Construction method and application of gene model for prognosis of triple negative breast cancer
A triple-negative breast cancer and construction method technology, applied in the field of gene model construction, can solve the problems of loss of function, inability to benefit, and median overall survival of less than one year, and achieve the effect of good predictive ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
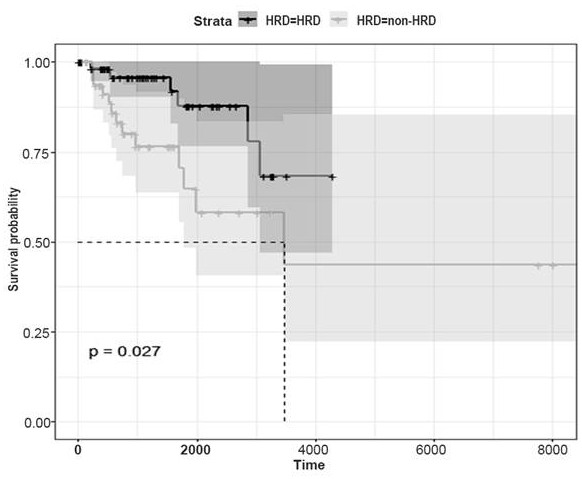
[0050] In this study, the clinical characteristics and RNA-sequencing data of 123 TNBC tumor samples were collected from the TCGA database, and the HRD grouping was determined based on the genomic data, and then a prognostic model of the HRD transcriptome was constructed and compared with the genomic HRD ratings for comparison.
[0051] 1 Data collection and preprocessing
[0052] S1. Obtain triple-negative breast cancer sample data from the database as a training set:
[0053] TCGA (The Cancer Genome Atlas) BC mRNA expression profiles (TCGA-BRCA cohort) and associated clinical information were downloaded from the Genome Data Commons data portal (https: / / portal.gdc.cancer.gov / ). Data were normalized and processed using R (version 3.6.0) software. The HRD information of tumor samples can be obtained from HRD related data (see HRD score: PMID: 29617664, TCGA_DDR_Data_Resources.xlsx format: DDR footprint), tumor samples are divided into HRD tumor samples (HRD score ≥ 42) and no...
Embodiment 2
[0067] Example 2: Validation of the predictive performance of the risk scoring model:
[0068] 2.1 Compared with the verification set:
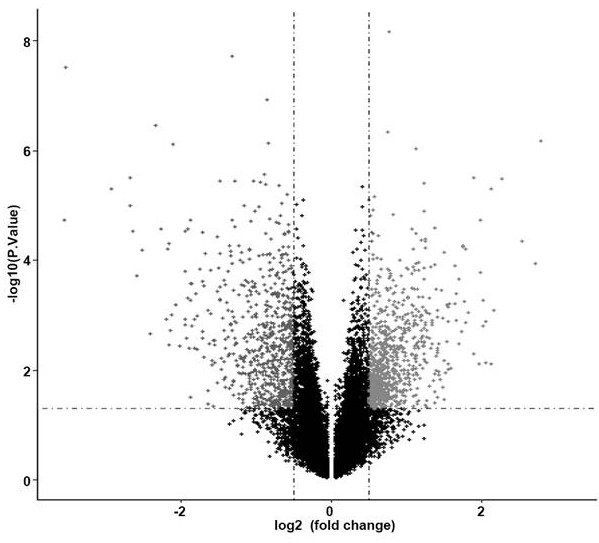
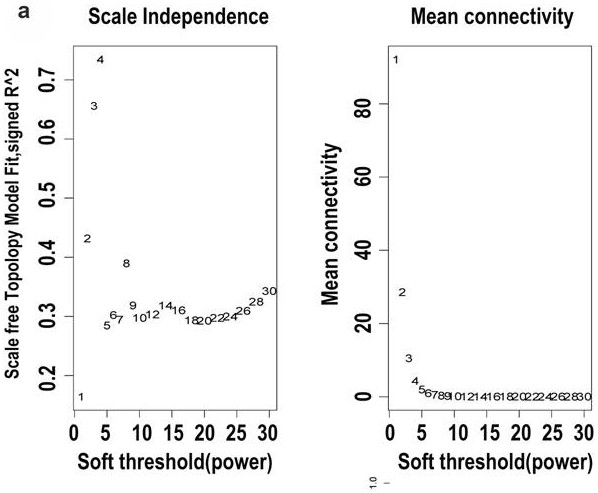
[0069] According to the gene expression level and the risk coefficient of each gene, the risk score of each patient is calculated, and the median risk score of each model is used as the threshold, and the samples are divided into high-risk and low-risk groups to draw the Kaplan-Meier curve and model The risk factor linkage map of the evaluation, compared with the low-risk group, judges the prognosis difference between the high-risk group and the low-risk group, and Kaplan-Meier survival analysis shows that the prognosis of the high-risk group is significantly poorer (P Figure 6a , Figure 6b ). The GSEGSE103091 data set was obtained from the Gene Expression Omnibus (GEO) database as a verification set, and the risk scoring model of the present invention was used for survival analysis, and the Kaplan-Meier curve was drawn ( Figure 10), the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com