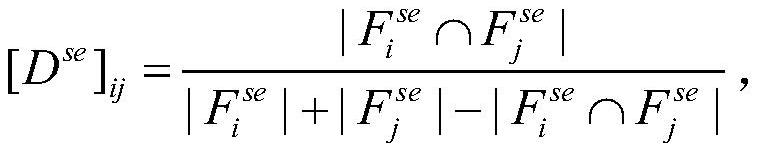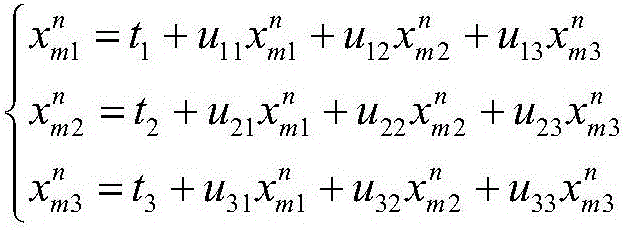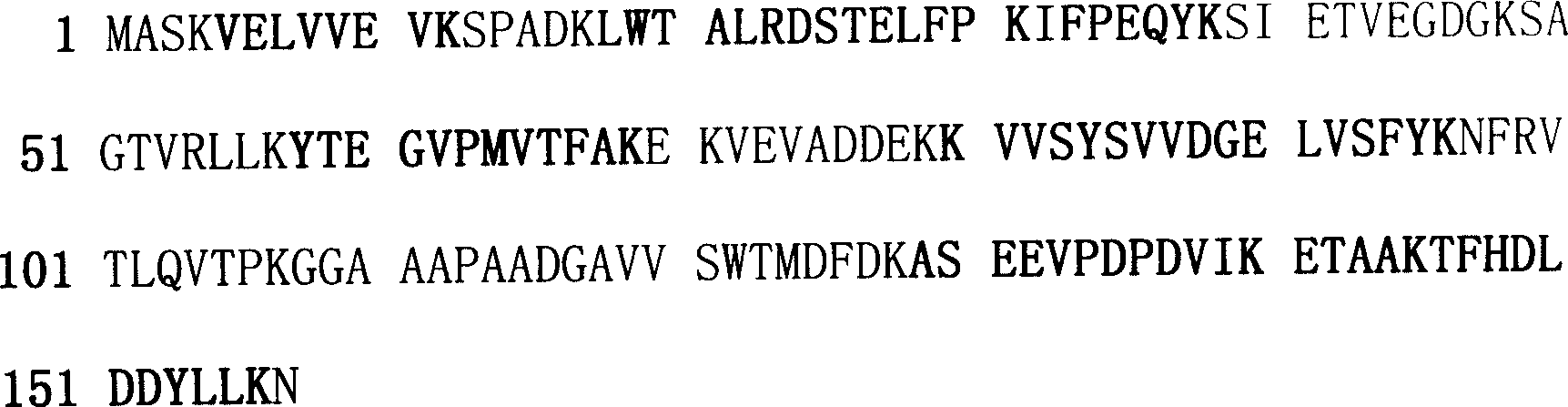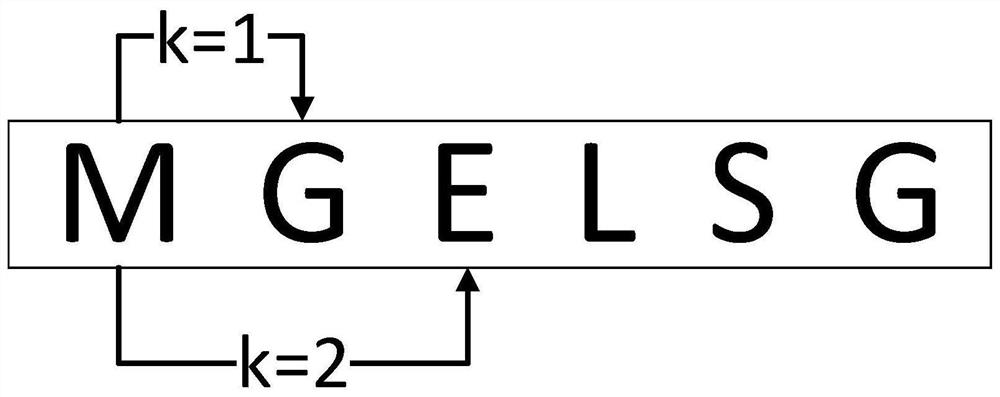Patents
Literature
37 results about "Protein database" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
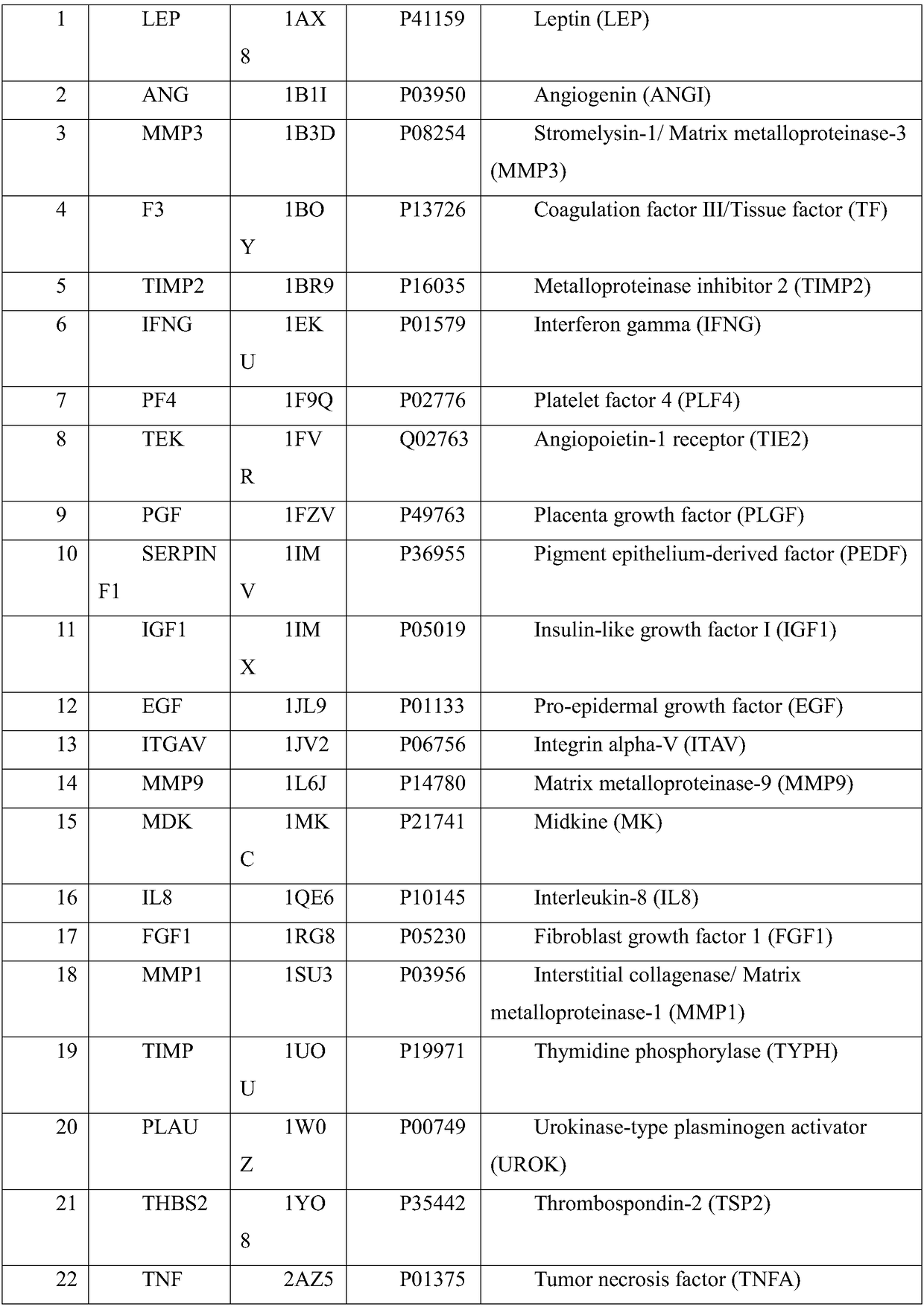
Method and device for detecting mutated proteins
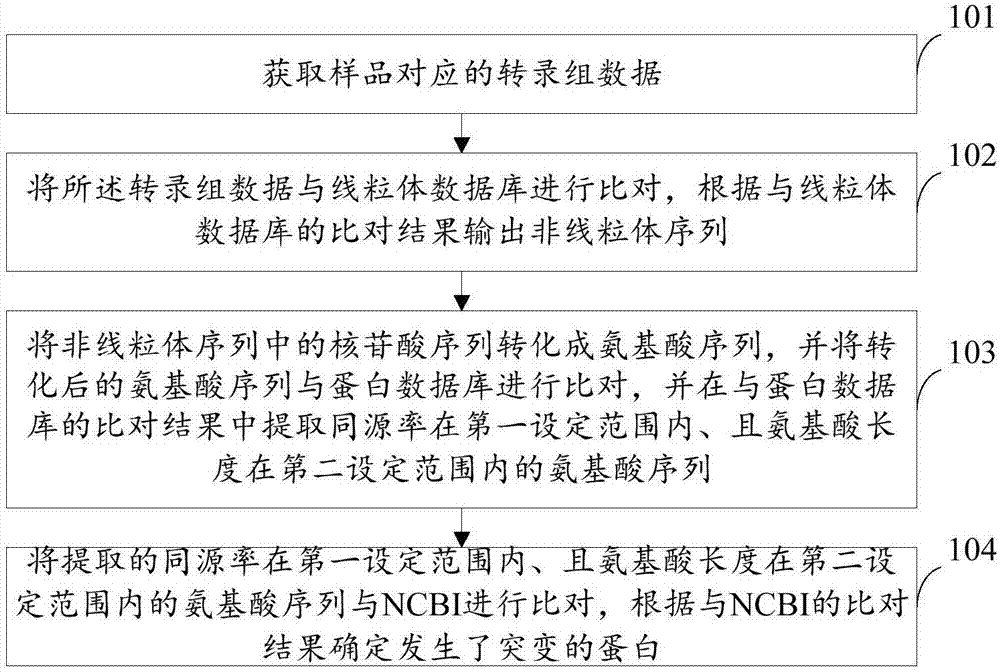
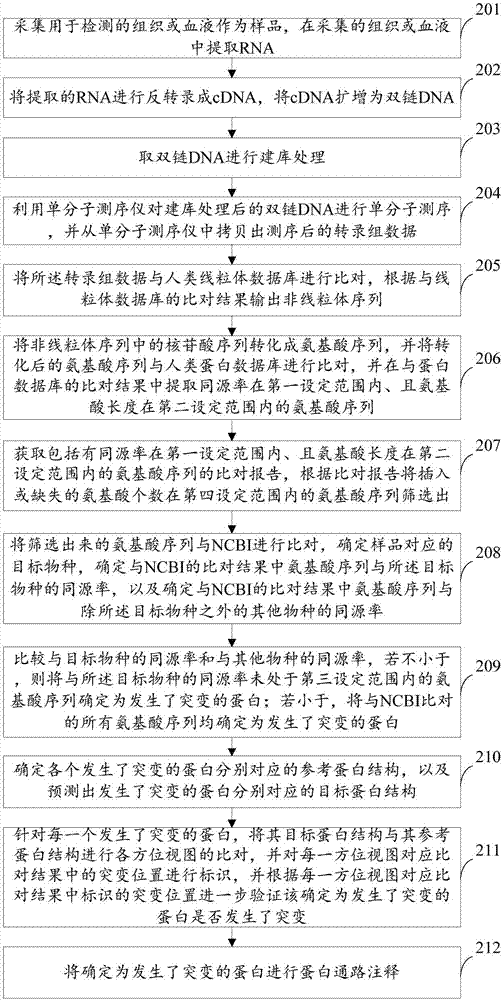
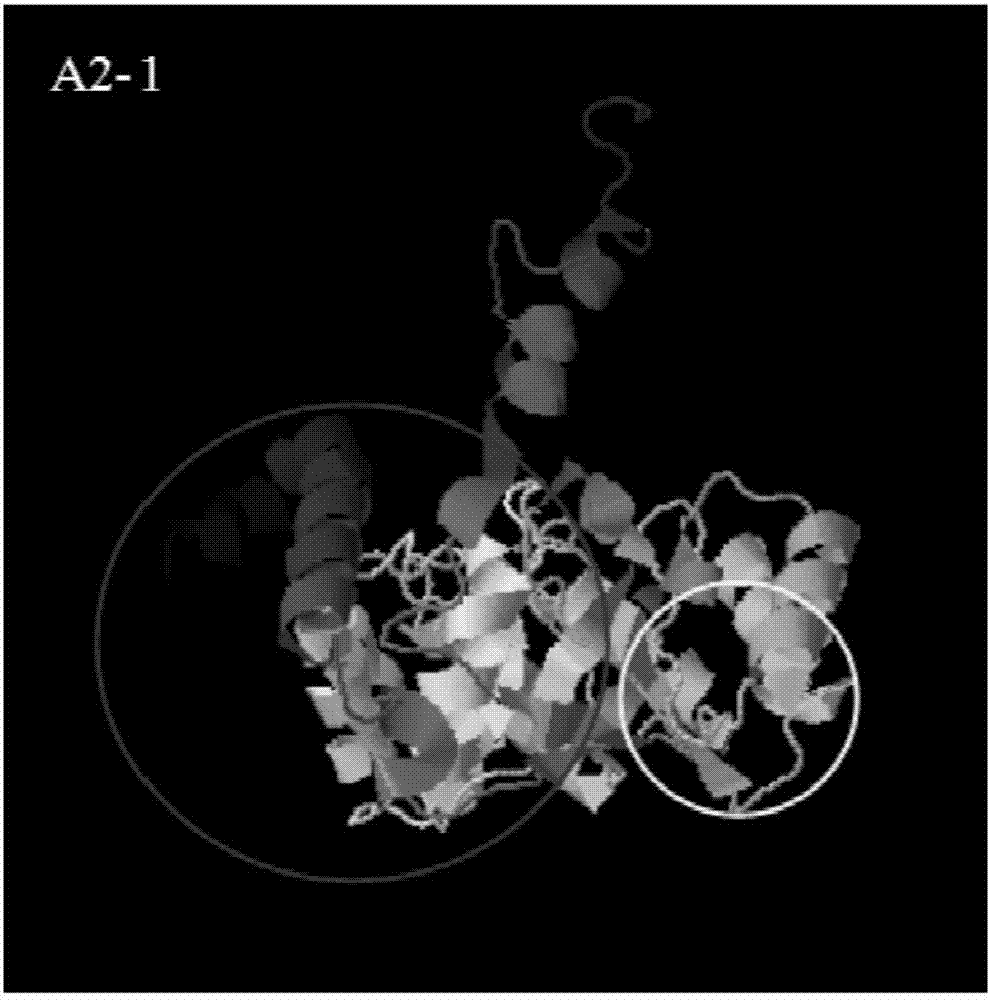
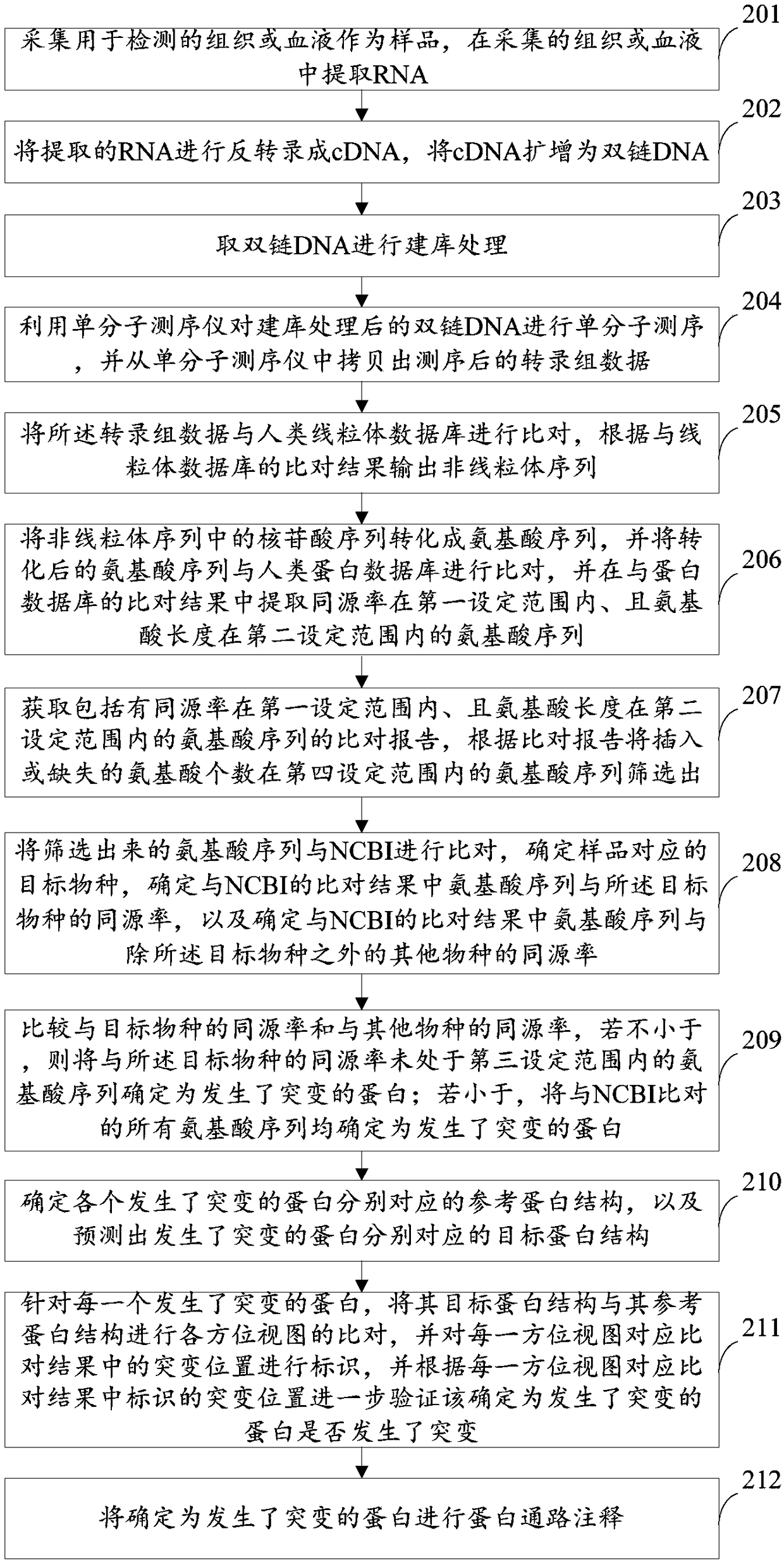
The invention provides a method and a device for detecting mutated proteins. The method includes acquiring transcriptome data corresponding to samples; comparing the transcriptome data to mitochondrion databases and outputting non-mitochondrion sequences according to comparison results of the transcriptome data and the mitochondrion databases; transforming nucleotide sequences in the non-mitochondrion sequences into amino acid sequences, comparing the transformed amino acid sequences to protein databases and extracting amino acid sequences with homogenous rates in first set ranges and amino acid lengths in second set ranges from comparison results of the transformed amino acid sequences and the protein databases; comparing the extracted amino acid sequences with the homogenous rates in the first set ranges and the amino acid lengths in the second set ranges to NCBI (national center of biotechnology information) and determining the mutated proteins according to comparison results of the extracted amino acid sequences and the NCBI. According to the scheme, the method and the device have the advantage that the mutated proteins in the samples can be detected by the aid of the method and the device.
