Patents
Literature
165 results about "Overall survival" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
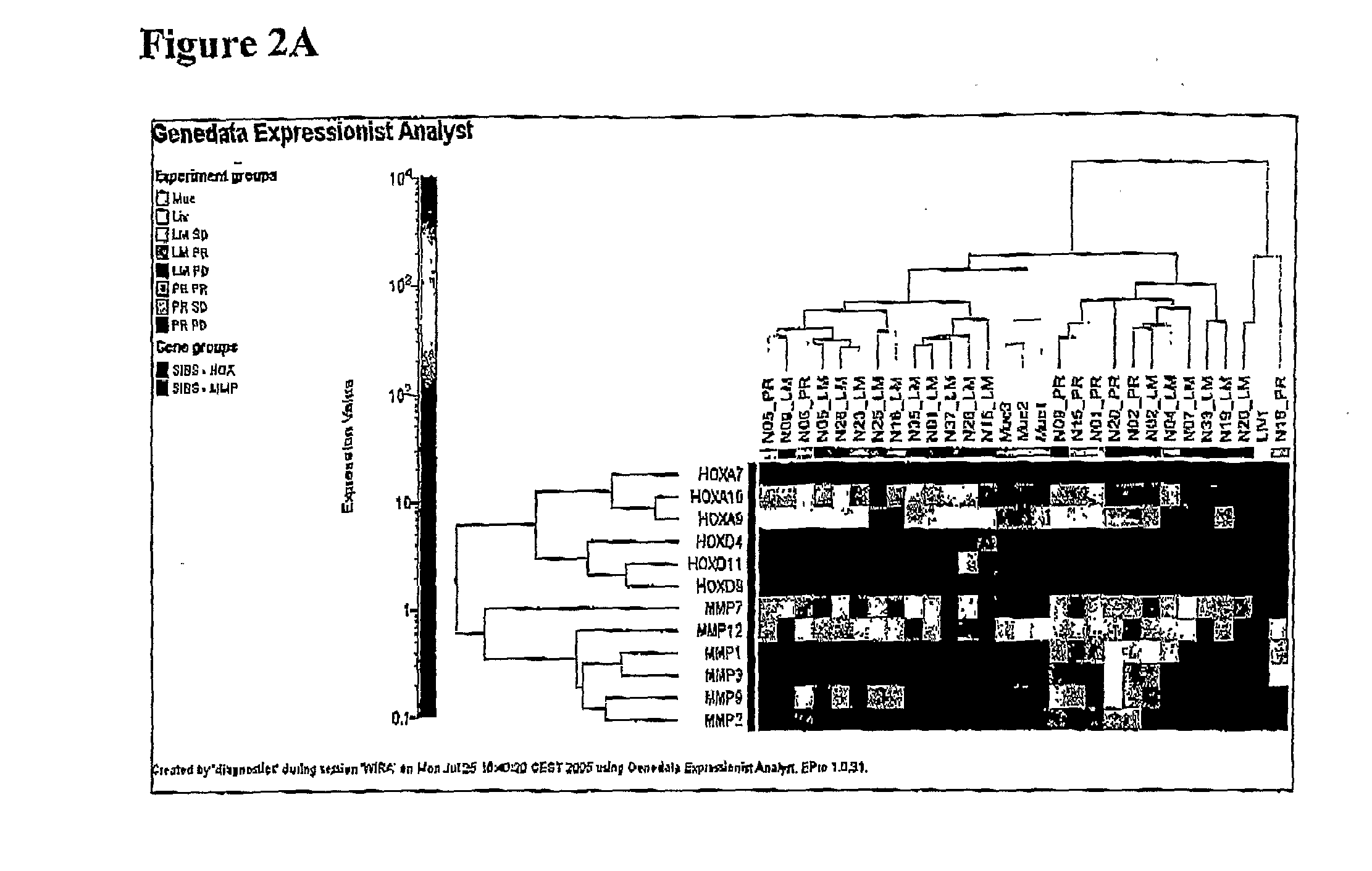
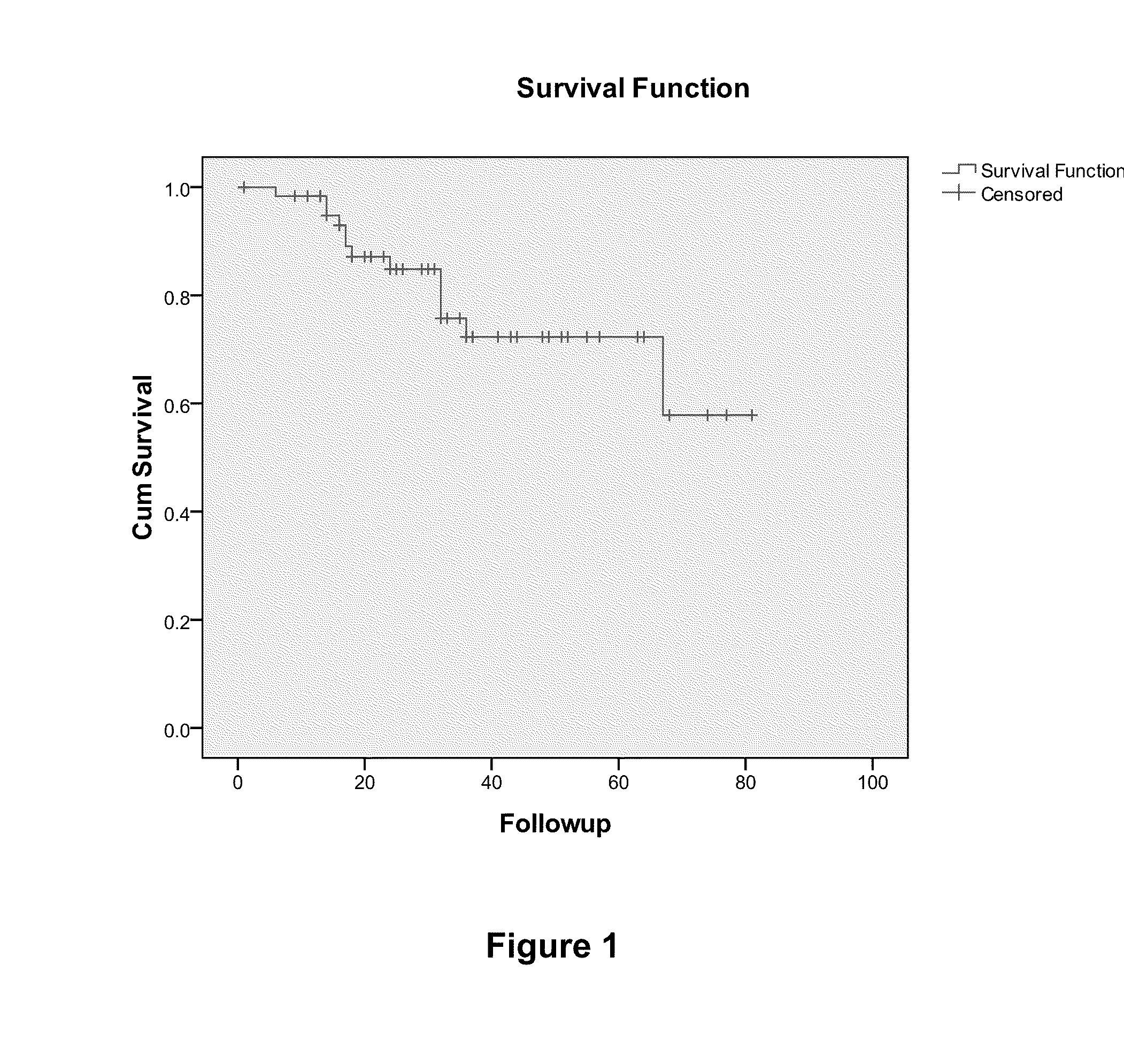
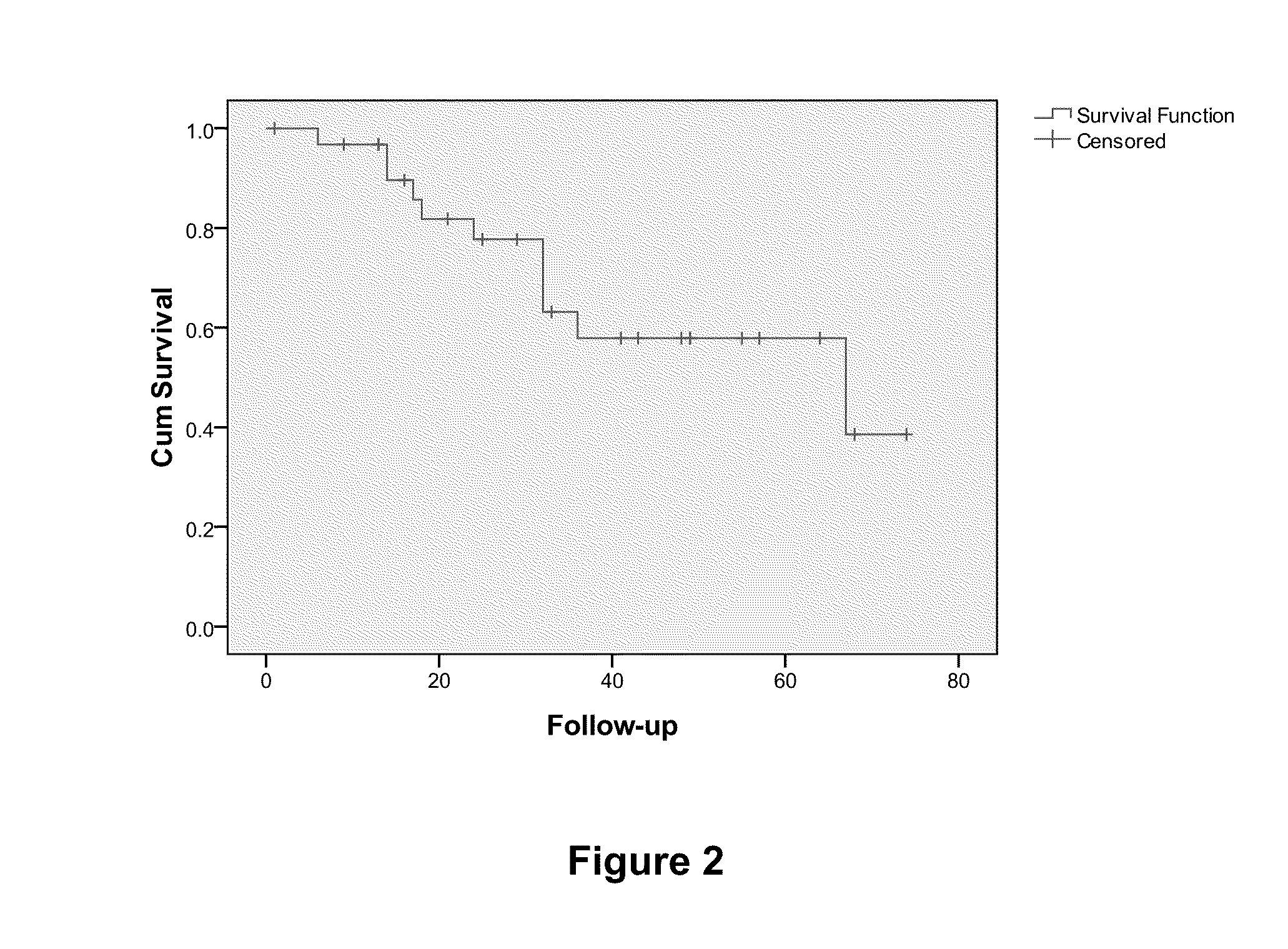
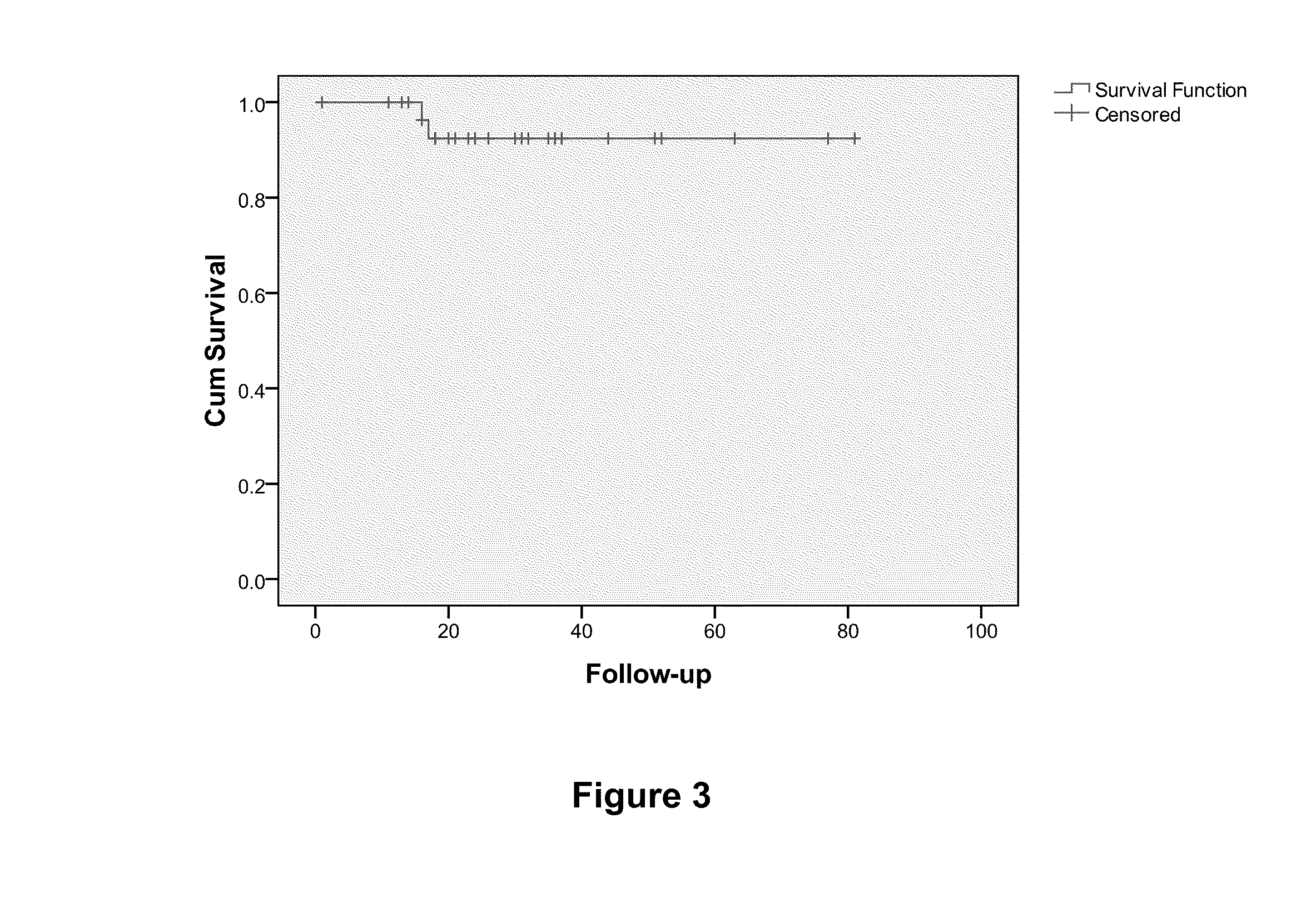
Overall Survival (OS) Overview. Overall survival, or OS, or sometimes just “survival” is the percentage of people in a group who are alive after a length of time—usually a number of years.
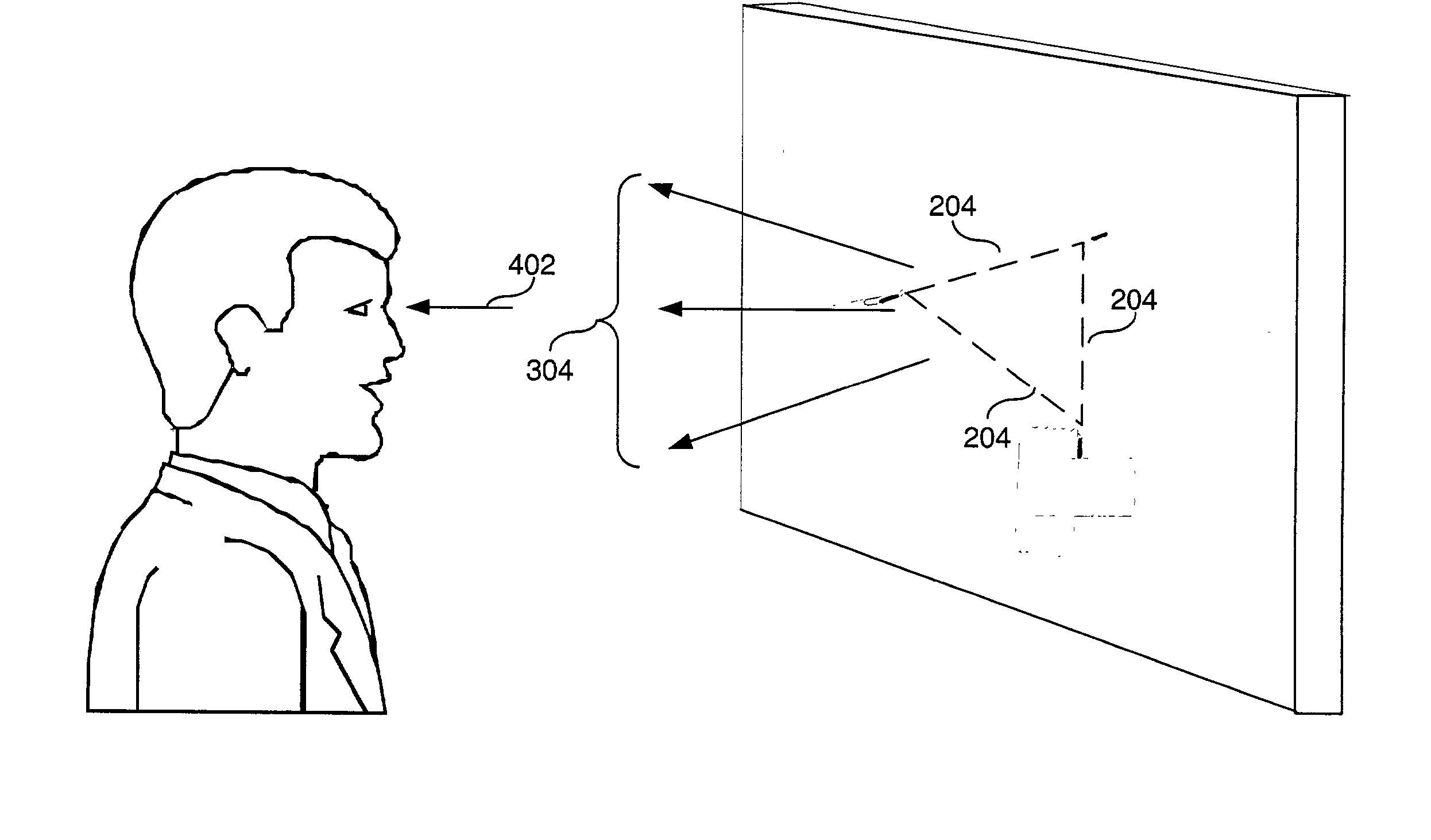
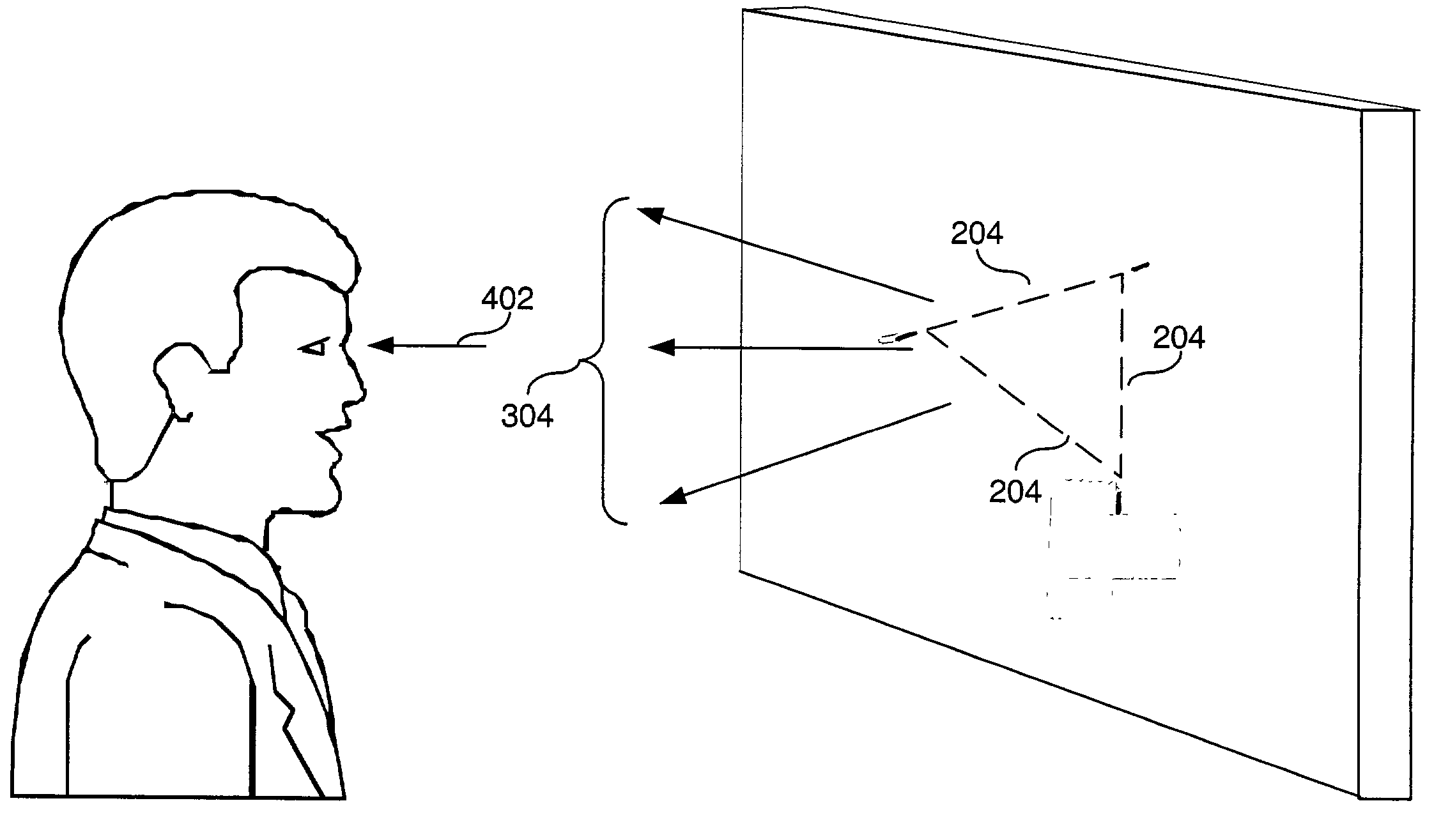
Apparatus, method and computer program product to facilitate ordinary visual perception via an early perceptual-motor extraction of relational information from a light stimuli array to trigger an overall visual-sensory motor integration in a subject
ActiveUS20040049124A1Increase probabilityEasy extractionSurgeryVaccination/ovulation diagnosticsOptical flowSensorimotor integration
An apparatus, method and computer program product is presented to address early visual-sensory motor perception of a subject, where the method comprising the steps of: (1) controlling photic energetic parameters and / or photic perceptual attributes to trigger pre-attentive cuing or increase reactivity in magnocellular activity towards transient visual stimuli of the subject; and (2) generating a optical field comprising optical events based on said photic energetic parameters and said photic perceptual attributes, wherein said optical field transforms into a simple optical flow in the perceptual visual field of said subject. The photic energetic parameters may comprise light array energetic features, including one or more of wavelength, amplitude, intensity, phase, polarization, coherence, hue, brightness, and saturation.
