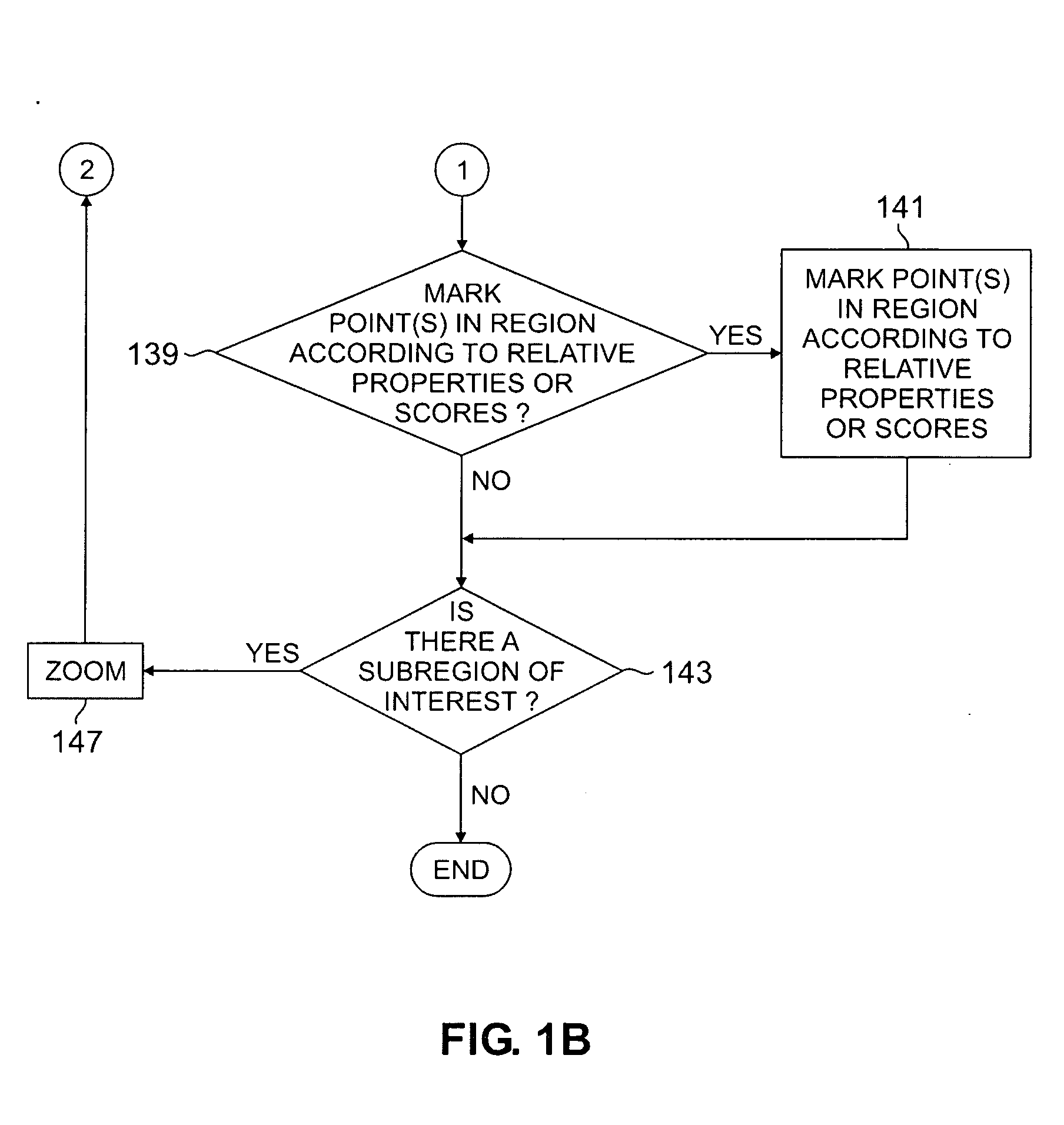
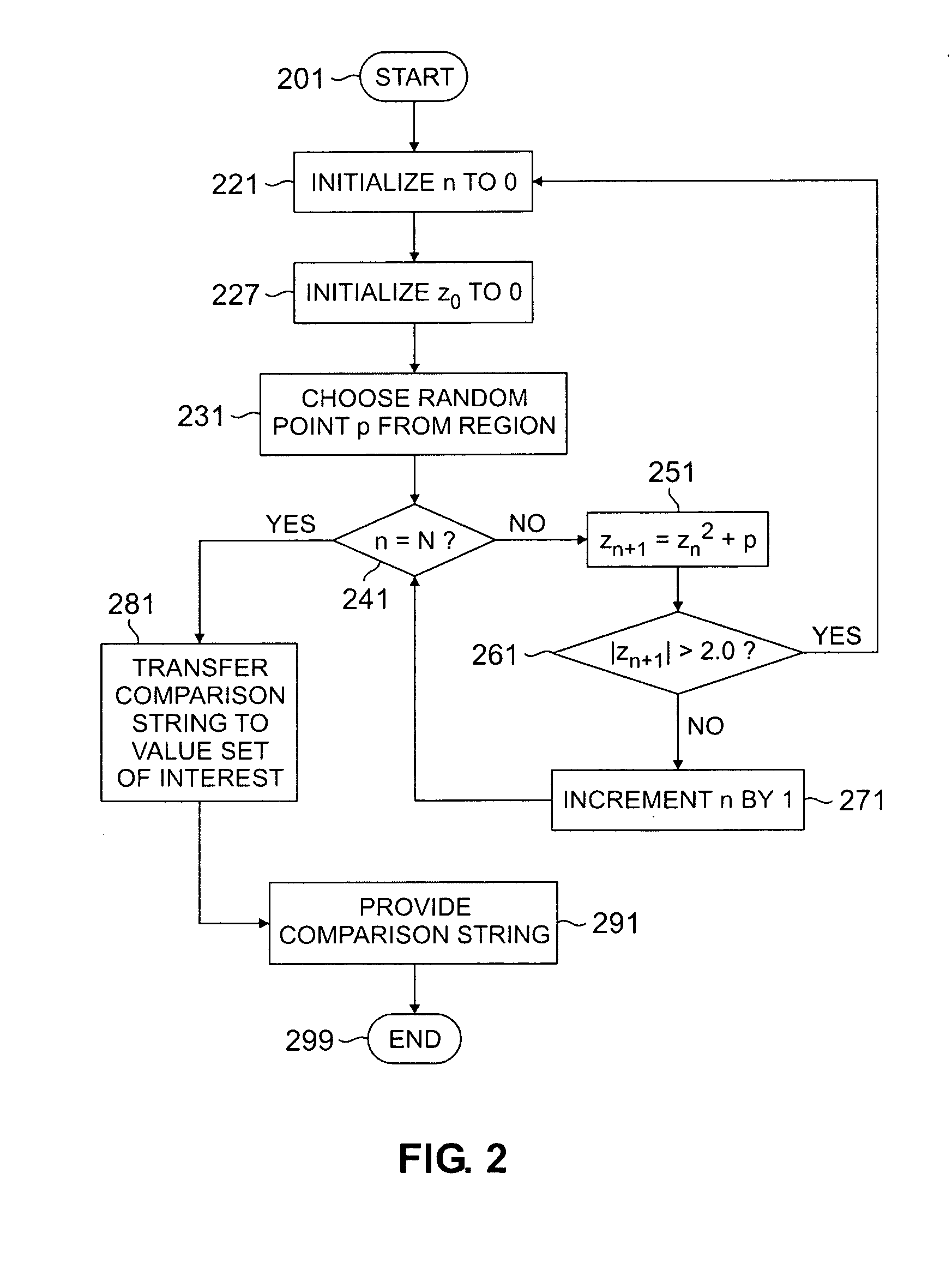
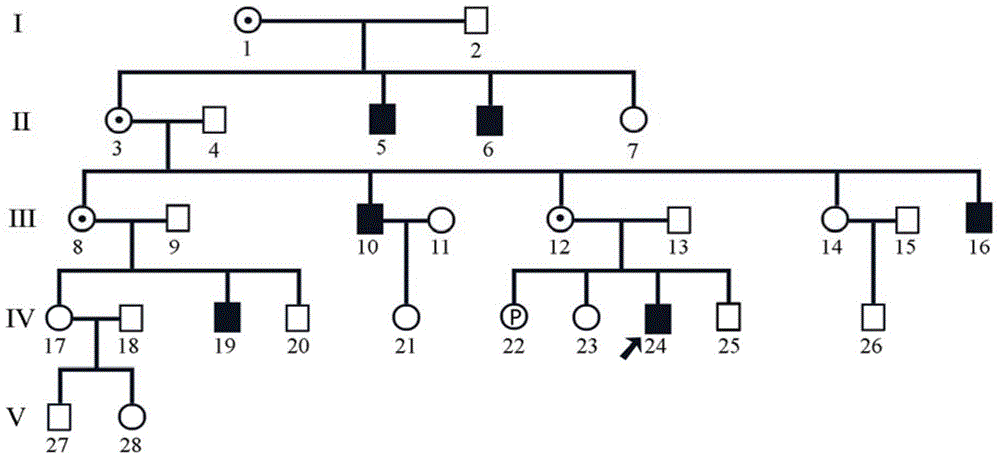
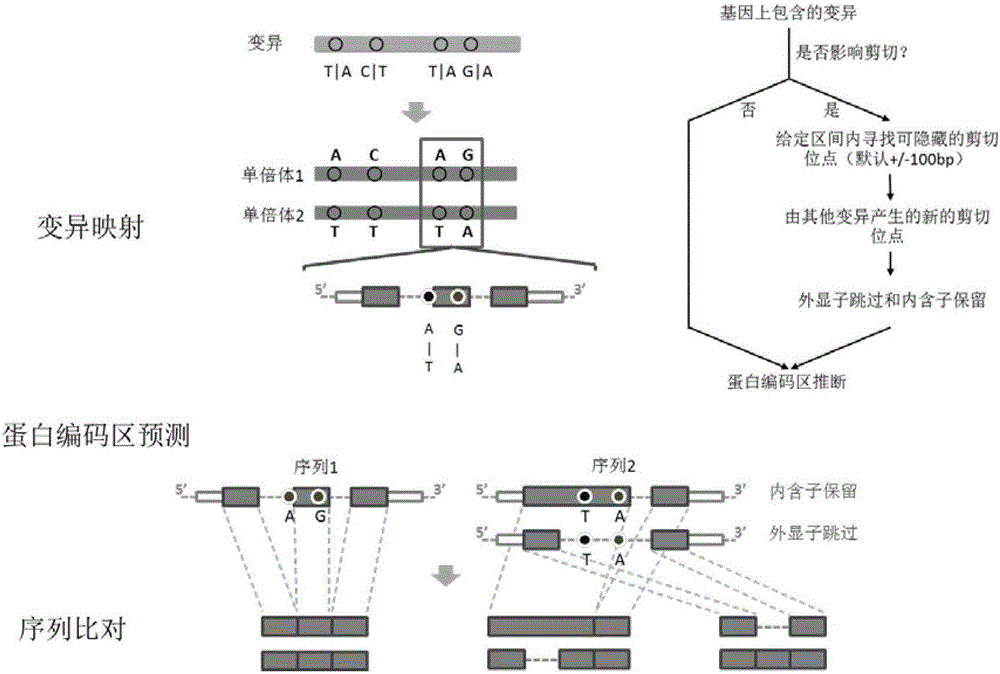
Patents
Literature
54 results about "Gene model" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
In TAIR, a Gene Model is defined as any description of a gene product from a variety of sources including computational prediction, mRNA sequencing, or genetic characterization.
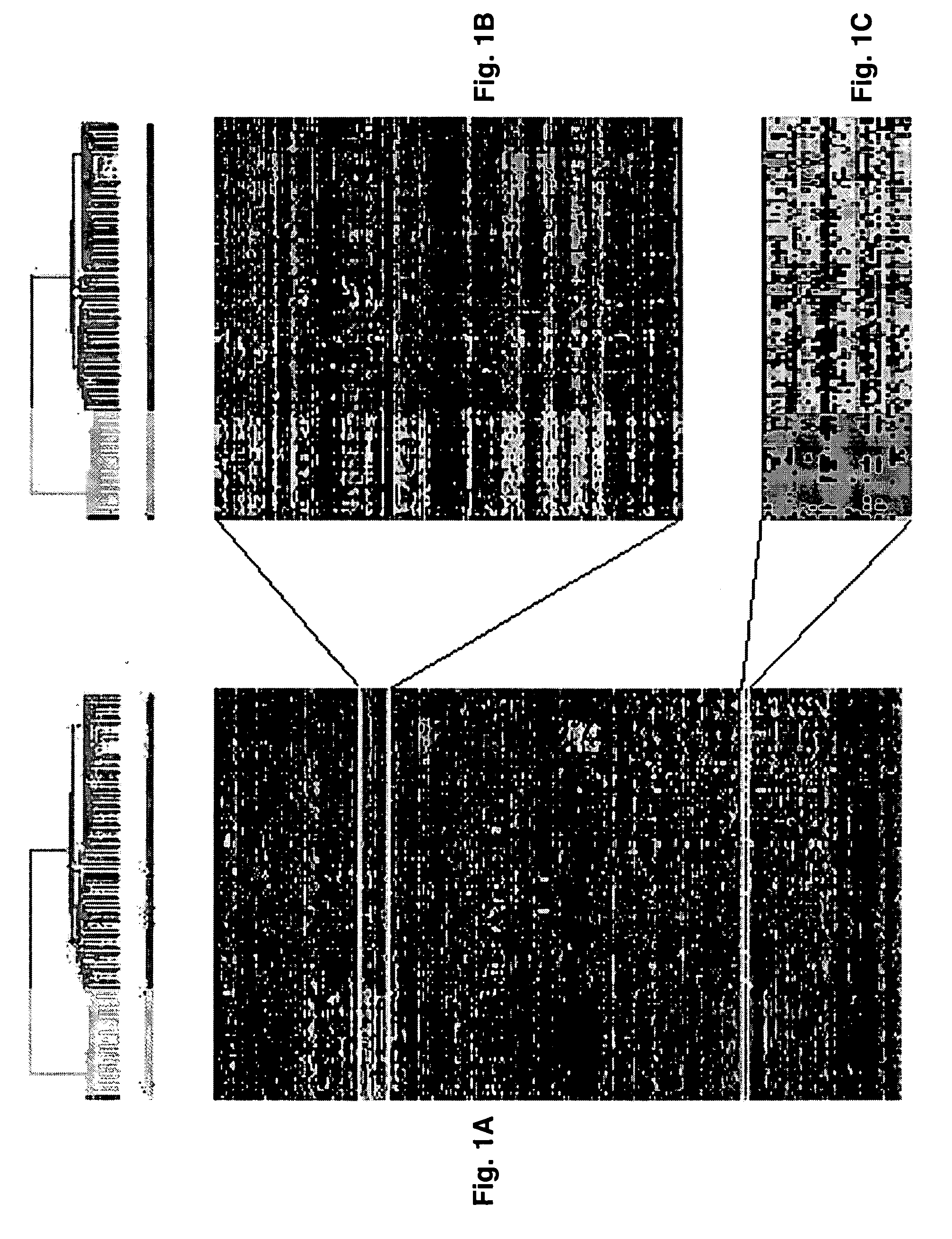
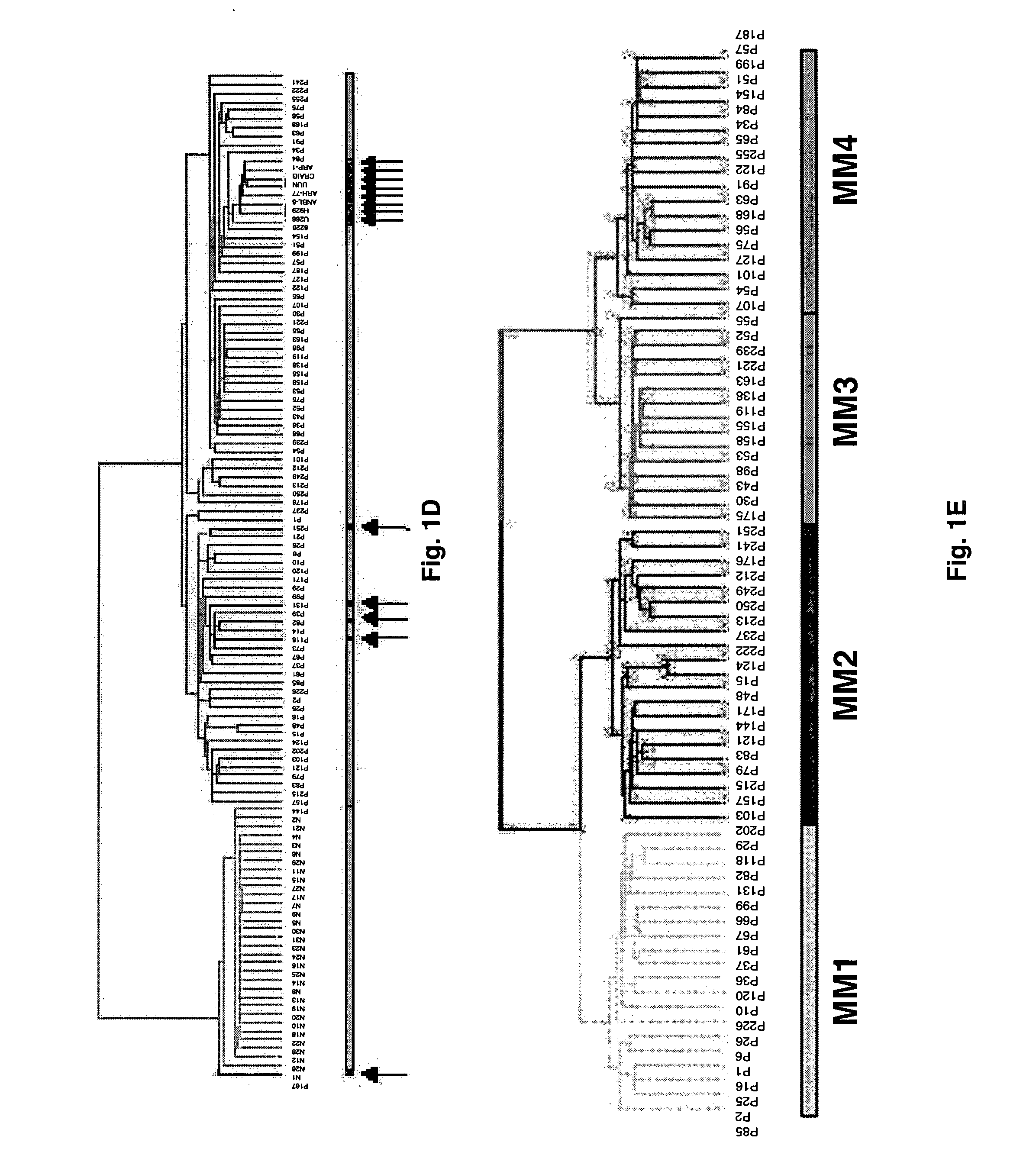
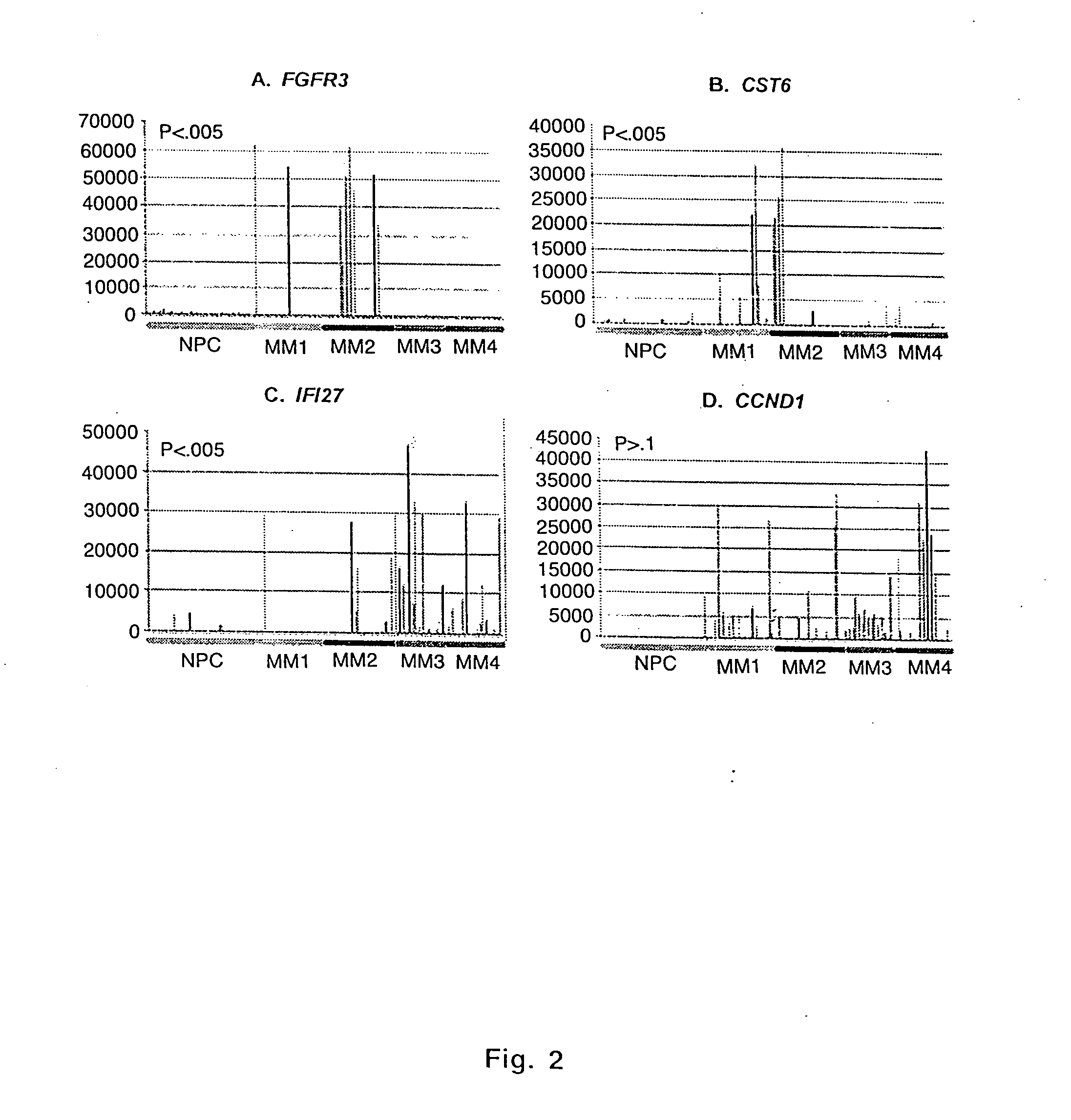
Diagnosis, prognosis and identification of potential therapeutic targets of multiple myeloma based on gene expression profiling
InactiveUS20050112630A1Microbiological testing/measurementBiological testingDevelopmental stageTumor specific
Gene expression profiling reveals four distinct subgroups of multiple myeloma that have significant correlation with various clinical characteristics. Diagnosis for multiple myeloma (and possibly monoclonal gammopathy of undetermined significance) based on differential expression of 14 genes, as well as prognosis for the four subgroups of multiple myeloma based on the expression of 24 genes are established. A 15-gene model that classifies myeloma into 7 groups is also reported. Gene expression profiling also allows placing multiple myeloma into a developmental schema parallel to that of normal plasma cell differentiation. Development of a gene expression- or developmental stage-based classification system for multiple myeloma would lead to rational design of more accurate and sensitive diagnostics, prognostics and tumor-specific therapies for multiple myeloma.
Owner:UNIV OF ARKANSAS FOR MEDICAL SCI THE
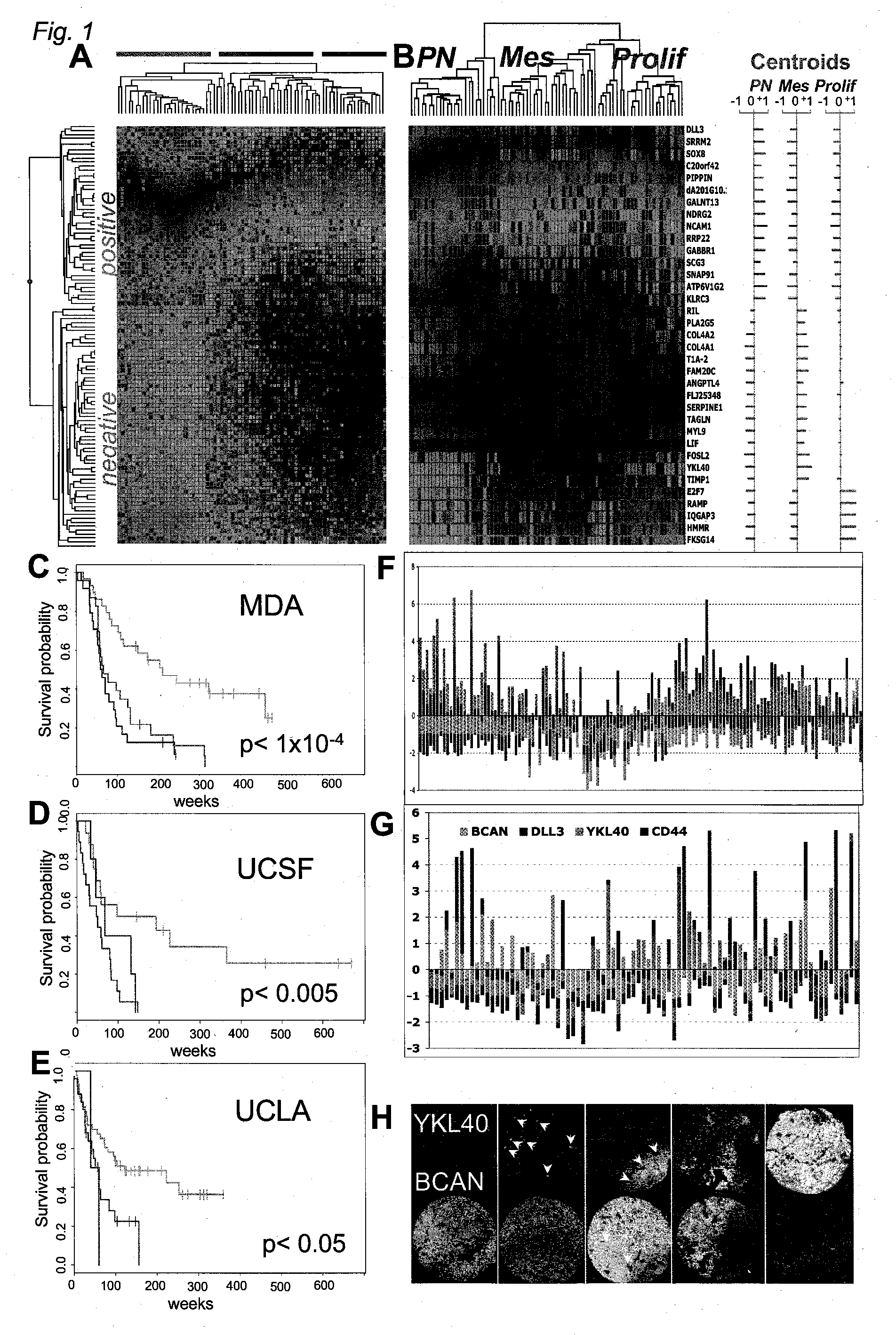
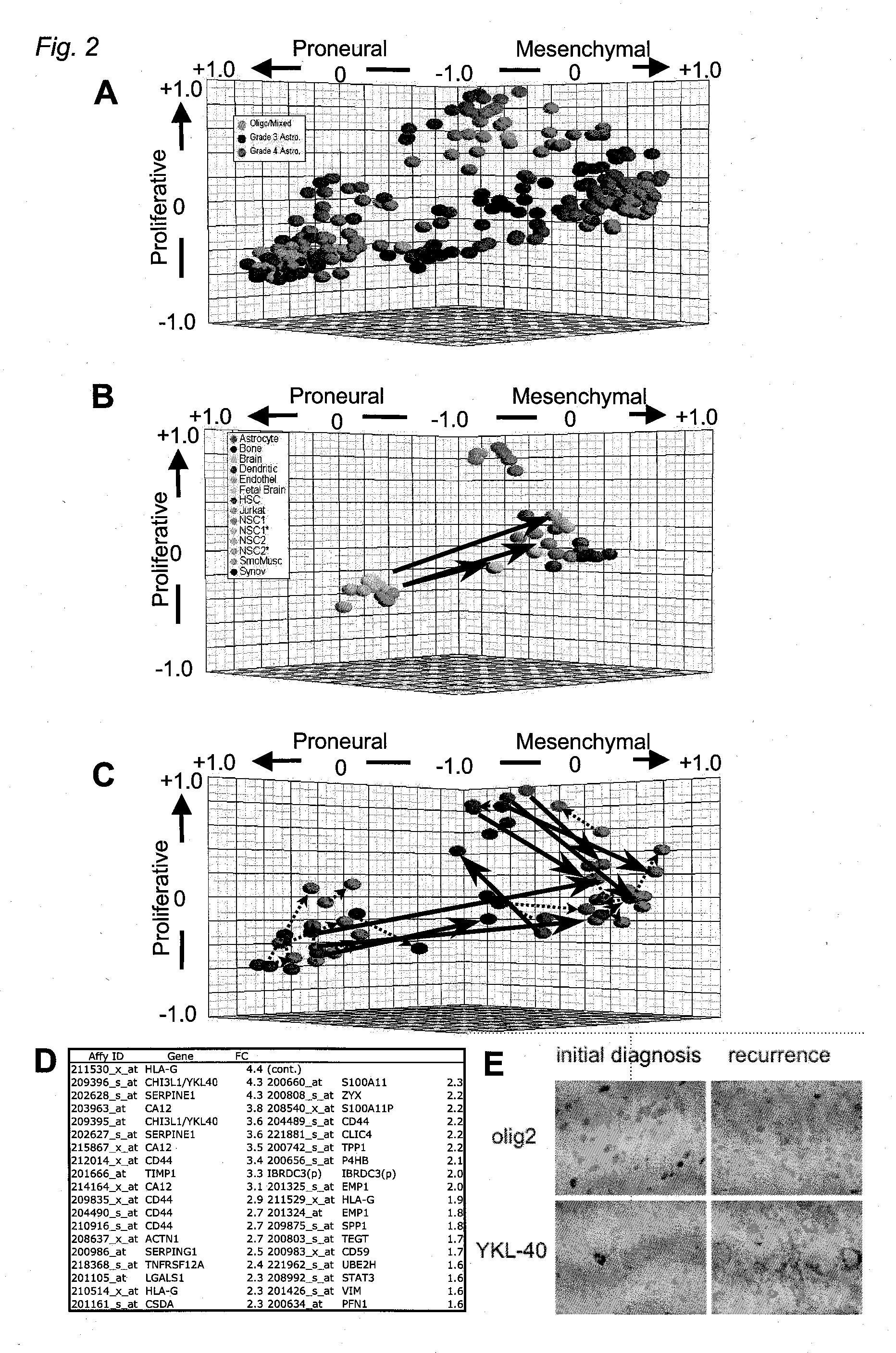
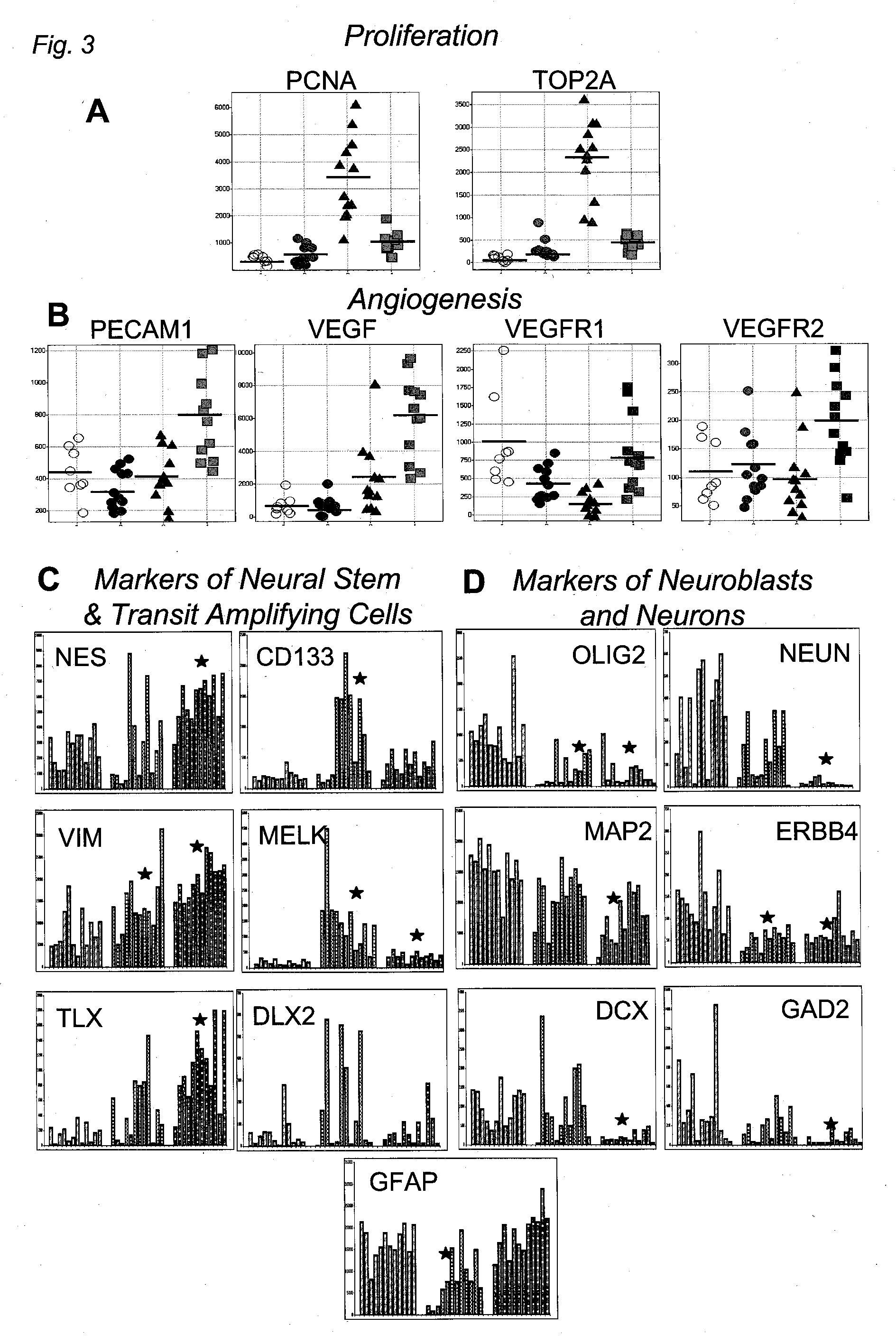
Method for Diagnosing, Prognosing and Treating Glioma
InactiveUS20070141066A1Long median patient survivalShort overall survivalHeavy metal active ingredientsCompound screeningAbnormal tissue growthAnti mitotic
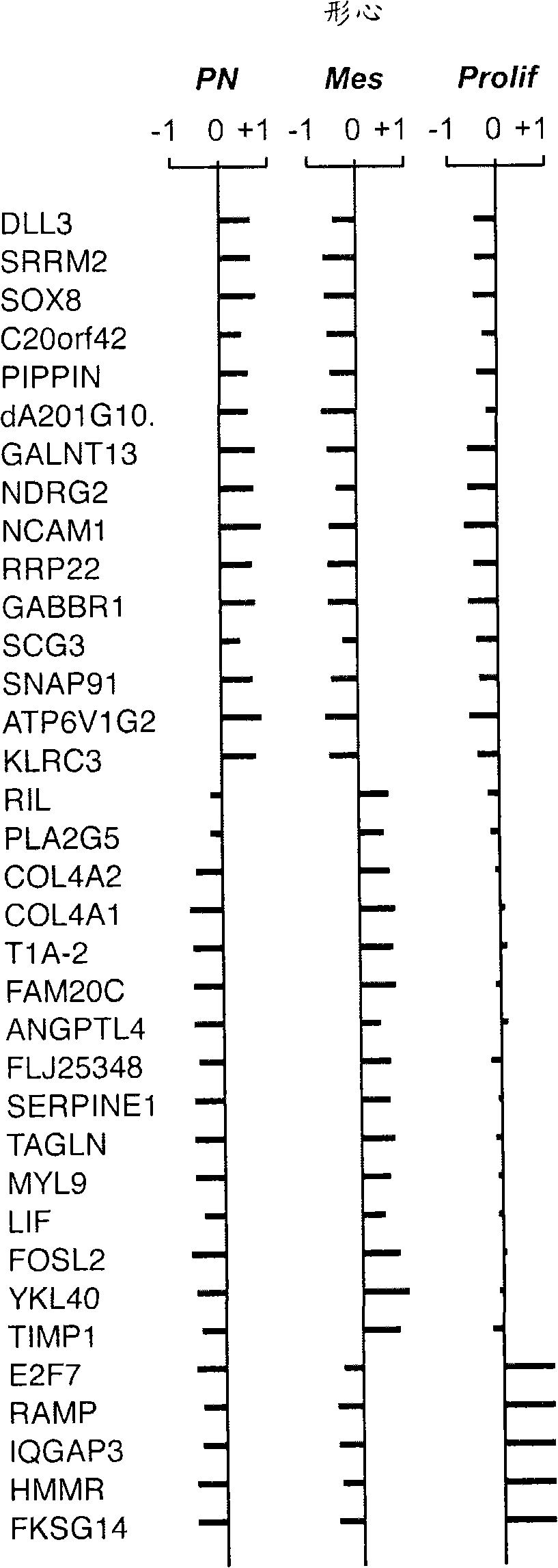
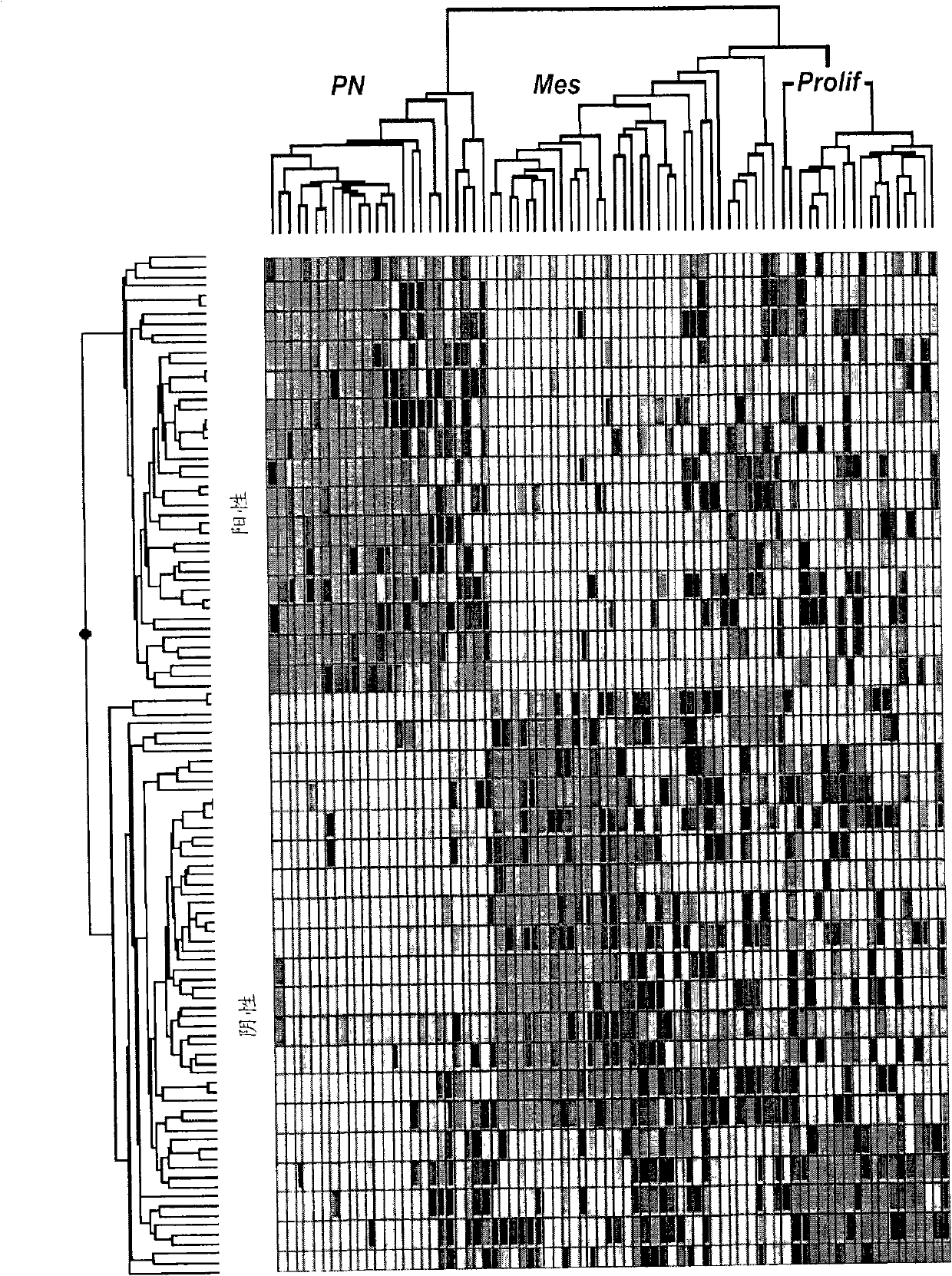
The invention provides generally a method of monitoring, diagnosing, prognosing and treating glioma. Specifically, the invention provides for three (3) prognostic subclasses of glioma, which are differentially associated with activation of the akt and notch signaling pathways. Tumor displaying neural or proneural PN lineage markers (including notch pathway elements) show longer median patient survival, while the two remaining tumor markers Prolif and Mes are associated with shortened survival. Tumors classified in this manner may also be treated with the appropriate PN- Prolif- or Mes-therapeutic corresponding to the subclassification in combination with anti-mitotic agents, anti-angiogenic agents, Akt antagonists, and neural differentiation agents. Alternatively, the invention also provides for method of prognosing and diagnosing glioma with a two-gene model based on the expression levels of PTEN and DLL3.
Owner:GENENTECH INC
Diagnosis, prognosis and identification of potential therapeutic targets of multiple myeloma based on gene expression profiling
