Patents
Literature
139 results about "Alternative splicing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
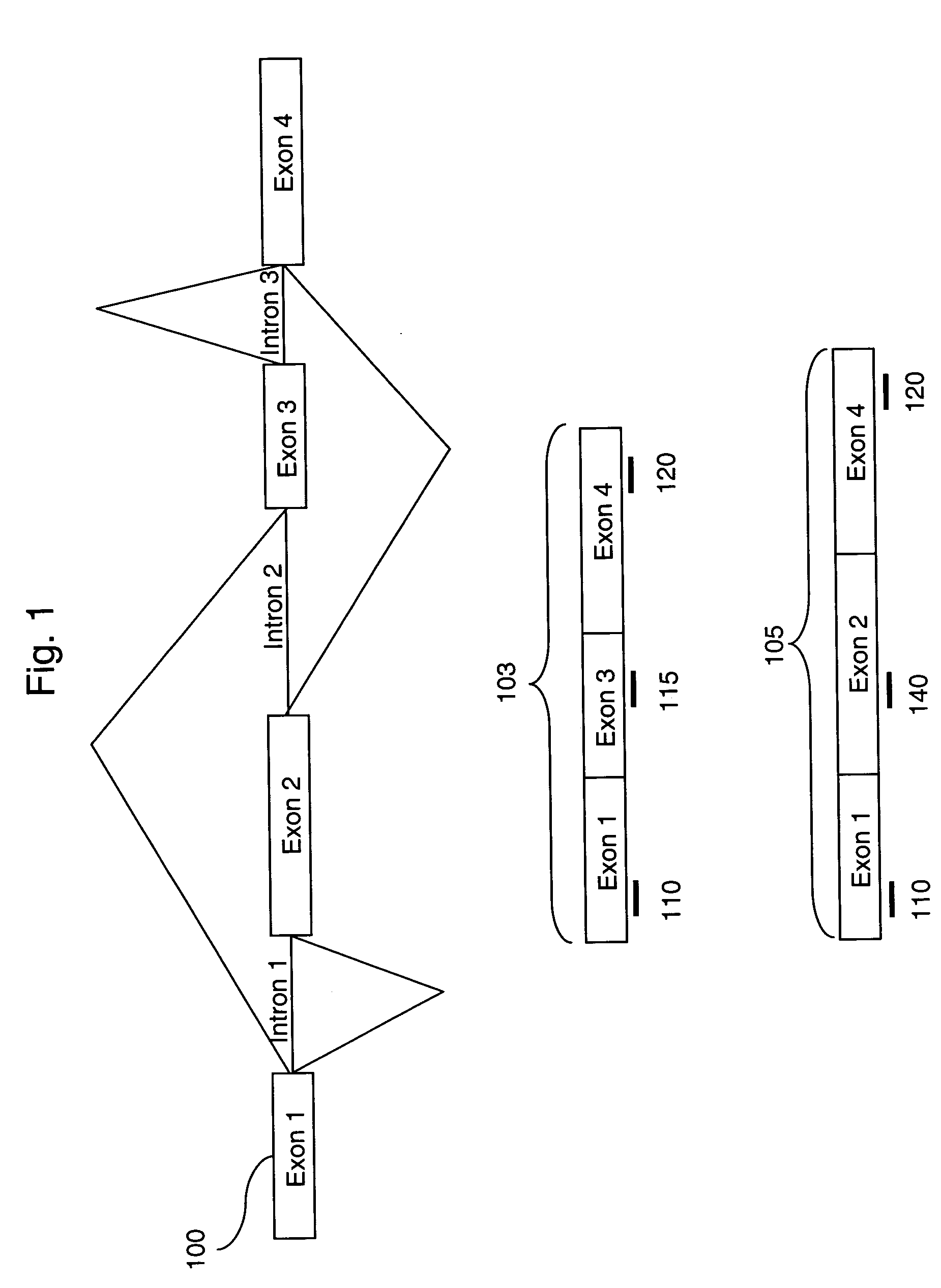
