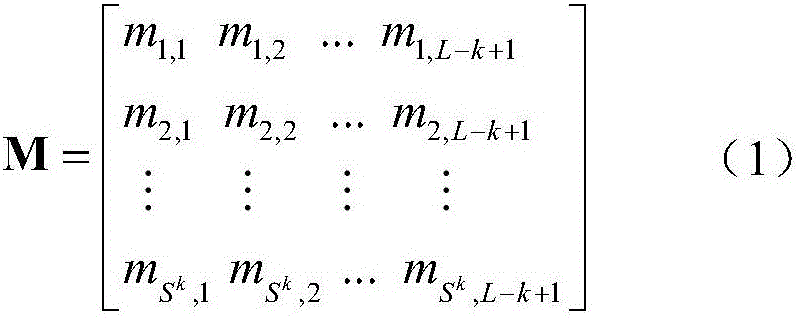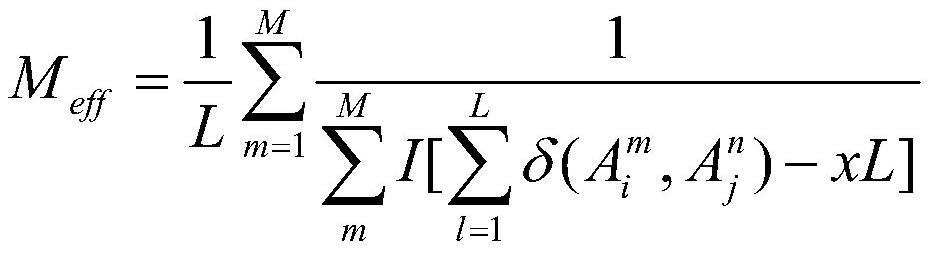Patents
Literature
71 results about "Multiple sequence alignment" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
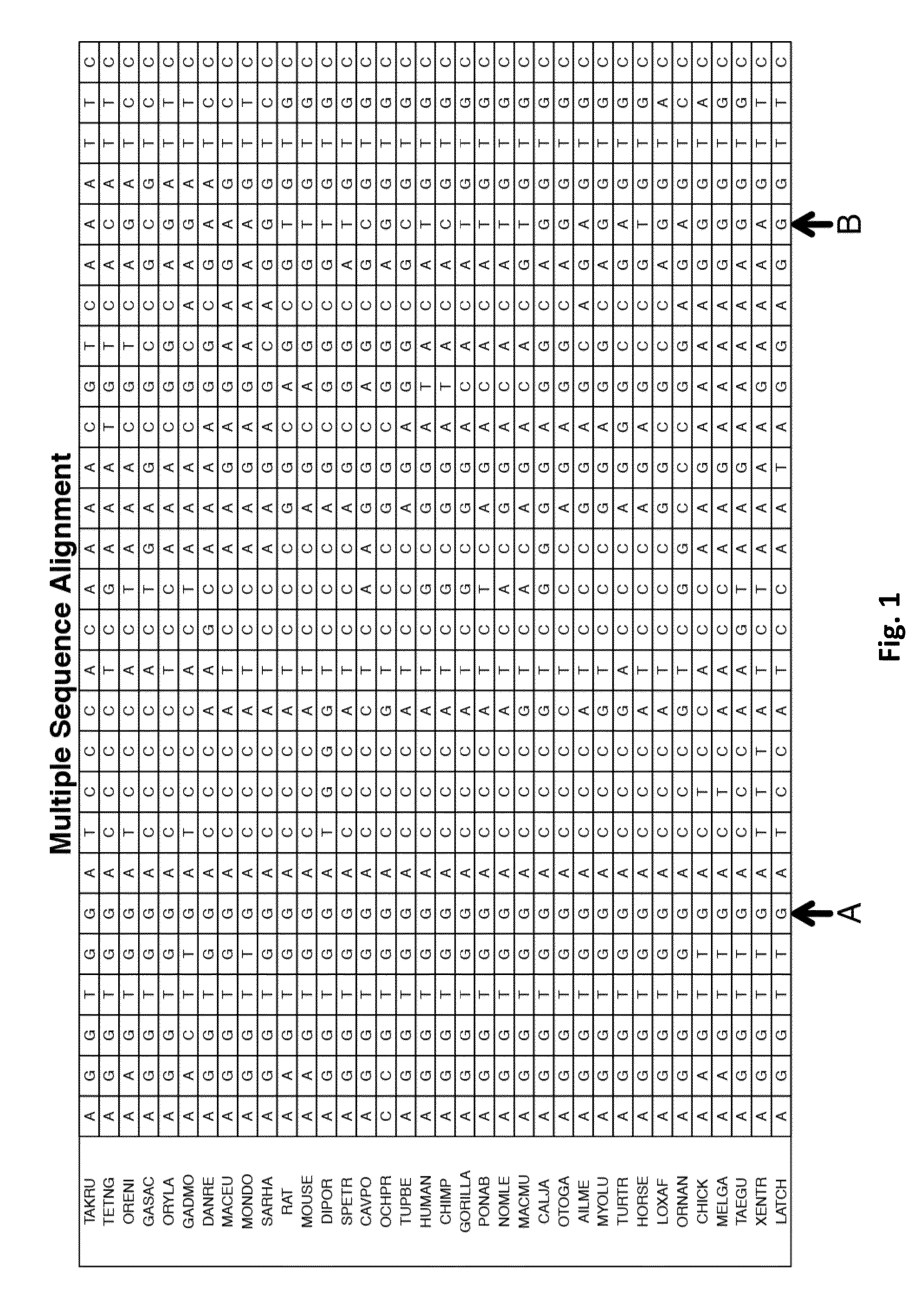
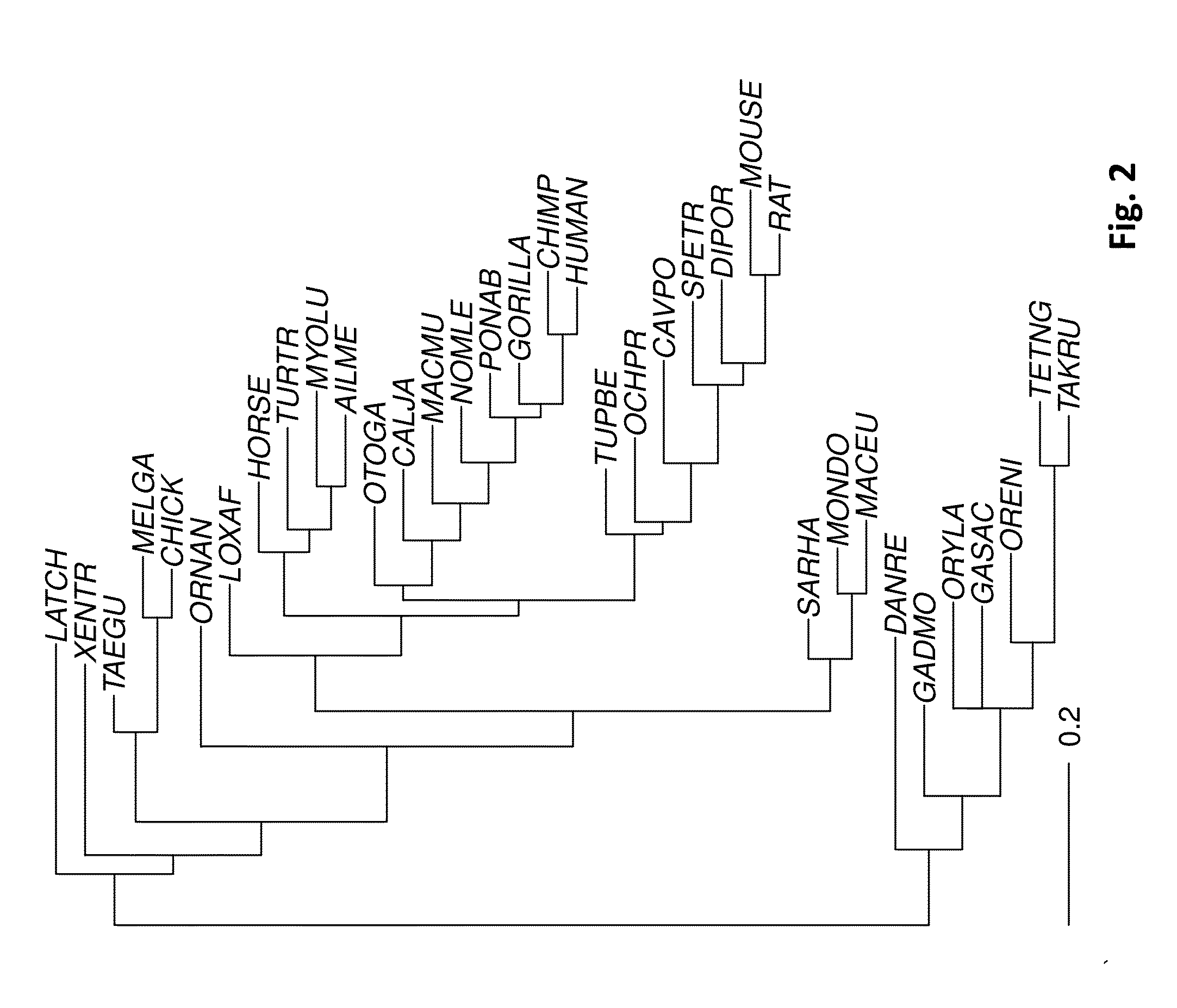
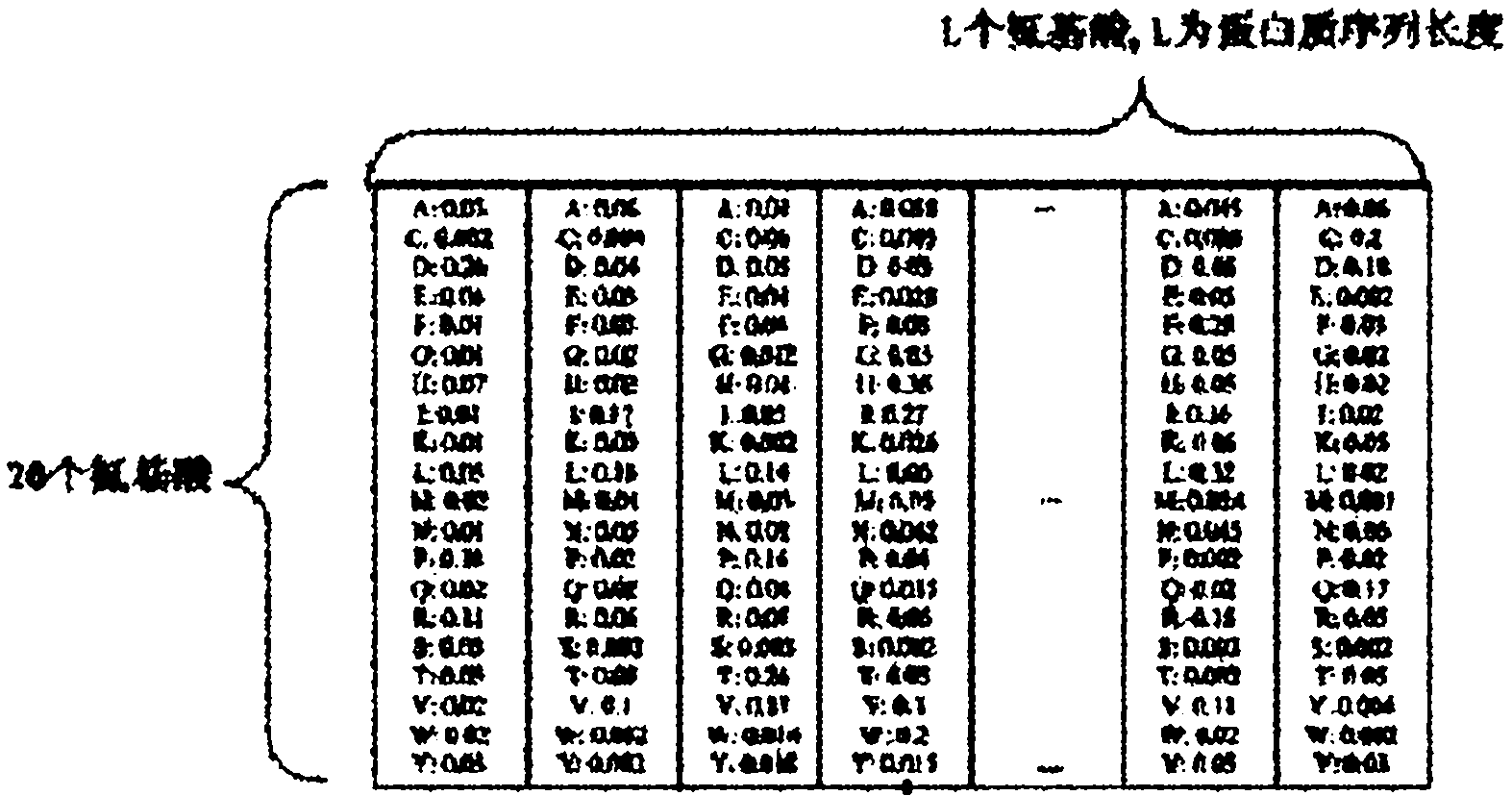
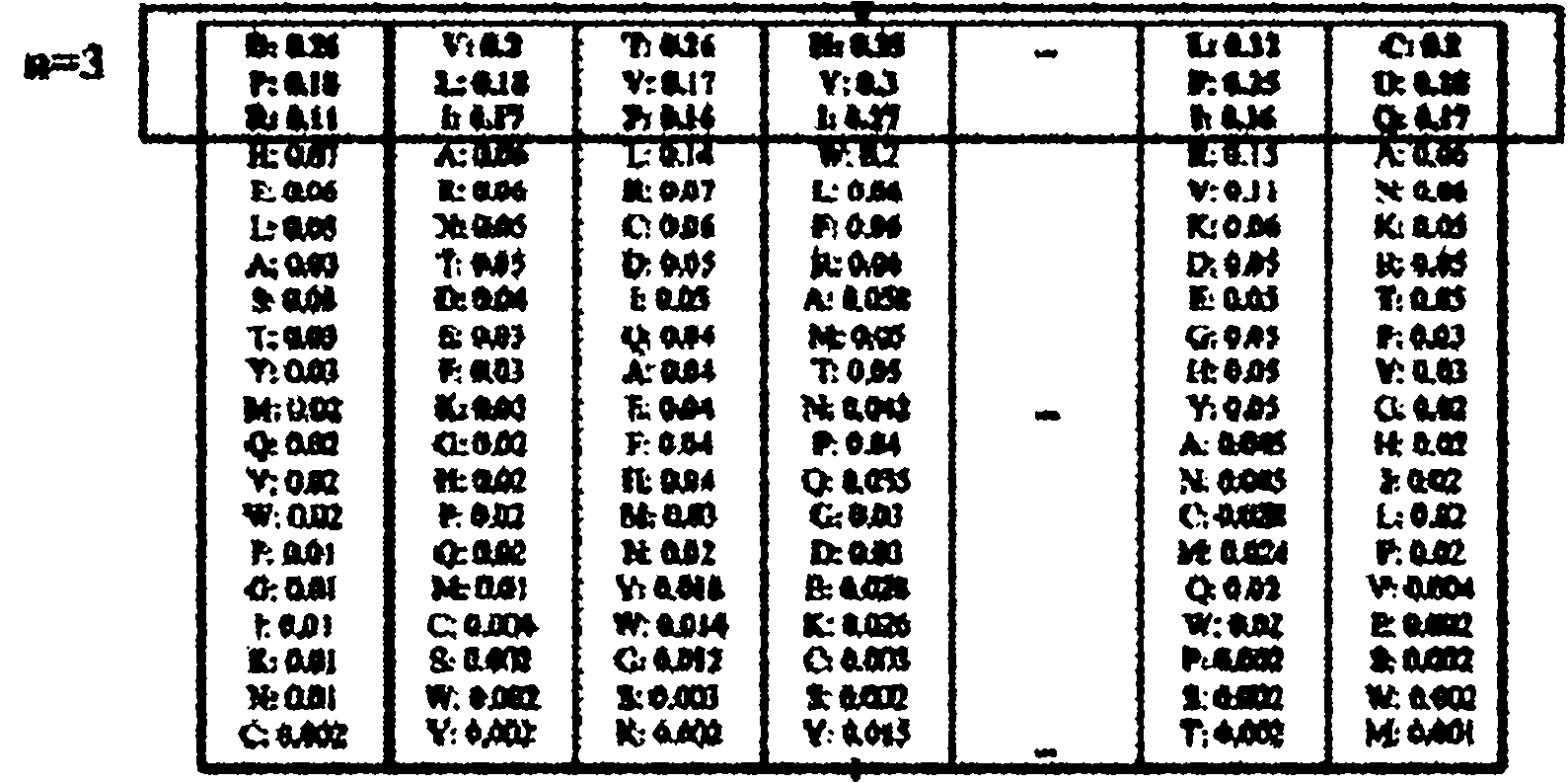
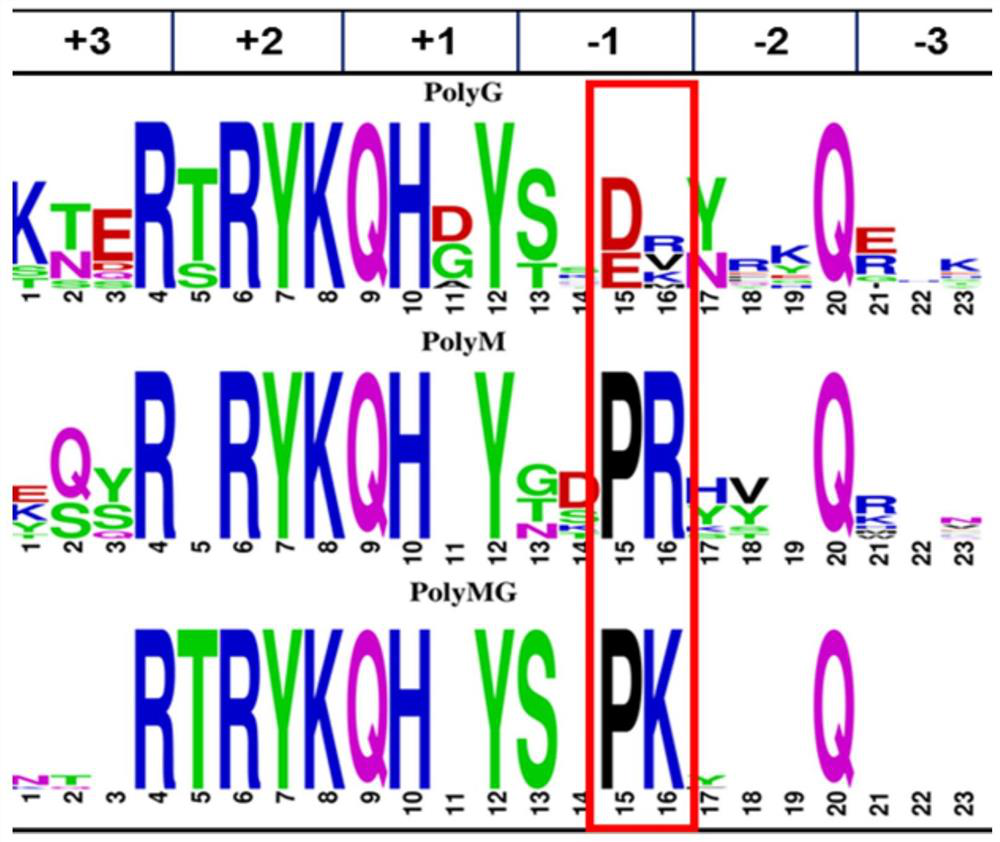
A multiple sequence alignment (MSA) is a sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. In many cases, the input set of query sequences are assumed to have an evolutionary relationship by which they share a linkage and are descended from a common ancestor. From the resulting MSA, sequence homology can be inferred and phylogenetic analysis can be conducted to assess the sequences' shared evolutionary origins. Visual depictions of the alignment as in the image at right illustrate mutation events such as point mutations (single amino acid or nucleotide changes) that appear as differing characters in a single alignment column, and insertion or deletion mutations (indels or gaps) that appear as hyphens in one or more of the sequences in the alignment. Multiple sequence alignment is often used to assess sequence conservation of protein domains, tertiary and secondary structures, and even individual amino acids or nucleotides.