Owner:天津市湖滨盘古基因科学发展有限公司
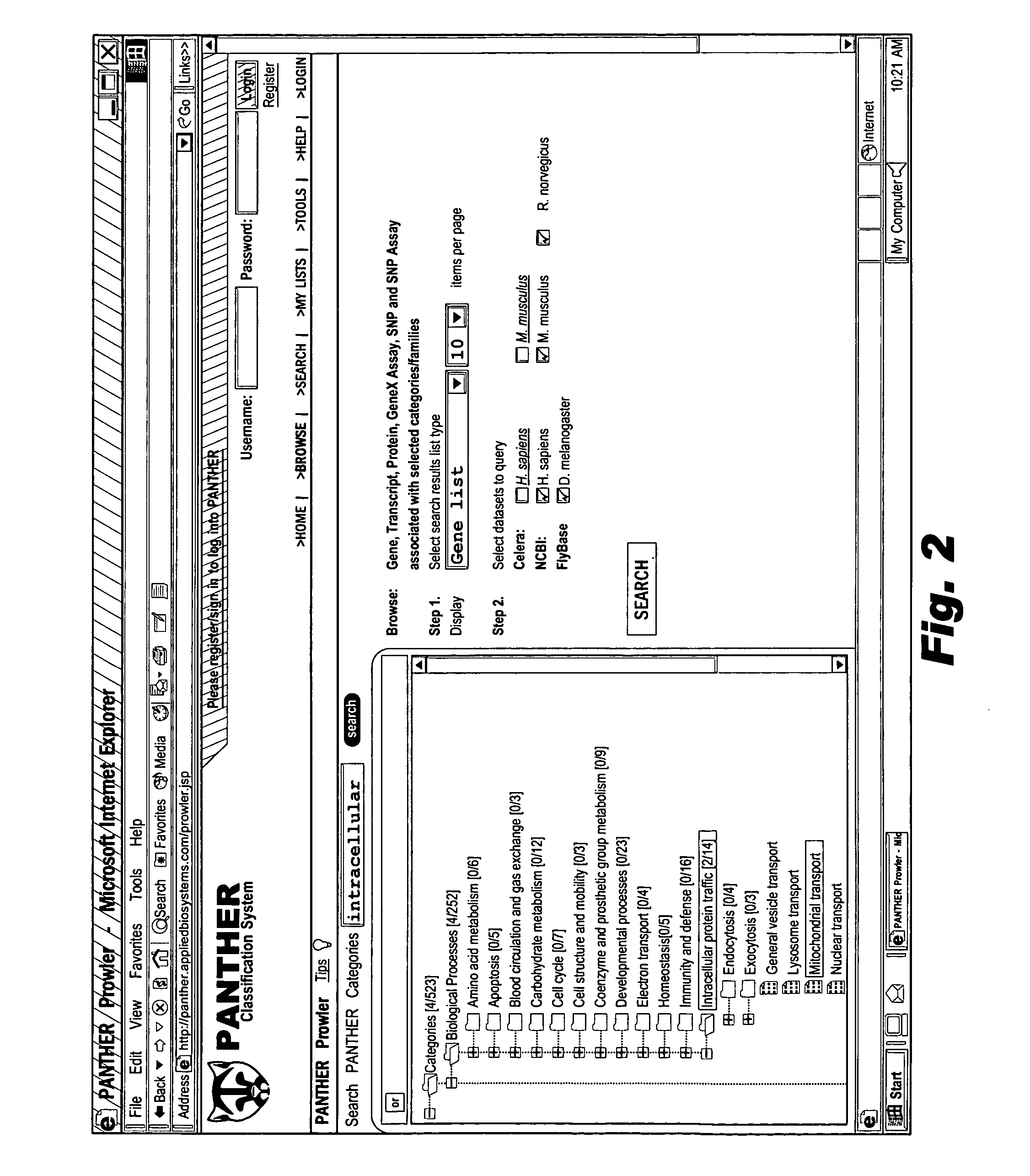
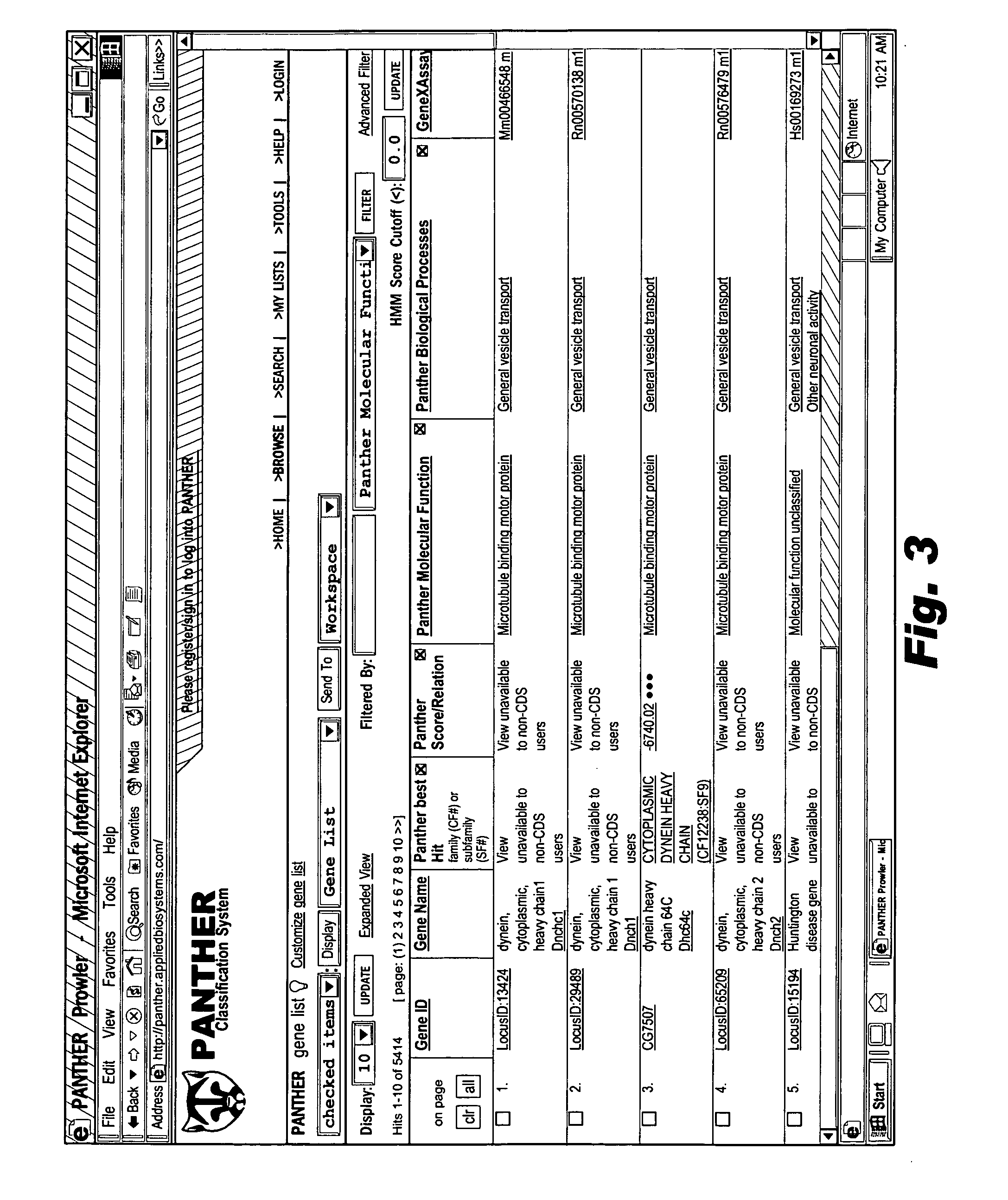
Browsable database for biological use
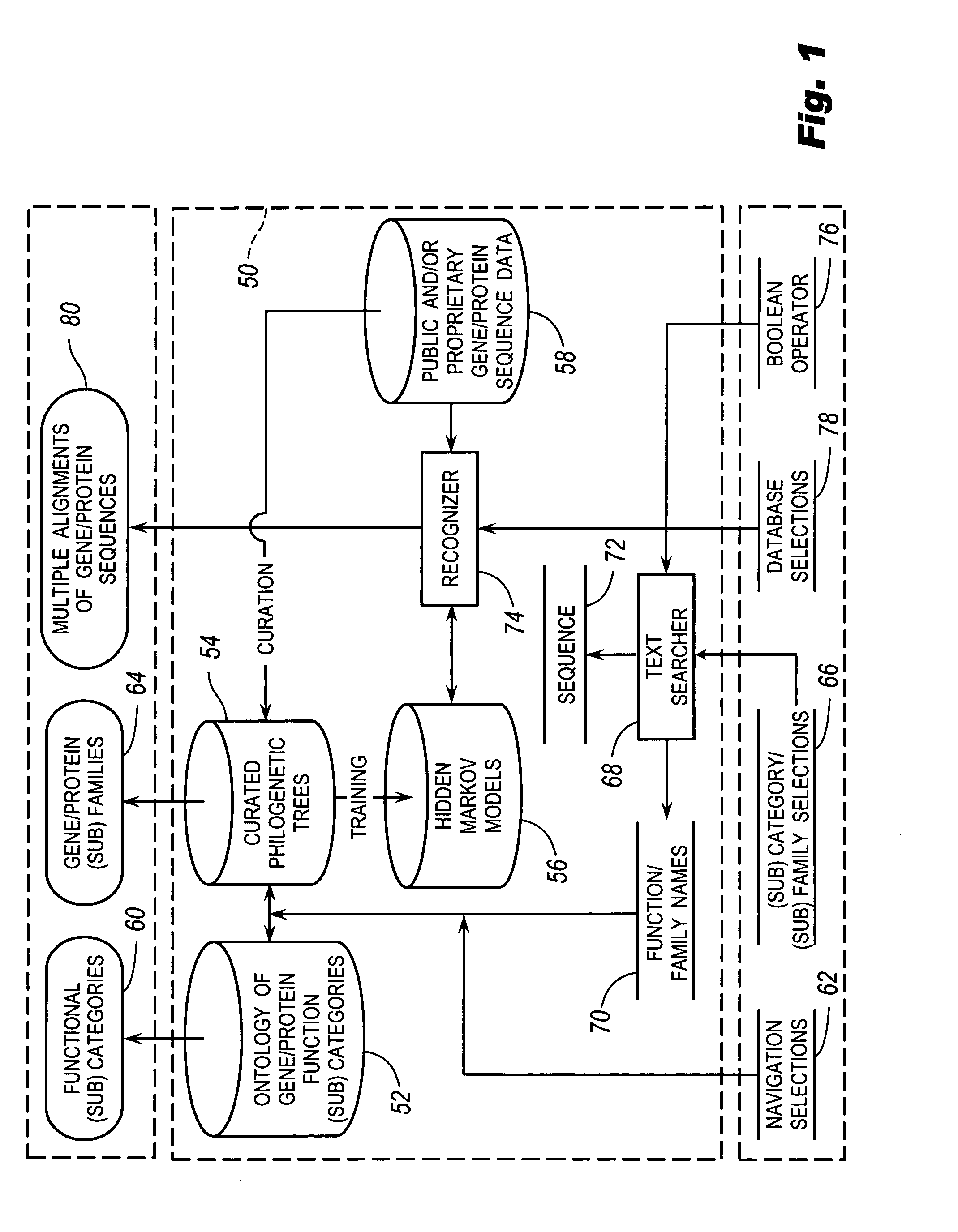
The browsable database can allow for high-throughput analysis of protein sequences. One helpful feature may be a simplified ontology of protein function, which allows browsing of the database by biological functions. Biologist curators may have associated the ontology terms with Hidden Markov Models (HMMs), rather than individual sequences, so that they can be applied to additional sequences. To ensure accurate functional classification, HMMs may be constructed not only for families, but for curator-defined subfamilies, whenever family members have divergent functions or nomenclature. Multiple sequence alignments and phylogenetic trees, including curator-assigned information, can be available for each family. Various versions of the browsable database may include training sequences from all organisms in the GenBank non-redundant protein database, and the HMMs can be used to classify gene products across the entire genomes of human, and Drosophila melanogaster.
Owner:APPL BIOSYSTEMS INC
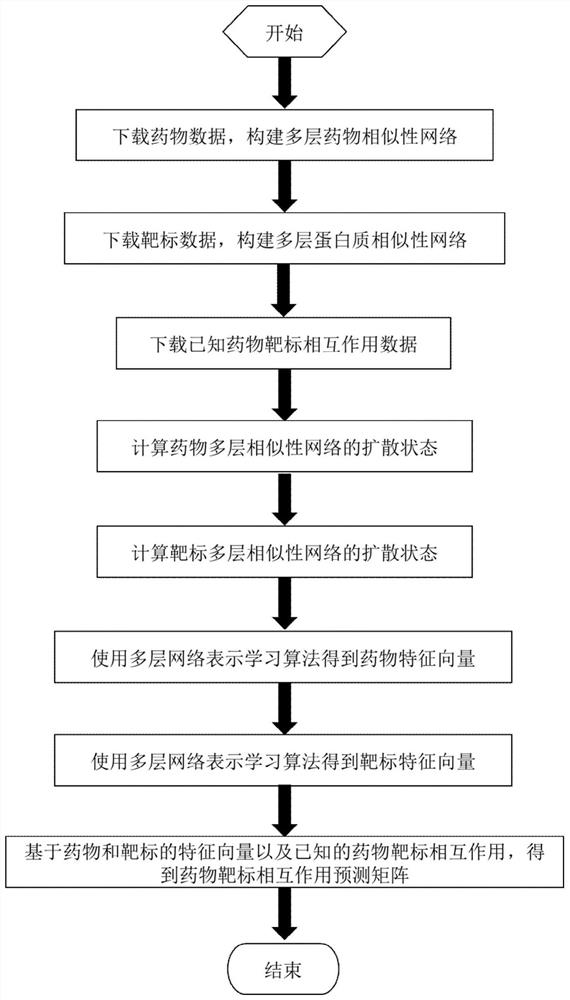
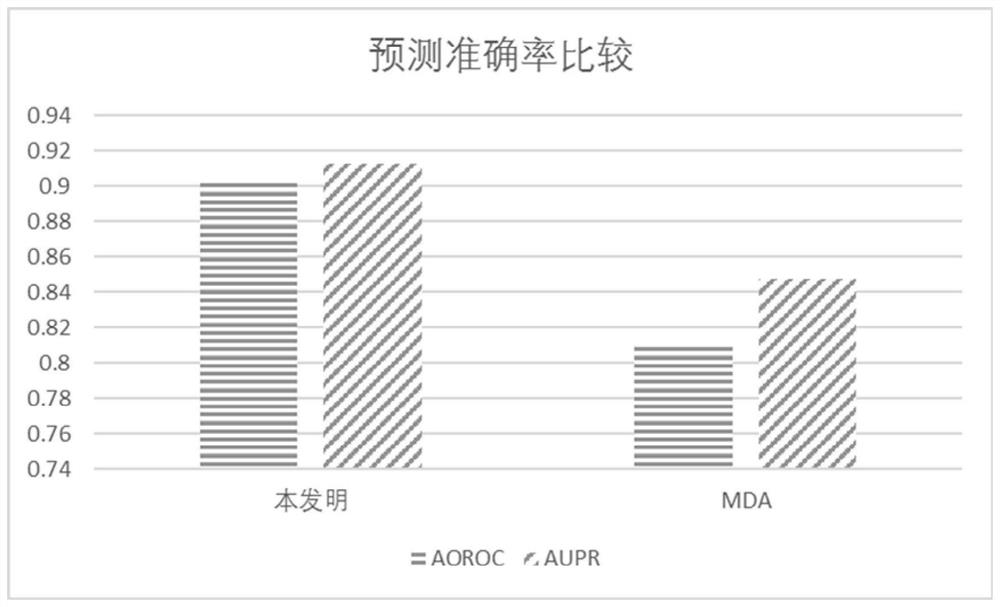
Drug target interaction prediction method based on multilayer network representation learning
PendingCN111785320AImprove forecast accuracyAvoid the disadvantage of being biasedBiostatisticsInstrumentsPharmaceutical drugProtein Feature
The invention discloses a drug target interaction prediction method based on multilayer network representation learning, and mainly solves the problem of low prediction accuracy in the prior art. Themethod comprises the following steps: downloading data from a drug and protein database, and respectively constructing multilayer similarity networks of drugs and proteins; calculating diffusion states of the two similarity networks respectively, and integrating the diffusion states respectively to obtain feature vectors of drugs and proteins; taking known drug target interaction data as supervision information, putting the drug and protein feature vectors into the same drug target space, and respectively obtaining projection matrixes of the drug and the protein by using a bilinear function; obtaining a prediction score matrix of drug target interaction according to the two projection matrixes and ranking the prediction score matrix; regarding eight top-ranked unknown drug target pairs aspotential drug target interactions. According to the method, the prediction accuracy of drug target interaction is improved, and the method can be used for predicting candidates of drug target pairs.