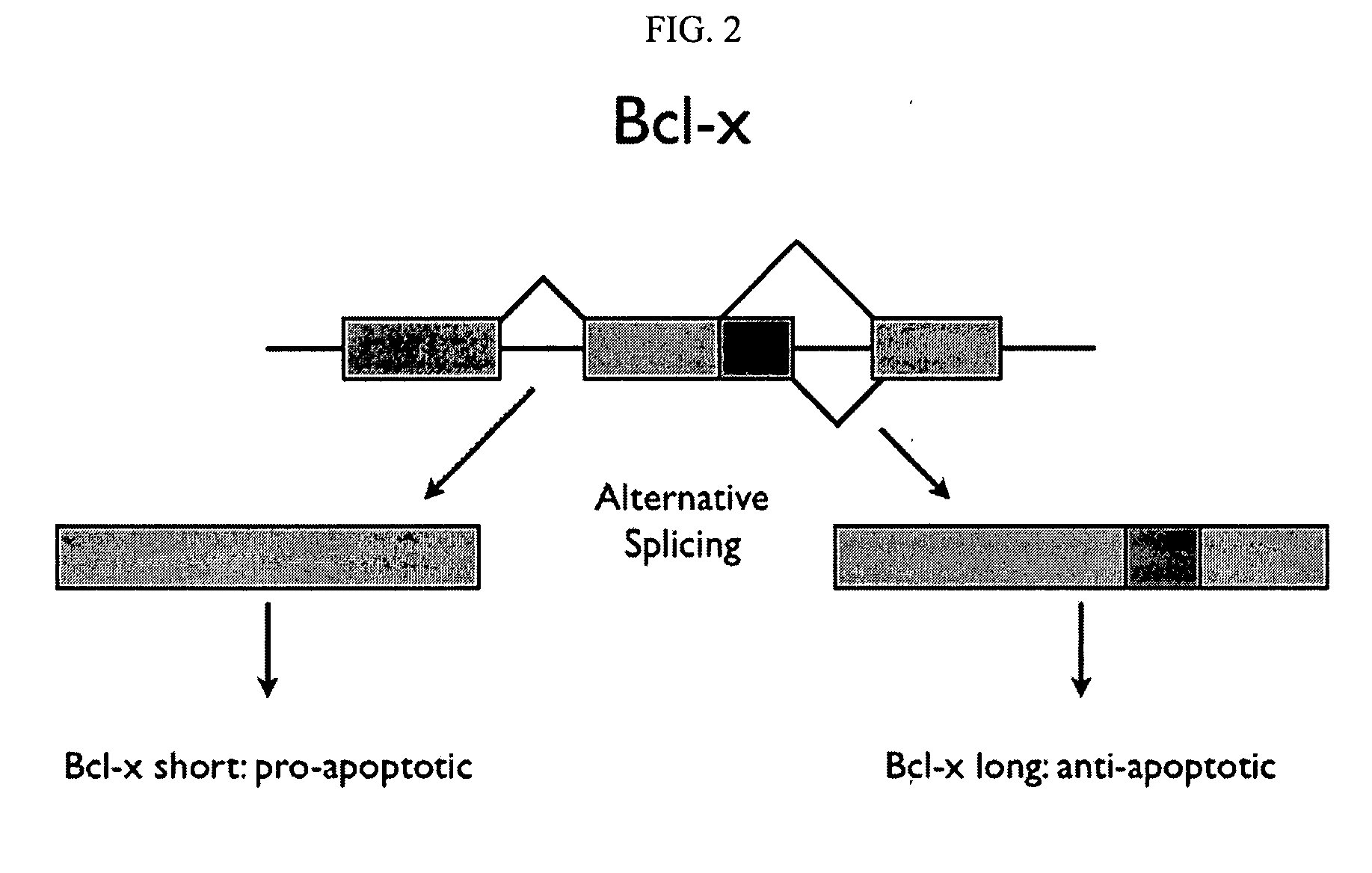
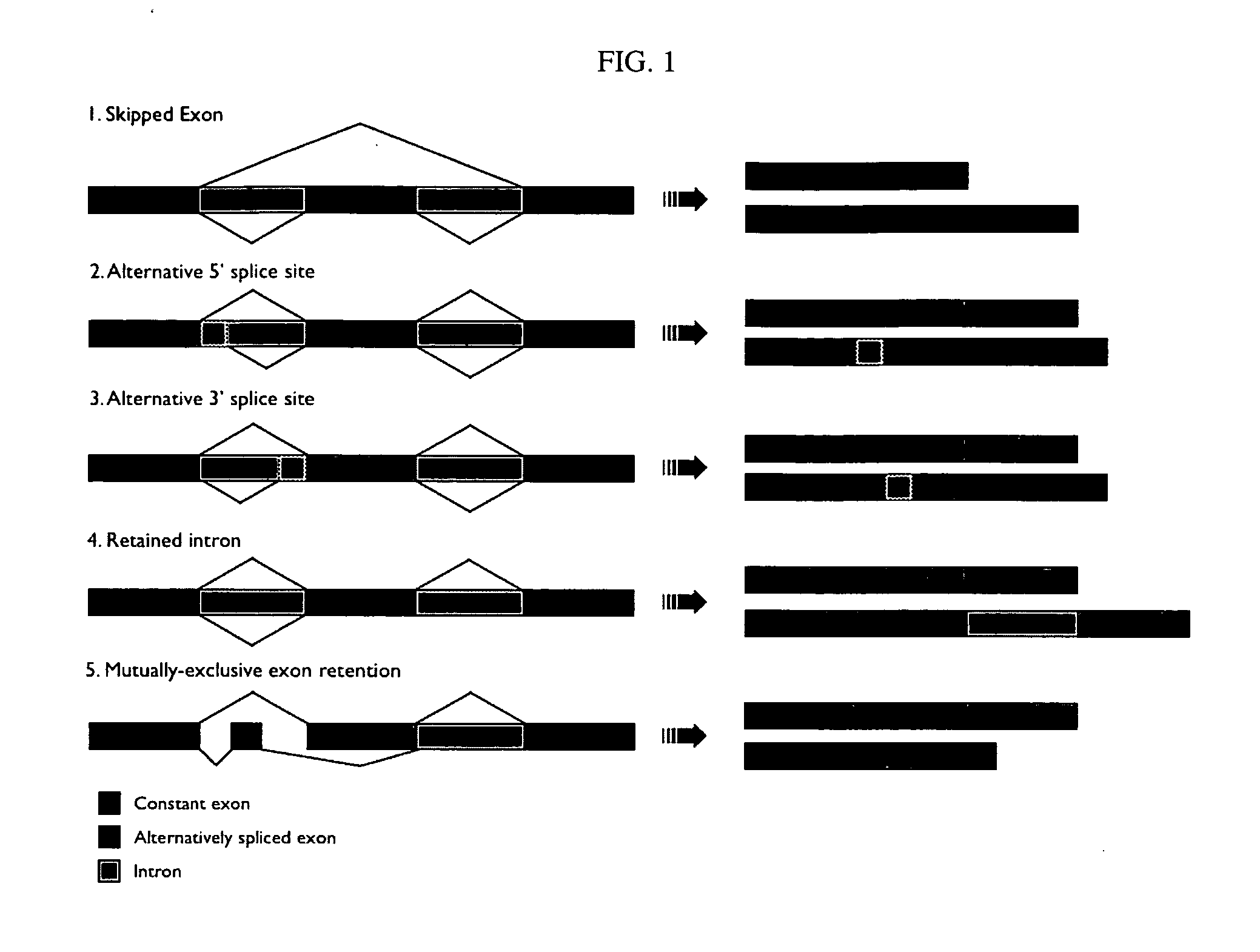
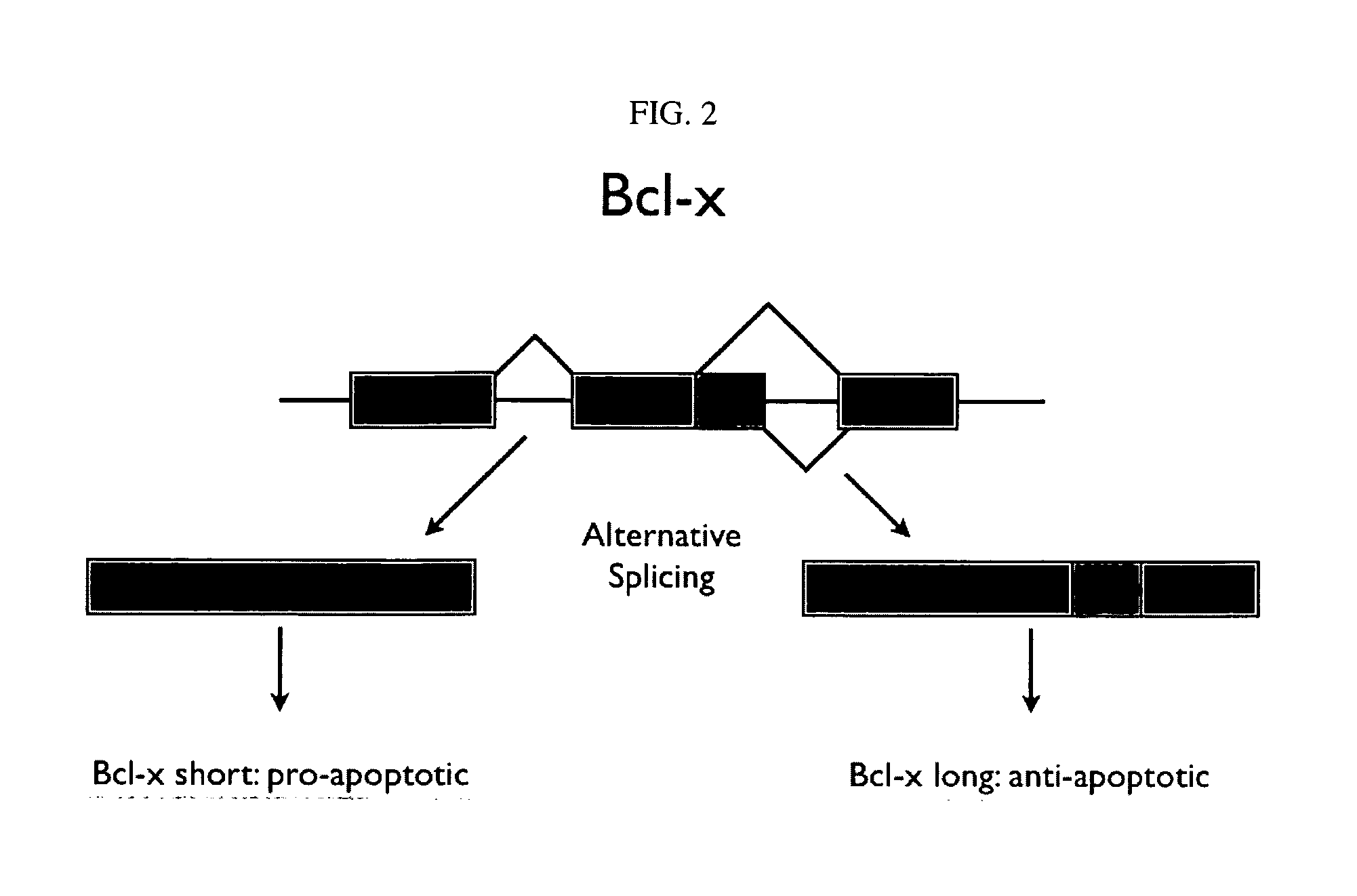