Browsable database for biological use
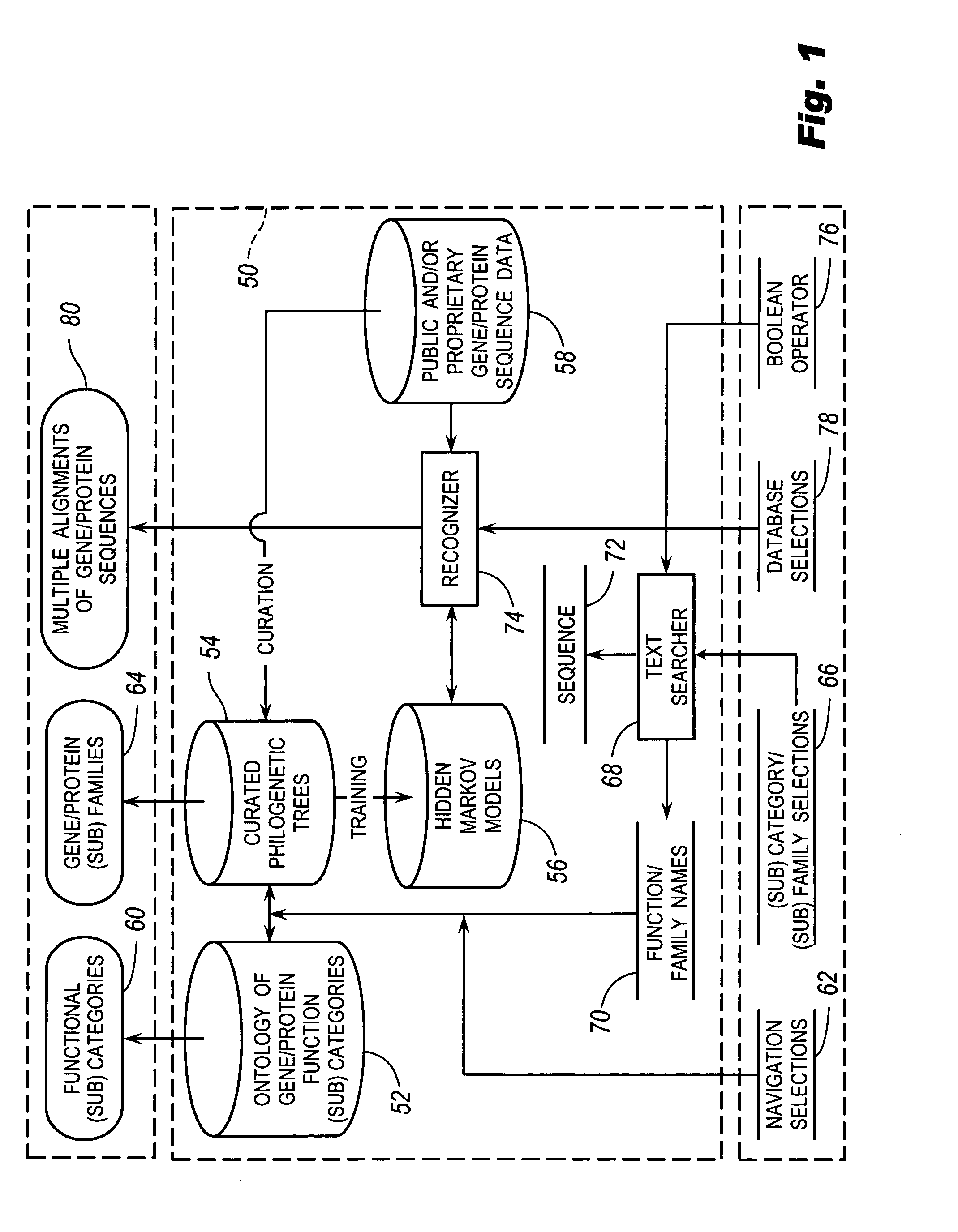
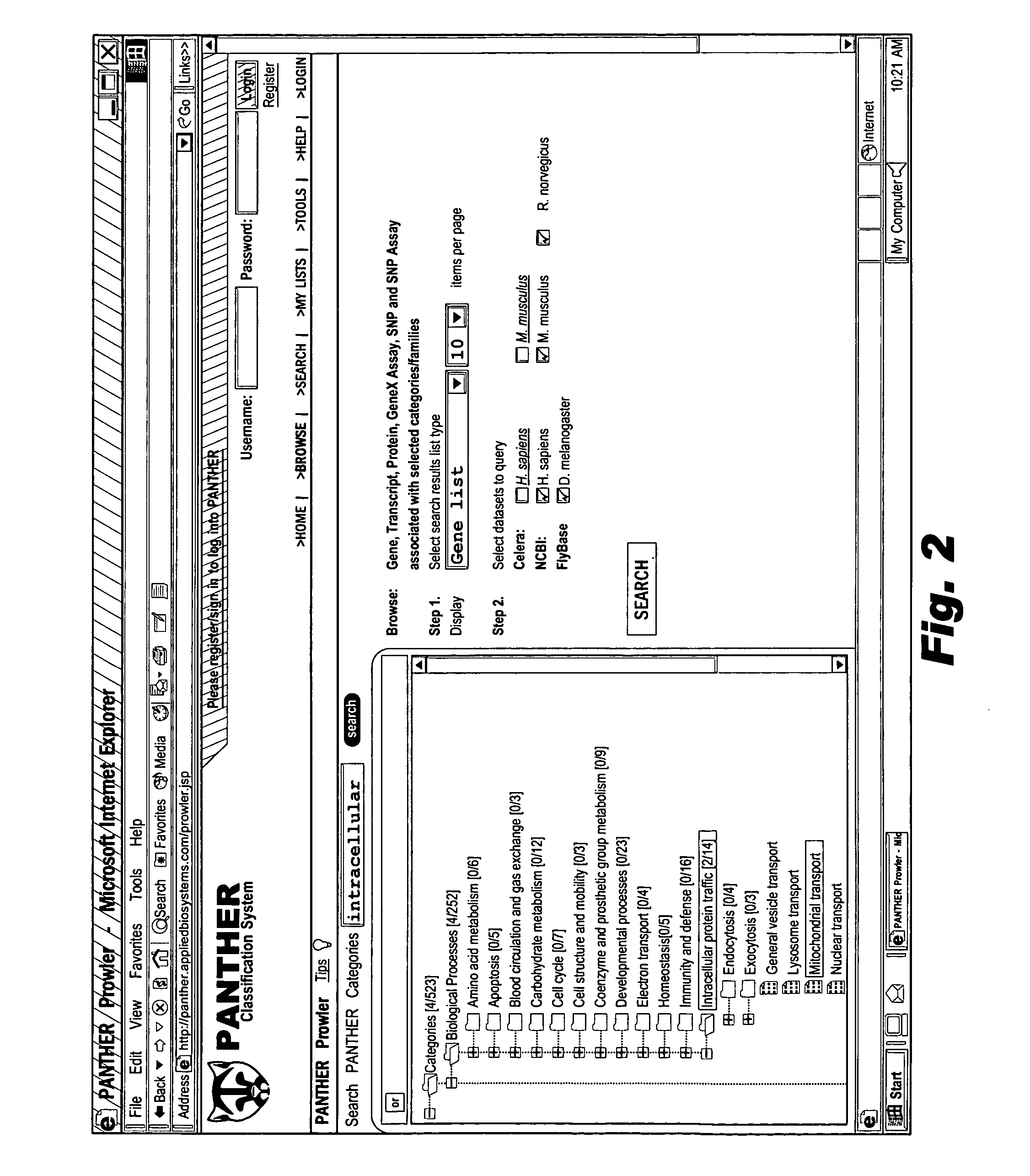
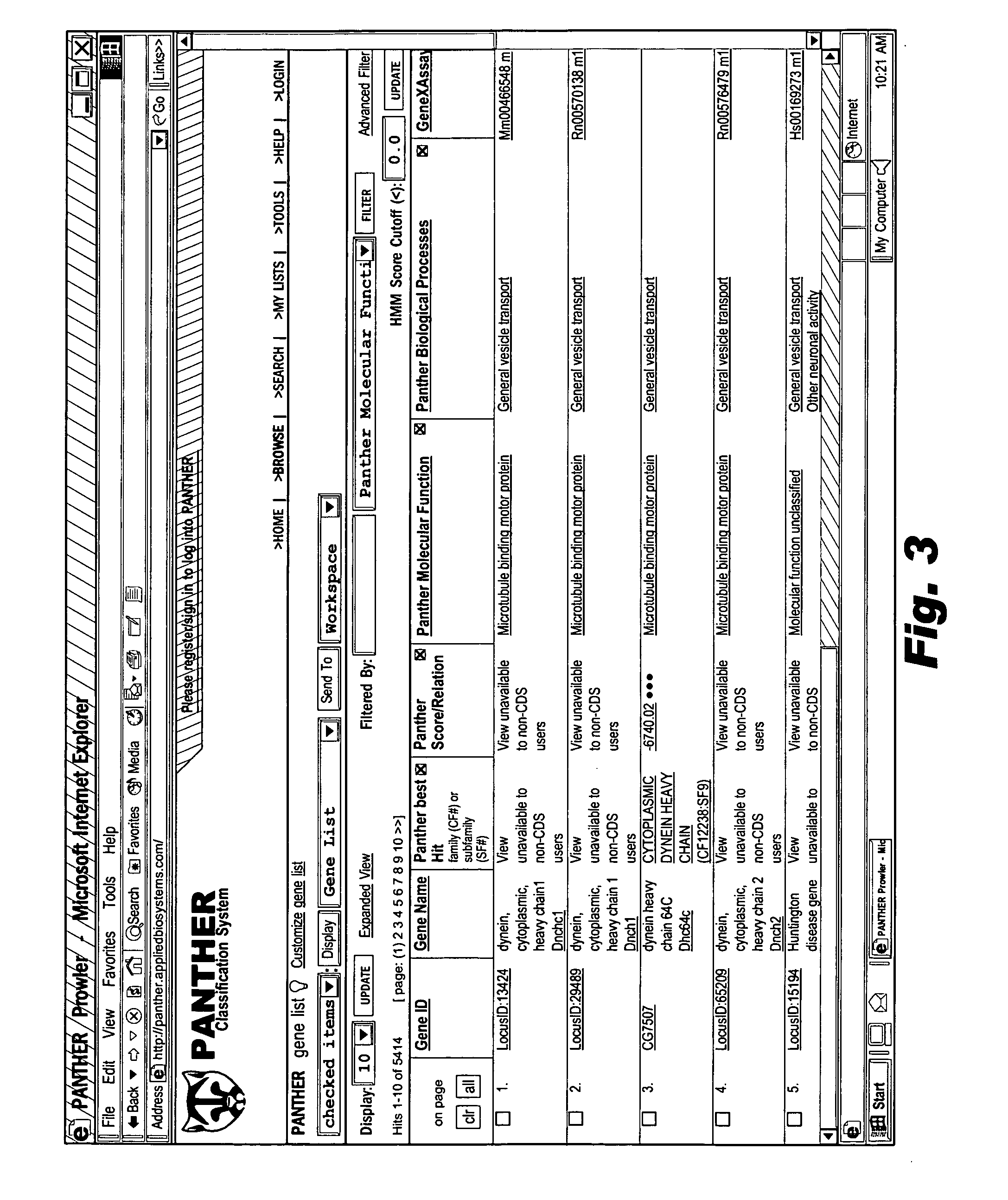
The browsable database can allow for high-throughput analysis of protein sequences. One helpful feature may be a simplified ontology of protein function, which allows browsing of the database by biological functions. Biologist curators may have associated the ontology terms with Hidden Markov Models (HMMs), rather than individual sequences, so that they can be applied to additional sequences. To ensure accurate functional classification, HMMs may be constructed not only for families, but for curator-defined subfamilies, whenever family members have divergent functions or nomenclature. Multiple sequence alignments and phylogenetic trees, including curator-assigned information, can be available for each family. Various versions of the browsable database may include training sequences from all organisms in the GenBank non-redundant protein database, and the HMMs can be used to classify gene products across the entire genomes of human, and Drosophila melanogaster.
Owner:APPL BIOSYSTEMS INC
Method of application classification in Tor anonymous communication flow
ActiveCN104135385AReduce loadImplement application classificationData switching networksTraffic capacitySequence alignment algorithm
The invention discloses a method of application classification in Tor anonymous communication flow, which mainly solves the problem of acquisition of upper-layer application type information in the Tor anonymous communication flow and relates to the correlation technique, such as feature selection, sampling preprocessing and flow modeling. The method comprises the following steps of: firstly, defining a concept of a flow burst section by utilizing a data packet scheduling mechanism of Tor, and serving a volume value and a direction of the flow burst section as classification features; secondly, preprocessing a data sample based on a K-means clustering algorithm and a multiple sequence alignment algorithm, and solving the problems of over-fitting and inconsistent length of the data sample through the manners of value symbolization and gap insertion; and lastly, respectively modeling uplink Tor anonymous communication flow and downlink Tor anonymous communication flow of different applications by utilizing a Profile hidden Markov model, providing a heuristic algorithm to establish the Profile hidden Markov model quickly, during specific classification, substituting features of network flow to be classified into the Profile hidden Markov models of different applications, respectively figuring up probabilities corresponding to an uplink flow model and a downlink flow model, and deciding the upper-layer application type included by the Tor anonymous communication flow to be classified through a maximum joint probability value.
Owner:南京市公安局
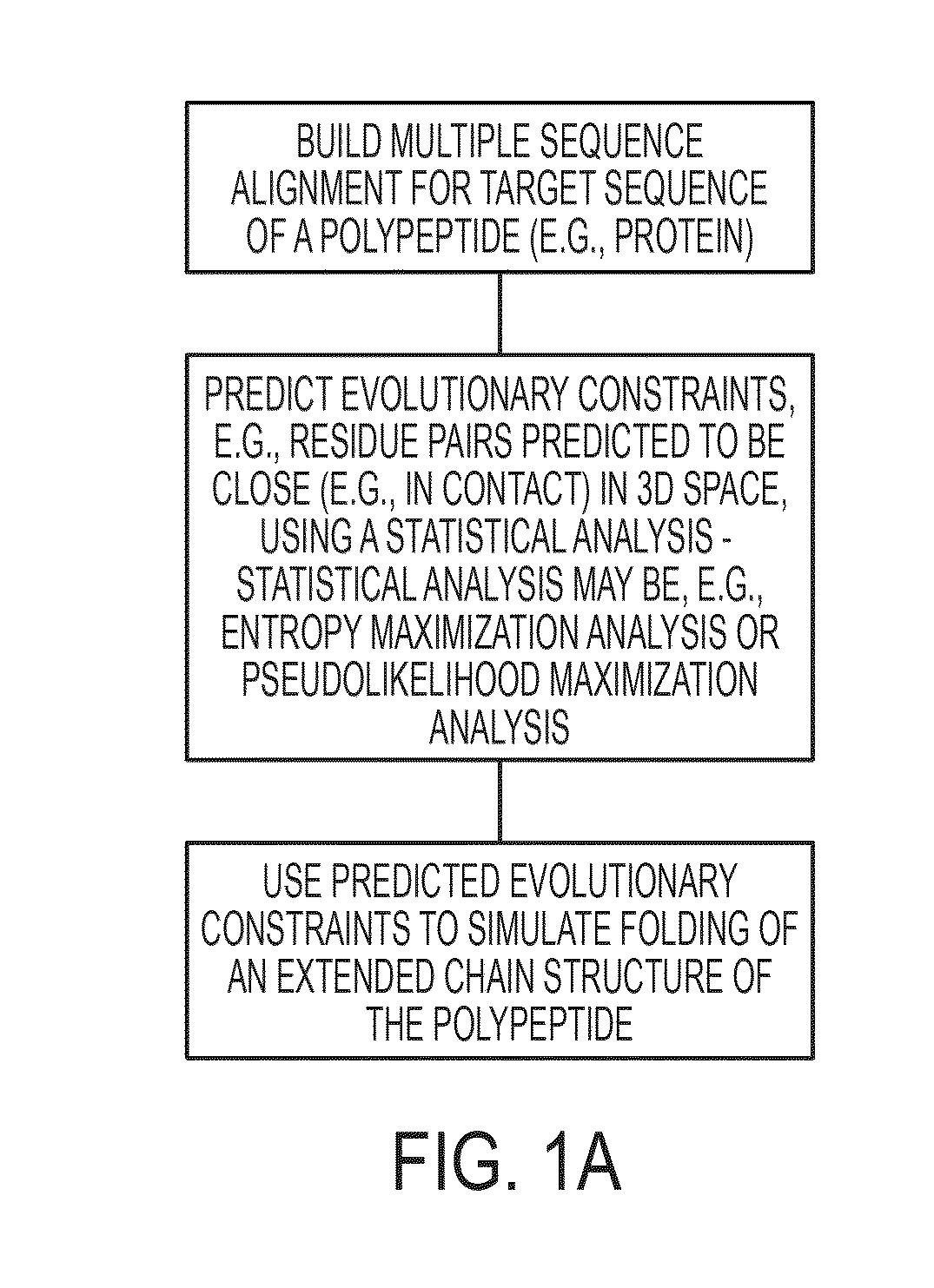
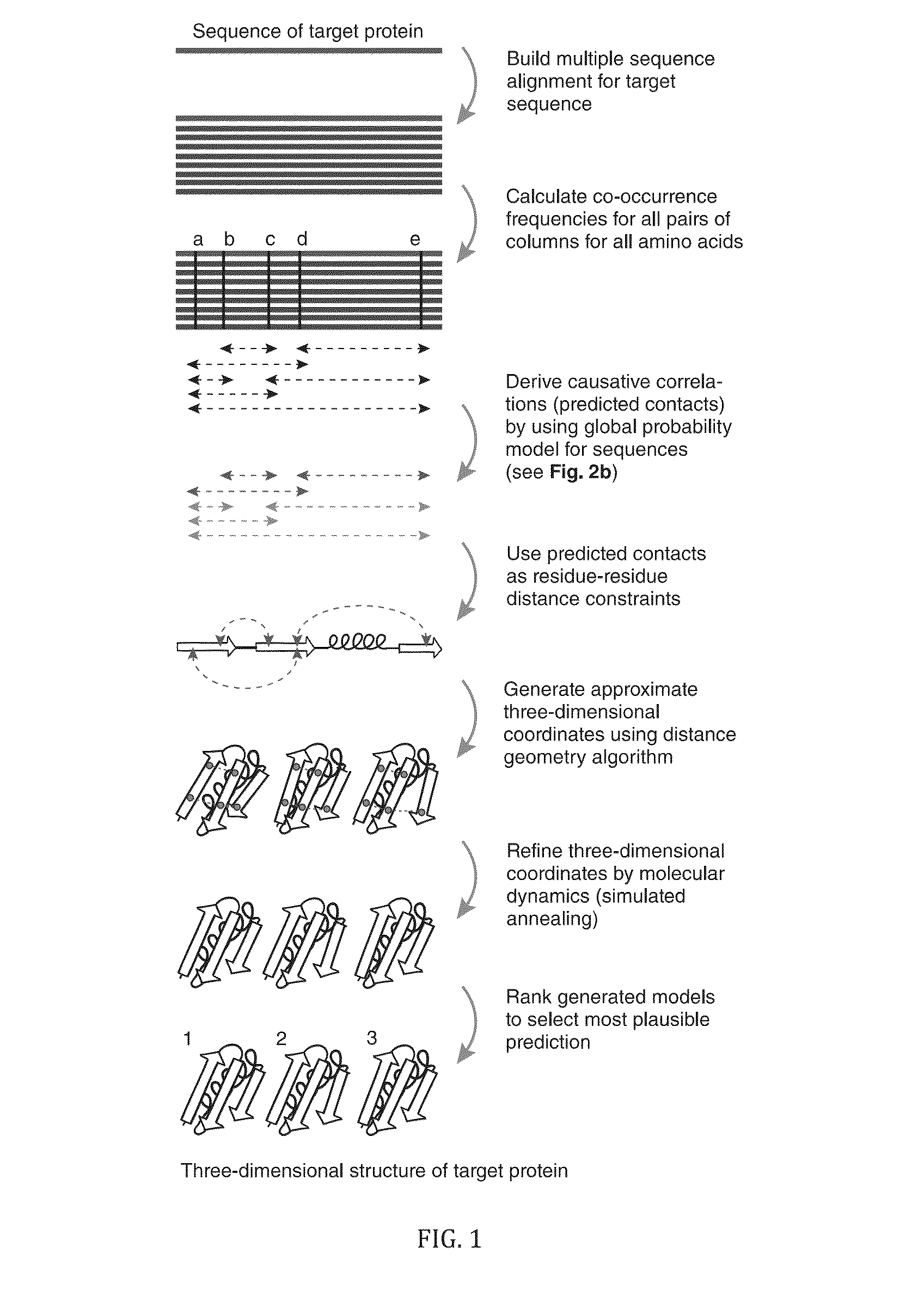
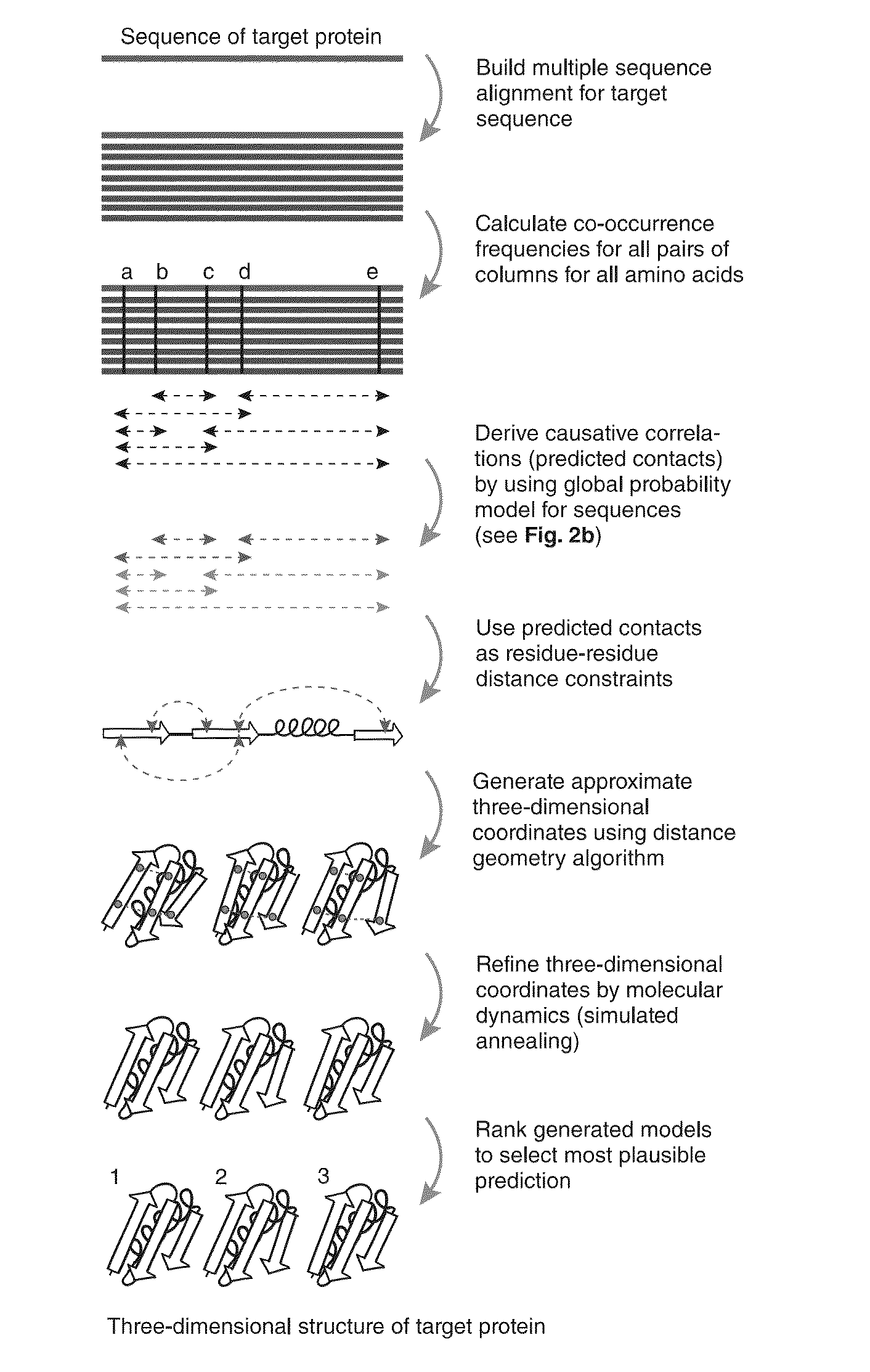
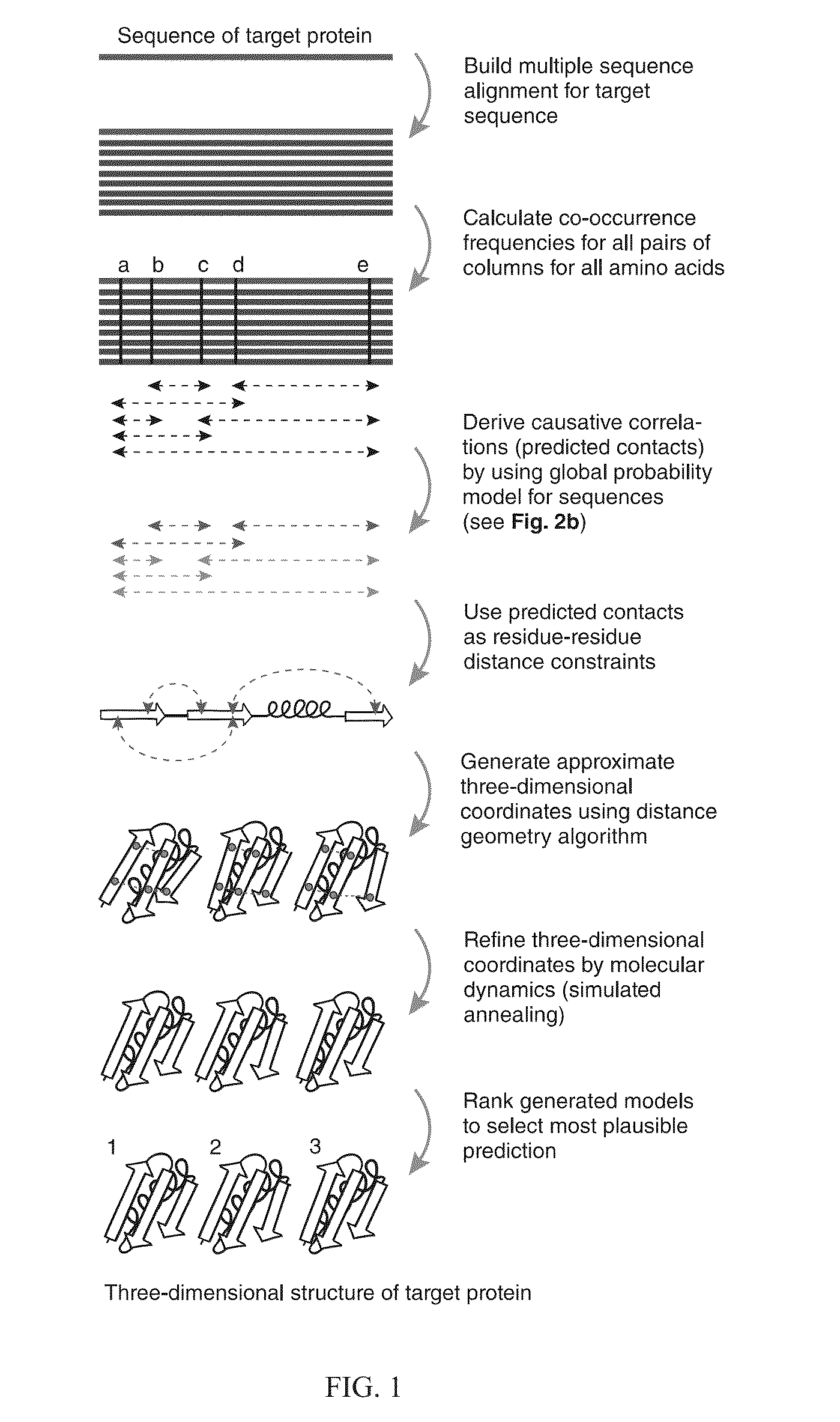
Methods and apparatus for predicting protein structure
InactiveUS20130303387A1Remove constraintsLibrary screeningAnalogue computers for chemical processesChain structureProtein structure
The present invention relates to a method for predicting three-dimensional structure of a protein from its sequence. Three-dimensional structure may be determined by: (a) generating a multiple sequence alignment for a candidate protein having a known sequence; (b) identifying a covariance matrix between all pairs of sequence positions in the multiple sequence alignment; (c) inverting the covariance matrix and identifying predicted evolutionary constraints using a statistical model of the candidate protein; and (d) simulating folding of an extended chain structure of the candidate protein using the predicted constraints.
