A method for predicting indirect target genes of microRNAs using literature mining
A target gene prediction, target gene technology, applied in biochemical equipment and methods, biological testing, special data processing applications, etc., can solve the problems of false positives, inaccurate differential expression, and true negatives in prediction results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
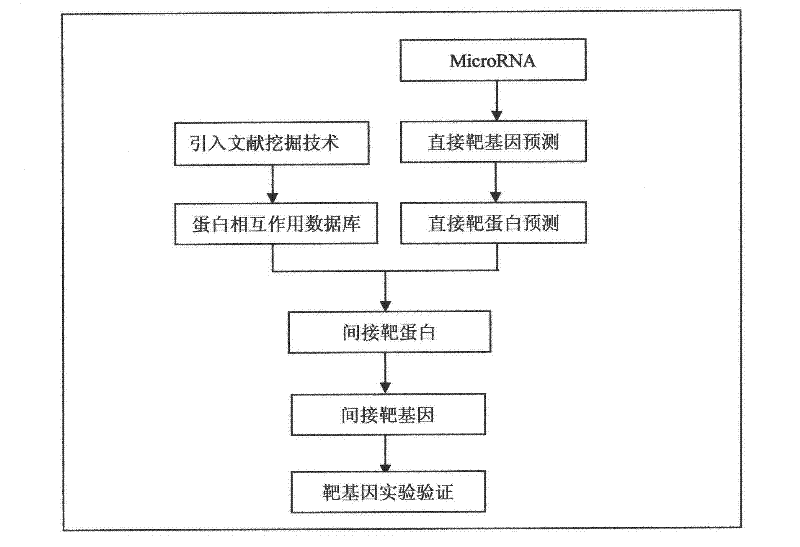
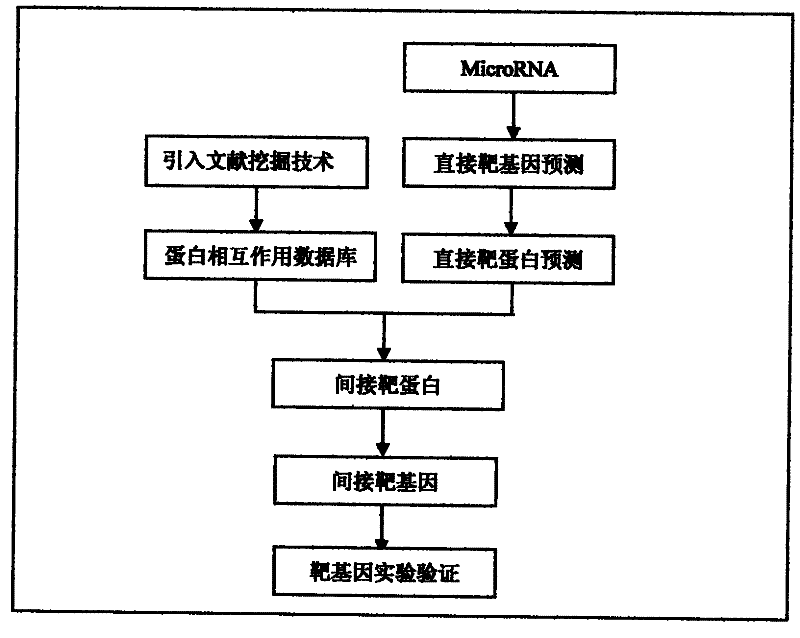
[0014] The method of the present invention will take the prediction of the target gene of mir-122 as an example to introduce the specific implementation method of the present invention.
[0015] 1. First use microT 3.0 (http: / / diana.cslab.ece.ntua.gr / microT / ), miRanda v5 (http: / / microma.sanger.ac.uk / targets / ), TargetScan 5.1 (http: / / www.targetscan.org / ), PicTar vertebrate 2007 (http: / / pictar.mdc-berlin.de / ) and other 4 softwares were used for target gene prediction, and a result set of mir-122 direct target genes was obtained, with a total of 935 genes;
[0016] 2. Predict the expression protein of the above 935 target genes, and obtain the direct protein result set of mir-122.
[0017] 3. Use literature mining technology to build a protein-protein interaction database. The protein-protein interaction data were downloaded from the MIPS database (http: / / mips.helmholtz-muenchen.de / proj / ppi / ) and the pathway data in the kegg database to construct a database of protein-protein i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com