InactiveUS20080293578A1Microbiological testing/measurementAnalogue computers for chemical processesGene targetsGene model
Provided herein is a method for gene expression profiling multiple myeloma patients into distinct subgroups via DNA hybridization and hierarchical clustering analysis of the hybridization data where the results may further be used to identify therapeutic gene targets. Also provided is a method for controlling bone loss in an individual via pharmacological inhibitors of DKK1 protein. In addition provided herein is a method for diagnosing multiple myeloma using a 15-gene model that classifies myeloma into groups 1-7.
Owner:BIOVENTURES LLC
Gene expression profiling based identification of DKK1 as a potential therapeutic targets for controlling bone loss
Gene expression profiling reveals four distinct subgroups of multiple myeloma that have significant correlation with various clinical characteristics. Diagnosis for multiple myeloma (and possibly monoclonal gammopathy of undetermined significance) based on differential expression of 14 genes, as well as prognosis for the four subgroups of multiple myeloma based on the expression of 24 genes are established. A 15-gene model that classifies myeloma into 7 groups is also reported. Gene expression profiling also allows placing multiple myeloma into a developmental schema parallel to that of normal plasma cell differentiation. Development of a gene expression- or developmental stage-based classification system for multiple myeloma would lead to rational design of more accurate and sensitive diagnostics, prognostics and tumor-specific therapies for multiple myeloma.
Owner:UNIV OF ARKANSAS FOR MEDICAL SCI THE
Method for studying cellular chronomics and causal relationships of genes using fractal genomics modeling
This present invention relates to methods of manipulation, storage, modeling, visualization and quantification of datasets. One application of the present invention is related to developing point-models of datasets represented by the various points in a multi-dimensional map. The invention can be adapted to genomic analysis by Fractal Genomics Modeling (FGM) for developing single point gene models which can be used for studying cellular chronomics and causal relationships of genes. Using FGM, evidence of genes that govern fundamental clocking cycles in cell development and tissue differentiation of an organism can be produced. This clocking mechanism and the FGM methods used to produce its genetic components and function are described in this disclosure.
Owner:HEALTH DISCOVERY CORP
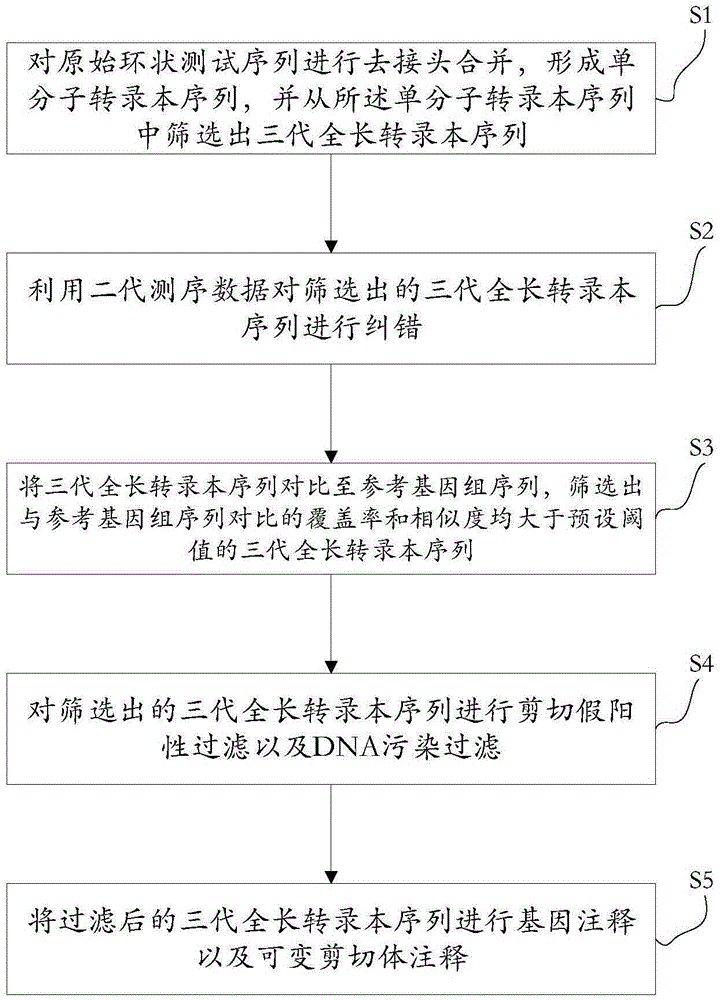
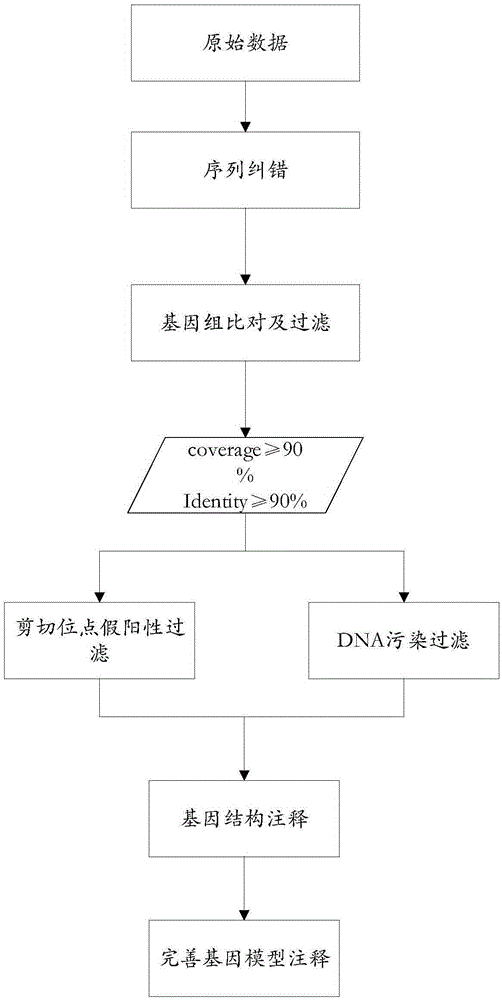
Method for detecting variable spliceosome in third generation full-length transcriptome
ActiveCN105389481AEfficient access to shear structuresPerfect commentSequence analysisSpecial data processing applicationsReference genome sequenceGene model