Owner:SLOAN KETTERING INST FOR CANCER RES +1
Newcastle disease virus/avian influenza virus H9 subtype/infectious bronchitis virus triplex fluorescence quantification detection reagent and detection method
InactiveCN105671204AHigh sensitivityStrong specificityMicrobiological testing/measurementMicroorganism based processesFluorescenceQuarantine
The invention relates to a newcastle disease virus / avian influenza virus H9 subtype / infectious bronchitis virus triplex fluorescence quantification detection reagent and a detection method and belongs to the technical field of animal quarantine. A newcastle disease virus M gene coding region specific sequence, an avian influenza virus H9 subtype H gene coding region specific sequence and a chicken infectious bronchitis virus M gene coding region specific sequence are selected as target regions, and on the basis of multi-sequence comparison, primer and probe design is conducted. The length of primers is about 20 basic groups, the GC content is 50-60%, a two-stage structure and repeatability do not exist in the primers, no complementary sequence exists between the primers or in the primers, and the melting temperature (Tm value) difference between the primers is smaller than 5 DEG C. In order to guarantee universal use of a newcastle disease virus probe, the length of the probe is only 13 basic groups, the probe is modified by LAN, and the Tm value of the probe is increased. The lengths of the other two virus probes are both about 25 basic groups, and the Tm values are about 5 DEG C higher than those of the primers.
Owner:山东省动物疫病预防与控制中心 +1
Methods and apparatus for predicting protein structure
InactiveUS20130304432A1Remove constraintsComputation using non-denominational number representationSequence analysisChain structureProtein structure
The present invention relates to a method for predicting three-dimensional structure of a protein from its sequence. Three-dimensional structure may be determined by: (a) generating a multiple sequence alignment for a candidate protein having a known sequence; (b) identifying a covariance matrix between all pairs of sequence positions in the multiple sequence alignment; (c) inverting the covariance matrix and identifying predicted evolutionary constraints using a statistical model of the candidate protein; and (d) simulating folding of an extended chain structure of the candidate protein using the predicted constraints.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE +1
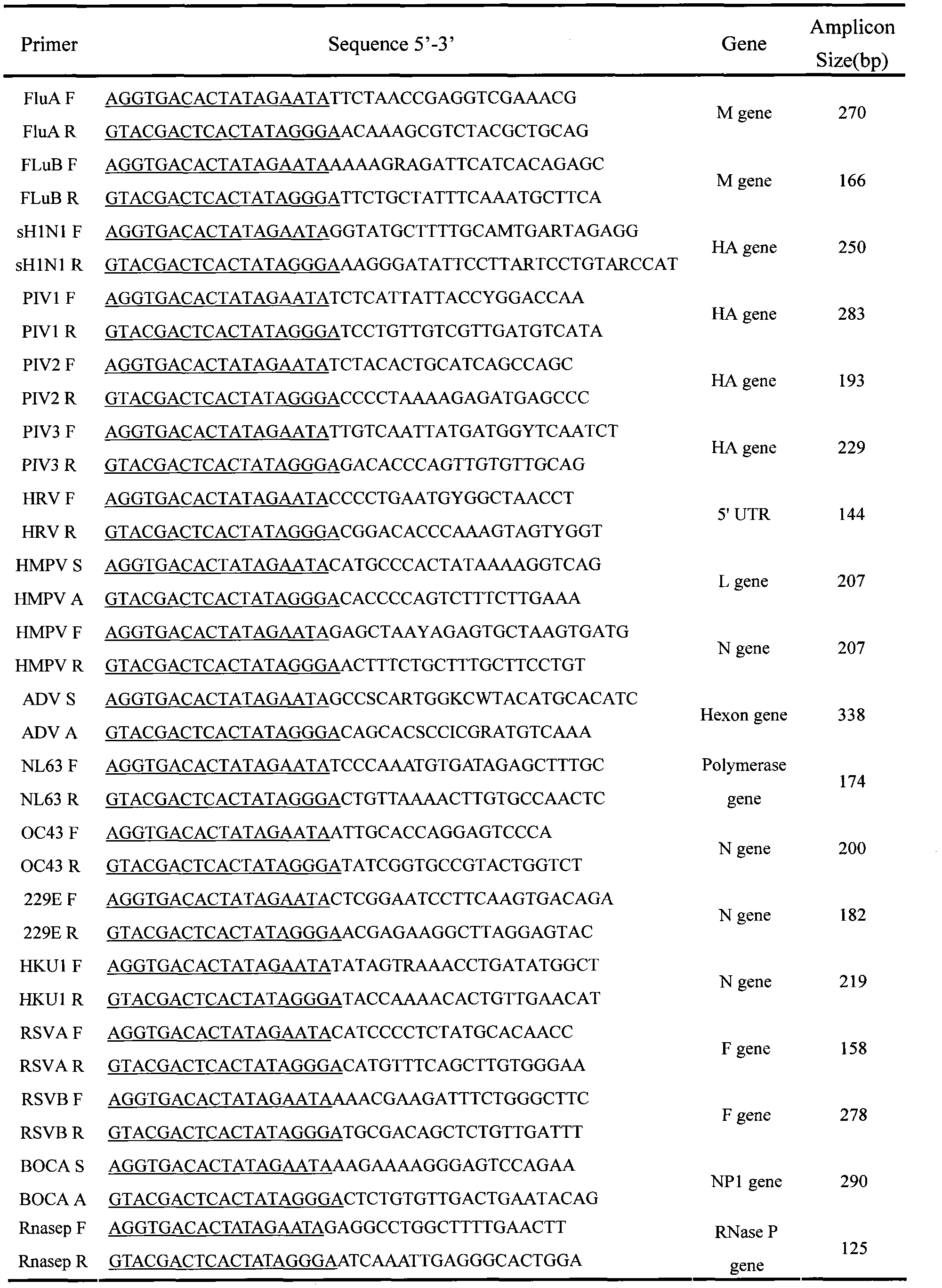
Application of GeXP multiplex gene expression genetic analysis system in genotyping of 16 common respiratory viruses
InactiveCN102839223AMicrobiological testing/measurementMicroorganism based processesDiseaseNucleotide
The invention belongs to the biotechnology application field, and relates to simultaneous detection and genotyping of infections of 16 respiratory viruses (including FluA, FluB, sH1N1, PIV1, PIV2, PIV3, RSVA, RSVB, HRV, HMPV, HBoV, CoV NL63, CoV OC43, CoV 229E, CoV HKU1 and Adv) of nasopharyngeal extract specimens of patients of respiratory-related diseases of all levels of disease prevention and control institutions and sentinel hospitals. Specifically, nucleotide sequences of representative strains of the 16 respiratory viruses are downloaded from NCBI; through literature review and multiple sequence alignment, pathogen relatively-conservative regions are determined, and multiplex specific primers are designed. Single-tube multiplex (18 stages) PCR detection is carried out for detecting the 16 respiratory virus conservative regions, and an entire reaction takes less than 2 hours. According to the invention, a defect that genotyping cannot be carried out with conventional single-tube multiplex fluorescent qualitative PCR detections can be overcome, and defects of complicated operations, long time, and high cost of conventional chip detection methods can be overcome. With the application provided by the invention, a novel idea is provided for respiratory virus genotyping technologies. With characteristics of high specificity, high sensitivity, and high speed, powerful technological support is provided for rapid and accurate screening and genotyping of the respiratory-disease-related viruses. The invention has important significance upon the researches of respiratory-tract patient infection pathogen spectrum of out nation, and upon molecular epidemiological investigations.
Owner:中国疾病预防控制中心病毒病预防控制所
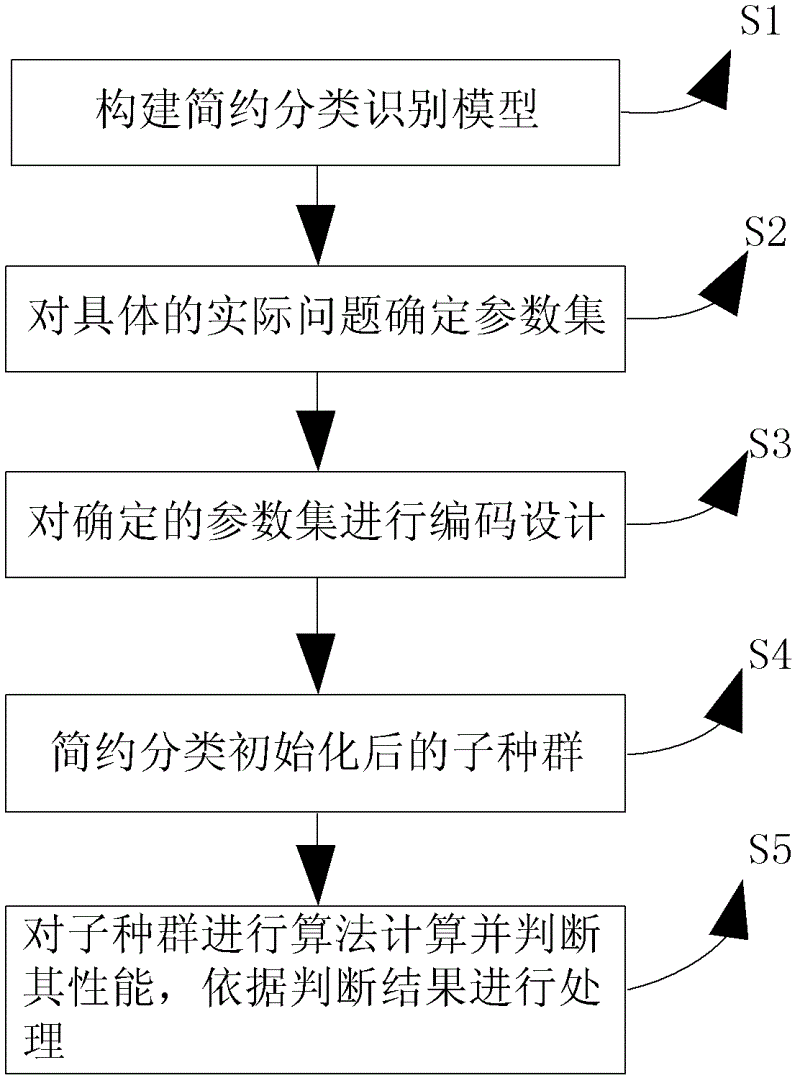
Processing method and processing device based on multiple sequence alignment genetic algorithm
InactiveCN102622535AParsimonious classificationImprove processing efficiencyGenetic modelsSpecial data processing applicationsLocal optimumSub populations
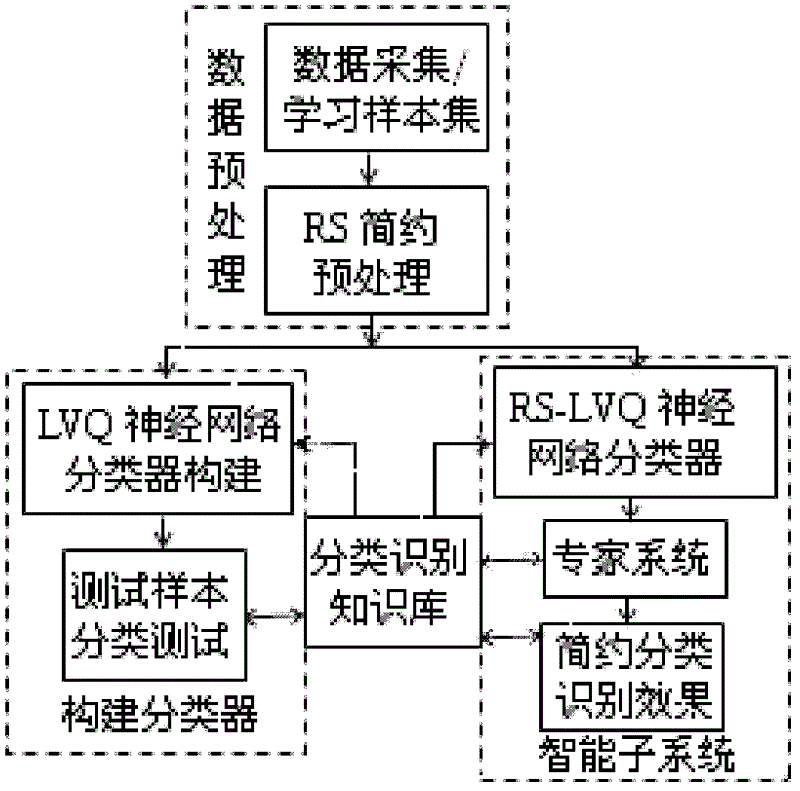
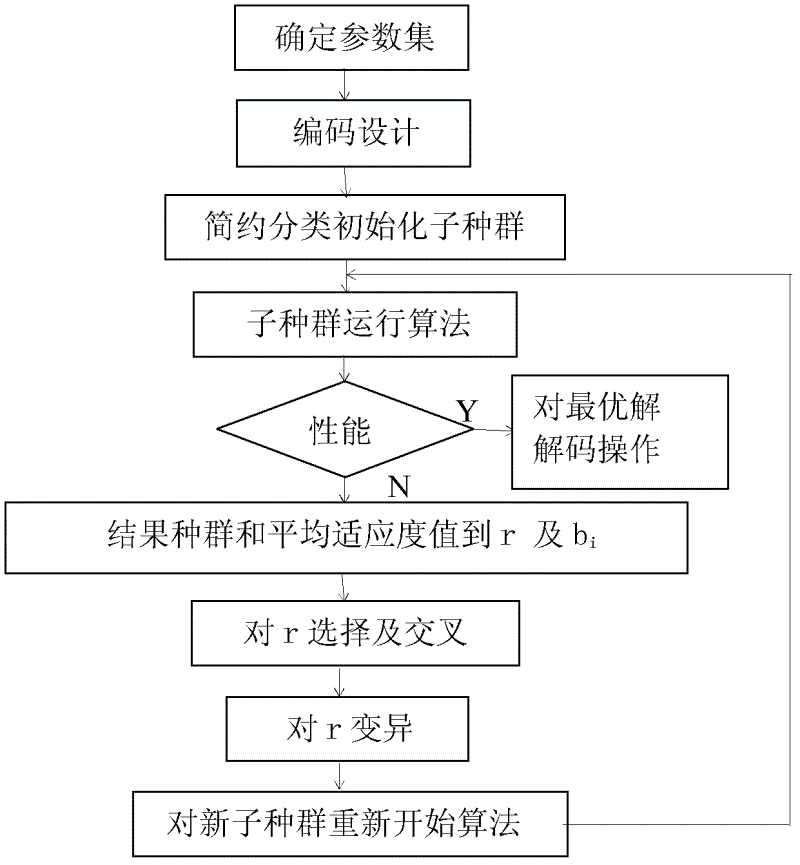
The invention provides a processing method and a processing device based on a multiple sequence alignment genetic algorithm. The processing method includes: building a brief classification identification model, determining a parameter set for special practical problems, performing code design on the determined parameter set, briefly classifying an initialized sub population, performing arithmetic calculation for the sub population and judging performance of the sub population, and processing the sub population according to judging results. The scheme is favorable for efficient comparison identification, overcomes shortcomings that the genetic algorithm is slow in convergence and apt to fall into local optimum and shortcomings that the existing attribute reduction algorithm is high in calculation complexity, not suitable for scale data reduction, not enough in describing of attribute collection and the like, and improves processing efficiency.
