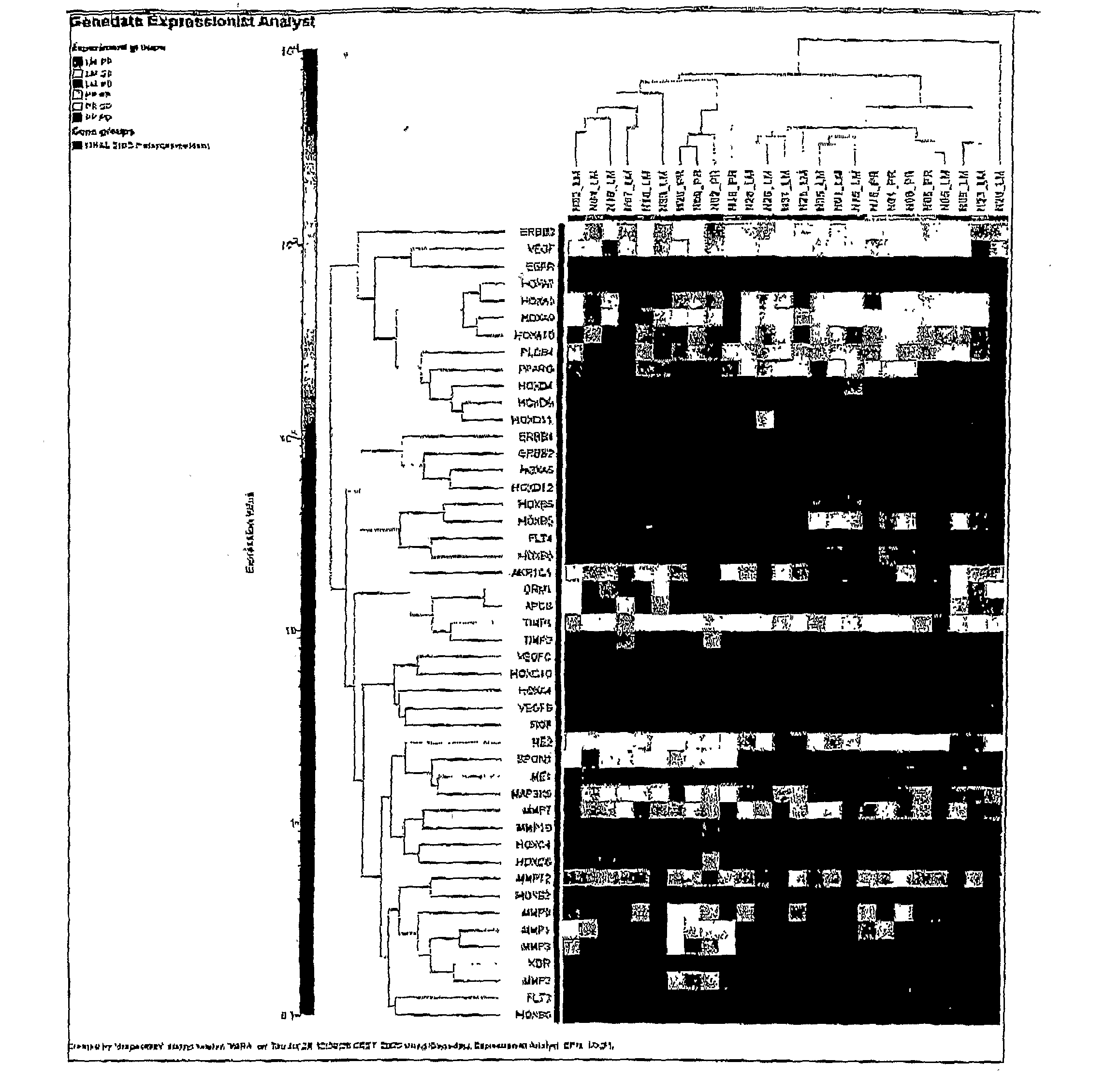
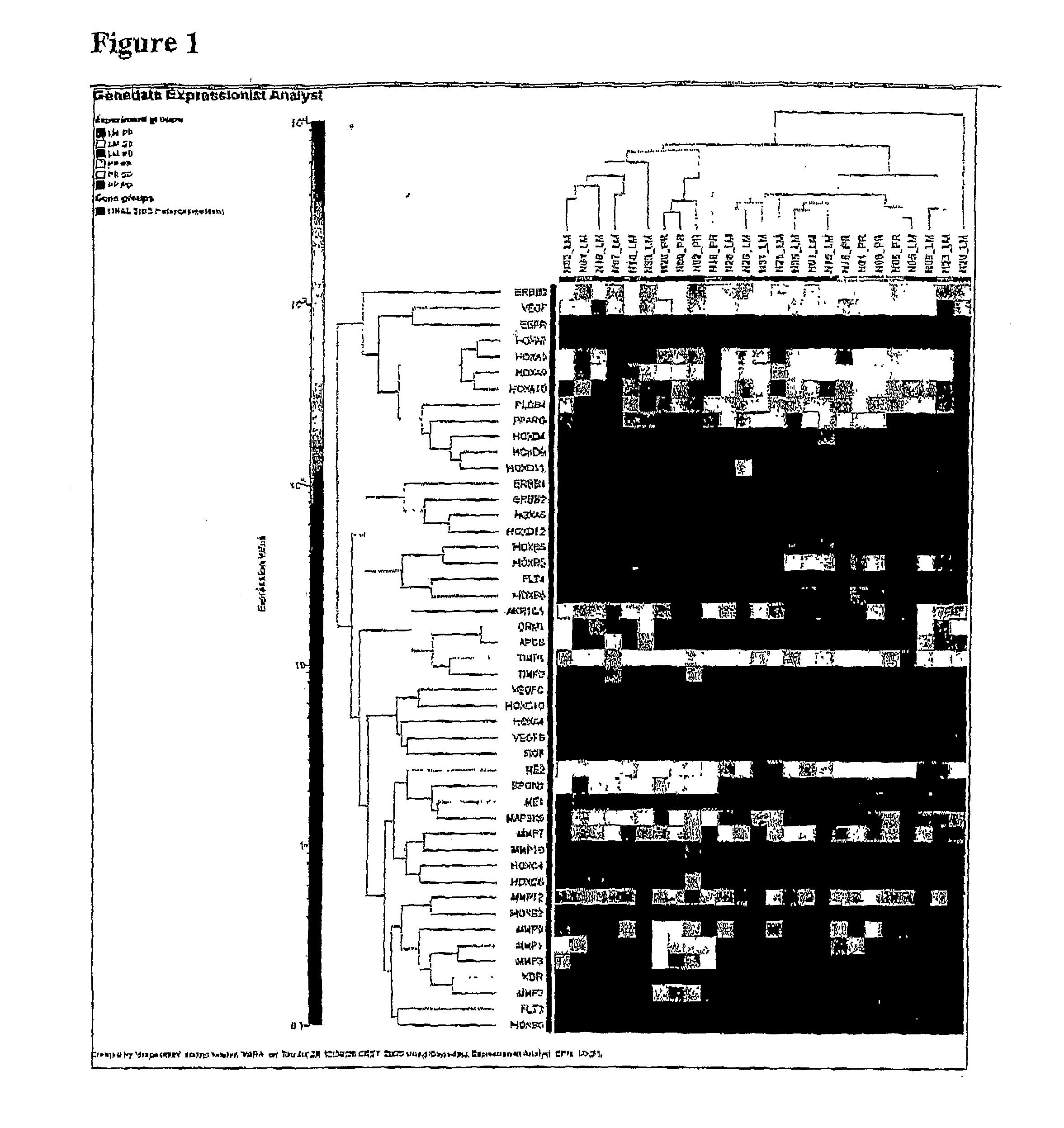
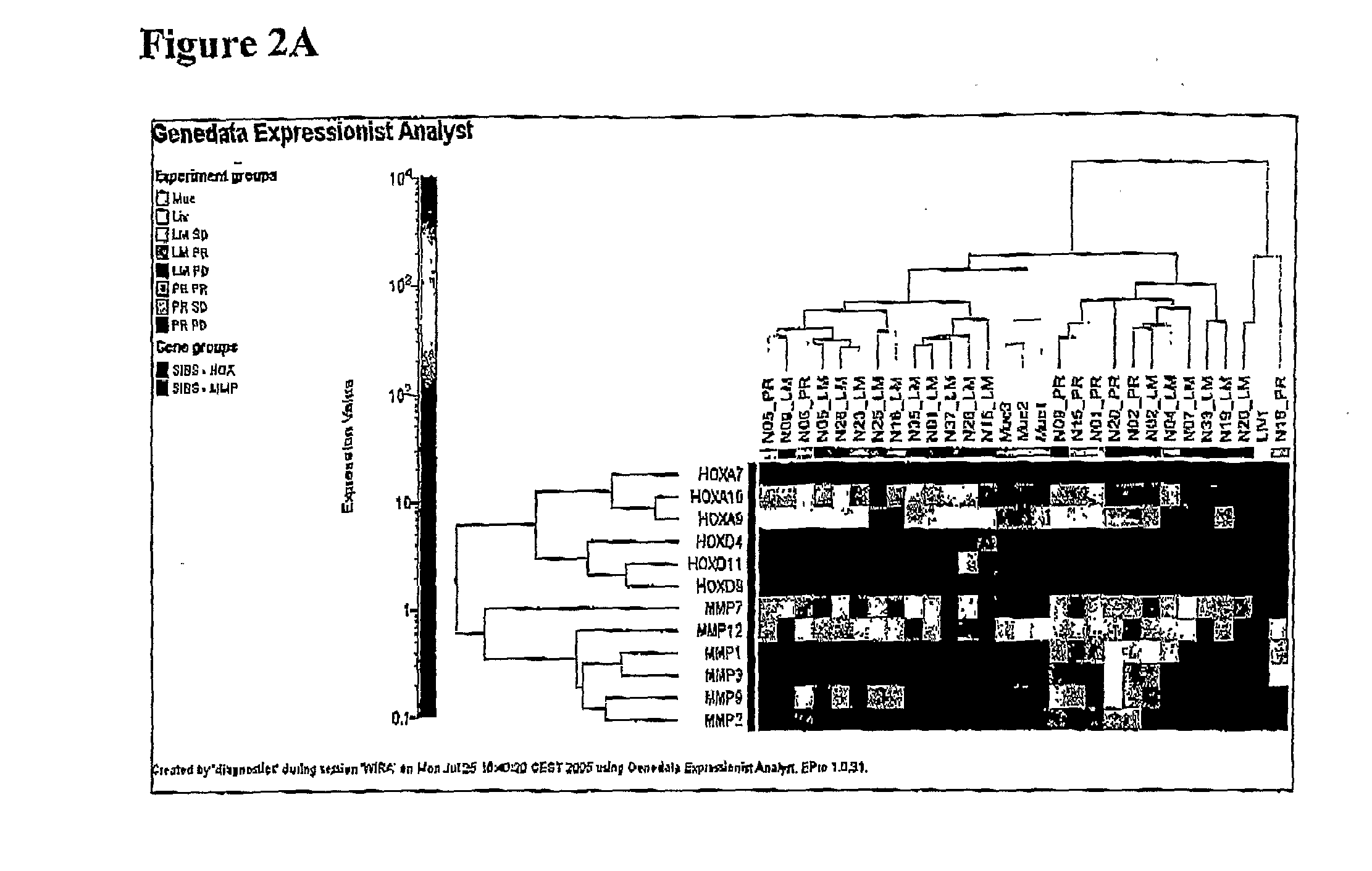
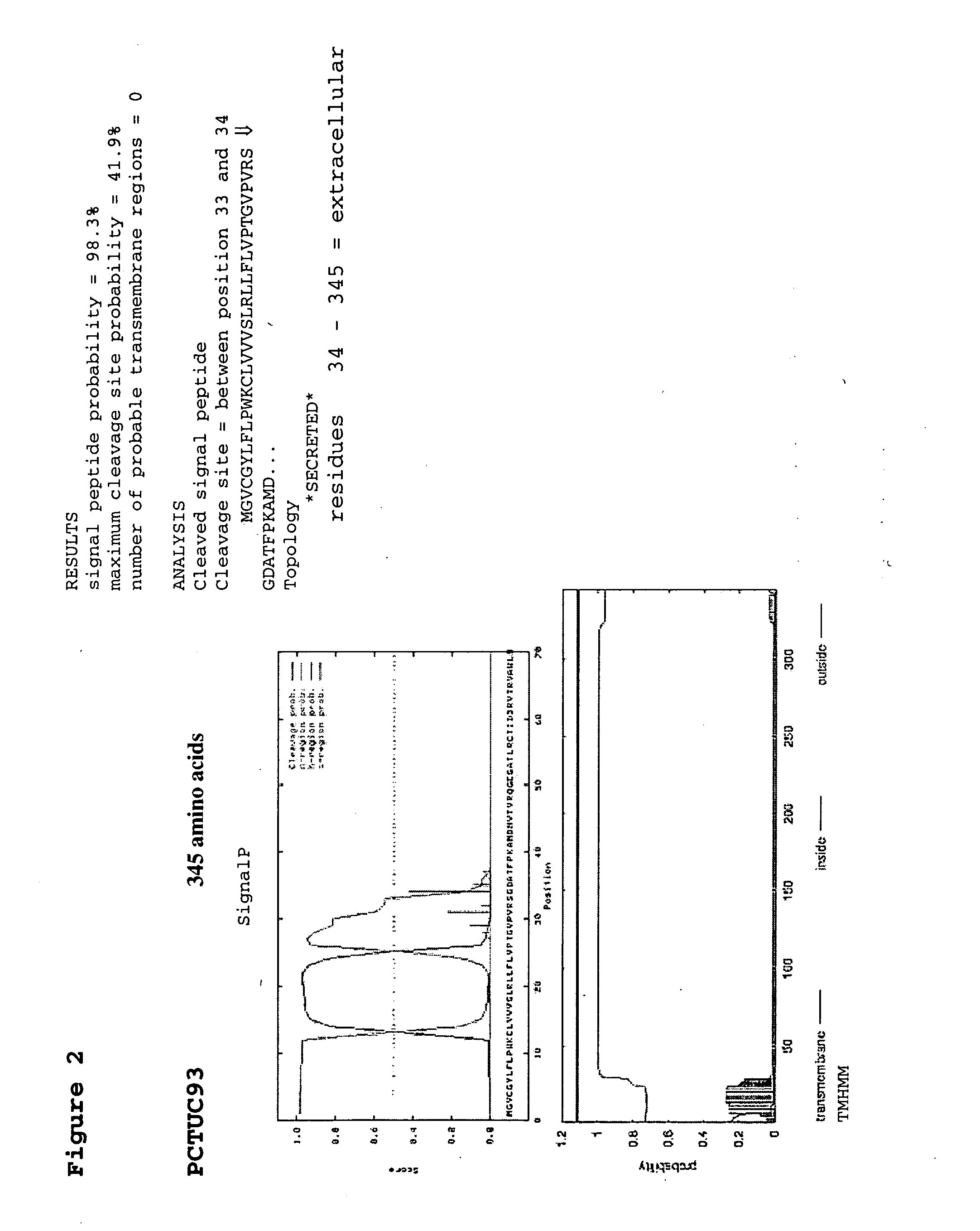
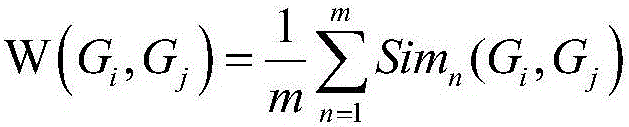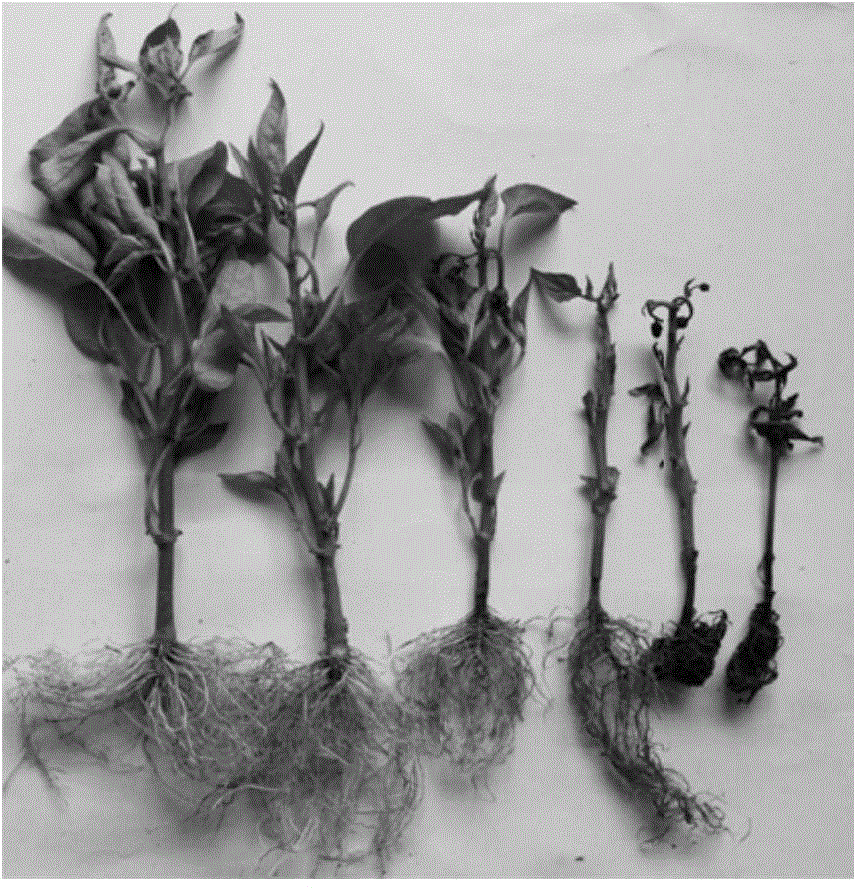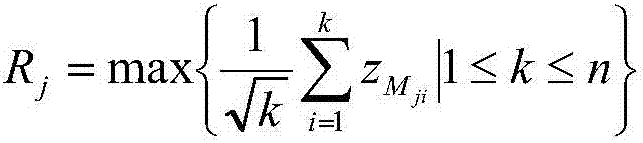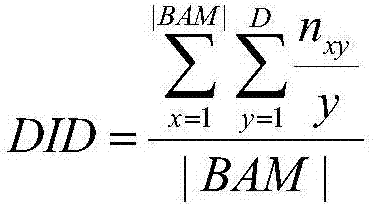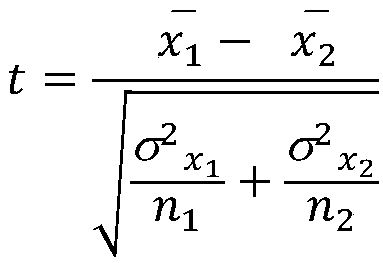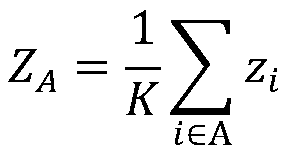Patents
Literature
317 results about "Differentially expressed genes" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Differential gene expression. 1. gene expression that responds to signals or triggers; a means of gene regulation, effects of certain hormones on protein biosynthesis.