The invention discloses a method for detecting a variable spliceosome in a third generation full-length transcriptome. The method comprises the following steps: merging original annular test sequences with joints removed to form a monomolecular transcript sequence, and screening a third generation full-length transcript sequence; comparing the third generation full-length transcript sequence with a reference genome sequence, and screening a third generation full-length transcript sequence having coverage and similarity with the reference genome sequence larger than preset thresholds; carrying out splicing false positive filtration and DNA contamination filtration on the screened third generation full-length transcript sequence; and carrying out gene annotation and variable spliceosome annotation on the filtered third generation full-length transcript sequence. An overlong read length of a third generation sequencing technology mentioned in the method disclosed by the invention is large enough to cover most RNA, the third generation full-length transcript sequence can be obtained by SMRT sequencing transcriptomes without being assembled, and a splicing structure of a gene can be effectively obtained by third generation transcriptome sequencing, and more perfect gene model annotation can be constructed.
Owner:嘉兴菲沙基因信息有限公司
Method for diagnosing, prognosing and treating glioma
The invention provides generally a method of monitoring, diagnosing, prognosing and treating glioma. Specifically, the invention provides for three (3) prognostic subclasses of glioma, which are differentially associated with activation of the akt and notch signaling pathways. Tumor displaying neural or proneural PN lineage markers (including notch pathway elements) show longer median patient survival, while the two remaining tumor markers Prolif and Mes are associated with shortened survival. Tumors classified in this manner may also be treated with the appropriate PN- Prolif- or Mes- therapeutic corresponding to the subclassification in combination with anti-mitotic agents, anti-angiogenic agents, Akt antagonists, and neural differentiation agents. Alternatively, the invention also provides for method of prognosing and diagnosing glioma with a two- gene model based on the expression levels of PTEN and DLL3.
Owner:GENENTECH INC
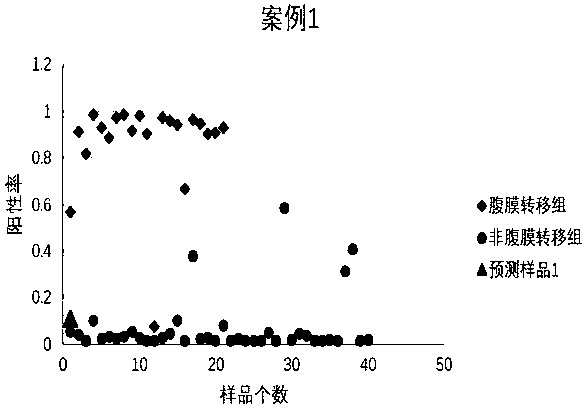
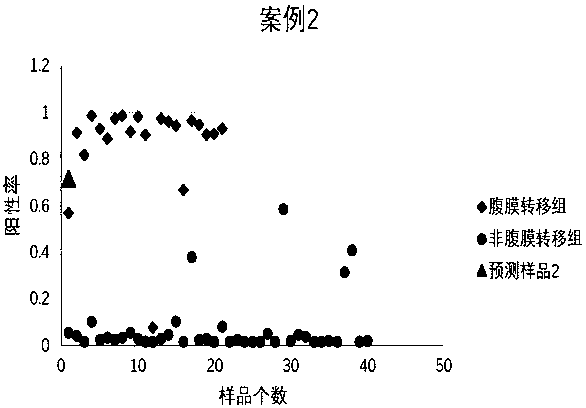
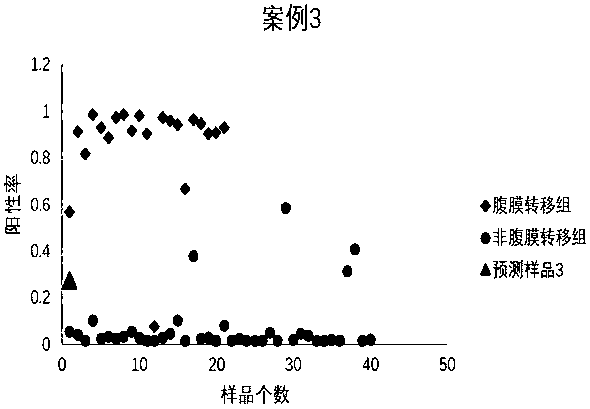
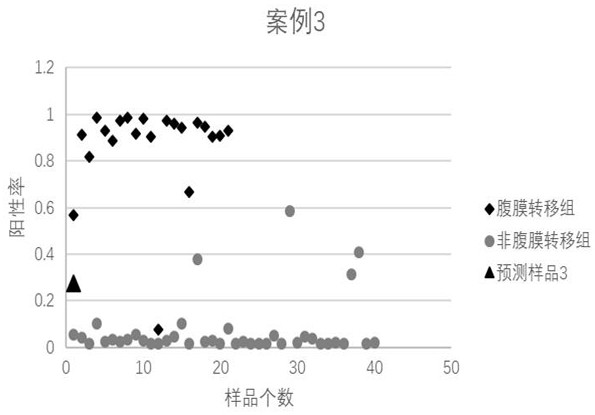
Peritoneal metastasis prediction model of gastric cancer based on 22 genes and application thereof
ActiveCN107586852AImprove survival rateImprove accuracyMicrobiological testing/measurementLymphatic SpreadRPS27A
The invention belongs to the field of genetic testing and provides a peritoneal metastasis prediction model of a gastric cancer based on 22 genes and application thereof, including PCLO, UGGT1, ZNF714, KIAA0825, COL23A1, MED1, NPAS2, TTC14, RPS27A, ASPH, ARHGEF12, SIK1, PAPPA, HHIPL1, MYO9B, ITPKB, ZNF862, MKNK1, MUC6, TRRAP, DUOX1 and KRTAP5-2; a risk of peritoneal metastasis is predicted effectively and specifically, according to SVM of a selected classifier and a positive judgment threshold value of 0.5. The application of the gene model of the invention is helpful for predicting the metastasis of patients with gastric cancer and has important value and significance for taking timely and effective clinical measures, making an individualized diagnosis and treatment plan and finally improving a survival rate of the patients with the gastric cancer.