Owner:XIDIAN UNIV
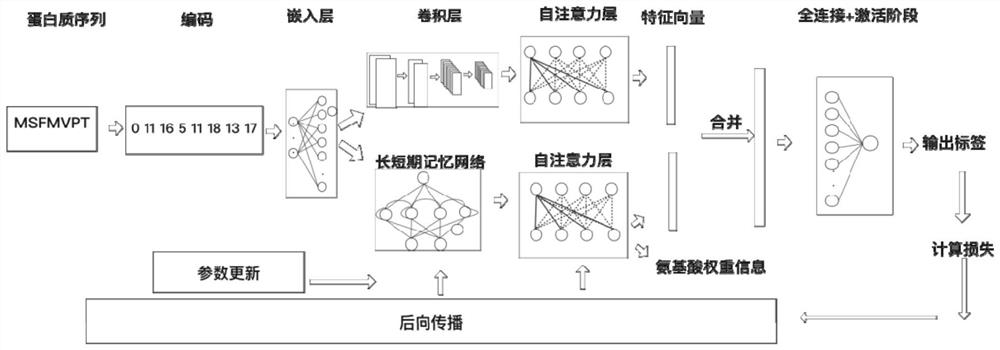
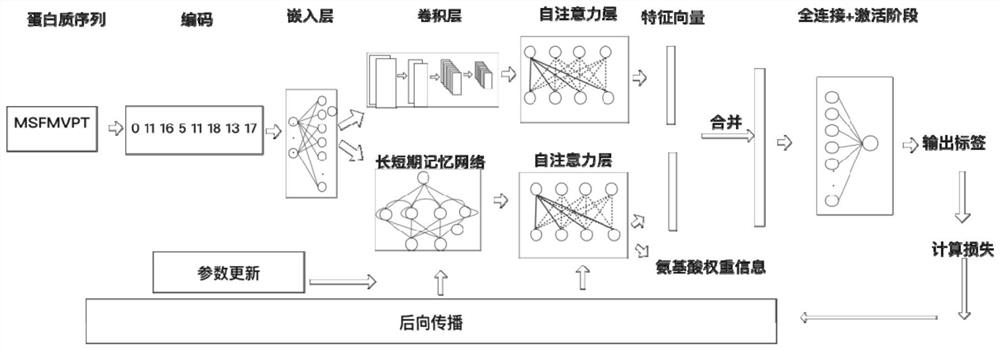
DNA binding protein identification and function annotation deep learning method based on self-attention mechanism
The invention discloses a DNA binding protein identification and function annotation deep learning method based on a self-attention mechanism. The method comprises the following steps: selecting a data set from a protein database, training and testing a constructed deep learning model by using the selected data set, and predicting whether protein can be combined with DNA by using the trained deeplearning model, wherein the deep learning model comprises a coding layer, an embedding layer, a long short-term memory neural network layer (LSTM), a convolutional neural network layer (CNN) and a self-attention layer (Self-Attention). Through a deep learning model based on a self-attention mechanism, whether the protein has a function of combining with DNA or not can be predicted according to primary sequence information of the protein, and an area with a combining function is found out.
Owner:TIANJIN UNIV
Lathyrus quinquenervius EST-SSR primer group developed based on transcriptome sequencing, method and application
InactiveCN107345256ALow costMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceSequence database
The invention provides a lathyrus quinquenervius EST-SSR primer group developed based on transcriptome sequencing, a method and application. The method comprises the steps of obtaining a set of all root, stem and leaf transcripts of two lathyrus quinquenervius varieties of different flower colors at a seedling stage to form an original sequence database; removing low-quality sequencing data with a joint and forming a high-quality filtered sequence database; splicing high-quality filtered sequences, reducing the transcripts and carrying out BLAST comparison on the transcripts and a reference protein database NR, reserving the optimal comparison result and determining the maximal sequence as a Unigene representative sequence; carrying out SSR locus scanning in Unigene by using MISA 1.0; and carrying out SSR primer design by adopting primer 3.0, screening SSR primers and carrying out fingerprint map construction on 43 parts of lathyrus quinquenervius materials. The lathyrus quinquenervius EST-SSR primer group is convenient, fast, accurate, low in cost, and a new thought is provided for the development of lathyrus quinquenervius EST-SSR primers and germplasm resources identification.
Owner:山西省农业科学院农作物品种资源研究所 +1
Method for reconstructing protein database and a method for screening proteins by using the same method
InactiveUS8296300B2Short screening timeEasy to handleGenetic modelsDigital computer detailsData miningDatabase
The present invention relates to a method for reconstructing protein database for identifying a protein and a method for screening a protein using the same, more precisely a method for reconstructing protein database, and a method for identifying a protein using the same. The method for reconstructing protein database and the method for identifying the protein of the invention are very useful for the investigation of endogenous proteins and their functions and interactions, and are further effectively used for the development of diagnostic and therapeutic agents for various diseases.
Owner:KOREA BASIC SCI INST
Method for constructing antibiotic resistance genbank
ActiveCN108491692ASpecial data processing applicationsBioinformaticsSequence annotationAntibiotic resistance genes
The invention discloses a bioinformatics method for constructing an antibiotic resistance genbank in the biotechnology field. The method comprises the following steps that: searching the protein sequence of a resistance gene in a GenBank; selecting a sequence with high accuracy as an initial sequence; adopting a Clustalw method for comparison; constructing a hidden Markov model, and searching a GenBank protein database to obtain all sequences which contain protein conserved sites; according to the E values of the sequences and the annotation information of the sequences in the GenBank database, removing the sequences which are highly homologous and do not conform to requirements; and after repeated sequences are removed, adding species annotation information; and integrating all protein sequences to finish database construction. By use of the method, the annotation information and the comparison similarity of a sequence can be comprehensively measured, and sequence collection speed andaccuracy can be improved. By use of the method provided by the invention, the construction of the antibiotic resistance genbank can be finished, and a basic data is provided for researching the primer design, the data analysis and the sequence annotation of the resistance gene.
Owner:RES CENT FOR ECO ENVIRONMENTAL SCI THE CHINESE ACAD OF SCI +1
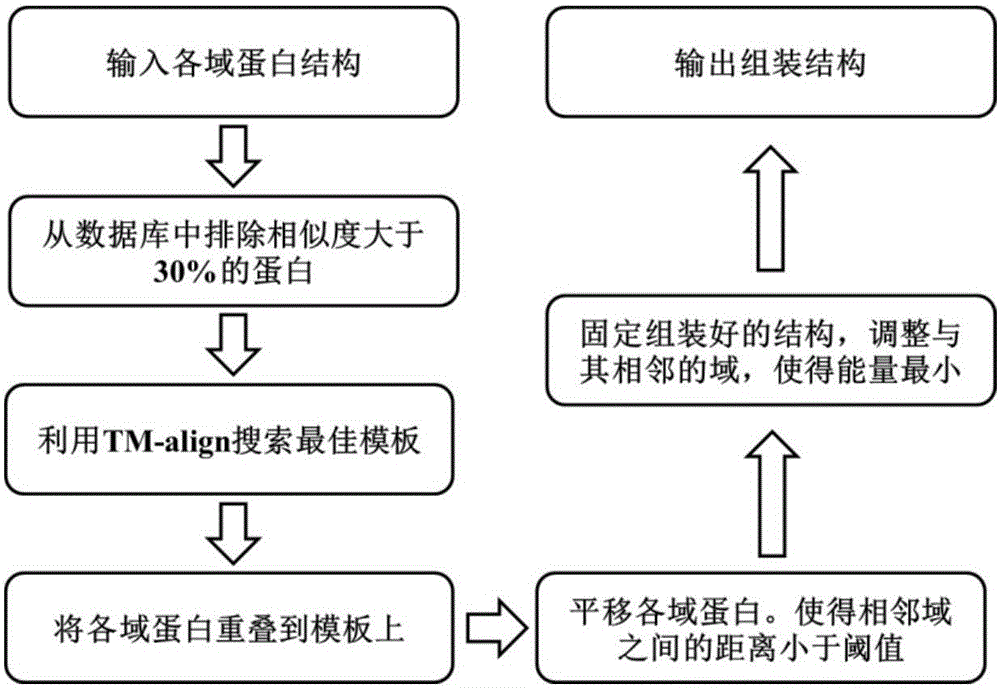
Template-based multi-domain protein structure assembling method
ActiveCN107180164AImprove forecast progressTo achieve the effect of guiding assemblySpecial data processing applicationsMolecular structuresProtein structureTemplate based
The invention discloses a template-based multi-domain protein structure assembling method. The method comprises the steps of firstly, comparing domain proteins once by utilizing a protein structure comparison tool TM-align according to the structures of the domain proteins, and finding out an optimal template from a multi-domain protein database; secondly, obtaining a rotation and translation matrix by utilizing a Kabsch method, overlapping the domain proteins to the template, and performing translation and rotation operations on the domain proteins, thereby enabling a distance between the domain proteins to be equal to a minimum allowed distance; thirdly, performing adjustment by performing random translation and rotation on an assembled structure, and measuring the quality of the assembled structure by utilizing a conflict factor between the domain proteins, an interactive atom quantity and a moving range of the assembled structure relative to the template; and in an assembling process, assembling the adjacent domain proteins in sequence, fixing the assembled structures, and when all the structures are assembled, outputting a final assembling result. The template-based multi-domain protein structure assembling method provided by the invention is relatively high in prediction precision.
Owner:ZHEJIANG UNIV OF TECH
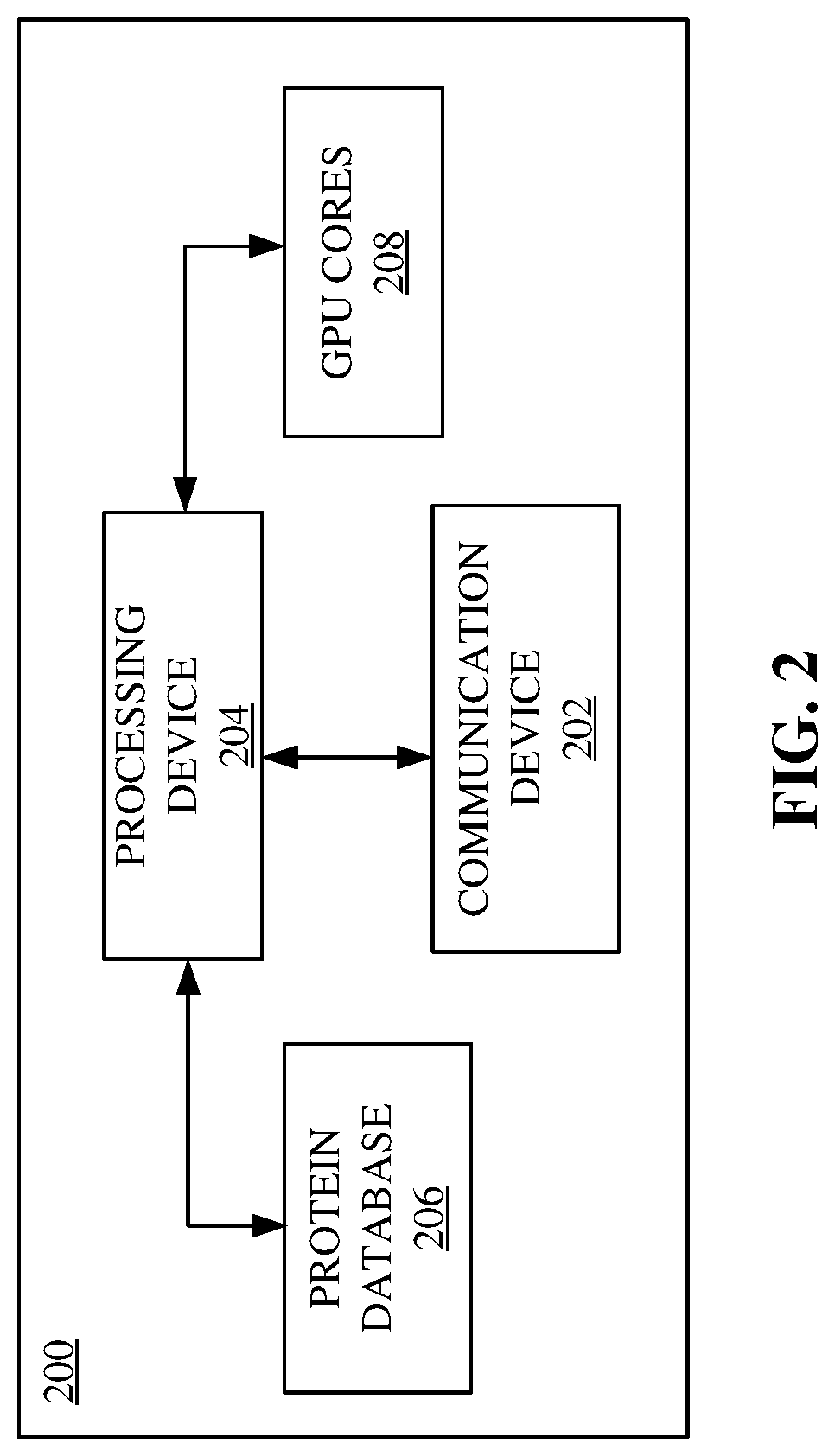
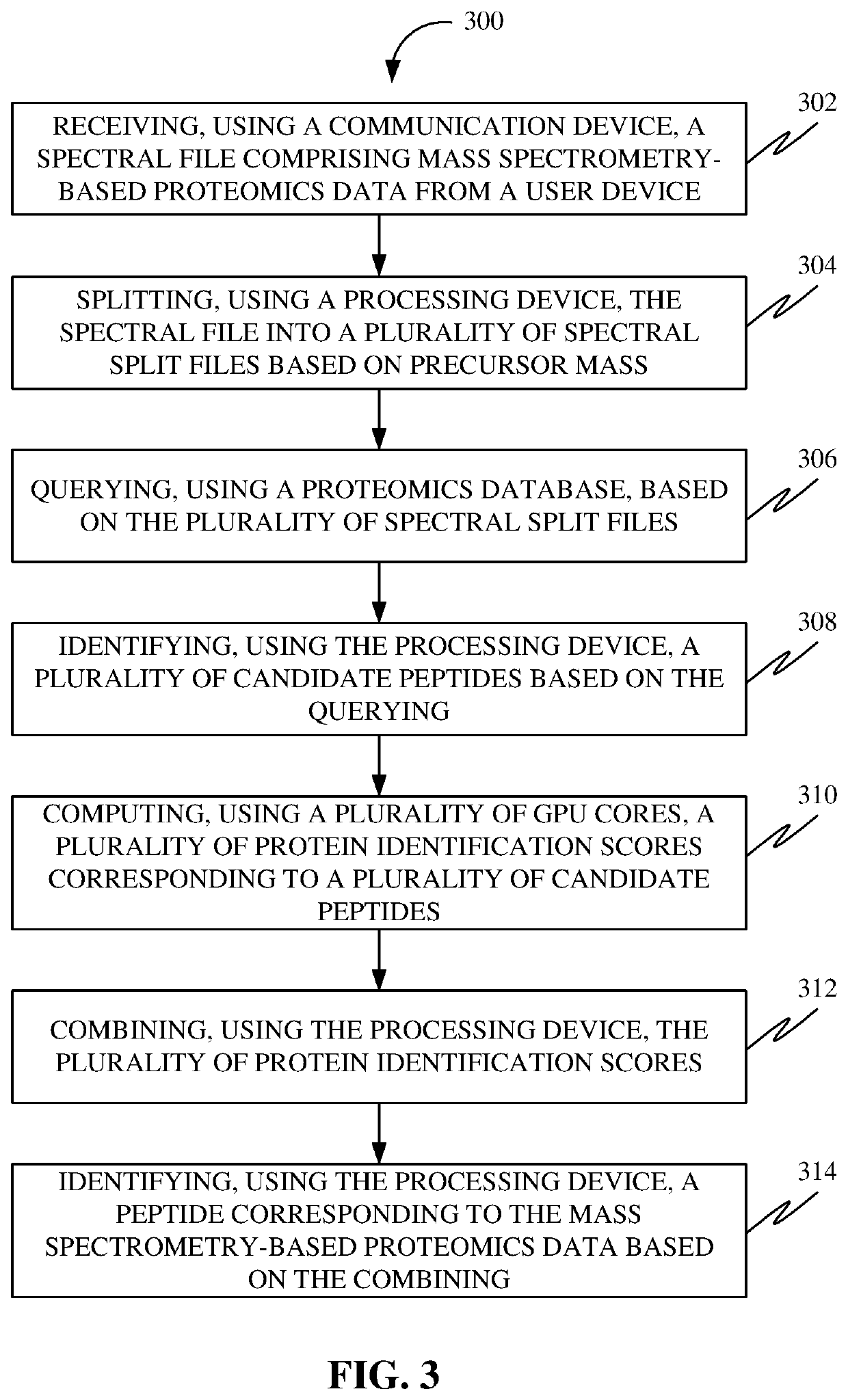
Methods, systems, apparatuses and devices for accelerating execution of a search query for peptide identification
A system for accelerating execution of a search query for peptide identification is disclosed. The system may include a communication device configured for receiving a spectral file including mass spectrometry-based proteomics data from a user device. Further, the system may include a processing device configured for splitting the spectral file into spectral split files based on precursor mass, identifying candidate peptides based on querying, combining protein identification scores, and identifying a peptide corresponding to the mass spectrometry-based proteomics data based on the combining. Further, the system may include a protein database configured for querying based on the plurality of spectral split files. Further, the system may include a plurality of GPU cores communicatively coupled to the processing device configured for computing the plurality of protein identification scores corresponding to candidate peptides.