Owner:SHANGHAI DIANJI UNIV
Lipase mutant with improved heat stability as well as preparation method and application thereof
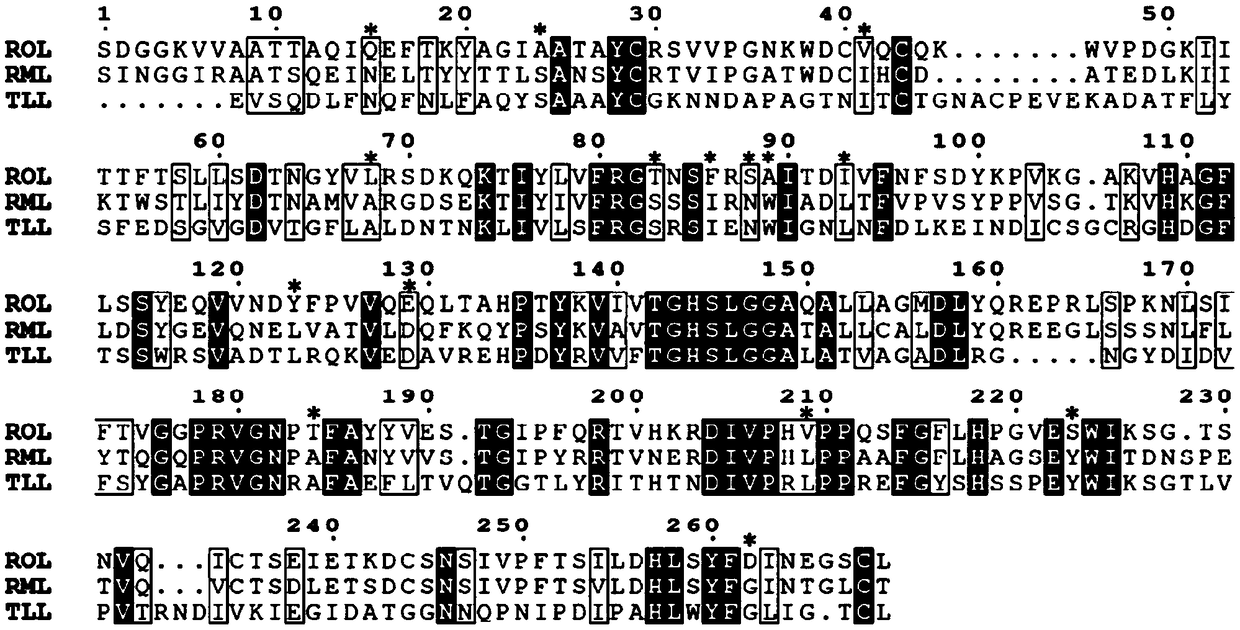
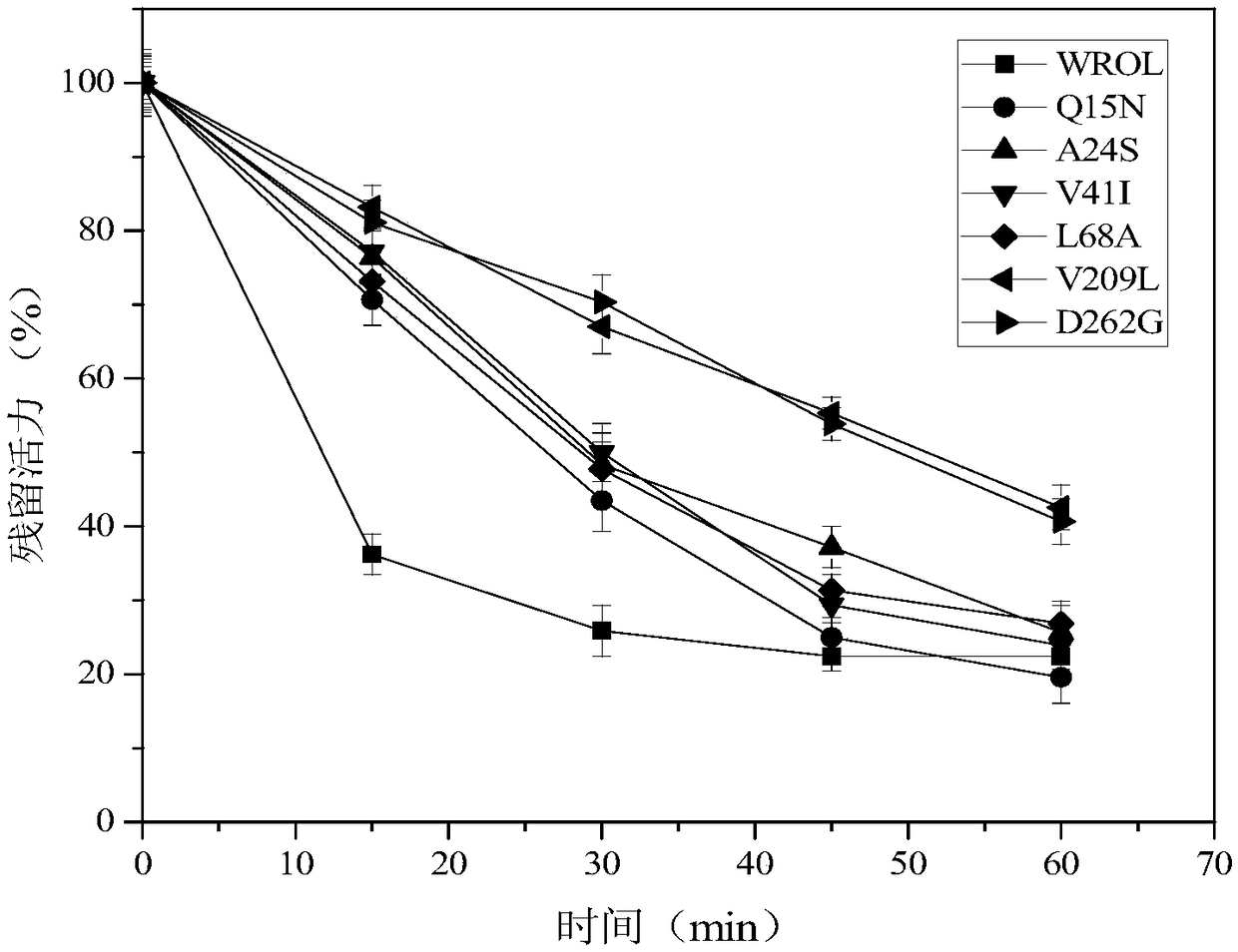
The invention discloses a lipase mutant with improved heat stability as well as a preparation method and application thereof, and belongs to the technical field of biology. The amino acid sequence ofthe lipase mutant is as shown in SEQ ID NO.1 or SEQ ID NO.3 or SEQ ID NO.5 or SEQ ID NO.7 or SEQ ID NO.9 or SEQ ID NO.11 or SEQ ID NO.13. Through multiple sequence alignment and the disulfide bond predicting result, rhizopus oryzaelipase ROL is subjected to heat stability transformation, so that the heat stability of the rhizopus oryzaelipase ROL is greatly improved; meanwhile, by virtue of a computer simulating technology, the heat stability improving principle is explained in molecular level. The heat stability of the lipase mutant provided by the invention is significantly improved; and through combination with the high Sn-1,3 selectivity of the lipase mutant, the lipase mutant has industrialized application value.
Owner:浙江容锐科技有限公司
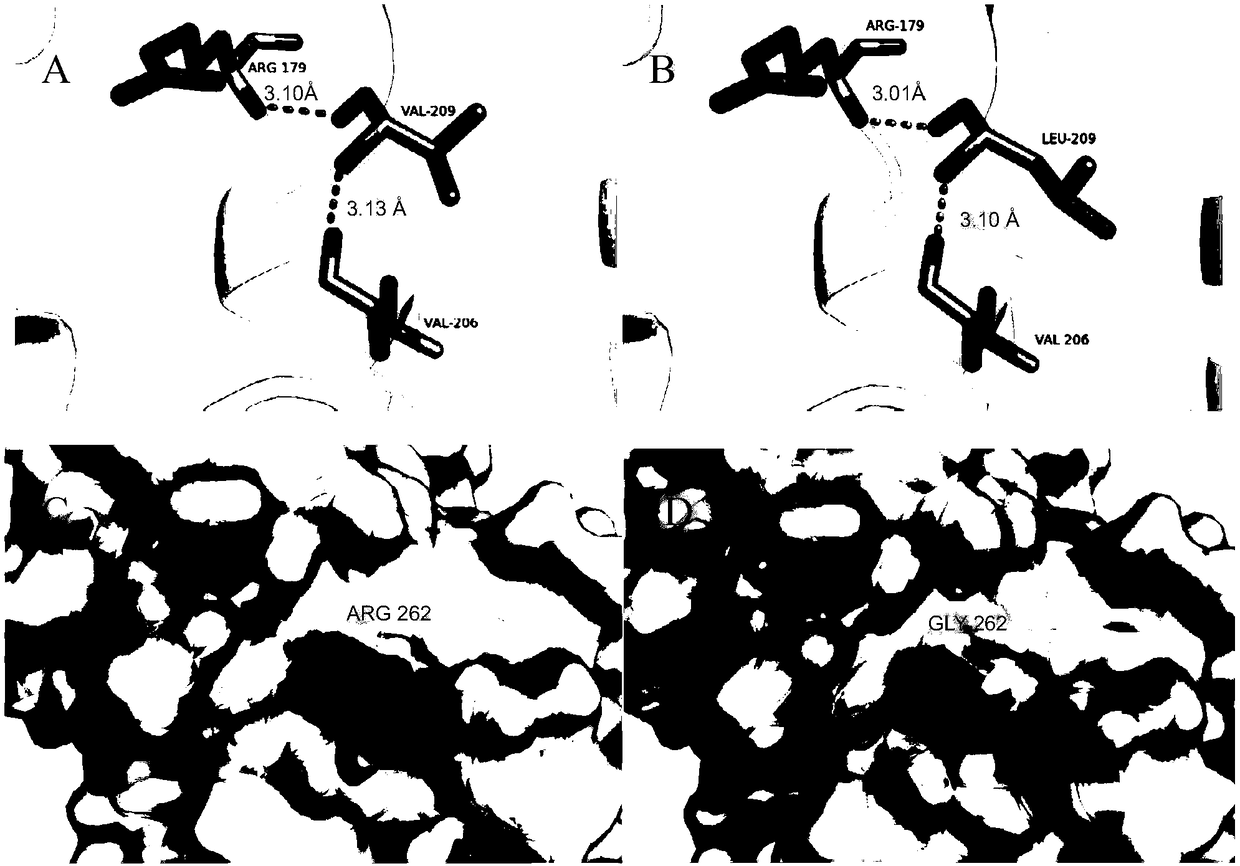
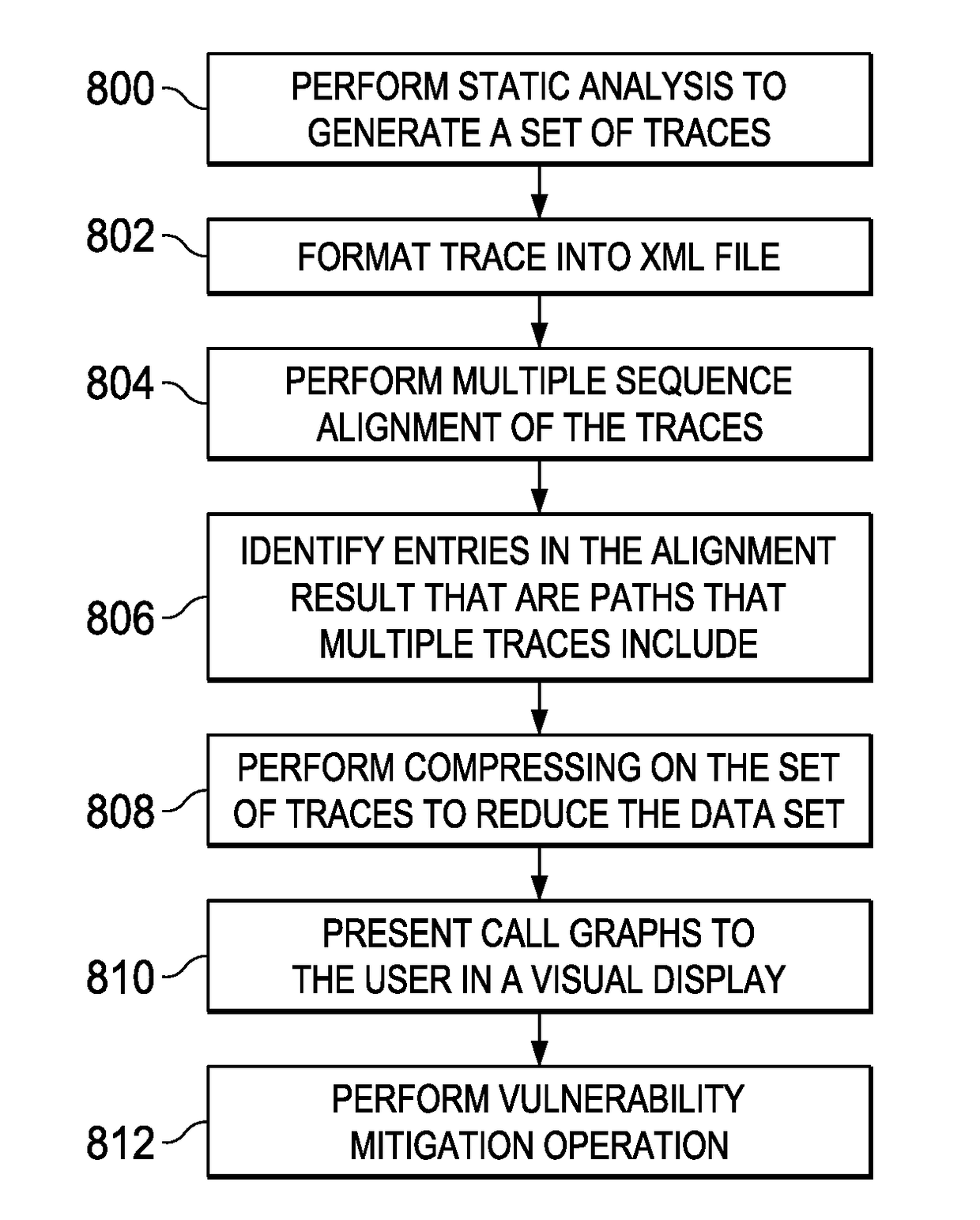
Using multiple sequence alignment to identify security vulnerability code paths
ActiveUS20170337123A1Enhanced security vulnerability determinationSimple processReverse engineeringSoftware testing/debuggingCode trackingMultiple sequence alignment
A static analysis tool is augmented to provide for enhanced security vulnerability determination from generated code traces. According to this disclosure, a multiple sequence alignment is applied to a set of traces generated by static analysis of application source code. The output of this operation is an alignment result that simplifies the traces, e.g., by representing many common nodes as a single node. In particular, the sequence alignment identifies entries in the alignment result that represent at least one code execution path that multiple traces in the set of traces include. A call graph can then be output that includes the at least one code execution path identified, and that call graph can also be simplified by applying a compression portions of the traces that are used to generate it. Using multiple sequence alignment and simplified call graphs enable a user to identify security vulnerabilities more efficiently.
Owner:IBM CORP
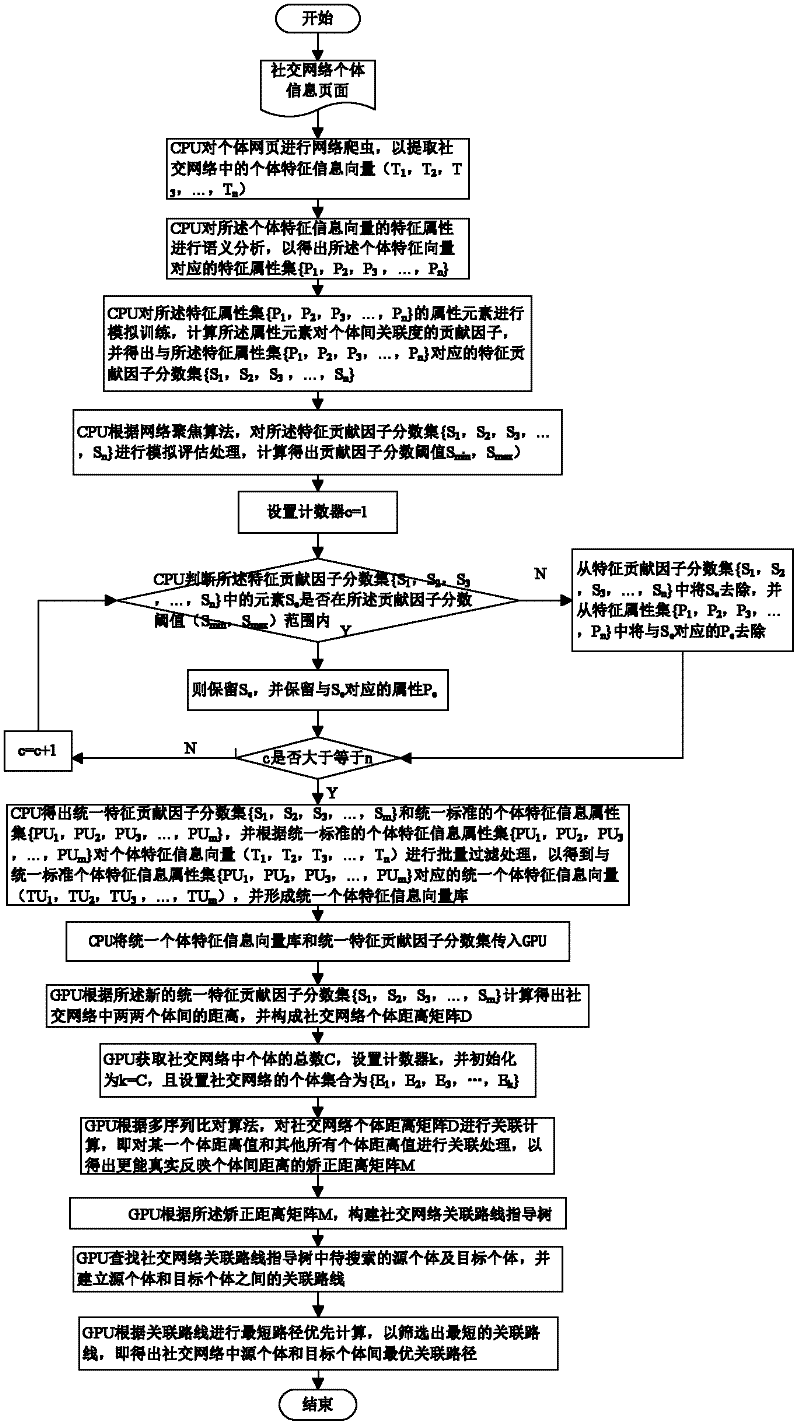
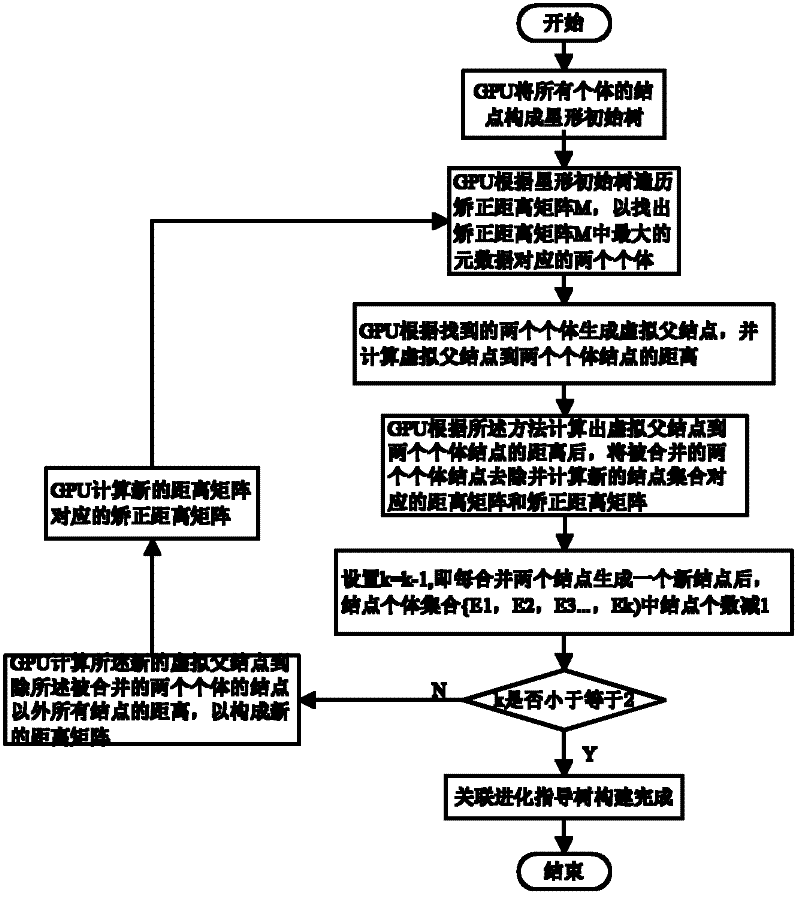
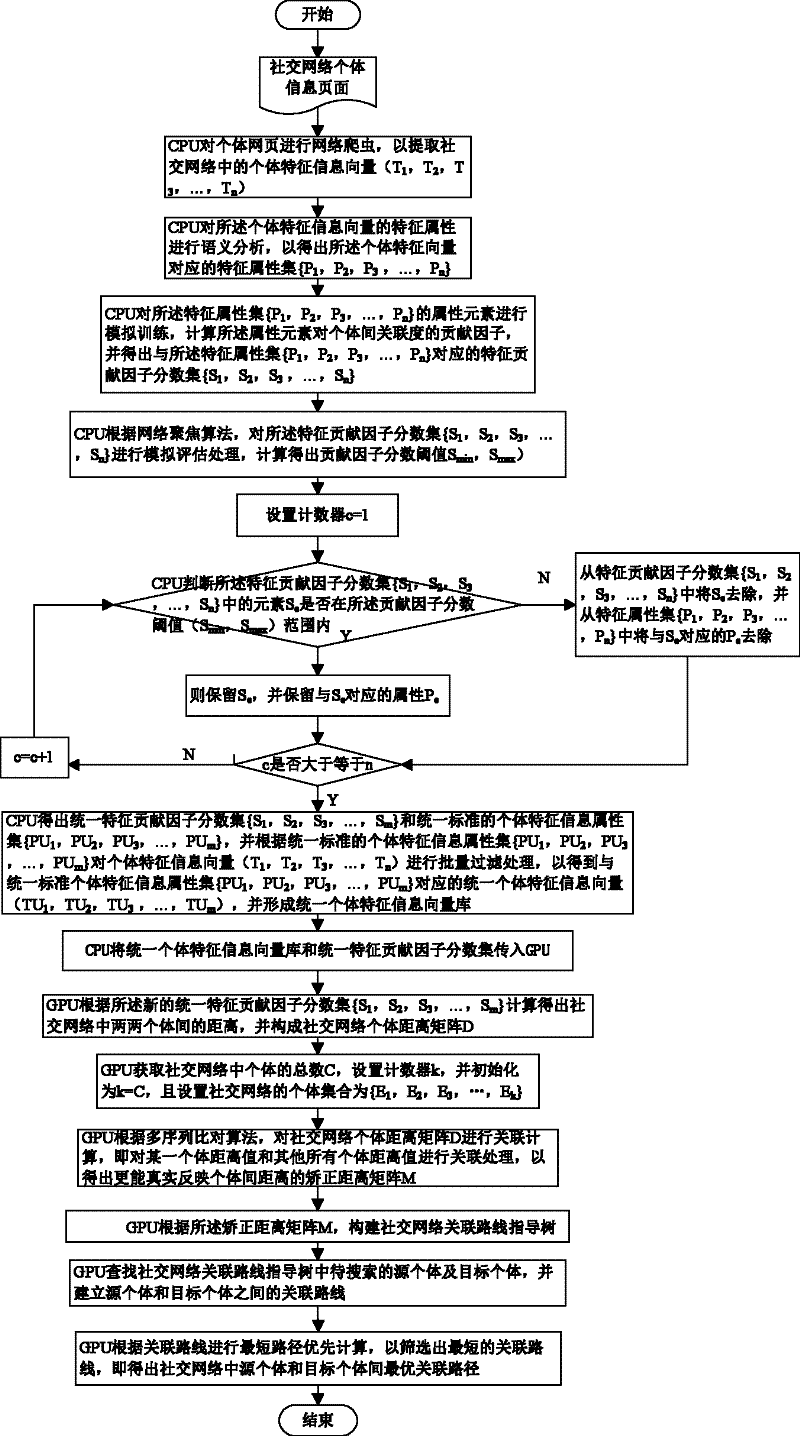
Social network association searching method based on graphics processing unit (GPU) multiple sequence alignment algorithm
InactiveCN102651030ASolve the large amount of dataSolve complexitySpecial data processing applicationsSearch problemDistance matrix
The invention discloses a social network association searching method based on a graphics processing unit (GPU) multiple sequence alignment algorithm. The method comprises the following steps that: a central processing unit (CPU) performs web crawler on an individual webpage so as to extract an individual characteristic vector from a social network; the CPU filters redundant characteristic information from the individual characteristic vector so as to generate a uniform individual characteristic information vector base; a GPU calculates an individual distance matrix and a correction distance matrix of the social network according to the uniform individual characteristic information vector base; the GPU establishes a social network association route guidance tree according to the correction distance matrix; and the GPU traverses the social network association route guidance tree so as to perform the optimal association route searching. By utilizing the advantage that the GPU is suitable for processing a large amount of dense data, associated searching problems which are solved by the the multiple sequence alignment algorithm are parallelized, complex and time-consuming operations, such as formation and traversing of the matrixes and the association route guidance tree, are finished by the GPU, and the problem of long time caused by a large amount of social network data and operation complexity is solved.
