingerprint and significant biology sing chart in the human activity or blood. The protein single protein fingerprint and the molecule sequence in the protein fingerprint can be used in the testing method of the reagent box of the body liquid part away from the body. The method is accurate, convenient and quick.
Owner:许洋
Charge repulsion induced single-chain collagen polypeptide probe and preparation method thereof
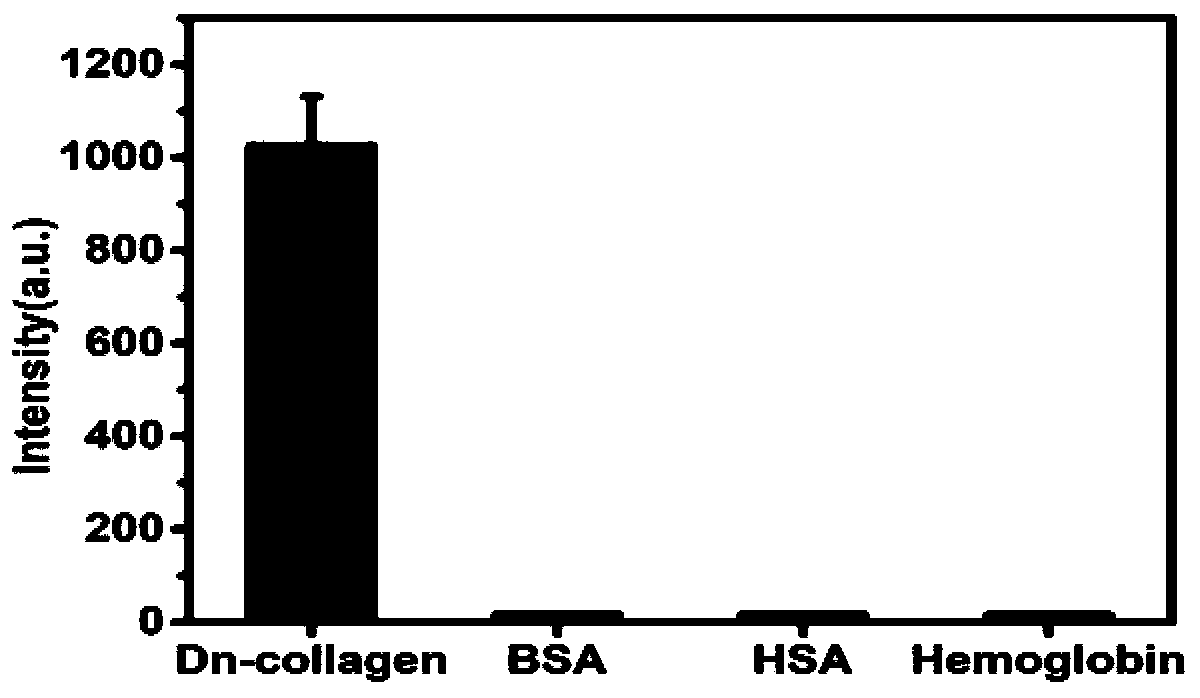
ActiveCN111057539AEasy to makeAvoid risk of injuryConnective tissue peptidesPeptide preparation methodsDiseaseSignalling molecules
The invention belongs to the technical field of biological detection, and particularly relates to a charge repulsion induced single-chain collagen polypeptide probe and a preparation method thereof. The single-chain collagen polypeptide probe provided by the invention contains a repetitive sequence of (Gly-Pro-Hyp)n, (Gly-Pro-Pro)n or (Gly-Hyp-Hyp)n in the middle, a fluorescence signal molecule ismodified on the sequence, and the C end of the sequence contains a plurality of charged amino acids. The novel polypeptide probe provided by the invention does not contain non-natural amino acid andis simple to prepare. The novel polypeptide probe has pH sensitivity, forms a triple helix structure under an acidic condition, can keep a stable single-chain structure under a physiological pH condition, has the capability of specifically binding lesion collagen without any pretreatment, and completely avoids the damage risk of pretreatment to a tissue sample. The novel polypeptide probe providedby the invention can be used for detecting the content of lesion collagen in vitro, and has a wide application prospect in early diagnosis and curative effect evaluation of collagen-related diseasessuch as tumors.
Owner:LANZHOU UNIVERSITY
Method for rapidly detecting bacillus coagulans and multiplex PCR reagent kit
ActiveCN105256054AExcellent screening performanceReduce economic lossMicrobiological testing/measurementBacteroidesMultiplex pcrs
The invention relates to the detection technology of bacillus and provides a method for rapidly detecting bacillus coagulans and a multiplex PCR reagent kit. The method is rapid and accurate in detection, high in sensitivity, easy to operate and low in cost. The reagent kit and method can specifically amplify coag genes and comK gene sequences in a bacillus coagulan genome, cannot obtain other bacterial gene segments through amplification and is high in specificity. Particularly for bacillus methylotrophicus and bacillus amyloliquefaciens with the high-similar molecular sequences as the bacillus coagulans, gene segments cannot be obtained through amplification. Whether bacillus coagulans exist in a sample to be detected can be accurately and effectively detected.