Owner:BRIGHTSTAR LEARNING LTD
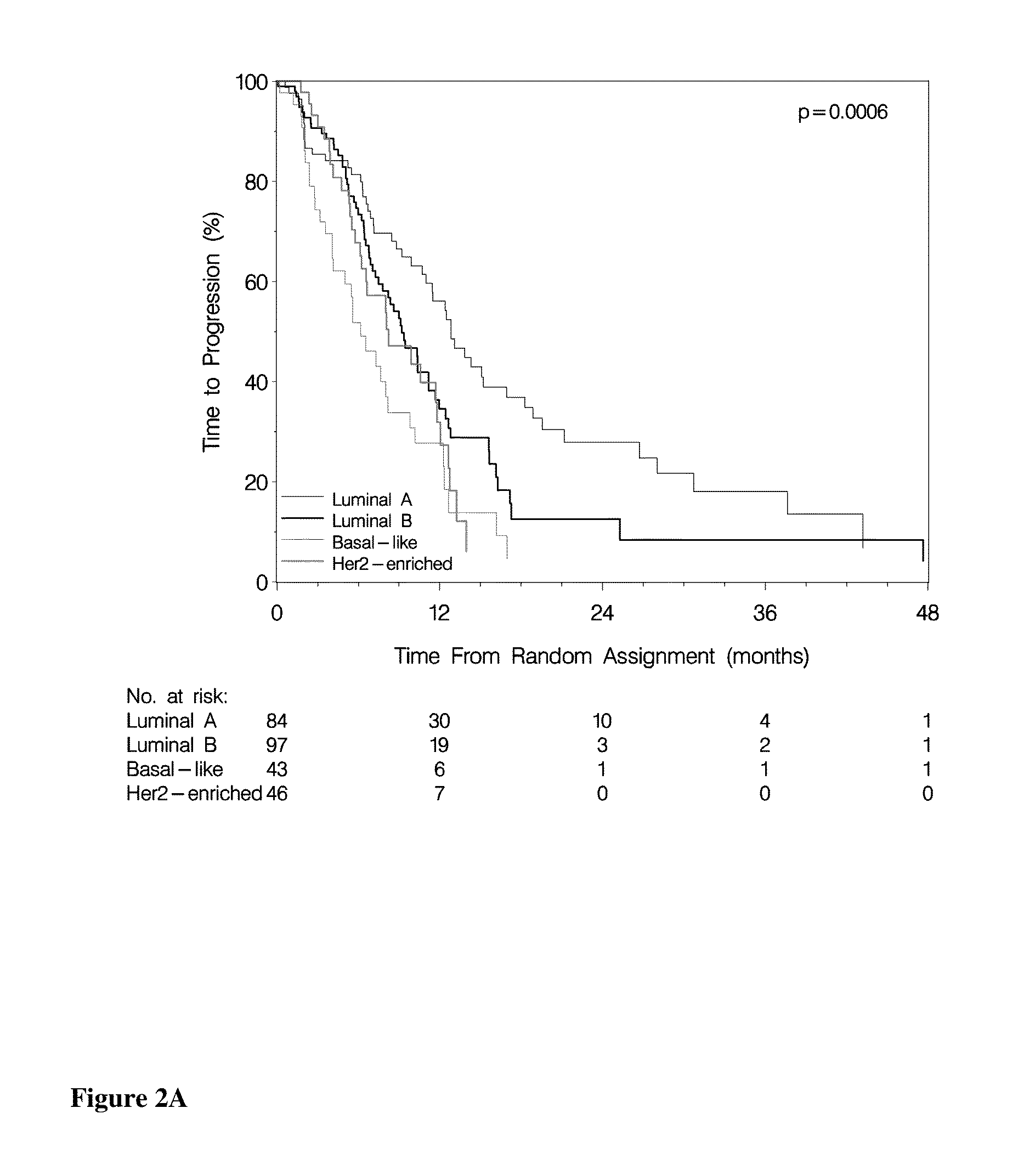
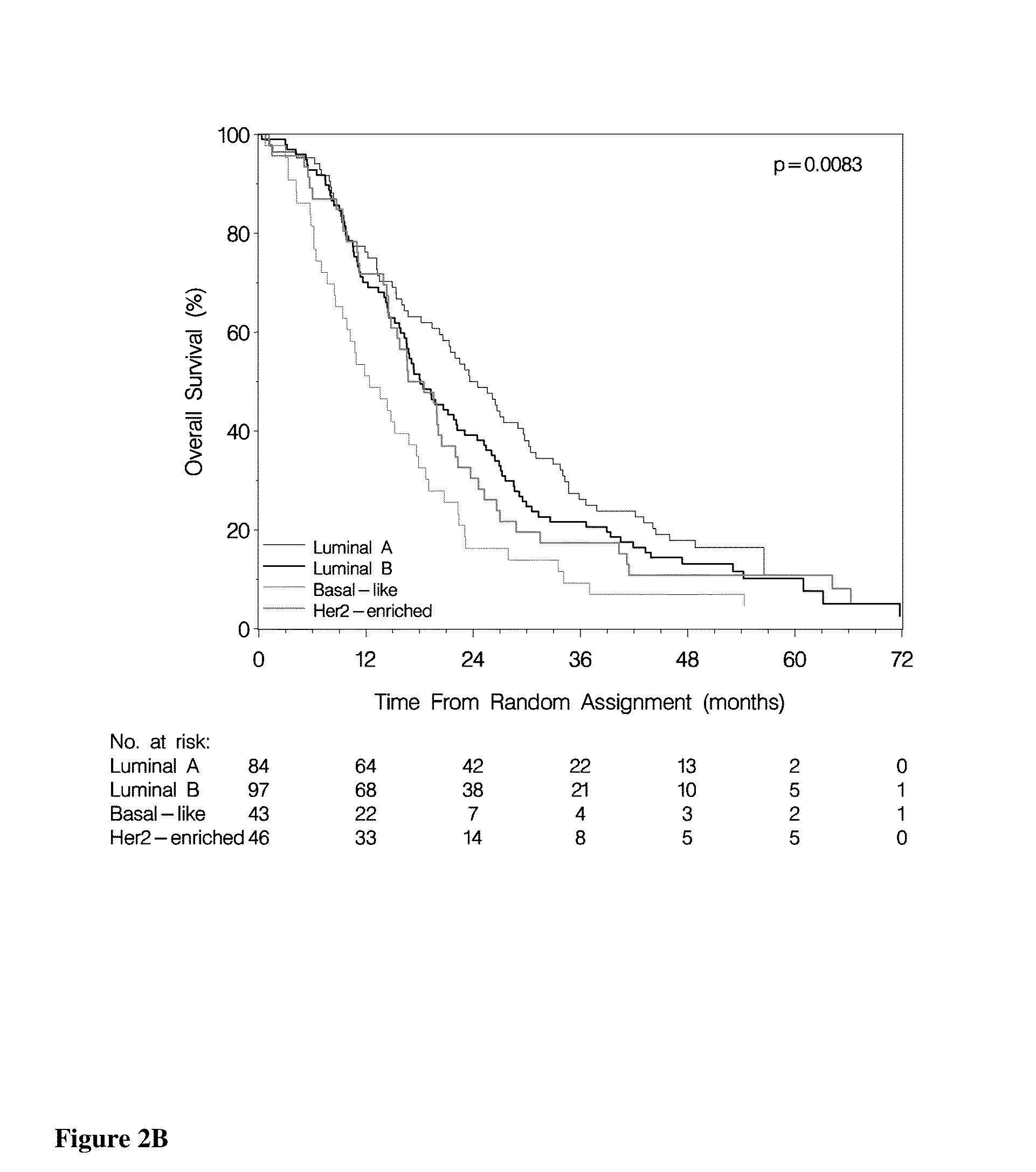
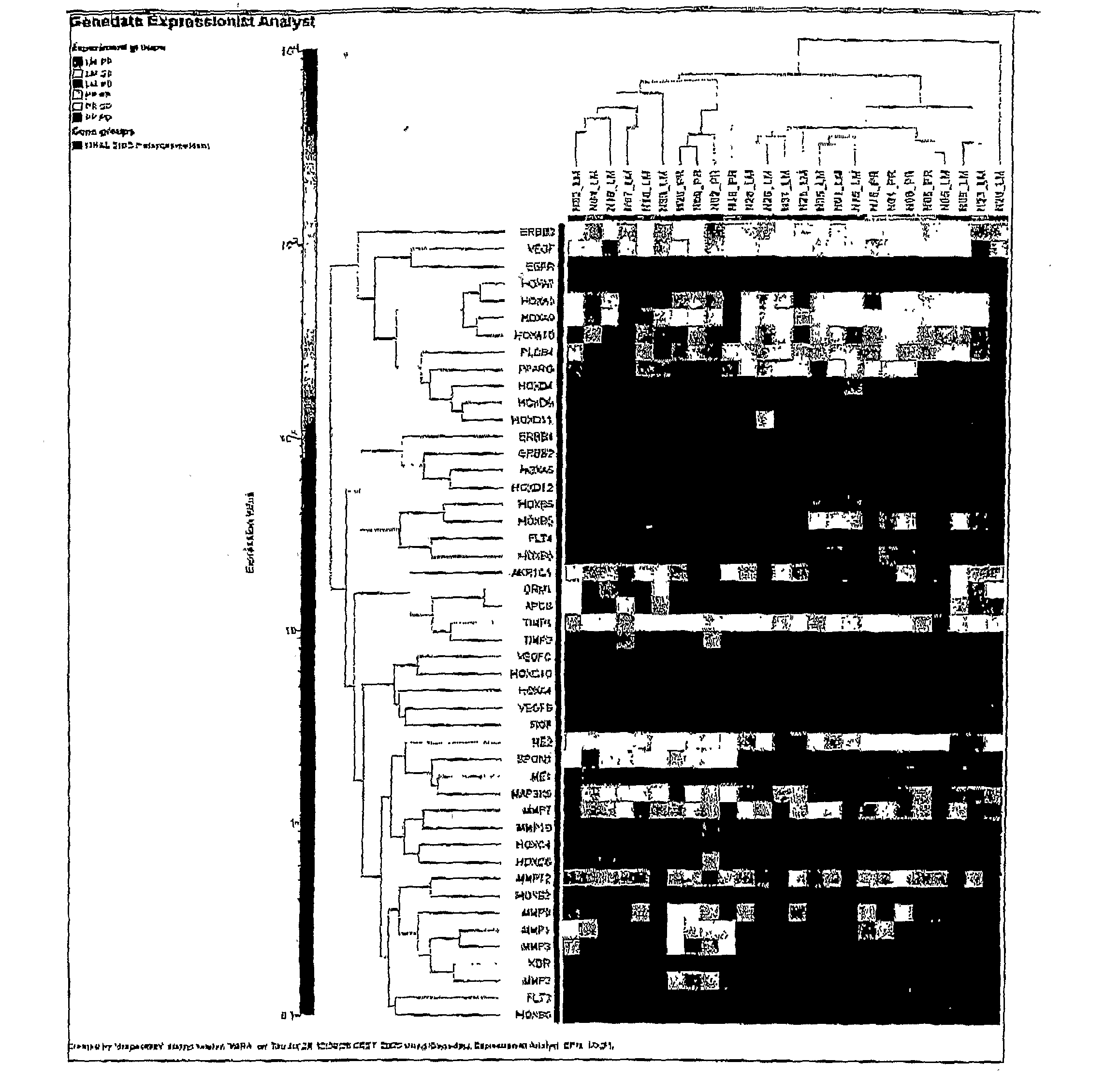
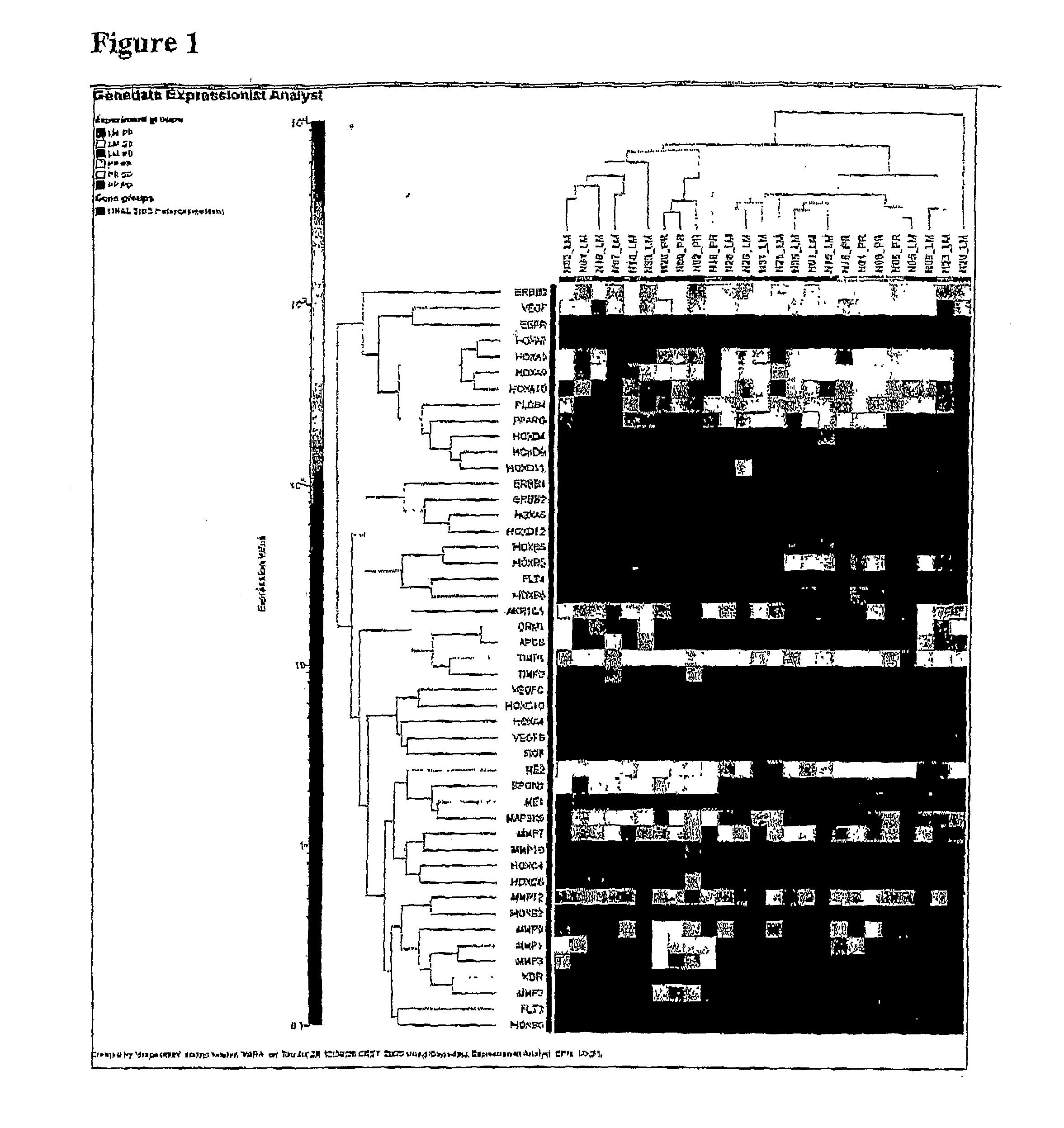
Methods of Treating Breast Cancer with Gemcitabine Therapy
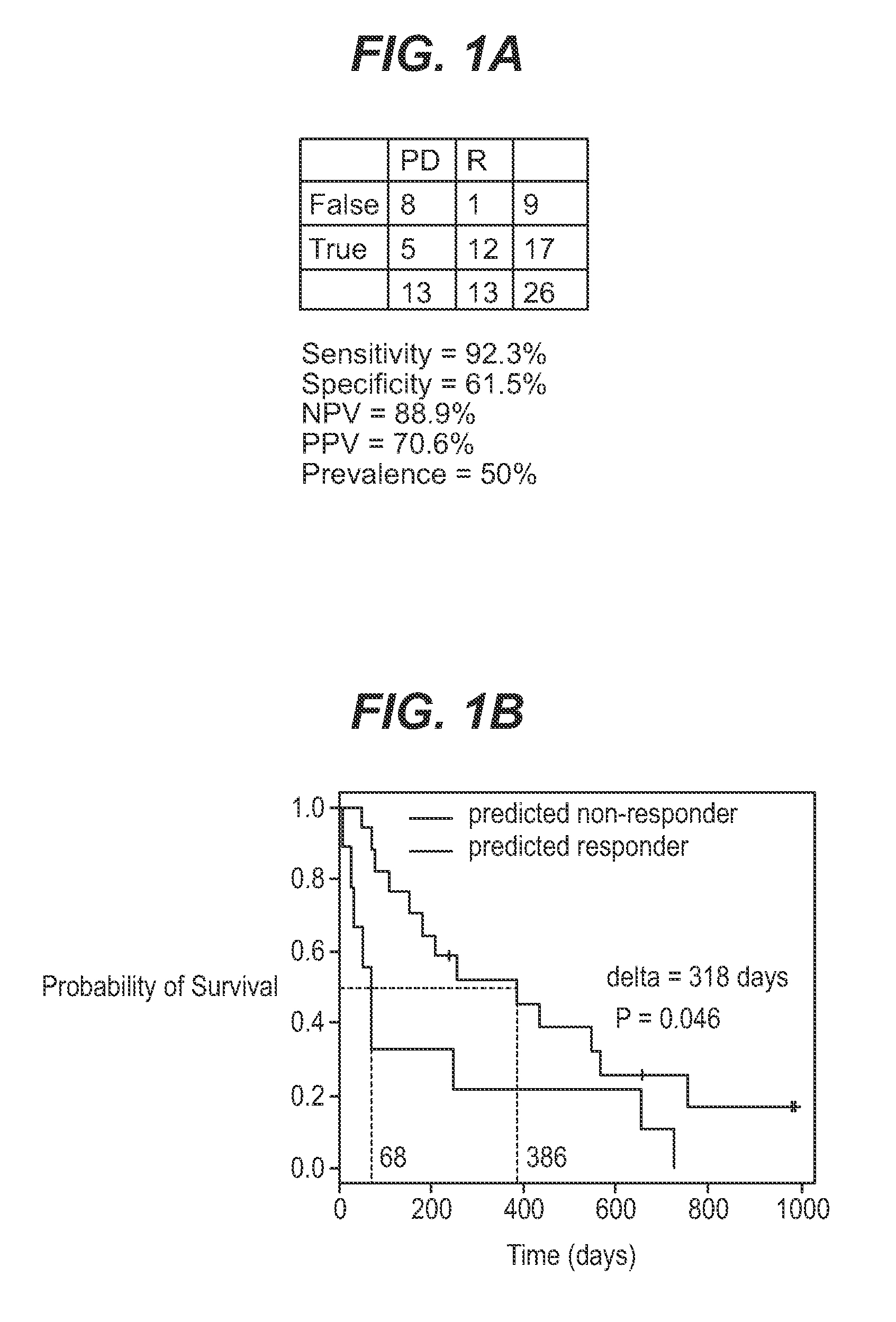
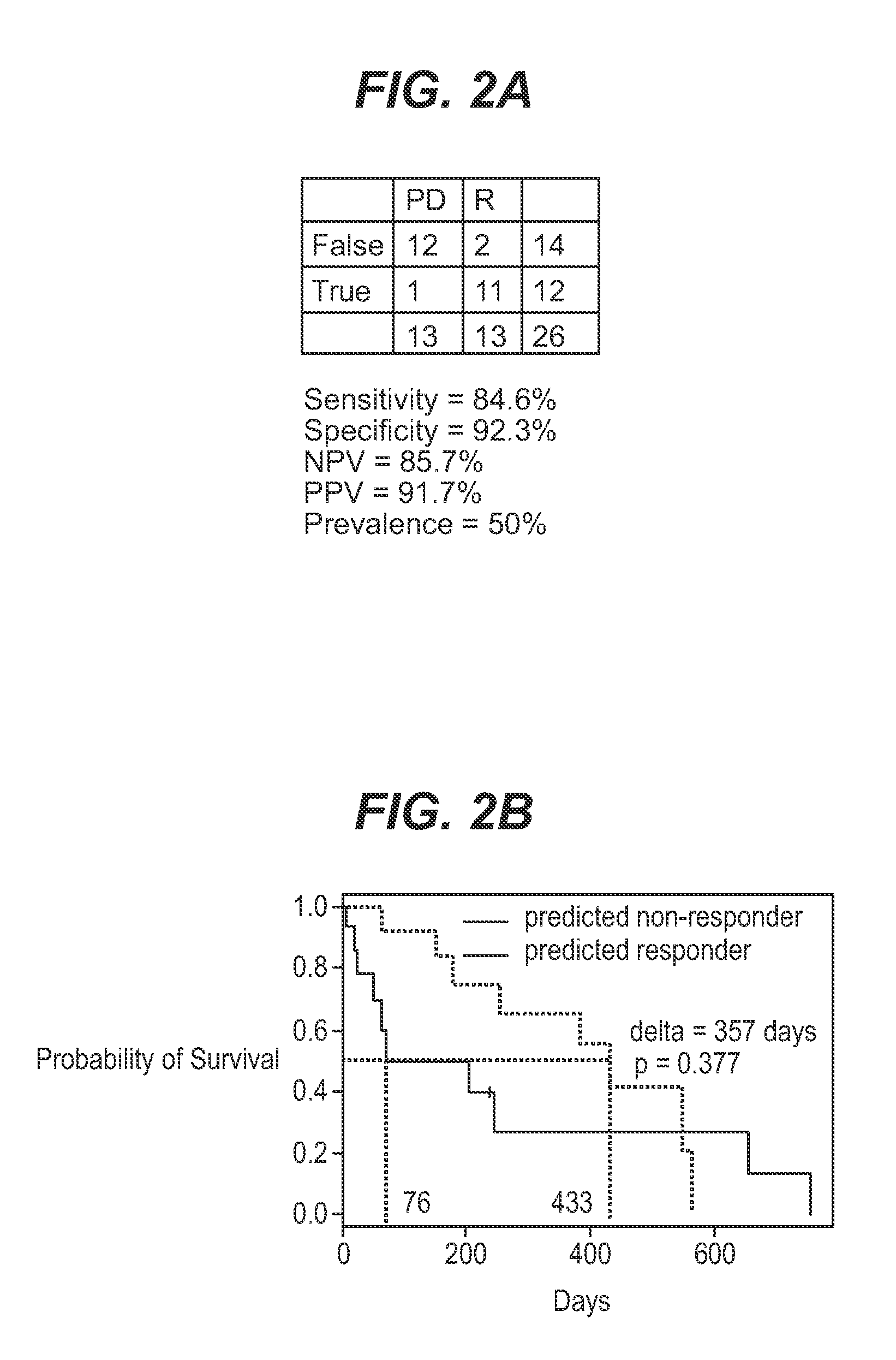
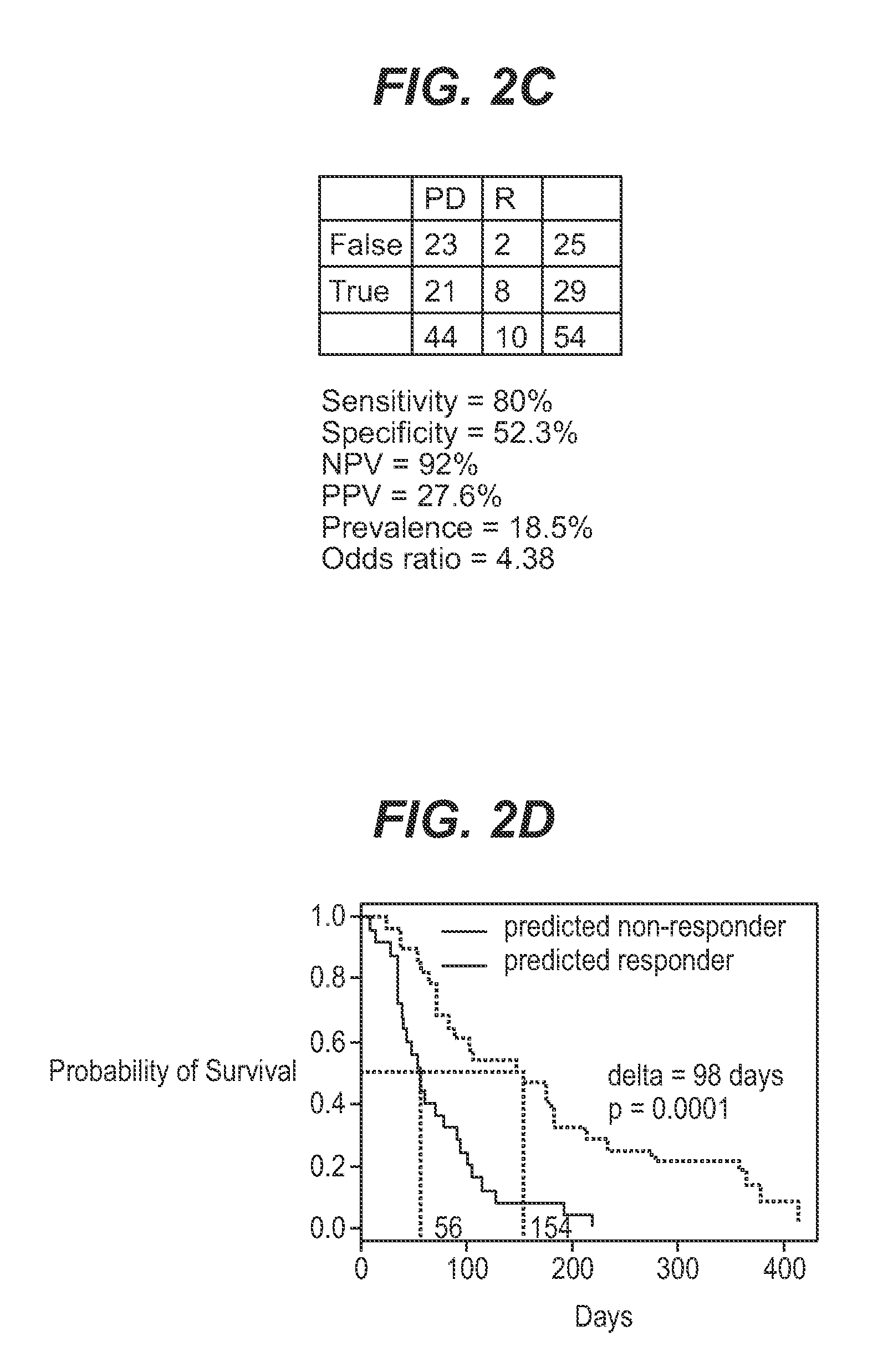
The application describes methods for predicting overall survival in subjects with breast cancer. The application also describes for screening subjects with breast cancer to determine if the breast cancer will be responsive to a breast cancer therapy including gemcitabine. The application further describes methods for treating subjects with breast cancer by screening them for the likelihood of the effectiveness of treating the cancer with a therapy including gemcitabine and administering the therapy in subjects when it is found that gemcitabine is likely to be effective.
Owner:NANOSTRING TECH INC +1
Methods and Kits for the Prediction of Therapeutic Success, Recurrence Free and Overall Survival in Cancer Therapies
InactiveUS20080305962A1Address bad outcomesLess aggressiveMicrobiological testing/measurementLibrary screeningTherapeutic antibodyAnticancer chemotherapy
The invention provides novel compositions, methods and uses, for the prediction, diagnosis, prognosis, prevention and treatment of malignant neoplasia and cancer. The invention further relates to genes that are differentially expressed in tissue of cancer patients versus those of normal “healthy” tissue. Differentially expressed genes for the identification of patients which are likely to respond to chemotherapy are also provided. The present invention relates to methods for prognosis the prediction of therapeutic success in cancer therapy. In a preferred embodiment of the invention it relates to methods for prediction of therapeutic success of combinations of signal transduction inhibitors, therapeutic antibodies, radio- and chemotherapy. The methods of the invention are based on determination of expression levels of 48 human genes which are differentially expressed prior to the onset of anti-cancer chemotherapy. The methods and compositions of the invention are most useful in the investigation of advanced colorectal cancer, but are useful in the investigation of other types of cancer and therapies as well.
Owner:SIEMENS HEALTHCARE DIAGNOSTICS INC
Prognostic biomarkers to predict overall survival and metastatic disease in patients with triple negative breast cancer
The present invention relates to a method for prognosing cancer in a subject with triple negative (TN) breast cancer, whose tumors lack expression of the estrogen receptor (ER), the progesterone receptor (PR) and normal (not amplified) levels of the human epidermal growth factor receptor 2 (HER2). Methods and biomarkers are disclosed that are useful for predicting the overall survival (OS) potential of cancer in a subject with triple negative breast cancer or for predicting metastatic disease in a subject with triple negative breast cancer. For example, the method comprises detecting in a sample from a subject one or more biomarkers selected from the group consisting of ANK3, CD24, EIF1, KLF6, KRAS, KRT1, MAP2K4, SDC4, SLC2A3, STK3, TFAP2C, and WRN. An increase or decrease in one or more biomarkers as compared to a standard is prognostic of OS of TN breast cancer. Likewise, in another example, the method comprises detecting in a sample from a subject one or more biomarkers selected from the group consisting of ANG, DICER1, EIF1, and MSH6. An increase or decrease in one or more biomarkers as compared to a standard is prognostic of metastasis of TN breast cancer.
Owner:VM INST OF RES
Methods of determining acute myeloid leukemia response to treatment with farnesyltransferase
InactiveUS7932036B1Improve accuracyHigh response rateSugar derivativesMicrobiological testing/measurementNewly diagnosedClinical study
We analyzed bone marrow from 67 patients from a phase 2 study of farnesyltransferase inhibition with tipifarnib (R115777, ZARNESTRA®), in older adults with previously untreated, poor-risk acute myeloid leukemia (AML) for N-Ras mutations, global gene expression, and / or quantitative PCR (qPCR) of specific genes. Microarray profiling identified a two-gene expression ratio (RASGRP1:APTX) which provided the greatest accuracy for predicting response to tipifarnib. We demonstrated that this classifier could predict response to tipifarnib in an independent set of 54 samples from relapsed or refractory AML, with a NPV and PPV of 92% and 28%, respectively (odds ratio of 4.4). Therefore, in both newly diagnosed and relapsed or refractory AML, this classifier improves the overall response rate by approximately 50% while maintaining a high NPV, and significantly improves patient overall survival. The two-gene classifier was also validated by qPCR in thirty AML samples from the same clinical study demonstrating a negative predictive value (NPV) and positive predictive value (PPV) of 81% and 50%, respectively (odds ratio of 4.3). These data indicate that a simple two-gene expression assay may have utility in diagnosing a population of AML patients who are more likely to respond to tipifarnib.