Owner:FUJIAN MEDICAL UNIV UNION HOSPITAL +1
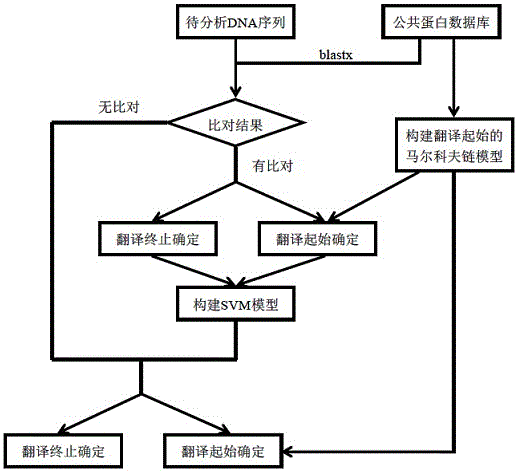
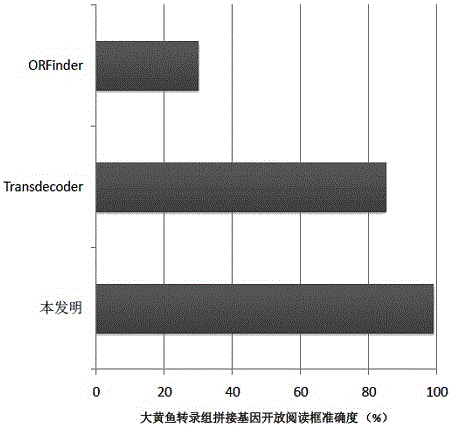
Method for analyzing structure of transcriptome gene sequence of non-model organism
ActiveCN106202998AEnrich the connotation of annotationsDefine biological functionBiostatisticsSpecial data processing applicationsStructural analysisNucleic acid sequence
The invention discloses a method for analyzing a structure of a transcriptome gene sequence of a non-model organism. The method comprises the following steps: (1) obtaining an optimal comparison result; (2) determining a protein coding mode and determining a translation termination position; (3) determining a coding starting position of the gene sequence; (4) classifying by utilizing a gene mode; (5) determining a nucleic acid sequence of a coding manner by utilizing a transcriptome sequence and training a nucleic acid sequence mode of a coding protein by utilizing a Markov chain; (6) determining a coding manner of a protein coding sequence of a gene which is not compared. The method disclosed by the invention is used for carrying out high-throughput structural analysis on a lot of gene sequences obtained by transcriptome sequencing of any non-model organism and function annotation of the transcriptome sequence is automatically finished in an analysis process; a Markov model and a support vector machine model are constructed by utilizing a compared high-reliability protein coding nucleic acid sequence, and a gene sequence which is not compared is analyzed, so that the credibility of the structural analysis of the sequence is higher.
Owner:JIMEI UNIV
Detection method of folate metabolism related gene
InactiveCN107254520AReduced activityLow costMicrobiological testing/measurementGel electrophoresisRelated gene
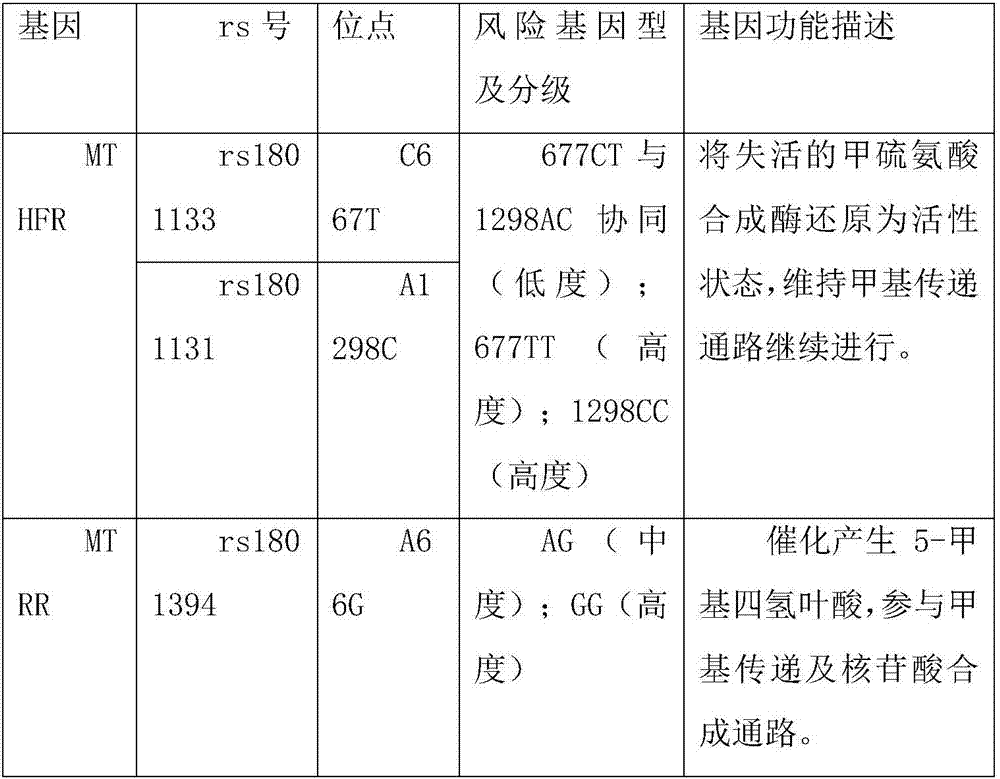
The invention discloses a detection method of a folate metabolism related gene, which includes steps of extracting DNA from oral mucosa cell of a detector; amplifying MTHFR gene C677T by SEQ NO 1 and SEQ NO 2; amplifying MTHFR gene A1298C by SEQ NO 3 and SEQ NO 4; amplifying MTHFR gene A66G by SEQ NO 5 and SEQ NO 6; extracting DNA from oral mucosa cell as a template to perform PCR reaction; performing sepharose gel electrophoresis of 1% of ordinary PCR amplified product on three pairs of primers; recycling a strip of a kit recycling item from the 1% of agarose gel electrophoretic band by sepharose gel; performing sequence testing reaction; purifying the sequence testing reaction product and denaturing; testing sequence by an upper 3730 sequence tester; analyzing three SNP gene models of the sequence testing result by Chromas software. The method has the advantages of high detection accuracy, and convenience of use.
Owner:安徽安龙基因科技有限公司
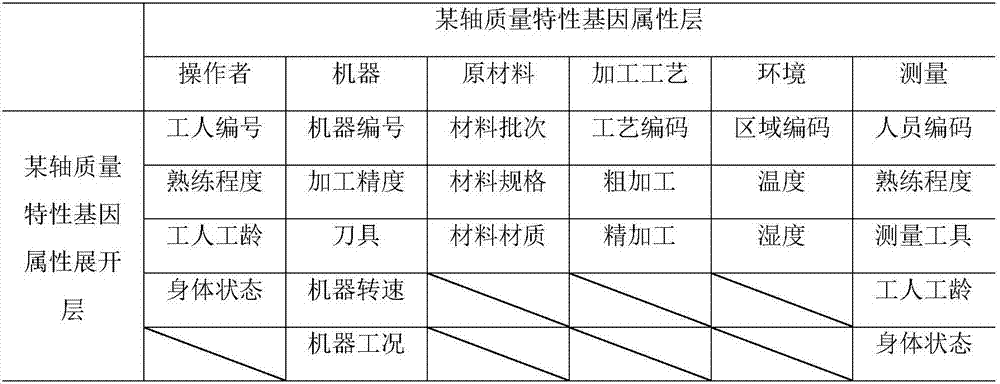
Mechanical and electrical product symbolic quality characteristic analysis and fault cause tracing method
InactiveCN107122907ASimple processResourcesManufacturing computing systemsGene modelInformation support
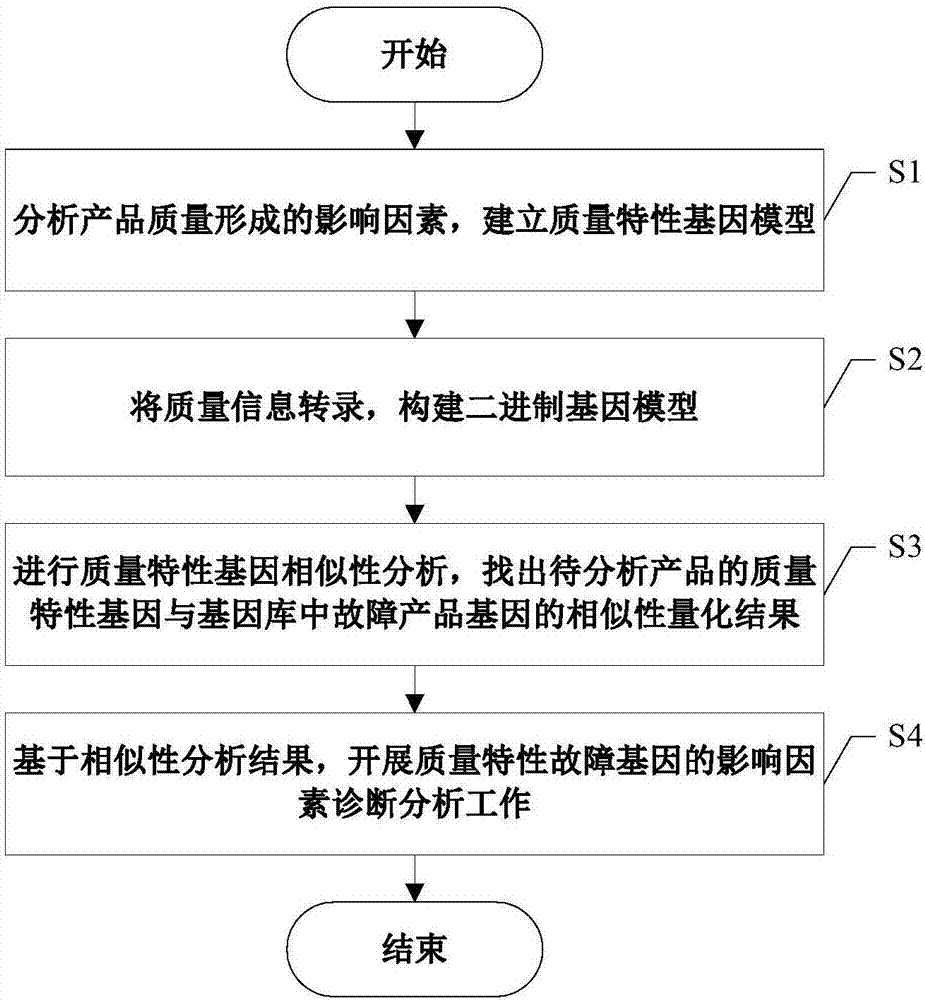
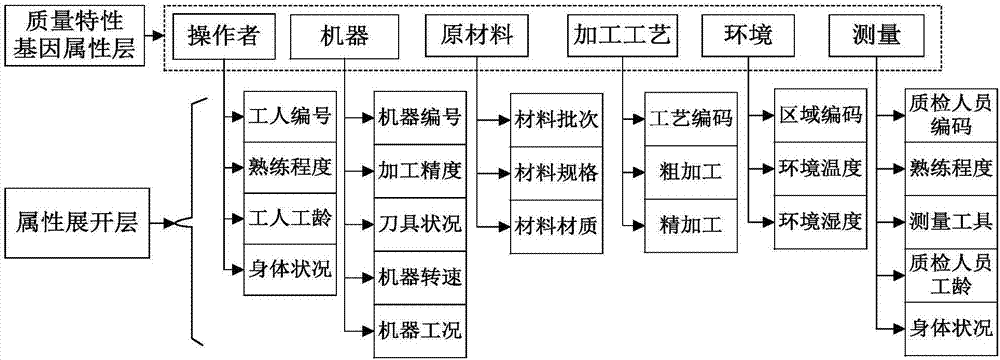
The invention discloses a mechanical and electrical product symbolic quality characteristic analysis and fault cause tracing method, takes a quality formation process of mechanical and electrical products as a breakthrough point, considers influence of operator, machine, material, the processing process, the environment and measurement (5M1E) factors on quality characteristics in particular and establishes a product quality characteristic gene model based on product quality characteristic influence factors. According to model input requirements, evaluation data of different product fault analysis personnel for fault products is collected, binary transcription of a quality characteristic influence degree is carried out according to an importance threshold, and a symbolic expression of a quality characteristic gene of the fault products is formed; based on the similarity principle, analysis on similarities between the fault product quality characteristic gene and gene database existing quality characteristic genes is developed, and necessary information support is provided for fault cause investigation and fault source tracking; lastly, through diagnosis analysis, causes of product faults are acquired.
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA
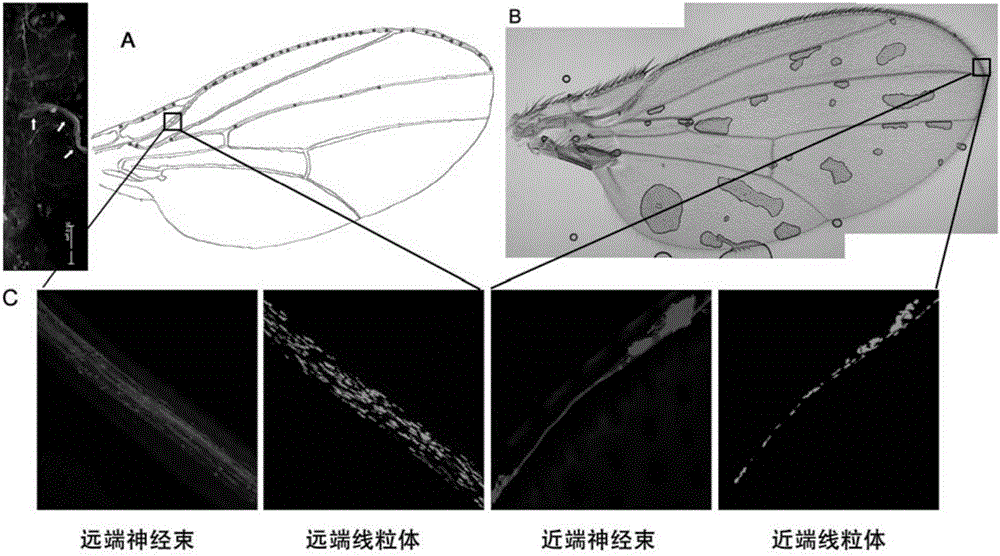
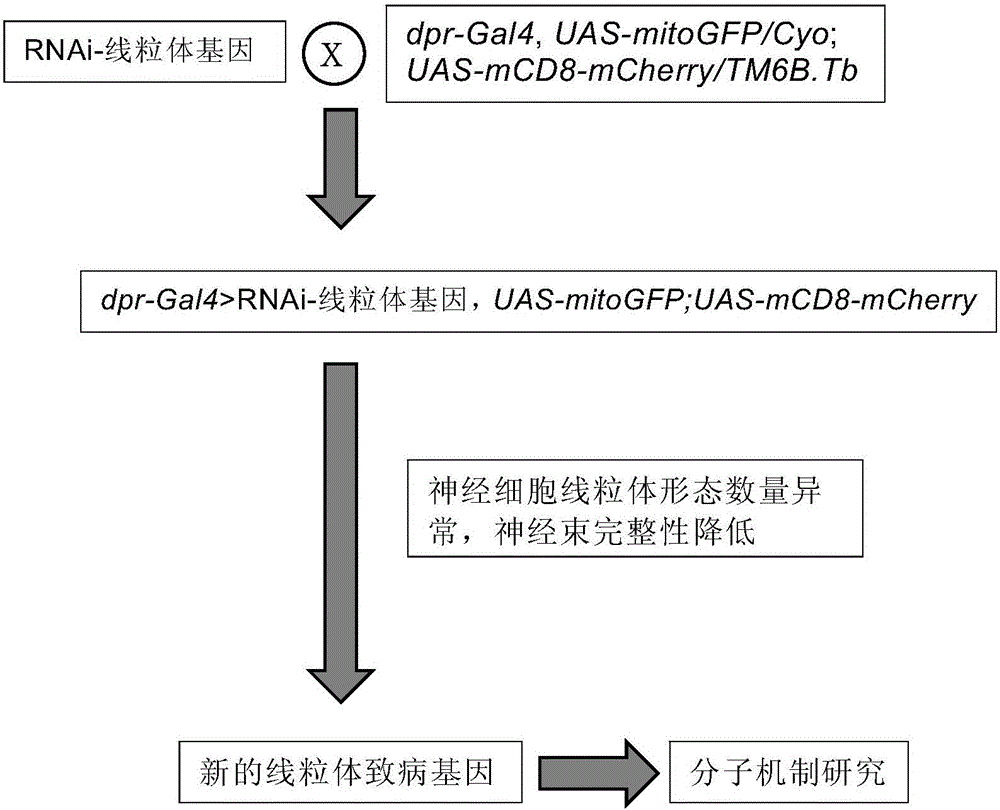
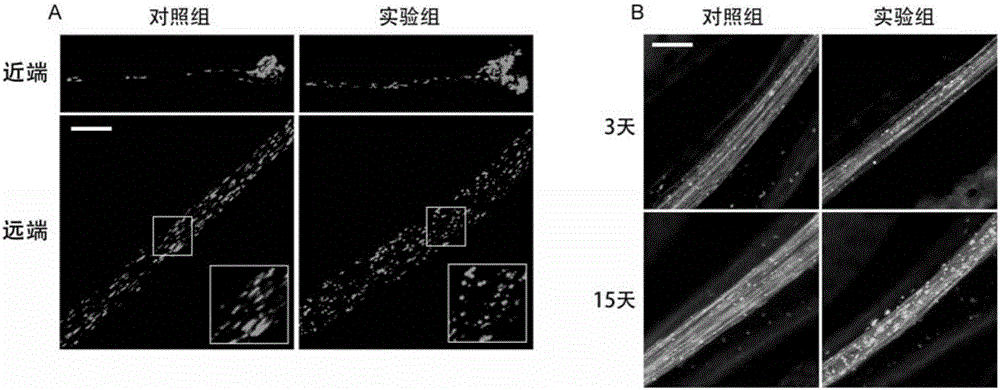
Building method of Drosophila melanogaster model for screening and researching mitochondria disease virulence gene, and application of same
The invention provides a building method of a Drosophila melanogaster model for screening and researching mitochondria disease virulence gene, and an application of the same. In the method, first fluorescent protein is represented by a mitochondria in a wing nerve cell in a Drosophila melanogaster model; second fluorescent protein is represented by in a wing nerve cell film in the Drosophila melanogaster model; the mitochondria is labeled via the first fluorescent protein positioned by the mitochondria; the nerve cell is labeled by the second fluorescent protein positioned by the nerve cell film; and research to mitochondria functions in a nervous system can be achieved in vivo. The Drosophila melanogaster transgene model built by the method can simply and conveniently screen and research mitochondria disease virulence genes; and demands for high flux, strong economic property and short time during mitochondria disease virulence gene screening can be met.