Method for determining relative abundance of nucleic acid sequences
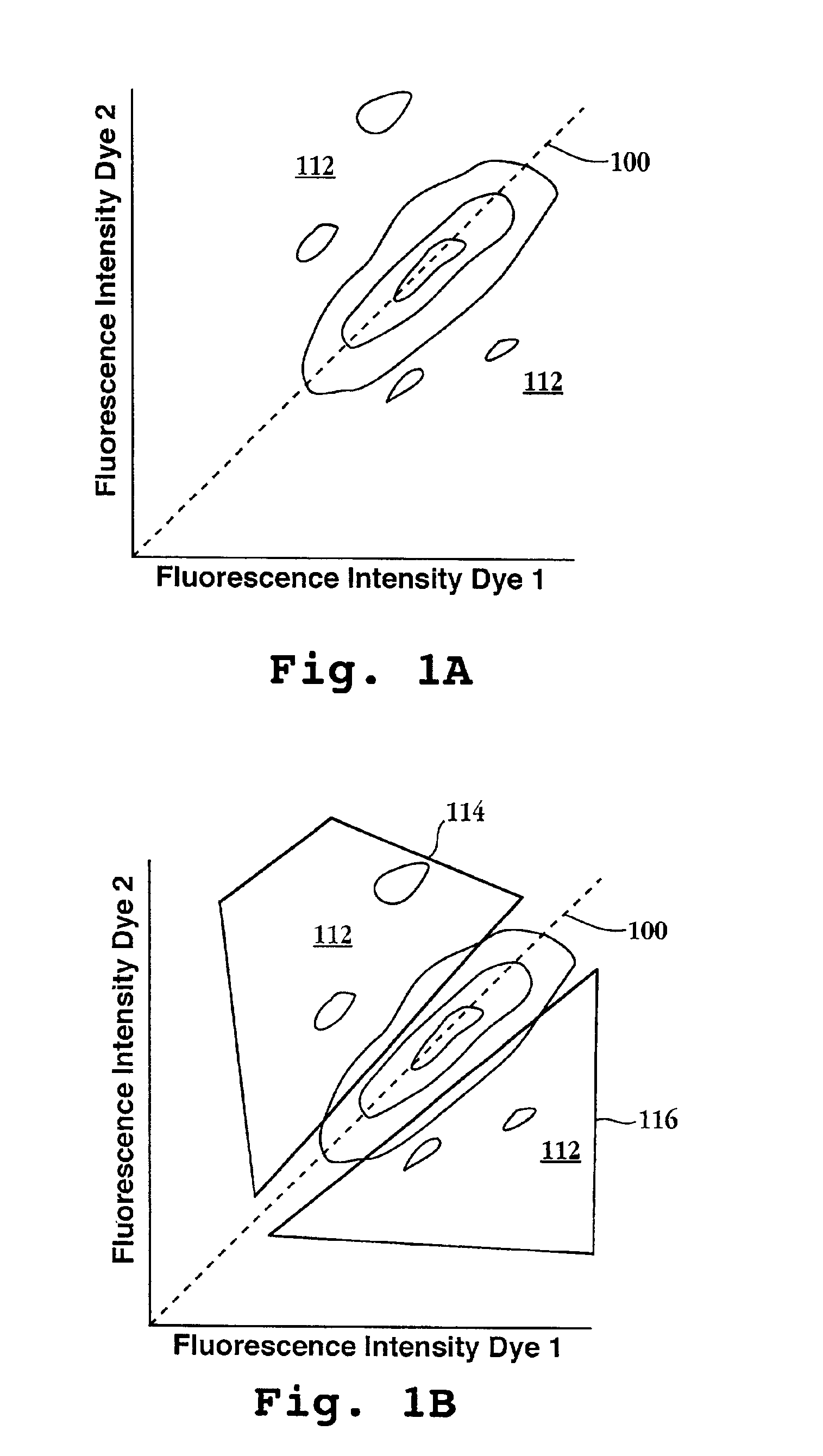
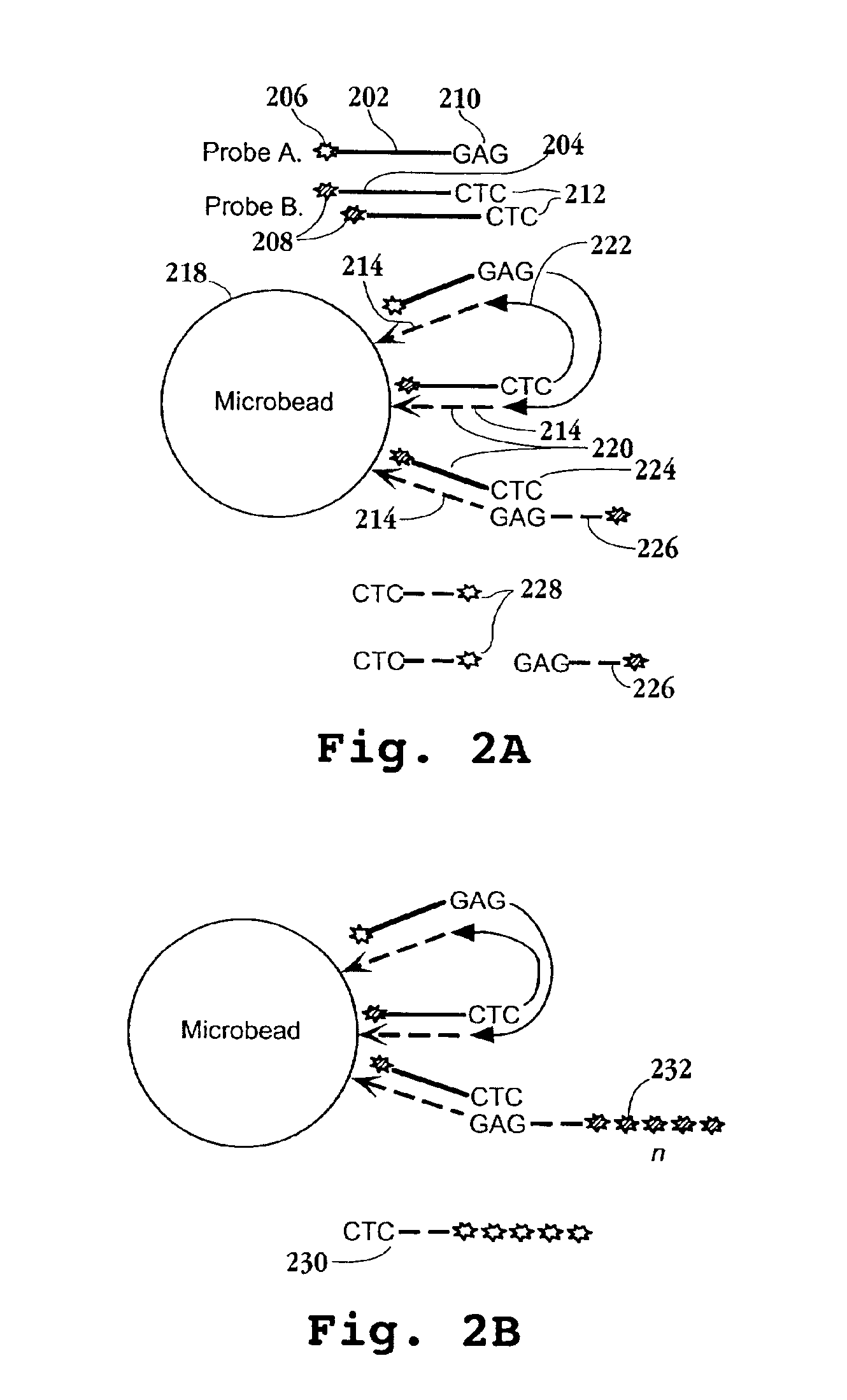
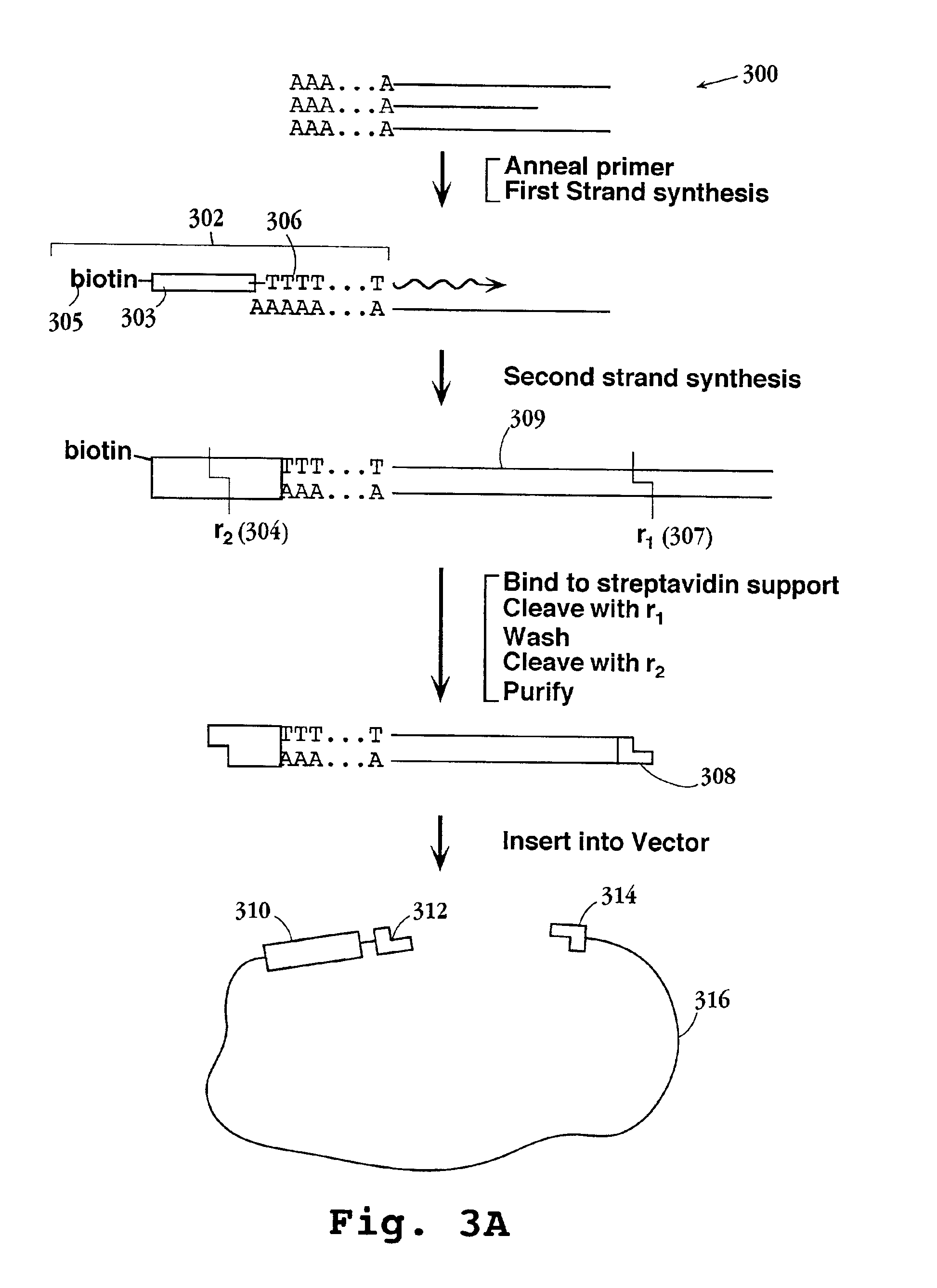
InactiveUS6897023B2Sugar derivativesMicrobiological testing/measurementHybridization probeFluorescence
Disclosed are methods for identifying nucleic acid sequences which are of different abundances in different nucleic acid source populations, e.g. differentially expressed genes or genomic variations among individuals or populations of individuals. In one embodiment, probes derived from the source nucleic acid populations are derivatized with a terminal sample ID (SID) sequence characteristic of that population. Upon competitive hybridization of the probes to a reference or index nucleic acid library containing all the sequences in the populations being compared, the SID tags remain single stranded, and those from the different sources are then annealed to one another. Unhybridized (remainder) SID sequences are then quantified. By labeling such remainder SID sequences with a fluorescent dye, FACS sorting of beads containing the hybridized probes can be carried out. The signal ratio upon which such sorting is based is enhanced compared to competitive hybridization using labeled probes without SID sequences.
Owner:THE MOLECULAR SCI INST +1
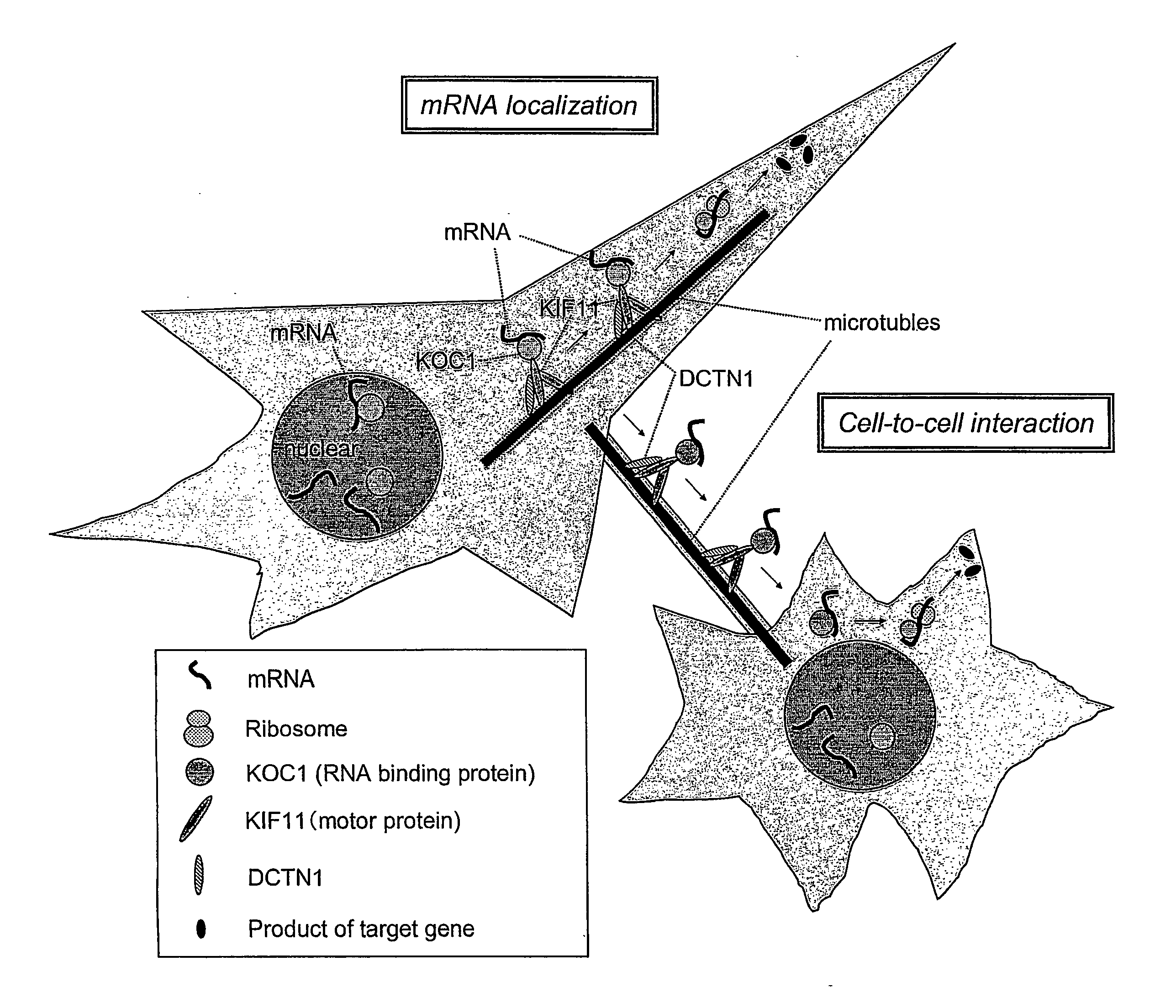
Method for Diagnosing Non-Small Cell Lung Cancer
InactiveUS20080050378A1Relieve symptomsReduced expression levelOrganic active ingredientsSugar derivativesOncologyDifferentially expressed genes
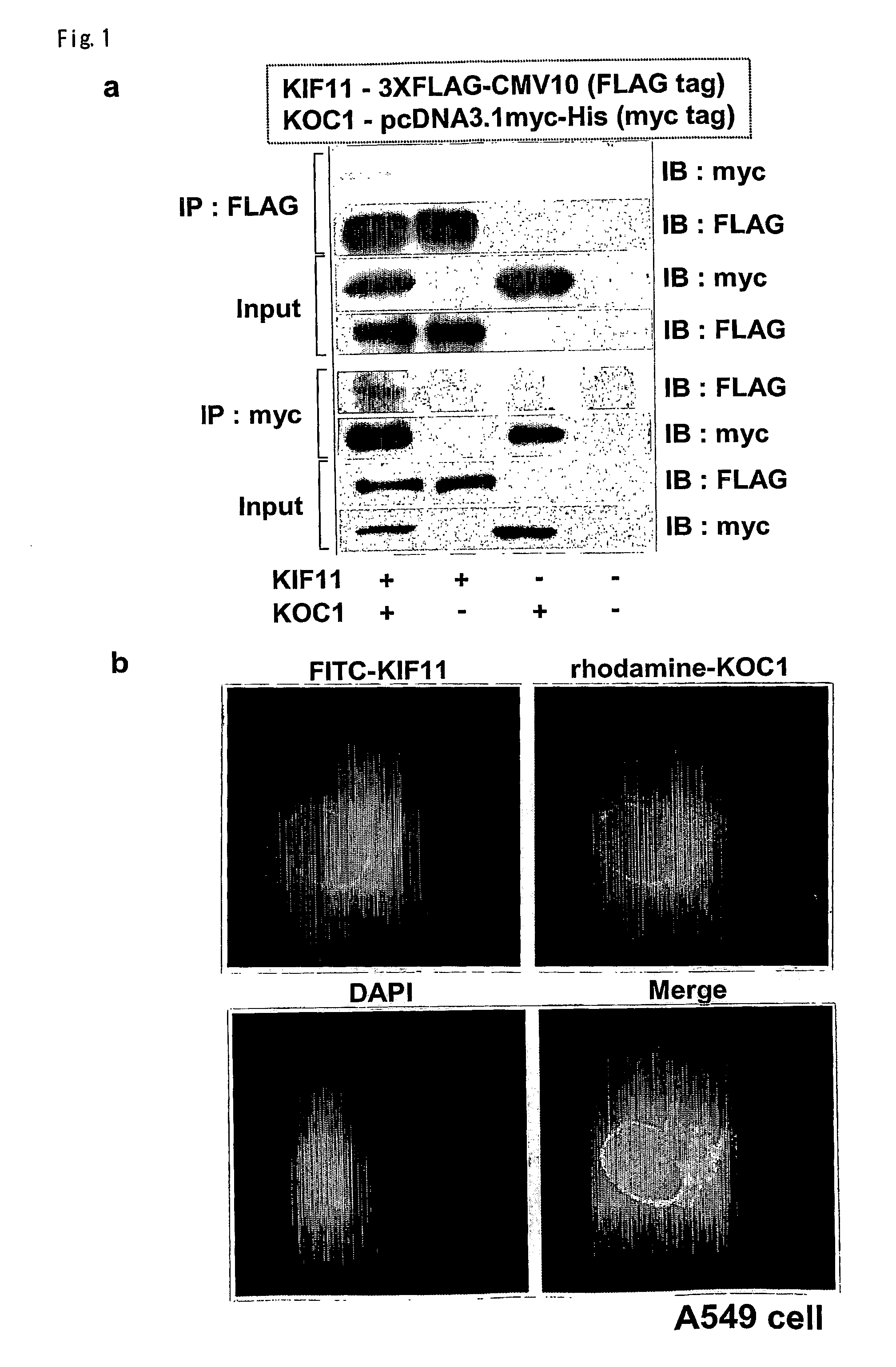
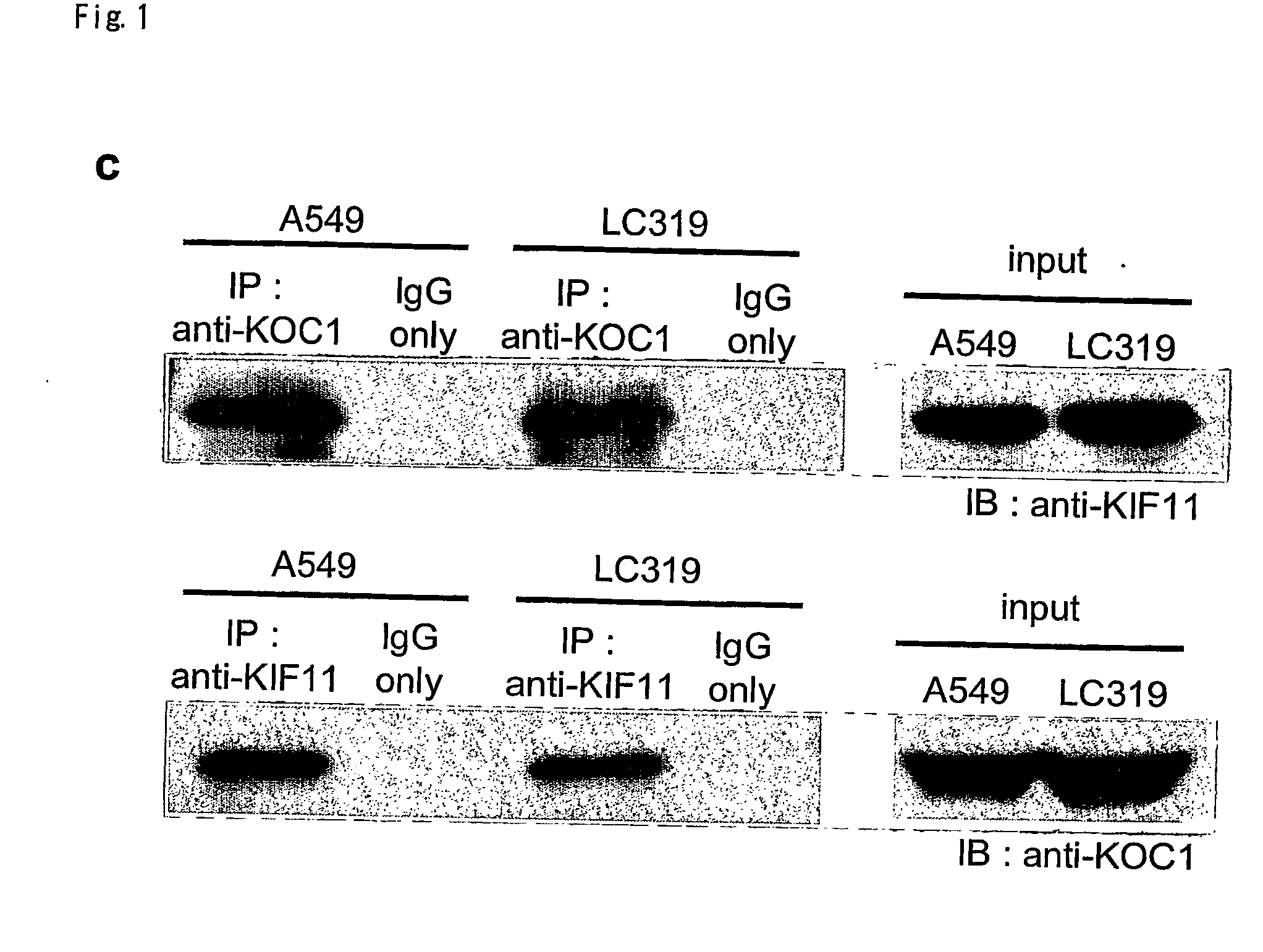
Disclosed are methods for detecting non-small cell lung cancer (NSCLC) using differentially expressed genes KIF11, GHSR1b, NTSR1, and FOXM1. Also disclosed are methods of identifying compounds for treating and preventing NSCLC, based on the interaction between KOC1 and KIF11, or NMU and GHSR1b or NTSR1.