Owner:JANSSEN DIAGNOSTICS LLC
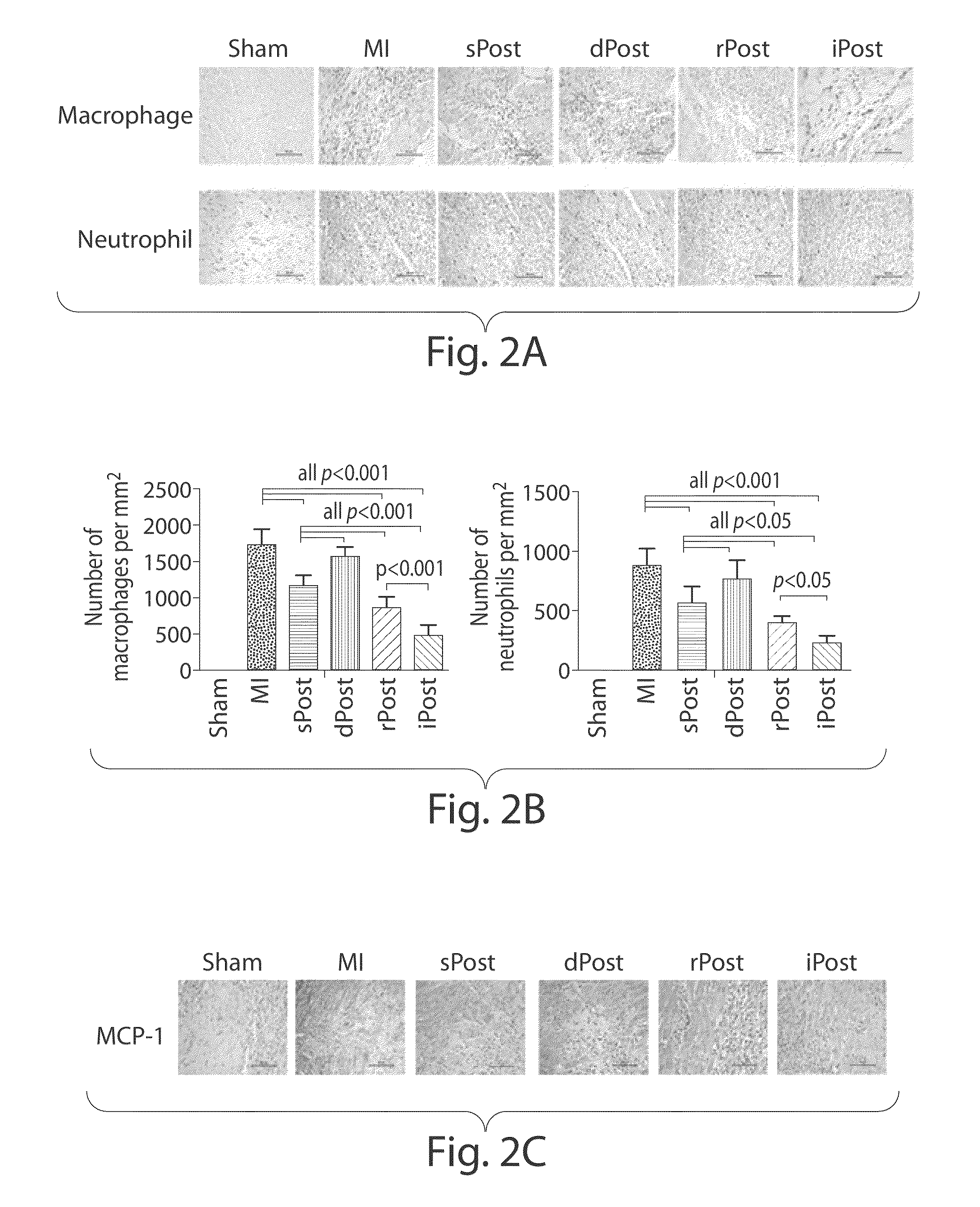
Use of remote ischemic conditioning to improve outcome after myocardial infarction
InactiveUS20110240043A1Promote resultsReduce riskOrganic active ingredientsDiagnosticsCardiac musclePost conditioning
The invention provides methods for reducing the incidence and / or severity and / or delaying the onset of heart dysfunction / failure and improving overall survival through the use of remote ischemic per-conditioning and post-conditioning.
Owner:HOSPITAL FOR SICK CHILDREN
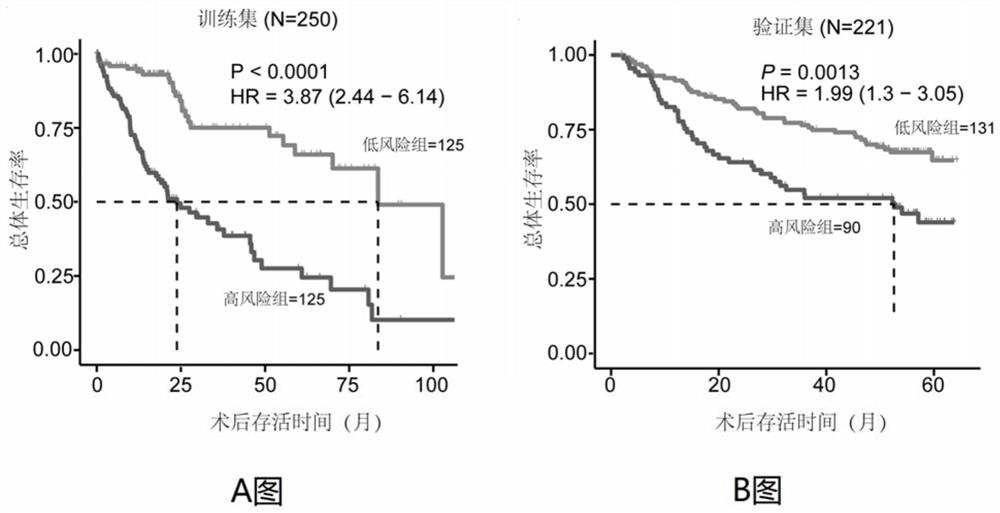
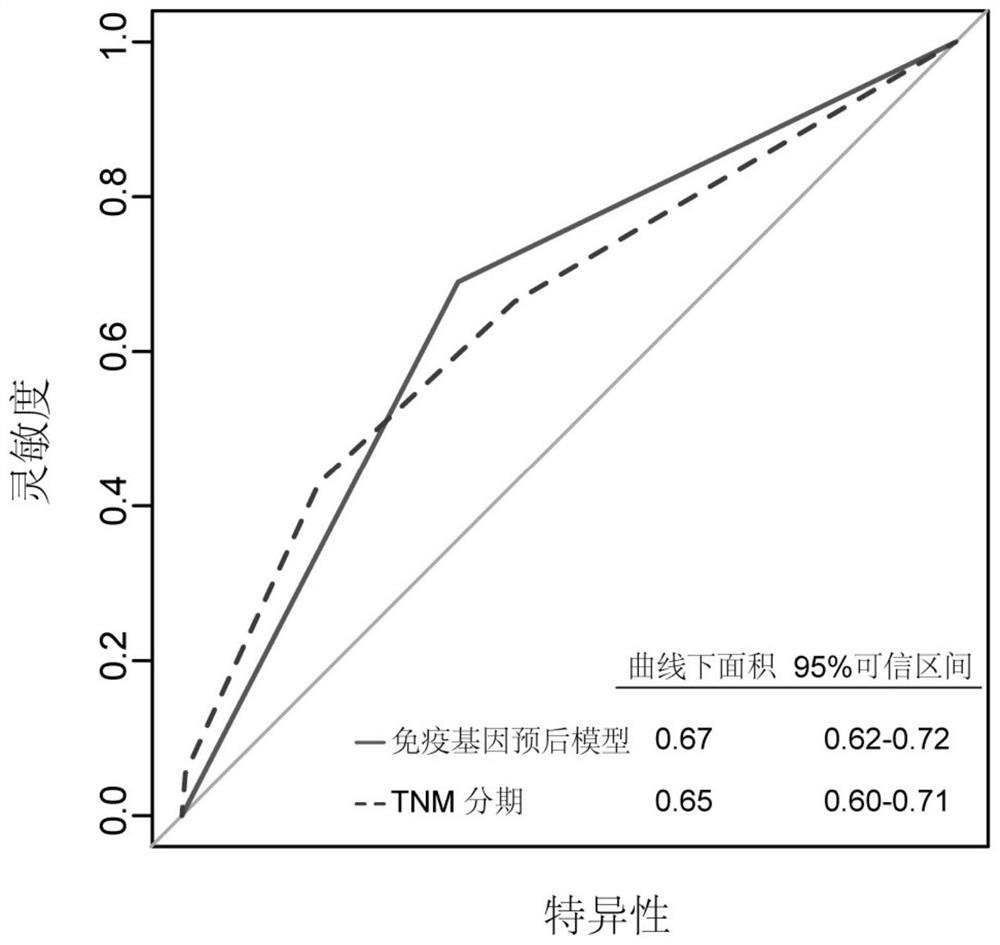
Immune gene prognosis model for predicting hepatocellular carcinoma tumor immune infiltration and postoperative survival time
ActiveCN112011616APromote the implementation of precision medicineObjective assessment of infiltrationMicrobiological testing/measurementBiostatisticsTNM staging systemMicroarray cgh
The invention relates to an immune gene prognosis model for predicting hepatocellular carcinoma tumor immune infiltration and postoperative survival time, and belongs to the technical field of biological medicines. The model can be used for evaluating the infiltration degree of immune cells in a tumor in clinical practice by detecting the expression levels of 22 specific immune related genes of ahepatocellular carcinoma patient, so that the model can be used for predicting hepatocellular carcinoma tumor immune infiltration in clinical practice and improve the prediction capability of the liver cancer immunotherapy response. The model can be used for judging the postoperative overall survival risk of a patient and guiding the formulation of a postoperative treatment strategy, and the corresponding microarray chip kit can realize the standardization and convenience of detection. Meanwhile, the immune gene prognosis model provided by the invention can increase the prediction accuracy andthe clinical net income of a hepatocellular carcinoma TNM staging system on the total survival time of three years and five years after operation. As a molecular marker for objectively and accuratelyevaluating the tumor immune state and poor prognosis risk of hepatocellular carcinoma, the model can realize accurate implementation of hepatocellular carcinoma immunotherapy and accurate prognosis prediction.
Owner:上海顿慧医疗科技发展有限公司
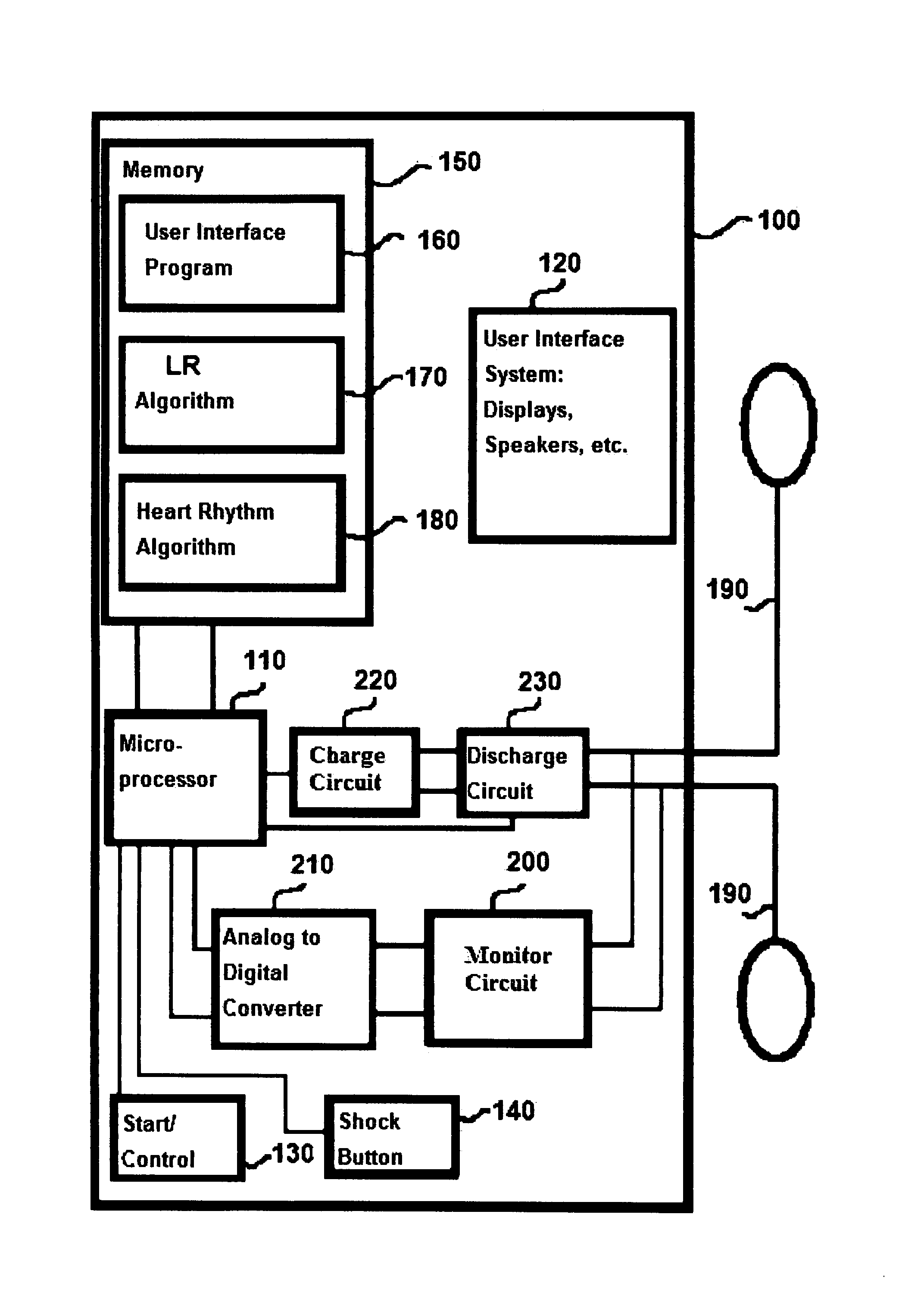
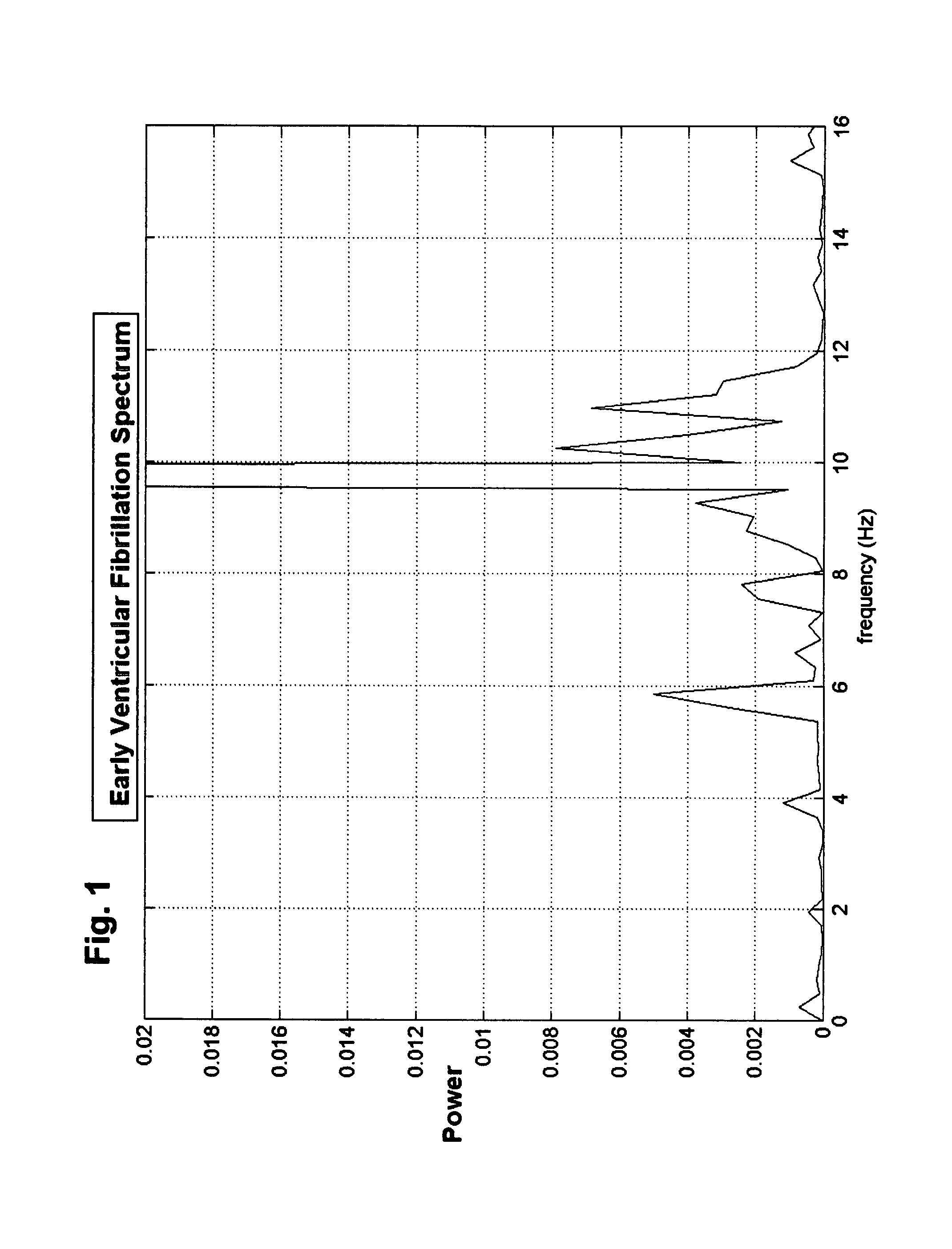
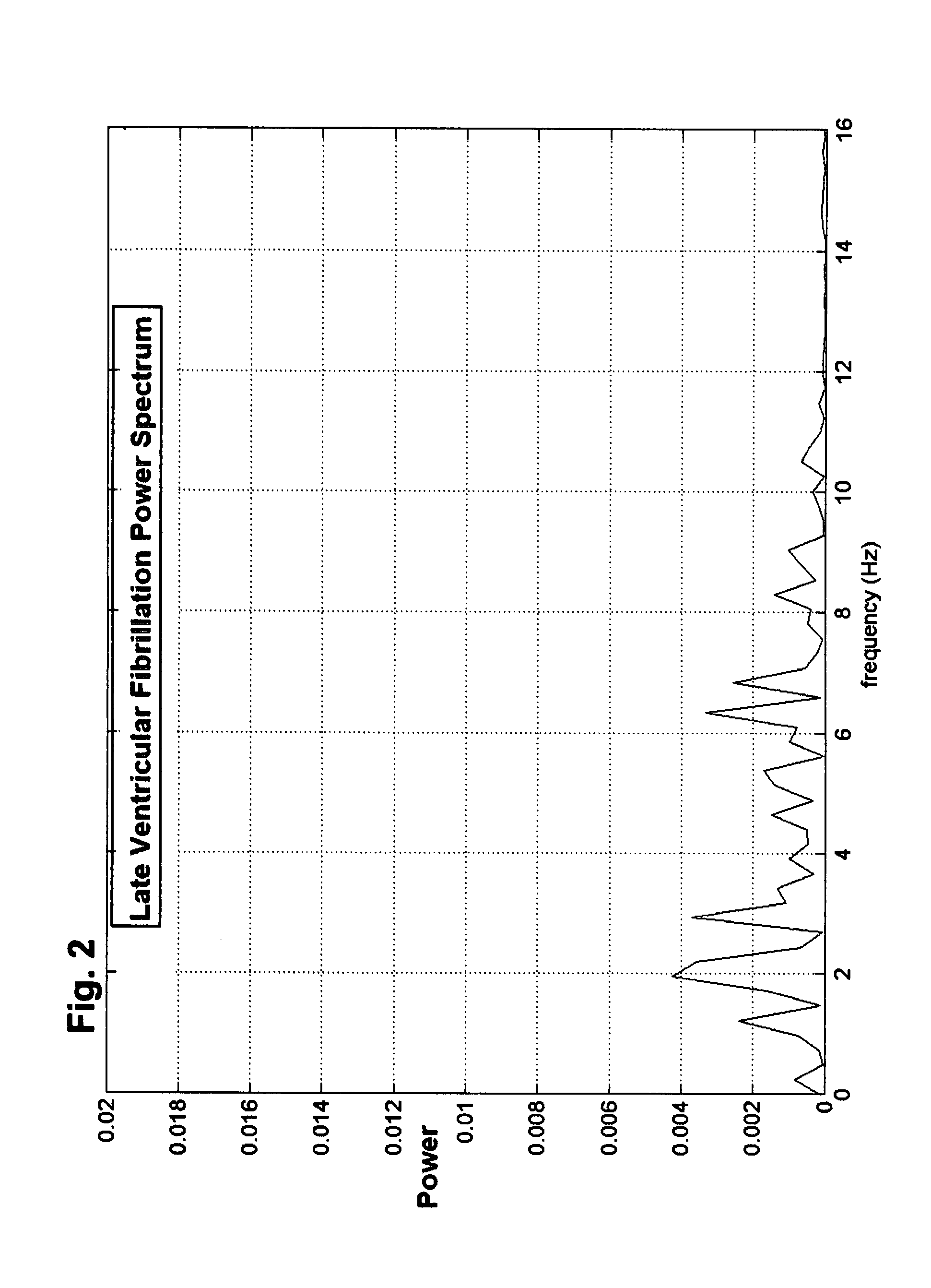
Methods and devices to guide therapy for ventricular fibrillation based on waveform analysis and survival benefit analysis
InactiveUS20050245974A1Easy to separateElectrocardiographyHeart defibrillatorsWaveform analysisLow frequency band
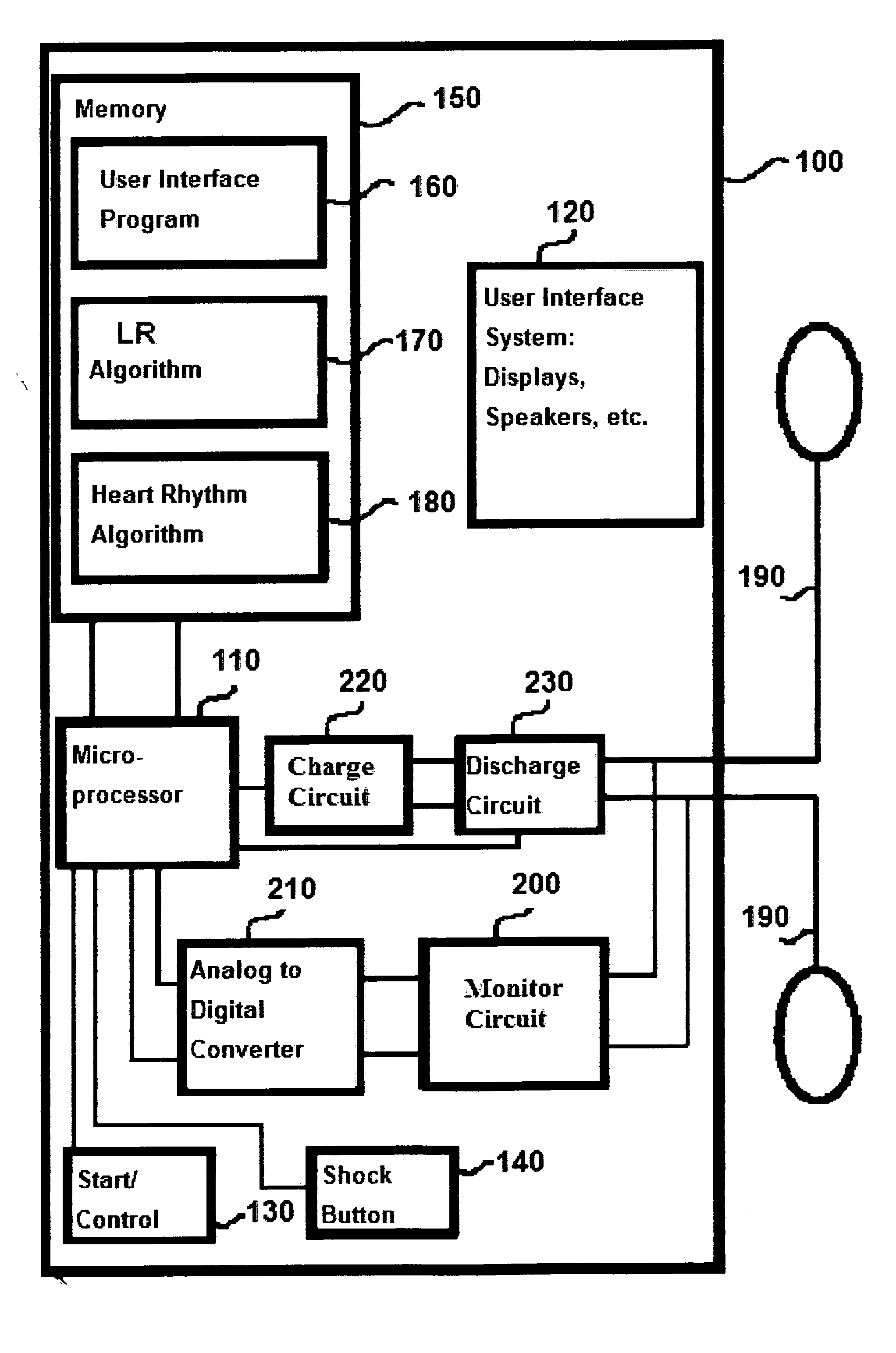
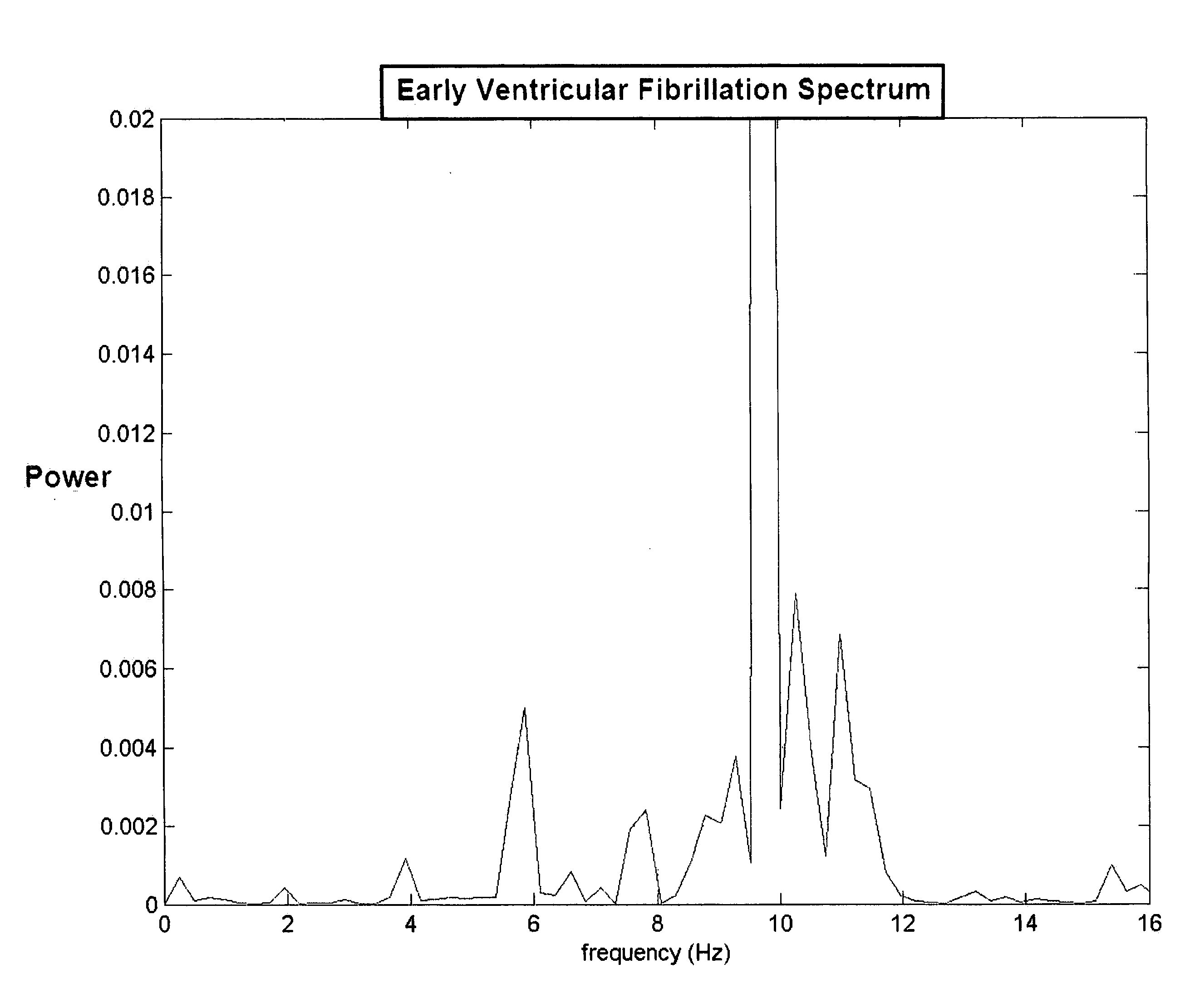
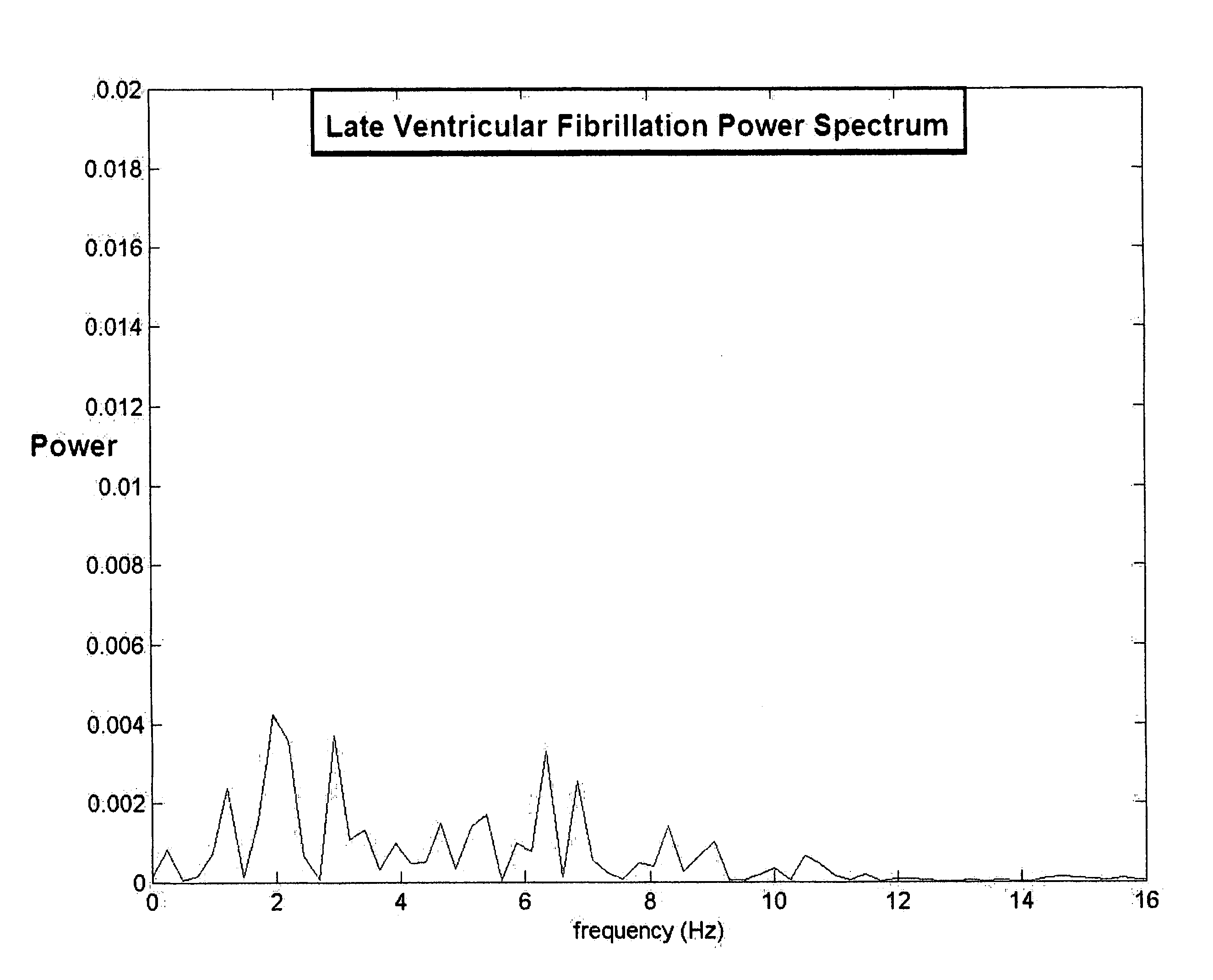
A method of determining a state of ventricular fibrillation from a waveform, includes: performing calculations on the Fourier transform of the waveform voltage values by using the quotient of the power in a high frequency band and of the power in a low frequency band; and determining the state of ventricular fibrillation by relating this ratio to the state of ventricular fibrillation. A defibrillation system for use in treatment of ventricular fibrillation includes at least one sensor to measure heart rhythm and at least one applicator to apply a defibrillation shock to a patient. The system further includes at least one processor which is adapted to calculate the ratio and also to obtain user input regarding survival of subgroups and average response times in the user's domain which allows calculation of an estimated maximum overall survival and selection of an optimum threshold for the ratio measure based on this maximum. The system further includes a user interface system in operative connection with the processor to provide information related to the ratio to a user.