Owner:TONGWEI
Method for improving soybean quality and DNA (Deoxyribonucleic Acid) molecule special for same
InactiveCN102127540AImprove qualityChange qualityVector-based foreign material introductionDNA/RNA fragmentationBiotechnologyWild type
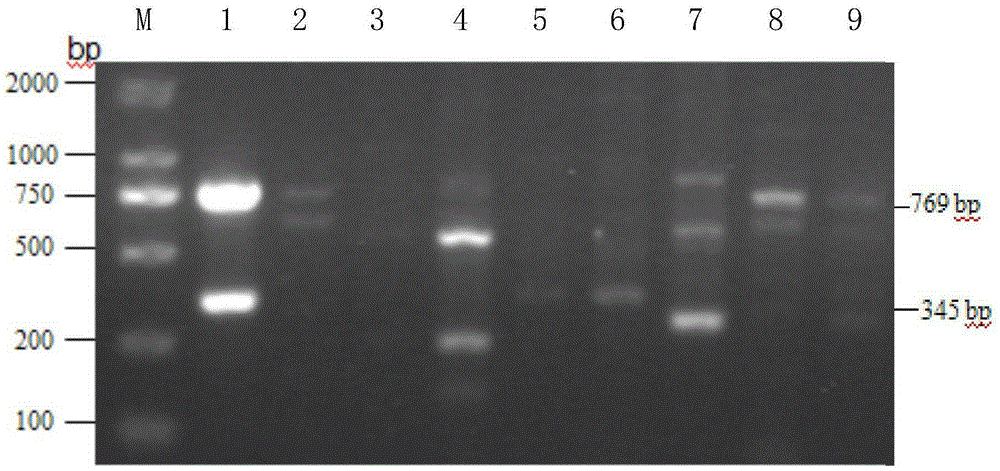
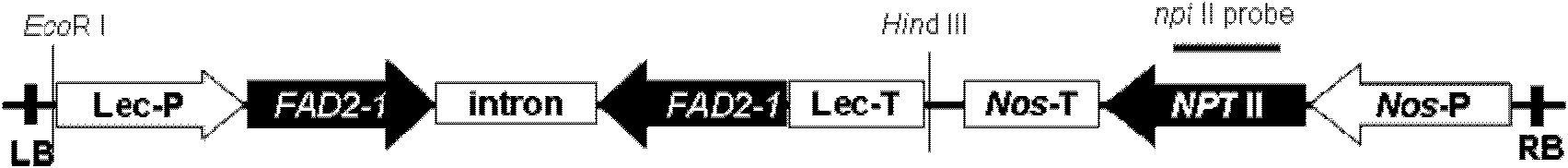
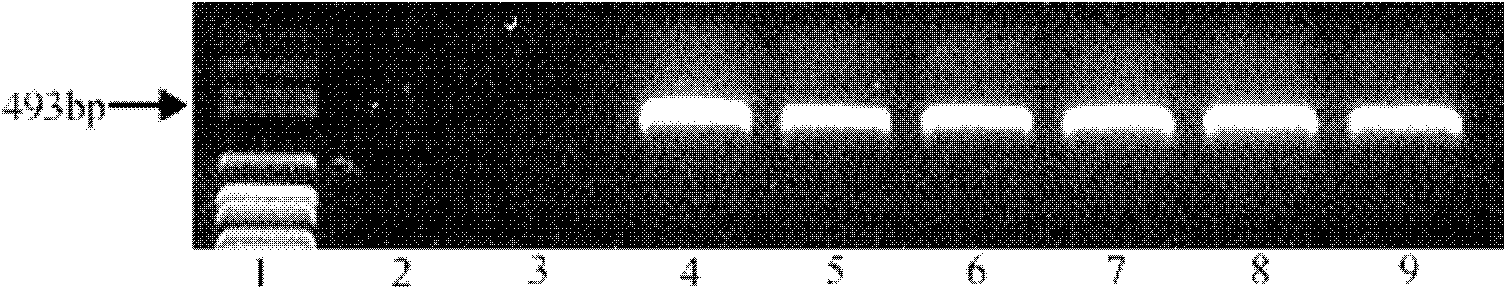
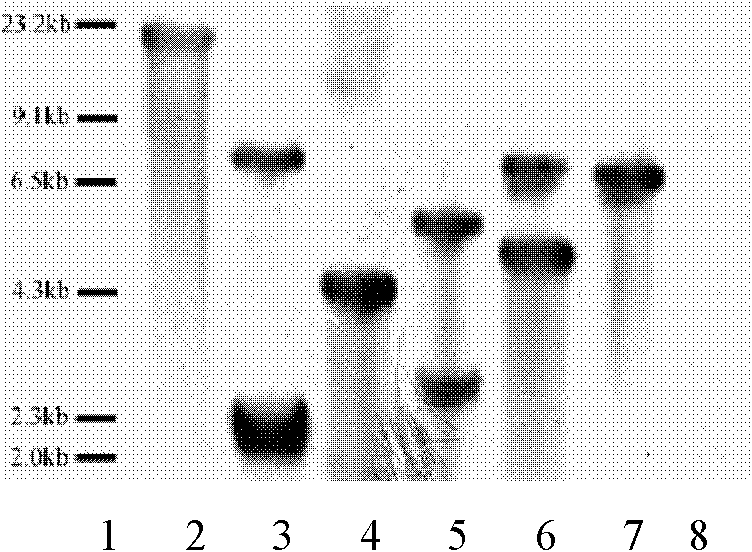
The invention discloses a method for improving soybean quality and a DNA (Deoxyribonucleic Acid) molecule special for the same, and provides a DNA molecule which is: (1) a DNA molecule shown as a sequence I in a sequence table or (2) a DNA molecule which is hybridized with the DNA sequence defined in (1) under the strict condition and has the same function as the DNA sequence defined in (1) or (3) a DNA molecule having at least 70 percent, at least 75 percent, at least 80 percent, at least 85 percent, at least 90 percent, at least 95 percent, at least 96 percent, at least 97 percent, at least 98 percent or at least 99 percent of homology and the same function as the DNA sequence defined in (1). An experiment in the invention indicates that a genetically engineered soybean seed of a transgenic soybean delta12-desaturase gene (FAD2-1) silent expression vector shows a high oleic acid phenotype, and transgenic soybean seed oil remarkably differs from wild type soybean in the aspects of oleic acid, linoleic acid, linolenic acid and palmitic acid.