Owner:ONCOTHERAPY SCI INC
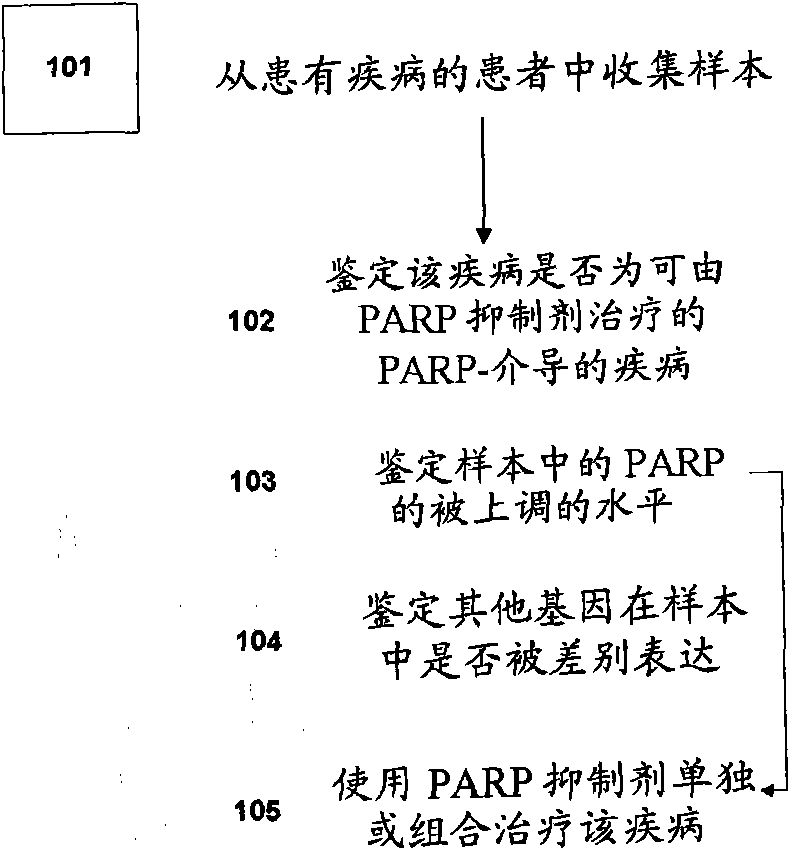
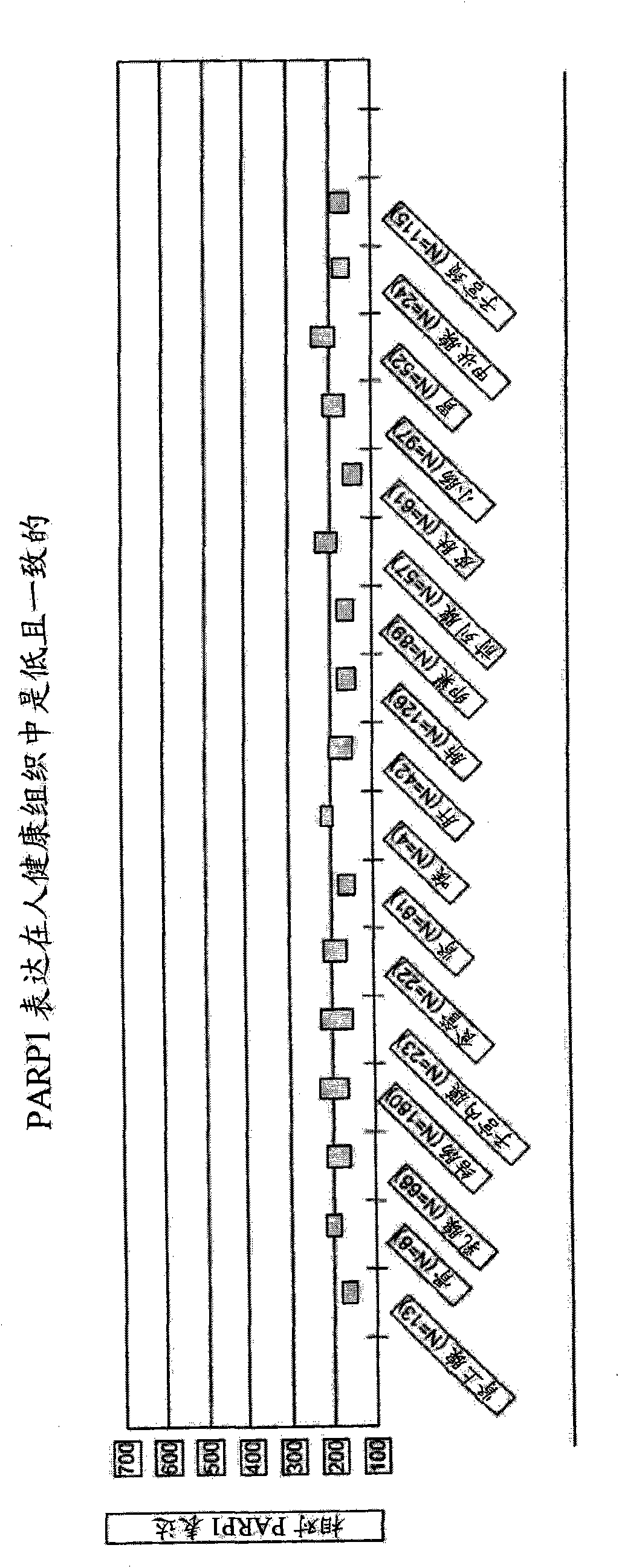
Methods of diagnosing and treating parp-mediated diseases
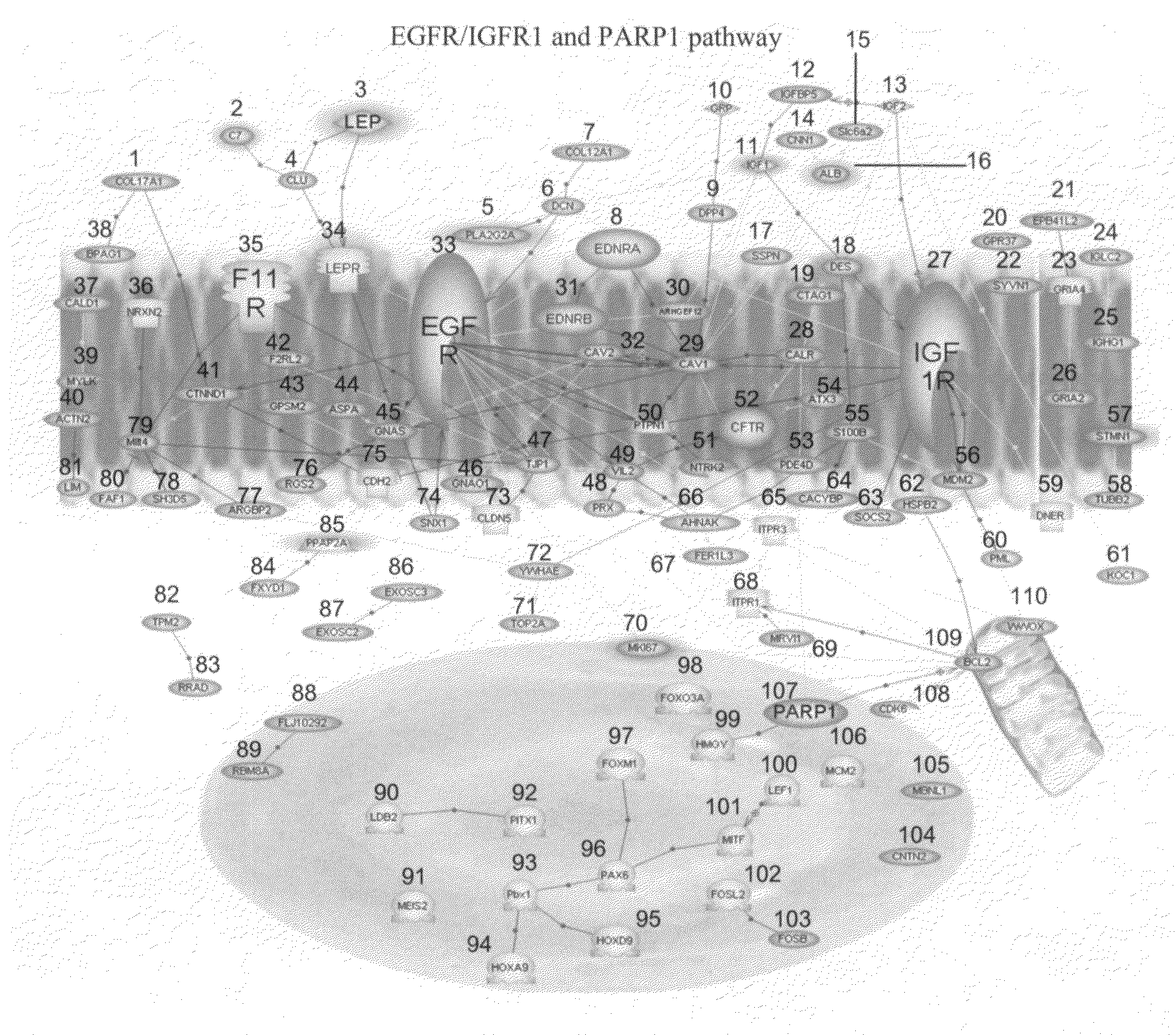
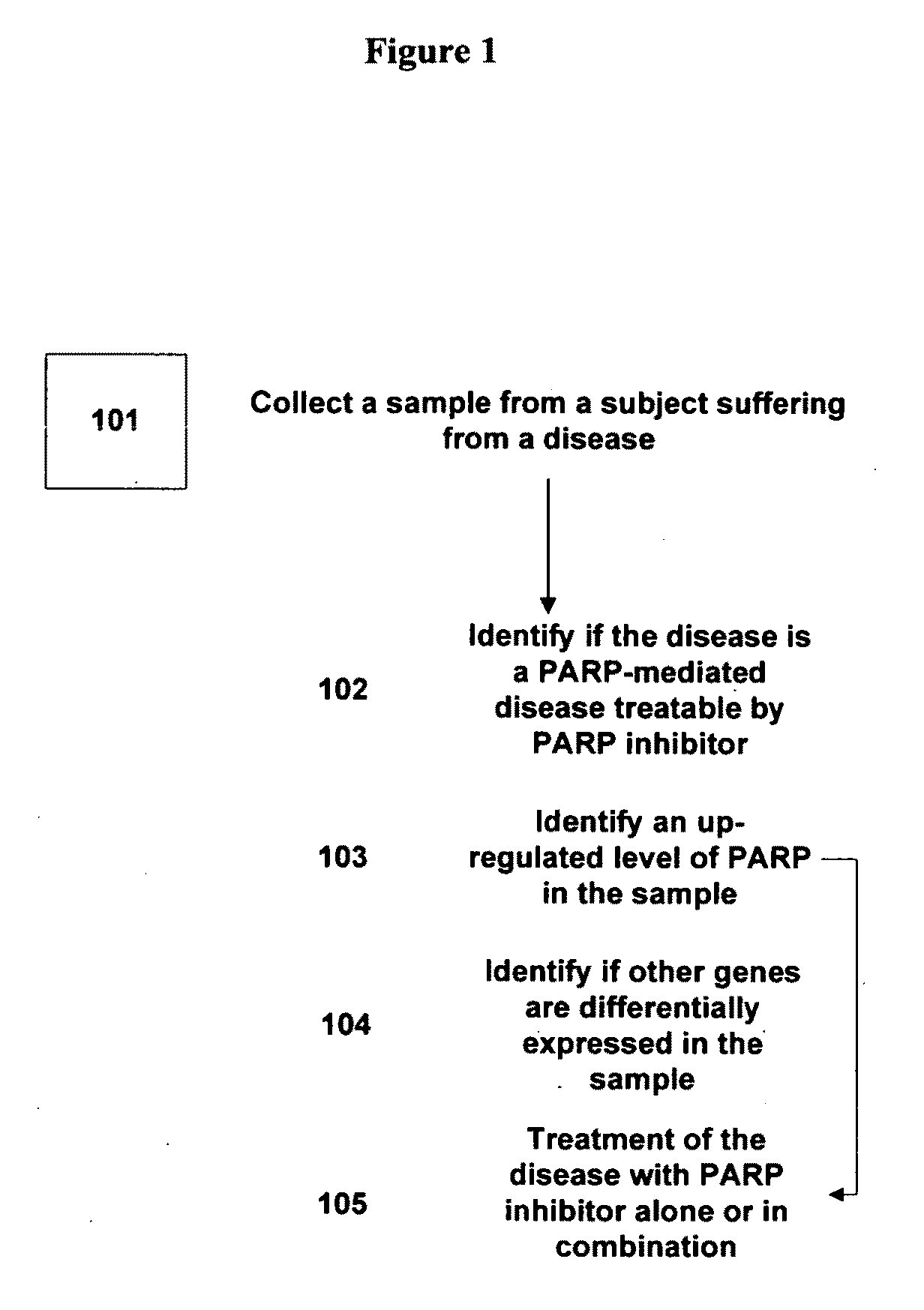
Disclosed are methods of identifying a disease treatable with modulators of differentially expressed genes in a disease, including at least PARP modulators, by identifying the level of expression of differentially expressed genes, including at least PARP, in a plurality of samples from a population, making a decision regarding identifying the disease treatable by modulators to the differentially expressed genes wherein the decision is made based on the level of expression of the differentially expressed genes. The method can further comprise treating the disease in a subject population with modulators of identified differentially expressed genes. The methods relate to identifying up-regulated expression of identified differentially-expressed genes in a disease and making a decision regarding the treatment of the disease. The level of expression of the differentially expressed genes in a disease can also help in determining the efficacy of the treatment with modulators to the differentially expressed genes.
Owner:BIPAR SCI INC
Proton-based transcriptome sequencing data comparison and analysis method and system
InactiveCN104657628AImprove accuracyImprove reliabilitySpecial data processing applicationsDifferentially expressed genesSingle-nucleotide polymorphism
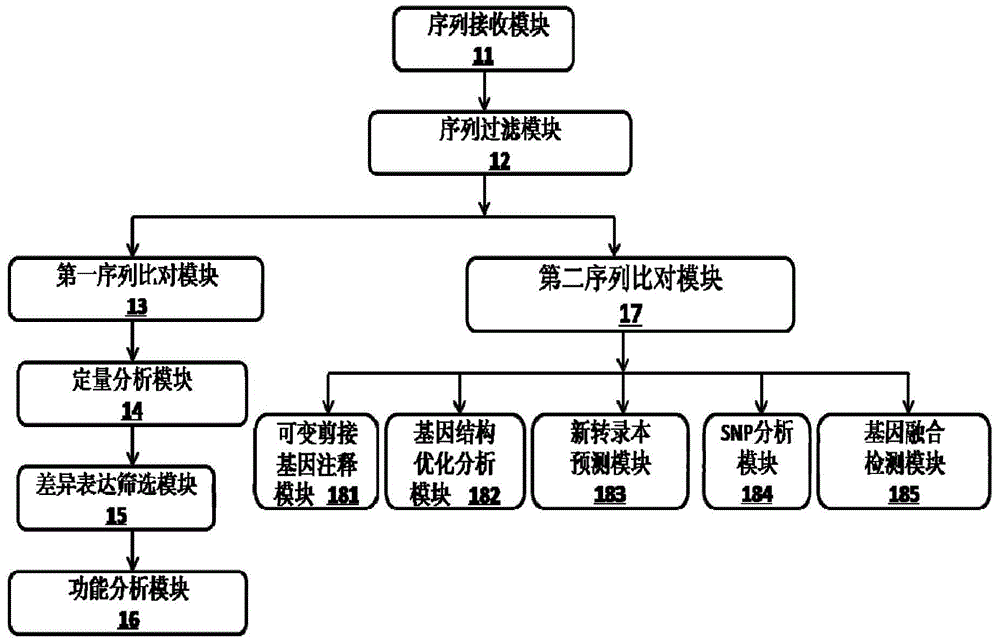
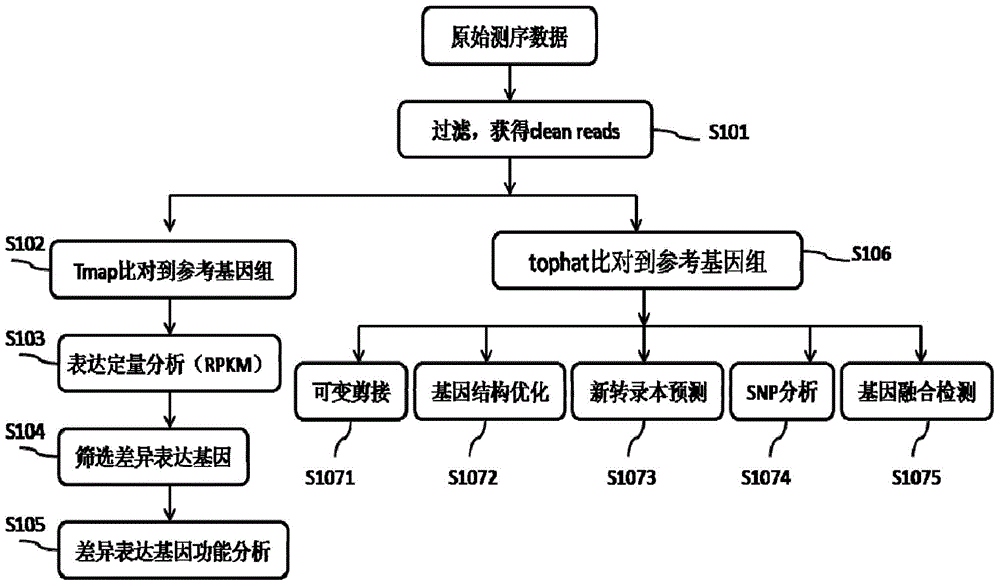
The invention provides a Proton-based transcriptome sequencing data comparison and analysis method and system. The method comprises the following steps: acquiring original sequencing data of at least two transcriptomes of a certain species by virtue of a Proton sequencing platform; filtering unqualified data to obtain clean reads; performing first-step analysis and second-step analysis, wherein the first-step analysis comprises the steps of comparing the clean reads with a reference genome of the species respectively, performing transcript quantitative analysis, screening significantly differently expressed genes and performing significantly differently expressed gene function analysis; the second-step analysis comprises the steps of comparing the clean reads to the reference genome of the species respectively, performing alternative splicing analysis, performing gene structure optimization analysis, performing new transcript prediction, performing SNP (Single Nucleotide Polymorphism) analysis and performing gene fusion detection. According to the method and the system, the transcriptome sequencing data comparison and analysis accuracy and reliability can be improved.
Owner:BGI TECH SOLUTIONS
Methods and Kits for the Prediction of Therapeutic Success, Recurrence Free and Overall Survival in Cancer Therapies
InactiveUS20080305962A1Address bad outcomesLess aggressiveMicrobiological testing/measurementLibrary screeningTherapeutic antibodyAnticancer chemotherapy
The invention provides novel compositions, methods and uses, for the prediction, diagnosis, prognosis, prevention and treatment of malignant neoplasia and cancer. The invention further relates to genes that are differentially expressed in tissue of cancer patients versus those of normal “healthy” tissue. Differentially expressed genes for the identification of patients which are likely to respond to chemotherapy are also provided. The present invention relates to methods for prognosis the prediction of therapeutic success in cancer therapy. In a preferred embodiment of the invention it relates to methods for prediction of therapeutic success of combinations of signal transduction inhibitors, therapeutic antibodies, radio- and chemotherapy. The methods of the invention are based on determination of expression levels of 48 human genes which are differentially expressed prior to the onset of anti-cancer chemotherapy. The methods and compositions of the invention are most useful in the investigation of advanced colorectal cancer, but are useful in the investigation of other types of cancer and therapies as well.
Owner:SIEMENS HEALTHCARE DIAGNOSTICS INC
Differentially expressed genes involved in cancer, the polypeptides encoded thereby, and methods of using the same
The invention relates nucleic acids and their encoded polypeptides, whose expression is modulated in cancer or tumor cells. The invention further relates to methods useful for treating or modulating cancer or tumors in mammals in need of such biological effect. This includes the diagnosis and treatment of oncological disorders. Additionally, the present invention further relates to the use of antibodies against the polypeptides of the present invention as diagnostic probes or as therapeutic agents as well as the use of polynucleotide sequences encoding the polypeptides of the present invention as diagnostic probes or therapeutic agents for the treatment of a broad range of pathological states.
Owner:PHARMACIA CORP
Methods of diagnosing and treating PARP-mediated diseases
Disclosed are methods of identifying a disease treatable with modulators of differentially expressed genes in a disease, including at least PARP modulators, by identifying the level of expression of differentially expressed genes, including at least PARP, in a plurality of samples from a population, making a decision regarding identifying the disease treatable by modulators to the differentially expressed genes wherein the decision is made based on the level of expression of the differentially expressed genes. The method can further comprise treating the disease in a subject population with modulators of identified differentially expressed genes. The methods relate to identifying up-regulated expression of identified differentially-expressed genes in a disease and making a decision regarding the treatment of the disease. The level of expression of the differentially expressed genes in a disease can also help in determining the efficacy of the treatment with modulators to the differentially expressed genes.