Owner:SHERMAN LAWRENCE DUANE
Apparatus, method and computer program product to facilitate ordinary visual perception via an early perceptual-motor extraction of relational information from a light stimuli array to trigger an overall visual-sensory motor integration in a subject
ActiveUS7309315B2Reinforce reciprocal relationshipFacilitates direct attainmentSurgeryVaccination/ovulation diagnosticsSensorimotor integrationOptical flow
An apparatus, method and computer program product is presented to address early visual-sensory motor perception of a subject, where the method comprising the steps of: (1) controlling photic energetic parameters and / or photic perceptual attributes to trigger pre-attentive cuing or increase reactivity in magnocellular activity towards transient visual stimuli of the subject; and (2) generating a optical field comprising optical events based on said photic energetic parameters and said photic perceptual attributes, wherein said optical field transforms into a simple optical flow in the perceptual visual field of said subject. The photic energetic parameters may comprise light array energetic features, including one or more of wavelength, amplitude, intensity, phase, polarization, coherence, hue, brightness, and saturation.
Owner:BRIGHTSTAR LEARNING LTD
Identification and Treatment of Aggressive Lung Cancer Tumors
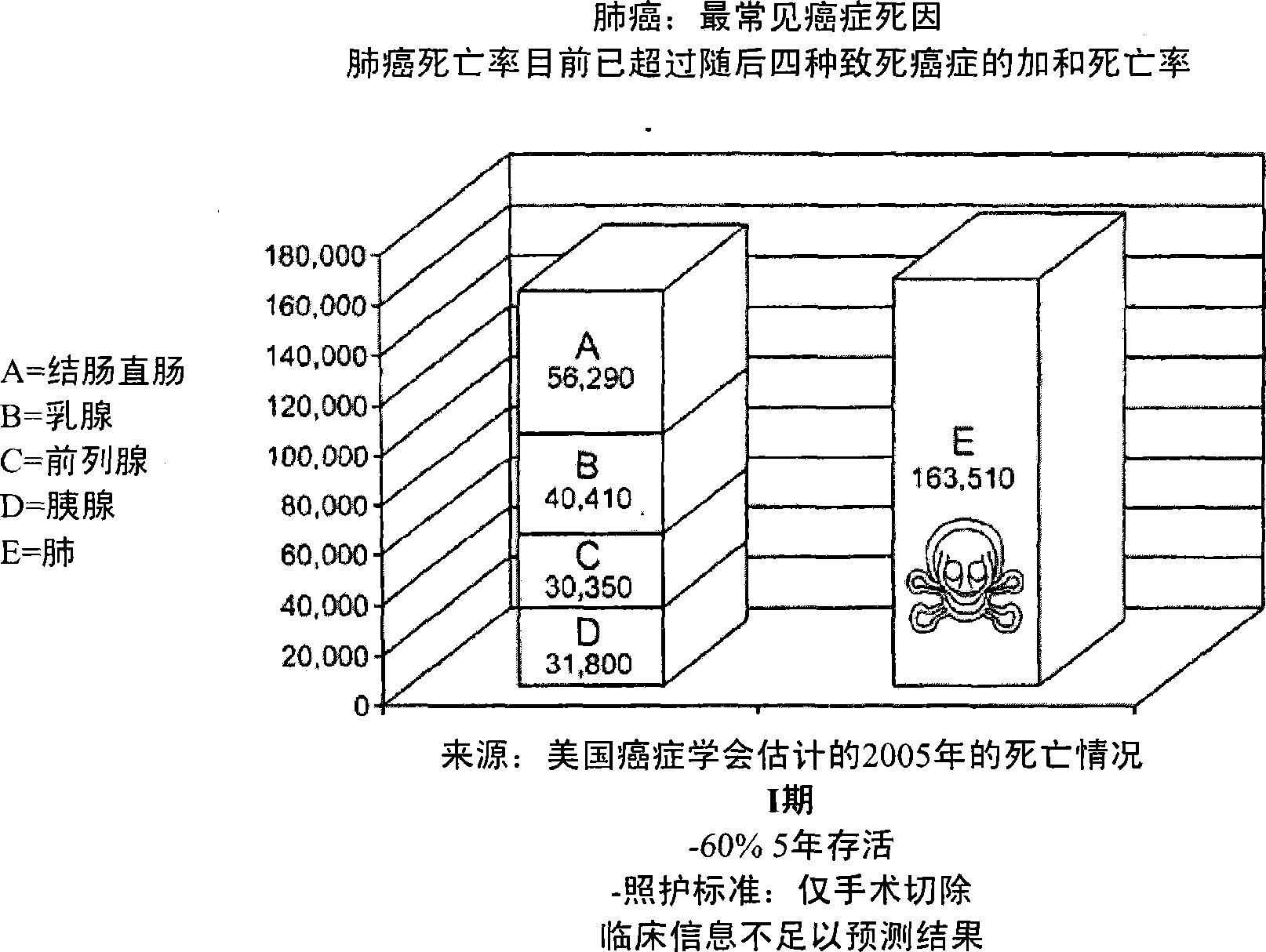
This invention relates to the identification and treatment of aggressive lung cancer tumors in patients. More particularly, it provides a method of identifying patients with non-small cell lung carcinoma (NSCLC) who have an aggressive node-negative (N0) tumor and a likelihood of a poor overall survival. The method comprises the step of determining if one or more of certain identified proteins are activated in tumor cells obtained from the patient's tumor, wherein the activation of one or more of the proteins indicates that the patient has an aggressive N0 tumor and is likely to have a poor overall-survival. The invention also provides a method for selecting a treatment for an NSCLC patient with an N0 tumor and a method for treating such patients. It further provides a kit for identifying an NSCLC patient with an aggressive N0 tumor and a likelihood of a poor overall survival and a pharmaceutical composition for treating such patients.
Owner:INST SUPERIORE DI SANITA +1
Methods and devices to guide therapy for ventricular fibrillation based on waveform analysis and survival benefit analysis
A method of determining a state of ventricular fibrillation from a waveform, includes: performing calculations on the Fourier transform of the waveform voltage values by using the quotient of the power in a high frequency band and of the power in a low frequency band; and determining the state of ventricular fibrillation by relating this ratio to the state of ventricular fibrillation. A defibrillation system for use in treatment of ventricular fibrillation includes at least one sensor to measure heart rhythm and at least one applicator to apply a defibrillation shock to a patient. The system further includes at least one processor which is adapted to calculate the ratio and also to obtain user input regarding survival of subgroups and average response times in the user's domain which allows calculation of an estimated maximum overall survival and selection of an optimum threshold for the ratio measure based on this maximum. The system further includes a user interface system in operative connection with the processor to provide information related to the ratio to a user.
Owner:SHERMAN LAWRENCE DUANE
Wireless sensor network clustering method based on K-means
ActiveCN107277889AAvoiding Cluster Imbalance ProblemsProlong survival timeNetwork topologiesHigh level techniquesMobile wireless sensor networkWireless mesh network
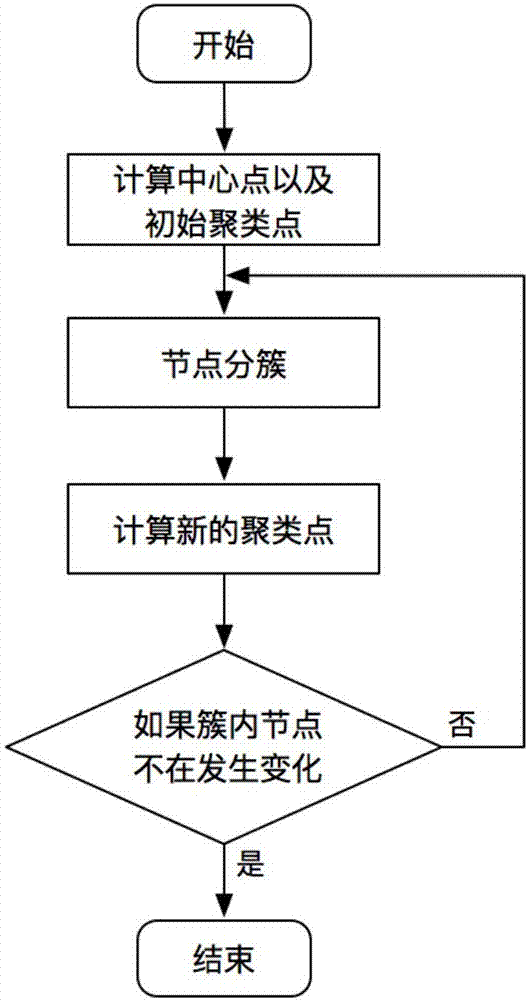
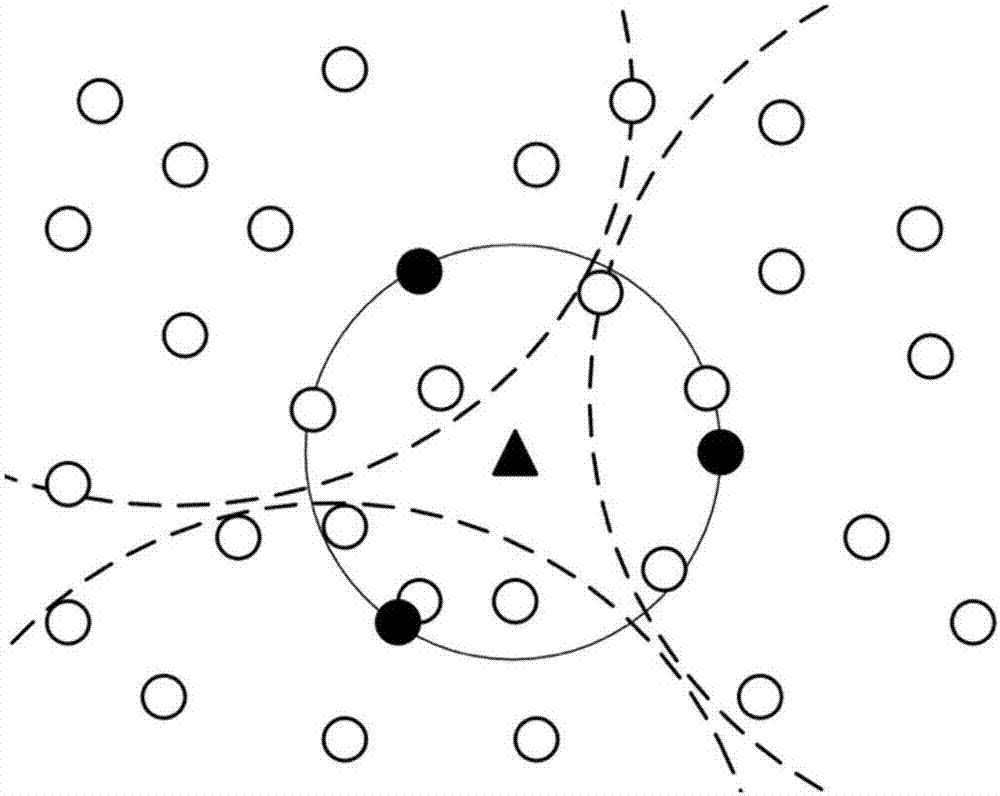
The invention proposes a wireless sensor network clustering method based on K-means. The method comprises the steps of (1) calculating center point position coordinates of each sensor in a network, and calculating coordinates of clustering points according to the center point position coordinates, (2) calculating a distance from each sensor to each clustering point in the network, and adding each sensor to a same cluster through selecting a clustering point with a nearest distance, wherein in the sensors added into the same cluster, a sensor with the smallest distance to the corresponding clustering point and energy higher than average energy of the sensors in the cluster is selected as the cluster head of the cluster, and (3) calculating the center point position coordinates of the sensors in the cluster for each cluster, taking the center point position coordinates as new cluster point coordinates, repeating the step (2) and the step (3) until the sensors in the cluster are not changed. According to the method, the power consumption of each sensor is reduced, and the overall survival time of a wireless sensor network is prolonged.
Owner:YANGZHOU UNIV
Methods of prognosis
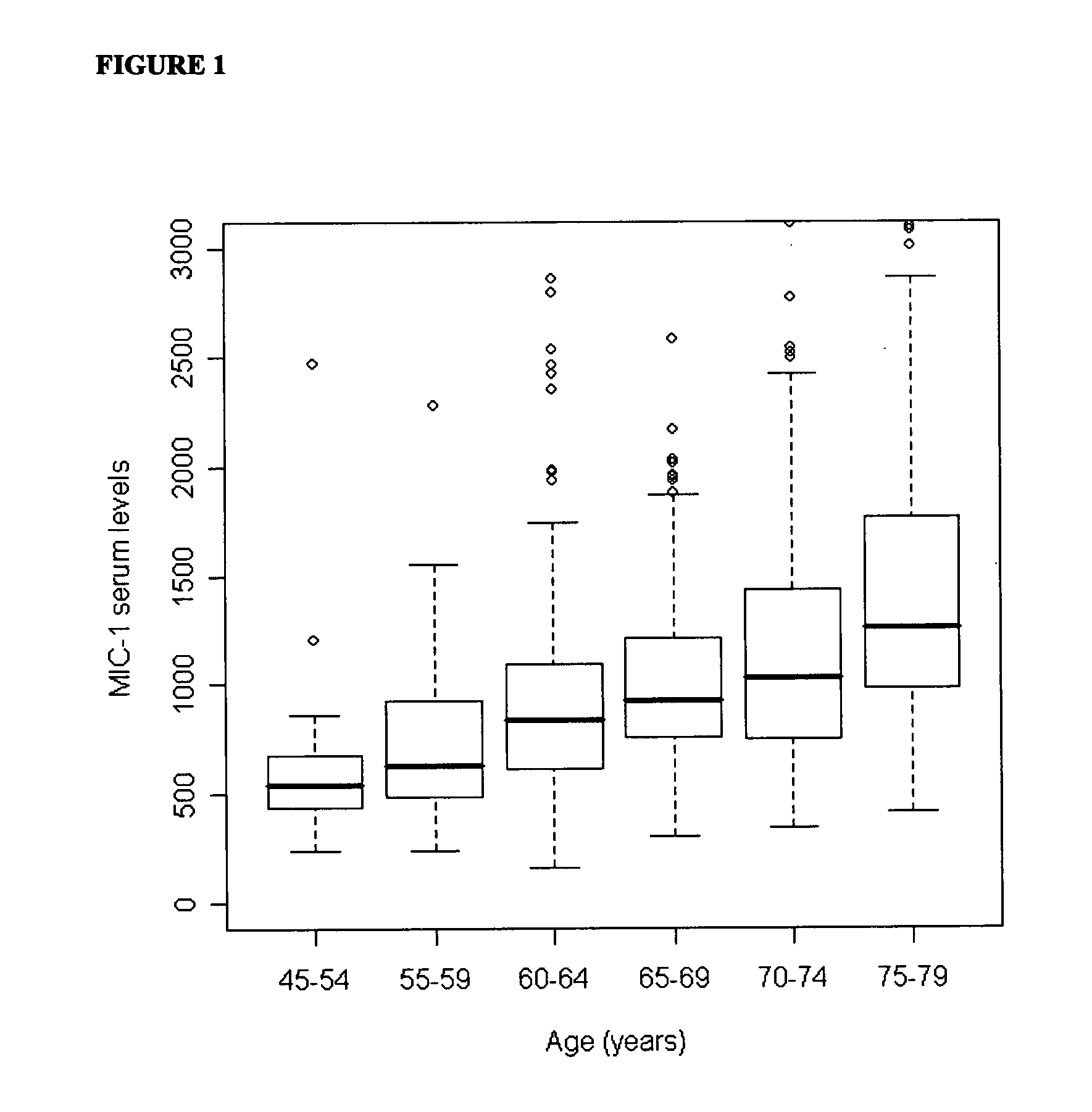
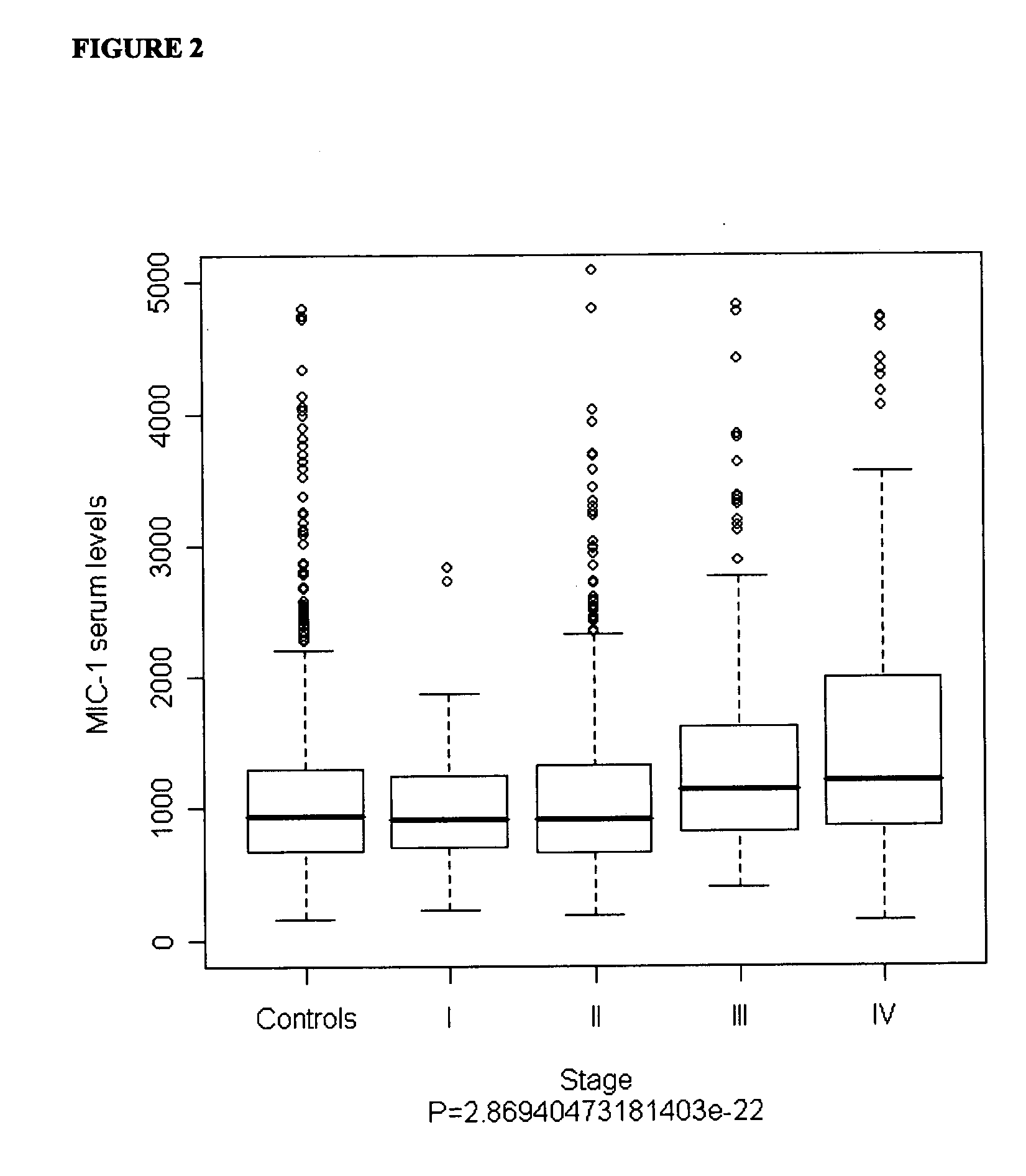
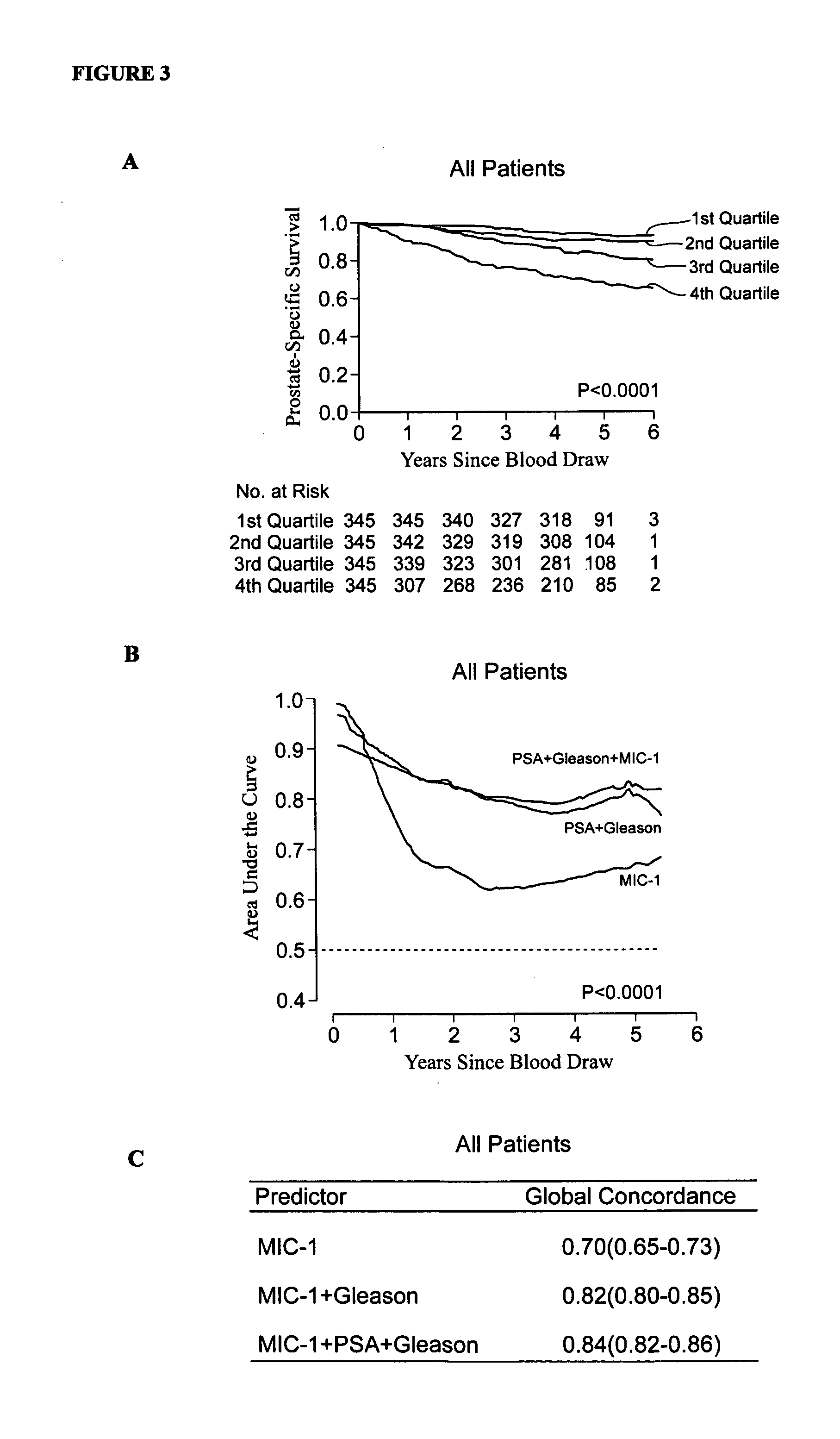
InactiveUS20110039284A1Raise the possibilityDisease diagnosisBiological testingMacrophage Inhibitory Cytokine 1Overall survival
The invention relates to the field of medical prognostics. In particular, the invention relates to methods for predicting prostate cancer progression and overall survival prognosis in a subject involving the detection of elevated amounts of macrophage inhibitory cytokine-1 (MIC-1) in a test body sample such as serum.