Owner:BRUKER SCI LLC
Compression and clustering-based batch protein homology search method
ActiveCN106022000AShorten the timeBiostatisticsSpecial data processing applicationsProtein DatabasesSequence clustering
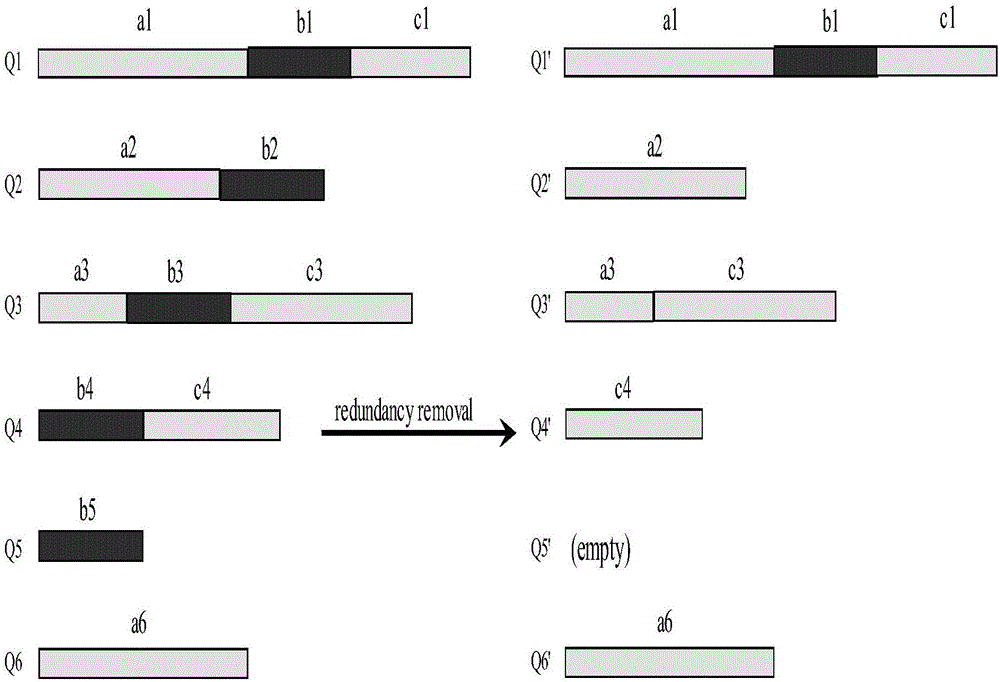
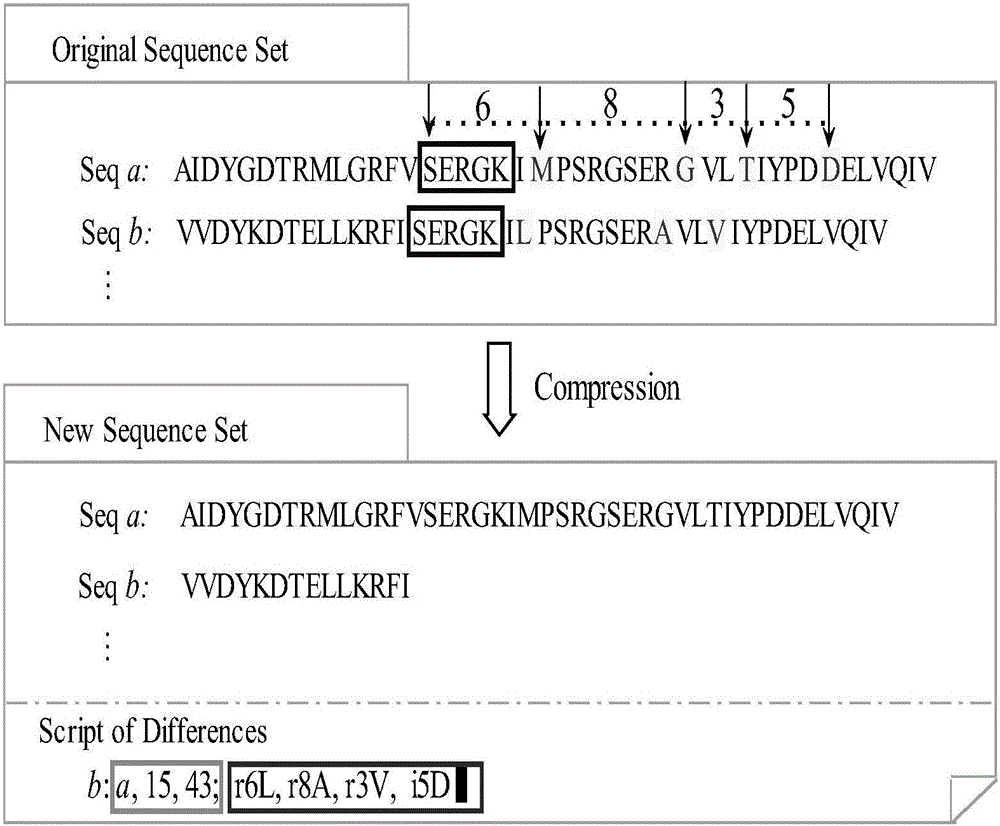
The invention discloses a compression and clustering-based batch protein homology search method and belongs to the cross field of computer application technologies and bio-technologies. The method comprises the steps of firstly performing compression operation on a query sequence and a protein database through redundancy analysis and redundancy removal processes by fully utilizing sequence similar information existent in a protein database sequence and the query sequence; secondly performing similar sub-sequence clustering on the compressed protein database; thirdly performing a search by utilizing a mapping principle based on the clustered database to discover potential results, and establishing an executable database according to the found potential result set; and finally performing a homology search in the executable database to obtain a final homology sequence. According to the method, the homology search is performed in the established executable database, so that the time for repeated sequence comparison and gapless expansion is greatly shortened.
Owner:DALIAN UNIV OF TECH
Screening method for inhibiting angiogenesis targets through celecoxib
The invention belongs to the field of treating tumor diseases, and particularly relates to a screening method for inhibiting angiogenesis targets through celecoxib. According to the method, by selecting protein expressed by 84 genes on a blood vessel formation polymerase chain reaction chip as a potential receptor protein database (PDB), the PDB and a UniProt database are used for screening 54 receptor protein structures, MGLTools is used for processing the receptor protein structures, after a molecular docking box is constructed, Autodock_vina is used as a molecular docking tool to calculatethe free energy of the binding site of the celecoxib and each protein receptor, and the protein receptors with the binding site free energy smaller than 9.0 kcal / mol are selected as potential targets.
Owner:ARMY MEDICAL UNIV
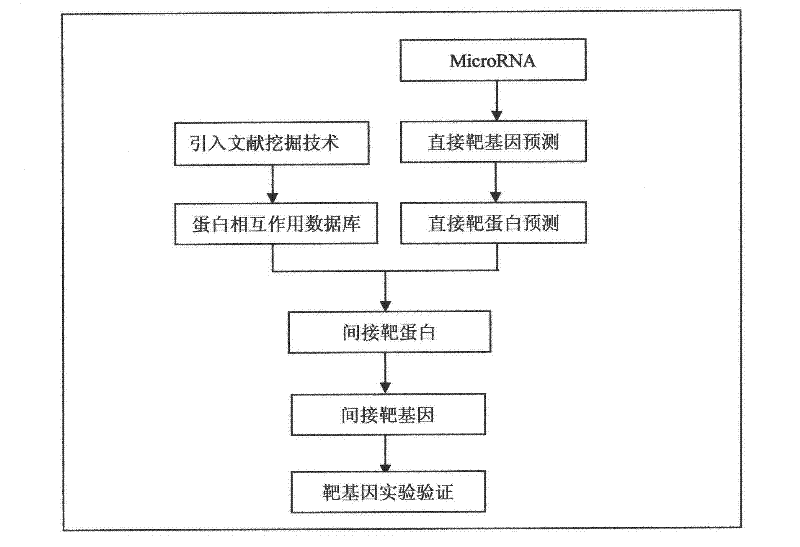
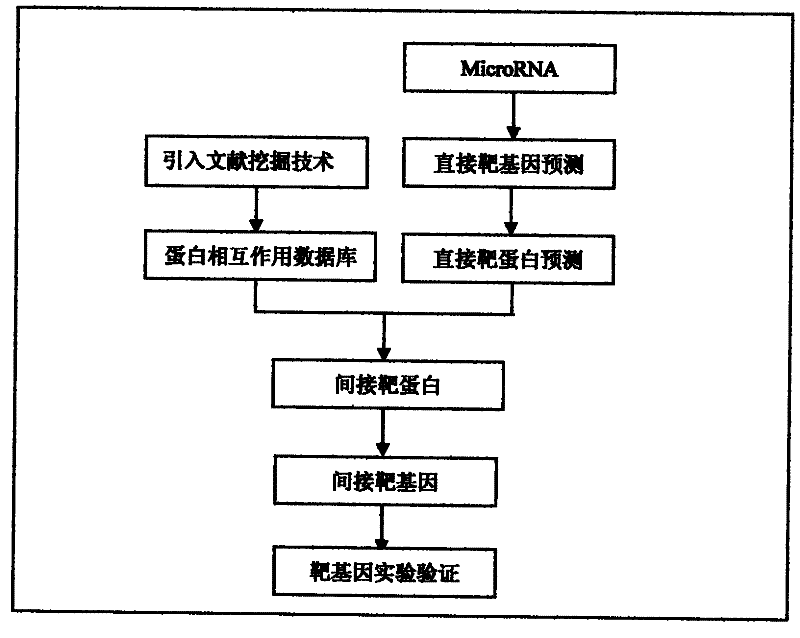
A method for predicting indirect target genes of microRNAs using literature mining
InactiveCN102268473AMicrobiological testing/measurementBiological testingPresent methodProtein target
The present invention designs a method for using literature mining technology to study the relationship between virus and human protein expression regulation, including the following main steps: step 1, predicting the direct target gene from microRNA; step 2, predicting the direct target protein from the above target gene; Step 3, use literature mining technology to construct a protein-protein interaction database; Step 4, use the constructed protein interaction database to screen out the indirect target protein and indirect target gene of microRNA from the direct target protein; Step 5, through the experiment Validation of indirect target genes of microRNAs. The feature of this method is that an "indirect" target gene model is proposed, and the literature mining technology is introduced to screen out the real microRNA target genes that are difficult to detect by conventional methods.
Owner:SHANGHAI CLUSTER BIOTECH
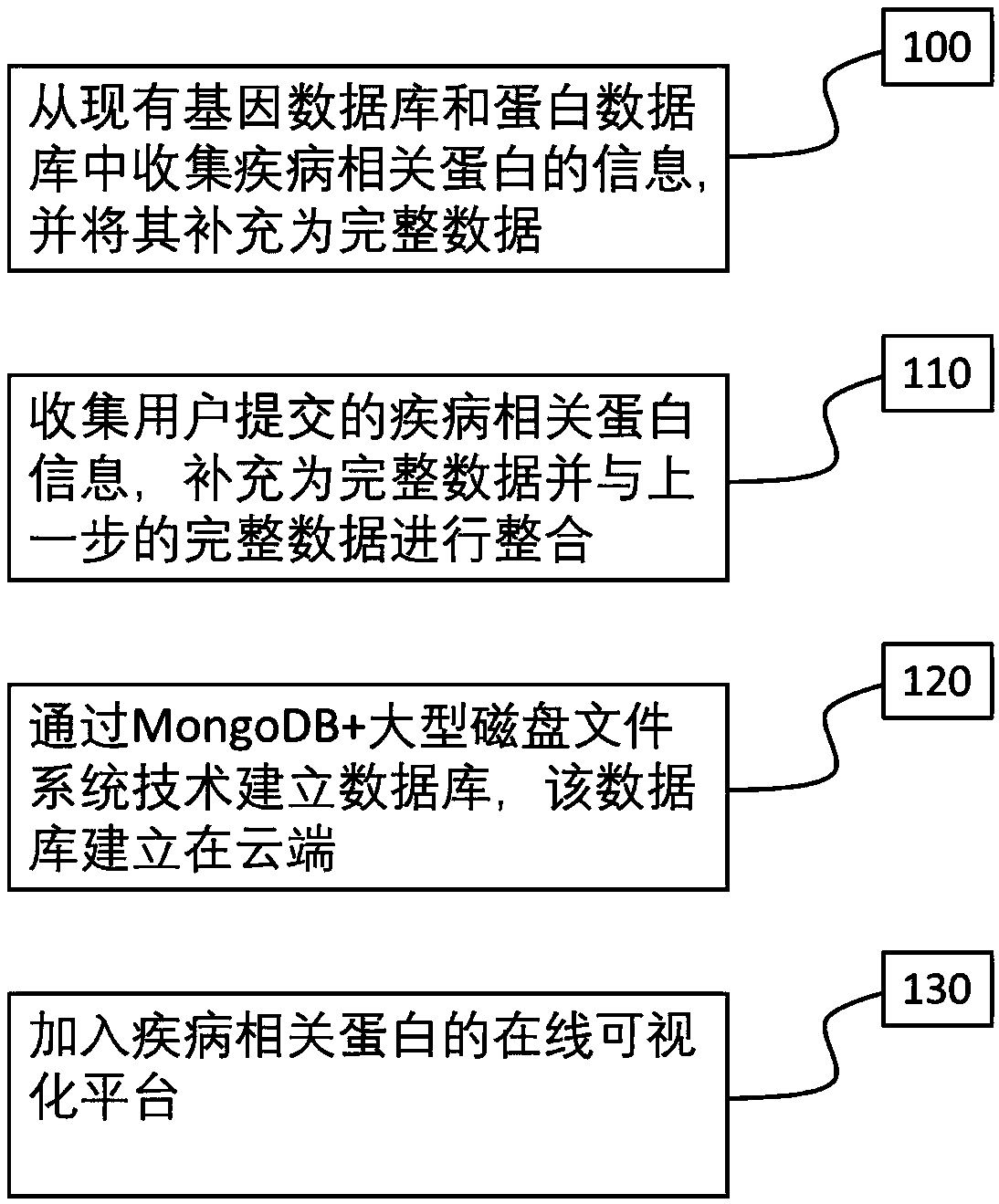
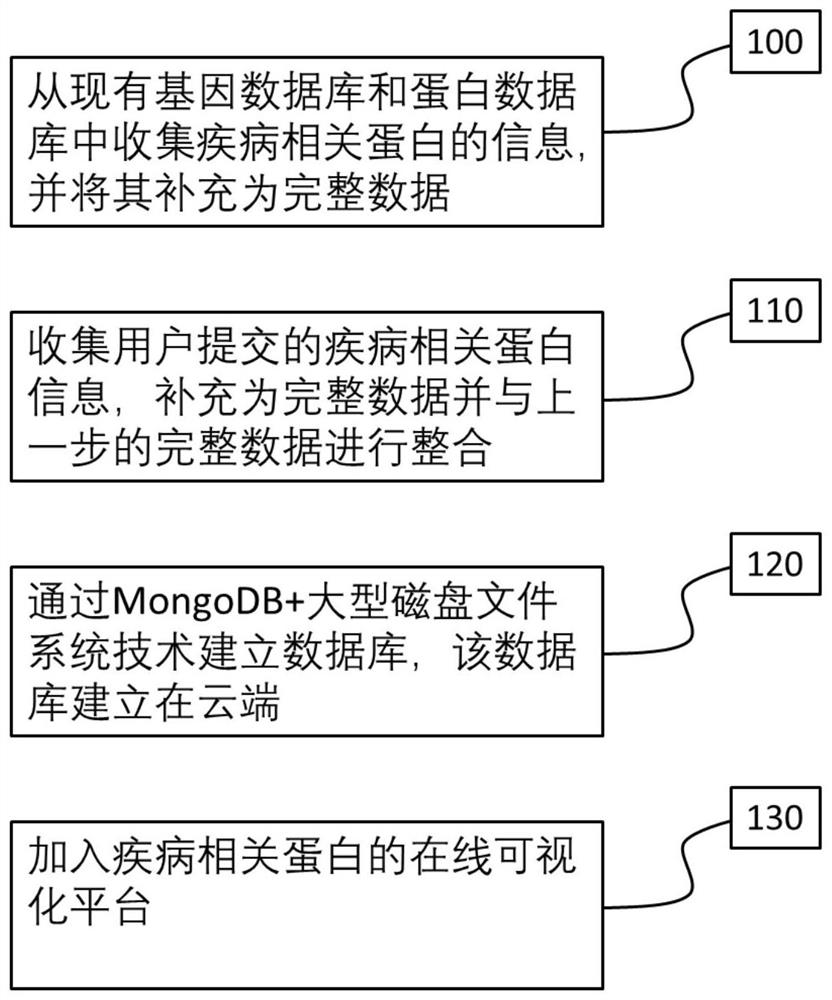
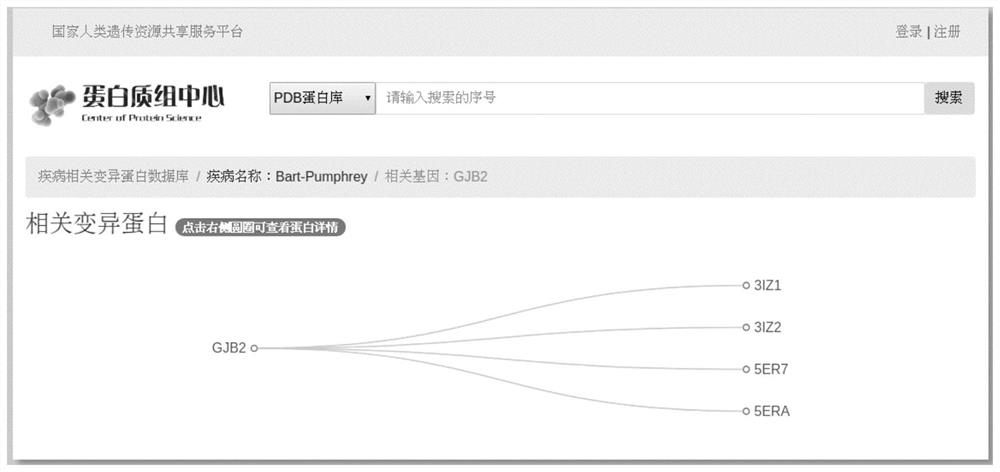
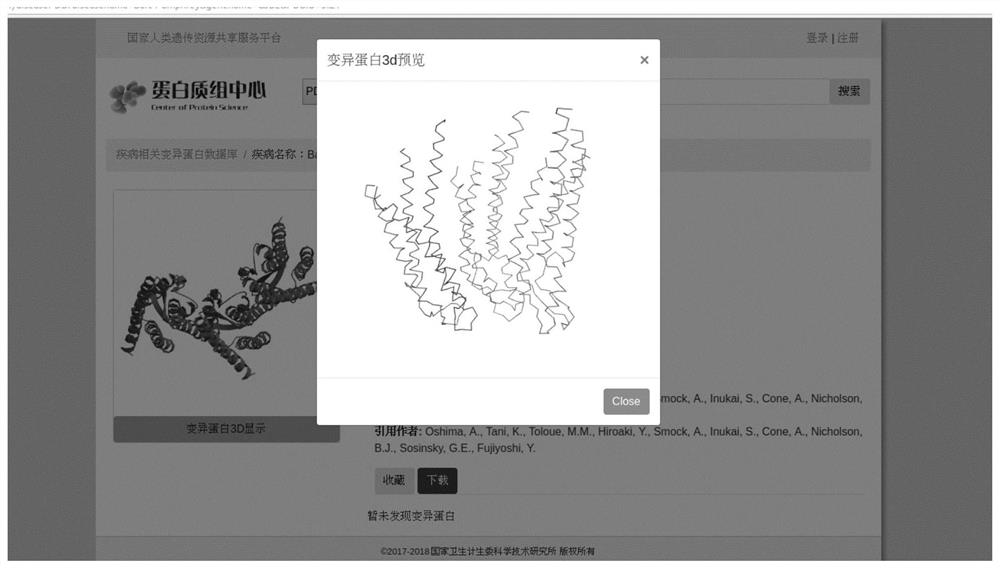
Disease Associated Protein Database
ActiveCN109086574AEasy to crawlTimely updateSpecial data processing applicationsDatabaseDisease cause
The invention relates to a disease-related protein database, a construction method and an application thereof. The database has the characteristics of extensive data collection of disease-related proteins, timely updating, including structural information, no local installation and fast access. At the same time, the construction method of the disease-related protein database and its application inscientific research are provided.