Owner:HUAZHONG UNIV OF SCI & TECH
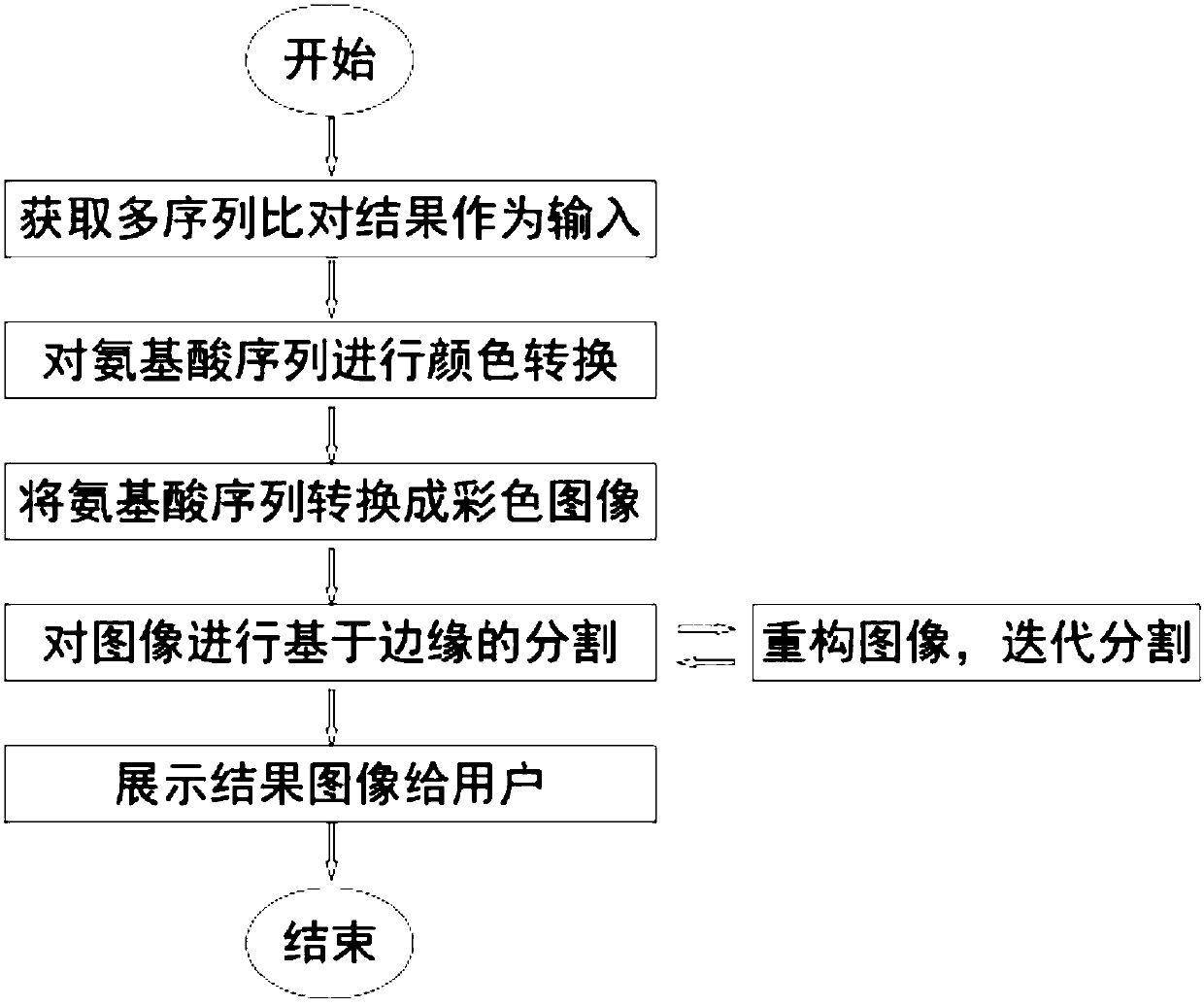
Multiple sequence alignment visualization method based on image processing
InactiveCN108052799AEliminate noise interferenceThe segmentation result is accurateImage enhancementImage analysisPattern recognitionColor transformation
The invention relates to a multiple sequence alignment visualization method based on image processing. The method includes following steps: S1, taking multiple amino acid sequences generated by a multiple sequence alignment algorithm as input; S2, respectively defining different colors for different types of amino acids, and performing color conversion on the amino acid sequences; S3, combining with image conversion to enable each amino acid in the amino acid sequences to correspond to one pixel in images, to enable color of each pixel to correspond to that of the corresponding amino acid andto convert multiple one-dimensional amino acid sequences into two-dimensional colored images; S4, utilizing an image segmentation method based on edge detection to segment converted images, and presenting segmented images to a user.
Owner:SUN YAT SEN UNIV
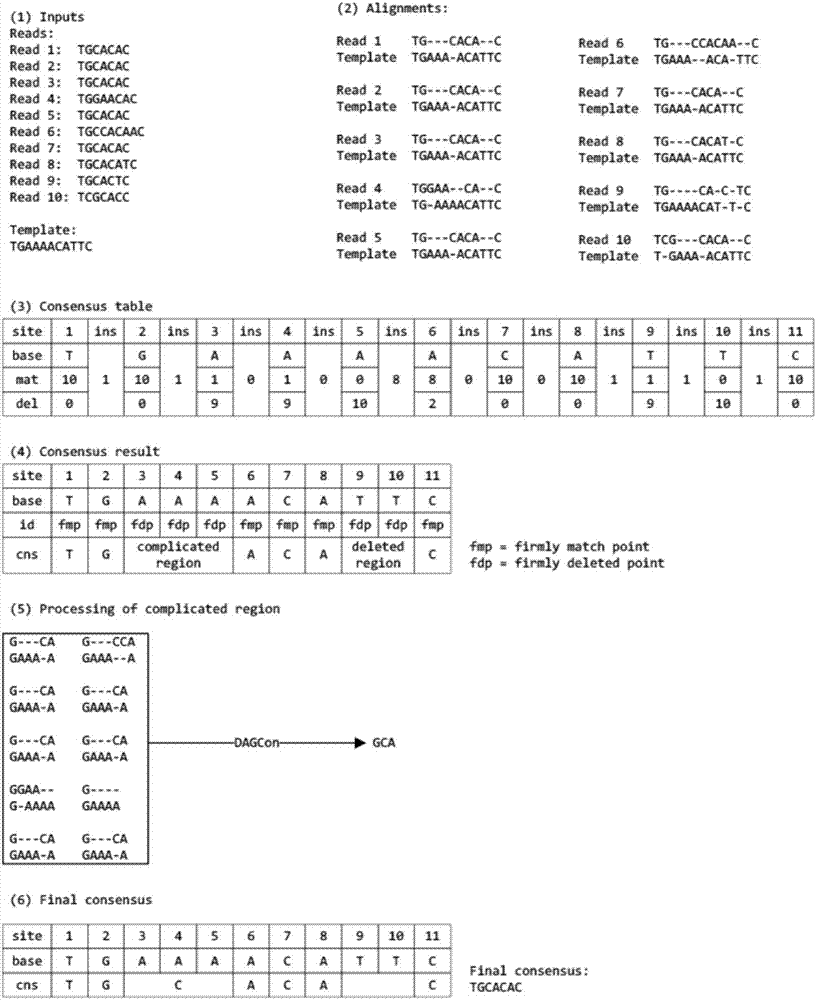
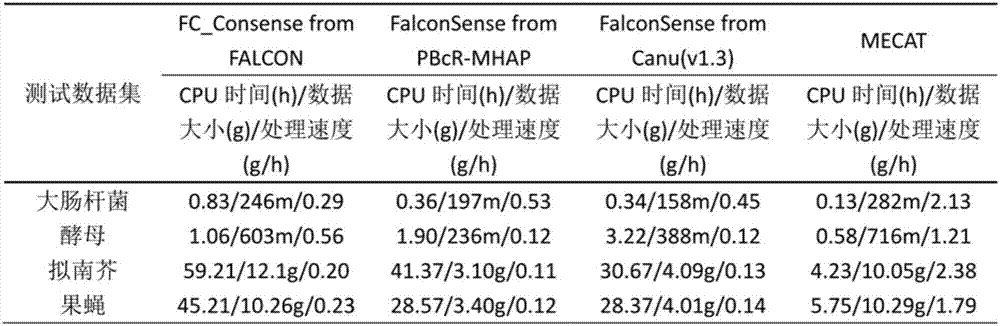
Third-generation sequencing sequence correction method based on local graph
InactiveCN107229842ASequence analysisSpecial data processing applicationsApplication softwareThird generation sequencing
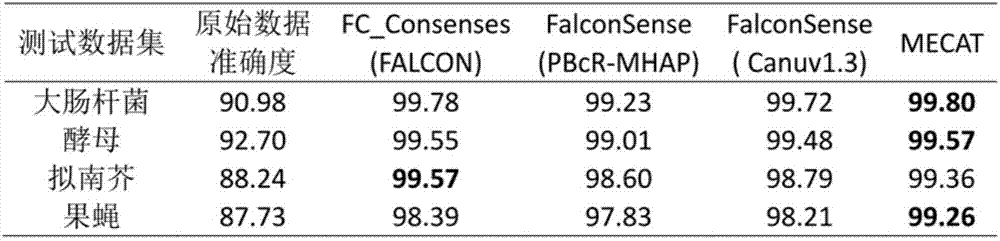
The invention relates to a third-generation sequencing sequence correction method based on a local graph and a system of the third-generation sequencing sequence correction method. The system comprises a two-two comparison module, a multi-sequence comparison module, a correction operation comparison module, a correction operation classification module, a consistent-area base position correction and complex-area local graph base sequence correction module and a module sequence correction division and defusion processing module, the two-two comparison module is connected with a single molecule real-time sequencing database and a nanopore sequencing database, and the single molecule real-time sequencing database and the nanopore sequencing database are input into the two-two comparison module separately. The precision of the third-generation sequencing sequence correction method and system can reach 99%, and the speed is 7-10 folds that of current application software.
Owner:ZHONGSHAN OPHTHALMIC CENT SUN YAT SEN UNIV
Evolutionary models of multiple sequence alignments to predict offspring fitness prior to conception
A system, device and method for receiving multiple aligned genetic sequences obtained from genetic samples of multiple organisms of one or more different species. A measure of evolutionary variation may be computed for one or more alleles at each of one or more aligned genetic loci. The aligned genetic loci in the multiple organisms may be derived from one or more common ancestral genetic loci or may be otherwise related. The measure of evolutionary variation may be a function of variation in alleles at corresponding aligned genetic loci in the multiple aligned genetic sequences. One or more likelihoods may be computed that an allele mutation at each of the one or more genetic loci in a simulated virtual progeny will be deleterious based on the measure of evolutionary variation of alleles at the corresponding aligned genetic loci for the multiple organisms.
Owner:ANCESTRY COM DNA
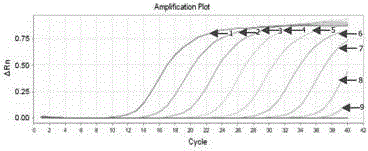
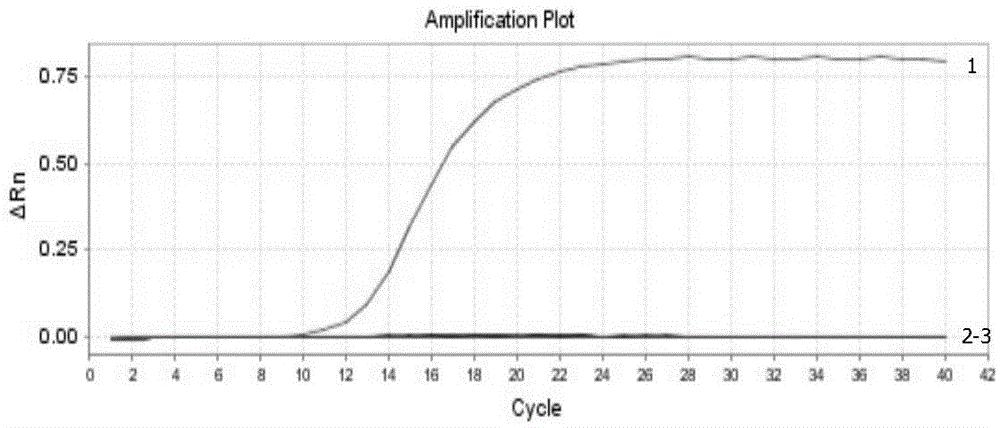
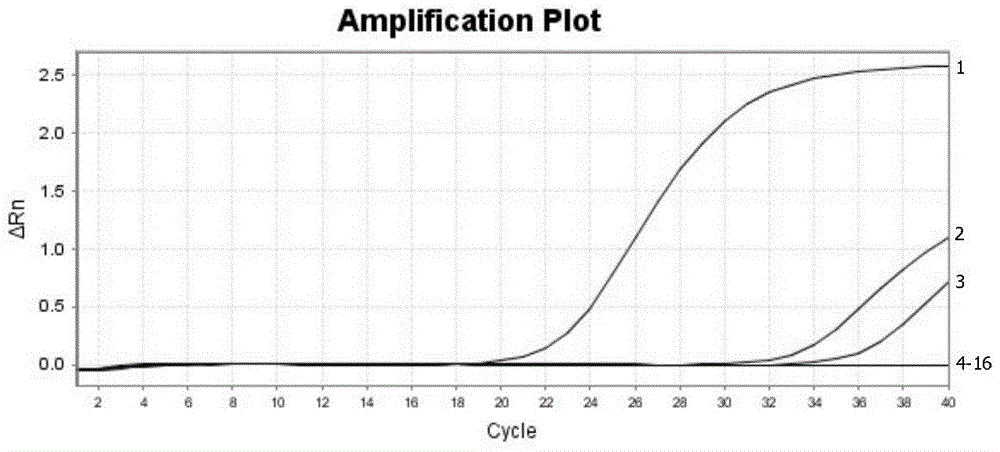
Real-time fluorescence RT-PCR detection kit for H1N1 type A swine influenza virus and application of detection kit
InactiveCN104450956AStrong specificityEasy to operateMicrobiological testing/measurementMicroorganism based processesFluorescenceTissue sample
The invention discloses a fluorescent quantitative RT-PCR detection kit for an H1N1 type A swine influenza virus, and an application of the detection kit. Through multiple sequence alignment, a primer and a probe with high specificity for detecting the H1N1 type A swine influenza virus is designed aiming at conservative gene segments of the H1N1 (2009) type A swine influenza virus, a Eurasian avian-like H1N1 swine influenza virus, a classical type H1N1 swine influenza virus and a human-derived H1N1 swine influenza virus, and is applied to real-time fluorescence RT-PCR detection. An experiment result proves that the specific PCR primer and TaqMan fluorescence probe disclosed by the invention are high in specificity when being used for detecting the H1N1 type A swine influenza virus; the sensitivity can reach 2.6*10<-5>ng; tissue samples such as nasal swabs, lungs and tracheas of to-be-detected swinery can be detected; the chick embryo allantoic fluid can also be detected; the detection kit is simple to operate and easy to popularize; basic operation and application are facilitated; and the detection kit can become a useful detection tool for diagnosis of H1N1 type A swine influenza virus diseases and epidemiological survey.
Owner:HARBIN VETERINARY RES INST CHINESE ACADEMY OF AGRI SCI
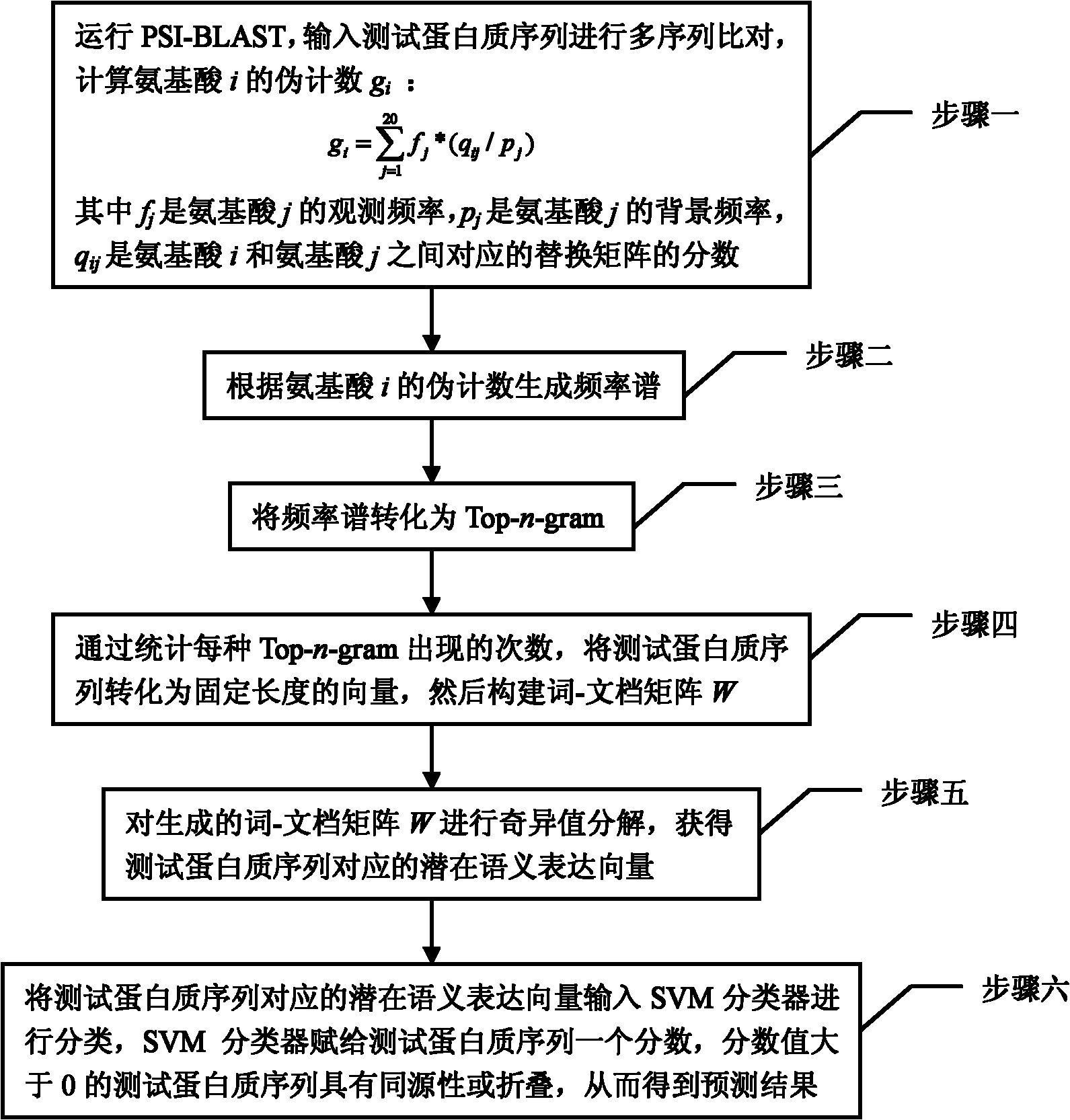
Remote protein homology detection and fold recognition method based on Top-n-gram
InactiveCN102043910AImprove predictive performanceEmphasize the importanceSpecial data processing applicationsPattern recognitionFrequency spectrum
The invention discloses a remote protein homology detection and fold recognition method based on a Top-n-gram, and relates to a remote protein homology detection and fold recognition method. The method is used for solving a problem that a binary spectrum cannot find out an optimal threshold and cannot distinguish difference of frequency of occurrences of amino acid in the prior protein remote homology detection and fold recognition method, and comprises the following steps: 1, operating a PSII-BLAST, inputting a tested protein sequence for multiple sequence alignment, and calculating a pseudocount of an amino acid i; 2, generating a frequency spectrum; 3, transforming the frequency spectrum into the Top-n-gram; 4, obtaining a latent semantic expression vector corresponding to the tested protein sequence; 5, inputting the latent semantic expression vector corresponding to the tested protein sequence into an SVM sorter for sorting, and obtaining a forecasting result. The protein remotehomology detection and fold recognition method based on the Top-n-gram is used in the filed of protein homology detection and fold recognition.