Owner:ST VINCENTS HOSPITAL SYDNEY
Method of prognosis and stratification of ovarian cancer
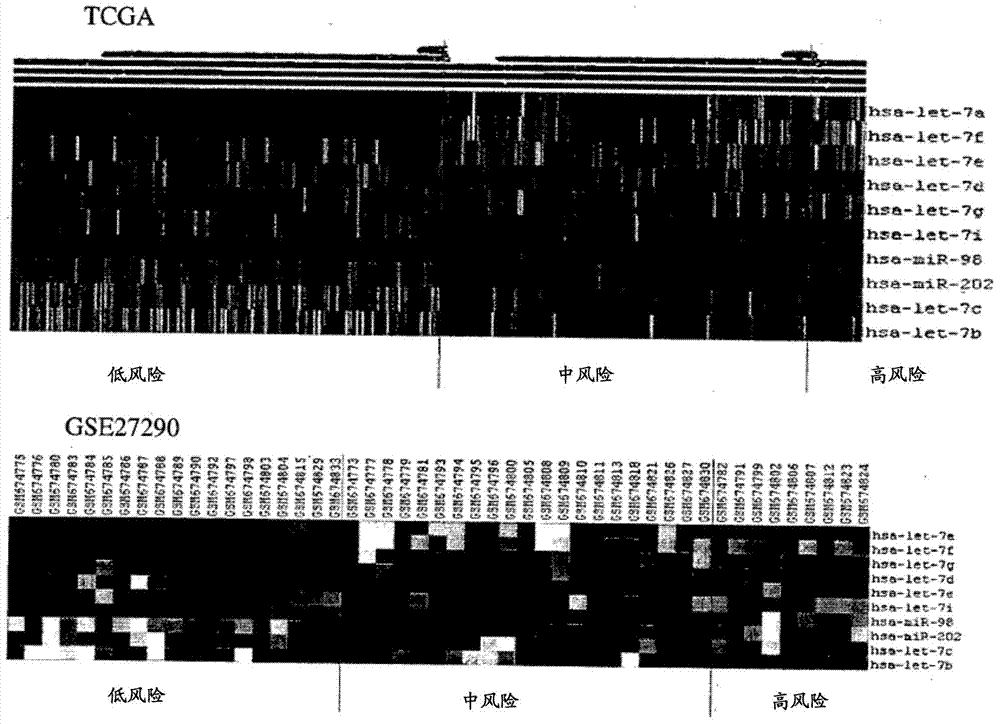
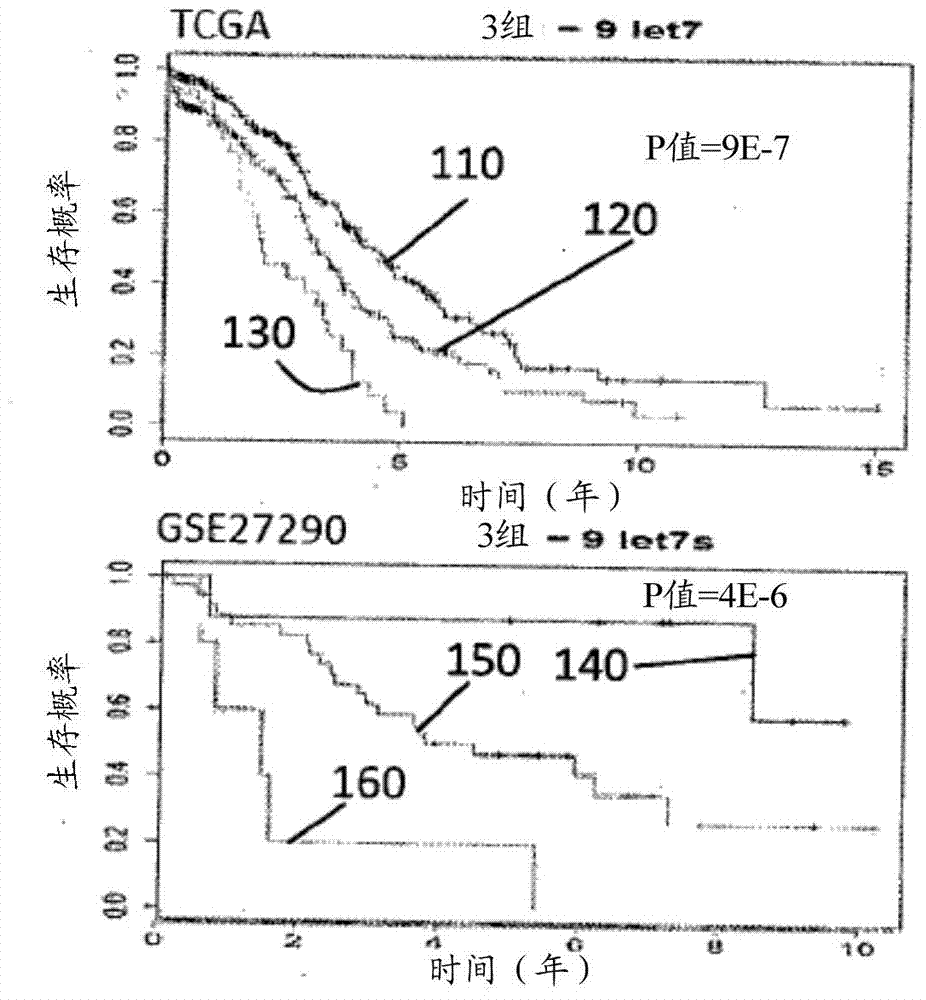
InactiveCN104854247AEasy to studyGenetic material ingredientsMicrobiological testing/measurementSurvival prognosisMicroRNA
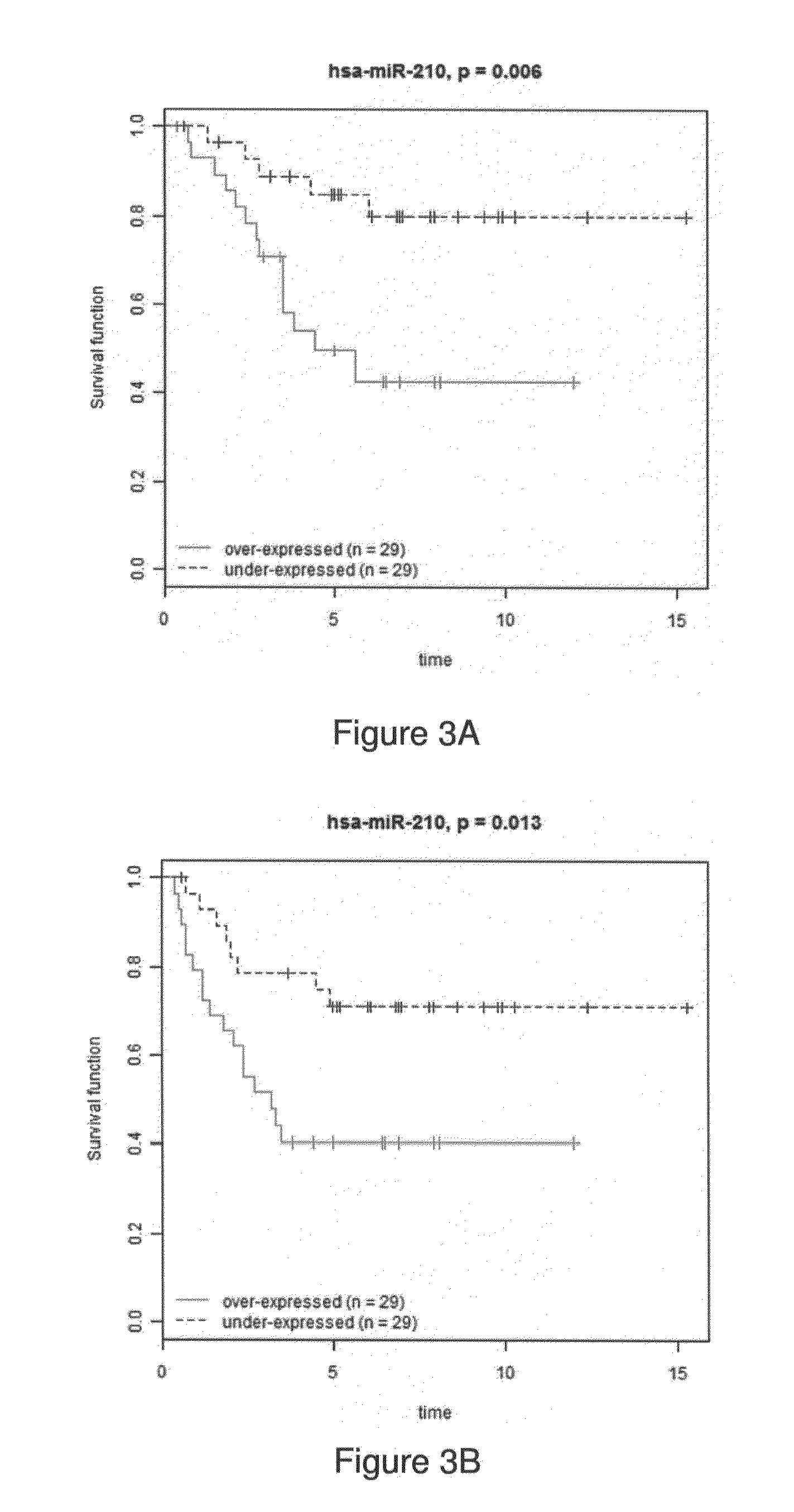
A method for the prognosis of overall survival or prediction of therapeutic outcome for a patient suffering from epithelial ovarian cancer (EOC), comprising: a. providing a sample from the patient, b. determining the expression level of microRNA family lethal-7b (let-7b) in the sample; c. using the expression level of the let-7b to obtain the prognosis of overall survival or prediction of therapeutic outcome for the patient.
Owner:AGENCY FOR SCI TECH & RES
Wireless sensor network target tracking method for energy conservation
ActiveCN103476147AExtend the life cycleGuaranteed accuracyNetwork topologiesHigh level techniquesNetworked systemWireless sensor networking
The invention discloses a specific scheme of wireless sensor network target tracking for energy conservation, which is used for accurately tracking the state of a target under the conditions of energy consumption constraint, topology switching and the like. The working mode of each node in a network is dynamically planned according to the energy consumption of each node and changes of the external condition, so that a net topology is dynamically switched to reduce the network energy consumption. Each node in a tracking network tracks the state information of a target in real time by acquiring target data and performing information exchange with neighbor nodes. By adopting the specific scheme of wireless sensor network target tracking, the influence of dynamic changes of a network topological structure on the overall performance of the tracking network can be quantitatively analyzed, the highest switching frequency for guaranteeing normal work of the tracking network is determined, and on the premise of ensuring the network overall survival time, the robustness and tracking accuracy of the tracking network are improved, and a specific deployment scheme of a network system is provided.
Owner:上海圣剑科技发展有限公司
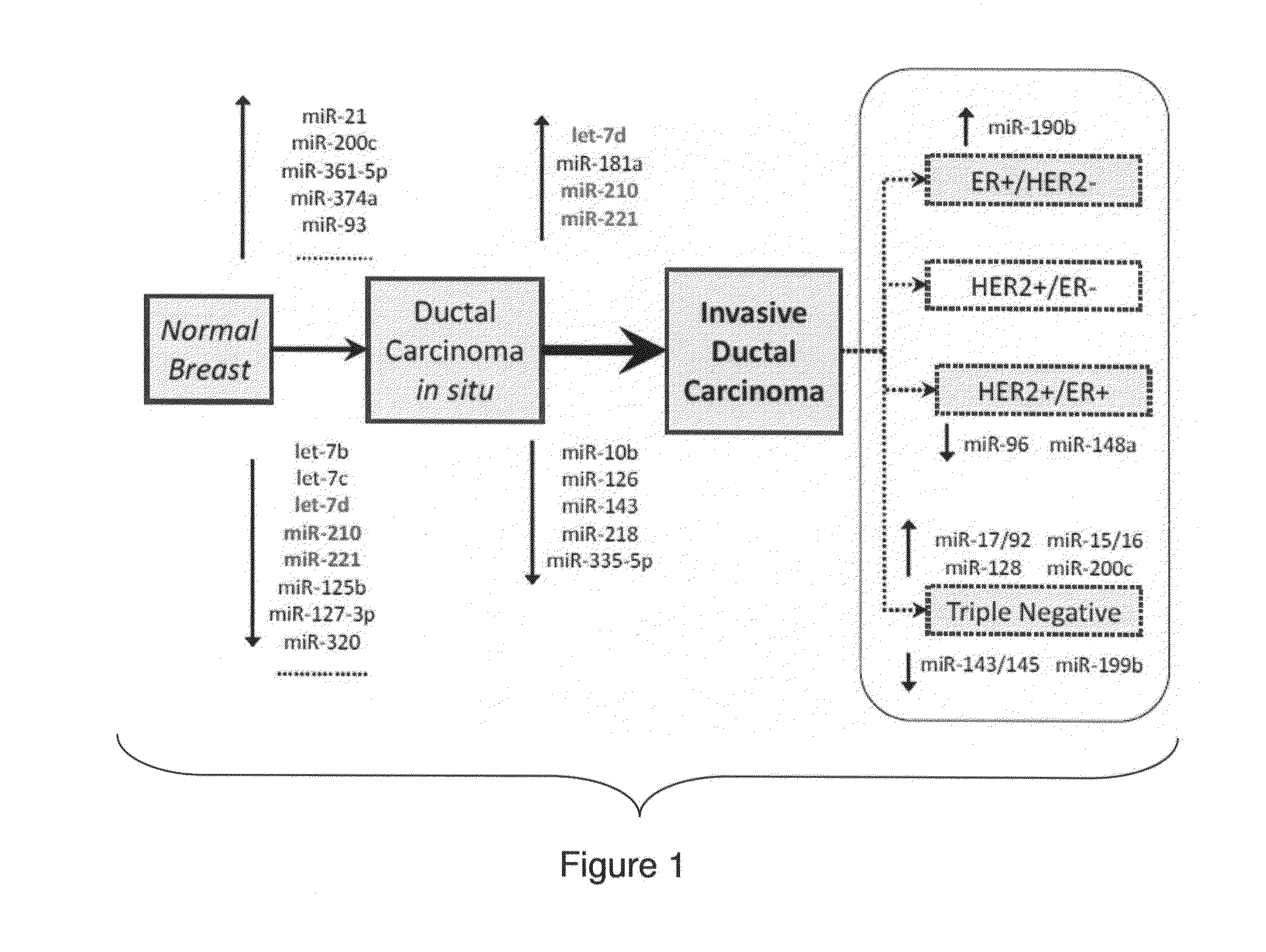
Breast Cancer Biomarker Signatures for Invasiveness and Prognosis
InactiveUS20130190386A1High expressionHigh riskDigital data processing detailsLibrary screeningLymphatic SpreadMicrorna profiling
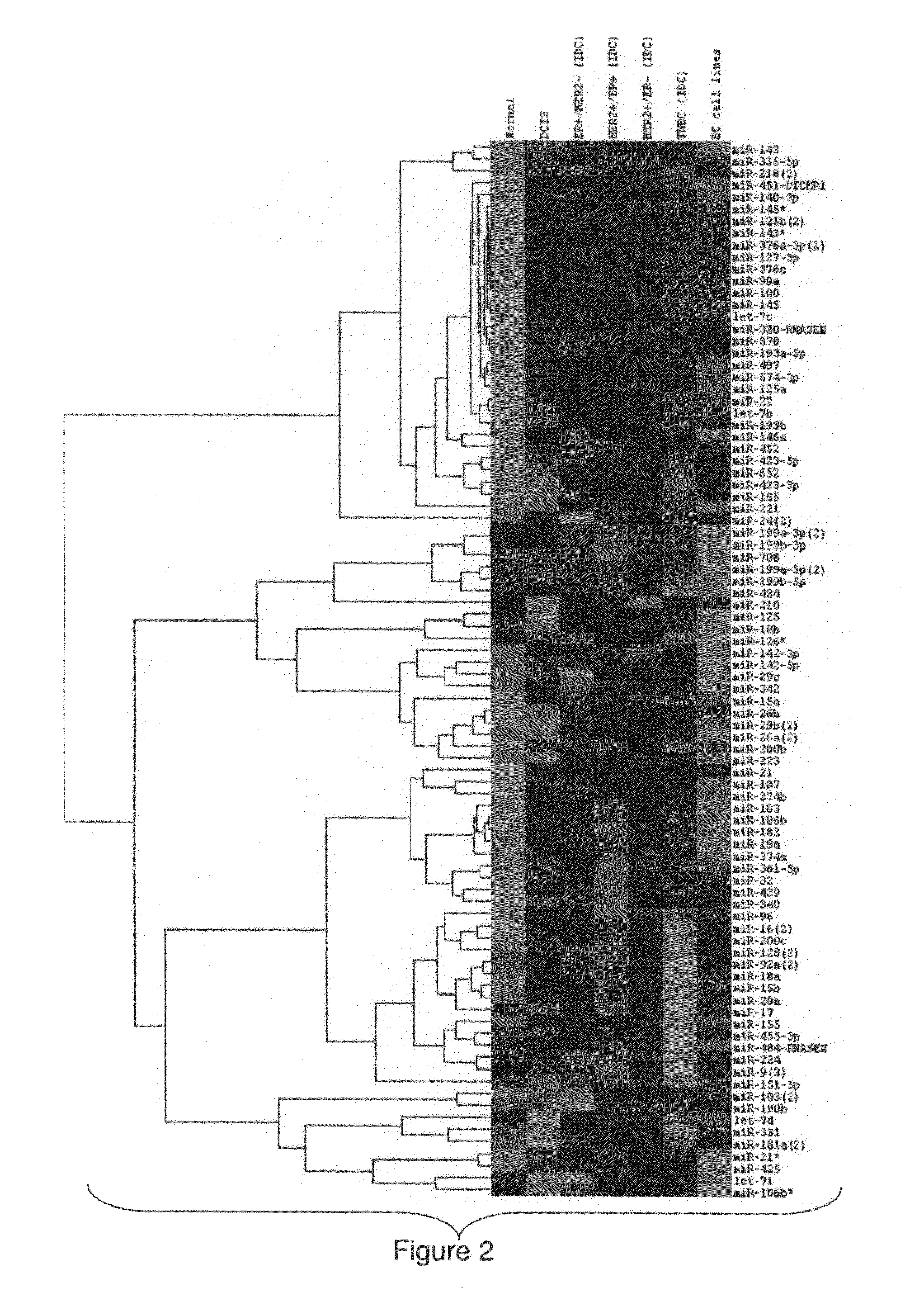
MicroRNA profiles transition from normal breast to ductal carcinoma in situ and transition to invasive ductal carcinoma (IDC) and methods of use thereof are described. Methods of diagnosis and prognosis using microRNA signatures to differentiate invasive from in situ carcinoma are described. Also described is the use of microRNA expression for predicting overall survival and time to metastasis.
Owner:THE OHIO STATES UNIV
Multigene prognostic assay for lung cancer
The present invention provides methods for providing a prognosis for lung cancer using a panel of eleven molecular markers that includes BAG1, BRCA1, CDC6, CDK2AP1, ERBB3, FUT3, IL11, LCK, RND3, SH3BGR, and WNT3A, which are differentially expressed in lung cancer. The eleven markers are related to patient prognosis to 5 -year overall survival outcomes, and are particularly useful in providing a prognosis for non-squamous NSCLC.
Owner:RGT UNIV OF CALIFORNIA +1
Markers and diagnostic reagents for the diagnosis or prognosis of lung cancer
The present invention relates to markers and diagnostic reagents for the diagnosis or prognosis of lung cancer. The present invention discloses for the first time that DDX56 is significantly increasedin patients with early disease progression (= 12 months) after treatment of two major subtypes of small cell lung cancer (lung squamous cell carcinoma, lung adenocarcinoma), and that the increased expression of DDX56 significantly affects the overall survival of the patients. Therefore, DDX56 can be used as a diagnostic marker for early recurrence of lung squamous cell carcinoma or lung adenocarcinoma.
Owner:CENT FOR EXCELLENCE IN MOLECULAR CELL SCI CHINESE ACAD OF SCI +1
Biomarker combination for molecular typing and/or prognosis prediction of muscle-invasive bladder cancer and application of biomarker combination
PendingCN109797221AMeet analysis needsEffectively assess the risk of adverse prognosisMicrobiological testing/measurementHybridisationGAS6Screening method
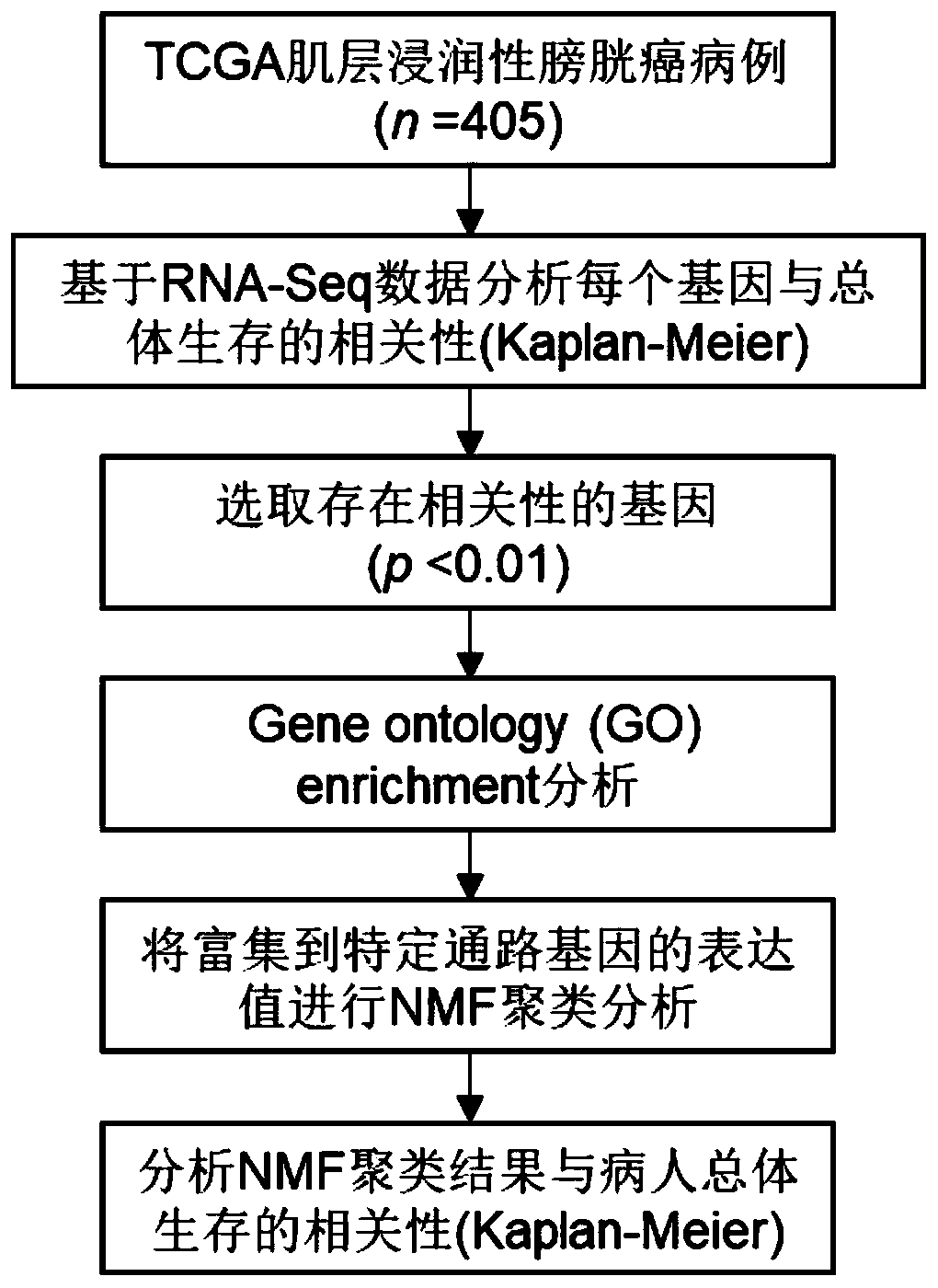
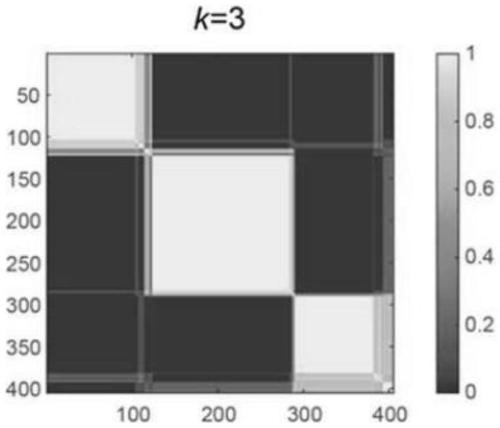
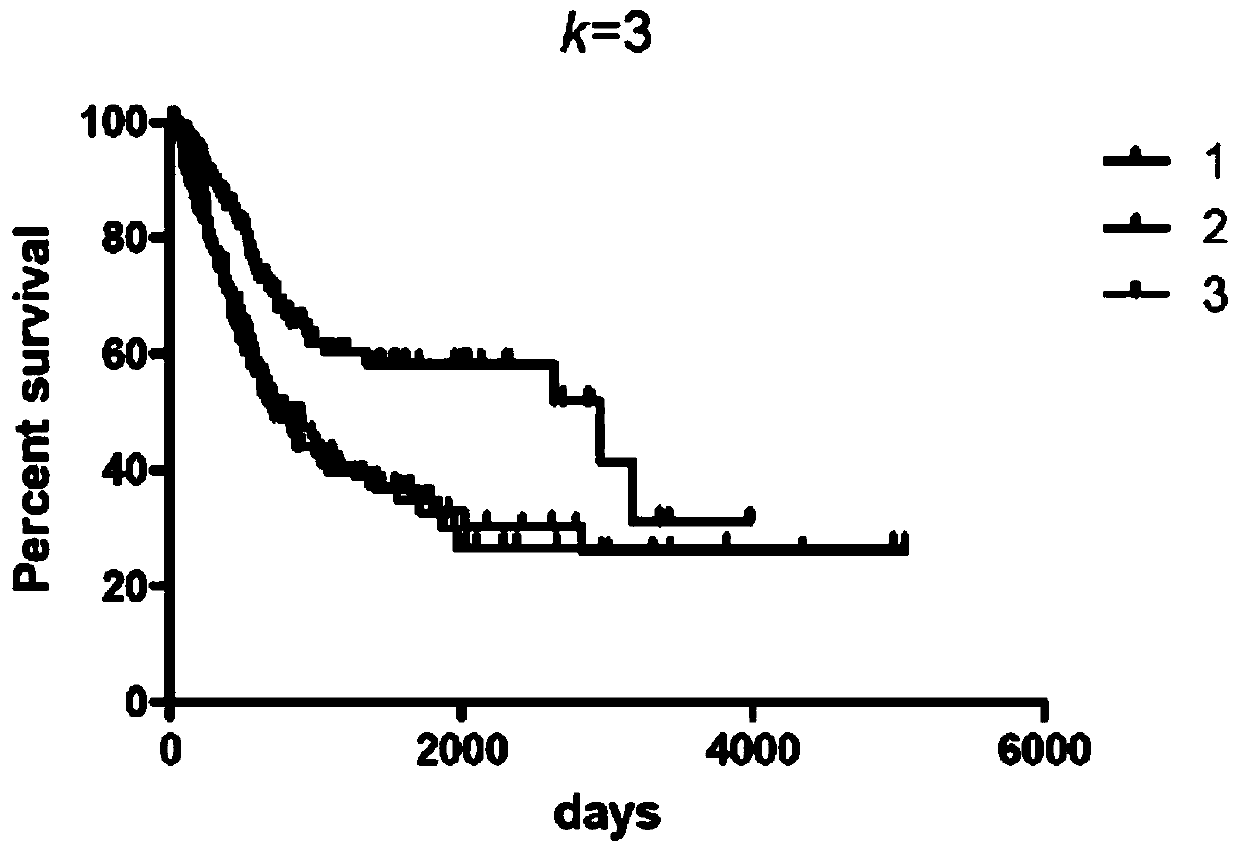
The invention relates to a biomarker combination for molecular typing and / or prognosis prediction of the muscle-invasive bladder cancer, and a screening method and application thereof. The biomarker combination comprises the following genes: FGF10, TP53INP1, DDR2, MYC, CDC73, IGF1, PLA2G1B, SKI, FN1, EGFR, PPARG, PDGFRA, PDGFD, GAS6, PDGFC, FNTB and CCNB1. Through non-negative matrix factorizationclustering analysis based on transcription data of the biomarker combination, the muscle-invasive bladder cancer can be classified to respectively correspond to different expression characteristic spectrums. The classification corresponds to significantly different overall survival statuses, thus being used for survival prognosis assessment. The transcription data analysis method adopted has theadvantages that the number of biomarker combinations is small, the analysis steps are simple, the requirements of large sample analysis are met, the requirements for the calculation ability are low, and the method is applicable to standardized transcription data; the transcription data can be a transcriptomic data sub set or a set of genetic transcription data for individual detection.