Owner:SHANGHAI INST OF ORGANIC CHEM CHINESE ACAD OF SCI
Method for efficiently and rapidly evaluating prognosis of II-stage colorectal cancer patient based on immune gene expression characteristic spectrum
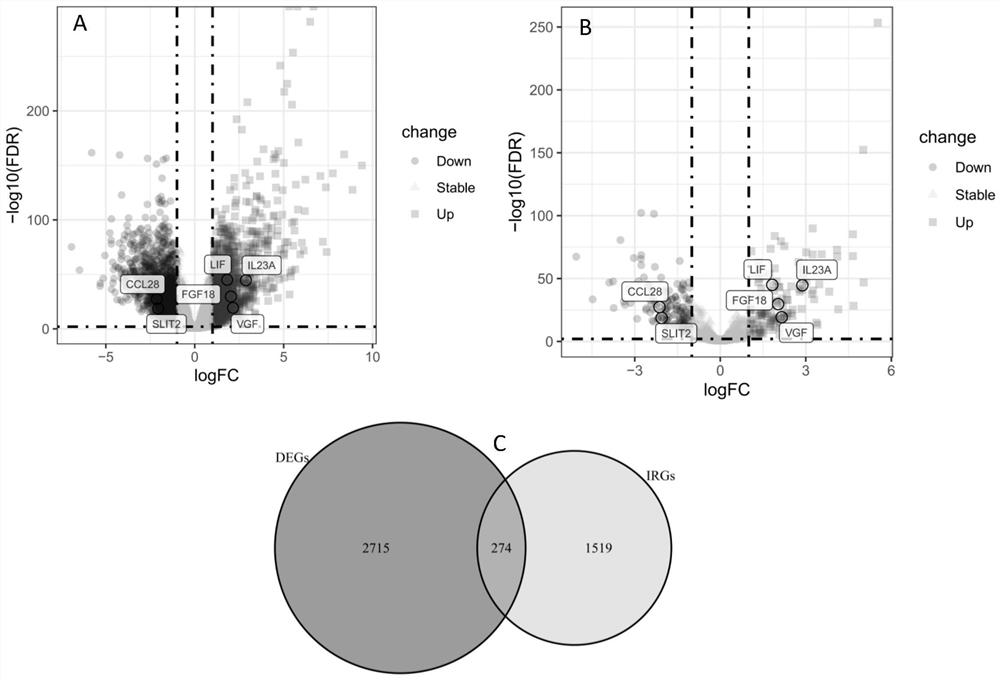
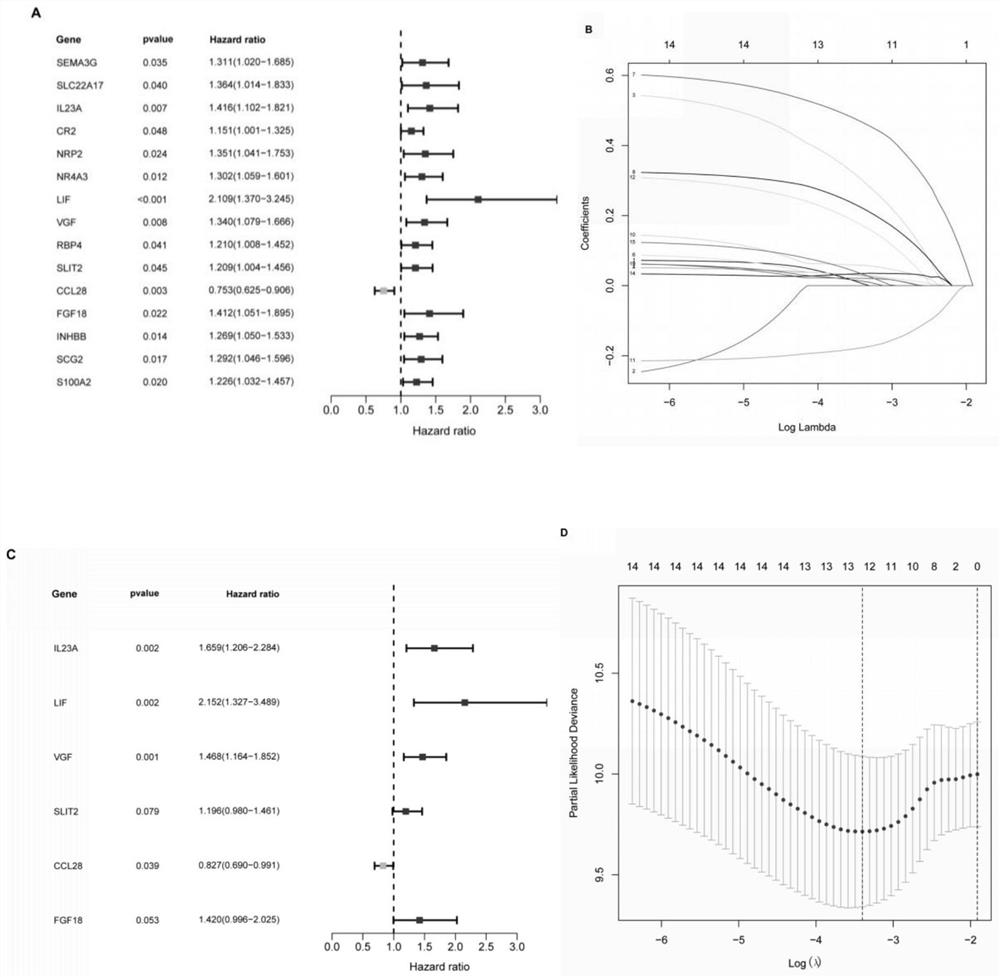
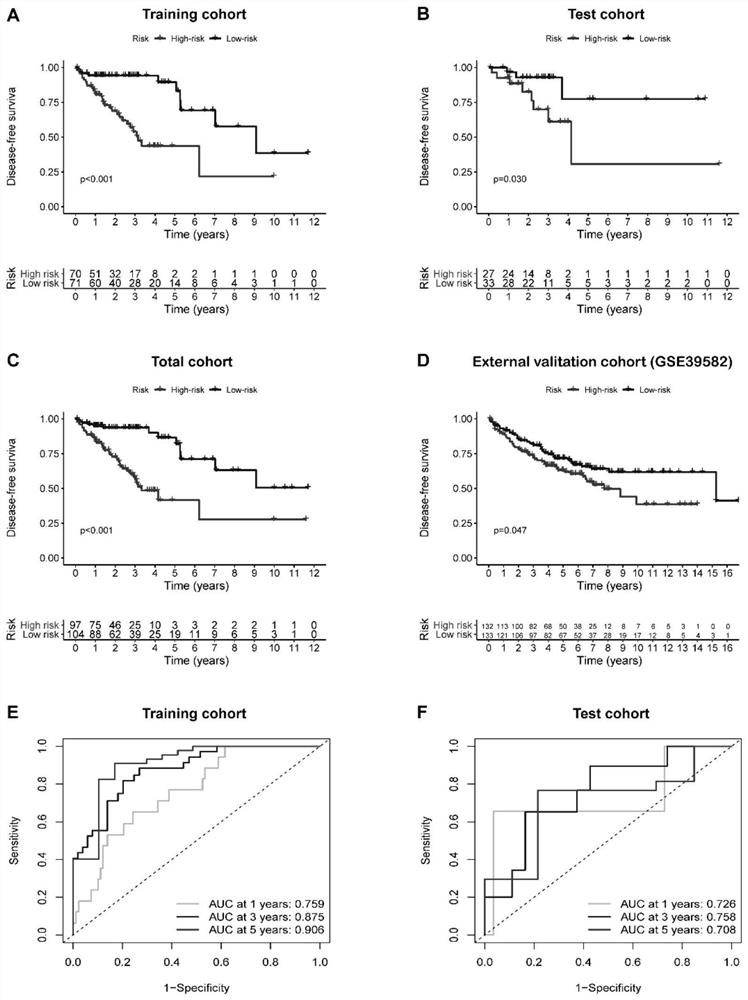
The invention discloses a method for efficiently and rapidly evaluating the prognosis of a patient with stage II colorectal cancer based on an immune gene expression characteristic spectrum and a construction method of an IRGCRC II prognosis model; the IRGCRC II prognosis model is adopted to evaluate the prognosis of the patient with stage II colorectal cancer, and the IRGCRC II prognosis model comprises CCL28, FGF18, IL23A, LIF, SLIT2 and VGF immune genes. According to the IRGCRC II prognosis model constructed by the invention, a gene model tool for evaluating the prognosis of the II-stage CRC patient is constructed on the basis of the content determination of six immune genes in the body of the II-stage colorectal cancer patient with high expression, and a new evaluation system is provided for evaluating the survival outcome of the II-stage colorectal cancer patient; and a new basis is provided for selection of treatment schemes such as whether the patient is subjected to chemical treatment or not.
Owner:THE SIXTH AFFILIATED HOSPITAL OF SUN YAT SEN UNIV
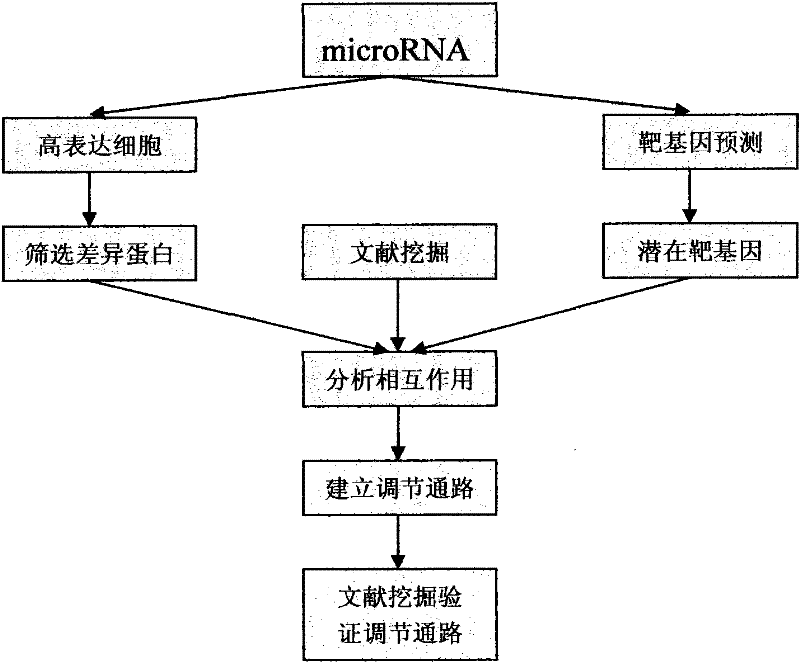
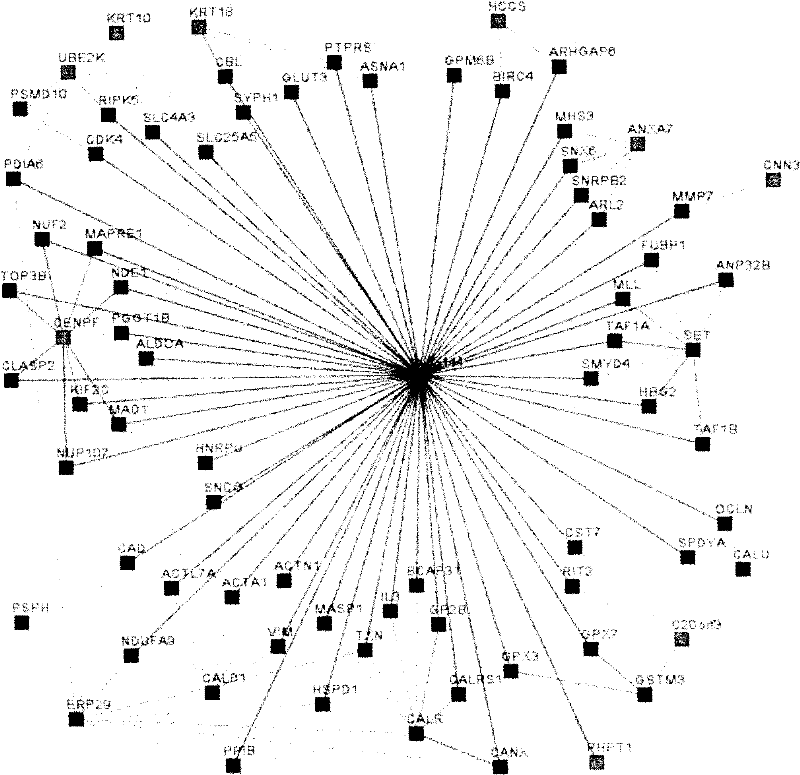
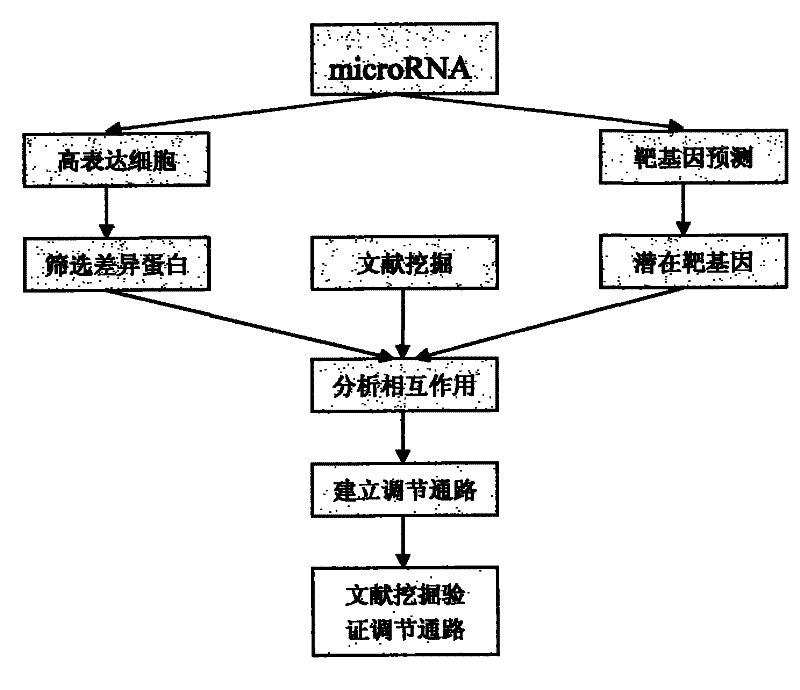
A method for studying the interaction between microRNA and protein
InactiveCN102268475AMicrobiological testing/measurementBiological testingExperimental validationGene model
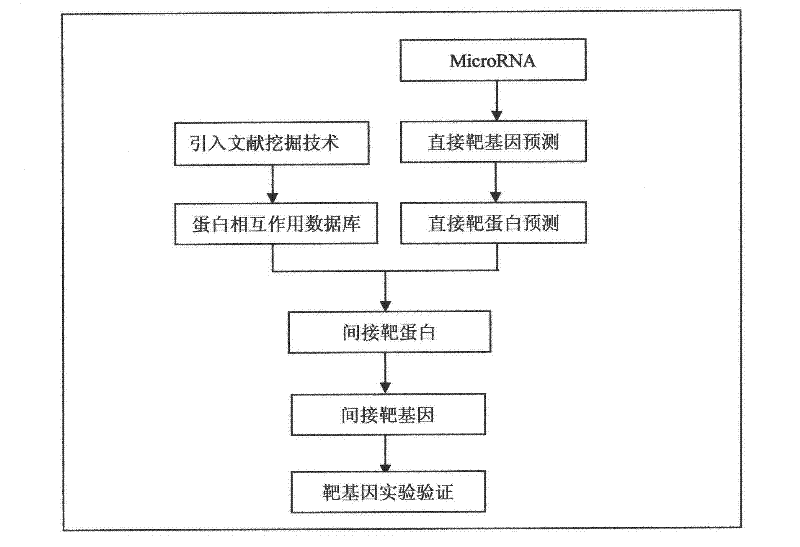
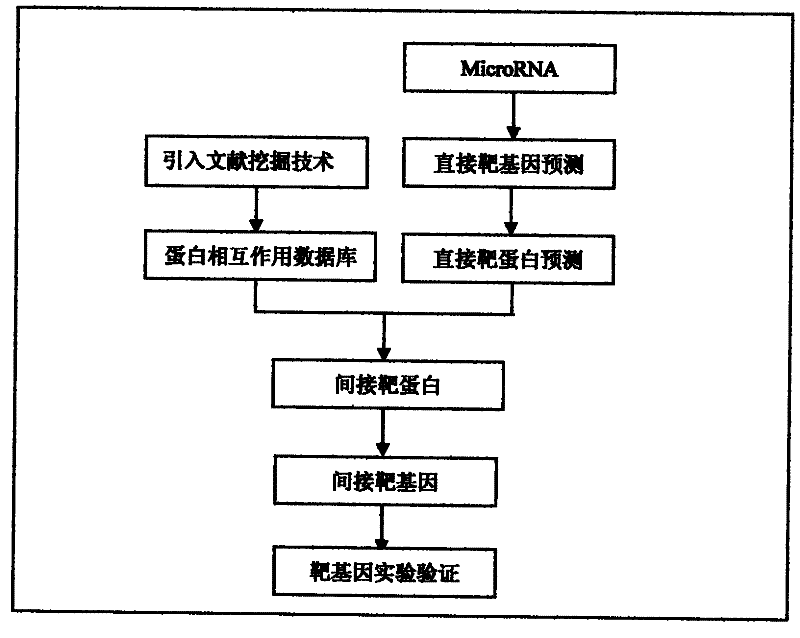
The present invention designs a method for studying the interaction between microRNA and protein, extracts the concept of an "indirect" target gene model, and conducts interaction research based on existing literature reports. The basic process is: Step 1, select microRNA For highly expressed cells, screen for differential proteins; step 2, prediction of microRNA target genes; step 3, introduce literature mining technology for differential protein and microRNA target genes, and build interaction network; step 4, integrate analysis results, and establish microRNA Regulatory pathways with proteins; step 5, introduce literature mining, search for literature reports that have been verified by experiments, and verify the above regulatory pathways.