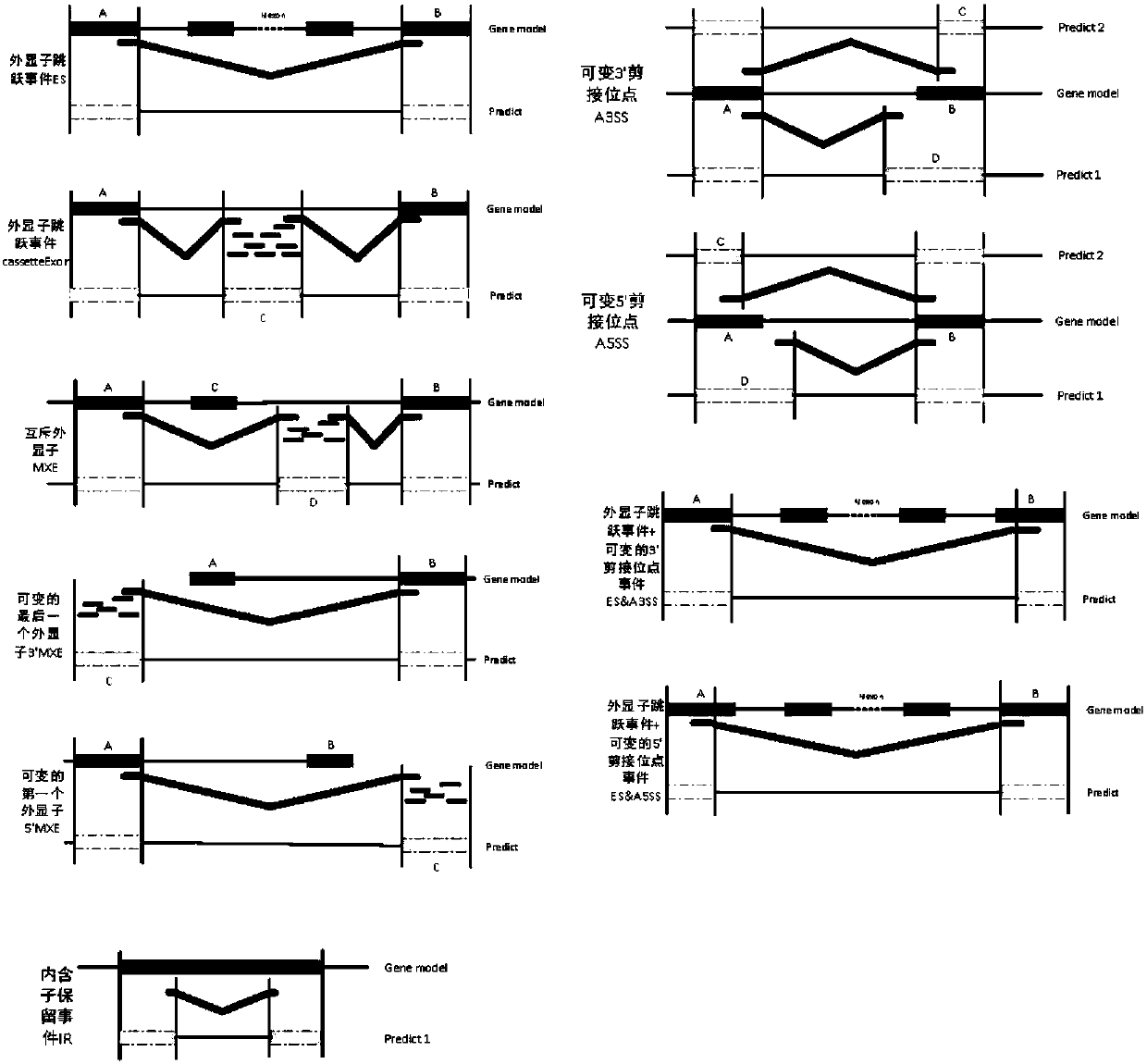
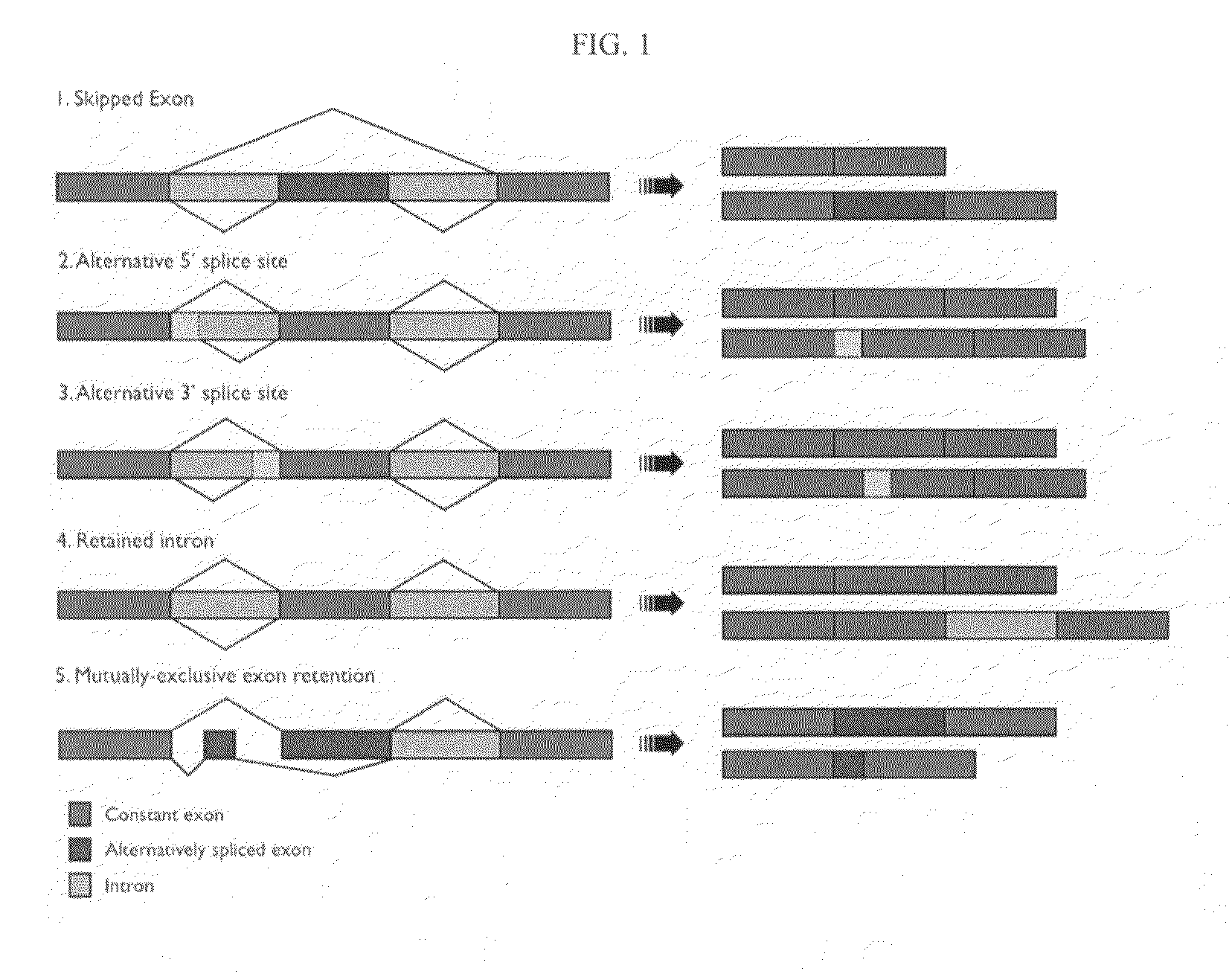
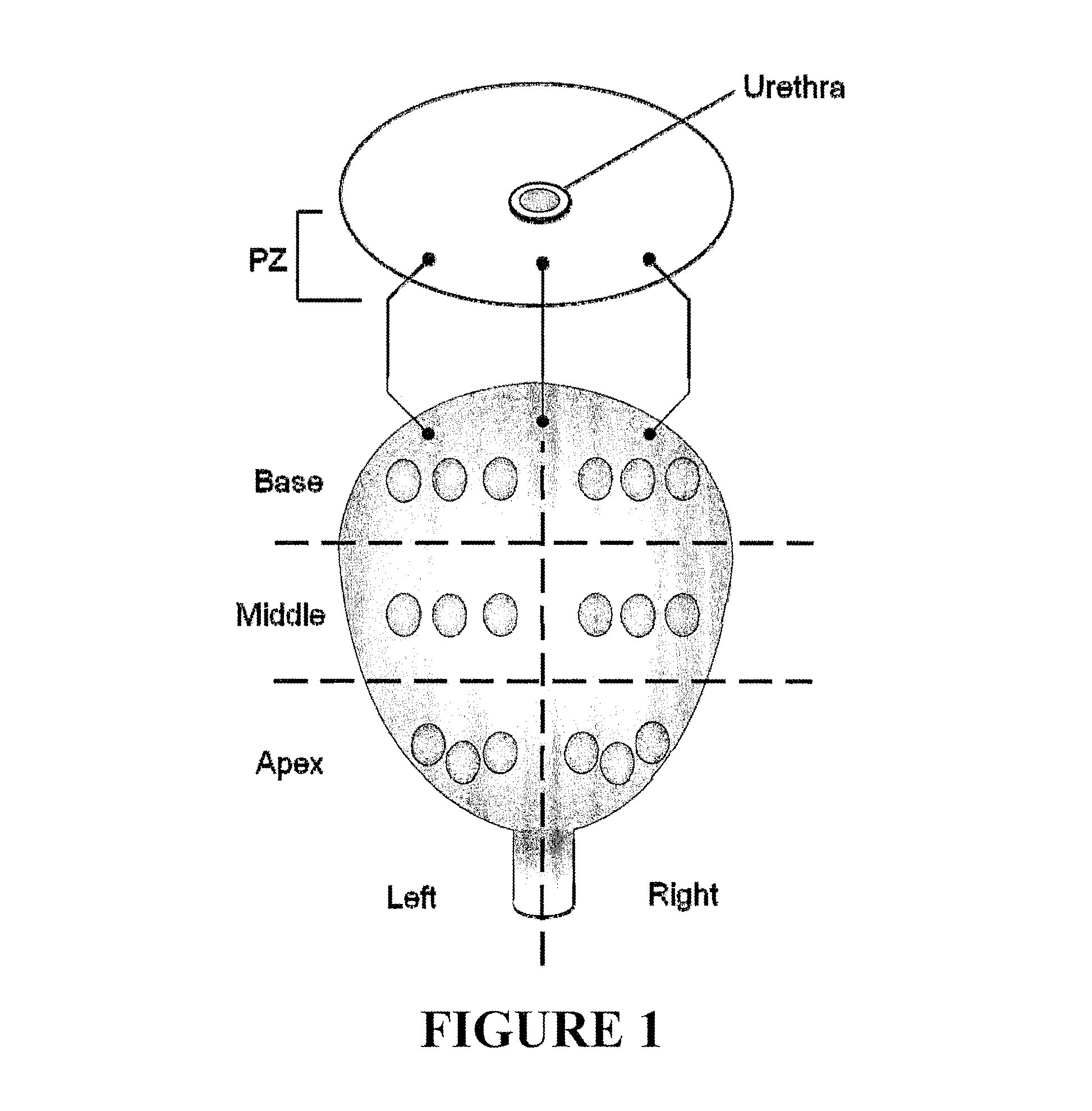