Owner:BIPAR SCI INC
Endothelial cell expression patterns
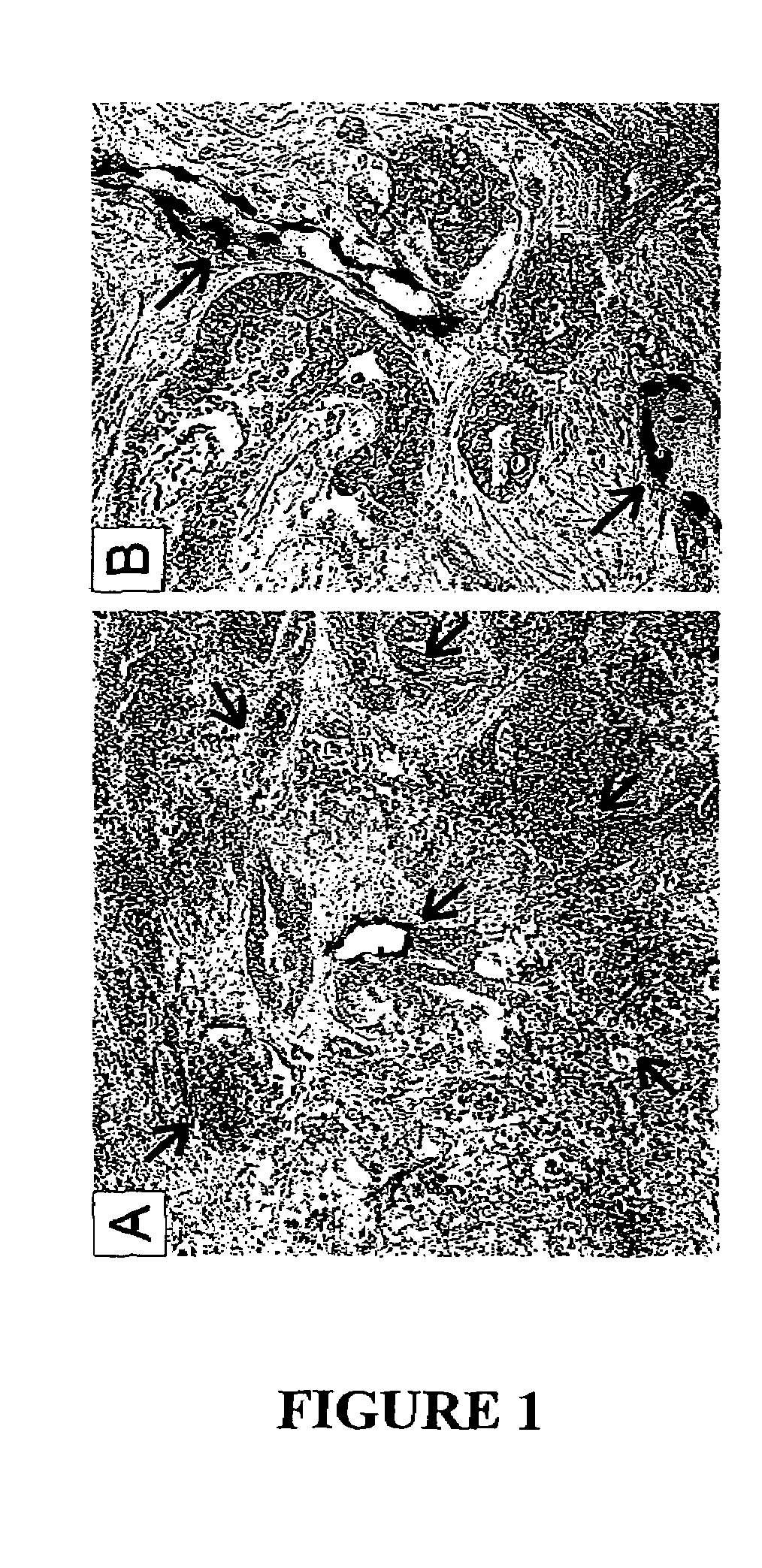
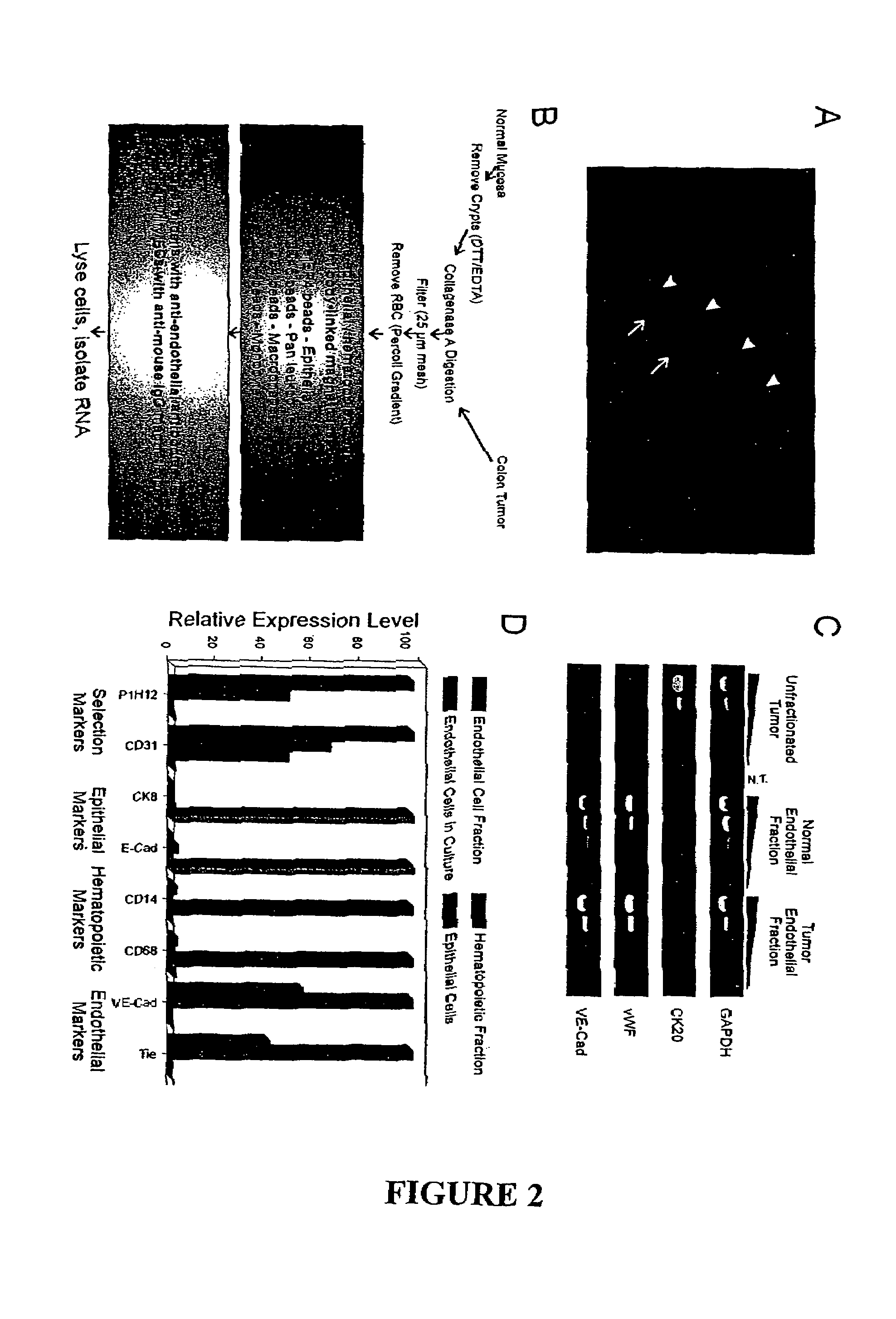
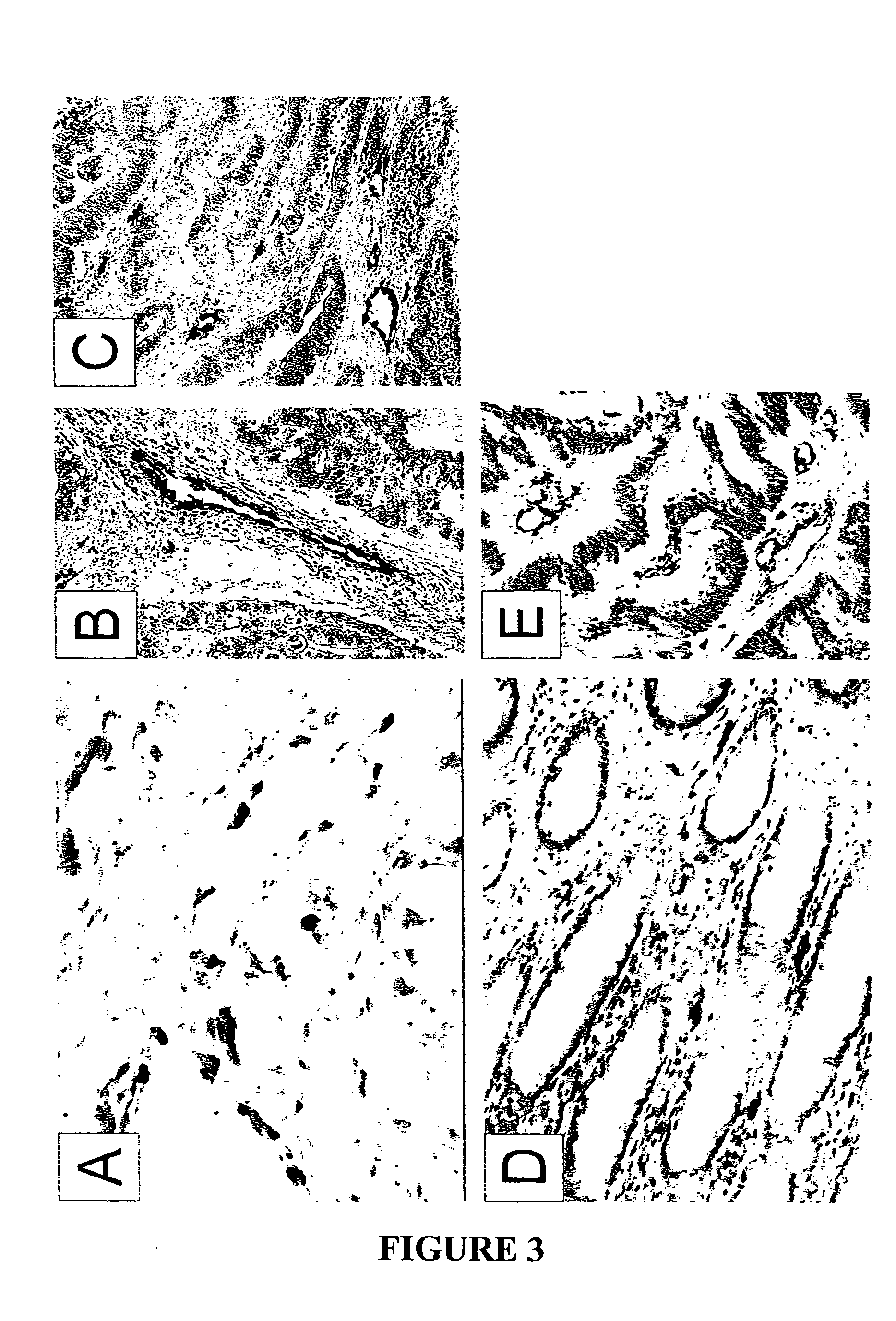
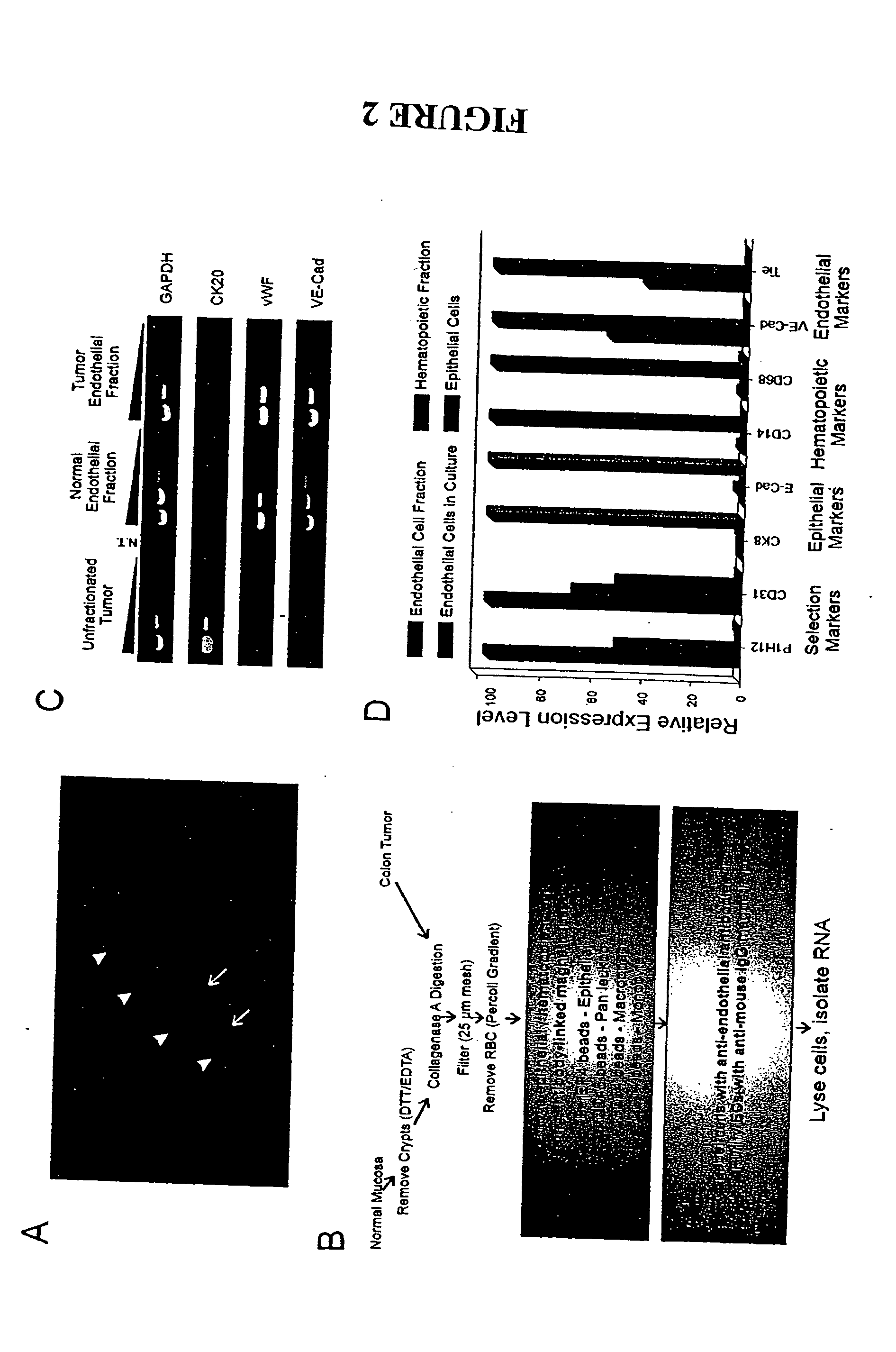
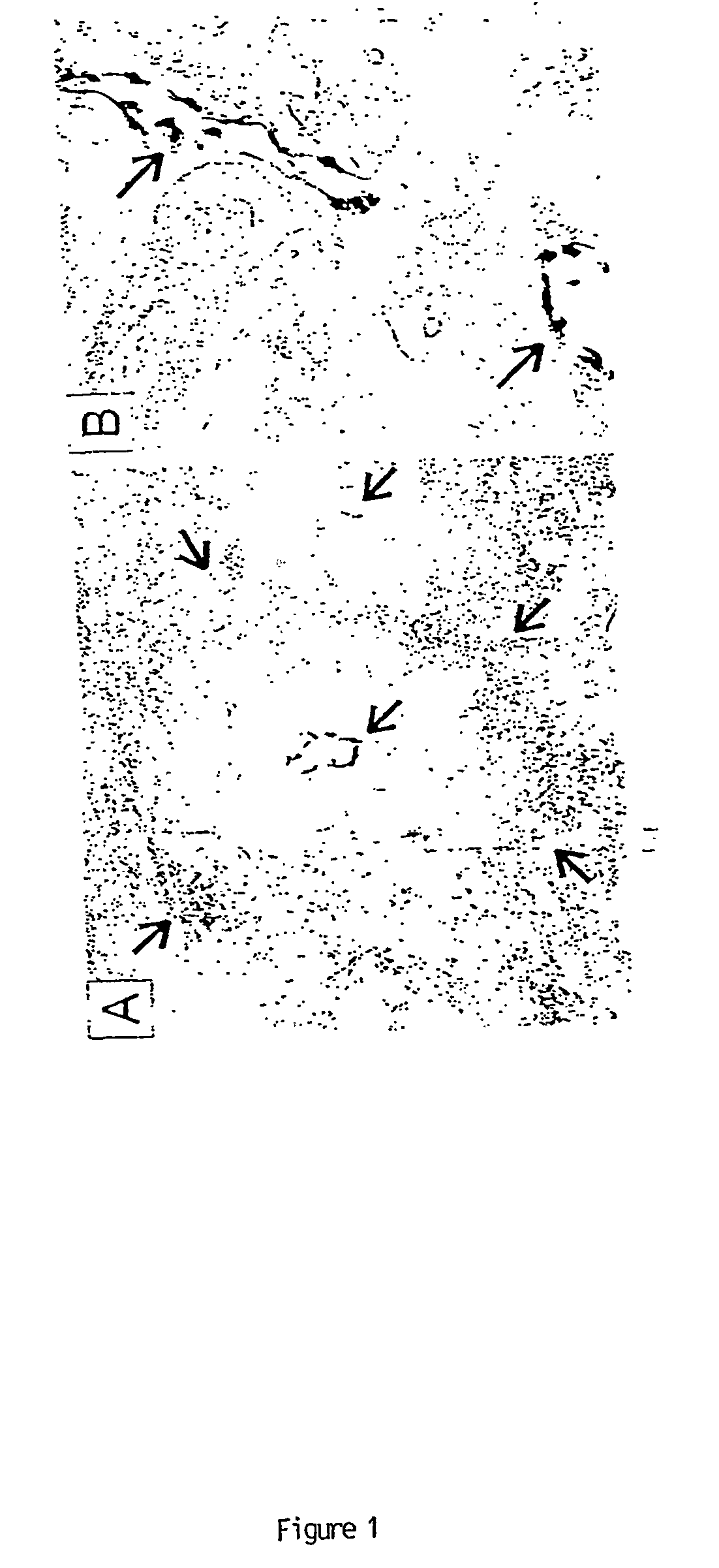
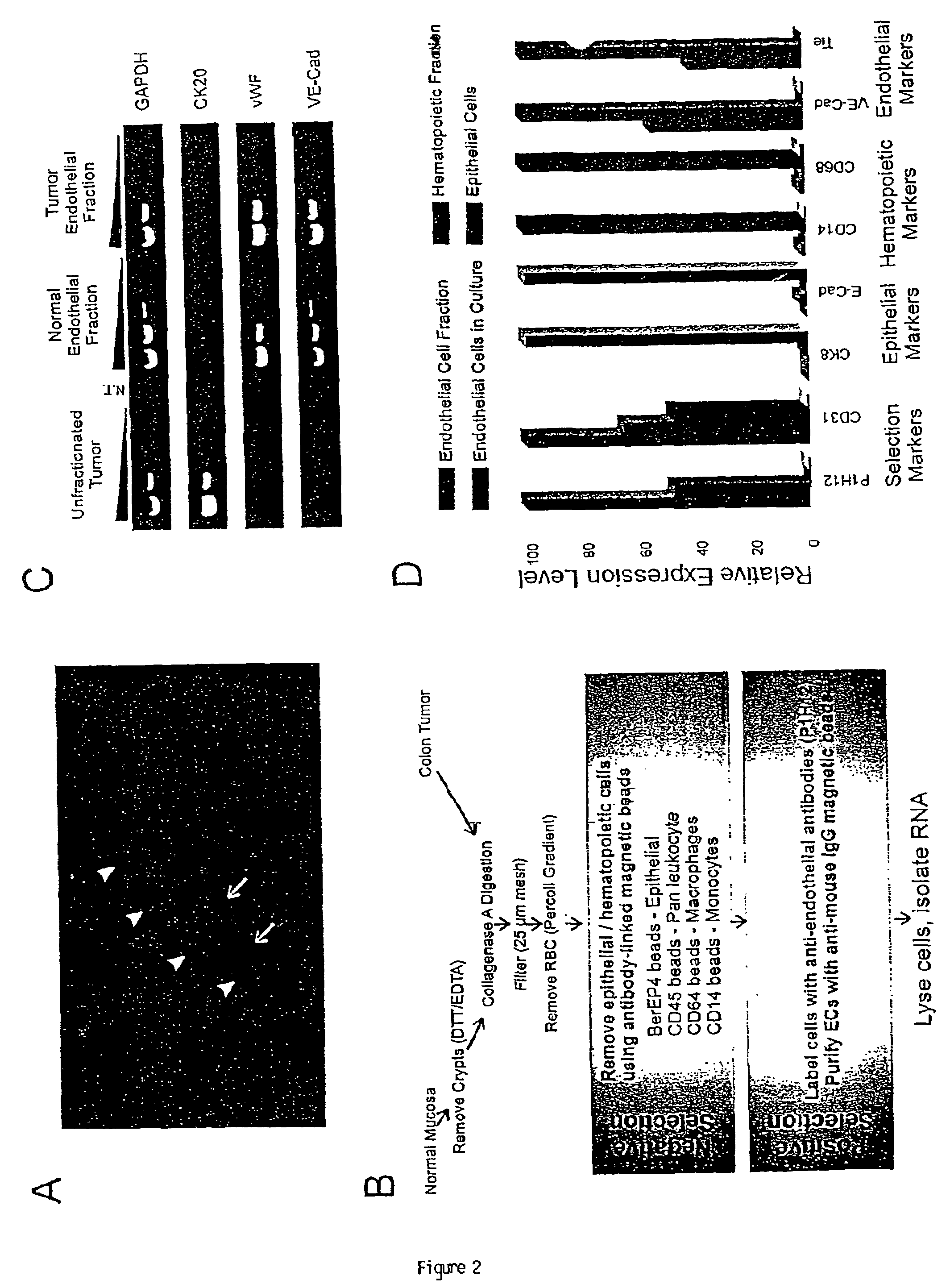
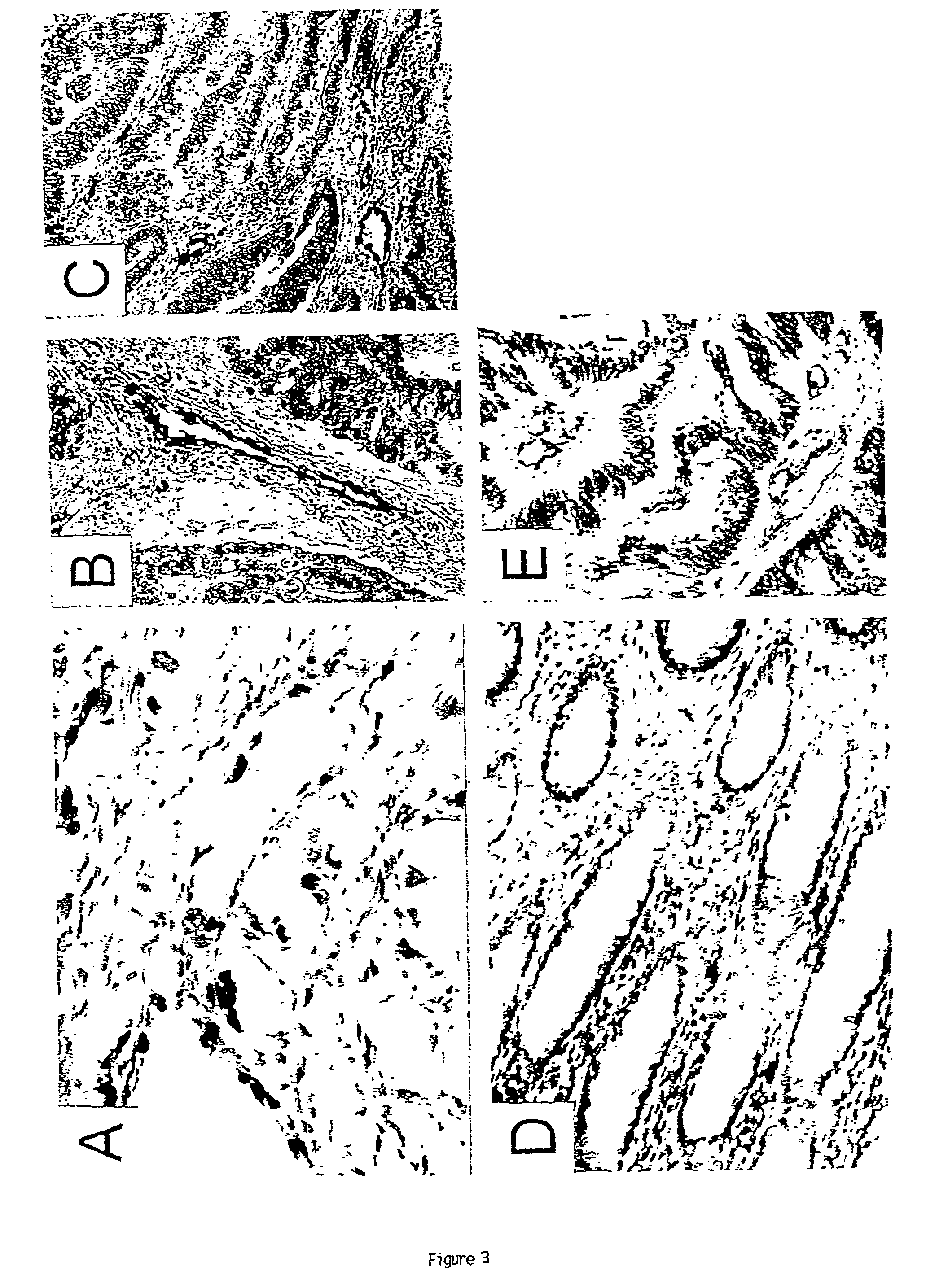
To gain a better understanding of tumor angiogenesis, new techniques for isolating endothelial cells (ECs) and evaluating gene expression patterns were developed. When transcripts from ECs derived from normal and malignant colorectal tissues were compared with transcripts from non-endothelial cells, over 170 genes predominantly expressed in the endothelium were identified. Comparison between normal- and tumor-derived endothelium revealed 79 differentially expressed genes, including 46 that were specifically elevated in tumor-associated endothelium. Experiments with representative genes from this group demonstrated that most were similarly expressed in the endothelium of primary lung, breast, brain, and pancreatic cancers as well as in metastatic lesions of the liver. These results demonstrate that neoplastic and normal endothelium in humans are distinct at the molecular level, and have significant implications for the development of anti-angiogenic therapies in the future.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
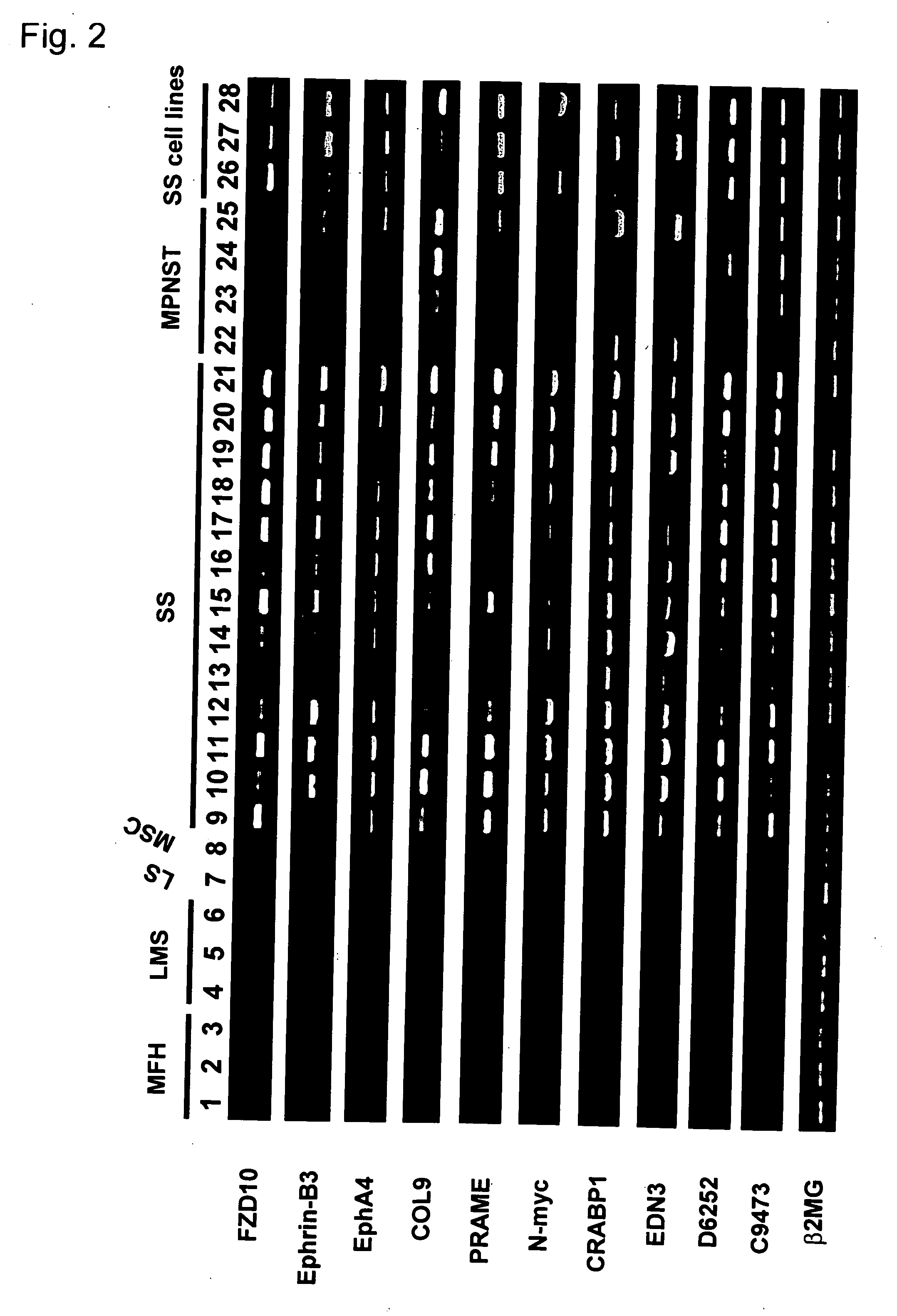
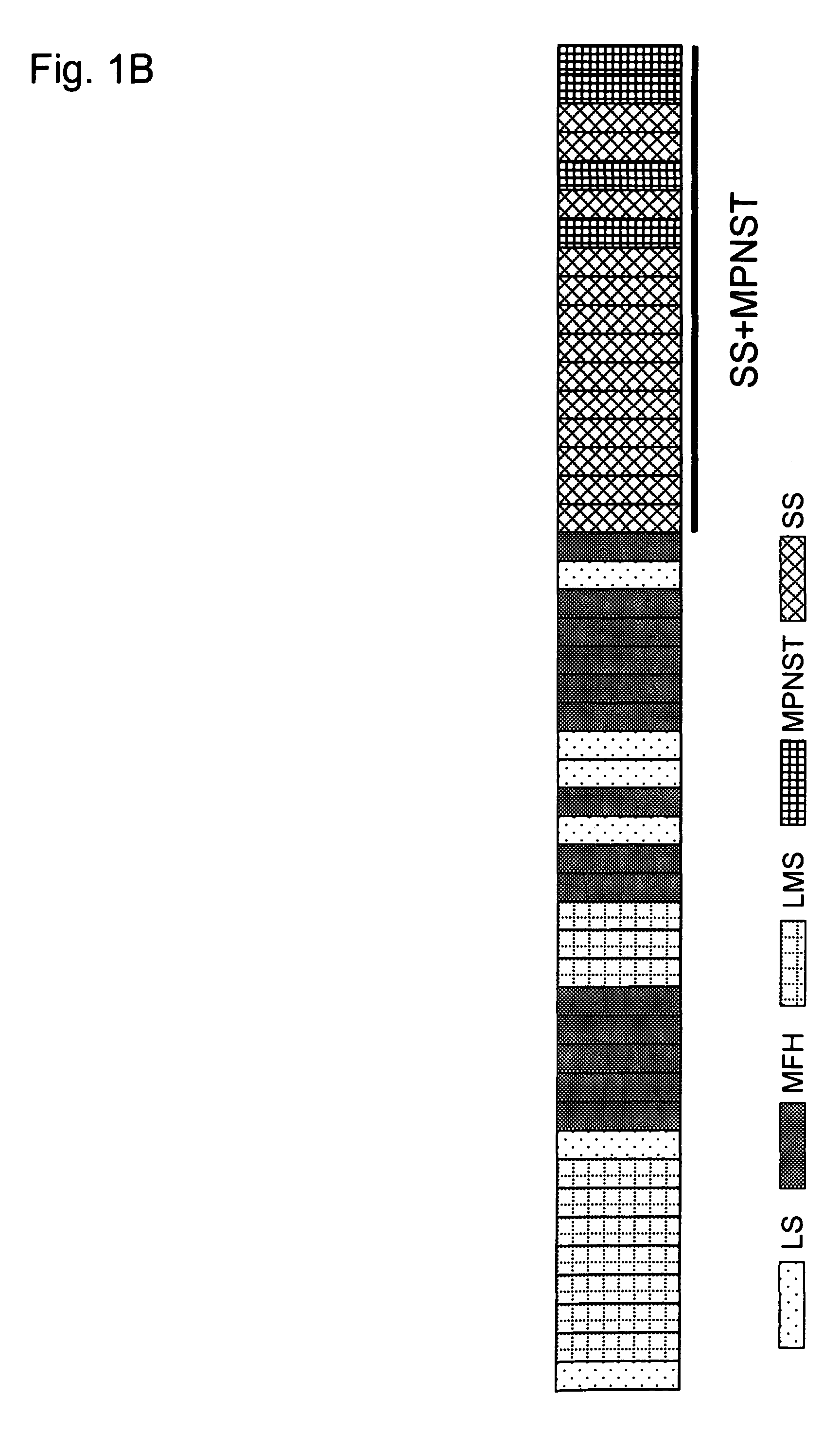
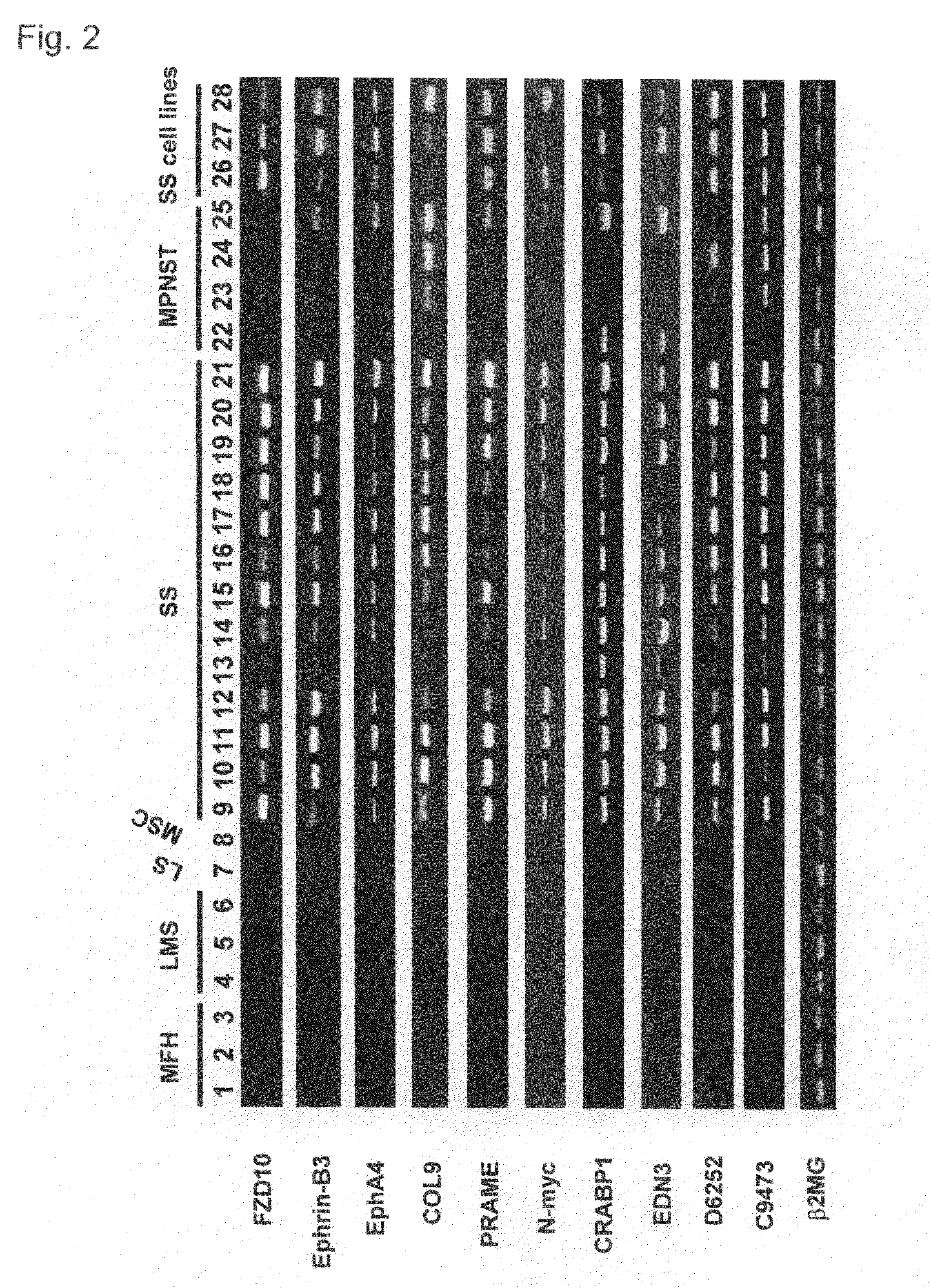
Method for treating synovial sarcoma
ActiveUS20050272063A1Prevent adverse side effectsMicrobiological testing/measurementImmunoglobulins against cell receptors/antigens/surface-determinantsSynovial sarcomaDifferentially expressed genes
Methods of detecting synovial sarcoma using differentially expressed genes are disclosed. Also disclosed are methods of identifying agents for treating synovial sarcoma. Further, a method for treating or preventing a disease that is associated with Frizzled homologue 10 (FZD10) in a subject is provided.
Owner:ONCOTHERAPY SCI INC
Methods and Kits for Predicting and Monitoring Direct Response to Cancer Therapy
InactiveUS20090203533A1Microbiological testing/measurementLibrary screeningCancer therapyDirect response