Owner:国家卫生健康委科学技术研究所
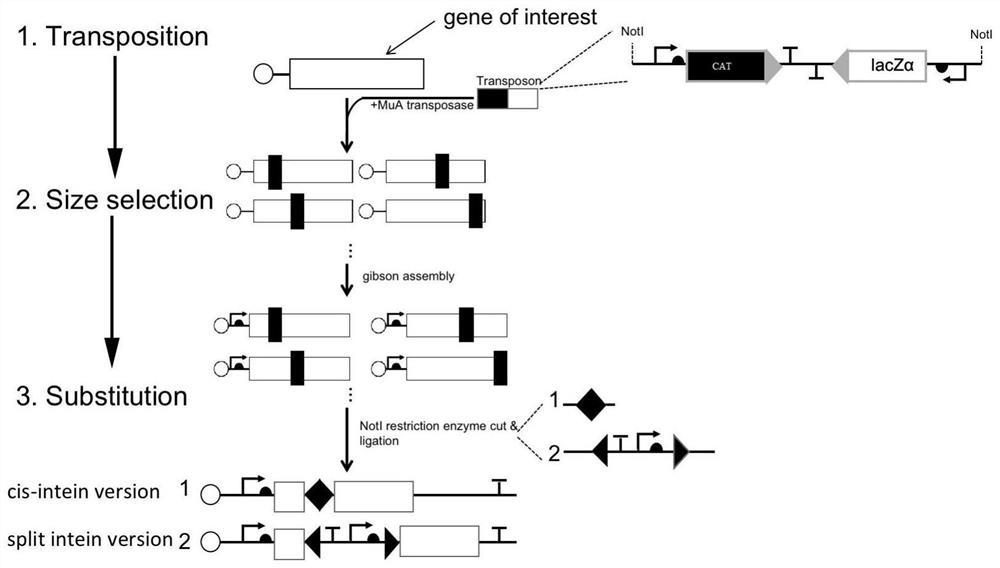
Method for screening splitting sites and application thereof
The invention provides a method for screening splitting sites and application of the method. The method comprises the following steps: S1, establishing a data writing a program by using a computer language, predicting that intein is subjected to splicing reaction after being embedded in every two adjacent amino acid residues in an amino acid sequence and is cut off, and forming a protein database by using the amino acid sequence formed by connecting adjacent peptide fragments; s2, performing an experiment: inserting intein sequence fragments into the gene fragments, then performing molecular cloning and translation to obtain peptide fragments, detecting whether the peptide fragments contain labeled amino acid sequences or not through mass spectrometry, and comparing the peptide fragments with the protein database to realize verification of the fracture sites. According to the method, a protein database is constructed through computer programming, then an experiment is carried out, and detection and verification of splitting sites are realized through mass spectrometry; final detection is achieved through mass spectrum, high-throughput screening is replaced, and the method is expanded to search for splitting sites of any active protein.
Owner:SHENZHEN INST OF ADVANCED TECH
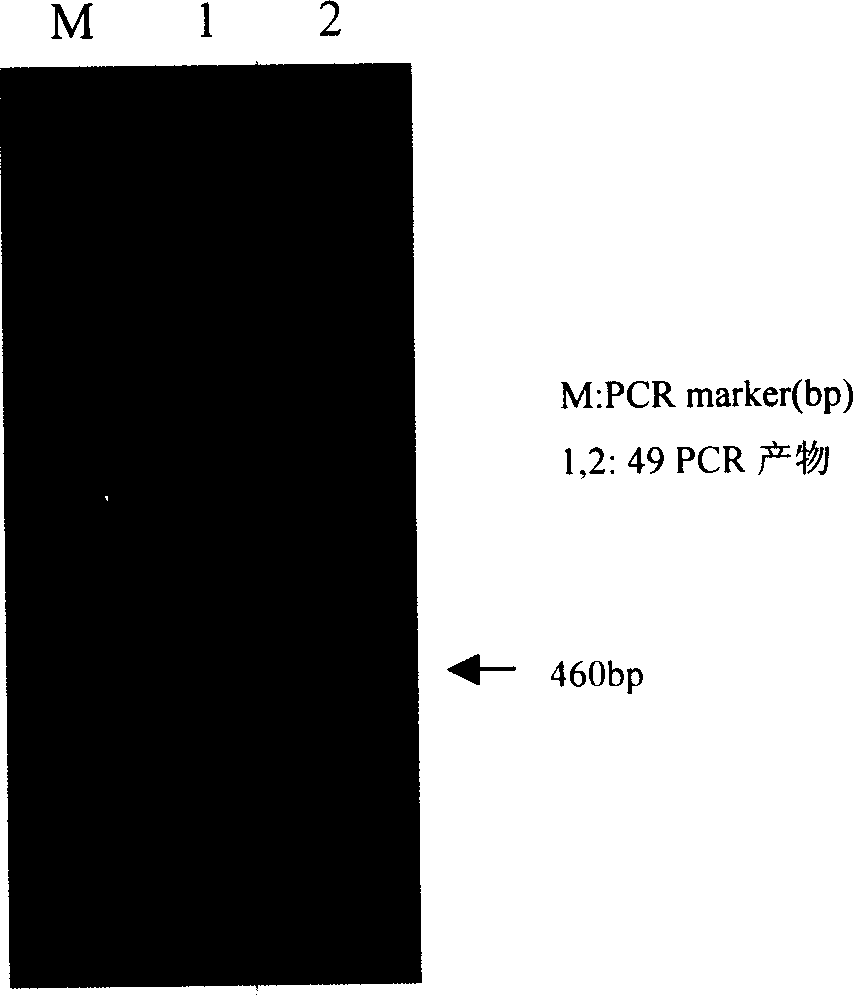
Salt-resistant gene in wheat
The invention obtains the specific expression protein of wheat salt tolerant mutant, and finds by mass spectrum identification, Web protein database search and bio-information technique that the protein has high homology with one assumed protein on the 4th rice chromosome. Accordingly, it designs primer, amplifies by PCR to obtain the corresponding wheat salt tolerance gene with 153 amino acids and 462bp length; clones the gene to construct pronucleus expression vector to express out a specific protein with molecular weight as 16.8KD and improve the salt tolerance of escherichia coli obviously. This invention settles foundation for wheat salt tolerance mechanism.
Owner:HEBEI NORMAL UNIV
Method For Using Protein Databases To Identify Microorganisms
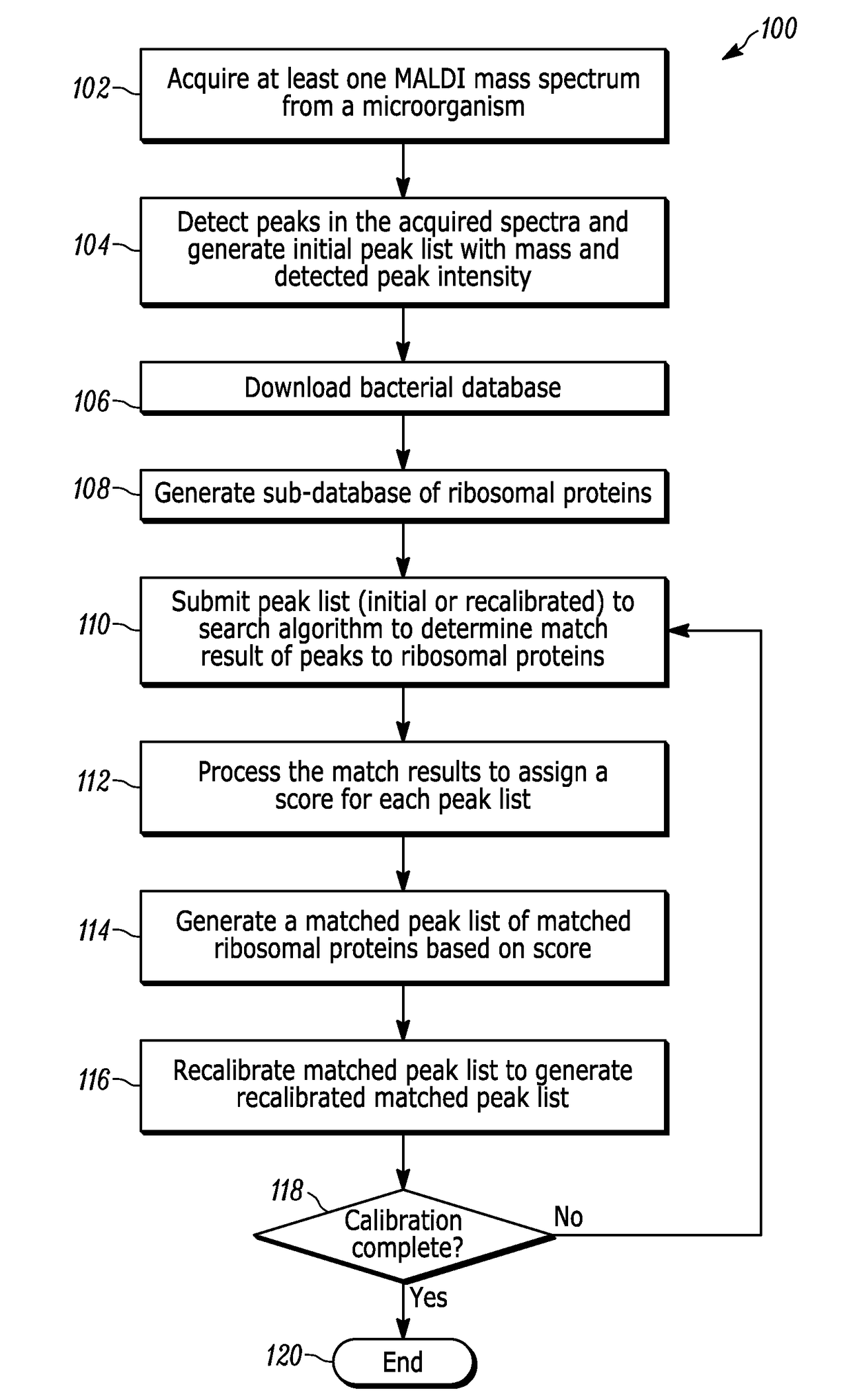
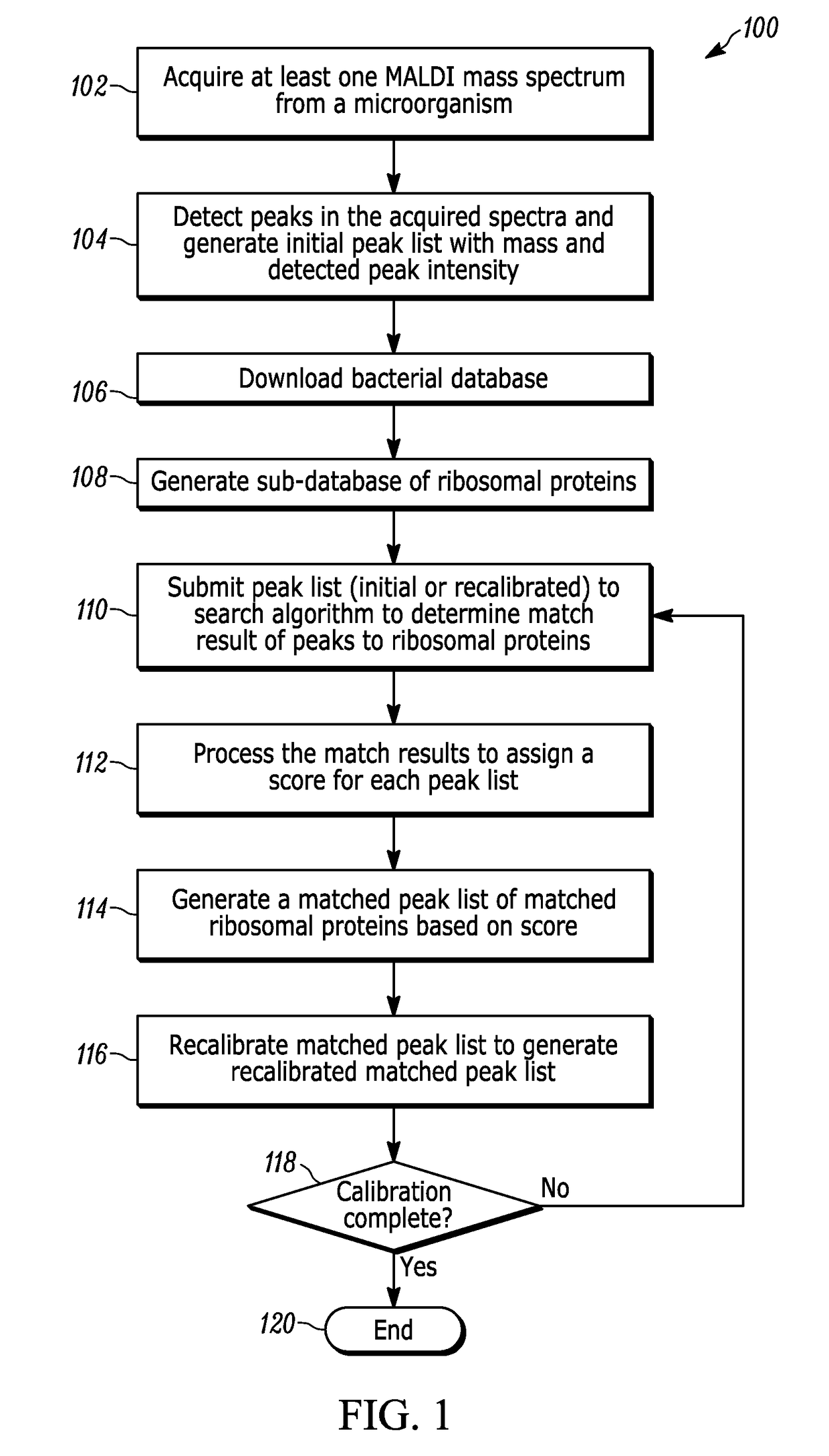
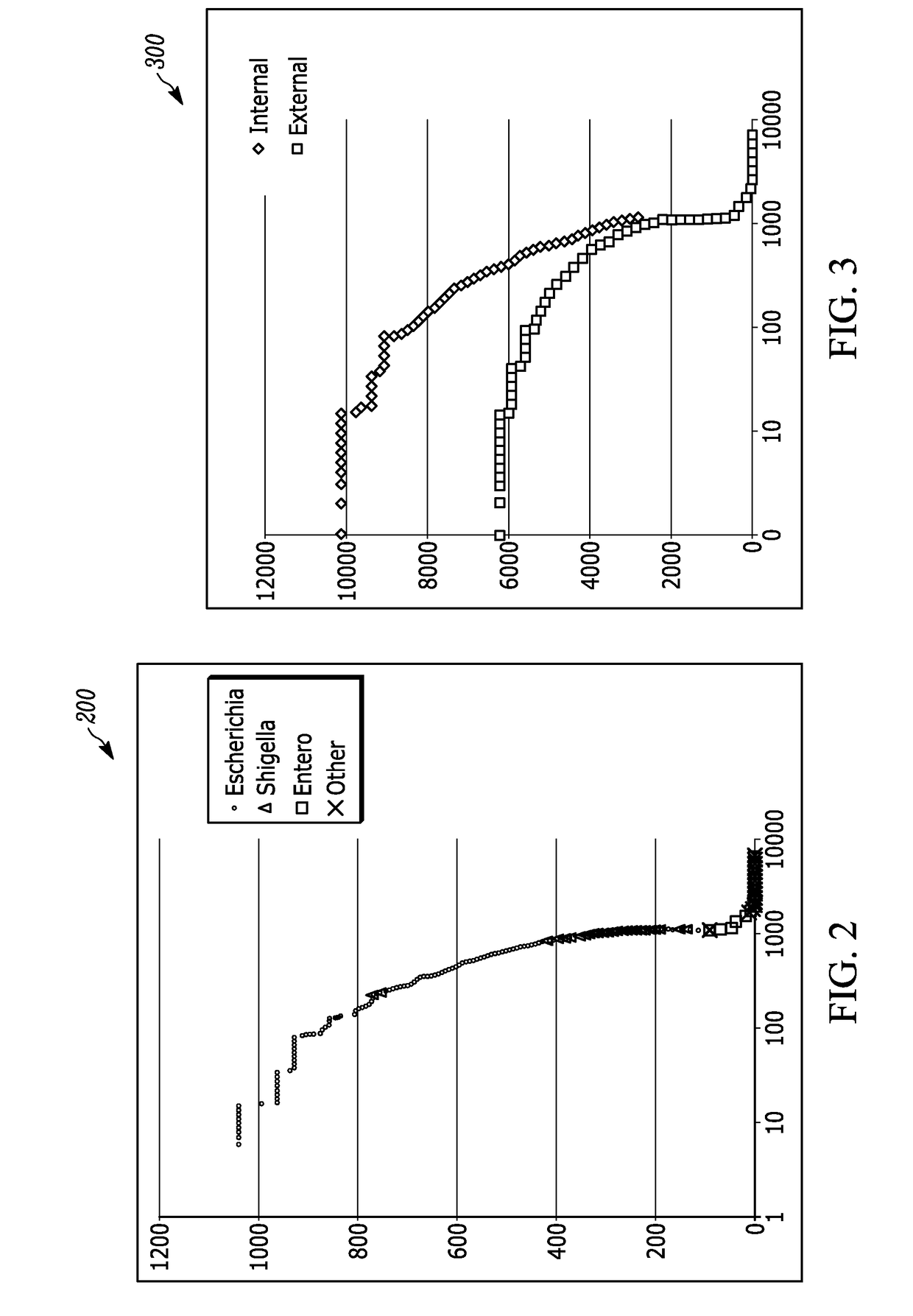
A method for identifying microorganisms by MALDI-TOF mass spectrometry includes acquiring a MALDI mass spectrum of a microorganism, detecting peaks in the acquired MALDI spectrum, generating a peak list comprising mass and intensity from the detected peaks in the acquired MALDI spectrum, acquiring a database of protein sequences deduced from DNA sequences for microorganisms, generating a sub-database of ribosomal proteins from the protein sequences and their masses in the database, matching masses of the detected peaks in the acquired MALDI spectrum to masses of the ribosomal proteins in the generated sub-database, scoring the matches obtained above for each represented microorganism, generating a peak list of accurate masses of matched ribosomal proteins, recalibrating the peak list comprising mass and intensity with the peak list of accurate masses of matched ribosomal proteins, identifying a microorganism with the highest score, and repeating until a desired improvement in the recalibrated peak list or a validated identification is achieved.
