Optimizing evidence theory based K nearest-neighbor alpha-helix prediction method
An evidence theory and nearest neighbor technology, applied in the field of bioinformatics and pattern recognition, can solve the problems of complex structure of membrane protein, inability to predict sequence, inability to predict, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
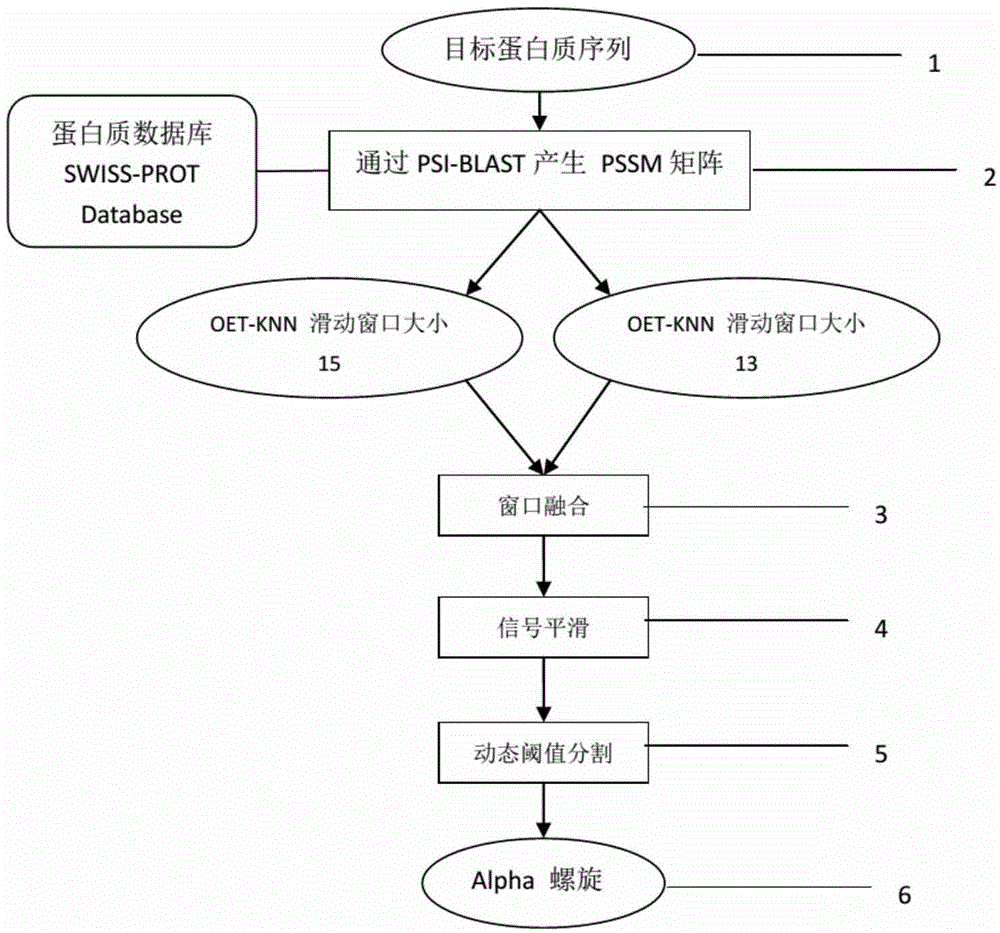
[0041] Such as figure 1 As described, this embodiment includes the following steps:
[0042] 1) Search the SWISS-PROT protein database according to the protein sequence to obtain the target amino acid sequence, as shown in Seq ID No.1.