Owner:INST OF BOTANY CHINESE ACAD OF SCI
Pig myostatin gene promoter and its applications
ActiveCN102725408AFix stability issuesResolve locationBacteriaGenetic material ingredientsAgricultural scienceHuman cell
A pig myostatin gene promoter and its applications are disclosed in the present invention. The DNA fragment provided in the present invention which is derived from pigs is anyone of the following DNA molecules 1)-4): 1) the DNA molecule of SEQ ID NO:2 shown in the Sequence Listing; 2) the DNA molecule of SEQ ID NO:3 shown in the Sequence Listing; 3) the DNA molecule which hybridizes to the DNA sequence in 1) or 2) under strict conditions and has promoter function; 4) the DNA molecule which has more than 90% homology with the DNA sequence in 1) or 2) and has promoter function. The experiments in the present invention prove that the promoter provided in the present invention can drive the expression of firefly luciferase reporter gene. The promoter can also be built into the reporter vector and then the vector is transfected into cultured swine and human cells, and the activity and efficiency of the promoter are identified by accurate quantitative methods through reporter gene test.
Owner:INST OF ANIMAL SCI & VETERINARY HUBEI ACADEMY OF AGRI SCI
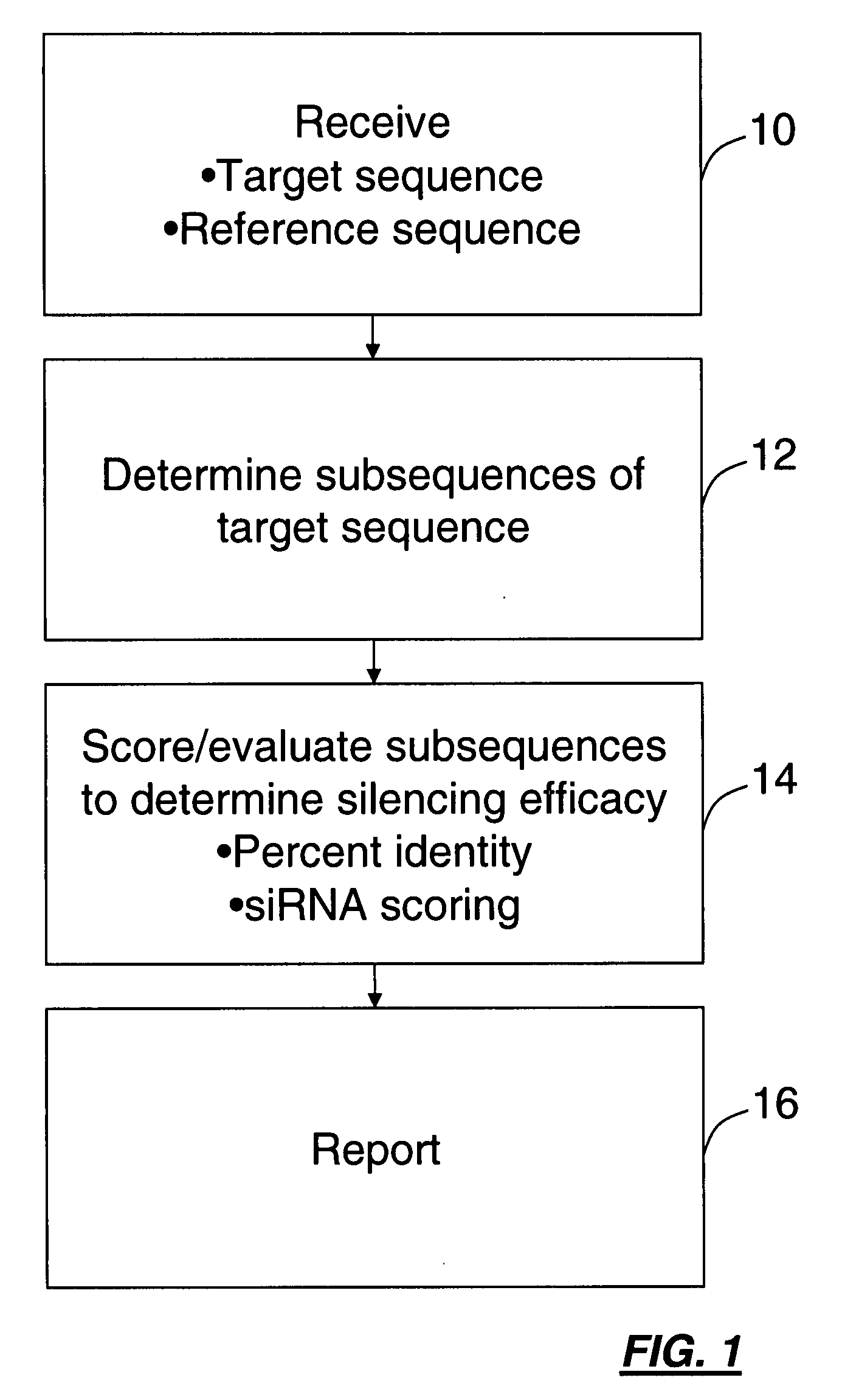
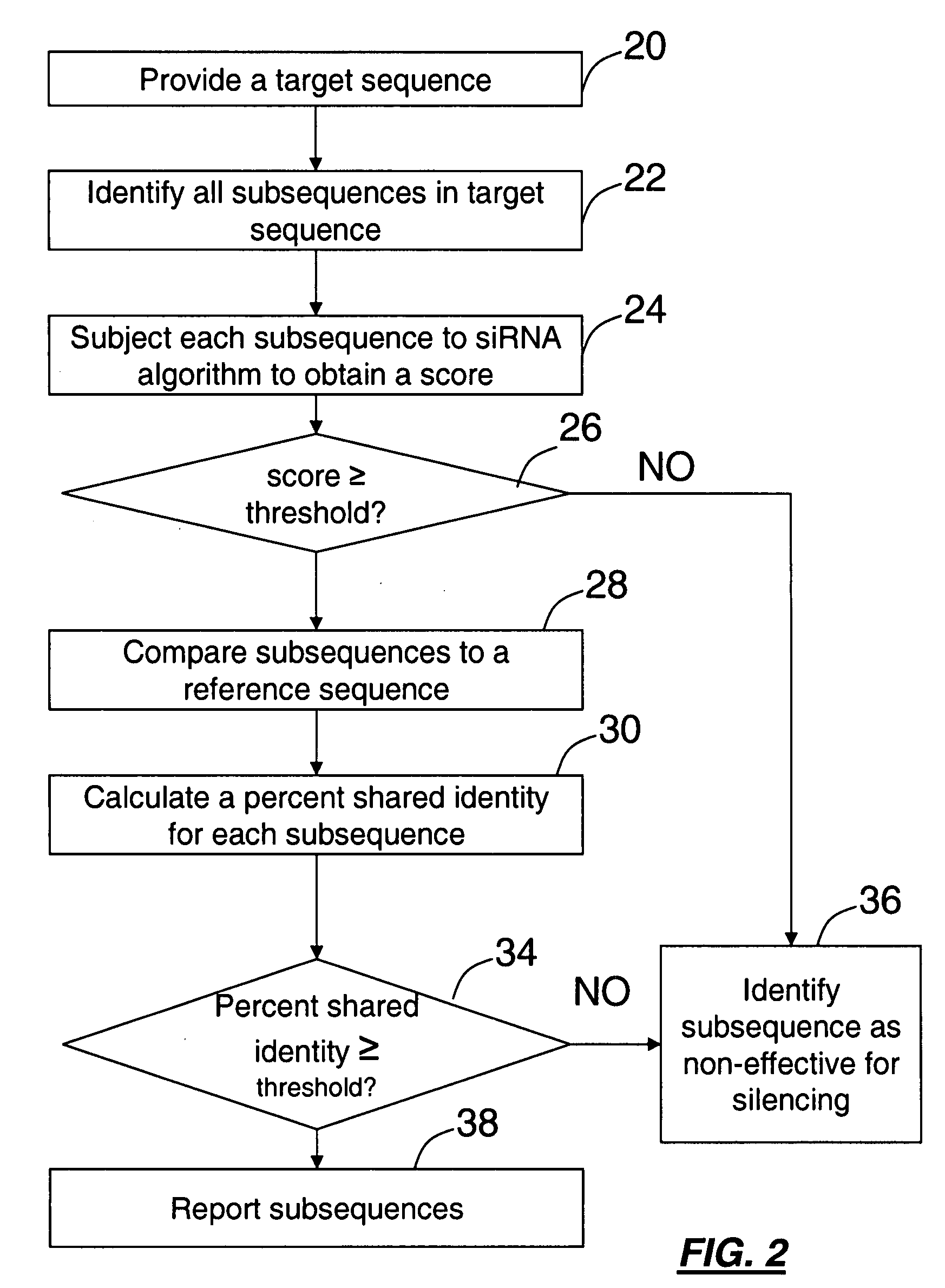
Computational method for choosing nucleotide sequences to specifically silence genes
A method for identifying subsequences in a polynucleotide sequence for specifically silencing a target gene is provided. The method is described for identifying sequences effective in silencing a target gene or a series of genes, but not others. Subsequences can be identified and scored using comparisons based on percent sequence identity with respect to a target reference sequence and siRNA algorithm analysis. The resulting subsequences may be ranked based on score, percent sequence identity. The identification of subsequences may be performed using a sliding window to identify all subsequences of a set length within the sequence. A user interface may be provided for displaying the results to a user.