The invention provides novel compositions, methods and uses, for the prediction, diagnosis, prognosis, prevention and treatment of malignant neoplasia and breast cancer. The invention further relates to genes that are differentially expressed in breast tissue of breast cancer patients versus those of normal “healthy” tissue. Differentially expressed genes for the identification of patients which are likely to respond to chemotherapy are also provided.
Owner:SIEMENS HEALTHCARE DIAGNOSTICS GMBH
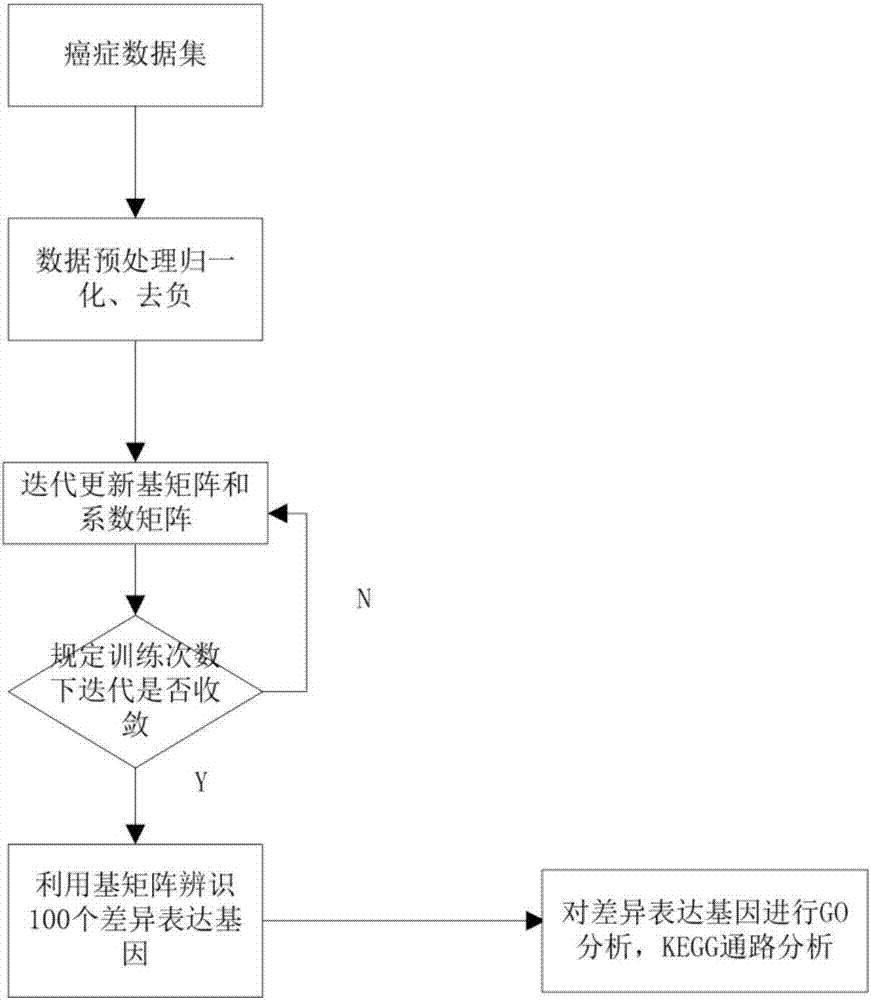
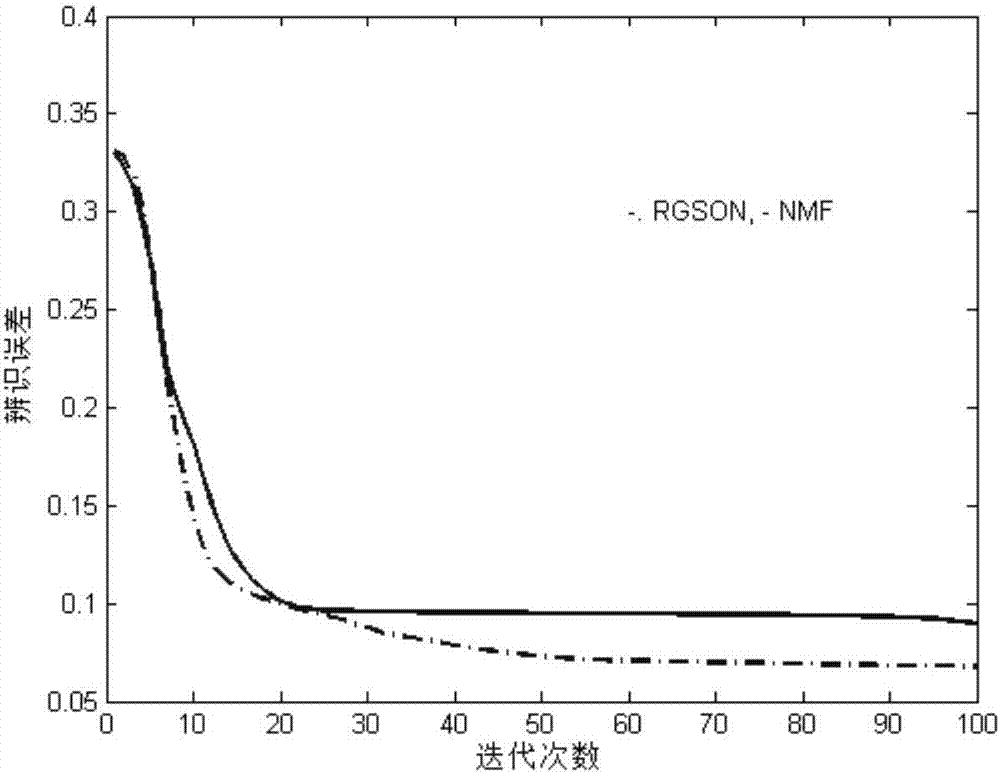
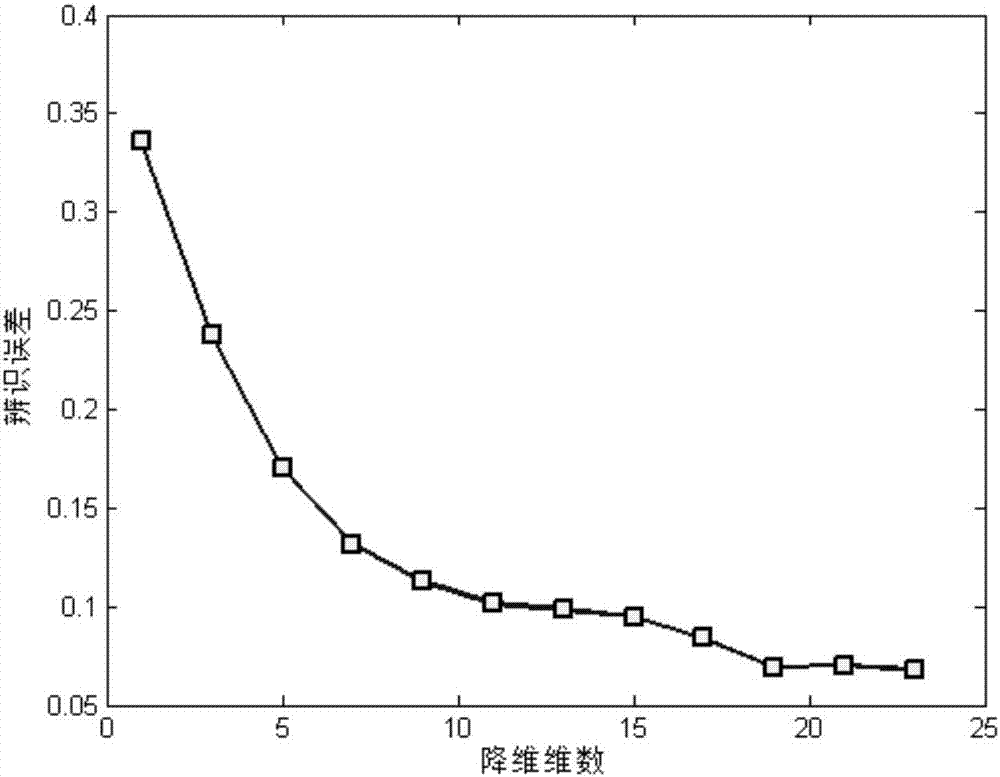
Differentially expressed gene identification method based on combined constraint non-negative matrix factorization
ActiveCN107016261AEffective decomposition resultsEfficient Sparse Decomposition ResultsSpecial data processing applicationsData setAlgorithm
The invention discloses a differentially expressed gene identification method based on combined constraint non-negative matrix factorization. The method comprises the following steps of 1, representing a cancer-gene expression data set with a non-negative matrix X, 2, constructing a diagonal matrix Q and an element-full matrix E, 3, introducing manifold learning in the classical non-negative matrix factorization method, conducting orthogonal-constraint sparseness and constraint on a coefficient matrix G, and obtaining a combined constraint non-negative matrix factorization target function, 4, calculating the target function, and obtaining iterative formulas of a basis matrix F and the coefficient matrix G, 5, conducting semi-supervision non-negative matrix factorization on the non-negative data set X, and obtaining the basis matrix F and the coefficient matrix G after iteration convergence, 6, obtaining an evaluation vector (the formula is shown in the description), sorting elements in the evaluation vector (the formula is shown in the description) from large to small according to the basis matrix F, and obtaining differentially expressed genes, 7, testing and analyzing the identified differentially expressed genes through a GO tool. The identification method can effectively extract the differentially expressed genes where cancer data is concentrated, and be applied in discovering differential features in a human disease gene database. The identification method has important clinical significance for early diagnosis and target treatment of diseases.