Owner:HARBIN INST OF TECH
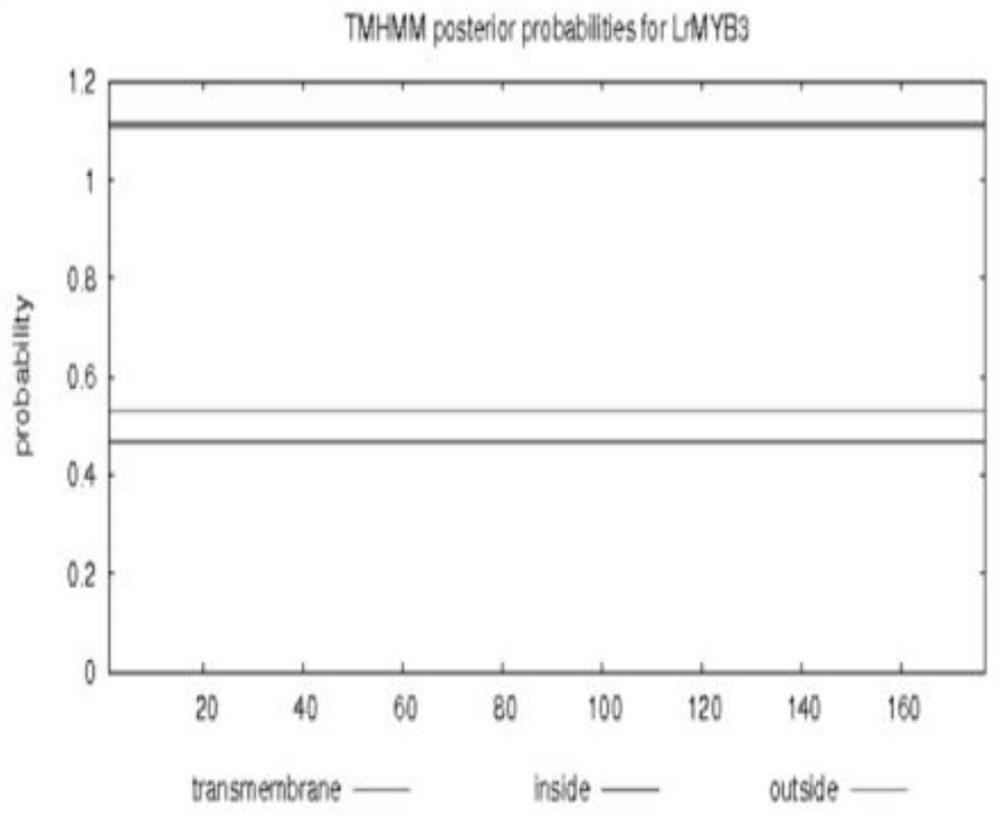
MYB transcription inhibition factor LrMYB3 related to lycium ruthenicum anthocyanin synthesis and application thereof
The invention discloses an MYB transcription inhibition factor LrMYB3 related to lycium ruthenicum anthocyanin synthesis and application thereof, and belongs to the technical field of gene engineering. According to transcriptome data of lycium ruthenicum, the MYB transcription inhibition factor LrMYB3 participating in anthocyanin synthesis is screened and cloned according to annotation results and expression quantity differences of MYB transcription factors, and belongs to a R2R3 type MYB transcription factor. Multiple sequence alignment and evolutionary tree analysis show that the transcription inhibition factor belongs to a FaMYB1-like transcription inhibition factor. QRT-PCR analysis shows that: the LrMYB3 is expressed in each tissue of the lycium ruthenicum, and the expression level is gradually increased along with ripening of the lycium ruthenicum. Subcellular localization and transcriptional activity detection experiments show that the LrMYB3 is a transcription factor which is localized in a cell nucleus and has no activation function.
Owner:WOLFBERRY SCI INST NINGXIA ACAD OF AGRI & FORESTRY SCI
Methods and apparatus for predicting protein structure
InactiveUS20160210399A1Computation using non-denominational number representationSequence analysisChain structureProtein structure
The present invention relates to a method for predicting three-dimensional structure of a protein from its sequence. Three-dimensional structure may be determined by: (a) generating a multiple sequence alignment for a candidate protein having a known sequence; (b) identifying a covariance matrix between all pairs of sequence positions in the multiple sequence alignment; (c) inverting the covariance matrix and identifying predicted evolutionary constraints using a statistical model of the candidate protein; and (d) simulating folding of an extended chain structure of the candidate protein using the predicted constraints.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE +1
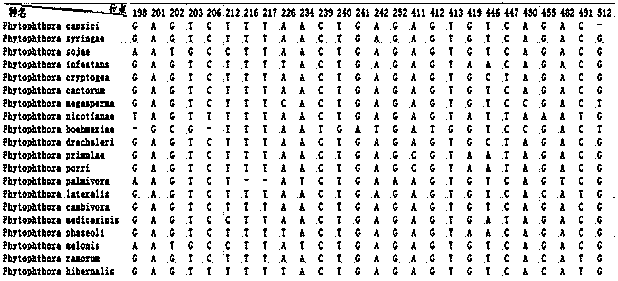
Method for detecting or auxiliarily detecting Phytophthora bacteria
InactiveCN104178576AEfficiently find base differencesThe result is accurateMicrobiological testing/measurementMicroorganism based processesBiotechnologyBase J
The invention provides a method for detecting or auxiliarily detecting Phytophthora bacteria, relating to the technical field of biology. The method comprises the following steps: carrying out multiple sequence comparison on DNA (deoxyribonucleic acid) bar code sequences of Phytophthora bacteria, detecting specific base sites of each variety, and meanwhile, obtaining a consistency sequence, wherein the specific base sites of each variety comprise differentiated base sites and SNP (single-nucleotide polymorphism) sites; comparing the DNA bar code sequence of the bacterium to be detected with the consistency sequence, and regulating the length to obtain a positioning sequence; and comparing the positioning sequence with the specific base sites of each variety, marking the degree of similarity between the bacterium to be detected and various bases, taking the maximum value, and determining that the bacterium to be detected belongs to the variety with the maximum value. The method can identify the Phytophthora bacteria, is simple to operate, and has the advantages of high accuracy, short time consumption and favorable specificity.
Owner:ANIMAL AND PLANT & FOOD DETECTION CENTER JIANGSU ENTRY EXIT INSPECTION AND QUARANTINE BUREAU +2
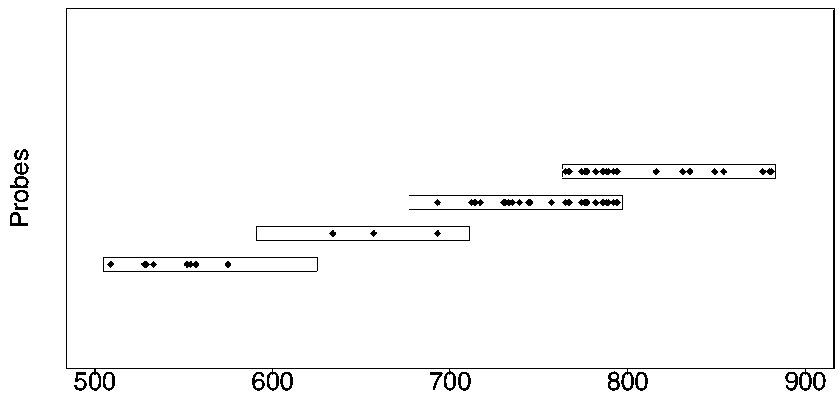
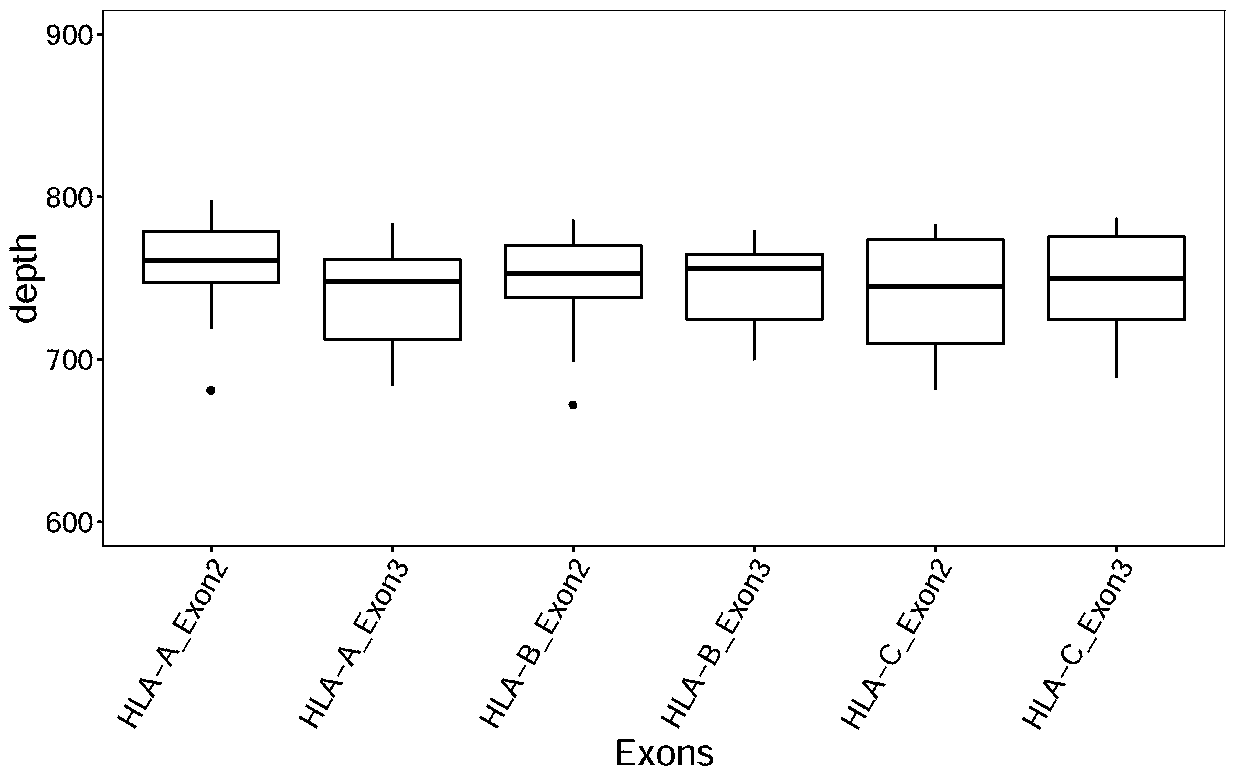
Design method of nucleic acid capture probe for HLA typing
ActiveCN110853708AEfficient captureImprove the effect of hybrid captureSequence analysisHybridisationRepresentative sequencesGene probe
The invention discloses a design method of a nucleic acid capture probe for HLA typing. The design method comprises the following steps: 1) constructing HLA-A, HLA-B and HLA-C sequence libraries; 2) performing multiple sequence alignment, and searching SNP sites on each gene Exon2 and Exon3 and upstream and downstream SNP sites; 3) selecting an area covering a set number of SNP sites as a probe design candidate area by using a sliding window separation algorithm; 4) performing clustering analysis to obtain a representative sequence of each probe design candidate region as a candidate probe; 5)repeating all the candidate probes to obtain capture probes of the three genes. The invention also discloses the probe designed by the method, and the sequence of the probe is shown as SEQ ID NO: 9-88. The nucleic acid capture probe is designed for all polymorphic sites of Exon2 and Exon3 regions with the GC proportion of 60% or above in the HLA gene, and the hybrid capture effect of the HLA geneprobe is improved.
Owner:上海仁东医学检验所有限公司
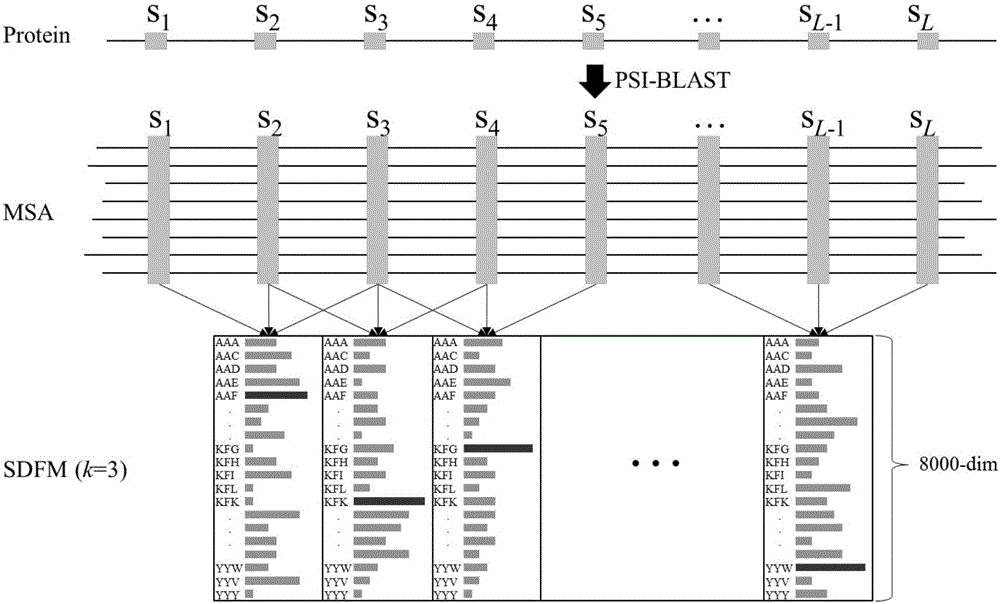
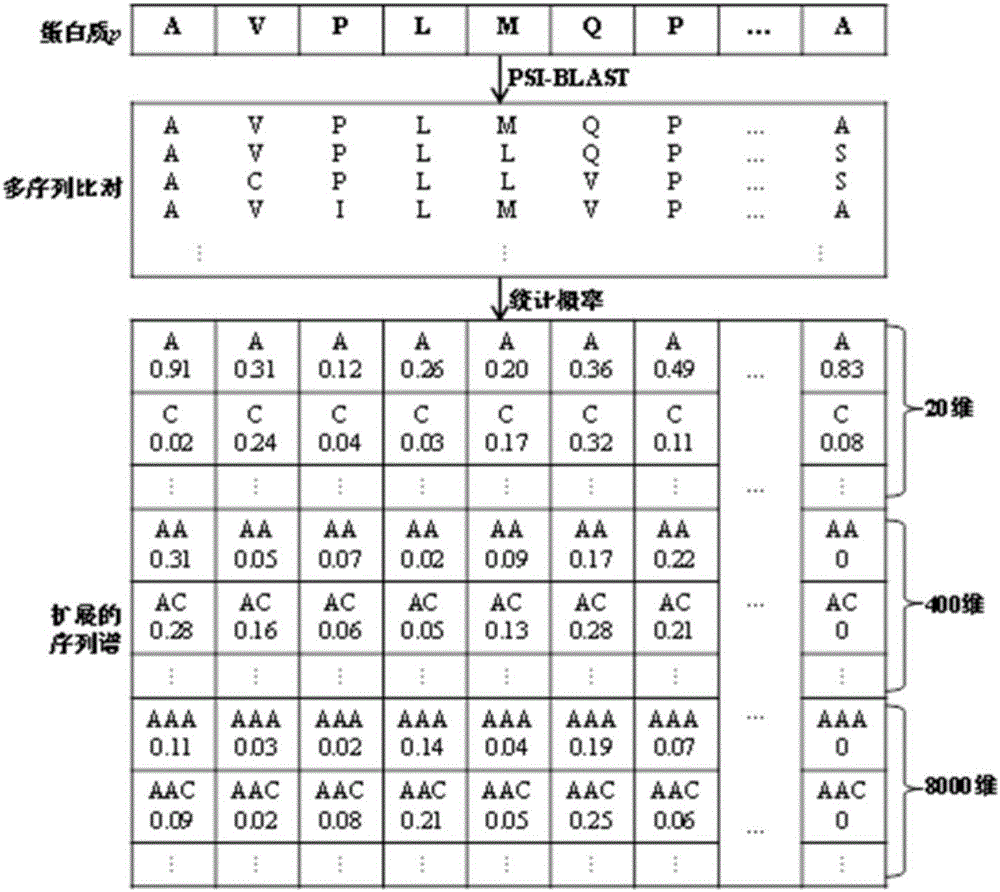
Sequence-order dependent frequency matrix-based biological sequence evolution information extraction method and application thereof
InactiveCN106529212ASequence analysisSpecial data processing applicationsSequence databaseComputer science
The invention provides a sequence-order dependent frequency matrix (SDFM)-based biological sequence evolution information extraction method. An SDFM is adopted for performing biological sequence evolution information extraction. The SDFM is obtained by adopting the following steps of for any biological sequence, firstly searching for a corresponding biological sequence database, and generating corresponding multiple sequence alignment (MSA); and secondly performing statistics on an occurrence frequency of each site biological sequence sub-string in the MSA to obtain the SDFM shown in a formula (1). According to the technical scheme, a dependency relationship of adjacent sites in a biological sequence is considered, and more more-accurate biological sequence evolution characteristics of functions, structures and the like can be extracted from multi-sequence alignment, so that probability distribution information subjected to statistics contains sequence site dependency relationship information.
Owner:HARBIN INST OF TECH SHENZHEN GRADUATE SCHOOL
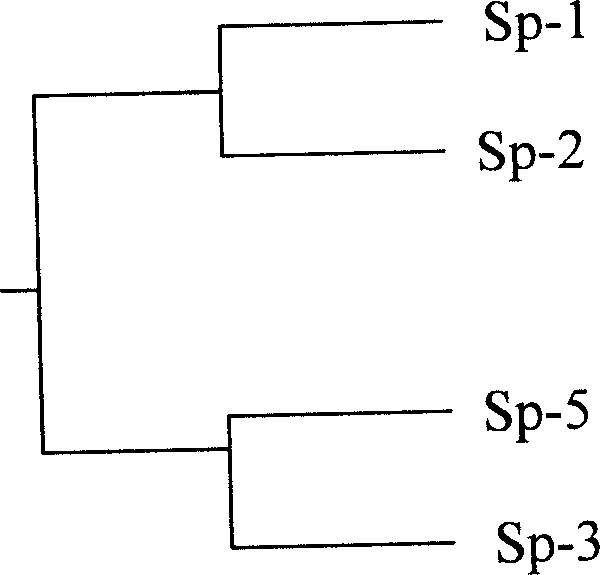
Method of discriminating good and bad production of spirurina strain
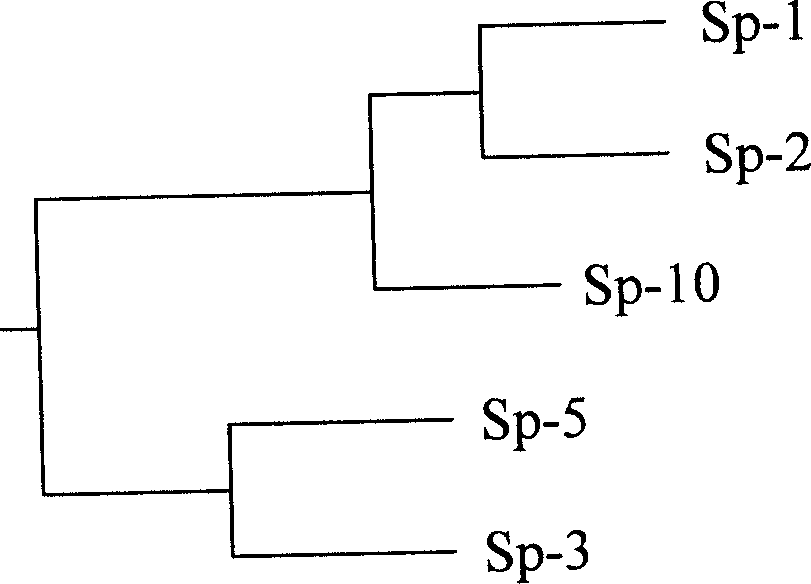
InactiveCN1904071ASimple and fastClear standardMicrobiological testing/measurementPhylogenetic treeReaction system
The present invention relates to a method for identifying spirulina line production character quality. Said method includes the following steps: selecting four plants of Sp-1, Sp-2, Sp-3 and Sp-5 from abtuse acron spirulina line; utilizing upstream primer PF:5'-CAATACATCTTCG CCGATTT-3' and downstream primer PR: 5'-CGTATTATCGGTAGTCATCGG-3'; in PCR reaction system making reaction: 94 deg.C 5 min, 94 deg.C 30s, 58 deg.C 1 min, 72 deg.C 2 min, 30 cycles; 72 deg.C 8 min; connecting PCR product recovered and purified by DNA gel onto PMD18-T carrier to screen male clone; extracting plasmid and making enzyme incision identification, sequencing the recombinant plasmid containing target fragment; making the cpc HID operon gene sequences of above-mentioned four plants and correspondent sequence of identified spirulina plant undergo the process of multiple sequence comparison and creating phylogenetic tree so as to identify spirulina line character quality.