Owner:SHANGHAI TENTH PEOPLES HOSPITAL
Prognostic biomarkers to predict overall survival and metastatic disease in patients with triple negative breast cancer
The present invention relates to a method for prognosing cancer in a subject with triple negative (TN) breast cancer, whose tumors lack expression of the estrogen receptor (ER), the progesterone receptor (PR) and normal (not amplified) levels of the human epidermal growth factor receptor 2 (HER2). Methods and biomarkers are disclosed that are useful for predicting the overall survival (OS) potential of cancer in a subject with triple negative breast cancer or for predicting metastatic disease in a subject with triple negative breast cancer. For example, the method comprises detecting in a sample from a subject one or more biomarkers selected from the group consisting of ANK3, CD24, EIF1, KLF6, KRAS, KRT1, MAP2K4, SDC4, SLC2A3, STK3, TFAP2C, and WRN. An increase or decrease in one or more biomarkers as compared to a standard is prognostic of OS of TN breast cancer. Likewise, in another example, the method comprises detecting in a sample from a subject one or more biomarkers selected from the group consisting of ANG, DICER1, EIF1, and MSH6. An increase or decrease in one or more biomarkers as compared to a standard is prognostic of metastasis of TN breast cancer.
Owner:VM INST OF RES
Gene combination and application thereof in preparation of reagent for predicting prognosis of patient treated by immune checkpoint inhibitor
ActiveCN110229894AImproved prognosisReduce financial burdenMicrobiological testing/measurementDNA/RNA fragmentationWilms' tumorBiology
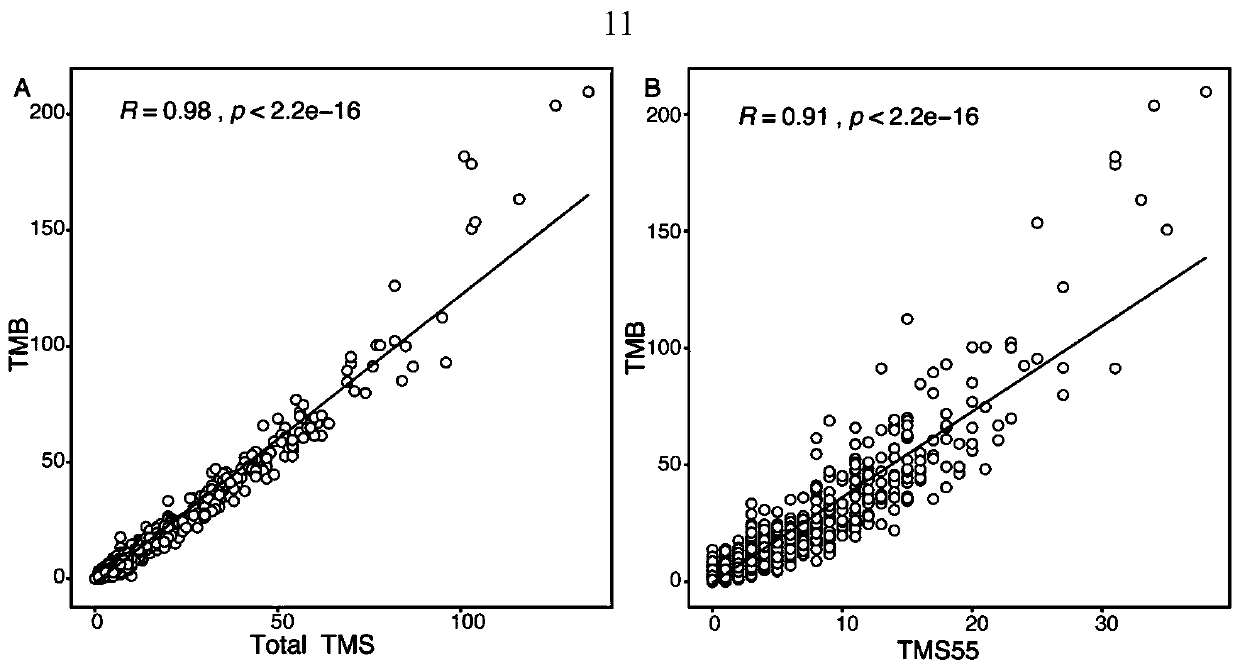
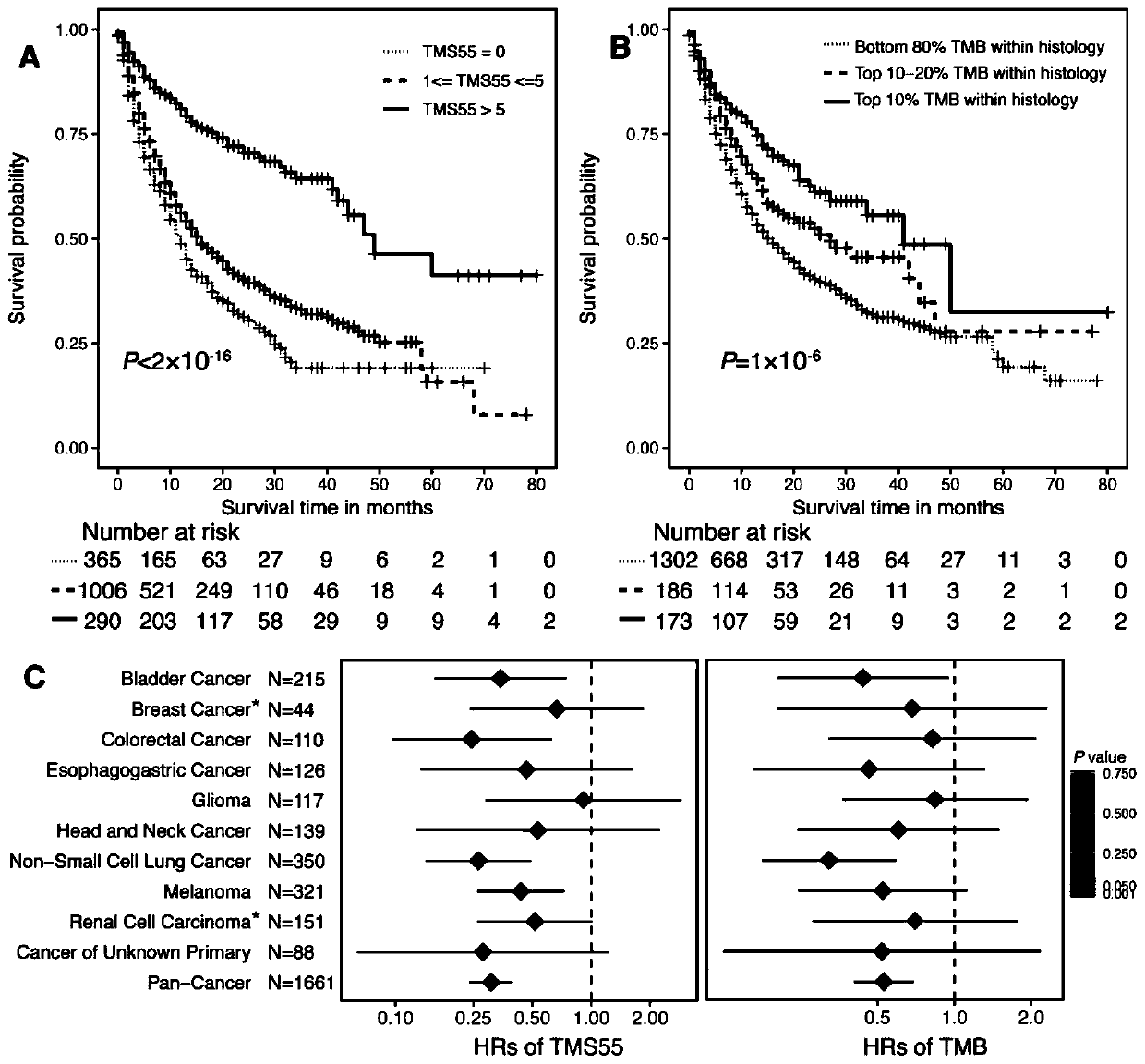
The invention provides a gene combination and application thereof in preparation of a reagent for predicting prognosis of a patient treated with an immune checkpoint inhibitor. Based on the combination of 55 genes, the invention develops a tumor mutation score TMS32055 method through the gene combination, and TMS is defined as the number of genes containing non-synonymous mutations in a specific gene combination. TMS 55 is divided into 3 groups: TMS 55 is equal to 0, TMS 55 is greater than or equal to 1 and is less than or equal to 5, and TMS 55 is greater than 5. Compared with a group that TMS 55 is equal to 0, the patients of groups that TMS 55 is greater than or equal to 1 and is less than or equal to 5 and TMS 55 is greater than 5 have better prognosis and longer overall survival time.The TMS55 based on the 55-gene combination has higher prediction efficiency than TMB in predicting the overall survival after immunotherapy, and has uniform cutoff value. Therefore, the TMS55 can beused for predicting the prognosis of patients receiving immune checkpoint inhibition treatment, and is easier to popularize and apply in clinical practice.
Owner:WUHAN UNIV
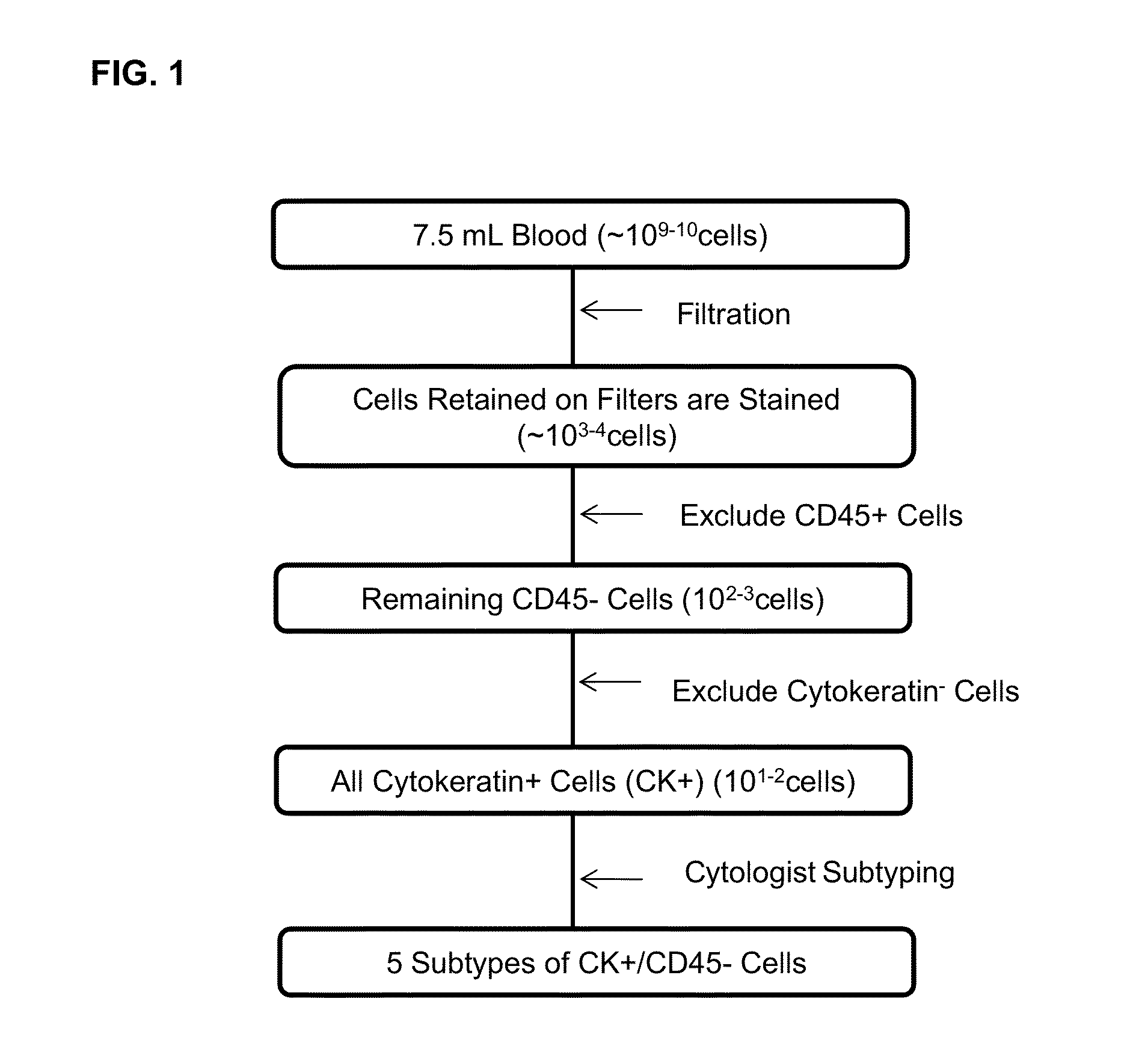
Methods for predicting overall survival of cancer patients based on numbers of circulating tumor cells
InactiveUS20160178630A1SurvivalReduced survivalSamplingBiological material analysisCirculating cancer cellOncology
Cytokeratin (CK)+ / CD45− / DAPI+ cells in the blood of cancer patients have been considered by most as circulating tumor cells (CTCs). Different methods of isolating CTCs results in a wide range of numbers and subgroups of CTCs from same patient. As a result, the clinical significance of the number of CTCs becomes cloudy. Provided herein is methodology to categorize CTCs into morphologically distinct subpopulations, and to use one of the subpopulations in methods for predicting overall survival of a patient having cancer.
Owner:CREATV MICROTECH
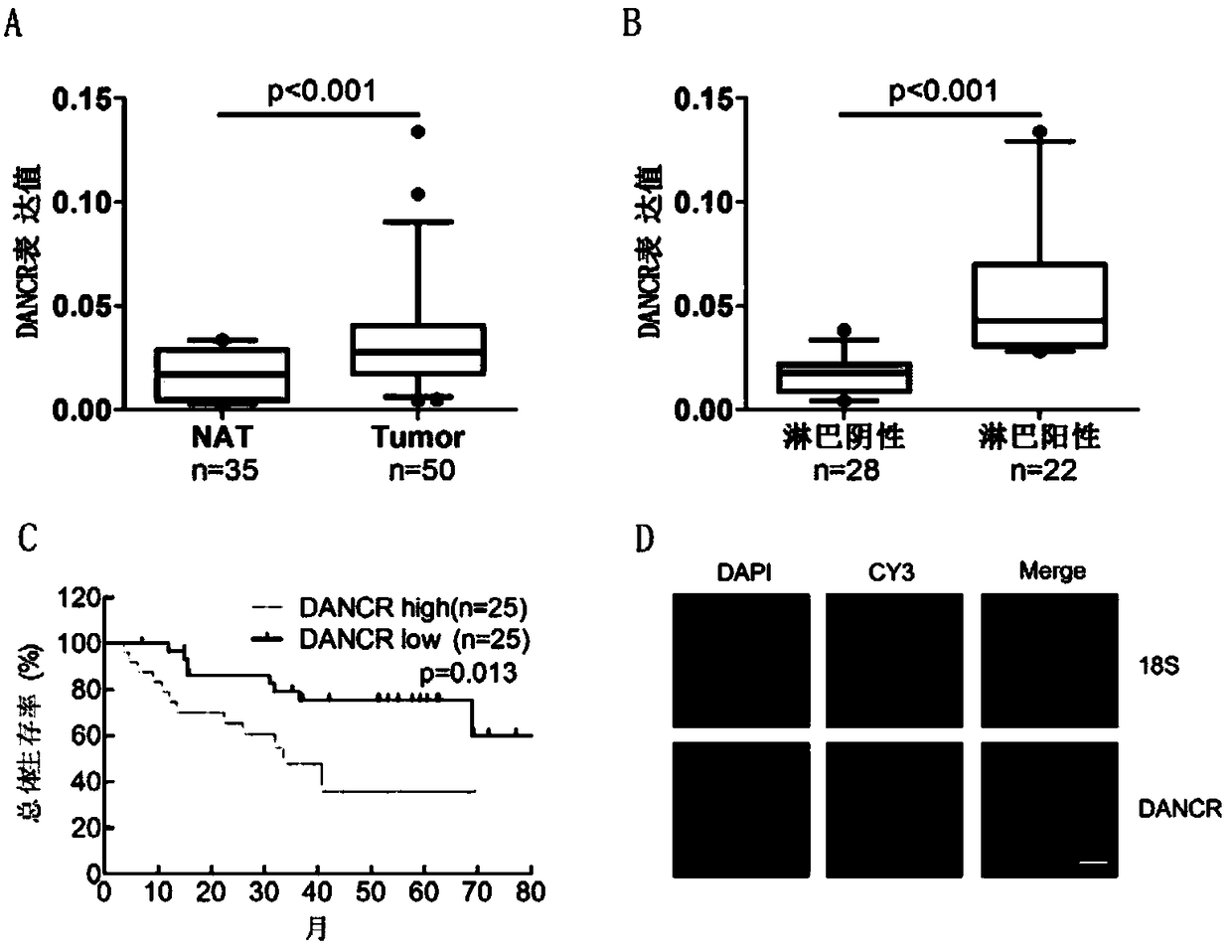
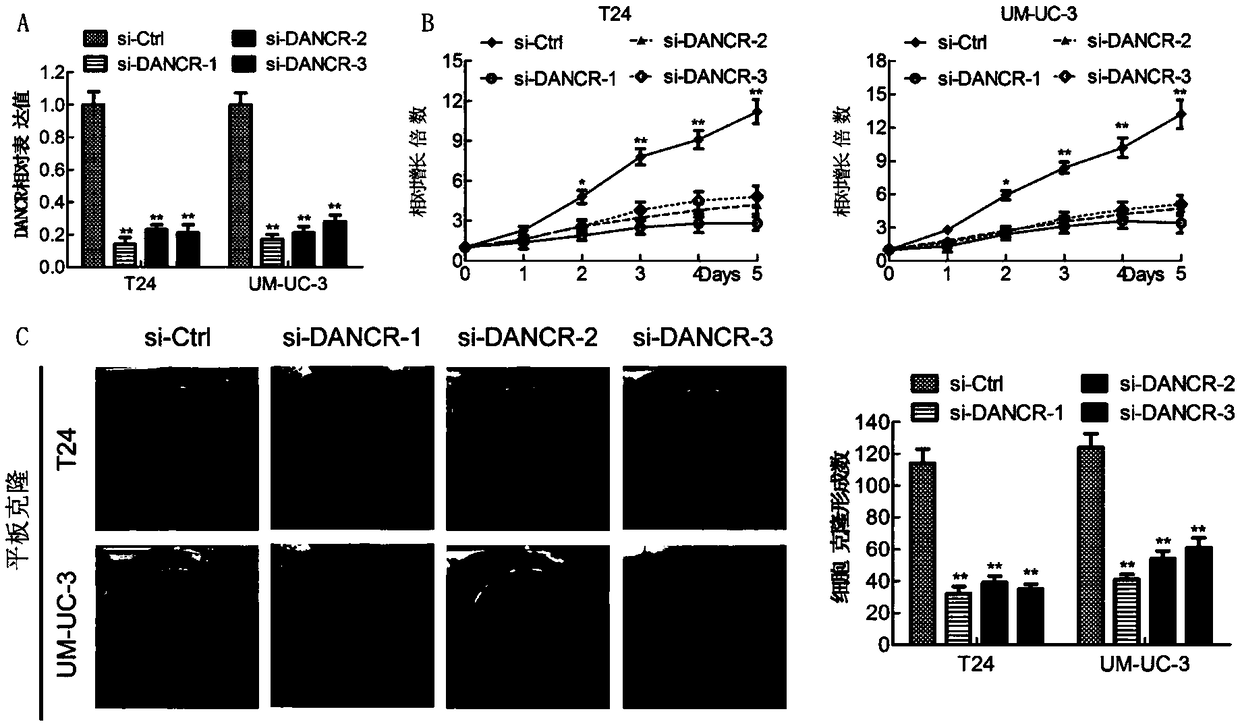
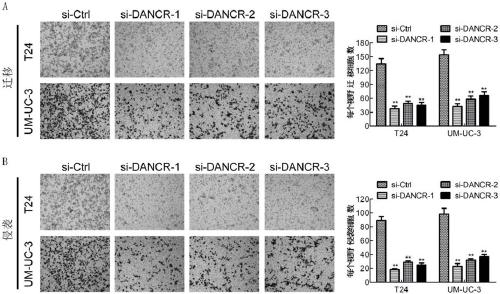
Molecular marker LncRNA DANCR for diagnosing and treating bladder cancer and application thereof
ActiveCN109371131APrevent proliferationInhibit migrationMicrobiological testing/measurementAntineoplastic agentsIn vivoCell migration
The invention discloses a molecular marker for diagnosing and treating the bladder cancer, namely LncRNA DANCR, and also discloses application of the molecular marker to screening or preparation of reagents used for diagnosing the bladder cancer diagnosing, bladder cancer lymphatic metastasis, bladder cancer prognosis relapse or bladder cancer clinical stages and application of the molecular marker to screening or preparation of drugs used for treating the bladder cancer. It is found that the LncRNA DANCR has high expression in bladder cancer cases with lymphatic metastasis, has negative correlation with overall survival prognosis of a patient and is an independent indicator for diagnosing the bladder cancer and judging bladder cancer development and survival prognosis; silencing of the LncRNA DANCR can restrain in-vitro proliferation, cell migration and invasion and in-vivo tumor formation and lymphatic metastasis of bladder cancer cells. It is verified that the LncRNA DANCR is an important cancerigenic factor of the bladder cancer for the first time, and can be used as a molecular marker for bladder cancer prognosis diagnosis and a new target spot for treatment.
