Molecule substitution label sequencing parallel detection method-oligomictic nucleic acid coding label molecule library micro-sphere array analysis
A technology of oligomeric nucleic acid and microsphere array, which is applied in the field of microsphere suspension array system, can solve the problems of uneven distribution, background deviation, poor repeatability, etc., and achieve the effect of good repeatability and high reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
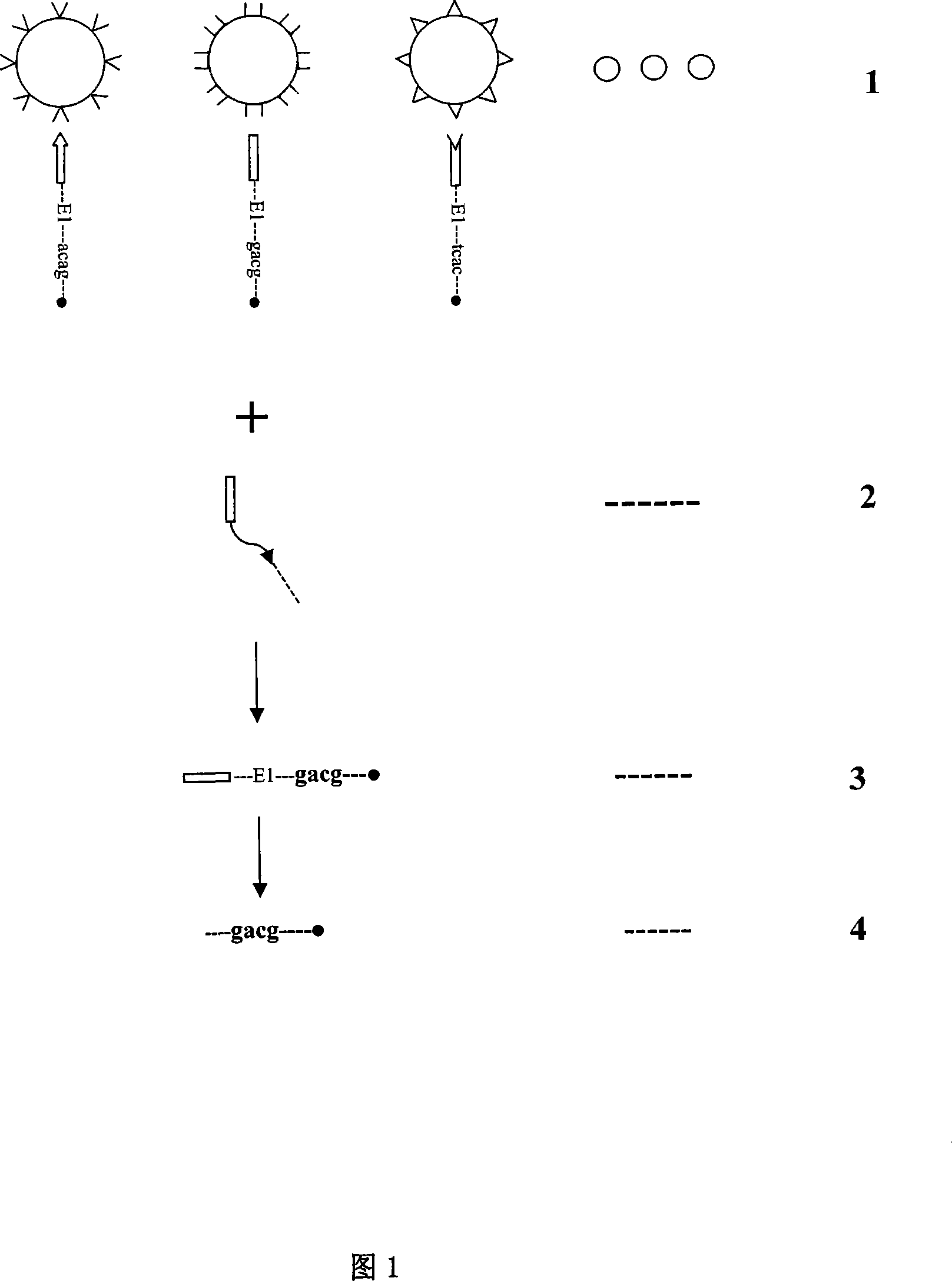
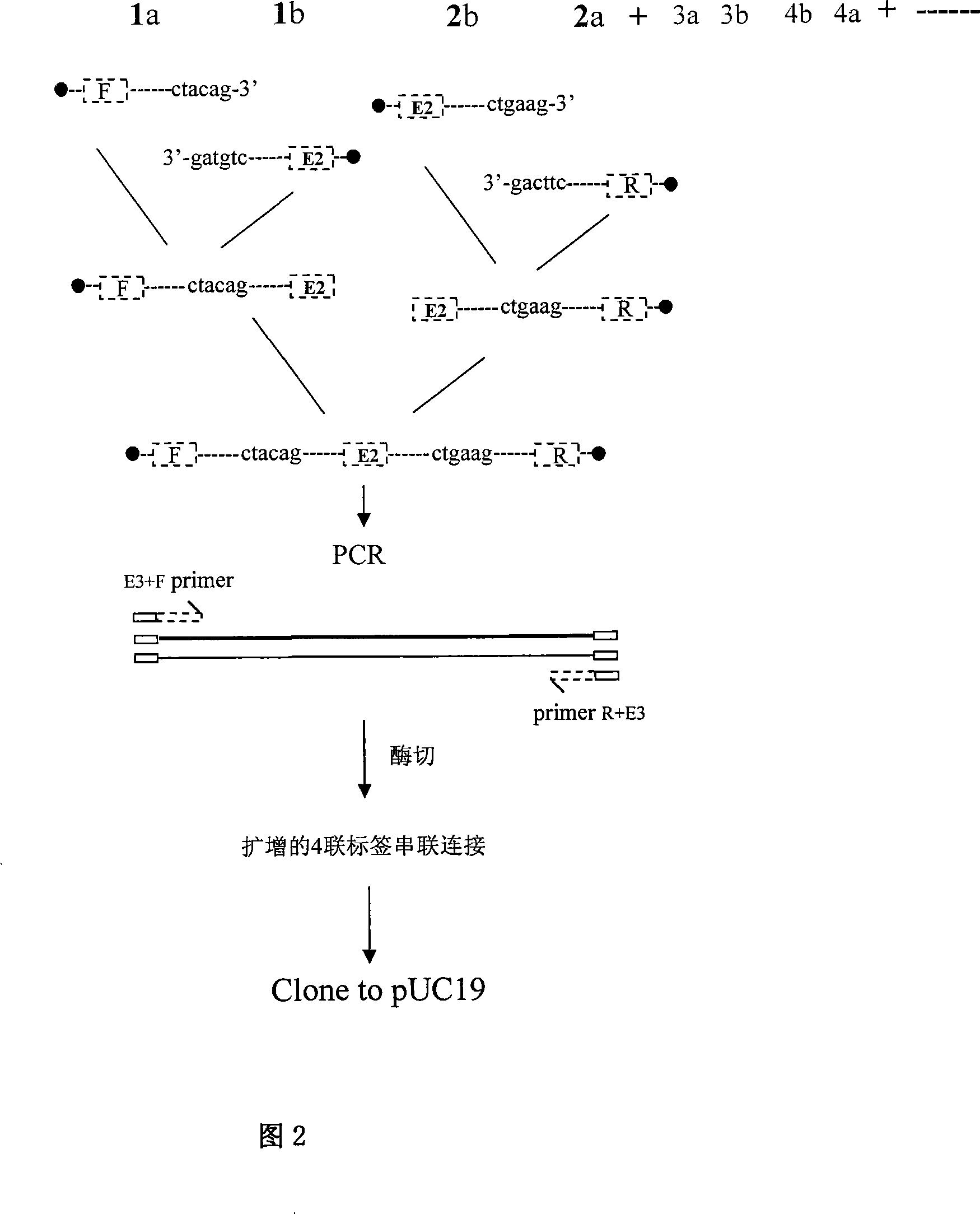
[0192] Example: Parallel detection of tumor suppressor genes pRB, p16, p53 and control Tubulin gene:
[0193] Oncogenes and tumor suppressor genes play the most critical roles in cell growth cycle regulation. In particular, the metabolism and inactivation of tumor suppressor genes pRB, p16, and p53 play the most important role in the mechanism of tumor cells losing growth control. Almost all tumors involve some change in them or in one of them. However, because the traditional research methods are single, the method of analyzing the research objects one by one is inevitable, and the whole real metabolic image cannot be restored.
[0194] This example constructs a human tumor suppressor gene pRB, p16, an indicator molecule library of p53, and adds the Tubulin (tubulin) gene of human cell structural protein as a constant control indicator molecule to jointly build a tagged pRB, p16, p53 and Tubulin indicating DNA adsorption to Seplarose CL-4B microsphere array library. The ov...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com