[0043] 2) Obtain the specific position scoring matrix PSSM as an amino acid feature through the PSI-BLAST sequence comparison tool;
[0044] 3) Extract feature vectors with sliding windows of size 13 and 15, respectively, and then perform fusion optimization;
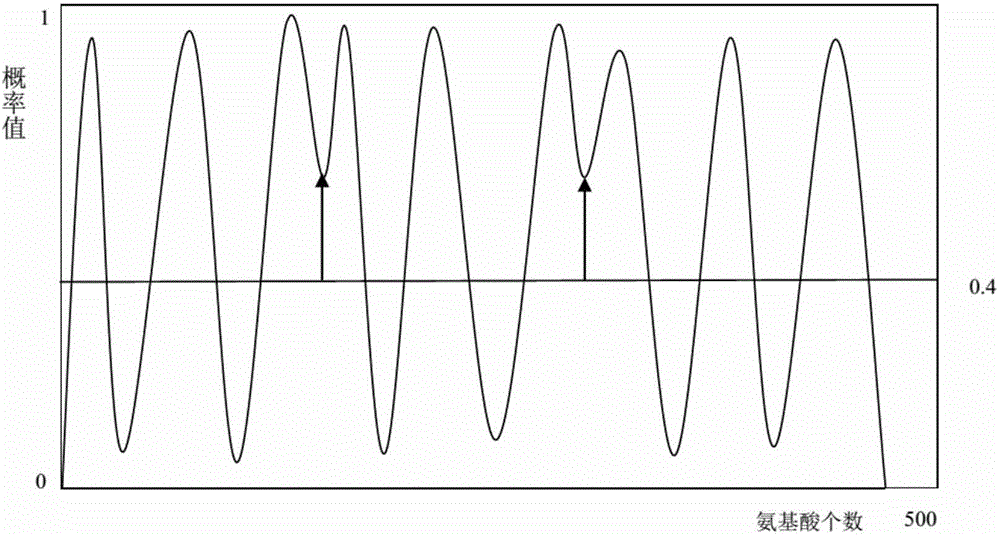
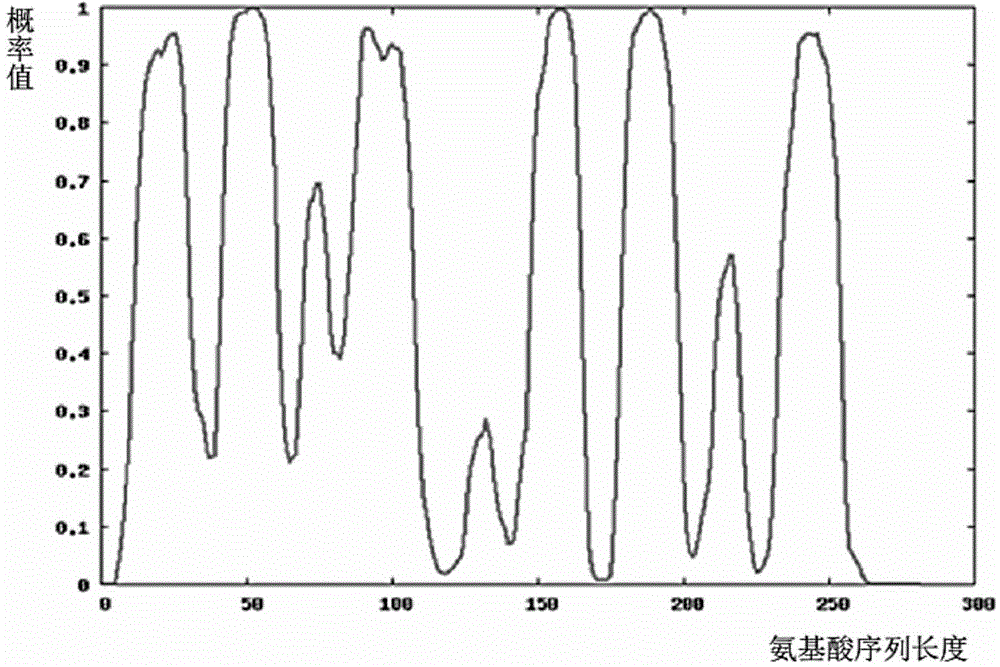
[0045] 4) Classifying the extracted feature vectors with the K-Nearest Neighbor Algorithm of Optimal Evidence Theory to obtain the prediction curve of amino acid sequence prediction probability;
[0046] 5) Use the median filtering method to smooth, remove noise, and reduce the burrs of the probability curve;
[0047] 6) Use dynamic threshold segmentation to obtain the division result of whether each amino acid in the target sequence belongs to the alpha helical transmembrane structu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com