Owner:SUN YAT SEN MEMORIAL HOSPITAL SUN YAT SEN UNIV
Methods of Prognosis for Non-Hodgkin Lymphoma
InactiveUS20120134986A1Predictive of survivalImprove the level ofBiocidePhosphorous compound active ingredientsBiologyTumor cells
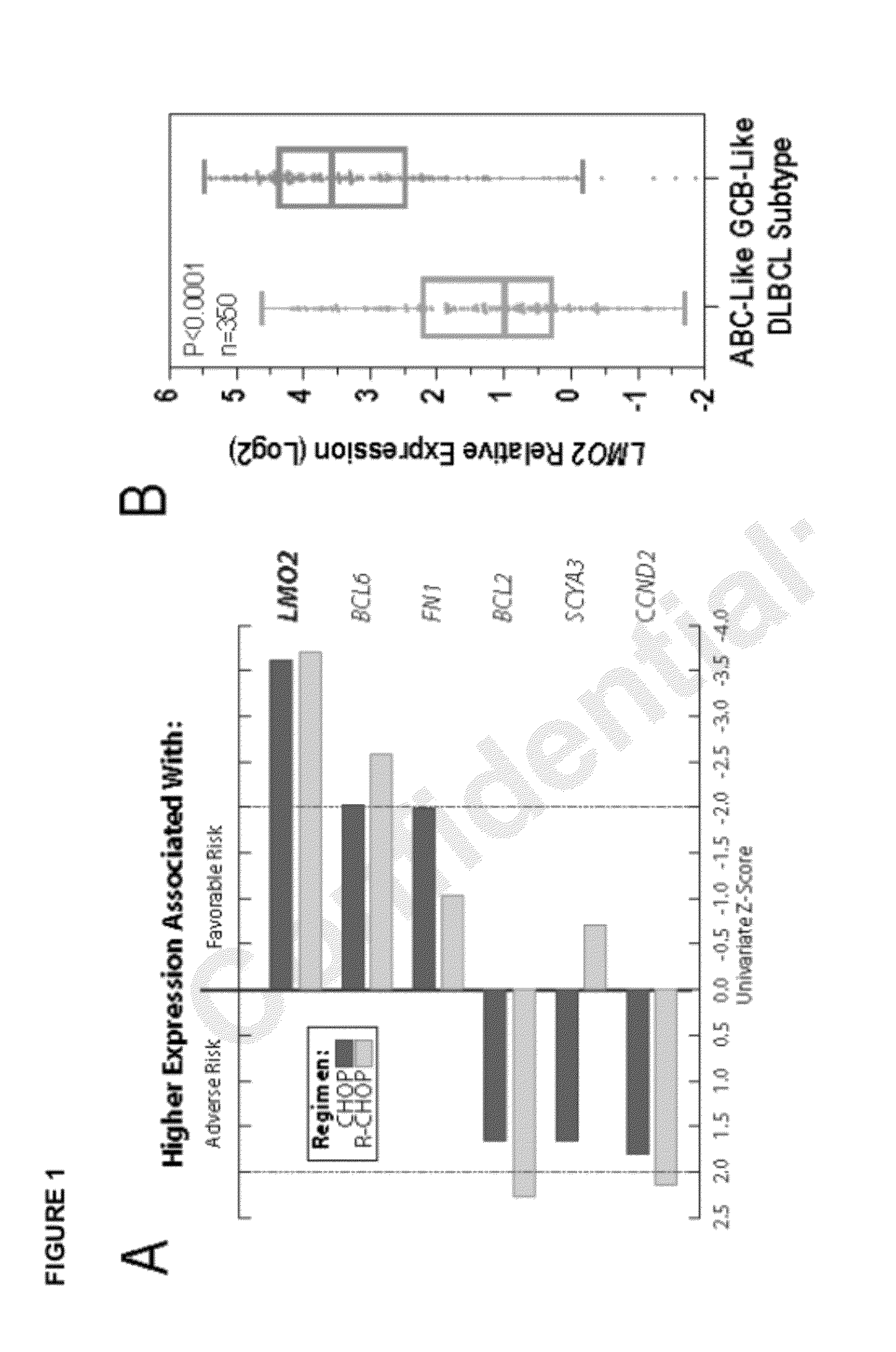
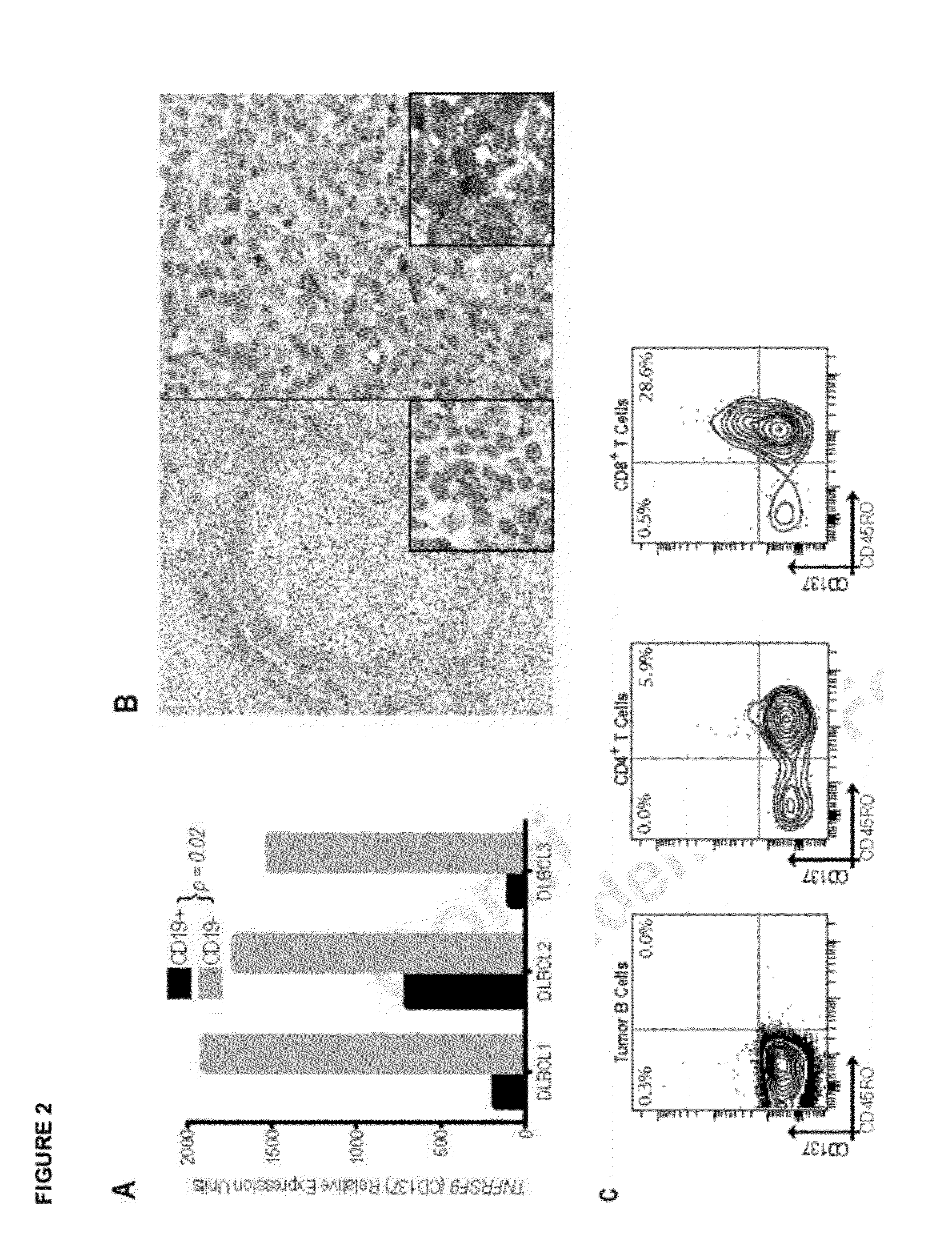
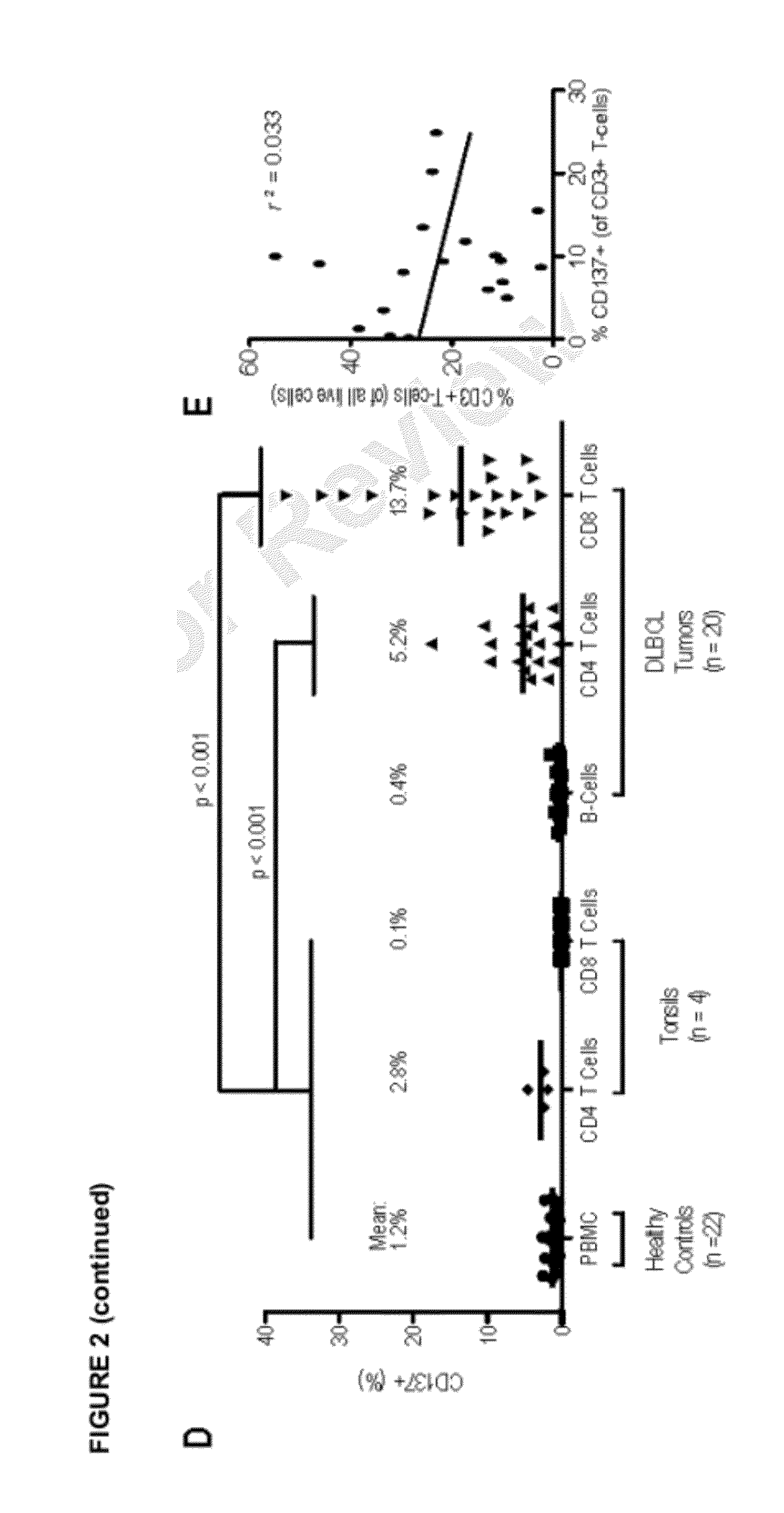
Measurement of a single gene expressed by tumor cells (LMO2) and a single gene expressed by the immune microenvironment (TNFRSF9), which determination may be referred to herein as a two gene score (TGS), powerfully predicts overall survival in patients with NHL, particularly overall survival in the context of anthracycline-based chemotherapy or co-treatment with anthracycline-based chemotherapy and anti-CD20 immunotherapy. It is shown herein that increased levels of LMO2 and TNFRSF9 correlate with a positive patient response and improved prognosis.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
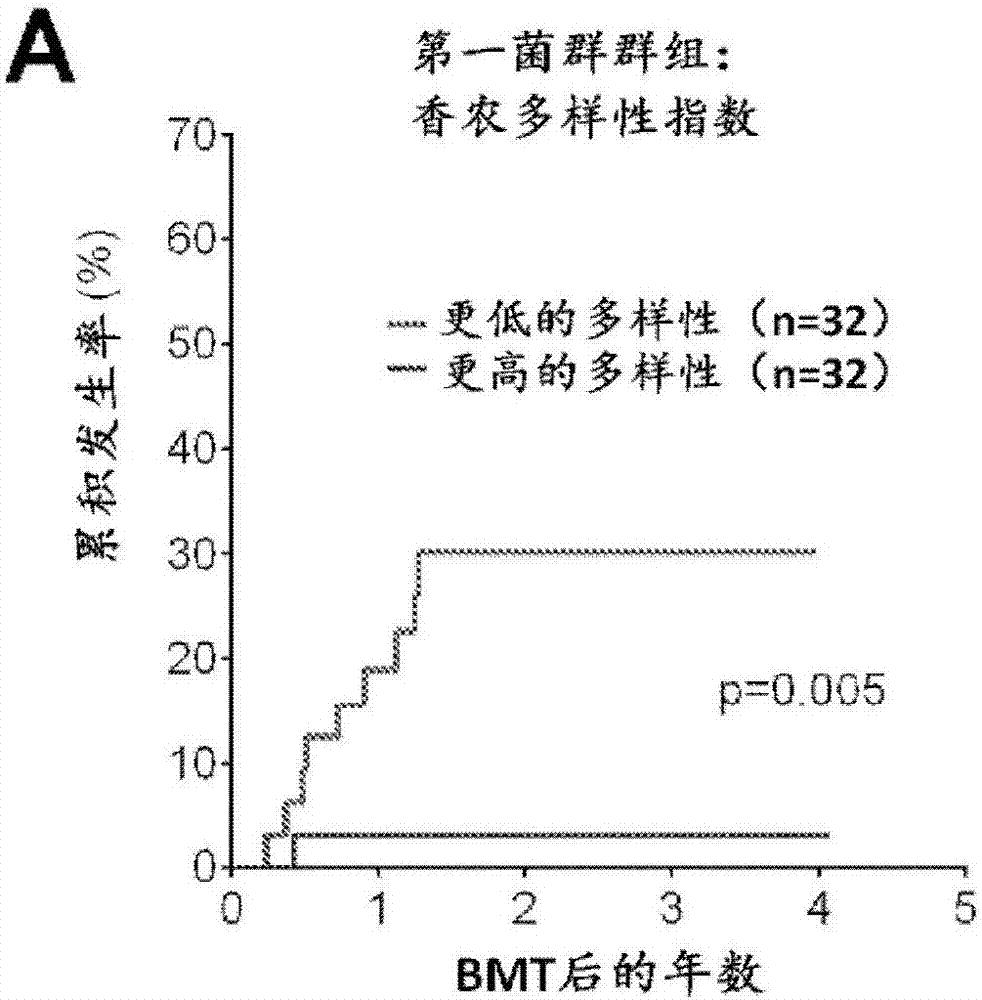
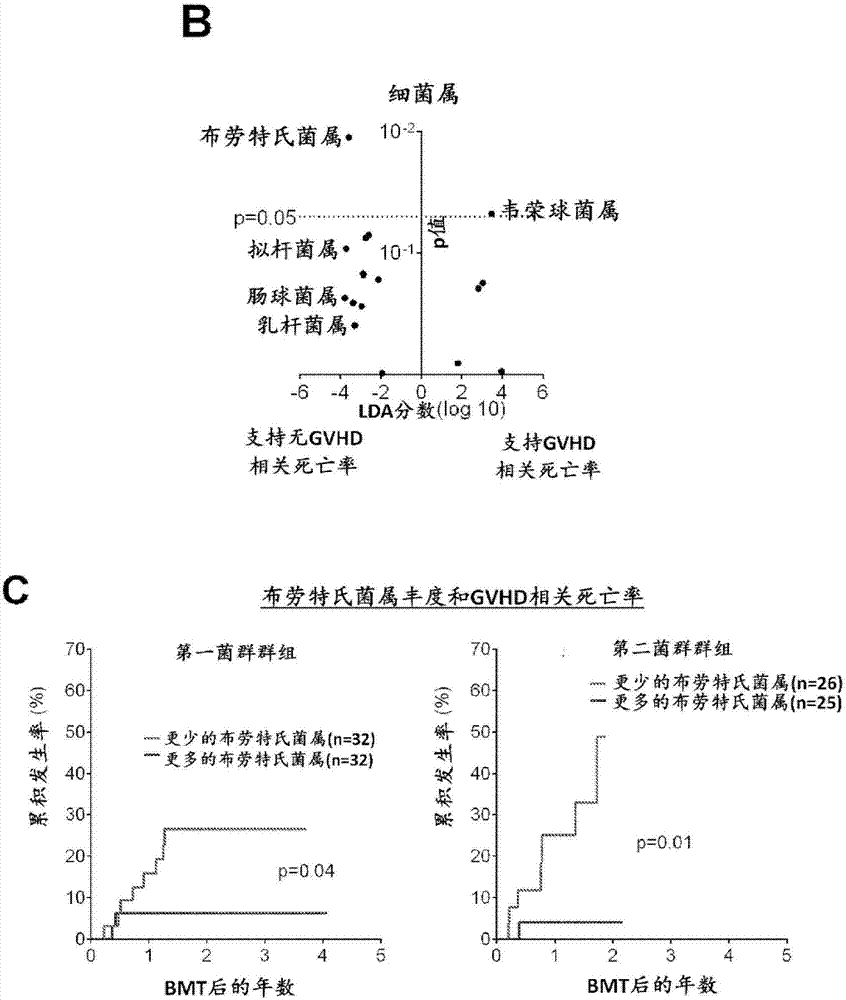
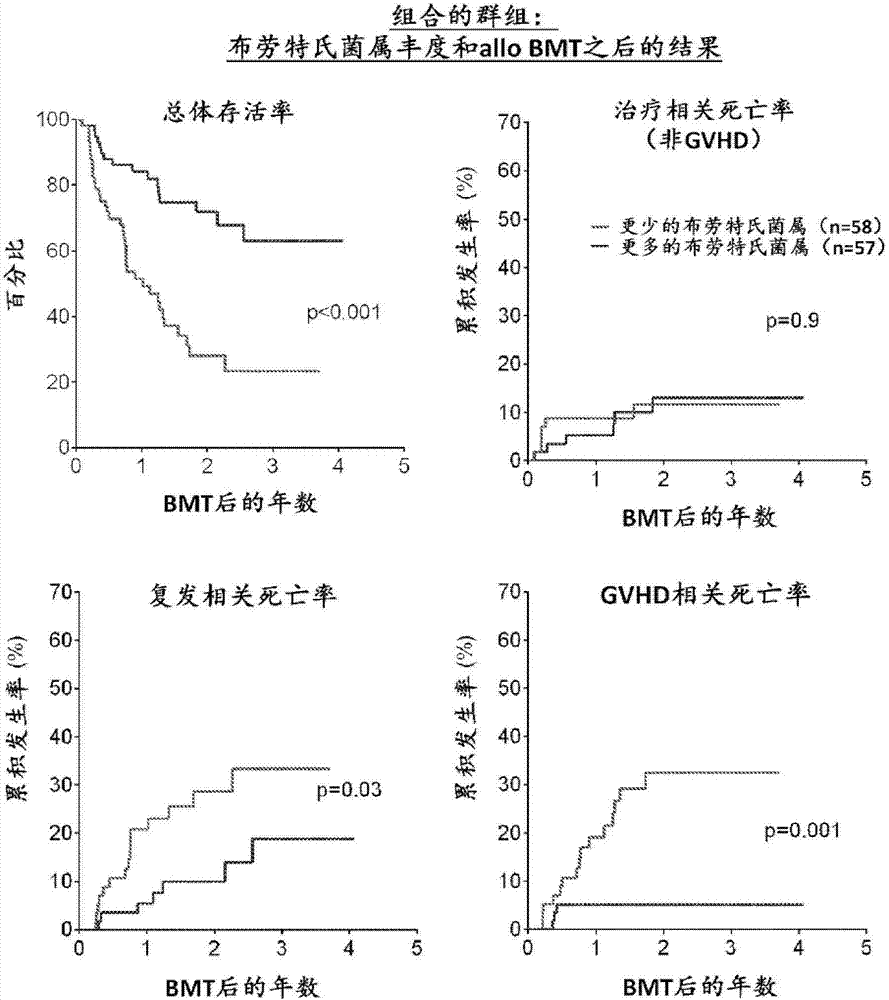
Intestinal microbiota and gvhd
PendingCN107249609AReduce abundanceRisk reduction or eliminationBacteria material medical ingredientsMicrobiological testing/measurementTreatment choicesSymbiotic bacteria
The present disclosure describes compositions and methods for increasing the abundance of commensal bacteria belonging to the order Clostridiales, including Blautia, Ruminococcus, Clostridium, Eubacterium, Holdemania and Dorea species, that are as-sociated with reduced lethal GVHD and improved overall survival following bone mar-row or hematopoietic stem cell transplant. The present disclosure, therefore, provides methods for reducing the likelihood, incidence or severity of GVHD by (1) avoiding the loss of endogenous beneficial species through antibiotic selection; (2) by administering a therapeutically effective amount of a composition comprising one or more Clostridiales associated with reduced GVHD to individuals who may lack or have lost those strains from their intestinal microbiota. Additionally, support for endogenous or reestablished Clostridiales related to reduced GVHD as a treatment option for reducing GVHD can also be provided in the form of nutritional supplementation, for example, sugars fermented by some species of Clostridiales with GVHD reducing activity.
Owner:MEMORIAL SLOAN KETTERING CANCER CENT
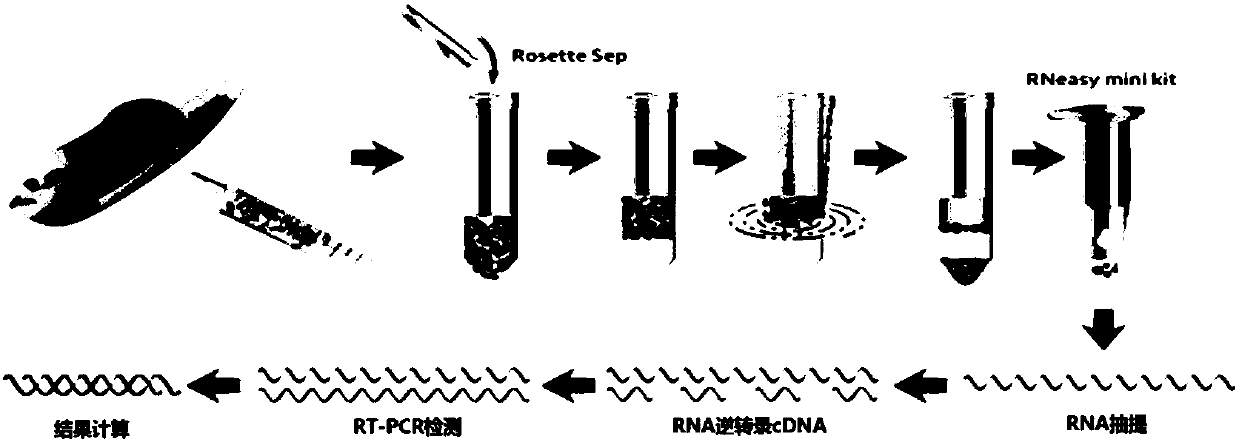
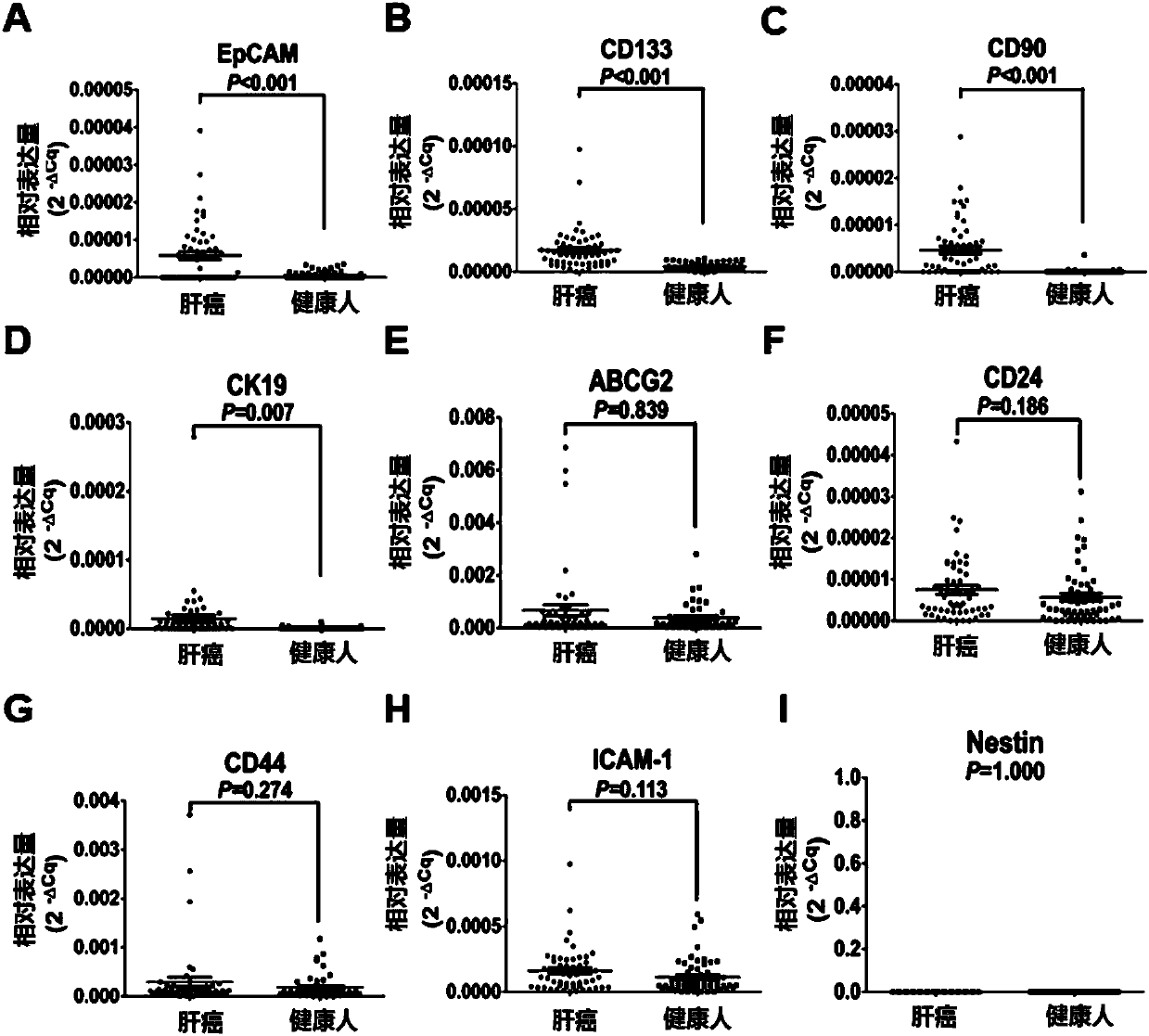
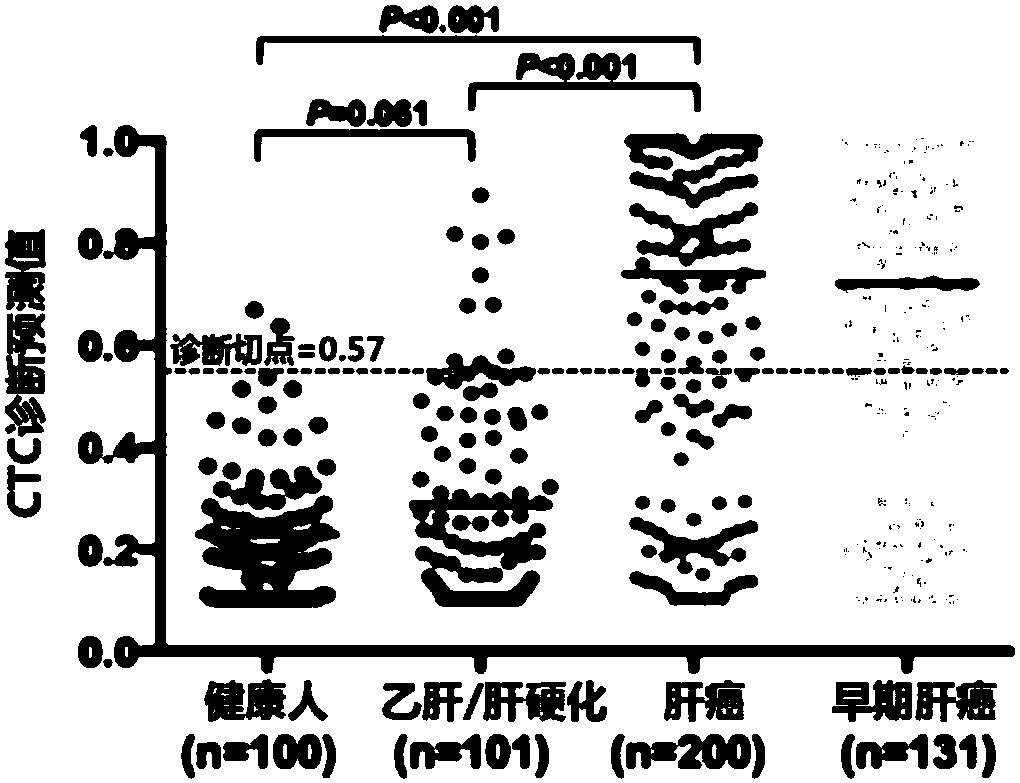
Joint detection kit for multiple markers on CTCs (circulating tumor cells) and application of kit
InactiveCN107586839AAvoid interferenceUse less bloodMicrobiological testing/measurementLymphatic SpreadHepatocellular carcinoma
The invention provides a joint detection kit for multiple stem cell markers on surfaces of CTCs (circulating tumor cells) and an application of the kit. The joint detection kit for multiple stem cellmarkers on surfaces of CTCs comprises 1) a peripheral blood CTC enrichment reagent; 2) an RNA extraction reagent; 3) an RNA reverse transcription reagent; 4) a fluorescence labeled Taqman probe and aspecific primer; 5) a qPCR reaction system reagent and a hepatocellular carcinoma prediction formula. The kit can be used for biopsy of minimally invasive fluids of tumor patients, early diagnosis andearly warning of tumor metastasis and relapse are realized, the anti-tumor therapy effect and the tumor progression condition are monitored, molecular subtyping based on CTCs and screening of specific sensitive drugs for the CTCs are realized, so that knowledge about tumor metastasis, relapse and drug resistance related mechanisms is improved, individualized medical treatment of tumor patients isguided, the overall survival of the tumor patients is significantly improved, and the kit has great clinical significance.