Owner:HANGZHOU HANGENE BIOTECH CO LTD
Endothelial cell expression patterns
InactiveUS20050142138A1Reduced activityHigh activitySenses disorderAntipyreticAbnormal tissue growthMolecular level
To gain a better understanding of tumor angiogenesis, new techniques for isolating endothelial cells (ECs) and evaluating gene expression patterns were developed. When transcripts from ECs derived from normal and malignant colorectal tissues were compared with transcripts from non-endothelial cells, over 170 genes predominantly expressed in the endothelium were identified. Comparison between normal- and tumor-derived endothelium revealed 79 differentially expressed genes, including 46 that were specifically elevated in tumor-associated endothelium. Experiments with representative genes from this group demonstrated that most were similarly expressed in the endothelium of primary lung, breast, brain, and pancreatic cancers as well as in metastatic lesions of the liver. These results demonstrate that neoplastic and normal endothelium in humans are distinct at the molecular level, and have significant implications for the development of anti-angiogenic therapies in the future.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Construction method for specific cancer differential expression gene regulation and control network
InactiveCN105740651AHigh precisionImprove calculation accuracyHybridisationSpecial data processing applicationsDifferentially expressed genesBioinformatics
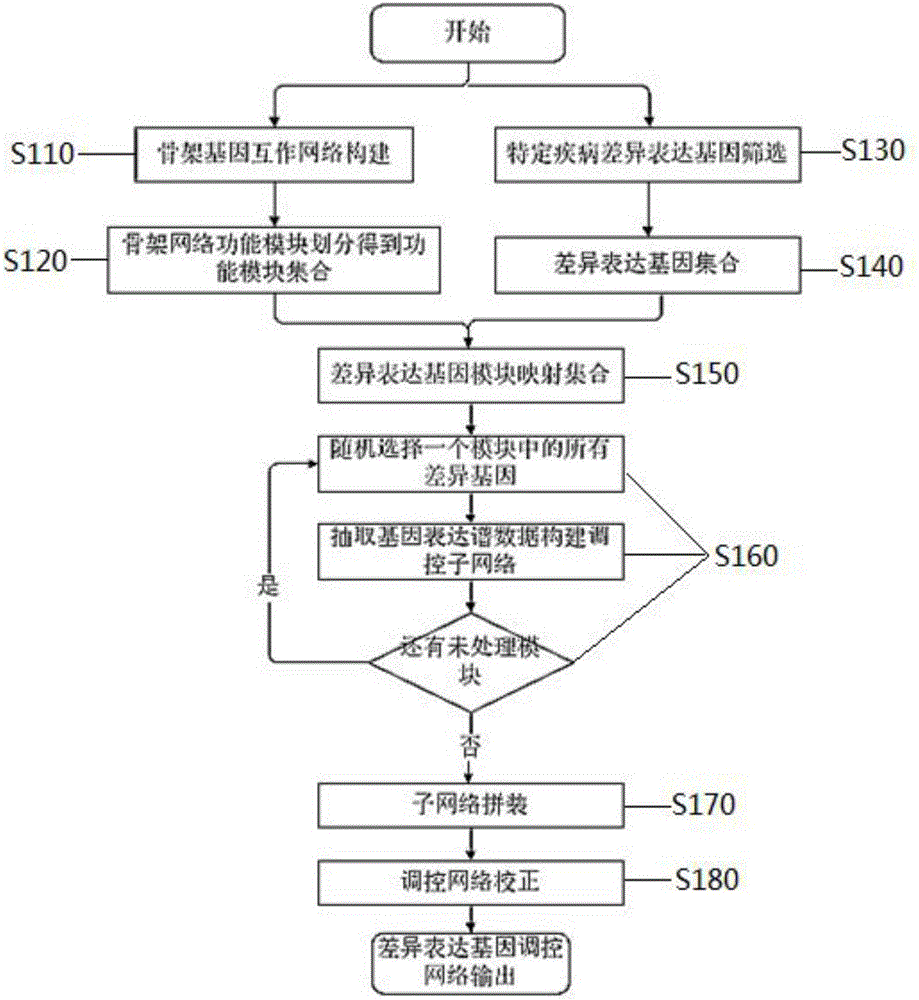
The invention discloses a construction method for a specific cancer differential expression gene regulation and control network.The method includes the following steps of firstly, constructing a framework gene interaction network according to function similarity weight numbers between genes; secondly, conducting module division on the framework gene interaction network through a segmenting method; thirdly, screening out differential expression genes through complete genome methylation data; fourthly, classifying the screened-out differential expression genes according to functions; fifthly, using all the differential expression genes mapped to each same function module as a function classification; sixthly, constructing the regulation and control network of all the genes in each function classification function; seventhly, conducting sub-network splicing under guidance of a framework network.The calculation complexity is greatly reduced, and high precision is achieved.
Owner:JILIN UNIV
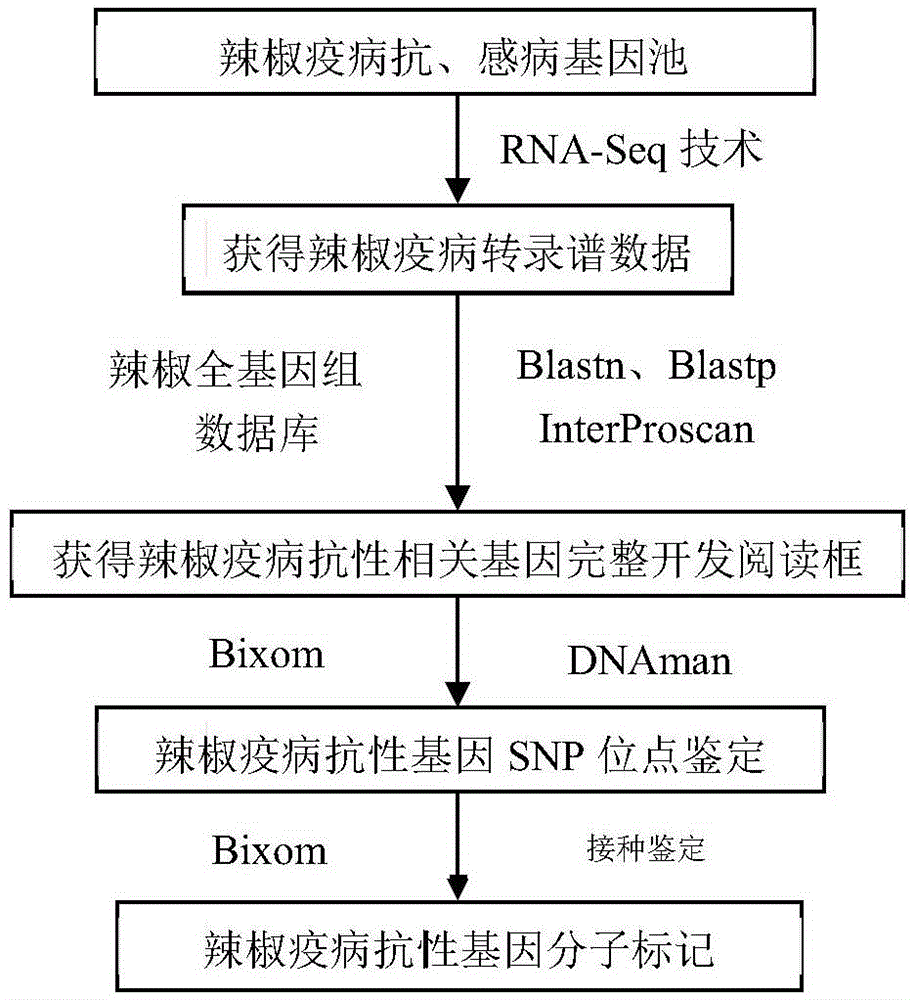
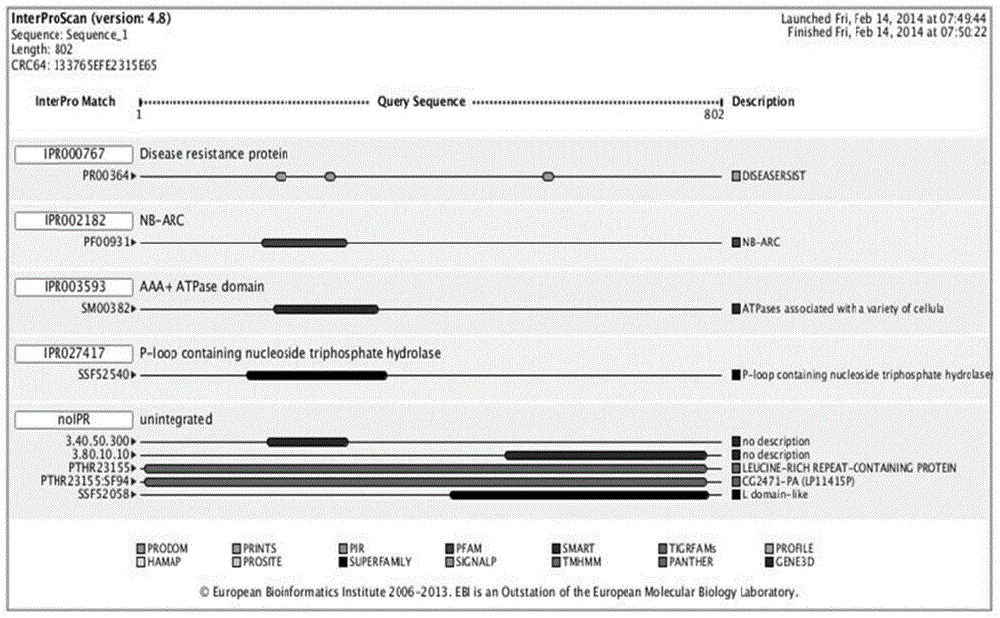
Method for obtaining capsicum phytophthora resistance candidate gene and molecular marker, and application
InactiveCN104560973AAccurate identificationLarge amount of data informationMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyData information
The invention relates to a method for obtaining a capsicum phytophthora resistance candidate gene and a molecular marker, and application. The method is used for obtaining the capsicum phytophthora resistance candidate gene by utilizing capsicum phytophthora transcriptome and whole-genome sequencing data information, differentially-expressed gene identification, bioinformatics analysis, molecular marker development and phytophthora inoculation identification and belongs to the technical field of capsicum biology. The method comprises the following steps: sequencing a phytophthora resistant and susceptible gene pool transcriptome obtained after phytophthora inoculation of an F2 population constructed by capsicum highly-resistant and highly-susceptible phytophthora materials, performing expression analysis and functional annotation on differential genes, extracting DNAs (Desoxvribose Nucleic Acid) of a capsicum phytophthora highly-resistant and highly-susceptible phytophthora material genome, performing primer design and PCR (Polymerase Chain Reaction) amplification, performing sequence difference analysis and SNP site identification, performing SNP specific primer design and validity verification, and performing other steps to efficiently obtain the capsicum phytophthora resistance candidate gene and the molecular marker. According to the method, the capsicum phytophthora resistance candidate gene can be accurately identified, and the effective molecular marker can be developed.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
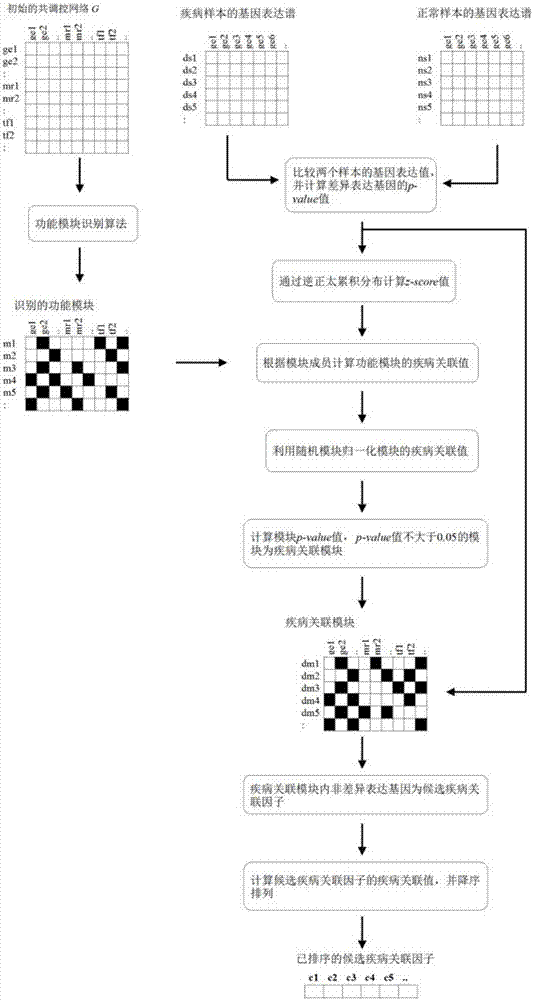
Method and system for recognizing disease related factor on basis of functional module
InactiveCN106874706AImprove accuracyImprove recognition accuracyBiostatisticsProteomicsDiseaseMedicine
The invention provides a method and a system for recognizing a disease related factor on the basis of a functional module. The method comprises the following steps: 1) recognizing the functional module which includes a transcription factor, a miRNA and a target gene on a human co-regulatory network; 2) acquiring a p-value value of a differential expression gene and confirming a disease related functional module according to the combination of the differential expression gene in the module; 3) taking the non-differential expression gene, the miRNA and the target gene in the disease related functional module as candidate disease related factors; and 4) calculating associated values of the candidate disease related factors and the disease and sequencing the candidate disease related factors according to the associated values. According to the method, various interaction relations are combined when the disease related factor is recognized; even under the condition of unknown transcription factor, miRNA and gene function, the method provided by the invention also can be used for forecasting and recognizing the disease related factor; the method provided by the invention is high in accuracy of forecasting the disease related factor.
Owner:HUNAN MICOME ZHONGJIN MEDICAL SCI & TECH DEV CO LTD
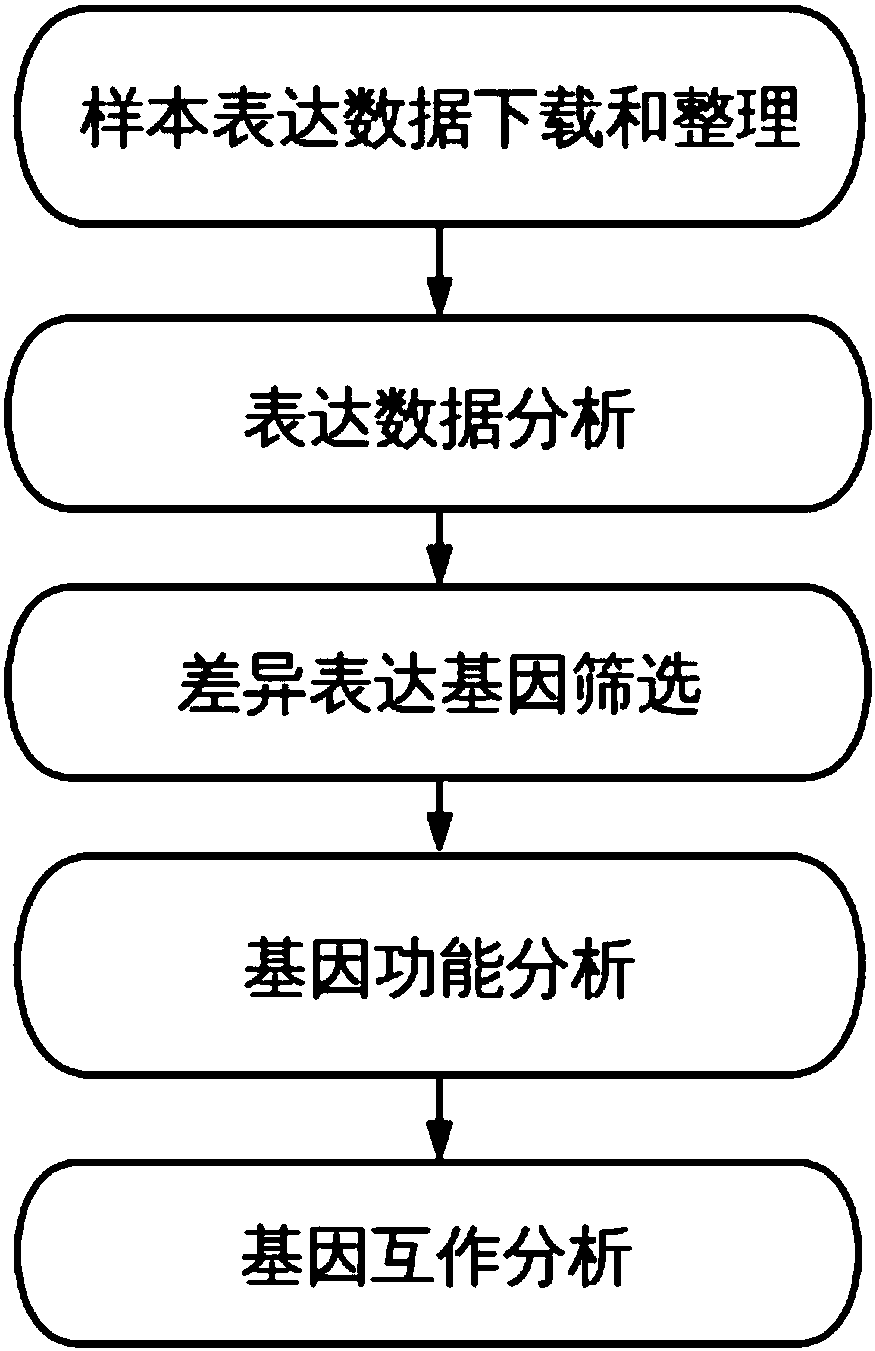
Method and system for discovering and integrating rectal cancer related genes by utilizing public data resources and analyzing functions of rectal cancer related genes, and application
ActiveCN107066835AReduce research costsImprove analysis efficiencyProteomicsGenomicsWeaknessDifferentially expressed genes
The invention discloses a method and a system for discovering and integrating rectal cancer related genes by utilizing public data resources and analyzing functions of the rectal cancer related genes, and an application. Based on the public data resources, open big data resources and diversified bioinformatics analysis means are reasonably used for performing analysis processing on mRNA expression data, and important genes related to complex diseases and functions of the important genes are identified. The method comprises the steps of sample data downloading and management; gene expression data analysis; difference expression gene screening; and gene function analysis and protein interaction analysis. According to the method and the system, the problems of weakness in integrating existing network resources, unfamiliarity with mRNA related most common databases and frontal analysis methods, incapability of independently finishing mRNA expression spectrum related bioinformatics analysis and the like can be solved; a plurality of risk pathways and genes related to the complex diseases such as the rectal cancer and the like can be discovered; and the method and the system are of important significance for biological targeted treatment of the complex diseases, biological drug research and development, pathogenesis explanation and risk prediction.