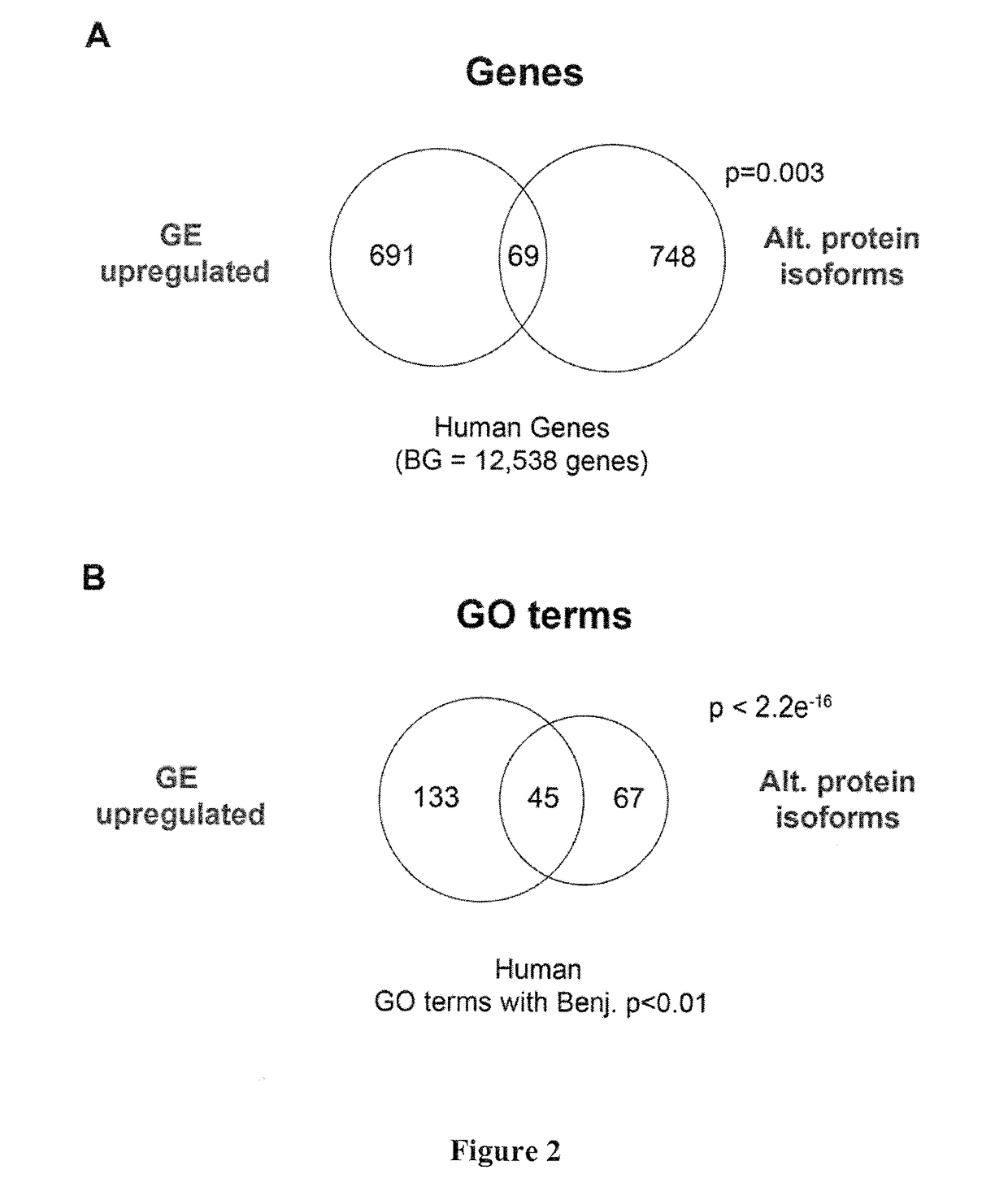
Alternative splicing, or differential splicing, is a regulated process during gene expression that results in a single gene coding for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions (see Figure). Notably, alternative splicing allows the human genome to direct the synthesis of many more proteins than would be expected from its 20,000 protein-coding genes.