Owner:ZHONGSHAN HOSPITAL FUDAN UNIV
Osteosarcoma prognosis marker and prognosis evaluation model
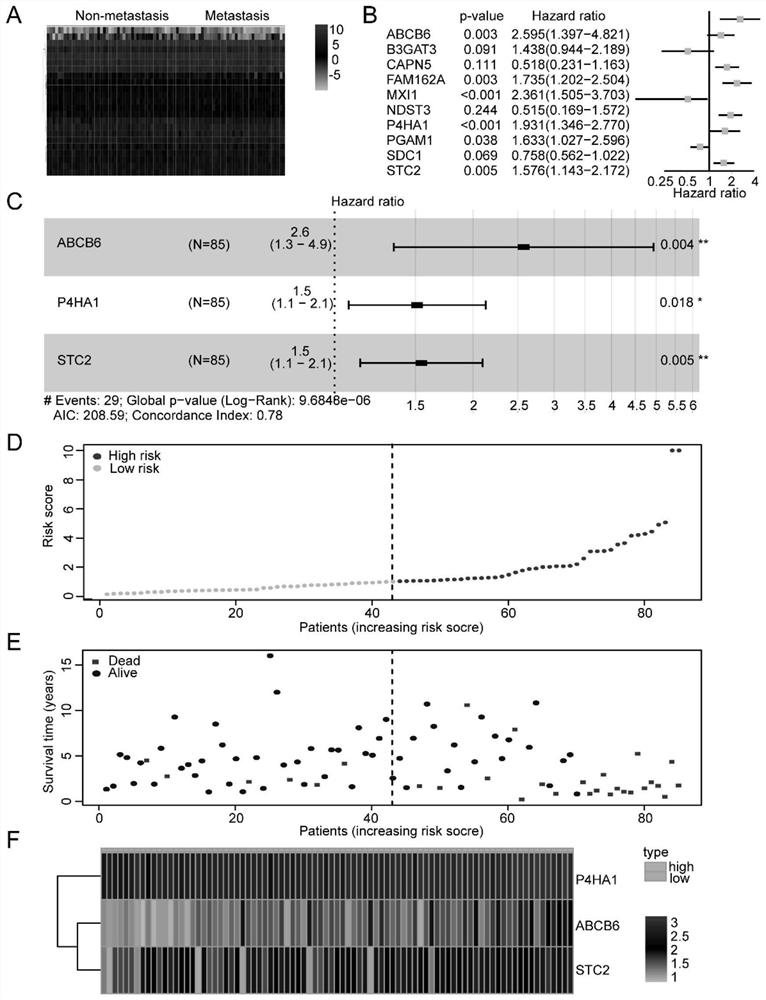
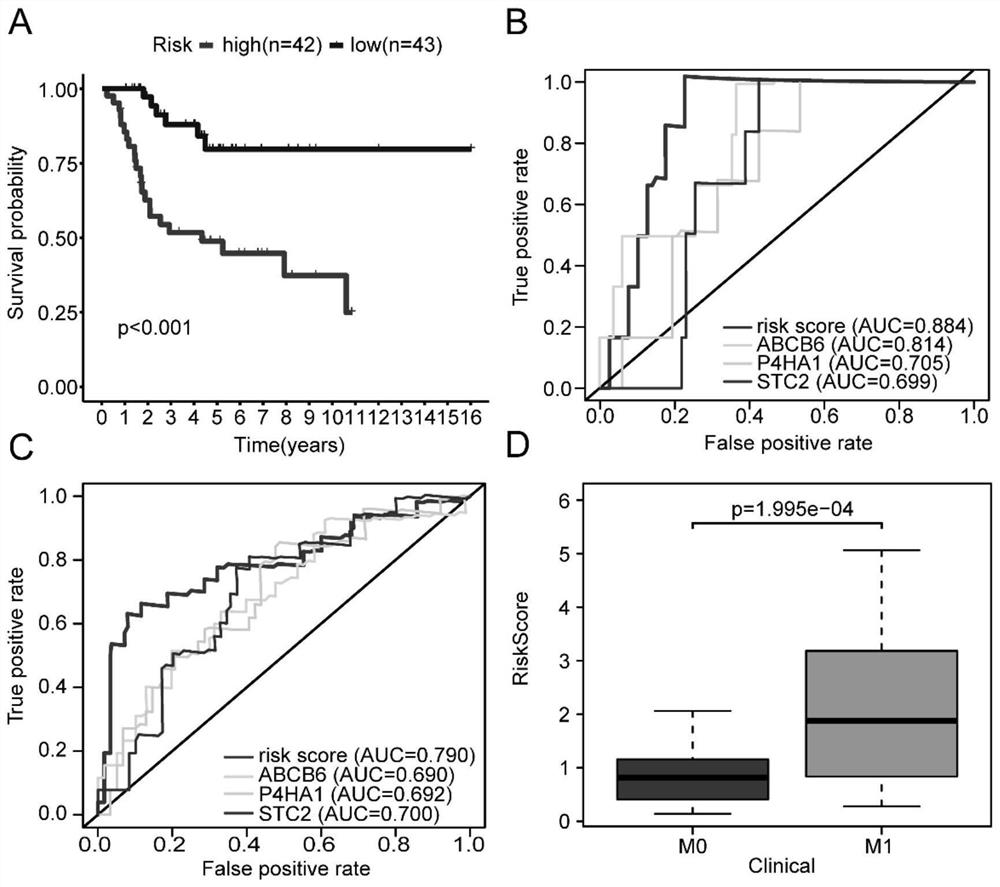
ActiveCN112063720APredict prognosisImprove 5-year survival rateMicrobiological testing/measurementMaterial analysisDiseaseOncology
The invention relates to an osteosarcoma prognosis marker and prognosis evaluation model. According to the osteosarcoma prognosis marker and prognosis evaluation model, overall survival time data of osteosarcoma patients in two independent databases of TARGET and GSE21257 are integrated, glycolysis-related genes P4HA1, ABCB6 and STC2 are screened and verified as osteosarcoma prognosis markers, anda prognostic risk assessment model is constructed based on the P4HA1, ABCB6 and STC2, and by a training set and a verification set, it is verified that the model has relatively high accuracy, and further analysis finds that the risk score of the model is also significantly related to metastasis and tumor immune microenvironment. The osteosarcoma prognosis marker and prognosis evaluation model areconducive to better predicting prognosis conditions of the osteosarcoma patients, and can bring great help for guiding clinical treatment of the osteosarcomas.
Owner:SHANGHAI FIRST PEOPLES HOSPITAL
Bad phosphorylation determines ovarian cancer chemo-sensitivity and patient survival
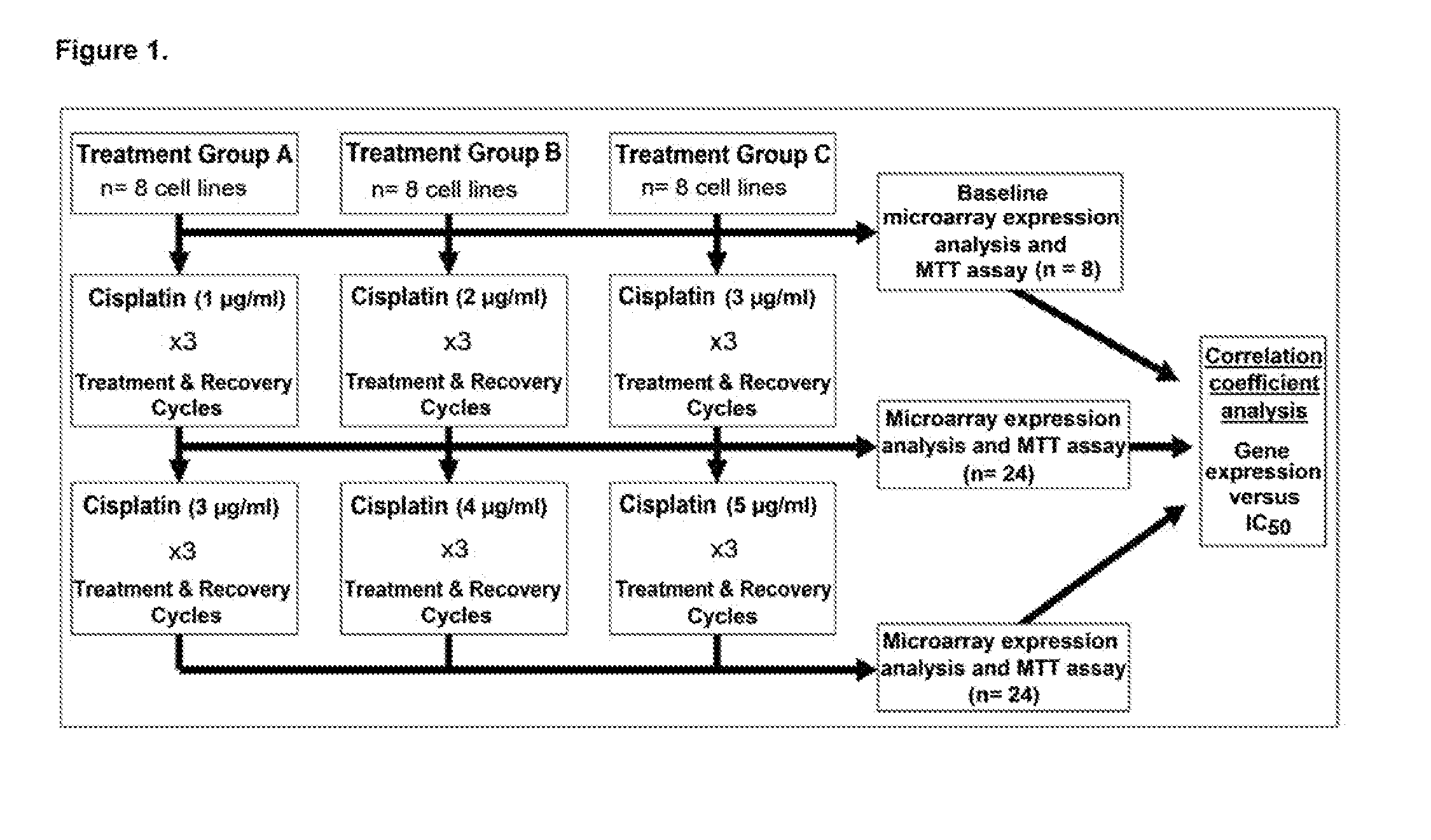
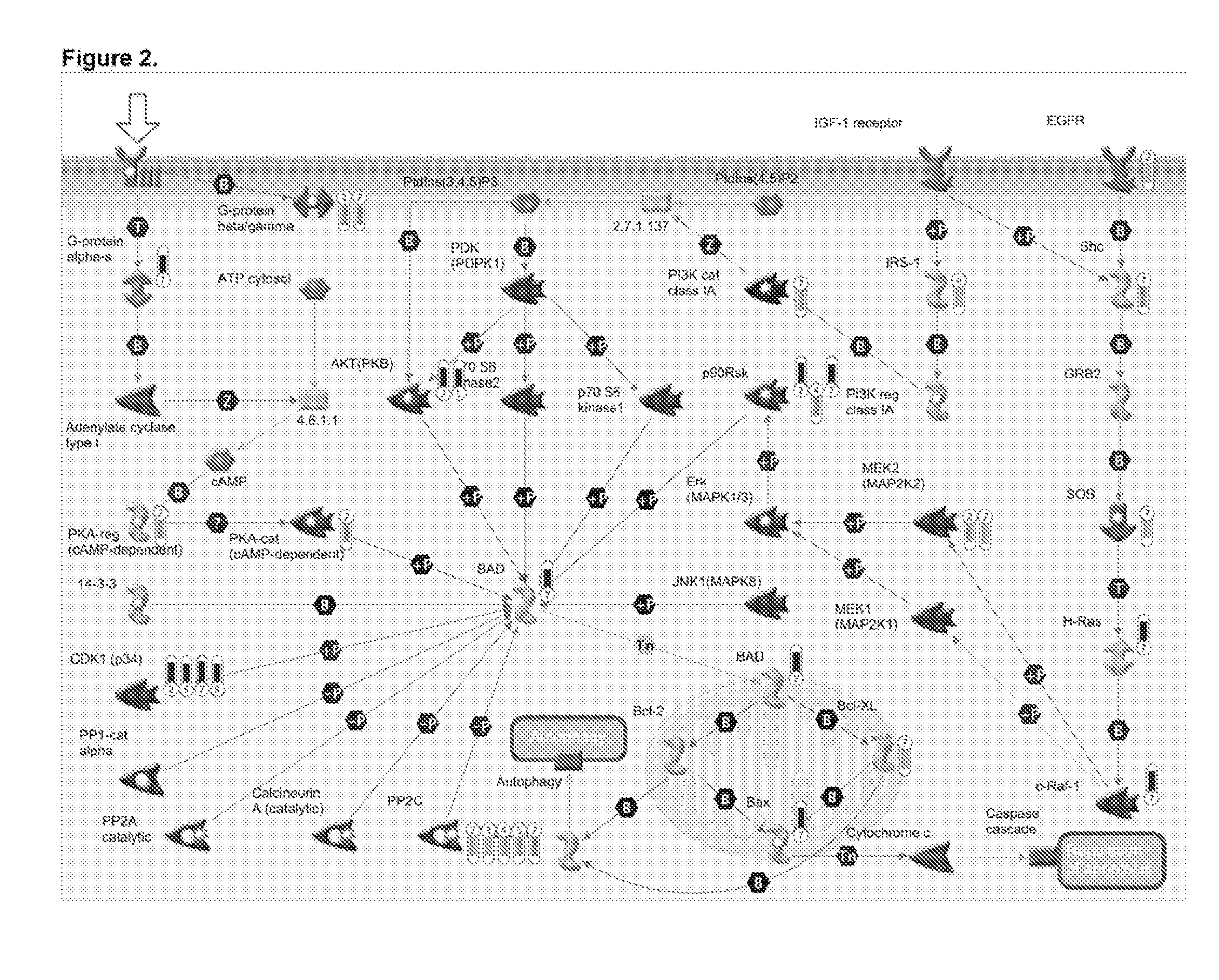
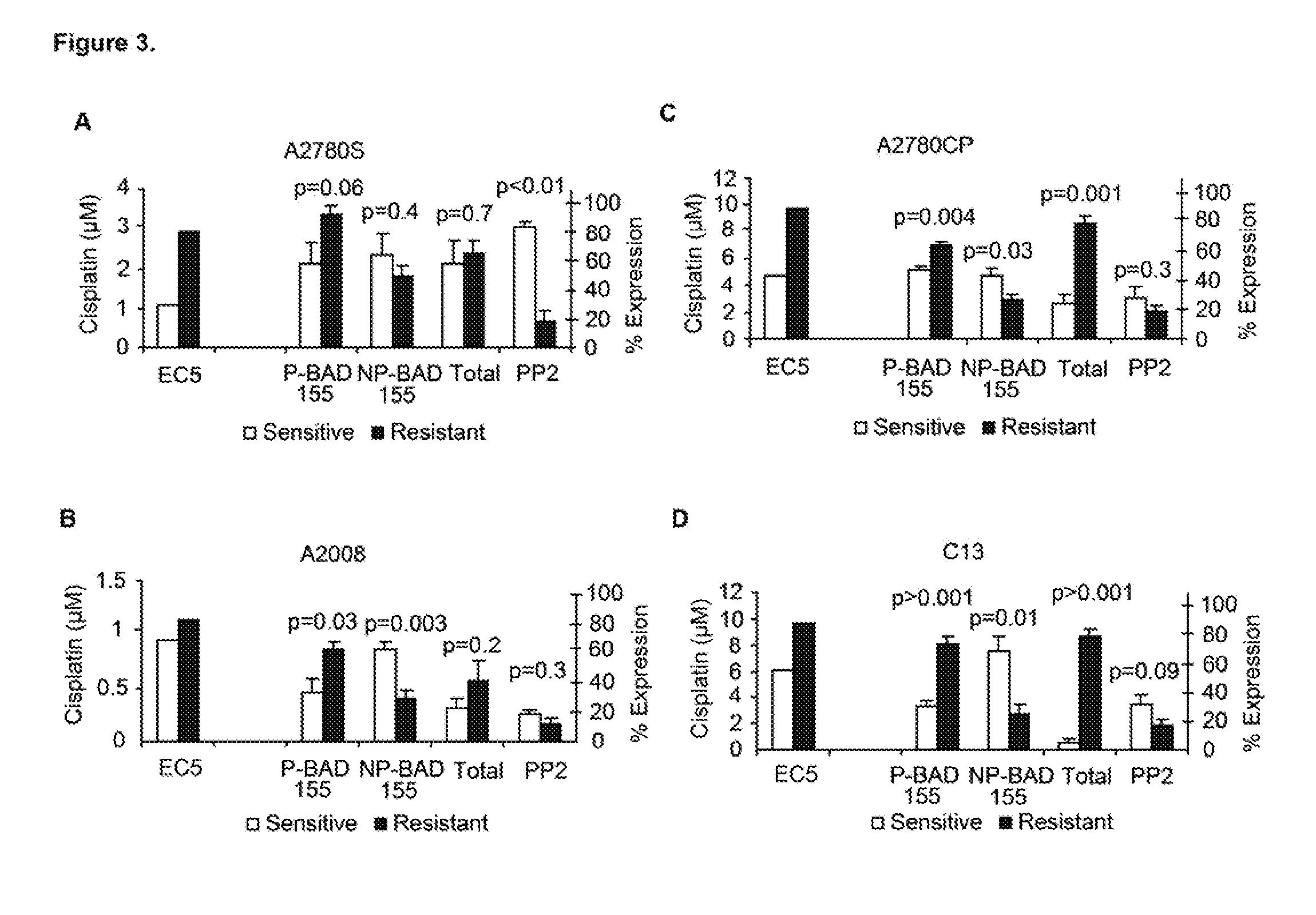
Despite initial sensitivity BAD-protein phosphorylation were evaluated in patient samples and cell lines as determinants of chemo-sensitivity and / or clinical outcome, and as therapeutic targets. Induced in-vitro OVCA cisplatin-resistance was associated with BAD-pathway expression. Expression of the pathway was also associated with resistance of 7 different cancers cell-types to 8 chemotherapeutic agents. Phosphorylation of the BAD-protein was associated with platinum-resistance in OVCA cells and primary OVCA specimens, and also overall patient survival. Targeted modulation of BAD-phosphorylation levels influenced cisplatin-sensitivity. A 47-gene BAD-pathway signature was associated in-vitro phospho-BAD levels and with survival of 838 patients with ovarian, breast, colon, and brain cancer. The survival advantage associated with both BAD-phosphorylation and also the BAD-pathway signature was independent of surgical cytoreductive status. The BAD apoptosis pathway influences human cancer chemo-sensitivity and overall survival. The pathway is useful as a biomarker of therapeutic response, patient survival, and therapeutic target.
Owner:H LEE MOFFITT CANCER CENT & RES INST INC
Treatment selection for lung cancer patients using mass spectrum of blood-based sample
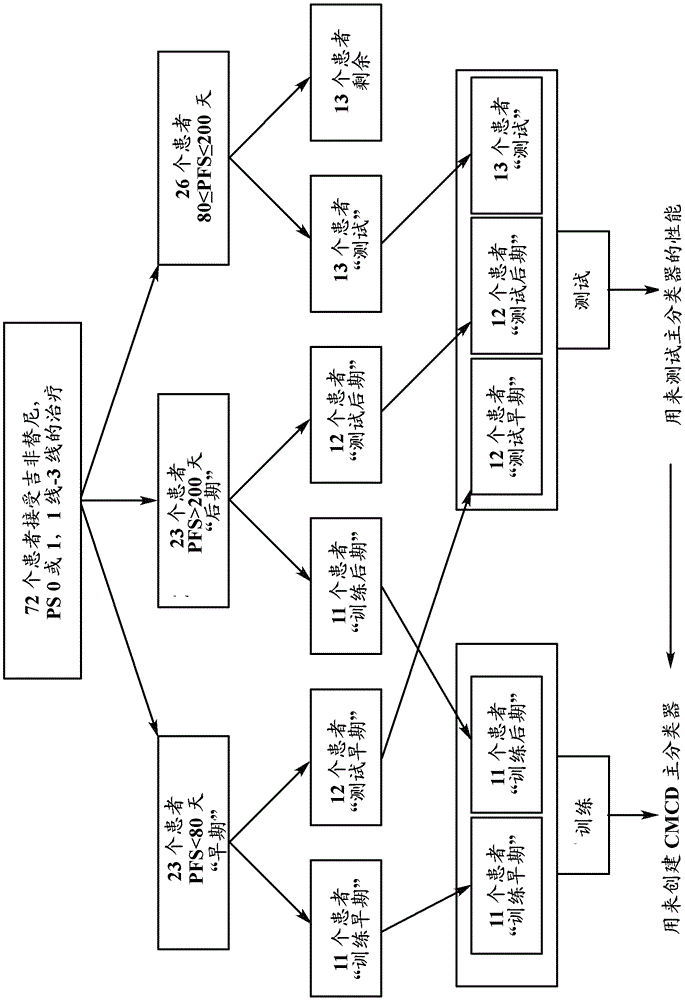
A test for predicting whether a non-small-cell lung cancer patient is more likely to benefit from an EGFR-I as compared to chemotherapy uses a computer-implemented classifier operating on a mass spectrum of a blood-based sample obtained from the patient. The classifier makes use of a training set which includes mass spectral data from blood-based samples of other cancer patients who are members of a class of patients predicted to have overall survival benefit on EGFRI-Is, e.g., those patients testing VS Good under the test described in U.S. Pat. No.7,736,905. The class-labeled group is further subdivided into two subsets, i.e., those patients which exhibited early (class label 'early') and late (class label 'late') progression of disease after administration of the EGFR-I in treatment of cancer.
Owner:BIODESIX
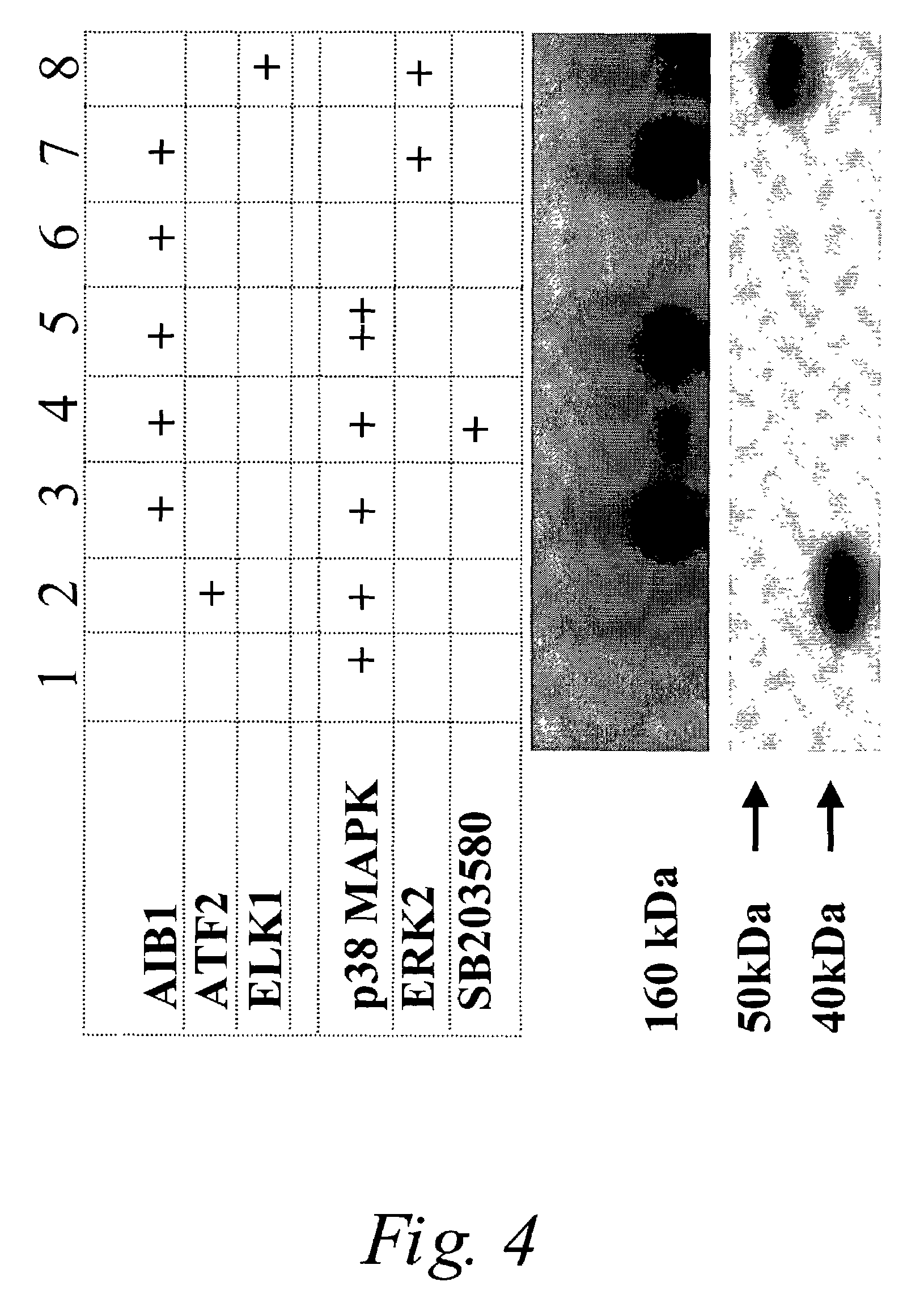
P38 MAPK pathway predicts endocrine-resistant growth of human breast cancer and provides a novel diagnostic and treatment target
InactiveUS7217533B2Avoid resistanceSamplingMicrobiological testing/measurementAdjuvantTreatment targets
Acquired and de novo endocrine resistance are major clinical problems in the management of breast cancer patients. Though the antiestrogen tamoxifen prolongs disease-free and overall survival in the adjuvant setting, and induces remissions in over half of the patients with estrogen receptor positive metastatic disease, all patients eventually acquire tamoxifen resistance. Furthermore, many of the resistant tumors actually appear to be stimulated by tamoxifen just as they are by estrogens. The present invention provides methods of predicting endocrine resistance comprising detecting the biological activity and / or expression of p38 MAPK and / or AIB1. The invention further provides methods of reducing, reversing, or preventing endocrine resistance comprising contacting a breast or prostate tumor with a p38 MAPK pathway inhibitor.
Owner:BAYLOR COLLEGE OF MEDICINE
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com