Owner:SOUTHEAST UNIV
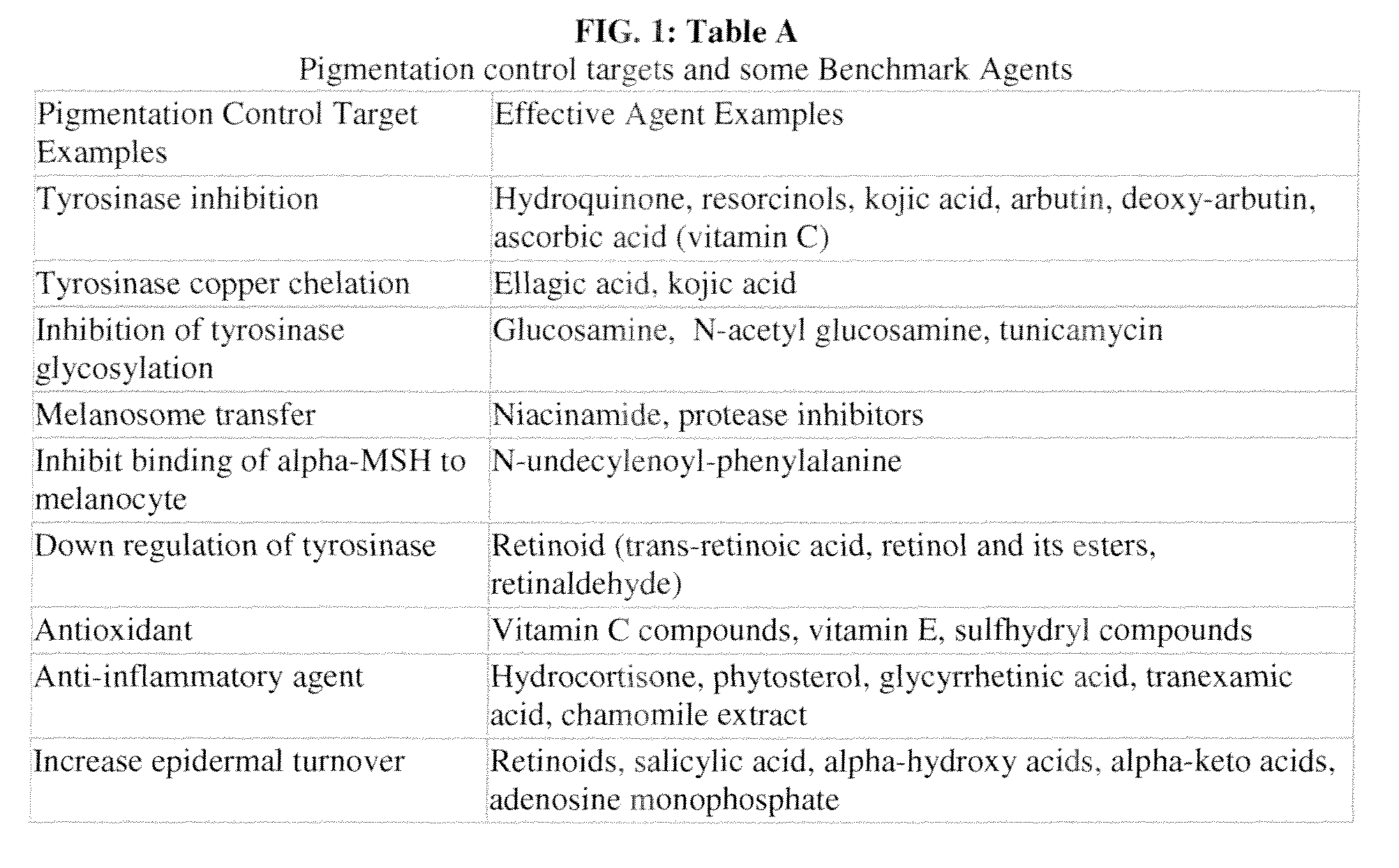
Method of Formulating a Skin-Lightening Composition
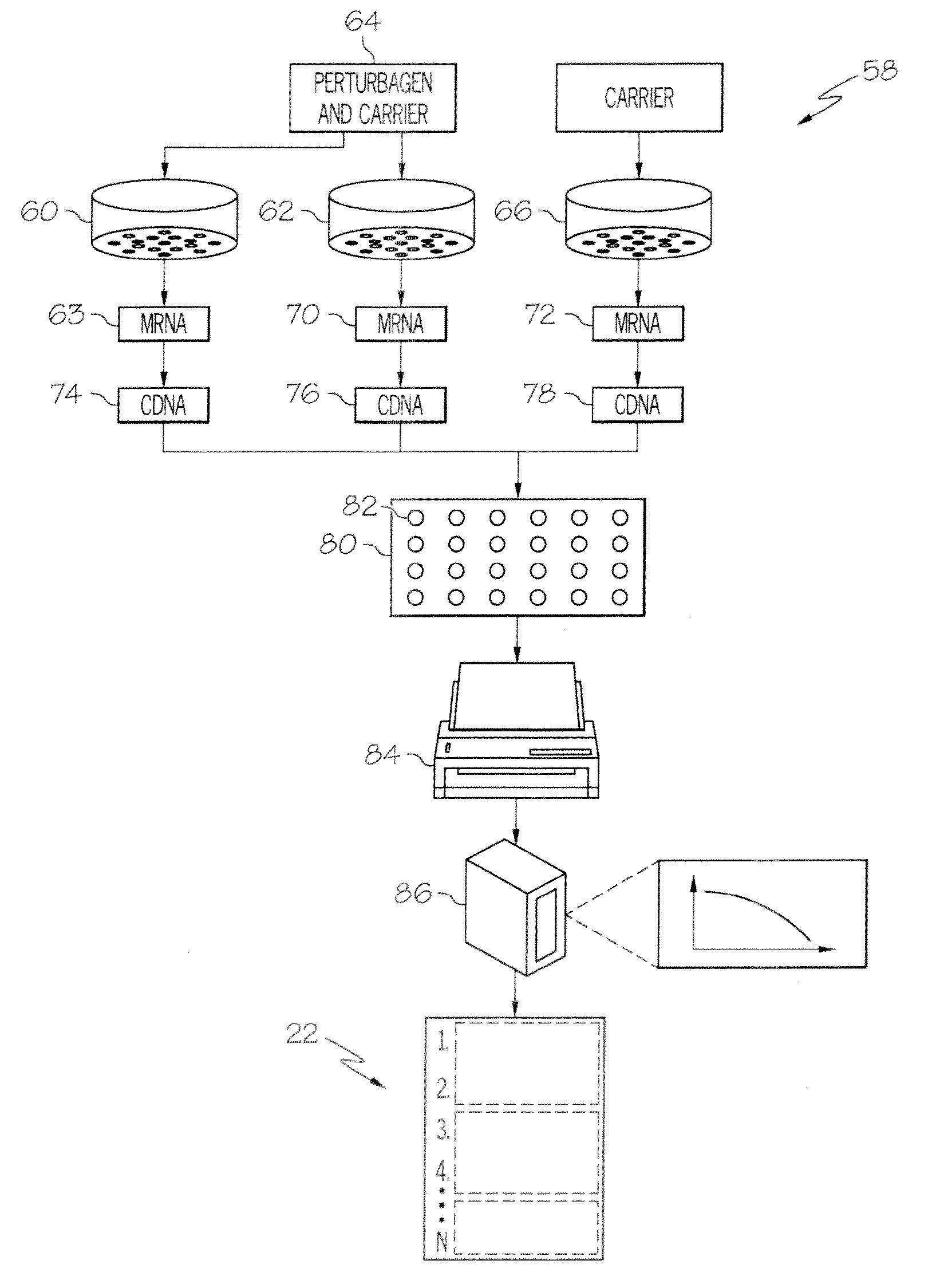
A method of formulating a skin-lightening composition by identifying actives with a data architecture and incorporating the actives into a skin care composition. The data architecture can be used to identify connections between perturbagens and genes associated with skin tone. A gene expression profile for a control human cell is compared to the gene expression profile a cell exposed to perturbagen to identify the genes which are differentially expressed. The differentially expressed genes are analyzed and arranged into an ordered list of identifiers, which along with other instances create a data architecture.
Owner:THE PROCTER & GAMBLE COMPANY
Method for treating synovial sarcoma
ActiveUS7803370B2Microbiological testing/measurementImmunoglobulins against cell receptors/antigens/surface-determinantsSynovial sarcomaDifferentially expressed genes
Methods of detecting synovial sarcoma using differentially expressed genes are disclosed. Also disclosed are methods of identifying agents for treating synovial sarcoma. Further, a method for treating or preventing a disease that is associated with Frizzled homologue 10 (FZD10) in a subject is provided.
Owner:ONCOTHERAPY SCI INC
Secreted and cytoplasmic tumor endothelial markers
InactiveUS20090233270A9Peptide/protein ingredientsGenetic material ingredientsAbnormal tissue growthHuman tumor
To gain a better understanding of tumor angiogenesis, new techniques for isolating endothelial cells (ECs) and evaluating gene expression patterns were developed. When transcripts from ECs derived from normal and malignant colorectal tissues were compared with transcripts from non-endothelial cells, over 170 genes predominantly expressed in the endothelium were identified. Comparison between normal- and tumor-derived endothelium revealed many differentially expressed genes, including a large nujber of genes that were specifically elevated in tumor-associated endothelium. Experiments with representative genes from this group demonstrated that most were similarly expressed in the endothelium of primary lung, breast, brain, and pancreatic cancers as well as in metastatic lesions fo the liver. Theses results demonstrate that neoplastic and normal endothelium in humans are distinct at the molecular level, and have significant implications for the development of anti-angiogenic.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Stochastic variable selection method for model selection
InactiveUS20050086010A1Facilitates proper identificationEasy to classifyBiostatisticsBiological testingDesign matrixModel selection
A method of identifying differentially-expressed genes includes deriving an analysis of variance (ANOVA) or analysis of covariance (ANCOVA) model for expression data associated with a number of genes; from the ANOVA or ANCOVA model, deriving a linear regression model defined at least in part by an observation vector representative of an observed subset of the gene-expression data, a design matrix of regressor variables, a vector of regression coefficients representing gene contribution to the observation vector, and a measurement error vector; and to the linear regression model, applying a hierarchical selection algorithm to designate a subset of the regression coefficients as significant regression coefficients, the selection algorithm representing at least one of the observation vector, the design matrix, and the measurement error vector as being hierarchically dependent on parameters having predetermined probabilistic properties, wherein the designated subset corresponds to a respective subset of the genes identified as differentially expressed.
Owner:CASE WESTERN RESERVE UNIV +1
Attenuated strain of Leishmania
Differentially expressed Leishmania genes and proteins are described. One differentially expressed gene (A2) is expressed at significantly elevated levels (more than about 10 fold higher) in the amastigote stage of the life cycle when the Leishmania organism is present in macrophages than in the free promastigote stage. The A2 gene encodes a 22 kD protein (A2 protein) that is recognized by kala-azar convalescent serum and has amino acid sequence homology with an S-antigen of Plasmodium falcilparum Vietnamese isolate VI. Differentially expressed Leishmania genes and proteins have utility as vaccines, diagnostic reagents, as tools for the generation of immunological reagents and the generation of attenuated variants of Leishmania.
Owner:MCGILL UNIV
Immortalized human Tuberous Sclerosis null angiomyolipoma cell and method of use thereof
InactiveUS20050266442A1Microbiological testing/measurementGenetically modified cellsDiseaseEpithelioid angiomyolipoma
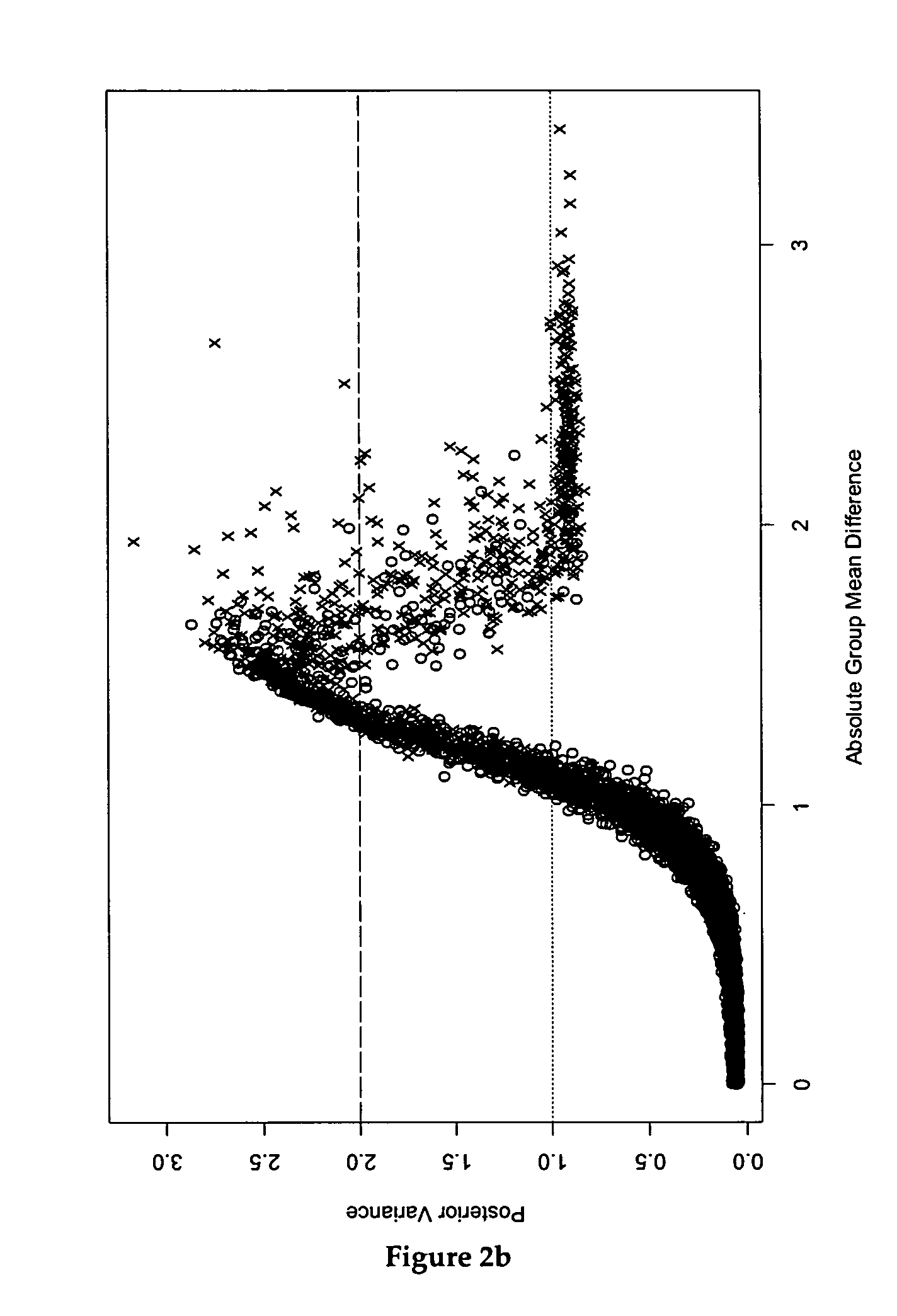
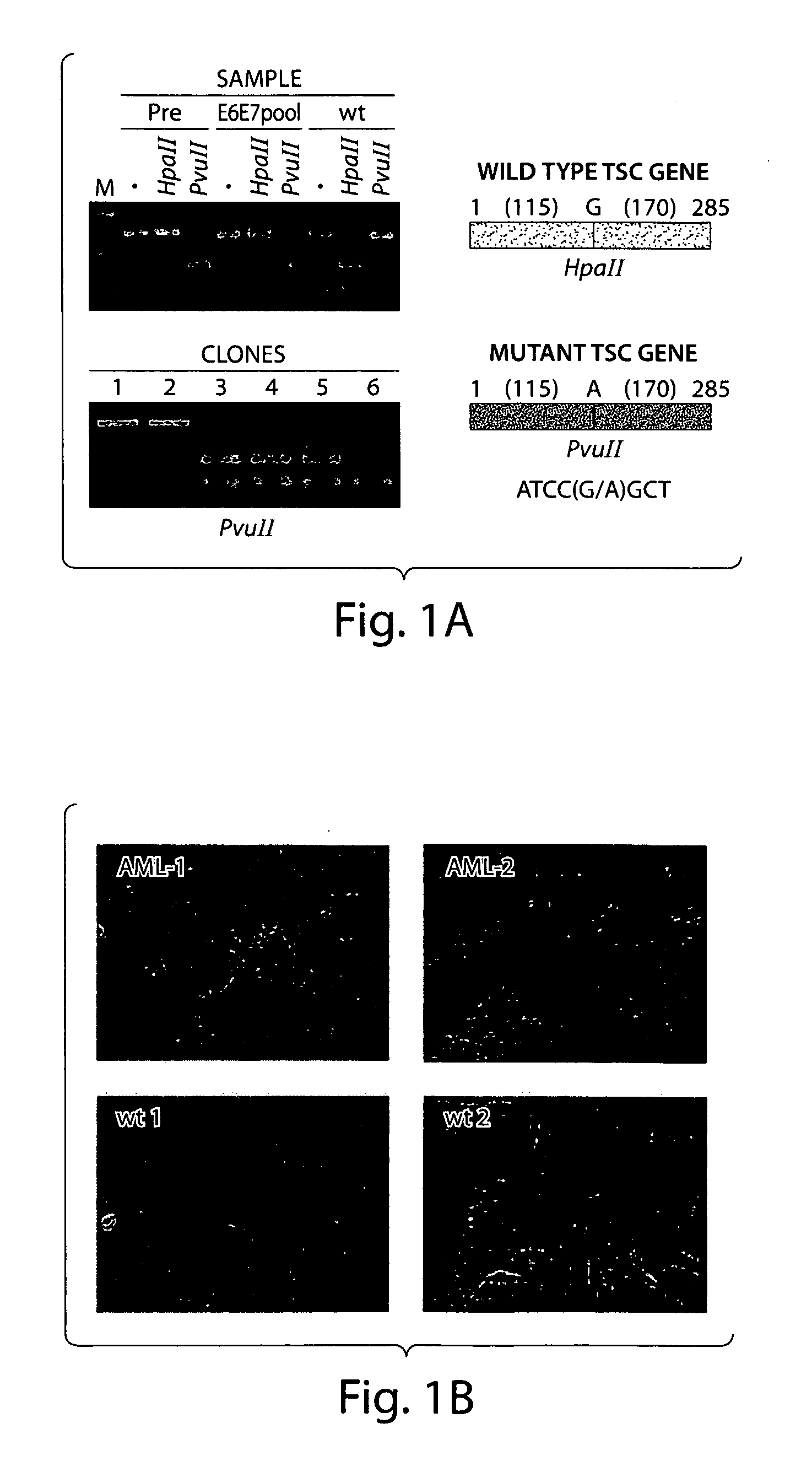
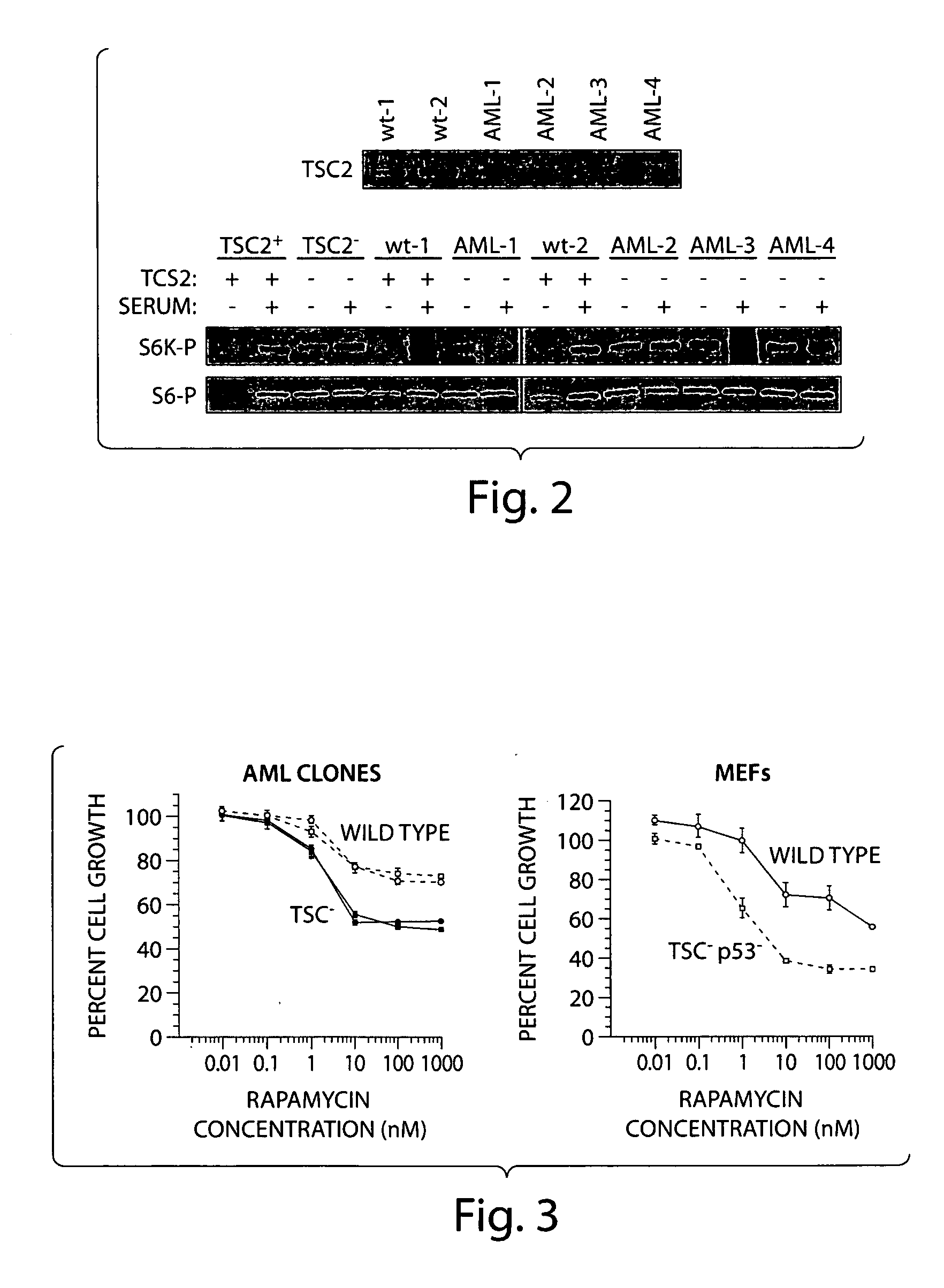
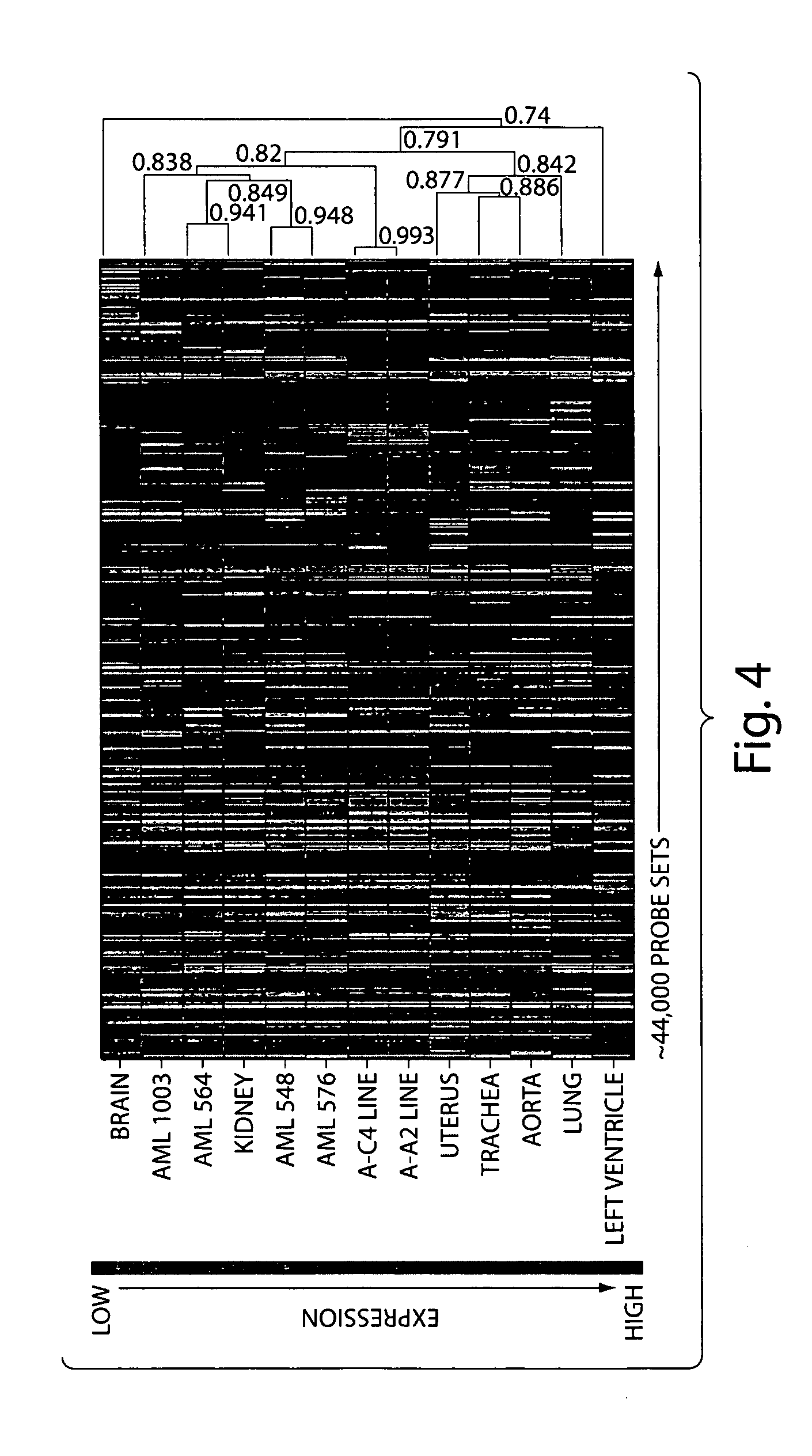
Disclosed are immortailized cells and cell lines that do not express the Tuberous Sclerosis Complex(TSC)-2 gene. Also sisclosed are methods of detecting TSC-related disorders using differentially expressed genes.