Owner:PIONEER HI BRED INT INC
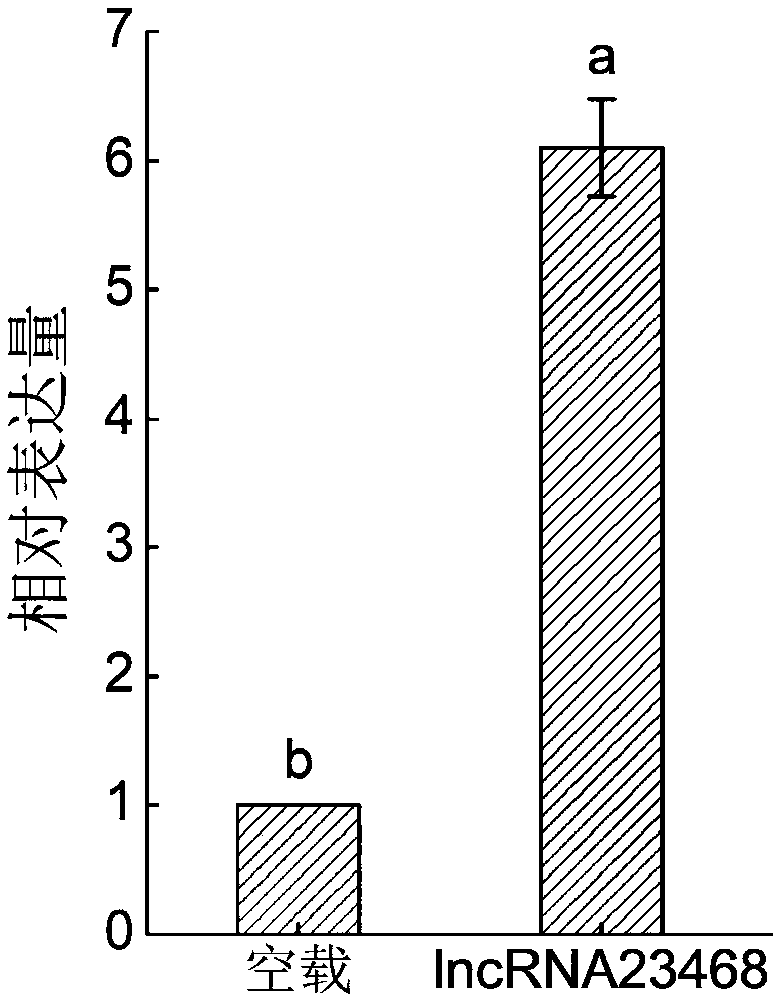
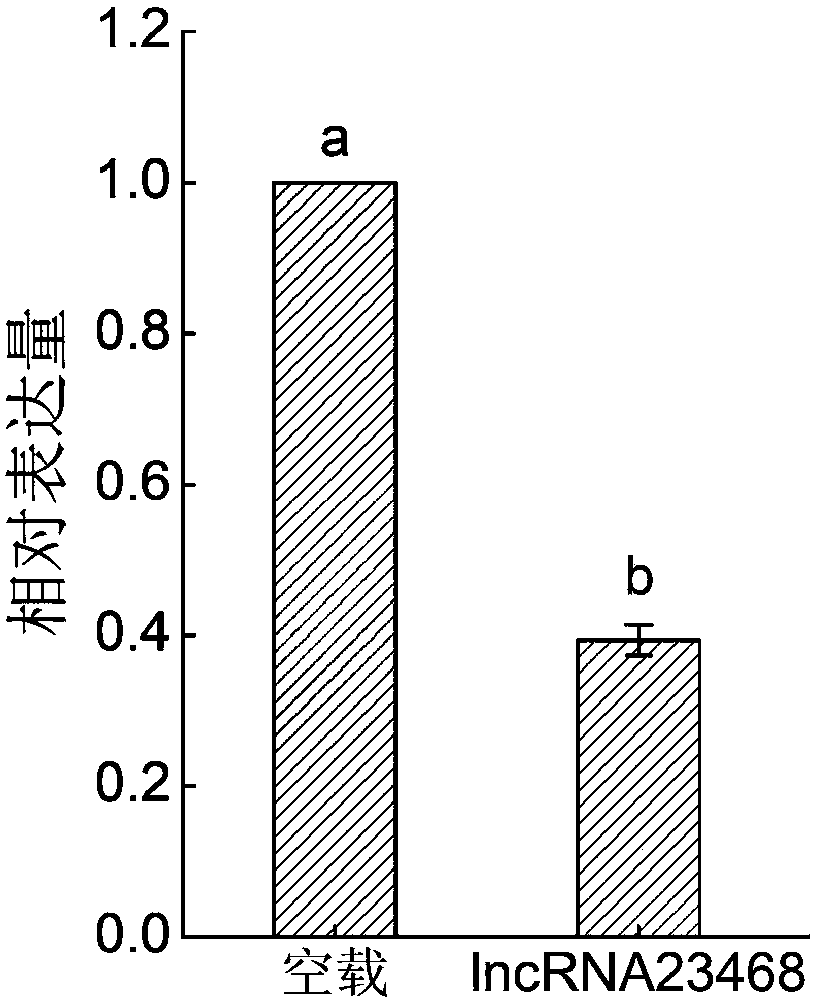
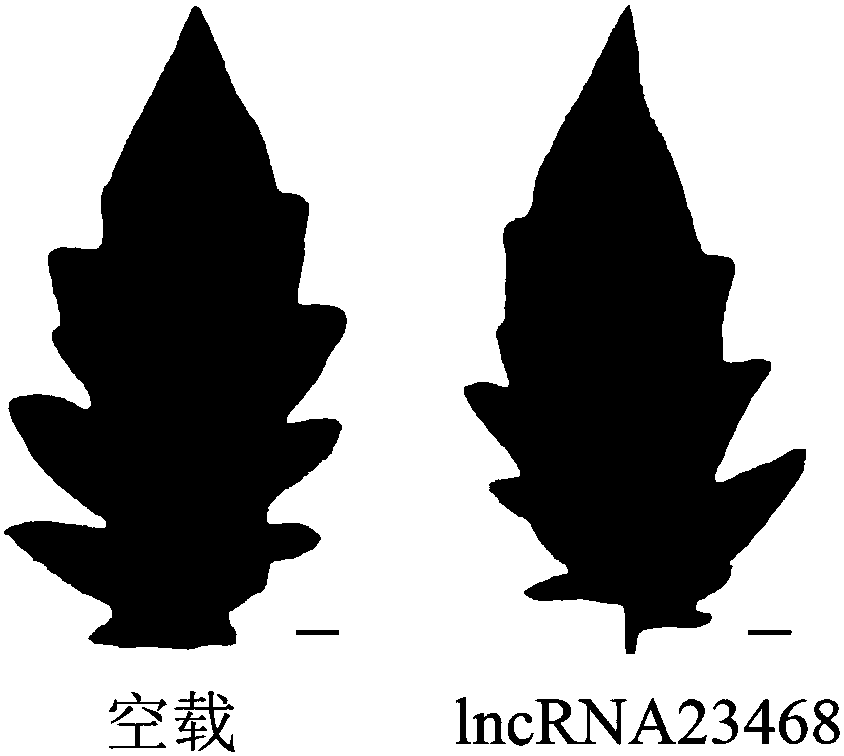
Long-chain non-coding RNA-lncRNA23468 of tomatoes, cloning method and application method thereof
ActiveCN108085318AProve silencingIncrease resistanceMicrobiological testing/measurementVector-based foreign material introductionEscherichia coliWild type
The invention provides long-chain non-coding RNA-lncRNA23468 for improving the resistance to late blight of tomatoes by silent miRNA482b, and provides a cloning method and an application method thereof. The DNA molecular sequence of the long-chain non-coding RNA-lncRNA23468 is shown as SEQ ID NO. 1; the cloning method comprises the steps of: adopting cDNA of wild type late-blight-resistant tomatoL3708 as a template to carry out PCR (Polymerise Chain Reaction) amplification, connecting an obtained