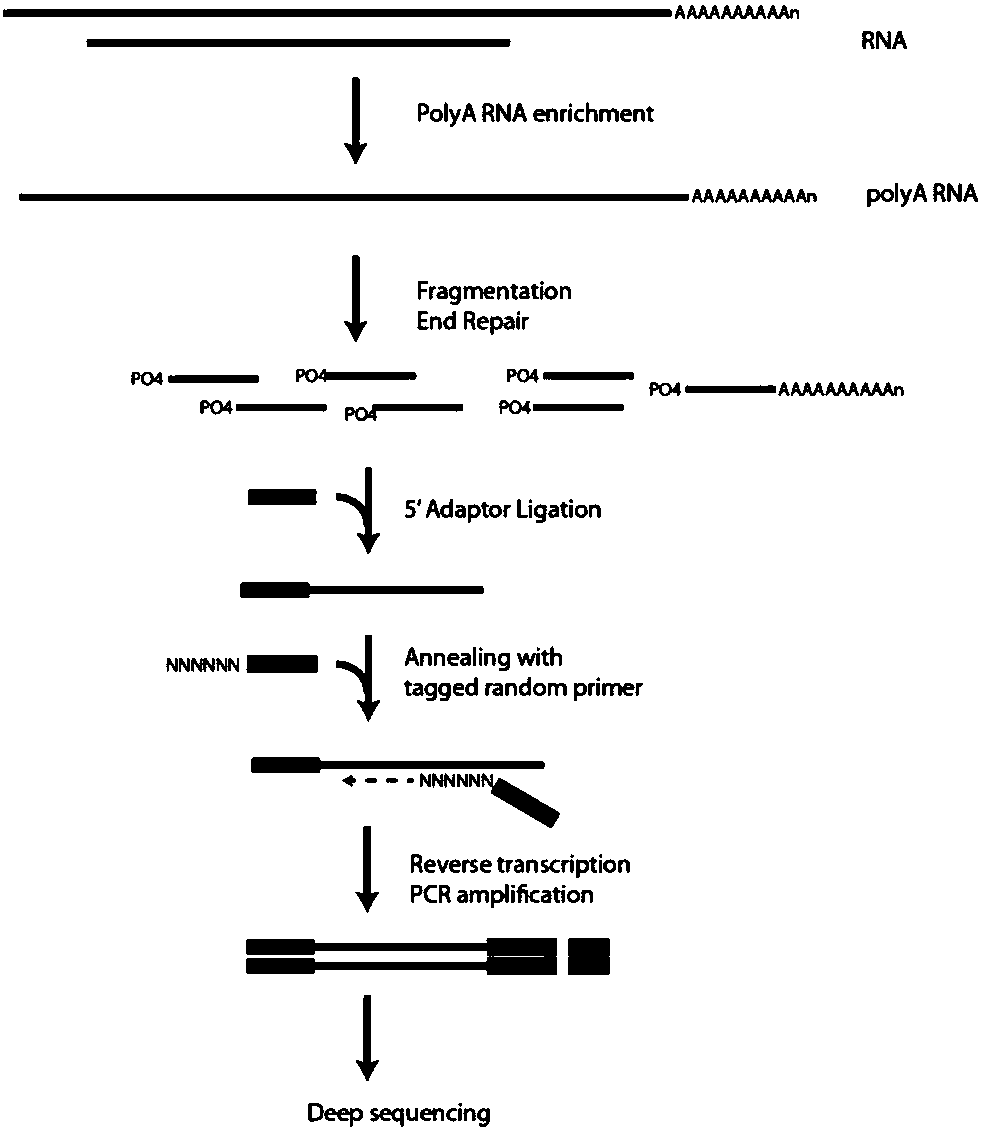
Methods of analysis of alternative splicing in human
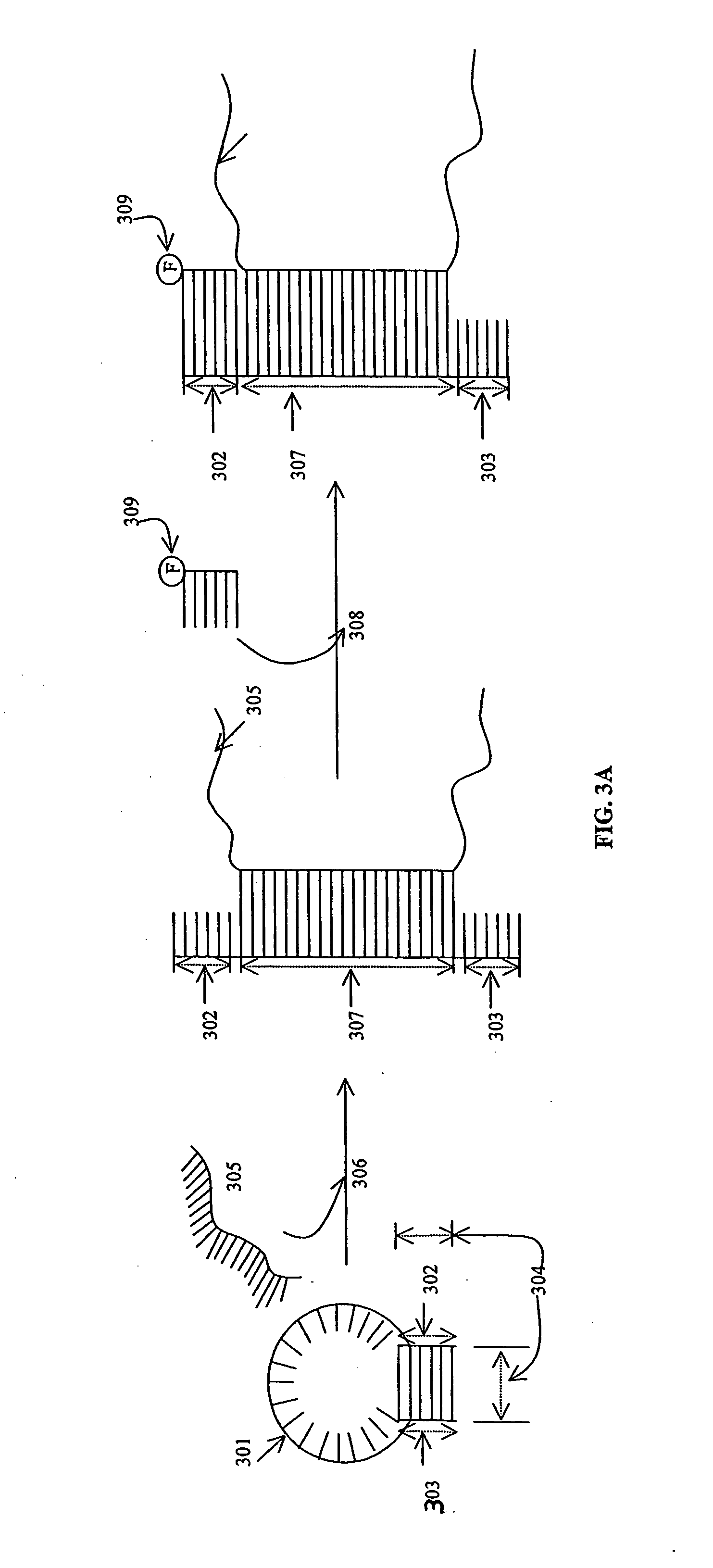