Owner:SHANGHAI CLUSTER BIOTECH
Gene detection kit for prognosis risk evaluation of patients with liver cancer and application
ActiveCN108611419ALow degree of biased amplificationLittle mutual interferenceMicrobiological testing/measurementDNA/RNA fragmentationGene modelMathematical model
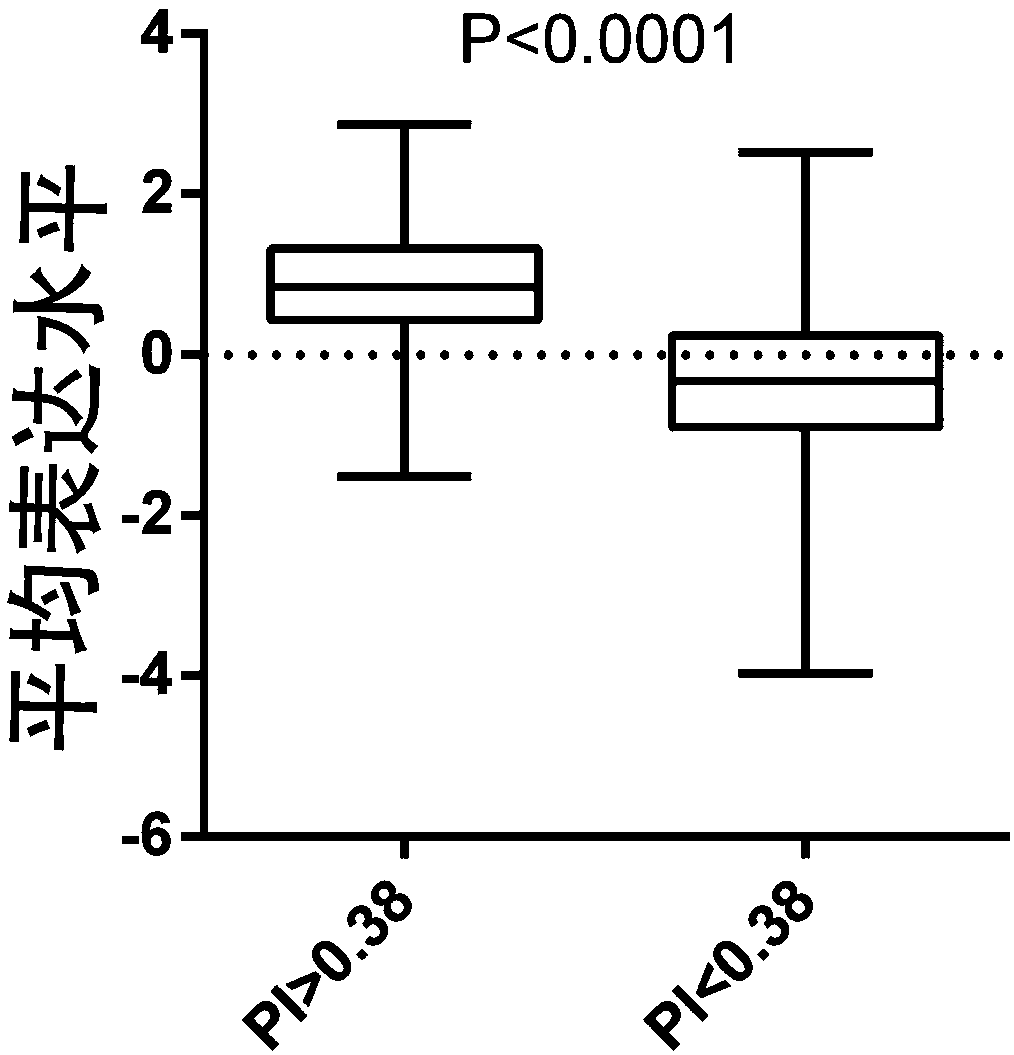
The invention relates to a gene detection kit for liver cancer grouping and prognosis risk evaluation and a mathematical model, and application thereof in liver cancer grouping and prognosis risk evaluation. The gene detection kit is characterized in that a liver cancer grouping and liver cancer prognosis risk evaluation model is built based on a second generation sequence detection technology anda TCGA (The Cancer Genome Atlas) database, and the result shows that liver cancer has two obviously-different prognosis subgroups, and a genetic group for prognosis risk evaluation of the liver cancer. According to the gene detection kit, a 6 gene model composed of CA9, CXCL5, G6PD, MMP12, MYBL2 and SLC1A5 is found, and the prognosis index is defined as PI=exp(p') / exp(1+p'), wherein p'=zCA9*1.64+zCXCL5*1.22+zMMP12*1.52+1.93*zMYBL12+0.76*zSLC1A5+2.43*zG6PD-3.86. The model is capable of recognizing and determining different liver cancer prognosis subgroups and reflecting the prognosis risk ofpatients with liver cancer.
Owner:MENGCHAO HEPATOBILIARY HOSPITAL OF FUJIAN MEDICAL UNIV
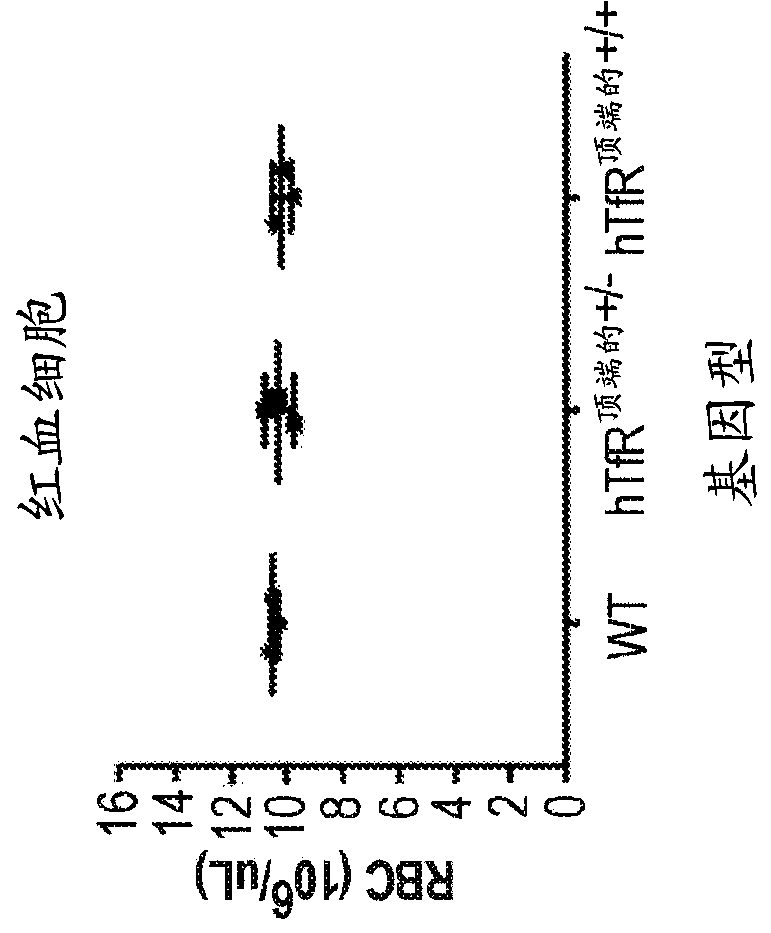
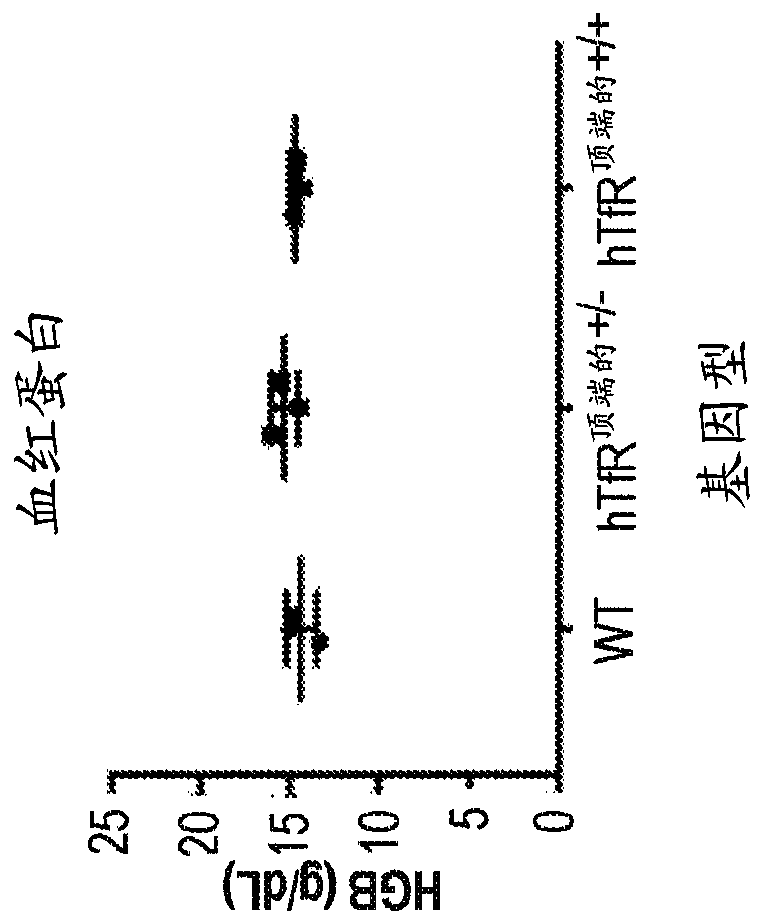
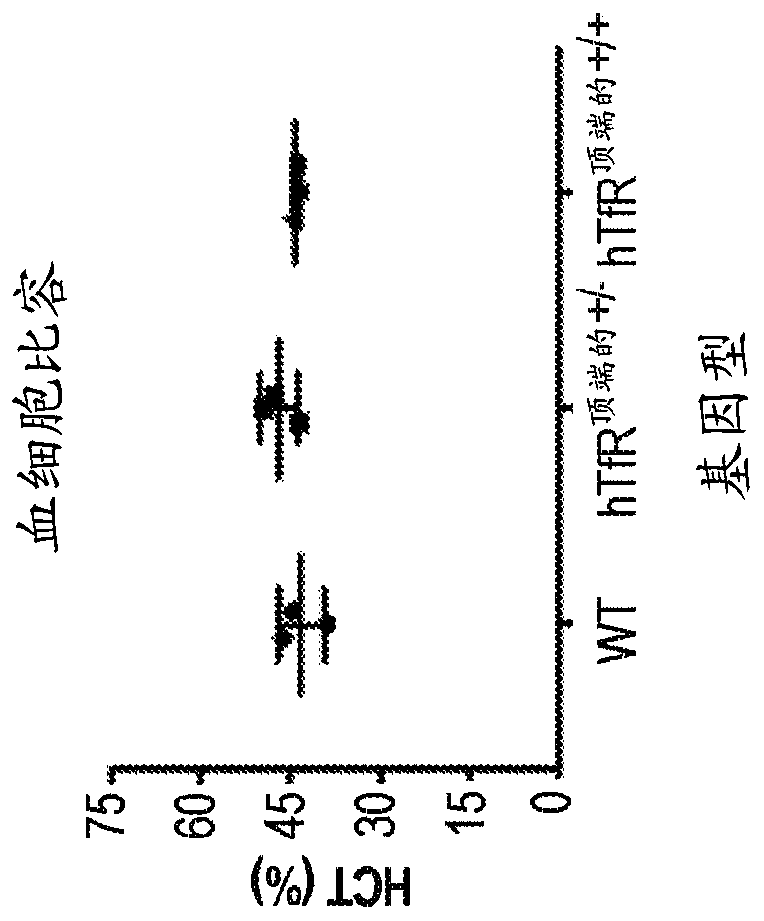
Transferrin receptor transgenic models
In some aspects, the present invention provides chimeric transferrin receptor (TfR) polynucleotides and polypeptides. In other aspects, this invention provides chimeric TfR transgenic animal models and methods of using the animal models to identify therapeutics that can cross the blood-brain barrier.