PCR product with pMD-19T cloning carriers, transforming escherichia coli DH5alpha, and choosing single colonies for sequencing. The long-chain non-coding RNA obtained by the method has the beneficial effects that after transient overexpression of the long-chain non-coding RNA in the tomatoes, theexpression quantity of the miR482b of the tomatoes is obviously reduced, and the resistance to the late blight is obviously improved; the long-chain non-coding RNA is used for silent miR482b and plays an important role in breeding the varieties of late-blight-resistant tomatoes.
Owner:DALIAN UNIV OF TECH
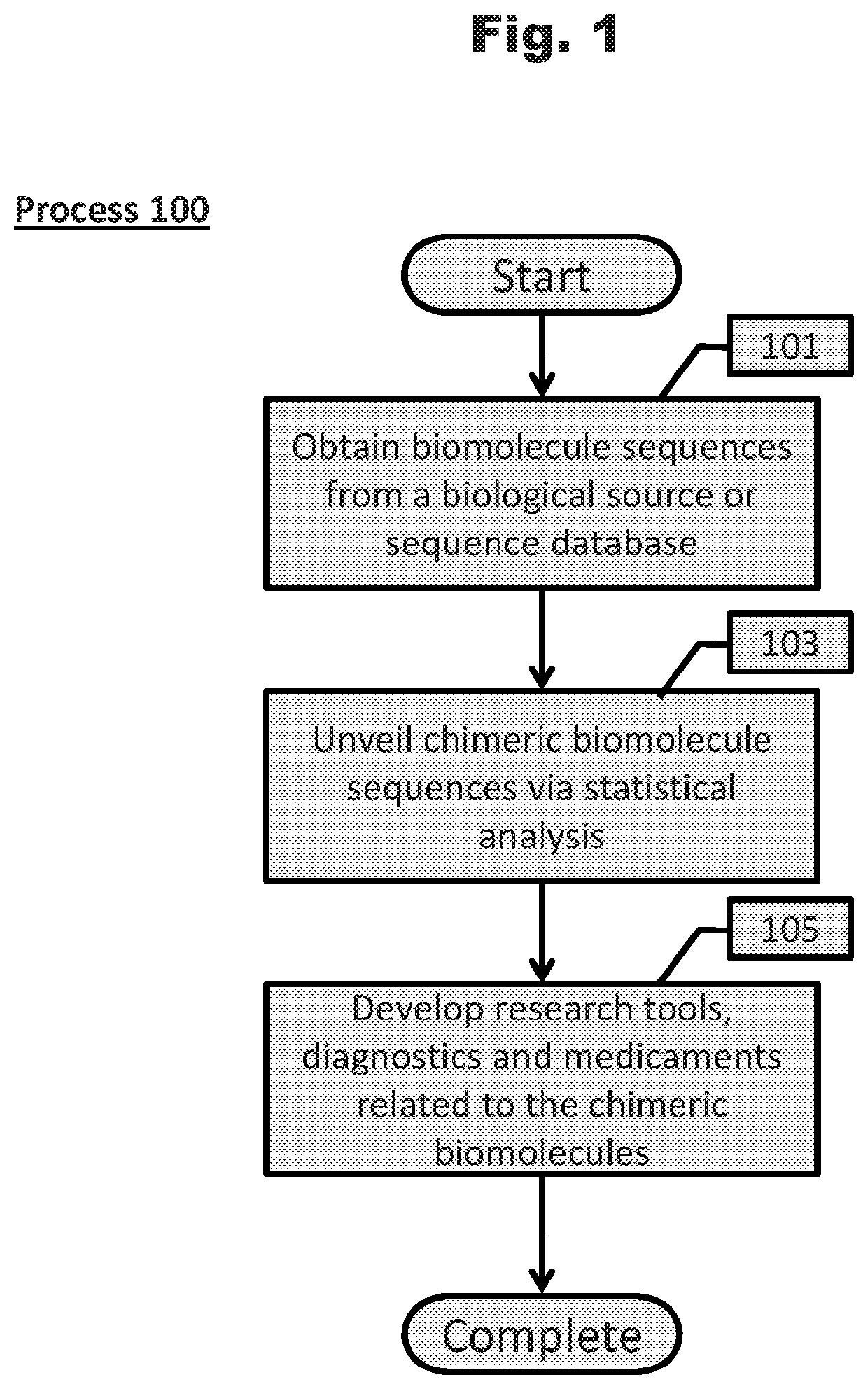
Accurate and Sensitive Unveiling of Chimeric Biomolecule Sequences and Applications Thereof
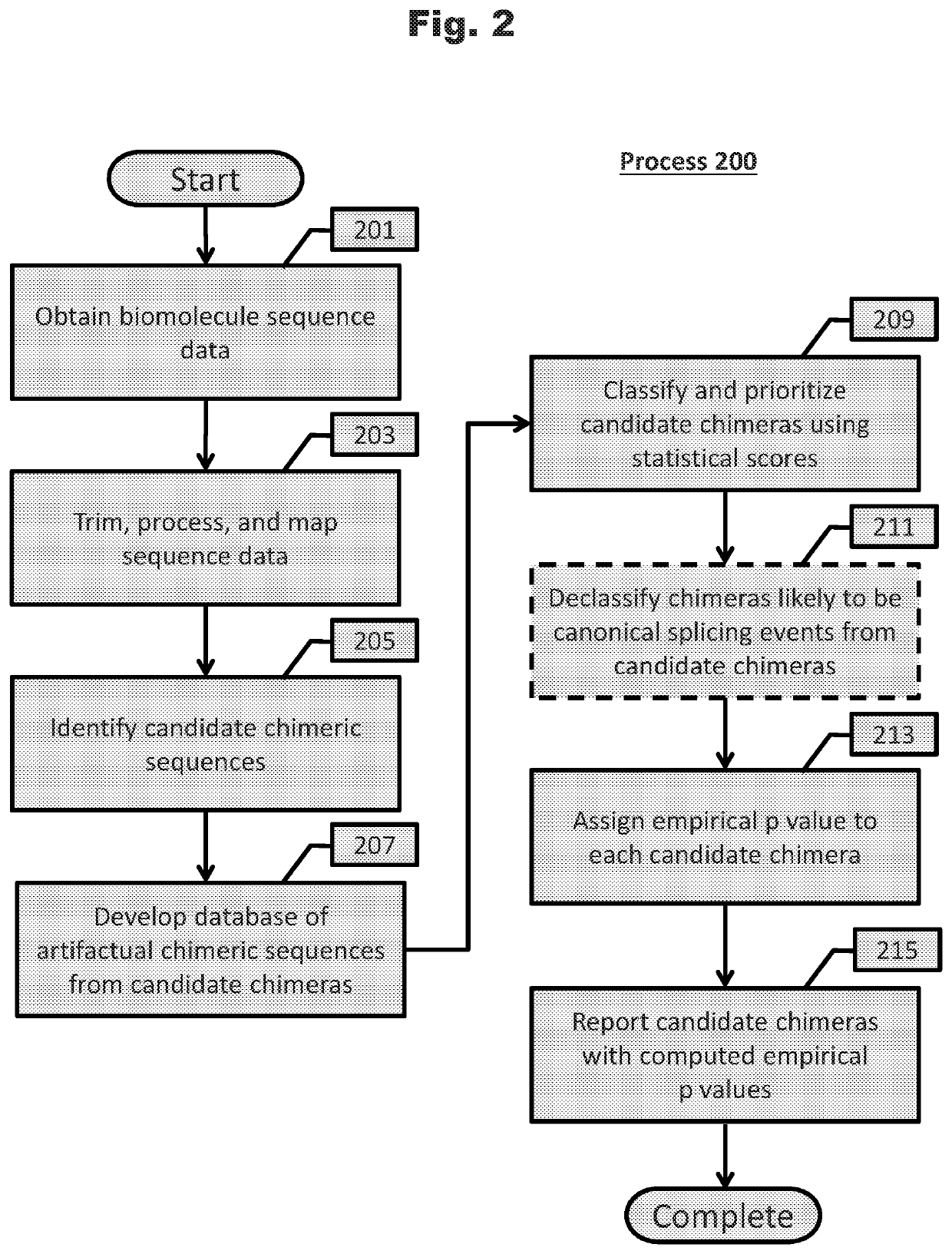
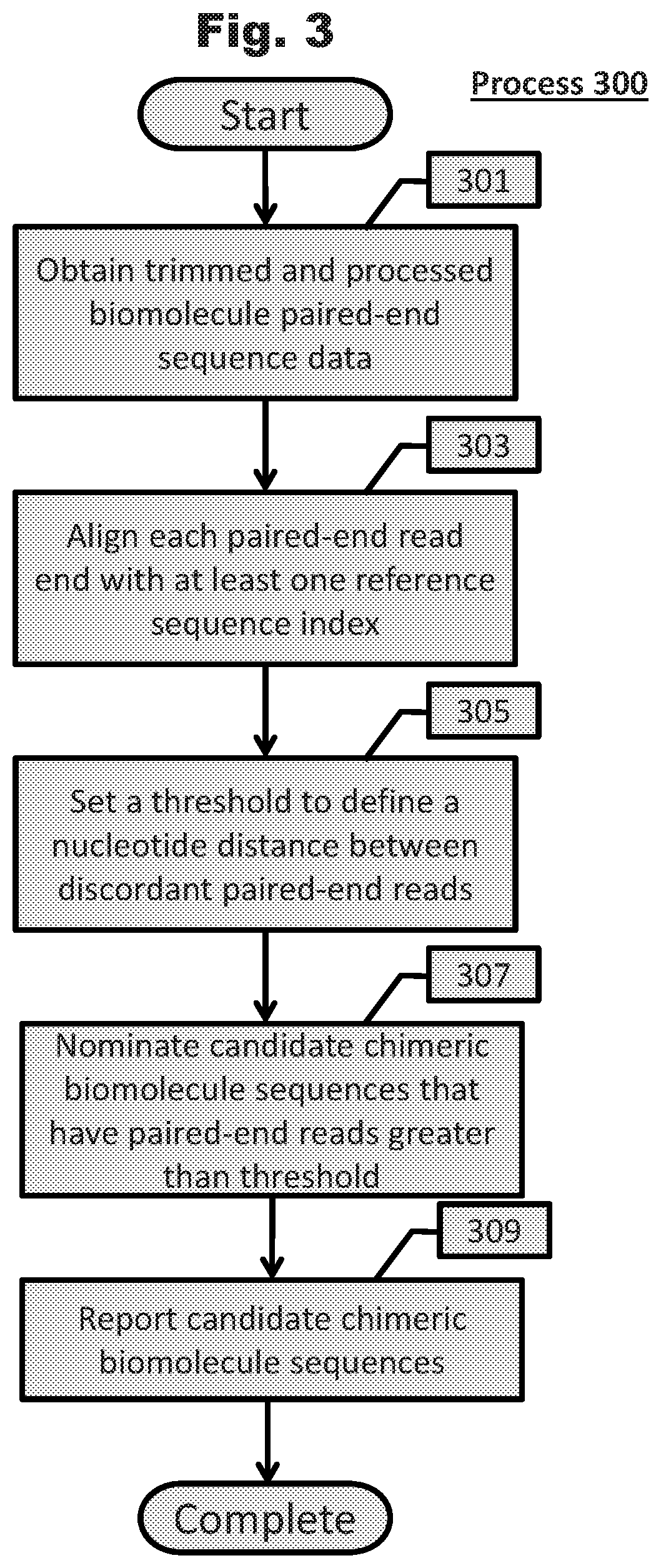