Owner:ZHEJIANG UNIV
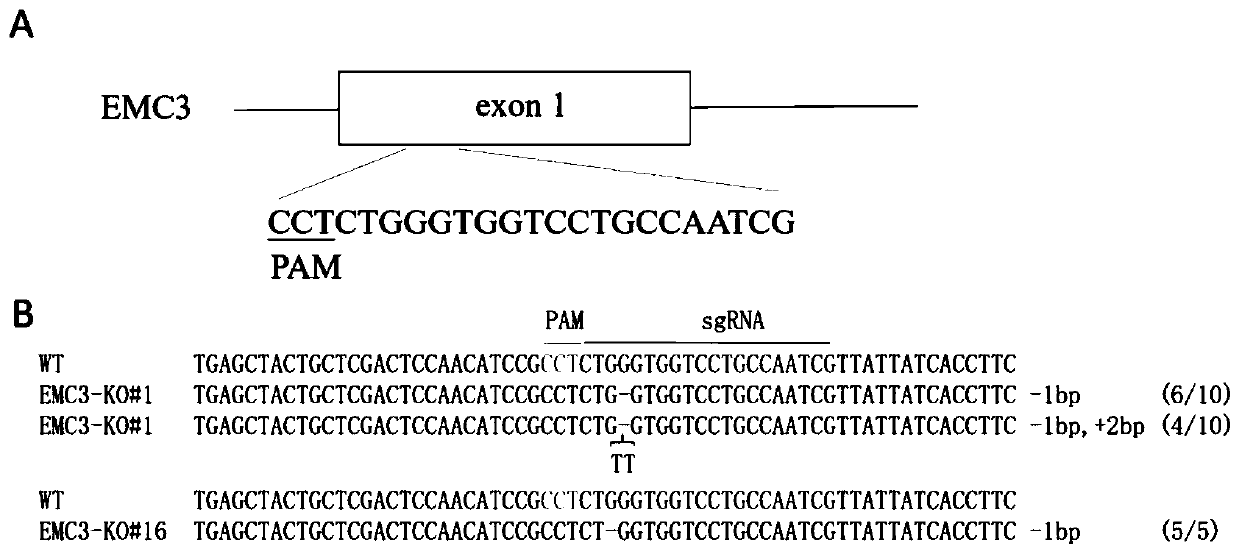
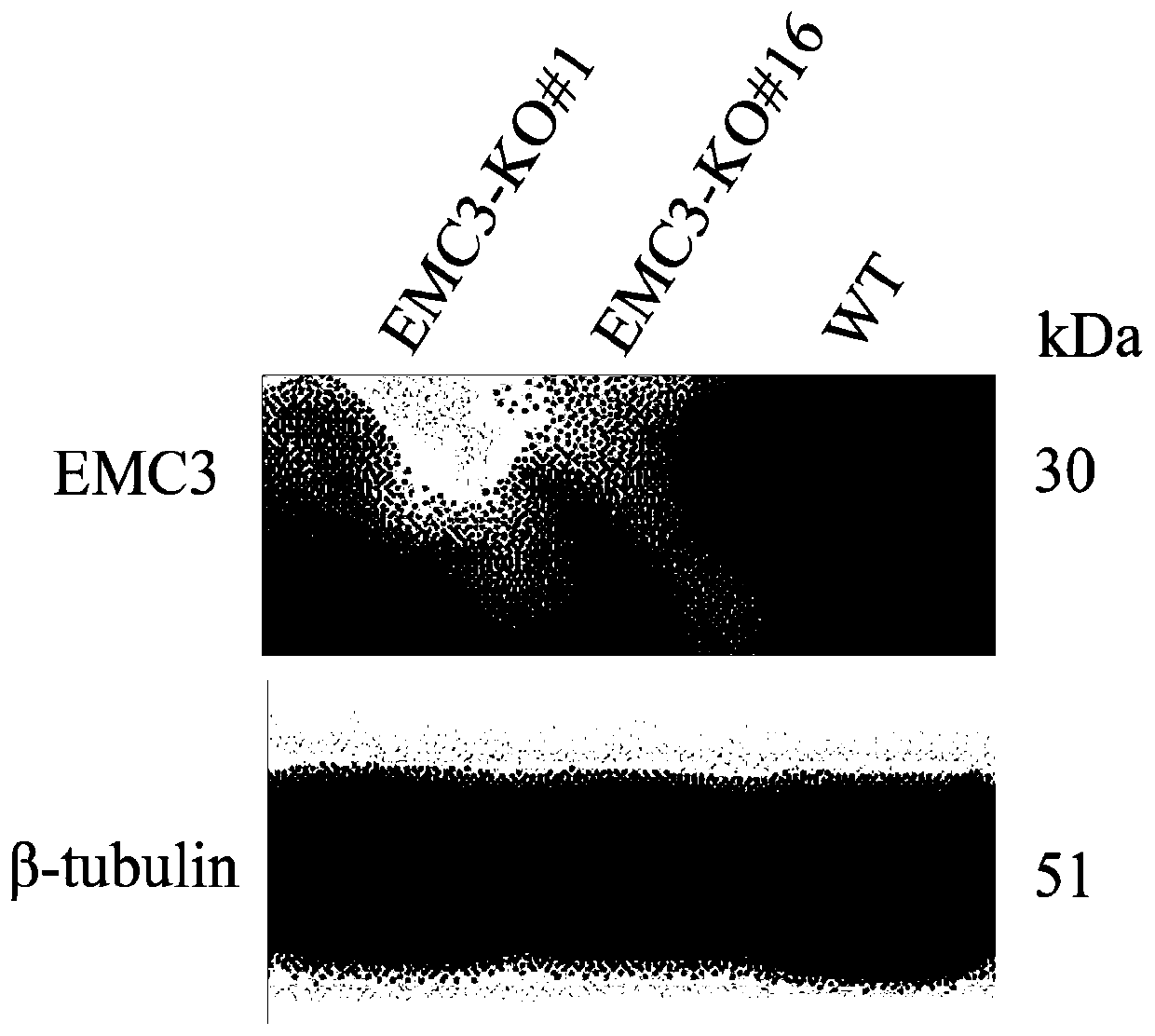
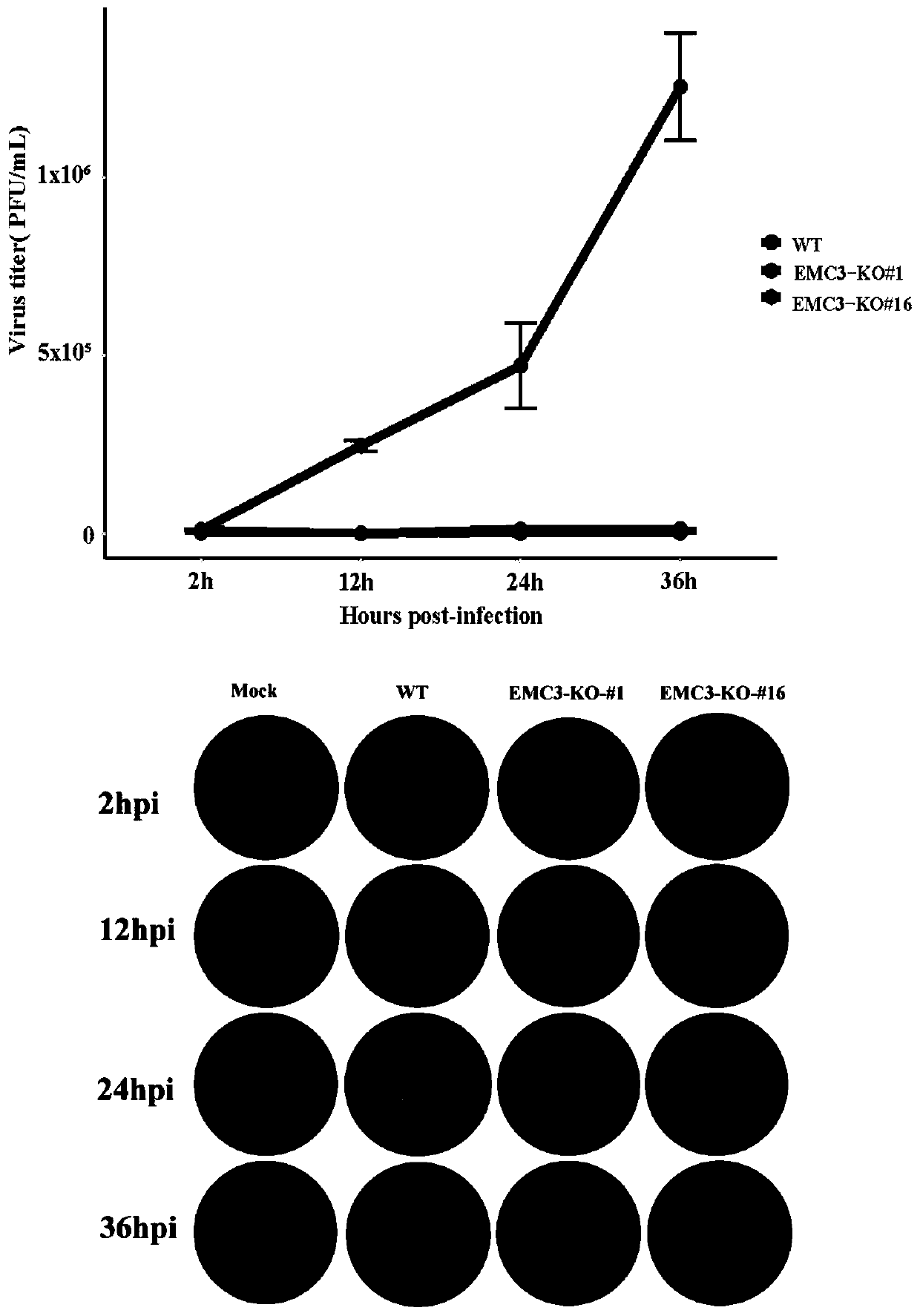
Application and fixed-point knockout method of EMC3 gene
InactiveCN110607280AImprove accuracyInhibition of replicationStable introduction of DNAAntiviralsNucleotideKidney cell
The invention belongs to the biology field, and particularly relates to application and a fixed-point knockout method of an EMC3 gene. Through targeted modification of the EMC3 gene by a gene editingtechnology and change of basic groups of a coding region to cause frame shift mutation of the EMC3 gene, an EMC3 gene knockout cell line is obtained. Experiments prove that by changing a nucleotide sequence of the EMC3 gene in a swine kidney cell (PK-15) to delete EMC3 protein expression, and proliferation of Japanese encephalitis virus (JEV) can be interfered significantly, so that host cells canbe effectively protected against JEV infection invasion induced death. Multiple sequence alignment analysis finds that the EMC3 gene sequence is highly conservative in pigs, people and mice. Therefore, the EMC3 gene can be used as a gene editing target for resisting Japanese encephalitis or used for developing anti-JEV drugs.
Owner:HUAZHONG AGRI UNIV
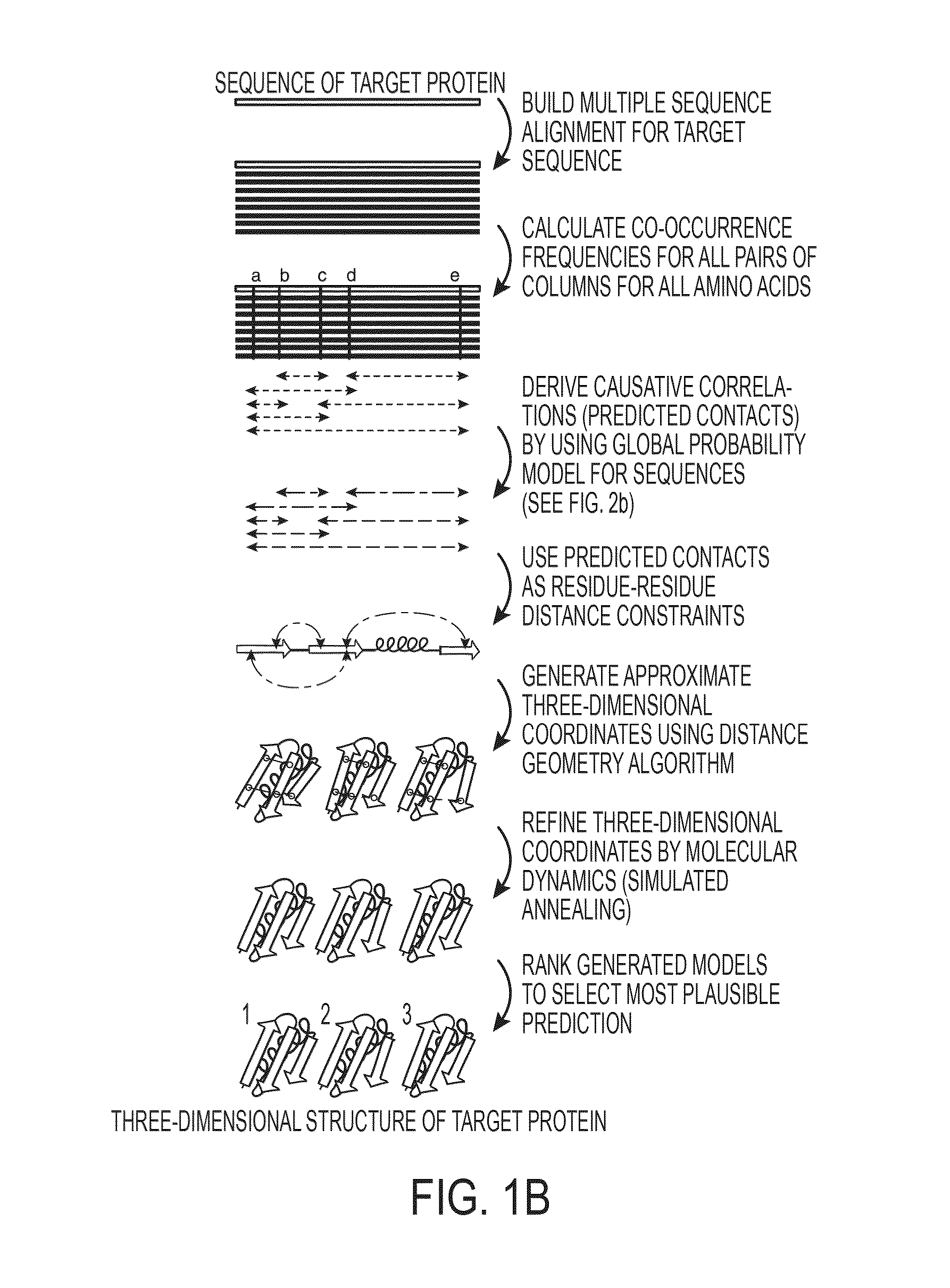
Macrogenome-based method for multiple-sequence alignment of proteins
InactiveCN113257337AEasy extractionIncrease the number of effective sequencesBiostatisticsInstrumentsProtein targetAlgorithm
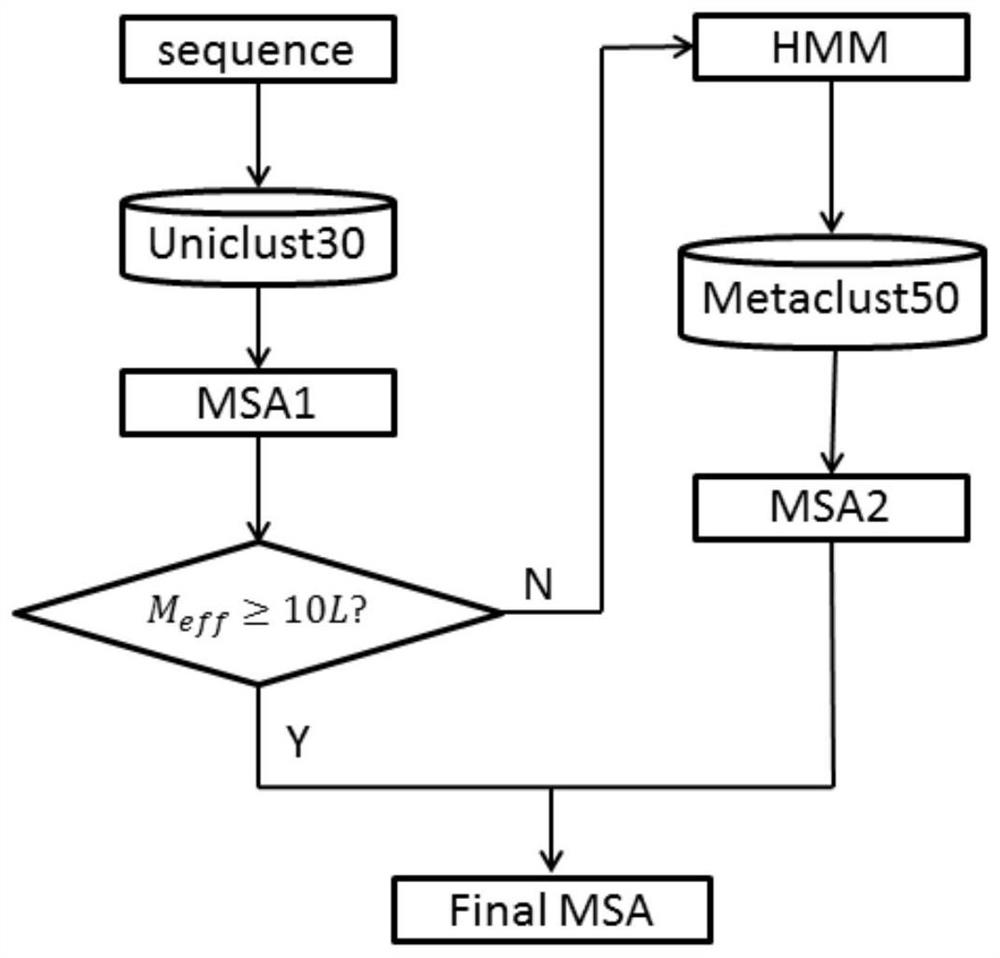
The invention relates to a macrogenome-based method for multiple-sequence alignment of proteins, which comprises the following steps: performing initial search on a UniClust30 database by using HHblits according to a sequence of a target protein, filtering out a sequence with residue gap exceeding 50% the target sequence length in searched MSA, and iterating the search process for three times; filtering obtained multi-sequence comparison files by using hhfilter to generate a sequence with a gap proportion exceeding 25% in the MSA, so as to obtain MSA1; calculating the valid sequence number Meff of MSA1, if Meff is larger than or equal to 10 L, ending the search, and taking the MSA1 as an output result of multi-sequence alignment; otherwise, constructing a hidden Markov model HMM of the MSA1, searching a Metaclust50 metagenome database to obtain multiple-sequence alignment MSA2, and combining the MSA1 with the MSA2 to obtain final MSA. The invention not only enhances the number and quality of the searched homologous sequences, but also improves the calculation efficiency.
Owner:ZHEJIANG UNIV OF TECH
Optimizing evidence theory based K nearest-neighbor alpha-helix prediction method
InactiveCN104537277AHelp predictOptimize forecast resultsCharacter and pattern recognitionSpecial data processing applicationsAlgorithmNear neighbor
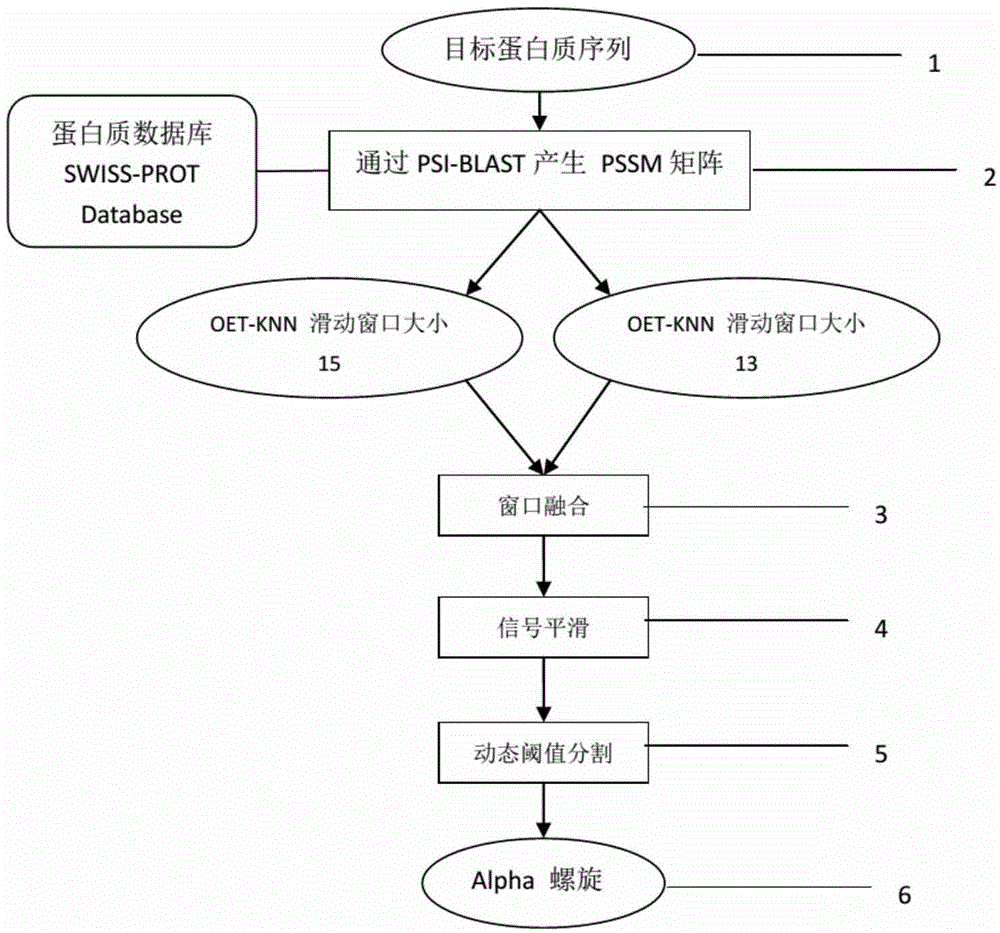
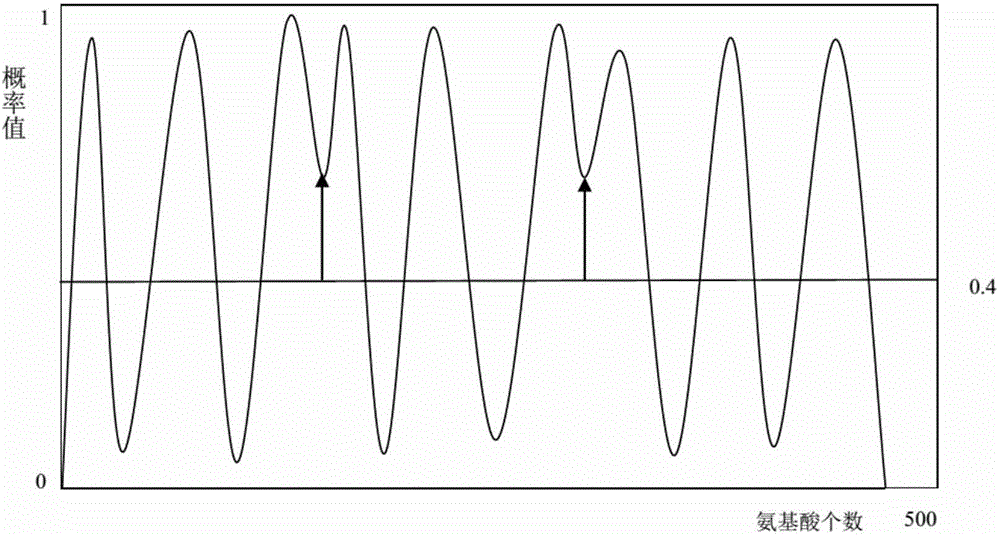
The invention discloses an optimizing evidence theory based K nearest-neighbor alpha-helix prediction method and relates to correlation techniques of mode recognition algorithms and computational biology. By means of the optimizing evidence theory based K nearest-neighbor alpha-helix prediction method, a membrane protein alpha-helix structure is accurately predicted when a protein sample with a high-resolution known structure is lacked. Multiple sliding window extraction feature vectors are fused to perform optimization by adopting a computational biology method including protein multiple sequence alignment and an OETPKNN algorithm, noise is smoothed by means of a median filtering method, then a prediction result is divided by means of a dynamic threshold method, and finally the membrane protein alpha-helix structure is obtained. By means of the optimizing evidence theory based K nearest-neighbor alpha-helix prediction method, the alpha-helix prediction accuracy is improved by higher than 20%, the tail end of an alpha-helix can be predicted, and a good effect is played on irregular alpha-helixes with the prediction length smaller than 15 amino acids.
