Methods and systems for controlling dna, RNA and other biological molecules passing through nanopores
A nanopore and molecular technology, applied in biochemical equipment and methods, nanotechnology for sensing, separation methods, etc., can solve the problems of not providing DNA movement, slowing down DNA translocation, and insufficient
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0044] Part I: Mechanical control of passage of DNA and other biomolecules through nanopores
[0045] The present disclosure provides methods for precisely controlling the movement of molecules, such as DNA, through a nanopore by mechanical means, or more specifically, by nanometer (nm) or sub-nanometer (nm) precision piezoelectric actuators or combinations of actuators, such as Methods for the movement of DNA through nanopores. Combined with electricity, this moves DNA into and out of the nanopore continuously or stepwise at a rate sufficient or slow enough for reliable base sensing, depending on the base sensing method chosen. For example: an ion current blocking base sensing approach using protein nanopores requires a sensing time of at least 1 ms (millisecond) per base.
[0046] Fundamentals - Balance of Forces
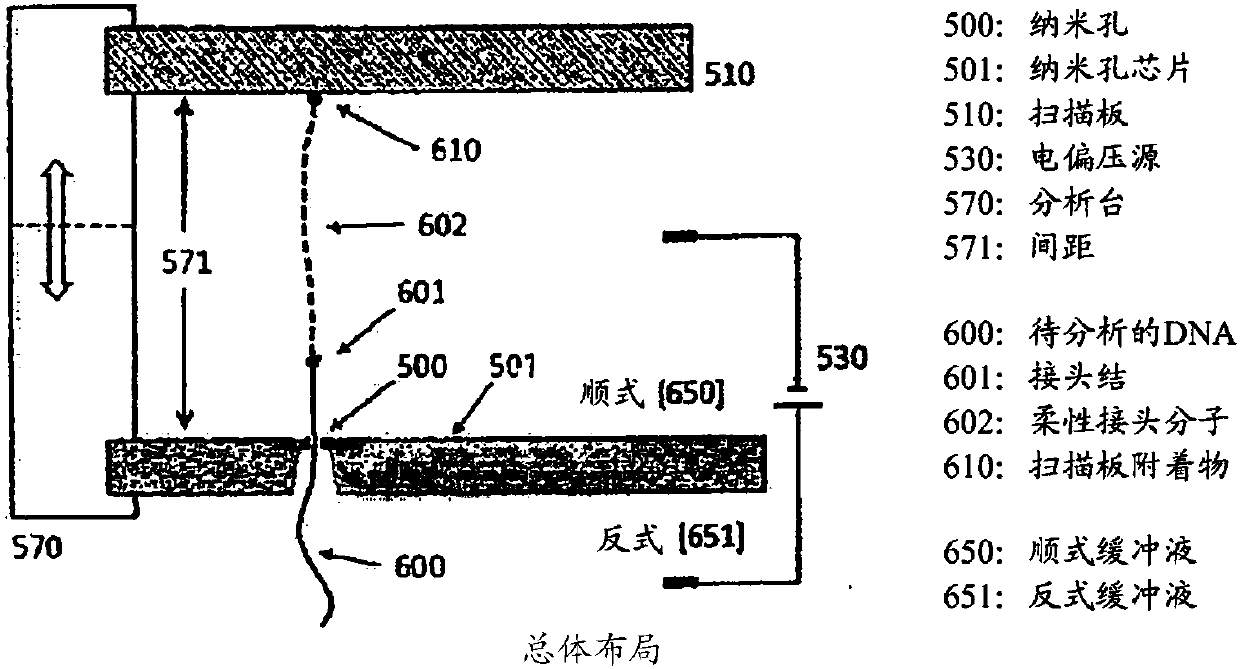
[0047] Basic principles such as figure 1 As shown, it uses single-stranded DNA (ssDNA) as an example.
[0048] The nanopore (500) is part of the nanopore chip...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com