Unveiling of chimeric biomolecule sequences and applications thereof are described. Generally, systems comprising statistical analysis are performed to unveil chimeric biomolecule sequences from sequencing data sets. Bloom filters and hierarchical bloom filter tree data structures can be constructed such that chimeric sequence unveiling systems are more efficient. Finally, chimeric sequences are used to develop research tools, diagnostics, and medicaments.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Molecular marker for diagnosis of diabetes and application thereof
PendingCN110452976AAddress riskAddressing Early Diagnosis IssuesMicrobiological testing/measurementDNA/RNA fragmentationForward primerThrombus
The invention belongs to the technical field of biology, and particularly to a molecular marker for diagnosis of diabetes and application thereof. For the problem that currently, no reliable biomarkercan perform risk prediction and early diagnosis on the diabetes, the following solution is proposed that the molecular marker comprises a kit and diagnostic primers, wherein the kit is internally provided with a chip, the chip comprises a gene chip, a gene primer and a molecular marker, the diagnostic primers comprise the forward primer and the reverse primer, and the forward primer and the reverse primer are both set to have RNA molecule sequences. Trough the detection of items related to a thrombus molecular marker, not only is further knowing of complications of DM vascular injuries and the pathogenesis of thrombosis facilitated, but also a basis is provided for using anticoagulant antiplatelet drugs for treatment, and the detection of molecules of type 2 diabetes patients has important clinical significance for the early prevention or intervention of thrombogenesis and vasculopathy.
Owner:淄博市中心医院
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com