Owner:SHANGHAI JIAO TONG UNIV
MYB transcription inhibition factor LrETC1 related to lycium ruthenicum anthocyanin synthesis and application thereof
ActiveCN113444731AMicrobiological testing/measurementPlant peptidesSequence databaseTranscription Repressor
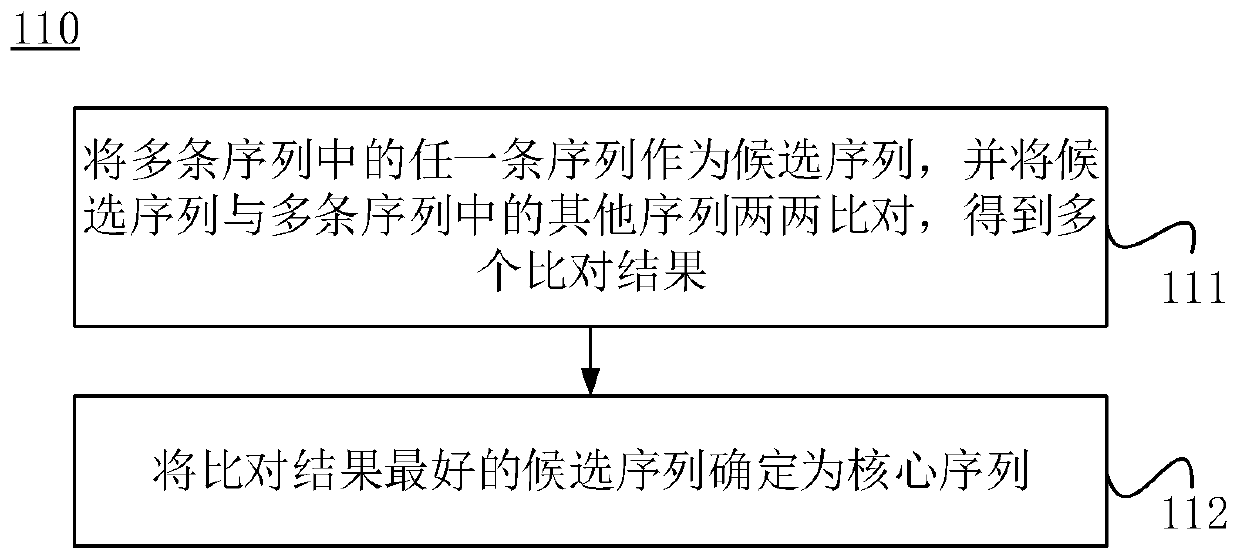
The invention discloses an MYB transcription inhibition factor LrETC1 related to lycium ruthenicum anthocyanin synthesis and application thereof, and belongs to the technical field of gene engineering. According to the invention, the MYB transcription inhibition factor LrETC1 participating in the anthocyanin synthesis is screened and cloned, the total cDNA of the gene is 240bp, and 79 amino acids are encoded; protein sequence database analysis shows that the transcription factor belongs to R3 type MYB; and multiple sequence alignment and evolutionary tree analysis show that the transcription inhibition factor belongs to AtCPC-like transcription inhibition factors, and is a transcription factor which is located in a cell nucleus and does not have an activation function. A LrETC1 transgenic arabidopsis thaliana strain is obtained through agrobacterium tumefaciens-mediated genetic transformation, compared with a wild type, seed coats of seeds are light brown, arabidopsis thaliana seedlings are free of pigment accumulation under the stress condition of high sucrose concentration, and the expression level of related structural genes is remarkably reduced.
Owner:WOLFBERRY SCI INST NINGXIA ACAD OF AGRI & FORESTRY SCI
Detection method of ranavirus in water
InactiveCN108728577AAvoid false negativesShorten the enrichment timeMicrobiological testing/measurementMicroorganism based processesRanavirusGiant salamander
The invention relates to a detection method of ranavirus in water and belongs to the field of virus detection. The detection method of the ranavirus in culture water is constructed in combination witha method for enriching viruses in water and a PCR technology. A method for enriching viruses in water with a NaCl-AlCl3.6H2O precipitation method is improved, and the enrichment time is shortened. Specific PCR primers are improved, multiple sequence alignment is performed on MCP (major capsid protein) of ranavirus including giant salamander ranavirus, rana tigrina ranavirus, common midwife toad virus and type-3 ranavirus, primers are designed for conservative areas of sequences, so that false negative of detection due to sequence variation of viruses in the evolutionary process is avoided, the blank of methods for detecting ranavirus in water on the market at present is effectively filled up, and the method has great social benefits and economic benefits.
Owner:SICHUAN AGRI UNIV
Alginate lyase mutant capable of relieving dependence of divalent metal ions and application of alginate lyase mutant
ActiveCN112921020AEliminate dependenciesImprove thermal stabilityBacteriaMicroorganism based processesLyaseDivalent metal ions
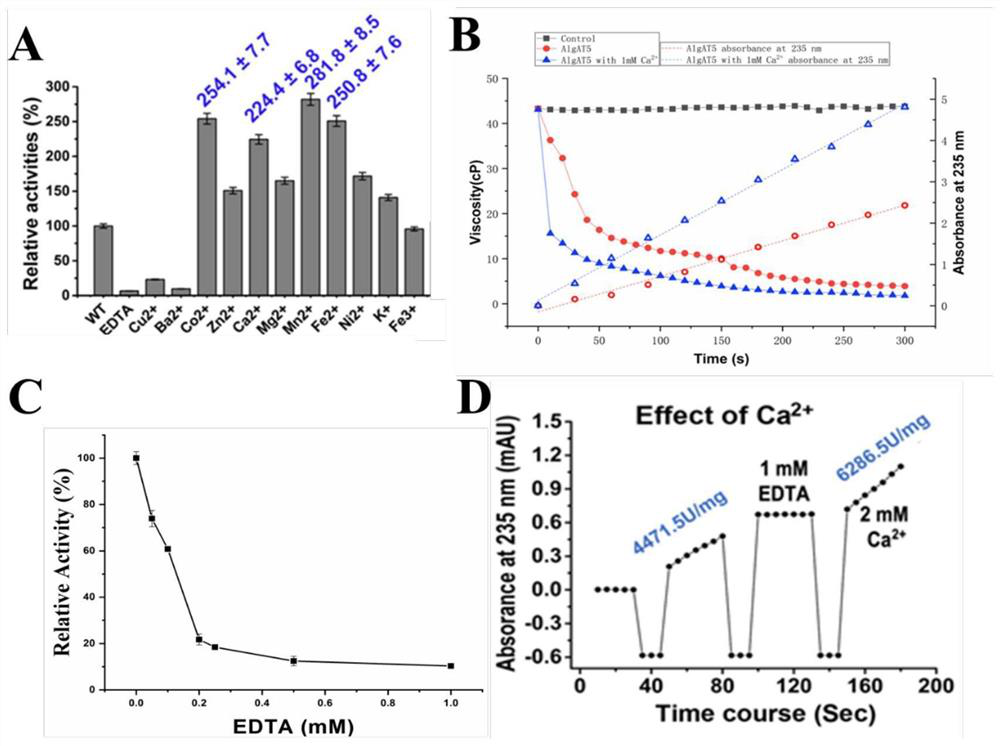
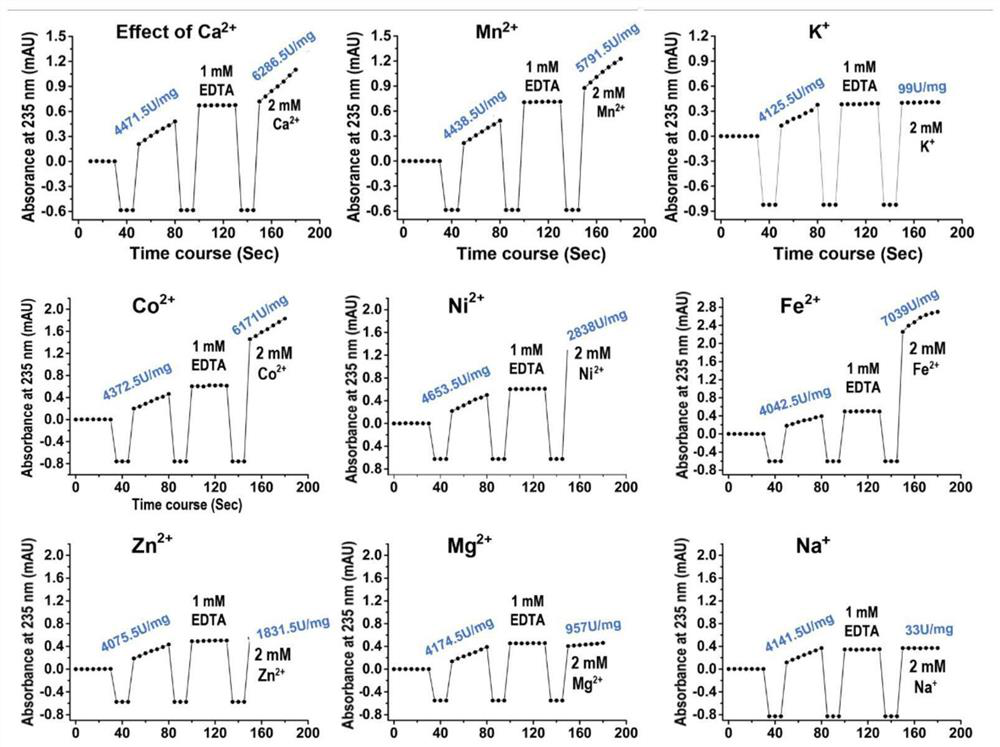
The invention relates to the technical field of protein engineering, in particular to an alginate lyase mutant capable of relieving dependence of divalent metal ions and application of the alginate lyase mutant. The mutant is characterized in that in an amino acid sequence of alginate lyase which takes PolyG as a specific substrate during enzymolysis in PL7 family alginate lyase, amino acid at least located at the same three-dimensional structure position as aspartic acid D146 in AlgAT5 or at the D146 position based on multiple sequence alignment of structure is positively charged amino acid, glycine, alanine, valine, leucine or isoleucine formed by mutating aspartic acid D or glutamic acid E. The beneficial mutations can provide theoretical and technical support for industrial development and application of alginate lyase, and promote green and efficient preparation of alginate oligosaccharide.
Owner:QINGDAO INST OF BIOENERGY & BIOPROCESS TECH CHINESE ACADEMY OF SCI
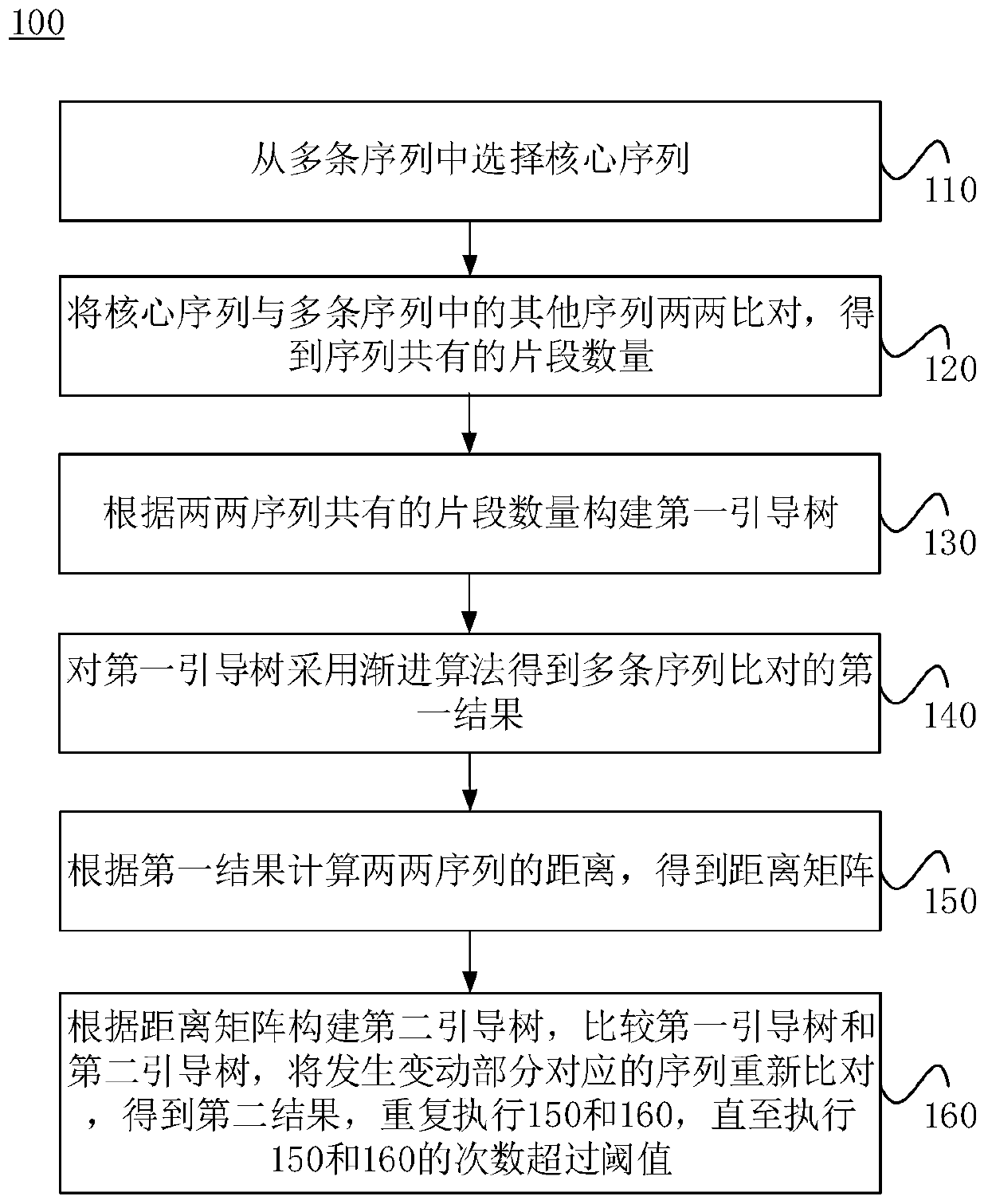
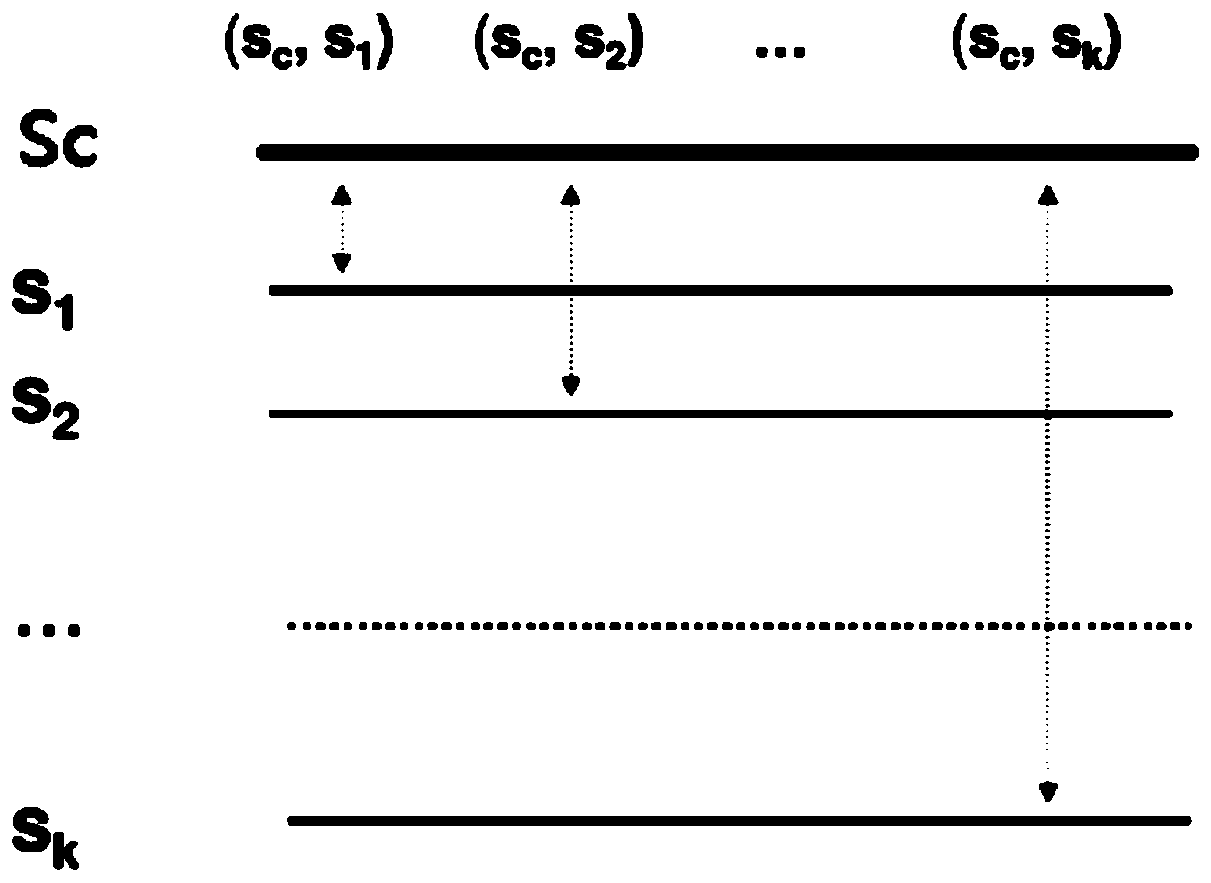
Method and system for optimizing multiple sequence alignment algorithms, and storage medium
ActiveCN109949867AShort timeReduce consumptionBiostatisticsSequence analysisSequence alignment algorithmDistance matrix
The invention relates to a method and a system for optimizing multiple sequence alignment algorithms, and a storage medium. The method comprises the steps of selecting a core sequence from multiple sequences; performing pairwise alignment on the core sequence and other sequences in the multiple sequences, and obtaining the number of common fragments of the sequences; constructing a first guiding tree according to the number of common fragments of the pairwise sequences; performing a progressive algorithm on the first guiding tree for obtaining a first result through alignment of multiple sequences; calculating the distance between the pairwise sequences according to the first result, and obtaining a distance matrix; constructing a second guiding tree according to the distance matrix, comparing the first guiding tree with the second guiding tree, performing re-alignment on the sequences which correspond with the changing part for obtaining a second result, and repeating processes of constructing the second guiding tree and comparing the first guiding tree with the second guiding tree until the number of comparison times exceeds a threshold, thereby shortening time consumption in sequence comparison, increasing processing process and reducing resource consumption.
Owner:INST OF SPECIAL ANIMAL & PLANT SCI OF CAAS
A polynomial method of constructing a non-deterministic (NP) turing machine
ActiveCN105190632AObvious featuresObvious advantagesKnowledge representationInference methodsPolynomial methodNon-deterministic Turing machine
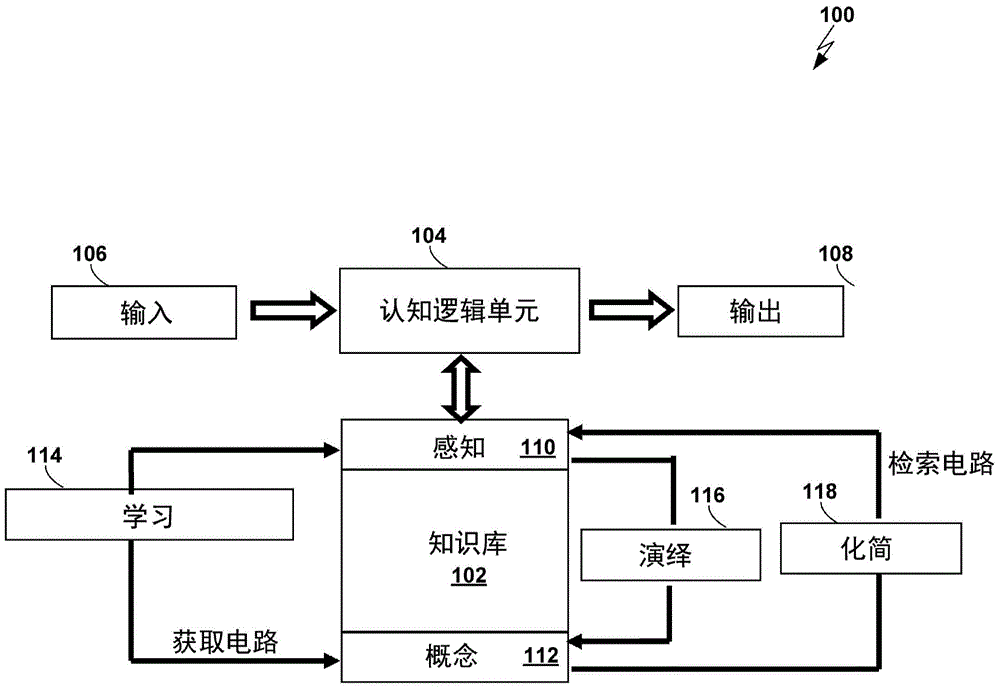
A nondeterministic Turning machine (NTM) performs computations using a spatial binary enumeration system, a three-dimensional relation system, a simulated-human logic system, and a bijective-set memory system. The NTM may be used to perform a variety of computational tasks, such as multiple sequence alignment, factorization, and other nondeterministic polynomial algorithms in polynomial time. The NTM may be constructed by a deterministic Turing machine (DTM) using the four systems listed above.
Owner:BEIJING BOSI BIOINTELLIGENCE TECH
Kit and method for rapidly detecting human bocavirus based on isothermal amplification
PendingCN112877477ARapid diagnosisStrong specificityMicrobiological testing/measurementMicroorganism based processesNucleotideA-DNA
The invention discloses a kit and a method for rapidly detecting a human bocavirus based on isothermal amplification. A nucleotide sequence of a conserved gene NP1 protein of the human bocavirus is compared through multiple sequences, a DNA fragment with the size of 270bp is determined as a targeted analysis sequence, the sequence is located between the 10th site and the 279th site of the NP1 gene, and a specific RPA primer is designed based on the sequence. Further, a recombinase polymerase isothermal amplification method suitable for rapidly and accurately detecting the human bocavirus is optimized and established. The detection limit of the method reaches 252ag which is higher than that of a traditional PCR detection technology, and the kit has the advantages of being high in sensitivity, high in specificity, easy to operate, rapid and the like and is suitable for clinical rapid detection of the human bocavirus.
Owner:NINGBO MUNICIPAL CENT FOR DISEASE CONTROL & PREVENTION
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com