Owner:VIRGIN INSTR CORP
Destructive beta-conglycinin beta subunit processing antigen area and screening method based on phage display technological orientation
The invention discloses a destructive beta-conglycinin beta subunit processing antigen area and a screening method based on phage display technological orientation. The amino acid sequence of the antigen area is as shown in SEQ ID NO. 15. By the aid of a series of bioinformatics software and reference of an analyzed beta-conglycinin three-dimensional crystalline structure of a PDB (protein database), the antigen area of ultrahigh-pressure destructive beta-conglycinin is researched by a phage display technology, a beta-conglycinin beta subunit and overlapping protein thereof are represented onthe surface of a phage, and an area with the beta-conglycinin beta subunit with antigenicity reduced by an ultrahigh-pressure is accurately positioned. A theoretical basis for screening processing methods is provided for food industry, and an application product for rapidly detecting the desensitization effect of processed food can be further developed.
Owner:HENAN UNIVERSITY OF TECHNOLOGY
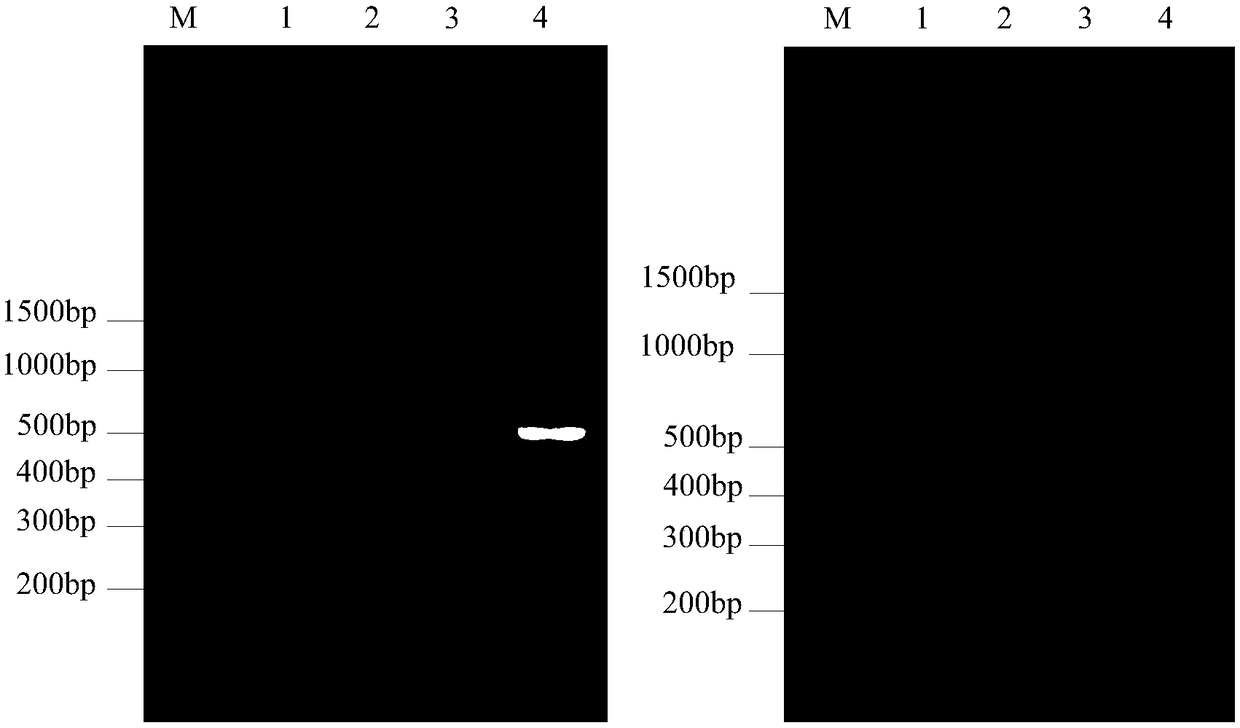
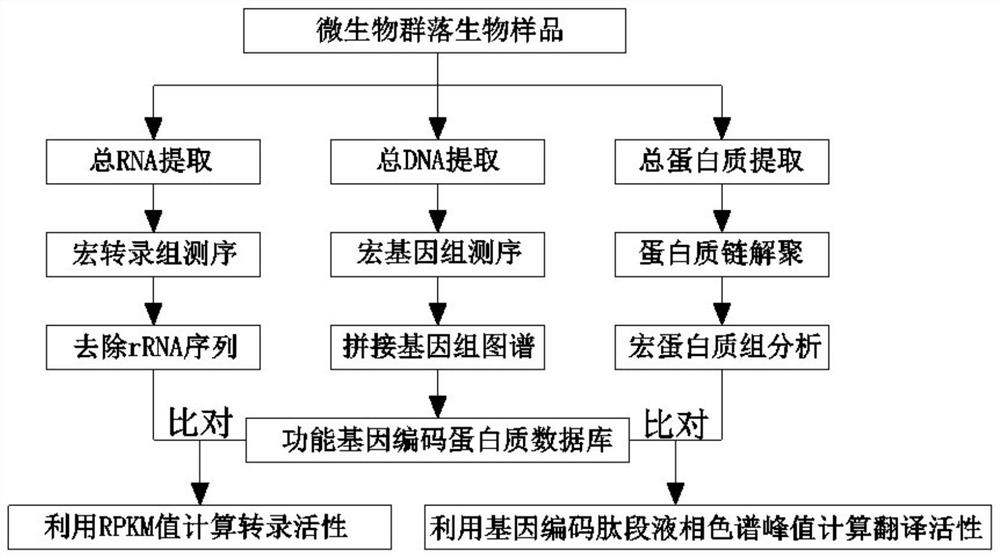
Method for analyzing transcription and translation activity of functional genes of microbial community
ActiveCN112342284AImprove integrityAdd depthComponent separationMicrobiological testing/measurementGenomic sequencingMicroorganism
The invention discloses a method for analyzing the transcription and translation activity of functional genes of a microbial community. The method comprises the following steps: extracting a total DNAsample, total RNA sample and total protein sample of the microbial community; carrying out metagenome sequencing, metatranscriptome sequencing and meta-protrinology analysis on the extracted samples;constructing a microbial community functional gene coded protein database by utilizing a metagenome sequencing result; analyzing the transcription activity of the functional genes of the microbial community on the basis of the protein database and an mRNA sequence data set generated by metatranscriptome sequencing; and analyzing the translation activity of the functional genes of the microbial community on the basis of a meta-protrinology analysis result and the protein database. According to the method, the unified microbial community functional genome protein database is constructed, and the expression quantities of transcription and translation of the functional genes of the community are analyzed according to the database; the differences of the expression levels of transcription andtranslation of the functional genes of the community are directly compared, thereby researching the post-transcriptional control process of the functional genes of the community.
Owner:PEKING UNIV
Identification of proteins using a physical parameter and accurate amino acid content
An unidentified purified protein can be uniquely identified by combining one or more physical parameters of the protein and an accurate amino acid content. The protein can be identified by input of the one or more physical parameters and the amino acid content into an interface that outputs data from a protein database.
Owner:US DEPT OF HEALTH & HUMAN SERVICES
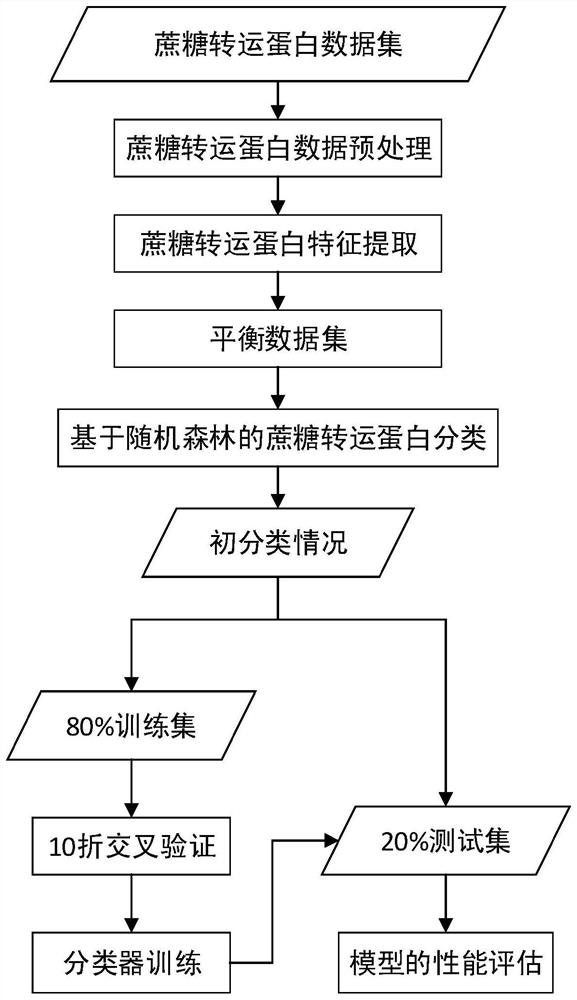
Random forest-based sucrose transporter identification method
PendingCN113902941ASolve classification problemsCharacter and pattern recognitionMachine learningData imbalanceData set
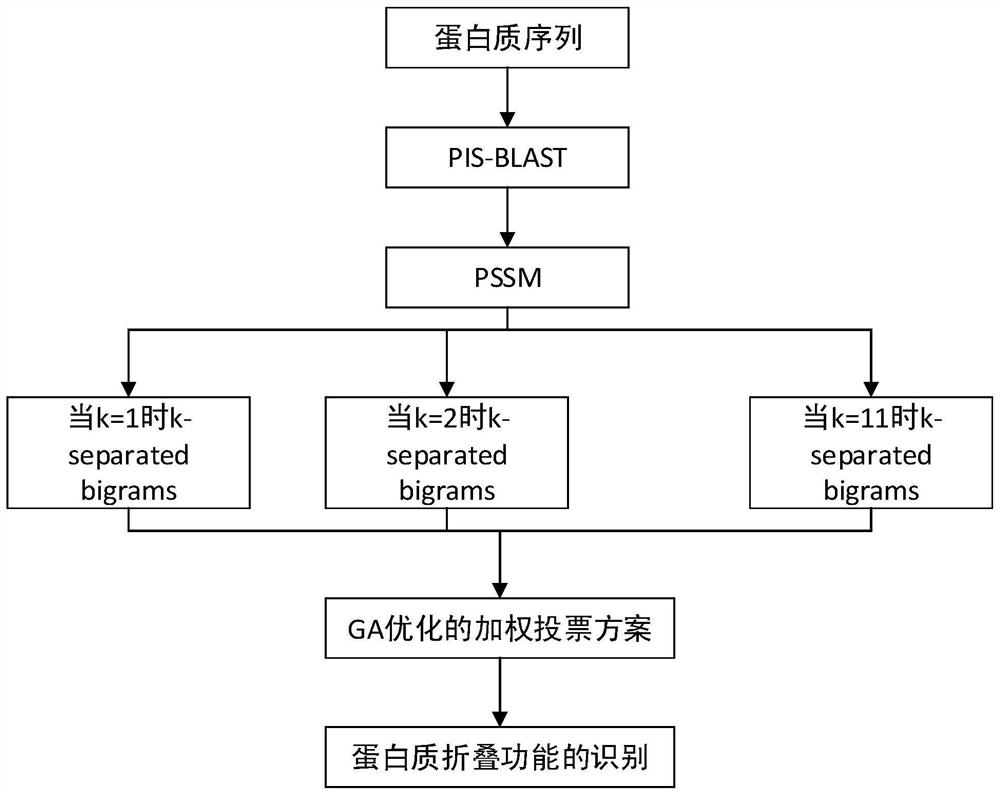
The invention relates to a random forest-based sucrose transporter identification method, which comprises the following steps of: firstly, acquiring initial data from a protein database, preprocessing the initial data, deleting sequences containing non-standard letters and sequences with too short lengths, and deleting sequences with the similarity greater than 60%; extracting different features according to physicochemical properties and evolutionary information of the protein, and taking each feature and the combined feature as feature input; then, due to the fact that the difference between the sample numbers of the positive example and the counter example is large, performing oversampling on the data set; and finally, under ten-fold cross validation, performing feature training on the oversampled training set by using a random forest, a support vector machine, stochastic gradient descent and naive Bayes, performing testing by using a test set, and analyzing a result. According to the method, a k-separated-bigrams-PSSM and random forest combined method is used, and a Borderline-SMOTE algorithm is introduced, so that the problem of data imbalance is solved, and the identification accuracy of the sucrose transporter is effectively improved.
Owner:NORTHEAST FORESTRY UNIVERSITY
Method for screening characteristic fragments of yak collagen through high-resolution mass spectrometry
InactiveCN113501861ARapid identificationEfficient technical supportConnective tissue peptidesBiological testingChromatographic separationResolution (mass spectrometry)
The invention discloses a method for screening characteristic fragments of yak collagen through high-resolution mass spectrometry. The method is characterized by comprising the following steps: (1) sample collection; (2) pretreatment; (3) performing enzymolysis; (4) high performance liquid chromatography separation: separating the enzymatic hydrolysate by adopting a high performance liquid chromatography system EasynLC with a nano-liter flow velocity; (5) mass spectrum identification: carrying out mass spectrum identification by using a Q-Exactive mass spectrometer after carrying out chromatographic separation on the enzymatic hydrolysate; and (6) characteristic fragment screening: retrieving a UniProt protein database in an original file of a mass spectrum identification result by using Mascot2.2 software, and screening to obtain the characteristic fragments of the yak collagen. According to the method, yak collagen subjected to enzymolysis is separated and identified through a high performance liquid chromatography-tandem mass spectrometry technology, data are compared and analyzed through Mascot2.2 software and a UniProt protein database, and yak collagen characteristic fragments which can be detected in an actual sample are screened out. The identified characteristic fragments can be used for detecting the authenticity and reliability of the yak-derived collagen, and an effective technical support is provided for the aspect of yak collagen component supervision.