Owner:THE ROTHBERG INST FOR CHILDHOOD DISEASES
Endothelial cell expression patterns
To gain a better understanding of tumor angiogenesis, new techniques for isolating endothelial cells (ECs) and evaluating gene expression patterns were developed. When transcripts from ECs derived from normal and malignant colorectal tissues were compared with transcripts from non-endothelial cells, over 170 genes predominantly expressed in the endothelium were identified. Comparison between normal- and tumor-derived endothelium revealed 79 differentially expressed genes, including 46 that were specifically elevated in tumor-associated endothelium. Experiments with representative genes from this group demonstrated that most were similarly expressed in the endothelium of primary lung, breast, brain, and pancreatic cancers as well as in metastatic lesions of the liver. These results demonstrate that neoplastic and normal endothelium in humans are distinct at the molecular level, and have significant implications for the development of anti-angiogenic therapies in the future.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
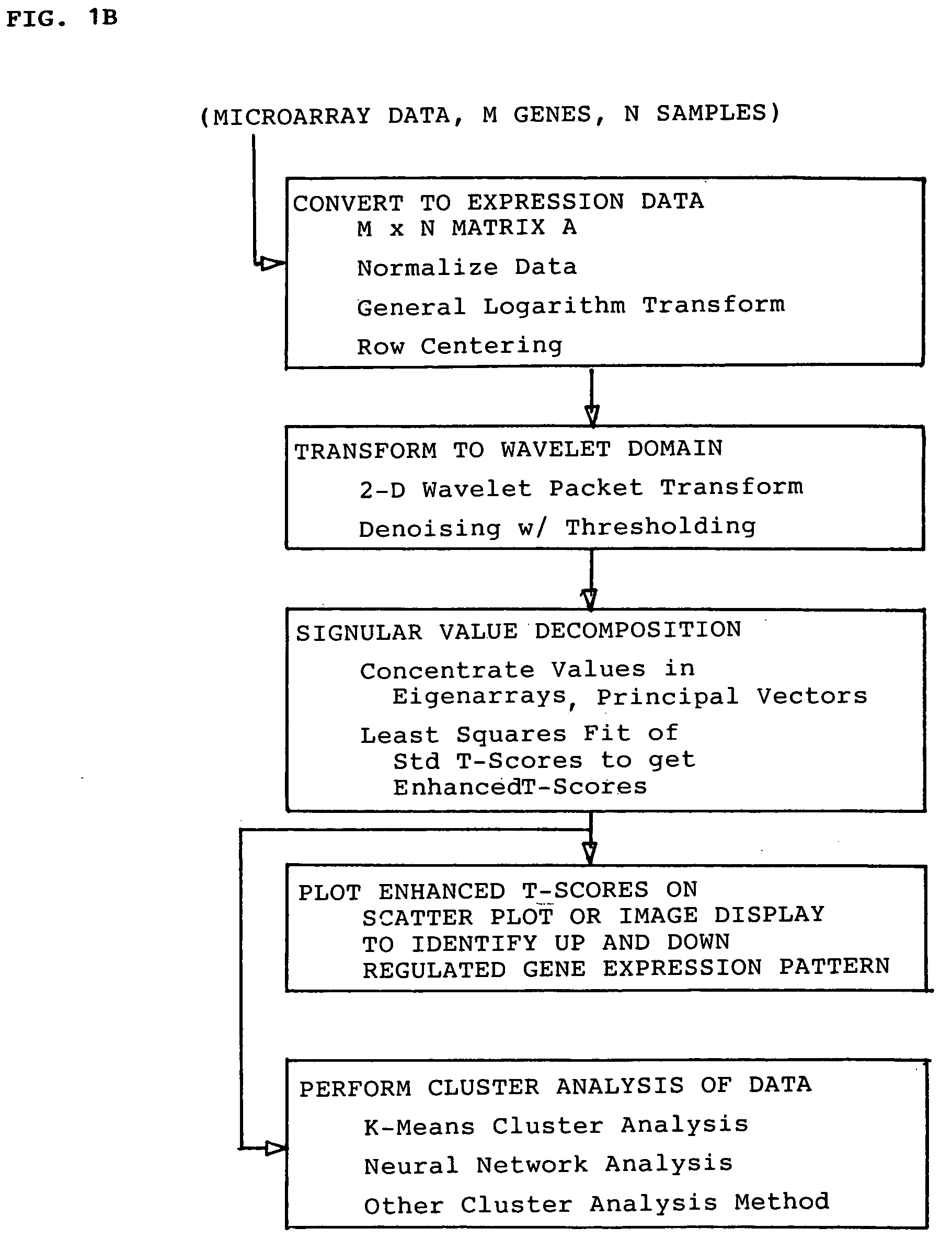
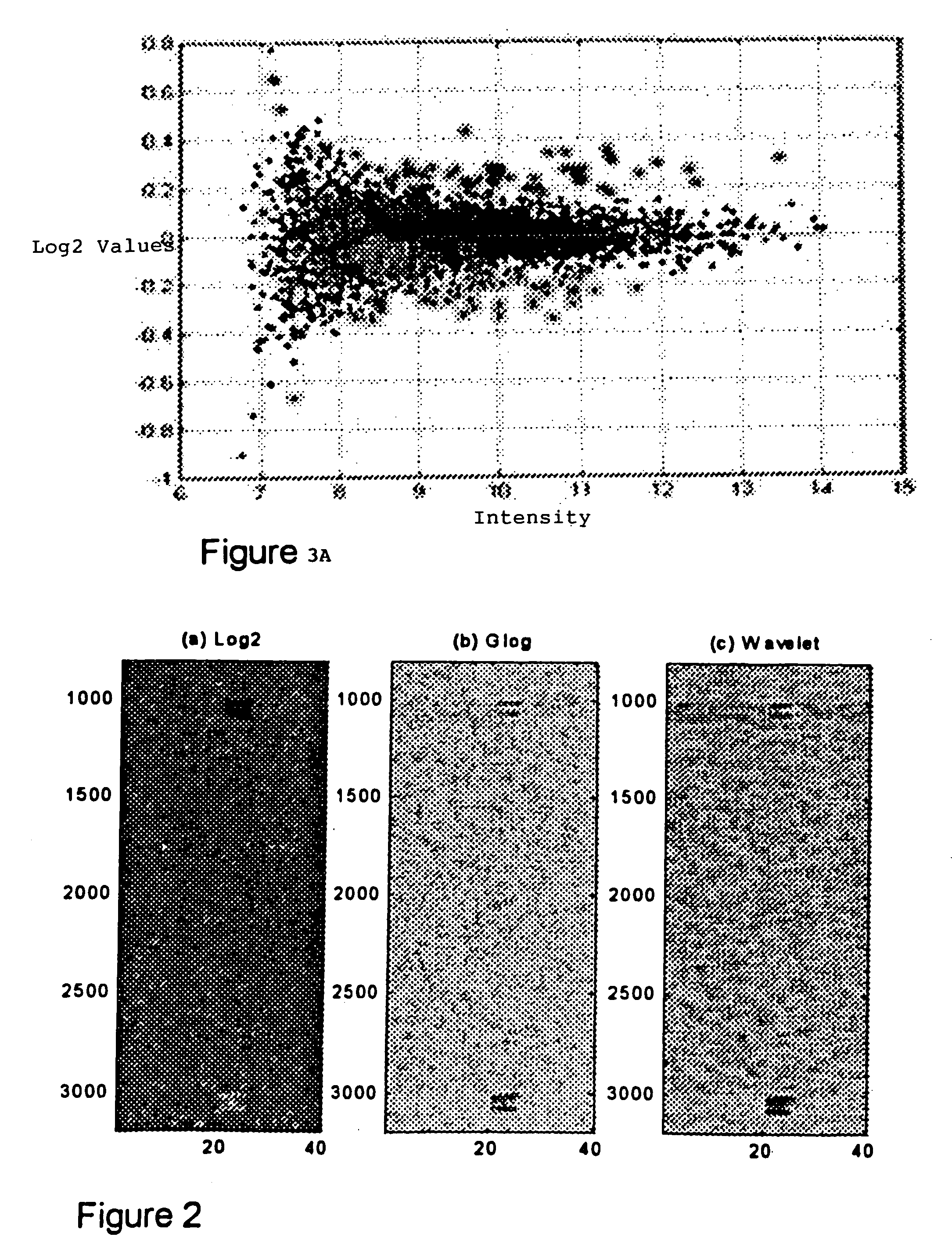
Methods for enhanced detection & analysis of differentially expressed genes using gene chip microarrays
InactiveUS20050181399A1Improve signal-to-noise ratioImprove concentrationMicrobiological testing/measurementBiological testingSingular value decompositionWavelet denoising
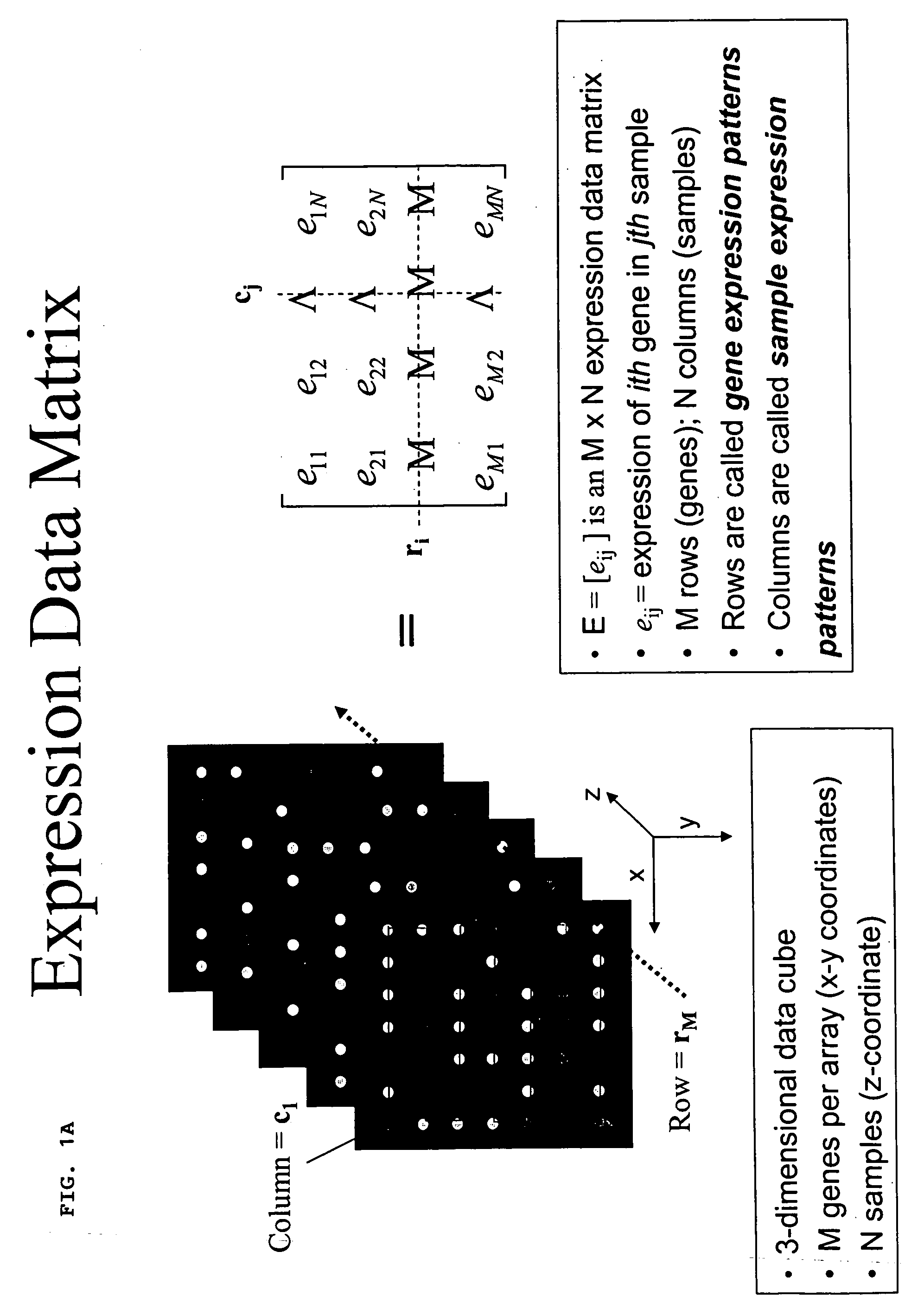
A method for enhanced detection and statistical analysis of differentially expressed genes in gene chip microarrays employs: (a) transformation of gene expression data into an expression data matrix (image data paradigm); (b) wavelet denoising of expression data matrix values to enhance their signal-to-noise ratio; and (c) singular value decomposition (SVD) of the wavelet-denoised expression data matrix to concentrate most of the gene expression signal in primary matrix eigenarrays to enhance the separation of true gene expression values from background noise. The transformation of gene chip data into an image data paradigm facilitates the use of powerful image data processing techniques, including a generalized logarithm (g-log) function to stabilize variance over intensity, and the WSVD combination of wavelet packet transform and denoising and SVD to clearly enhance separation of the truly changed genes from background noise. Detection performance can be assessed using a true false discovery rate (tFDR) computed for simulated gene expression data, and comparing it to estimated FDR (eFDR) rates based on permutations of the available data. Where a small number (N) of samples in a group is involved, a pair of specific WSVD algorithms are employed complementarily if N>5 and if N<6.
Owner:UNIV OF HAWAII
System for Identifying Connections Between Perturbagens and Genes Associated with a Skin Hyperpigmentation Condition
InactiveUS20130261024A1Microbiological testing/measurementOrganic compound librariesHypopigmentationSkin hyperpigmentation
A system for identifying connections between perturbagens and genes associated with a skin hyperpigmentation condition. The system includes a computer readable medium having a plurality of instances stored thereon, and a skin hyperpigmentation-relevant gene expression signature. Each instance includes an instance list of rank-ordered identifiers of differentially expressed genes, and the hyperpigmentation-relevant gene expression signature includes a gene expression signature list of identifiers representing differentially expressed genes associated with a hyperpigmentation condition or differentially expressed genes associated with a benchmark skin-lightening agent. The system also includes a programmable computer with computer-readable instructions that allow the computer to accessing the instances and a hyperpigmentation-relevant gene expression signature stored on the computer readable medium, comparing the hyperpigmentation-relevant gene expression signature to the plurality of the instances, and / or assigning a connectivity score to each of the plurality of instances.
Owner:THE PROCTER & GAMBLE COMPANY
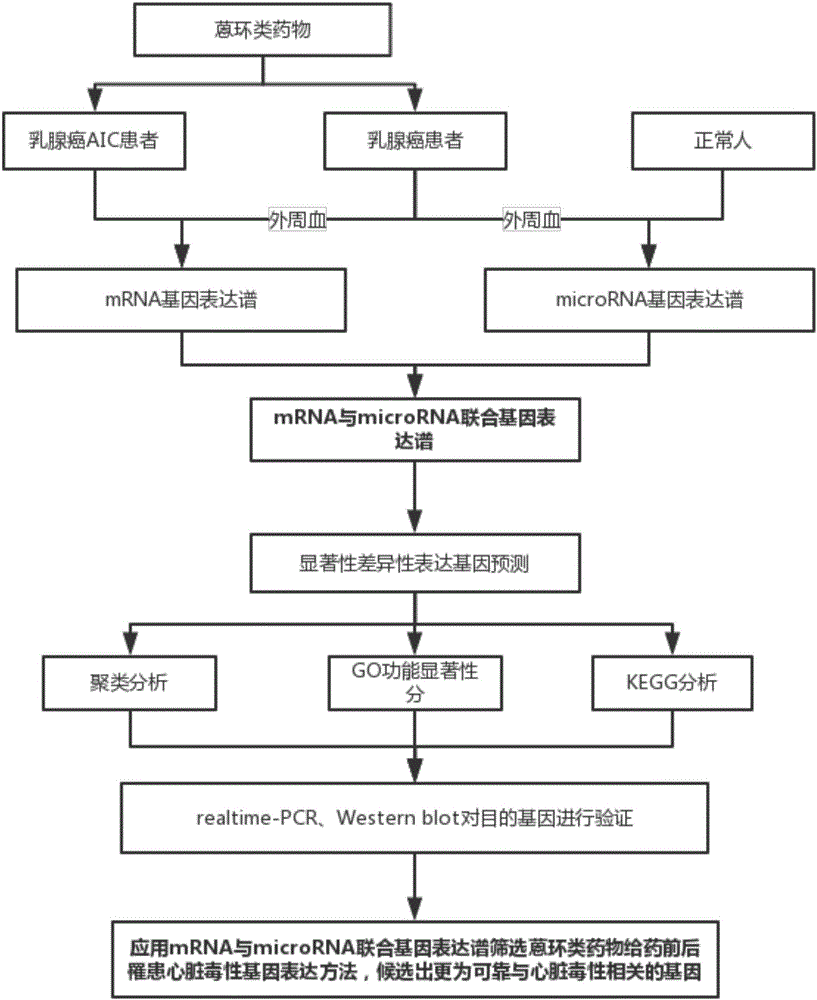
Method for screening anthracycline cardiotoxicity genes through mRNA expression profiles and competitive endogenesis RNA expression profiles jointly
The invention relates to a method for screening anthracycline cardiotoxicity genes through mRNA expression profiles and competitive endogenesis RNA expression profiles jointly. The method includes the steps that expression values of a large quantity of genes are detected through mRNA and competitive endogenesis RNA expression profile chips, and the correlations of all the genes are ordered; the obtained mRNA feature genes and the obtained competitive endogenesis RNA feature genes are combined; a gene pool is subjected to further gene selection with a genetic algorithm; differential expression genes are selected by comparing the data between different samples of mRNA and competitive endogenesis RNA joint expression profiles of breast cancer patients, breast cancer AIC patients and able-bodied persons; the differential expression genes are predicted, and the genes with the scores larger than 140 and free energy smaller than 20 are selected as reliable target genes; genes with obvious differences are selected from the target genes, and constructed breast-cancer anthracycline cardiotoxicity attacking target genes are verified through the residual RNA. By means of the method, more reliable potential gene markers related to anthracycline cardiotoxicity can be selected as a candidate.
Owner:JINZHOU MEDICAL UNIV
Method of Generating a Hyperpigmentation Condition Gene Expression Signature
ActiveUS20130261006A1Nucleotide librariesMicrobiological testing/measurementReference sampleHypopigmentation
A method of generating a hyperpigmentation condition gene expression signature for use in identifying connections between perturbagens and genes associated with a skin pigmentation condition. The method includes providing a gene expression profile for a reference sample of human skin cells not affected with a pigmentation condition; generating a gene expression profile for a sample of human skin cells from a subject exhibiting the hyperpigmentation condition; comparing the expression profiles to determine a gene expression signature that includes a set of differentially expressed genes; assigning an identifier to each gene constituting the gene expression signature and ordering the identifiers according to the direction of differential expression to create one or more gene expression signature lists; and storing the one or more gene expression signature lists on at least one computer readable medium.
Owner:THE PROCTER & GAMBLE COMPANY
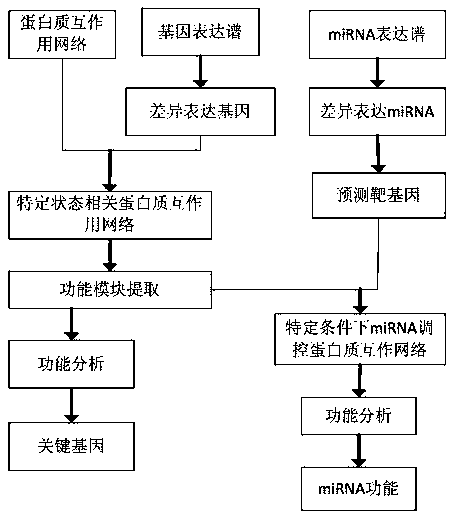
Method of miRNA function recognition based on multi-genomics
The invention relates to a miRNA function recognition method based on multi-genomics, and relates to a miRNA function recognition method based on multi-genome. The present invention aims to solve theproblem of low accuracy of miRNA function recognition in the prior art. The method of the invention comprises: analyzing differentially expressed genes through gene expression profiles; constructing disease-or drug-related protein networks; selecting functional modules of the constructed protein network related to disease or drug; performing enrichment analysis of the selected functional modules to determine the key genes in the functional modules; analyzing differentially expressed miRNAs by miRNA expression profiles to predict the target genes of differentially expressed miRNAs; performing construction of miRNA-regulated disease-or drug-related protein networks; performing functional enrichment analysis on the nodal genes of the constructed miRNA-regulated disease or drug-related proteinnetwork to identify the function of miRNA. The invention is used in the field of bioinformatics.
Owner:QIQIHAR UNIVERSITY
Method for diagnosing non-small cell lung cancers
InactiveCN1705753AOrganic active ingredientsMicrobiological testing/measurementBiologyDifferentially expressed genes
Disclosed are methods for detecting non-small cell lung cancer using differentially expressed genes. Furthermore, novel human genes whose expression is elevated in non-small cell lung cancer compared to no-cancerous tissues are provided. Also disclosed are methods of identifying compounds for treating and preventing non-small cell lung cancer.
Owner:ONCOTHERAPY SCI INC
Membrane Associated Tumor Endothelium Markers
InactiveUS20110104059A1Reduced activityHigh activitySenses disorderPeptide/protein ingredientsMolecular levelFhit gene
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com