Owner:DENALI THERAPEUTICS INC
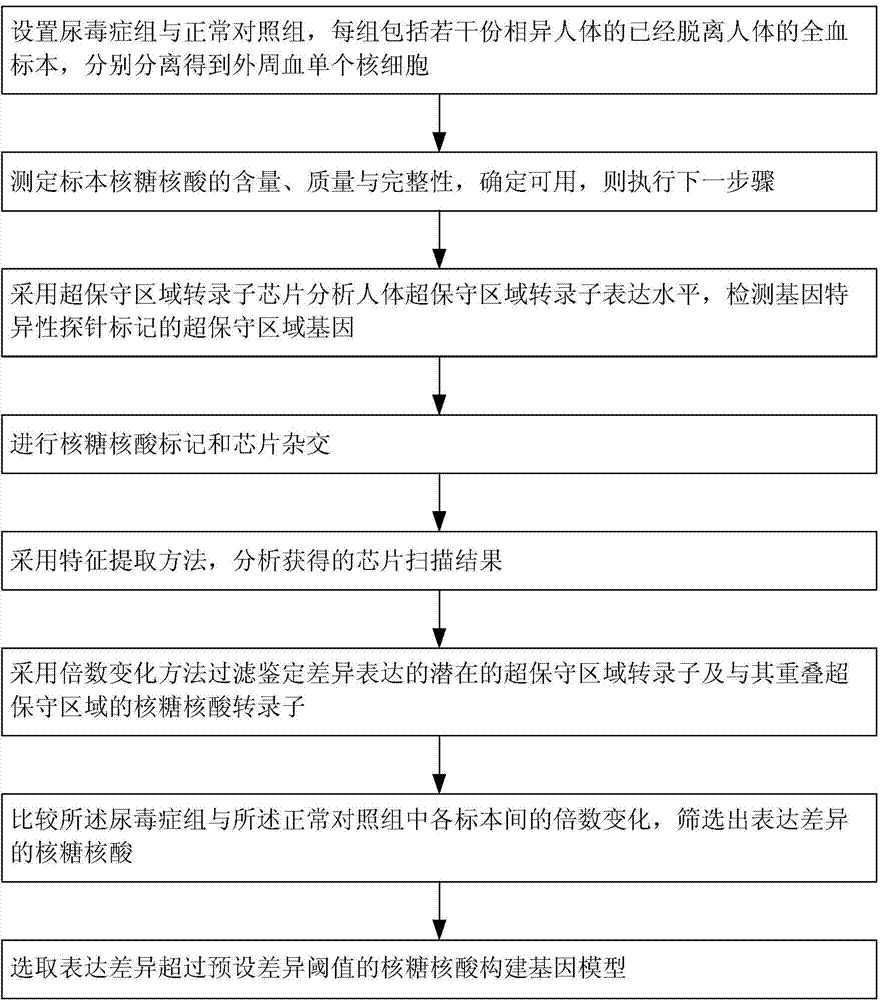
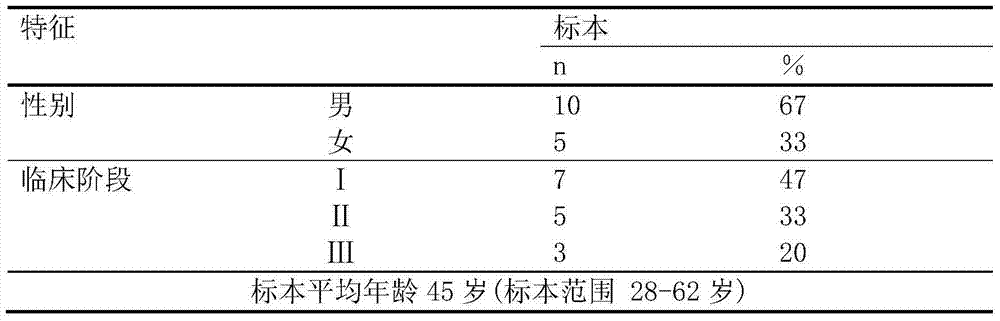
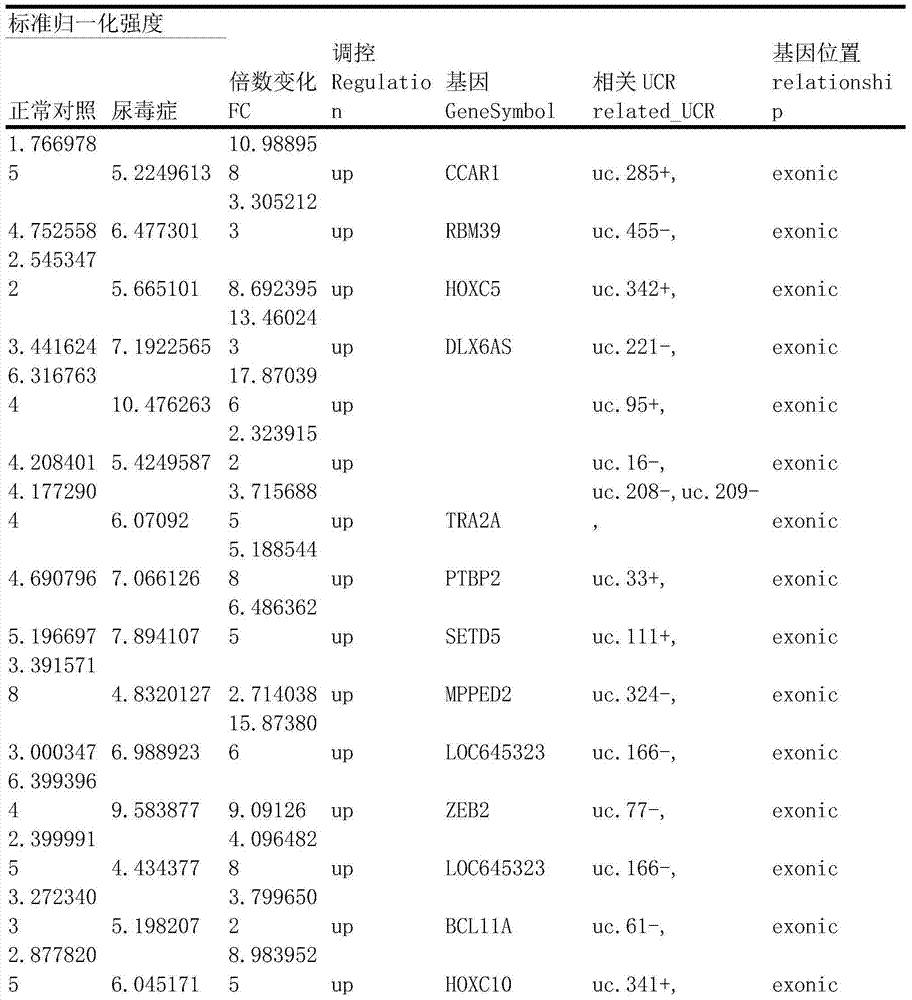
Method for constructing transcription expression gene model of ultra-conserved region of uremic peripheral blood mononuclear cell, and application of model
ActiveCN103937879AMicrobiological testing/measurementBlood/immune system cellsHuman bodyPeripheral blood mononuclear cell
The invention relates to a method for constructing a transcription expression gene model of an ultra-conserved region of a uremic peripheral blood mononuclear cell, and an application of the model. The construction method comprises the following steps: arranging a uremia group and a normal control group, wherein each of the above groups comprises a plurality of whole blood specimens separate from different human bodies, and respectively separating to obtain the uremic peripheral blood mononuclear cell; determining the ribonucleic acid content, the mass and the integrity of the specimens; analyzing the transcripton expression level of the human body ultra-conserved region by adopting an ultra-conserved region transcripton chip, and detecting a gene specificity probe marked ultra-conserved region gene; carrying out ribonucleic acid marking and chip hybridization; analyzing the obtained chip scanning result; filtering a latent ultra-conserved region transcripton and a ribonucleic acid transcripton of the ultra-conserved region overlapping with the ultra-conserved region transcripton for dientifyign differential expression; comparing the multiple change among the specimen in each of the uremia group and the normal control group, and screening expression difference ribonucleic acid; and selecting ribonucleic acid with the expression difference exceeding a preset difference threshold to construct the gene model.
Owner:中国人民解放军联勤保障部队第九二四医院
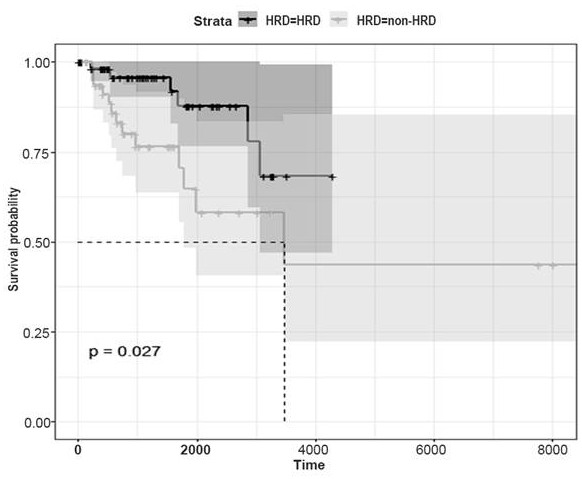
Construction method and application of gene model for prognosis of triple negative breast cancer
The invention discloses a construction method and application of a gene model for triple-negative breast cancer prognosis, and belongs to the technical field of biomedicine, and the construction method is characterized by comprising the following steps: acquiring sample data of triple-negative breast cancer patients from a database, screening out differential genes in combination with HRD scores, and constructing a gene model for triple-negative breast cancer prognosis on the basis of the differential genes. The key genes related to prognosis of the triple-negative breast cancer are screened out, the risk scoring model is constructed based on the key genes, the risk scoring model of HRD characteristics is developed, the prognosis value of the risk scoring model in triple-negative breast cancer patients is proved, and the risk scoring model is expected to be applied to a clinical intelligent decision-making system.
Owner:南京墨宁医疗科技有限公司
Endometrioid adenocarcinoma prognosis-related gene and protein as well as application thereof
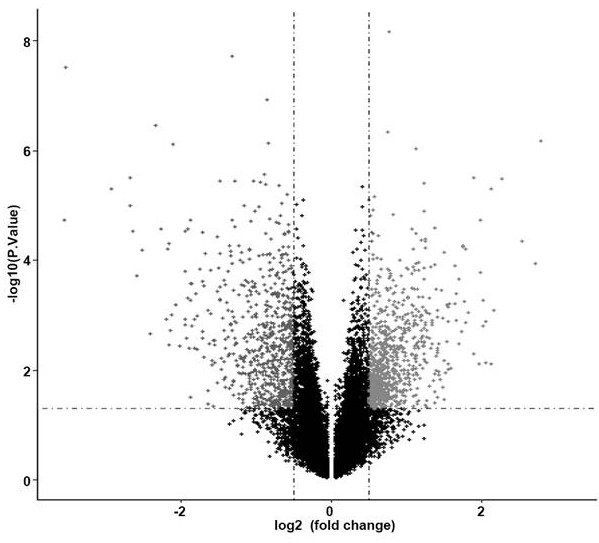
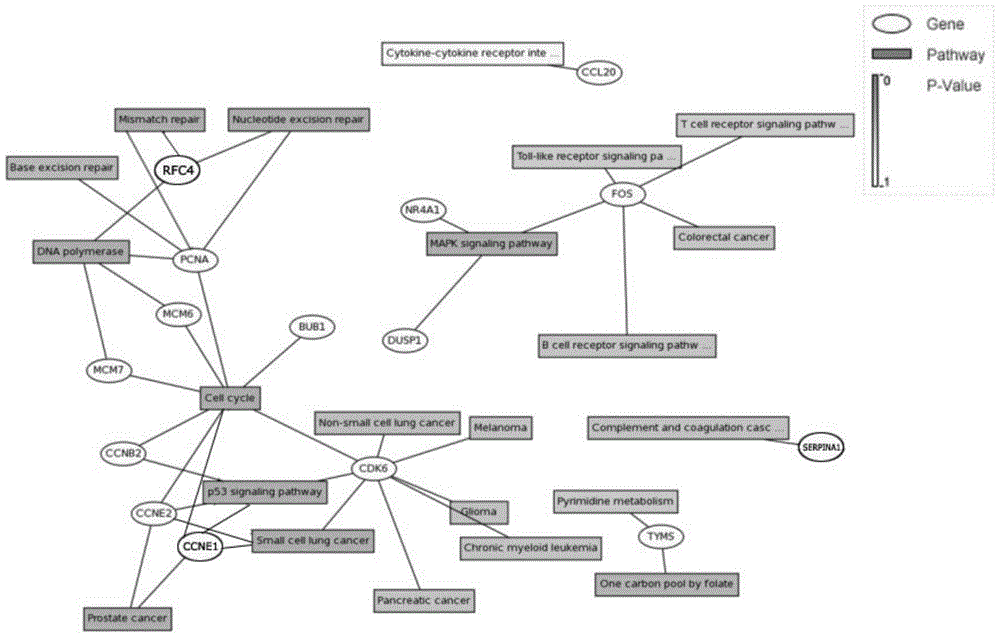
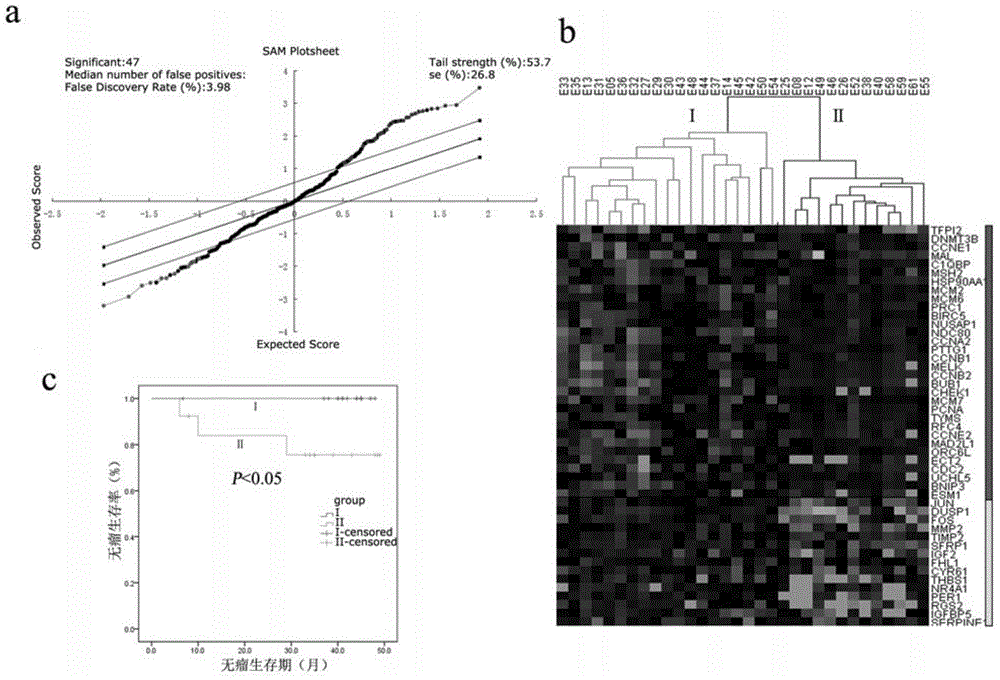
The invention discloses endometrioid adenocarcinoma prognosis-related gene and protein as well as an application thereof. The invention provides a kit a to assist grouping of patients population with endometrioid adenocarcinoma, comprising a substance for detecting mRNA level expression of 21 genes: PRC1, NUSAP1, MCM6, PIWIL2, TYMS, CCNB2, ESM1, BUB1, BARD1, MCM7, CDK6, PCNA, NDC80, CCNE2, RFC4, CCNE1, SERPINA1, FOS, DUSP1, CCL20, and NR4A1. The experiment found a gene model and a protein marker capable of performing prognostic prediction of disease-free survival time of endometrioid adenocarcinoma. The model has important clinical meaning for reflecting biological characteristics of endometrioid adenocarcinoma and prognostic prediction.
Owner:PEOPLES HOSPITAL PEKING UNIV
Process for aligning targeted nucleic acid sequencing data
Provided is a computer-implemented method of aligning RNA including receiving onto a data storage unit primer sequences and transcript sequences transcribable from a reference genome based on a gene model, generating target sequences to be amplified from a combination of the primer sequences and the transcript sequences, generating a modified reference genome based on the plurality of target sequences, aligning sequence reads generated from a test sample comprising RNA amplicon molecules to the of target sequences, and generating an alignment profile for the test sample based on the aligning.Also provided is a computer system for performing the foregoing method.
Owner:ILLUMINA INC
A method for predicting indirect target genes of microRNAs using literature mining
InactiveCN102268473AMicrobiological testing/measurementBiological testingPresent methodProtein target
The present invention designs a method for using literature mining technology to study the relationship between virus and human protein expression regulation, including the following main steps: step 1, predicting the direct target gene from microRNA; step 2, predicting the direct target protein from the above target gene; Step 3, use literature mining technology to construct a protein-protein interaction database; Step 4, use the constructed protein interaction database to screen out the indirect target protein and indirect target gene of microRNA from the direct target protein; Step 5, through the experiment Validation of indirect target genes of microRNAs. The feature of this method is that an "indirect" target gene model is proposed, and the literature mining technology is introduced to screen out the real microRNA target genes that are difficult to detect by conventional methods.
Owner:SHANGHAI CLUSTER BIOTECH
Method for constructing expression system of exogenous genes in campylobacter jejuni and shuttle plasmids thereof
The invention relates to a method for constructing an expression system of exogenous genes in a campylobacter jejuni. Under the condition of anaerobic environment and temperature controlled at 37 DEG C, the method comprises the steps of preparing promoters, constructing the shuttle plasmids, and establishing a bioluminescent transgene model of the campylobacter jejuni. By using electrotransformation to introduce the campylobacter jejuni ATCC33291 and ATCC35921 which are expressed under the selection pressure of kanamycin antibiotics, the method can observe bioluminescence without adding any substrate, visually expresses bioinformation, and has a wide research prospect.
Owner:NORTHWEST A & F UNIV
Pathogenic gene model of mental retardation and building method and application of pathogenic gene model
ActiveCN106191032AFacilitate understanding of pathogenesisFacilitate understanding of prenatal diagnosis and preventionMicrobiological testing/measurementDNA preparationNormal peoplePrenatal diagnosis
Owner:戴勇 +1
Method for determining influence of genetic variation on function based on genomic environment
The invention provides a method for determining the influence of genetic variations on function based on a genomic environment. According to the method, each gene serves as a unit, and the common influence of all variations on the gene is annotated. The method includes the following steps: 1, all the variations are mapped to all genes of a given genetic model according to the coordinate positions of the variations; 2, according to all the variations on all the genes, the individualized sequence of each gene is reconstructed; 3, all the individualized sequences are analyzed, and the influences of the variations on the genes are obtained. The method fully considers the genomic environment where the variations are located, a large quantity of annotation errors are avoided, and the accuracy of the annotation variation effect is remarkably improved.
Owner:PEKING UNIV
Transferrin receptor transgenic models
In some aspects, the present invention provides chimeric transferrin receptor (TfR) polynucleotides and polypeptides. In other aspects, this invention provides chimeric TfR transgenic animal models and methods of using the animal models to identify therapeutics that can cross the blood-brain barrier.