Owner:青海瑞肽生物科技有限公司
Disease-associated protein database
The invention relates to a disease-related protein database and its construction method and application. The database has the characteristics of extensive collection of existing disease-related protein data information, timely updates, structural information, no local installation and fast access. At the same time, the construction method of the disease-related protein database and its application in scientific research are provided.
Owner:国家卫生健康委科学技术研究所
Method and application of designing enzyme preparation feeding mode in fermentation process by computer
ActiveCN113637712BIncrease productionAdd lessMicrobiological testing/measurementBiofuelsBiotechnologyEconomic benefits
Owner:NANJING NORMAL UNIVERSITY
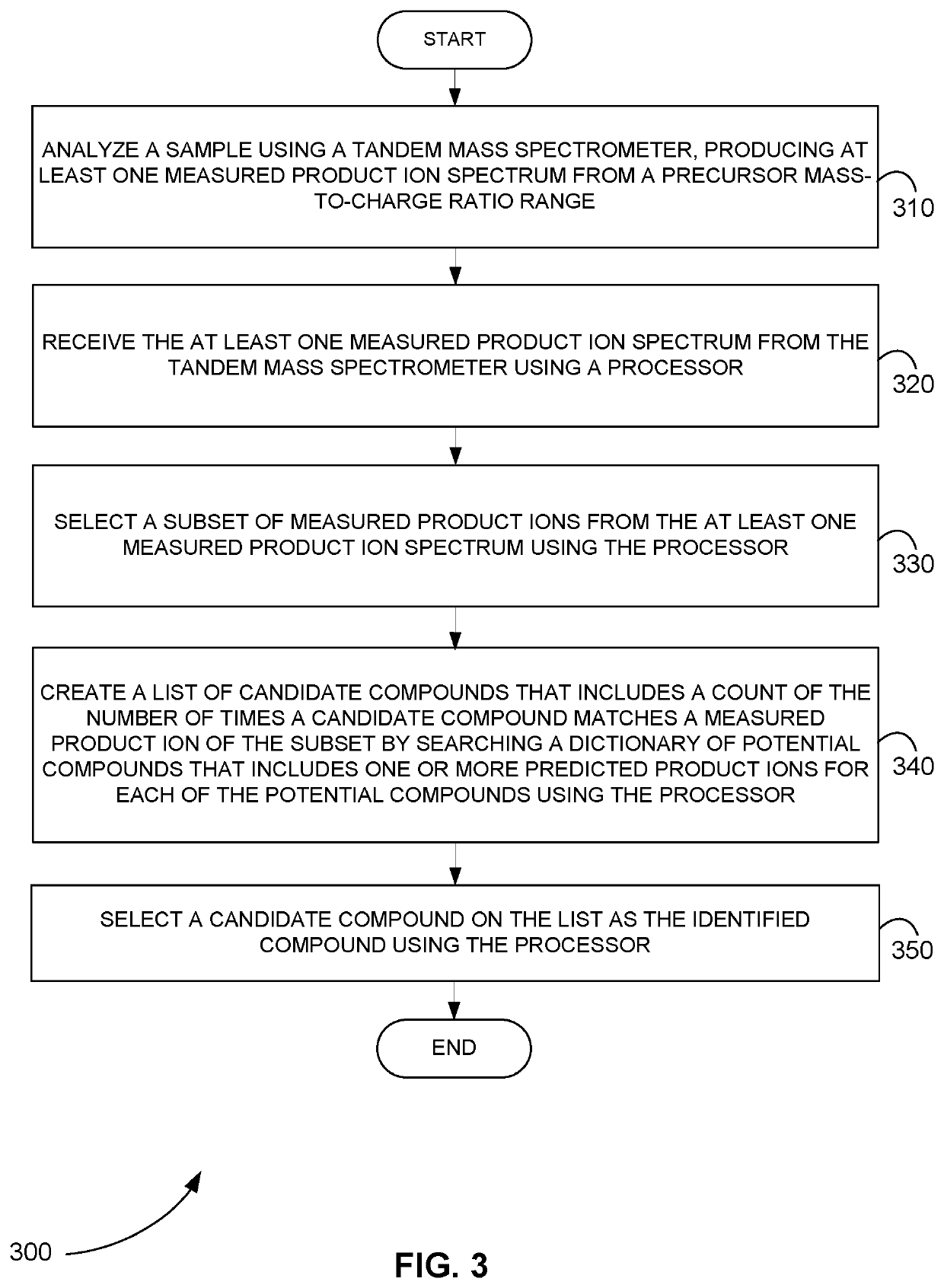
Systems and methods for identifying compounds from MS/MS data without precursor ion information
ActiveUS11456164B2Molecular entity identificationBiological testingChemical physicsChemical compound
Systems and methods are provided for identifying a precursor ion without using any a priori precursor ion information. In one method, a sample is analyzed using a tandem mass spectrometer, producing at least one measured product ion spectrum from a precursor mass-to-charge ratio range. The at least one measured product ion spectrum are received. A subset of measured product ions is selected from the at least one measured product ion spectrum. A list of candidate compounds is created by searching a dictionary of potential compounds that includes one or more predicted product ions for each of the potential compounds using the subset of measured product ions. A candidate compound on the list is selected as the identified compound. In another method, the measured product ions are assumed to correspond to shortened forms of the peptide and a protein database is searched for shortened forms of the peptide.
Owner:DH TECH DEVMENT PTE
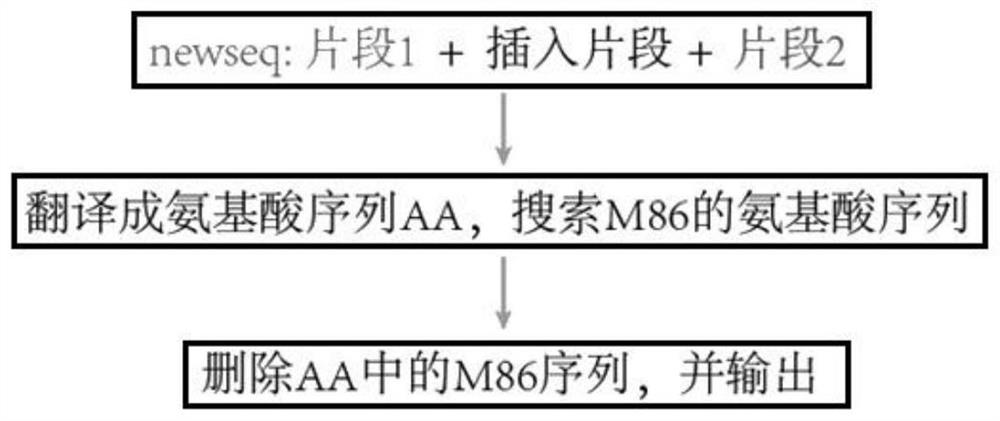
Method and device for detecting mutant protein
The invention provides a method and a device for detecting mutated proteins. The method includes acquiring transcriptome data corresponding to samples; comparing the transcriptome data to mitochondrion databases and outputting non-mitochondrion sequences according to comparison results of the transcriptome data and the mitochondrion databases; transforming nucleotide sequences in the non-mitochondrion sequences into amino acid sequences, comparing the transformed amino acid sequences to protein databases and extracting amino acid sequences with homogenous rates in first set ranges and amino acid lengths in second set ranges from comparison results of the transformed amino acid sequences and the protein databases; comparing the extracted amino acid sequences with the homogenous rates in the first set ranges and the amino acid lengths in the second set ranges to NCBI (national center of biotechnology information) and determining the mutated proteins according to comparison results of the extracted amino acid sequences and the NCBI. According to the scheme, the method and the device have the advantage that the mutated proteins in the samples can be detected by the aid of the method and the device.
Owner:天津市湖滨盘古基因科学发展有限公司
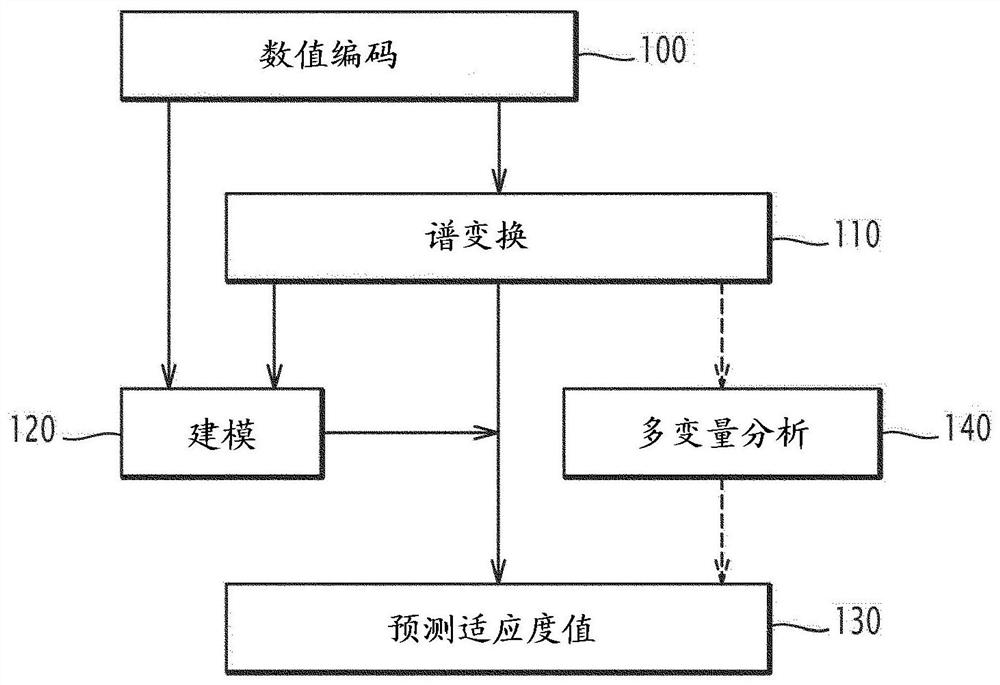
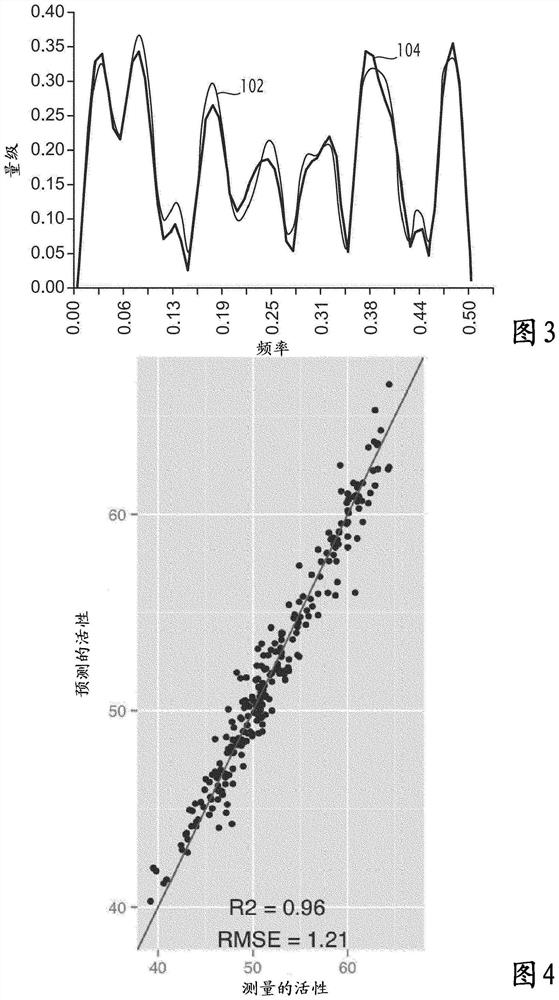
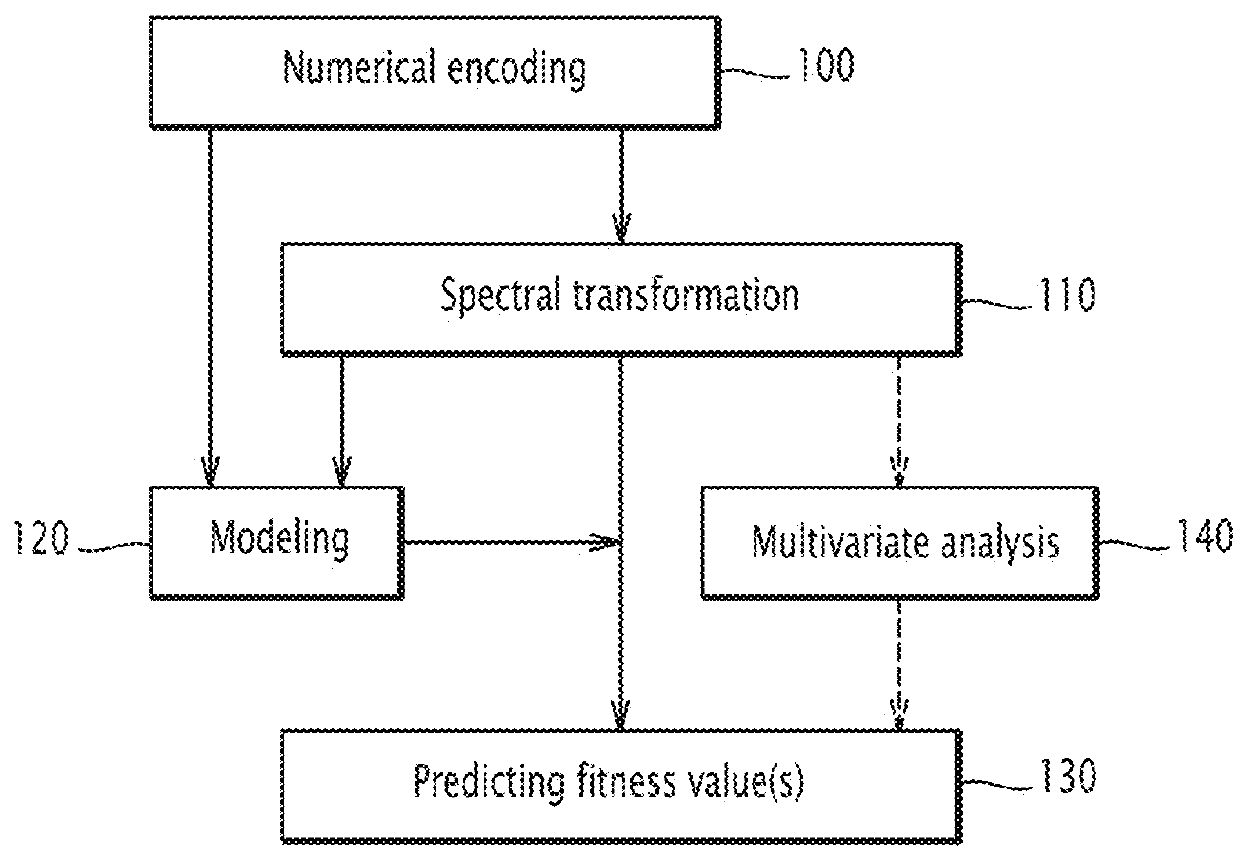
Method and electronic system for predicting at least one fitness value of protein, related computer program product
A method for predicting at least one fitness value of a protein is executed on a computer and comprises the steps of: encoding (100) an amino acid sequence of the protein into a numerical sequence according to a protein database, the numerical sequence comprising a value for each amino acid in the sequence; calculating (110) a protein profile from the sequence of values; and for each fitness: comparing (130) the calculated protein profile with protein profile values of a predetermined database containing protein profile values for different values of the fitness, predicting (130) the value of the fitness according to the comparing step.
Owner:PEACCEL
Method and electronic system for predicting at least one fitness value of a protein, related computer program product
A method for predicting at least one fitness value of a protein is implemented on a computer and includes the following steps: encoding the amino acid sequence of the protein into a numerical sequence according to a protein database, the numerical sequence comprising a value for each amino acid of the sequence; calculating a protein spectrum according to the numerical sequence; and for each fitness: comparing the calculated protein spectrum with protein spectrum values of a predetermined database, said database containing protein spectrum values for different values of said fitness, predicting a value of said fitness according to the comparison step.