Owner:DENALI THERAPEUTICS INC
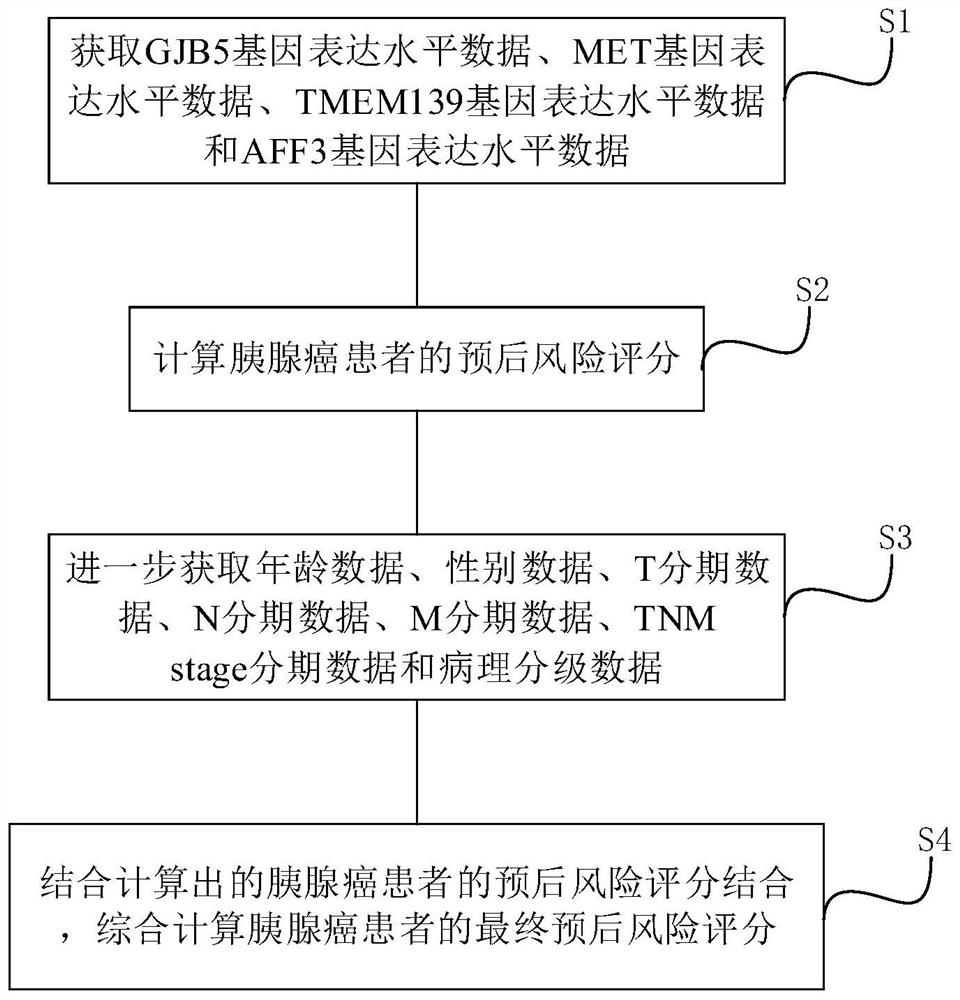
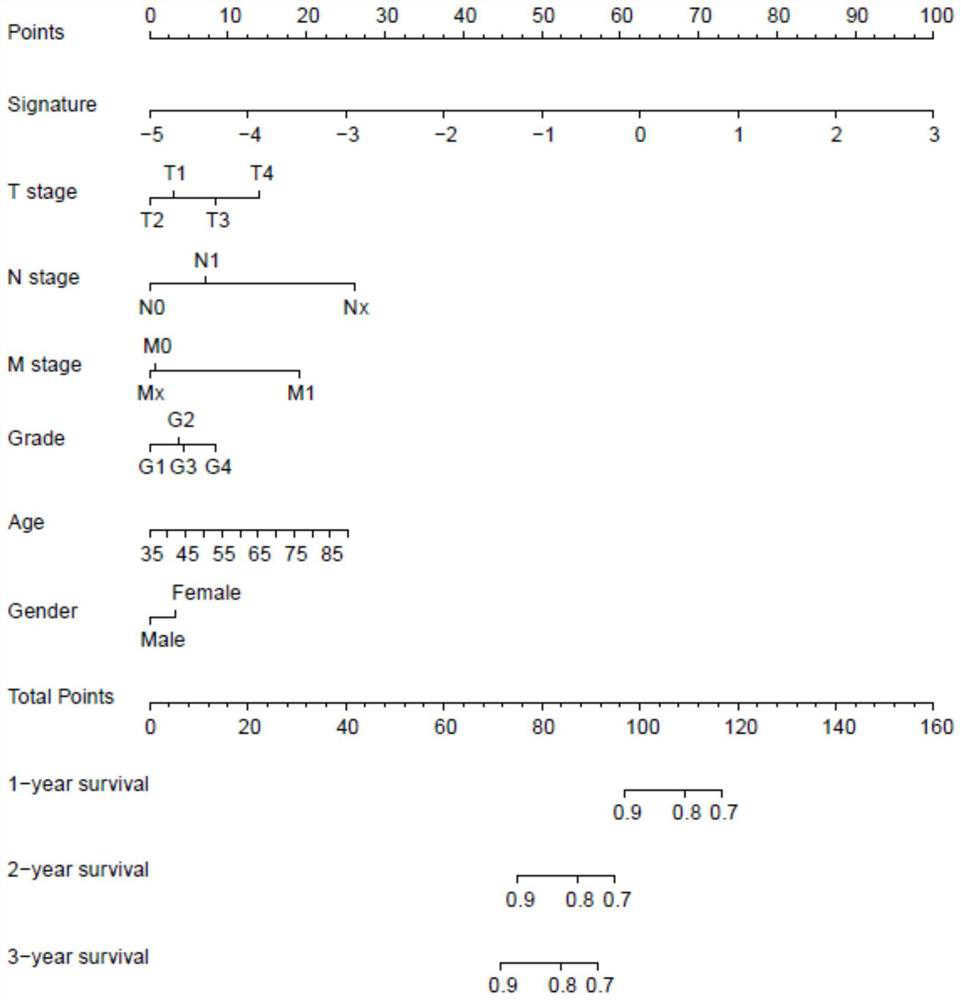
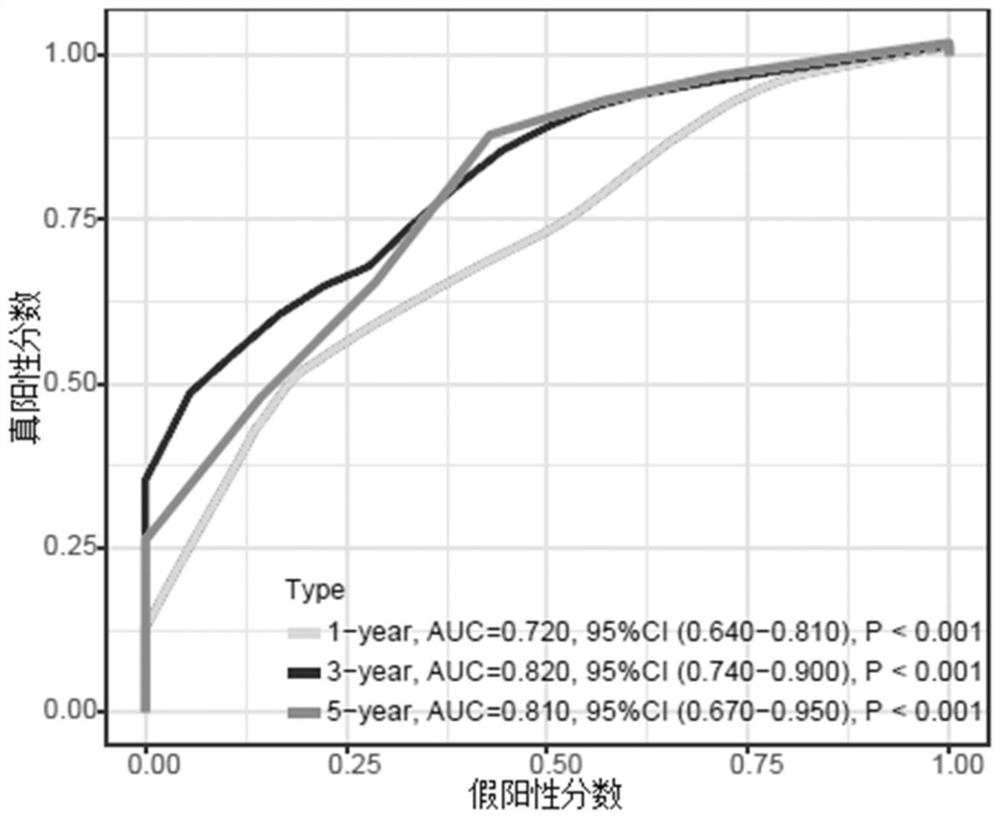
Pancreatic cancer prognosis risk prediction method and device established based on metabolic genes
PendingCN114373548AImproved prognosisImprove robustnessHealth-index calculationHybridisationPancreas CancersGene model
The invention relates to a pancreatic cancer prognosis risk prediction method and device established based on metabolic genes, and the method comprises the following steps: obtaining GJB5 gene expression level data, MET gene expression level data, TMEM139 gene expression level data and AFF3 gene expression level data, and calculating a prognosis risk score of a pancreatic cancer patient. Compared with the prior art, the four-gene model provided by the invention has relatively strong robustness, can exert stable prediction efficiency in data sets of different platforms, and has the advantages of excellent efficiency, small number of detected genes and the like.
Owner:FUDAN UNIV SHANGHAI CANCER CENT
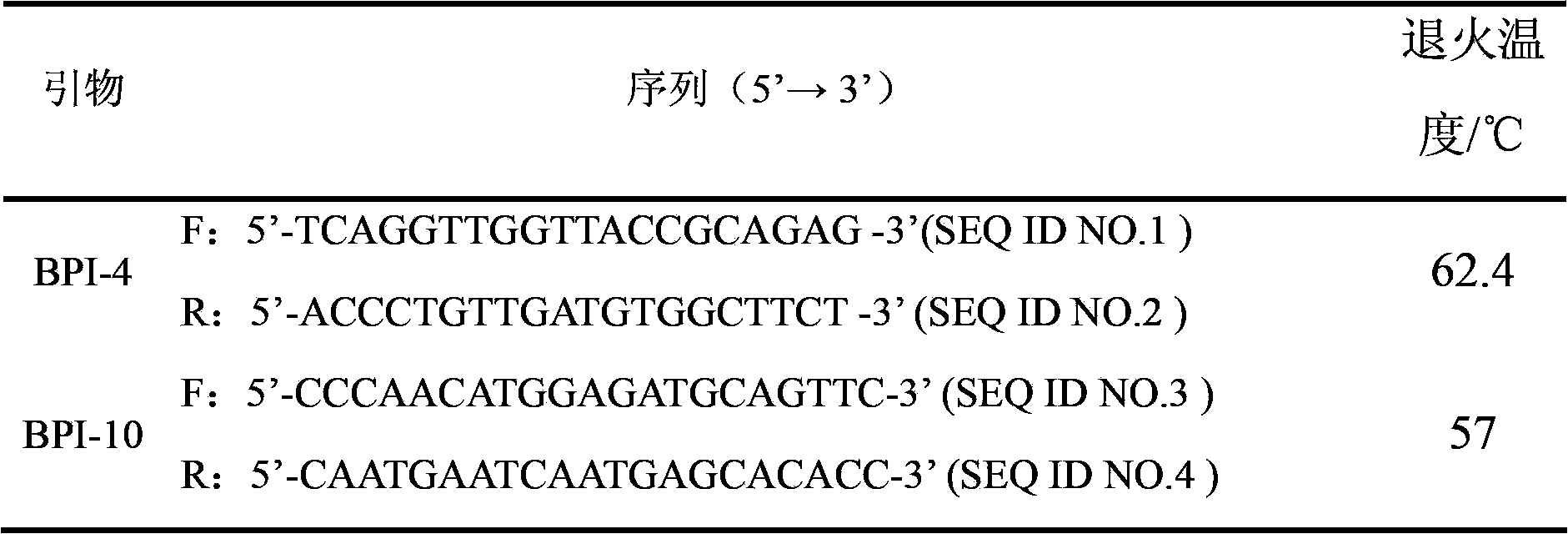
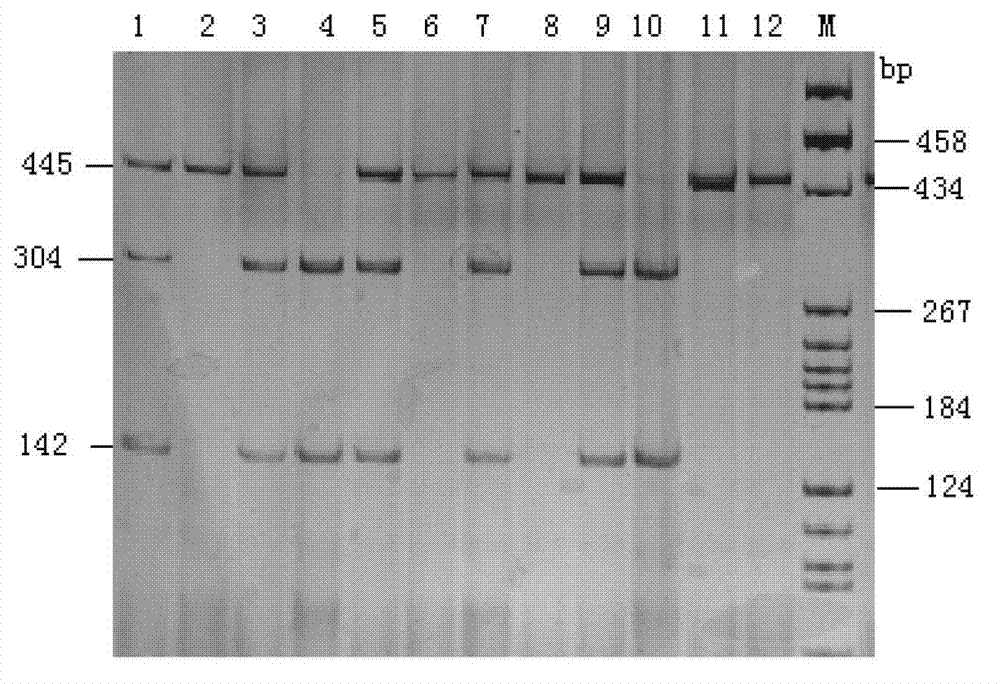
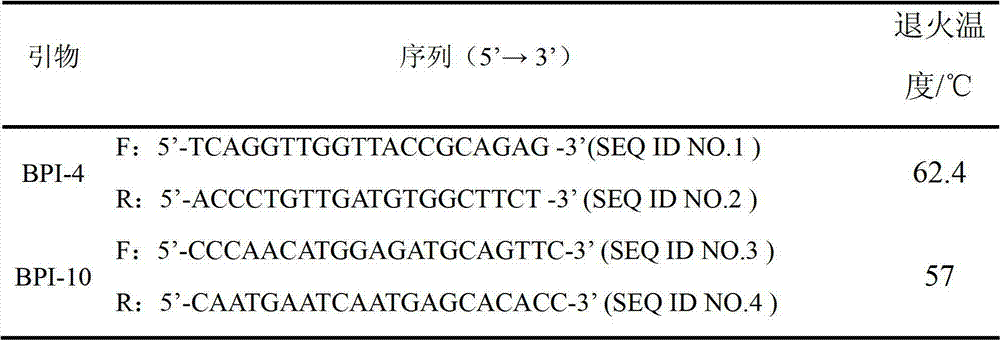
Application of bactericidal/permeability-increasing protein gene in serving as common disease resistance genetic marker of pigs
ActiveCN103173531BImprove disease resistanceEnhance immune responseMicrobiological testing/measurementSingle-strand conformation polymorphismEnzyme digestion
The invention relates to a genetic marker for screening the combined gene model of the fourth and tenth exon variation sites of a common disease resistance BPI (bactericidal / permeability increasing protein) gene of pigs, and belongs to the technical fields of genetic breeding and molecular marker assisted selection of pigs. The method comprises the following steps of: extracting DNA (deoxyribonucleic acid) of a pig genome of a sample to be tested, and performing PCR (polymerase chain reaction) amplification on the pig genome; analyzing the PCR product of the exon 4 through SSCP (single strand conformation polymorphism), and performing enzyme digestion on the PCR product of the exon 10 through restriction enzyme HpaII, wherein when the gene model of the pig body to be tested is a CDAB model, the pig body is strong in common disease resistance. According to the application, a molecular genetic marker with higher efficiency and accuracy is provided to molecular marker assisted selection of the pig disease resistance breeding work, and an effective molecular marker breeding means is provided for improving the common disease resistance and immune response capability of piglets. The detection method is simple in operation, low in cost and high in accuracy, can realize automatic direct detection, and takes a magnificent effect in pig breeding.
Owner:YANGZHOU UNIV
Application of bactericidal/permeability-increasing protein gene in serving as common disease resistance genetic marker of pigs
ActiveCN103173531AImprove disease resistanceEnhance immune responseMicrobiological testing/measurementSingle-strand conformation polymorphismEnzyme digestion
The invention relates to a genetic marker for screening the combined gene model of the fourth and tenth exon variation sites of a common disease resistance BPI (bactericidal / permeability increasing protein) gene of pigs, and belongs to the technical fields of genetic breeding and molecular marker assisted selection of pigs. The method comprises the following steps of: extracting DNA (deoxyribonucleic acid) of a pig genome of a sample to be tested, and performing PCR (polymerase chain reaction) amplification on the pig genome; analyzing the PCR product of the exon 4 through SSCP (single strand conformation polymorphism), and performing enzyme digestion on the PCR product of the exon 10 through restriction enzyme HpaII, wherein when the gene model of the pig body to be tested is a CDAB model, the pig body is strong in common disease resistance. According to the application, a molecular genetic marker with higher efficiency and accuracy is provided to molecular marker assisted selection of the pig disease resistance breeding work, and an effective molecular marker breeding means is provided for improving the common disease resistance and immune response capability of piglets. The detection method is simple in operation, low in cost and high in accuracy, can realize automatic direct detection, and takes a magnificent effect in pig breeding.
Owner:YANGZHOU UNIV
Prediction model of peritoneal metastasis of gastric cancer based on 22 genes and its application
ActiveCN107586852BImprove survival rateImprove accuracyMicrobiological testing/measurementRPS27AGene model
The invention belongs to the field of gene detection, and provides a gastric cancer peritoneal metastasis prediction gene model and its application, including PCLO, UGGT1, ZNF714, KIAA0825, COL23A1, MED1, NPAS2, TTC14, RPS27A, ASPH, ARHGEF12, SIK1, PAPPA, HHIPL1, MYO9B, ITPKB , ZNF862, MKNK1, MUC6, TRRAP, DUOX1, and KRTAP5‑2; the selected classifier SVM and the positive judgment threshold of 0.5 are effective and specific for predicting the risk of peritoneal metastasis. The application of the gene model of the present invention helps to predict the metastasis of gastric cancer patients, and is of great value and significance for timely taking effective clinical measures, formulating individualized diagnosis and treatment schemes, and finally improving the survival rate of gastric cancer patients.
Owner:FUJIAN MEDICAL UNIV UNION HOSPITAL +1
Gene model for judging prognosis of hepatocellular carcinoma patient, construction method and application
ActiveCN113539376AAvoid overtreatmentActive treatment planMicrobiological testing/measurementBiostatisticsCell-Extracellular MatrixGene model
The invention discloses a gene model for judging prognosis of hepatocellular carcinoma as well as a construction method and application of the gene model. The method comprises the following steps: comparing the data of a hepatocellular carcinoma patient sample with transcriptome data of a normal patient sample to obtain a gene with differential expression, integrating the gene with an extracellular matrix gene set, and shrinking through an LASSO-COX regression model to obtain a model with 18 genes. The model can be used for evaluating the prognosis of hepatocellular carcinoma patients and distinguishing and selecting hepatocellular carcinoma patients with poor prognosis, so that clinicians are guided to provide a more positive treatment scheme, and meanwhile, the hepatocellular carcinoma patients with low risk can be prevented from being over-treated. The gene model is beneficial to construction of a tissue chip based on extracellular matrix genes, and can be used for rapid prognosis evaluation of a hepatocellular carcinoma postoperative patient so as to realize clinical transformation.
Owner:ZHEJIANG UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com