Owner:PEACCEL
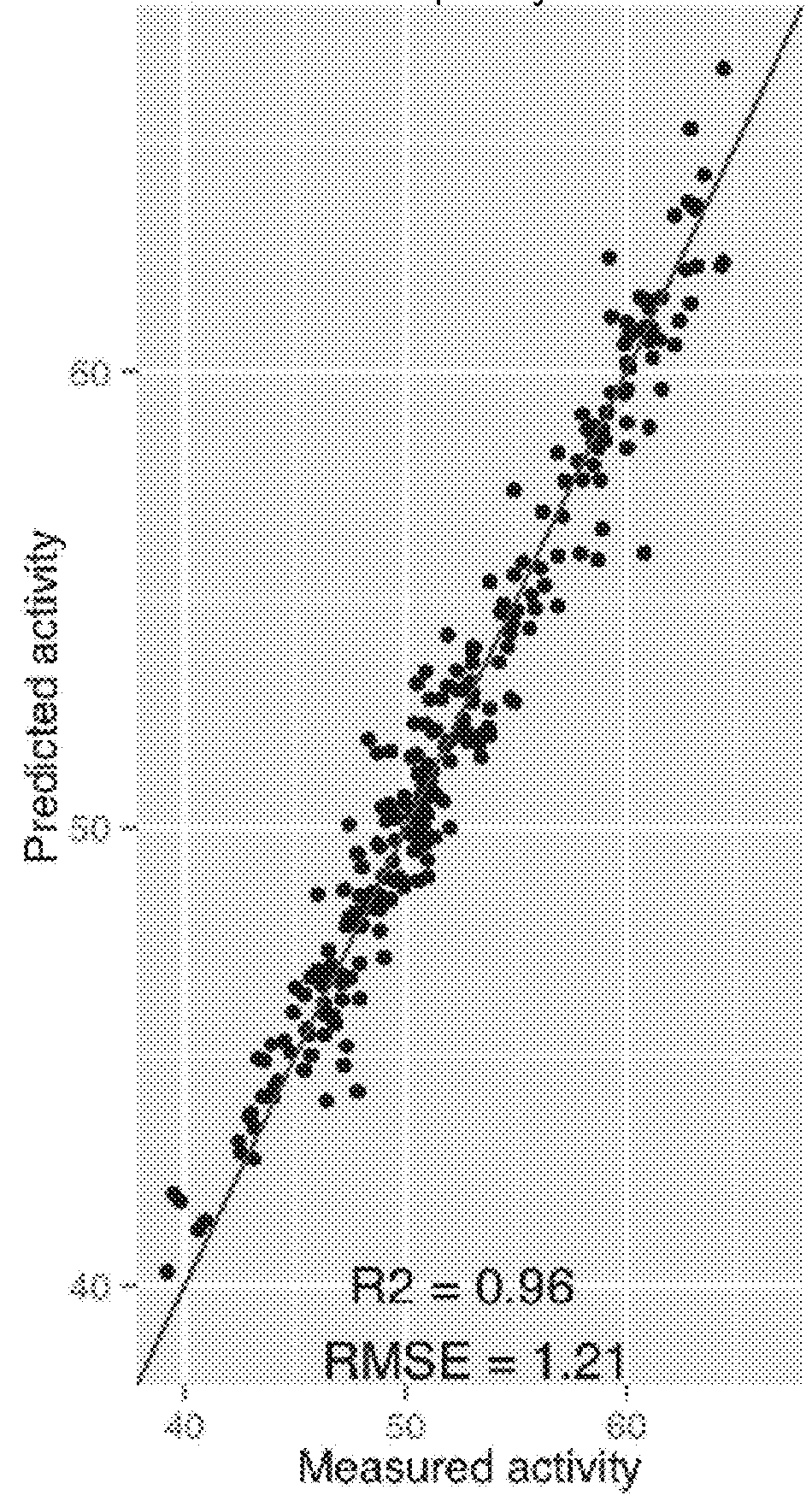
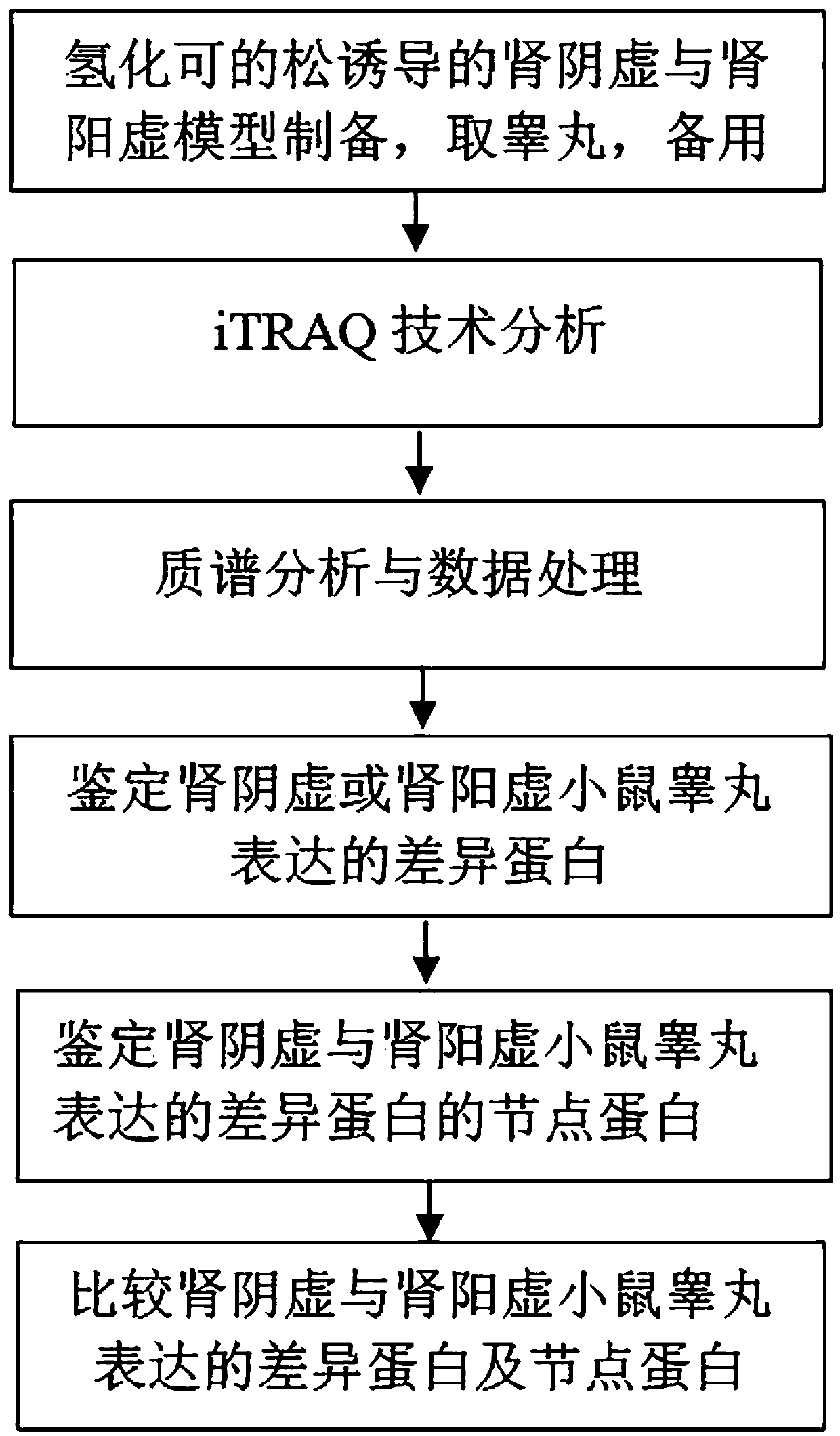
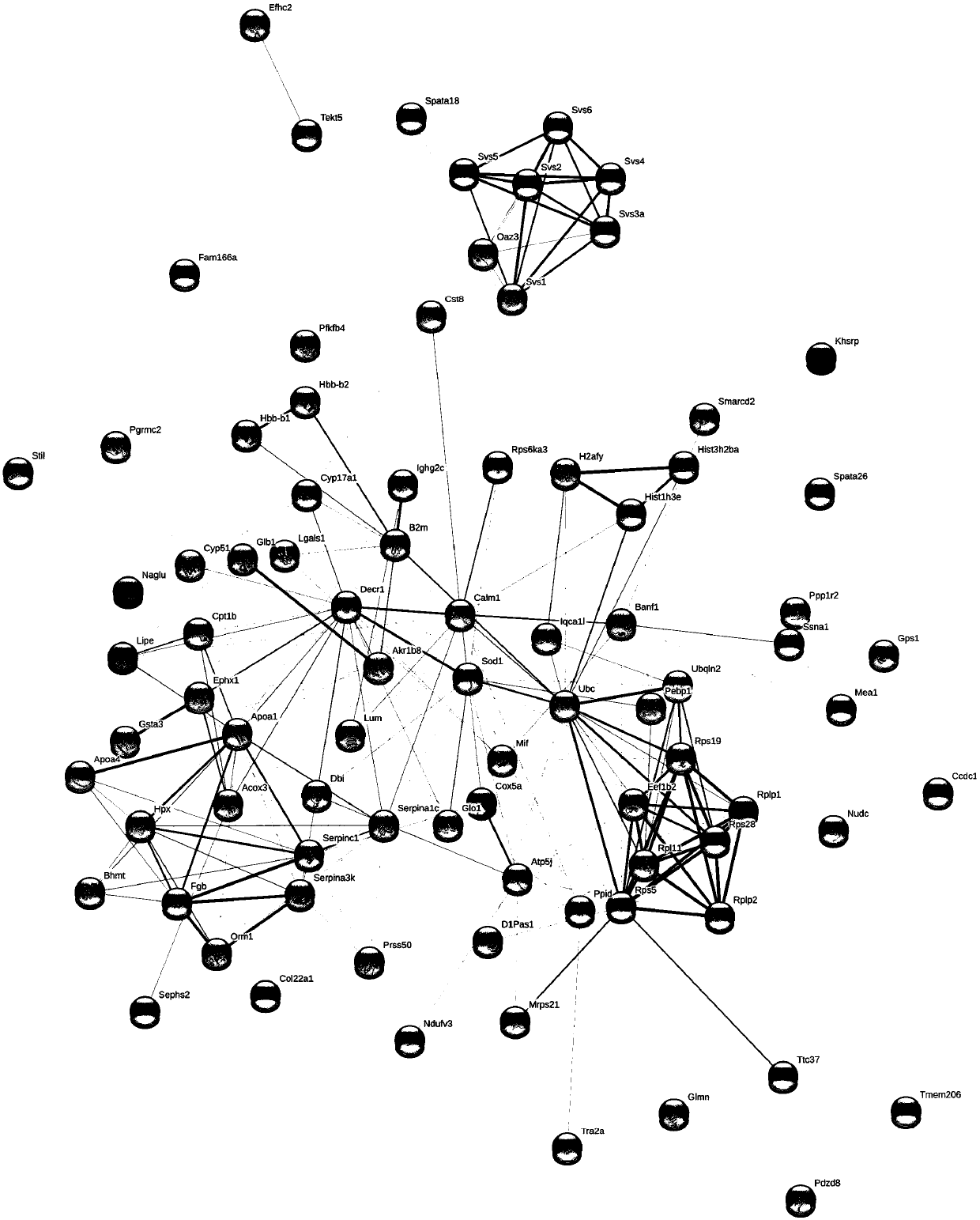
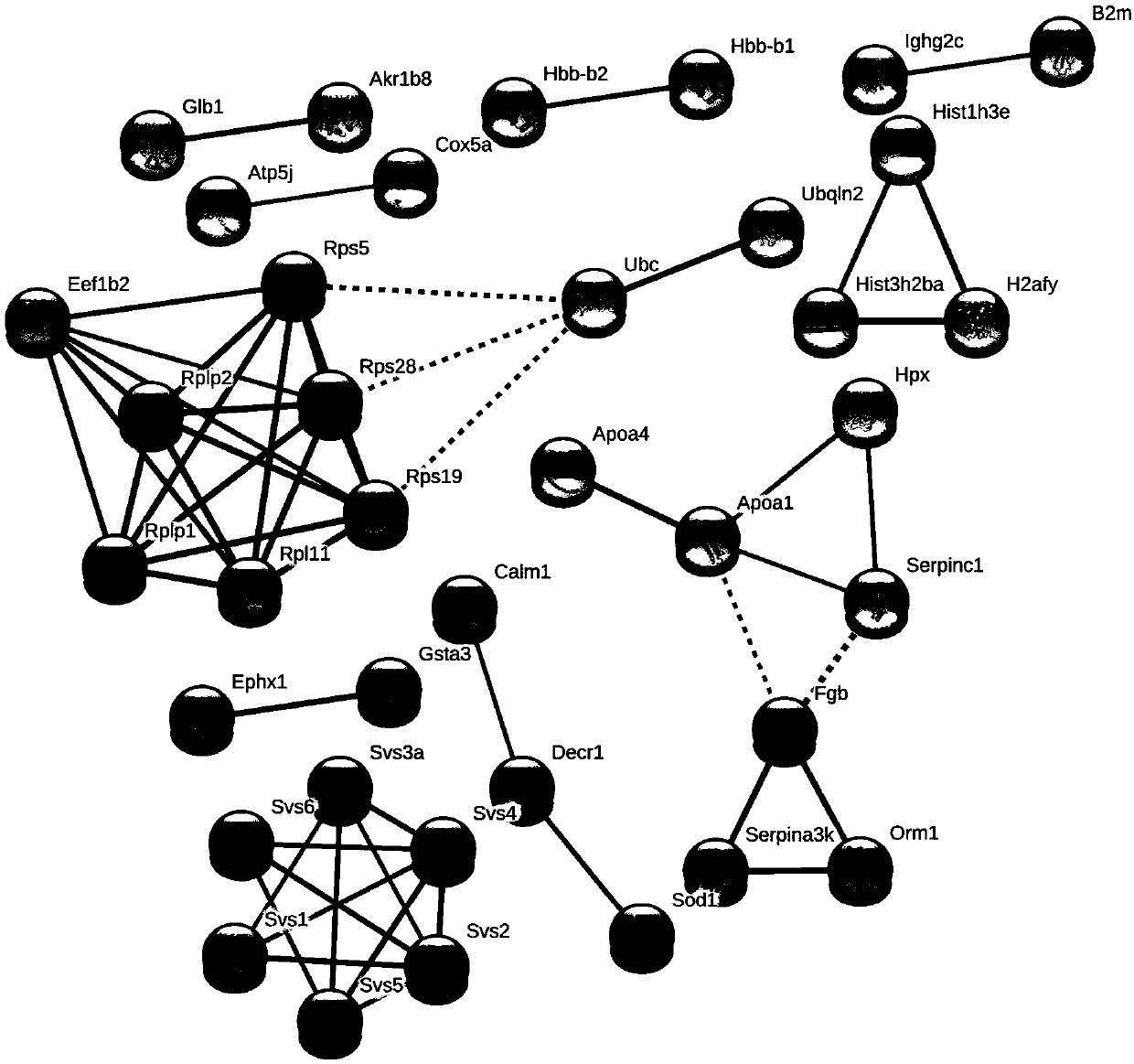
Differential protein database for expressing kidney deficiency testis and construction method thereof
The invention relates to a differential protein database for expressing kidney deficiency testis and a construction method thereof. The differential protein database for expressing kidney deficiency testis is obtained by selecting a mode of deficiency of kidney-yin and a mode of deficiency of kidney-yang induced by hydrocortisone and comparing differential proteins of a normal model, the mode of deficiency of kidney-yin and the mode of deficiency of kidney-yang for expressing testis by means of an iTRAQ technology, a protein interaction network and a clustering analyzing means. Essence of deficiency of the kidney is explained by means of the differential protein database constructed, and the differential protein database provides a basis for diagnosing and treating deficiency of the kidneyin the aspect of proteomics and has very important meaning in diagnosing and treating male reproductive dysfunction.
Owner:FOURTH MILITARY MEDICAL UNIVERSITY
A deep learning approach for DNA-binding protein identification and functional annotation based on self-attention mechanism
The invention discloses a deep learning method for DNA binding protein identification and functional annotation based on a self-attention mechanism. A data set is selected from a protein database, and the constructed deep learning model is trained and tested by using the selected data set, and then the trained deep learning model is used for training and testing. The deep learning model can predict whether protein can bind to DNA; wherein, the deep learning model includes coding layer, embedding layer, long short-term memory neural network layer (LSTM), convolutional neural network layer (CNN) and self-attention Layer (Self‑Attention). Through the deep learning model based on the self-attention mechanism, the present invention can predict whether it has the function of binding with DNA according to the primary sequence information of the protein, and find out the region with the binding function.
Owner:TIANJIN UNIV
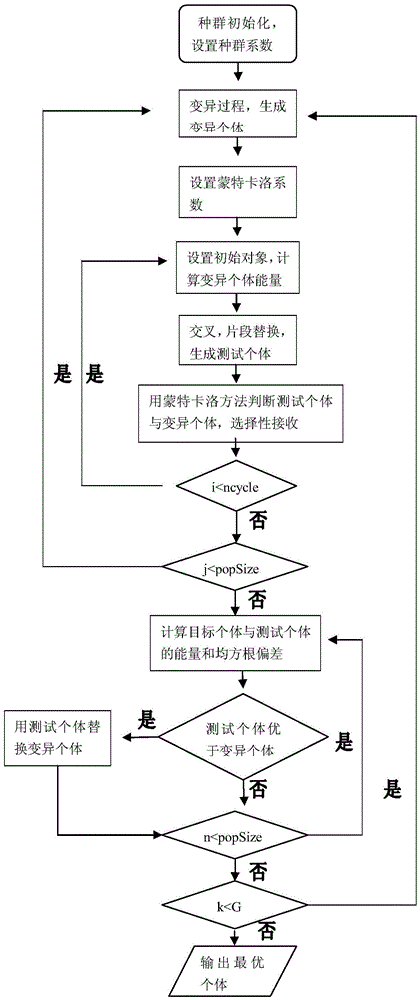
A 3D protein structure prediction method based on Monte Carlo local dithering and fragment assembly
ActiveCN103714265BReduce search space complexityFast convergenceSpecial data processing applicationsSide chainComputational physics
The invention discloses a method for predicting a protein three-dimensional structure based on Monte Carlo local shaking and fragment assembly. The method comprises the following steps that firstly, according to the difficult problem that search space of protein high-dimensional conformation space is complex, the effectiveness of fragment replacement is judged under a Rosetta force field model through the Monte Carlo statistical method according to a protein database configuration fragment bank; under a differential evolution group algorithm framework, the complexity of the search space is reduced through fragment assembly, meanwhile, false fragment assembly is removed through the Monte Carlo statistical method, and the conformation search space is gradually reduced through the diversity of an evolutionary algorithm, and therefore the searching efficiency is improved; meanwhile, a module with coarseness is adopted, a side chain is ignored, and cost of a search is effectively reduced. The method for predicting the protein three-dimensional structure based on Monte Carlo local shaking and fragment assembly can effectively obtain optimal local stable conformation and is high in predicting efficiency and good in convergence correctness.
Owner:ZHEJIANG UNIV OF TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com