InactiveUS20050244851A1Bioreactor/fermenter combinationsBiological substance pretreatmentsChemical biologyNucleic acid sequencing
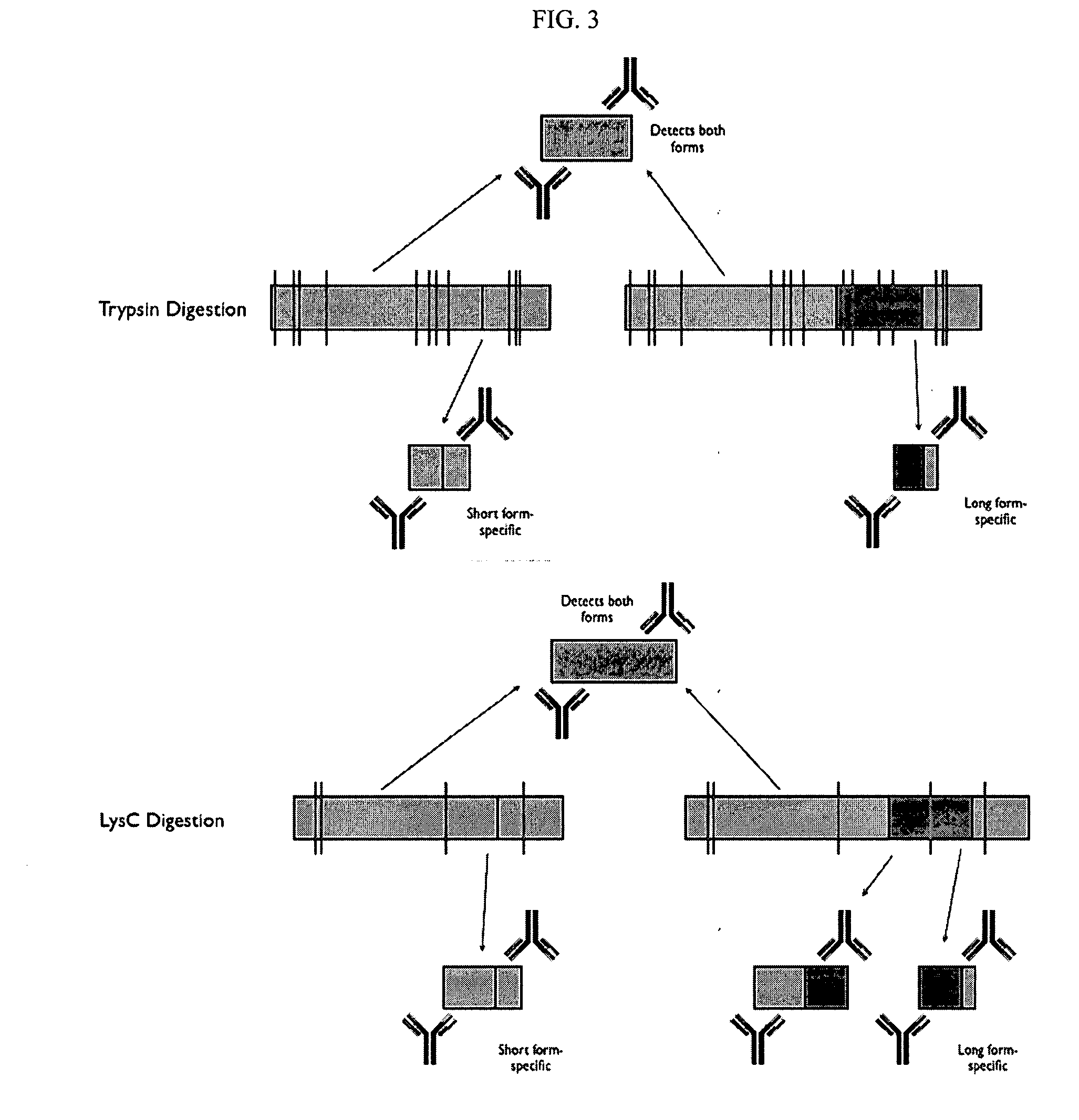
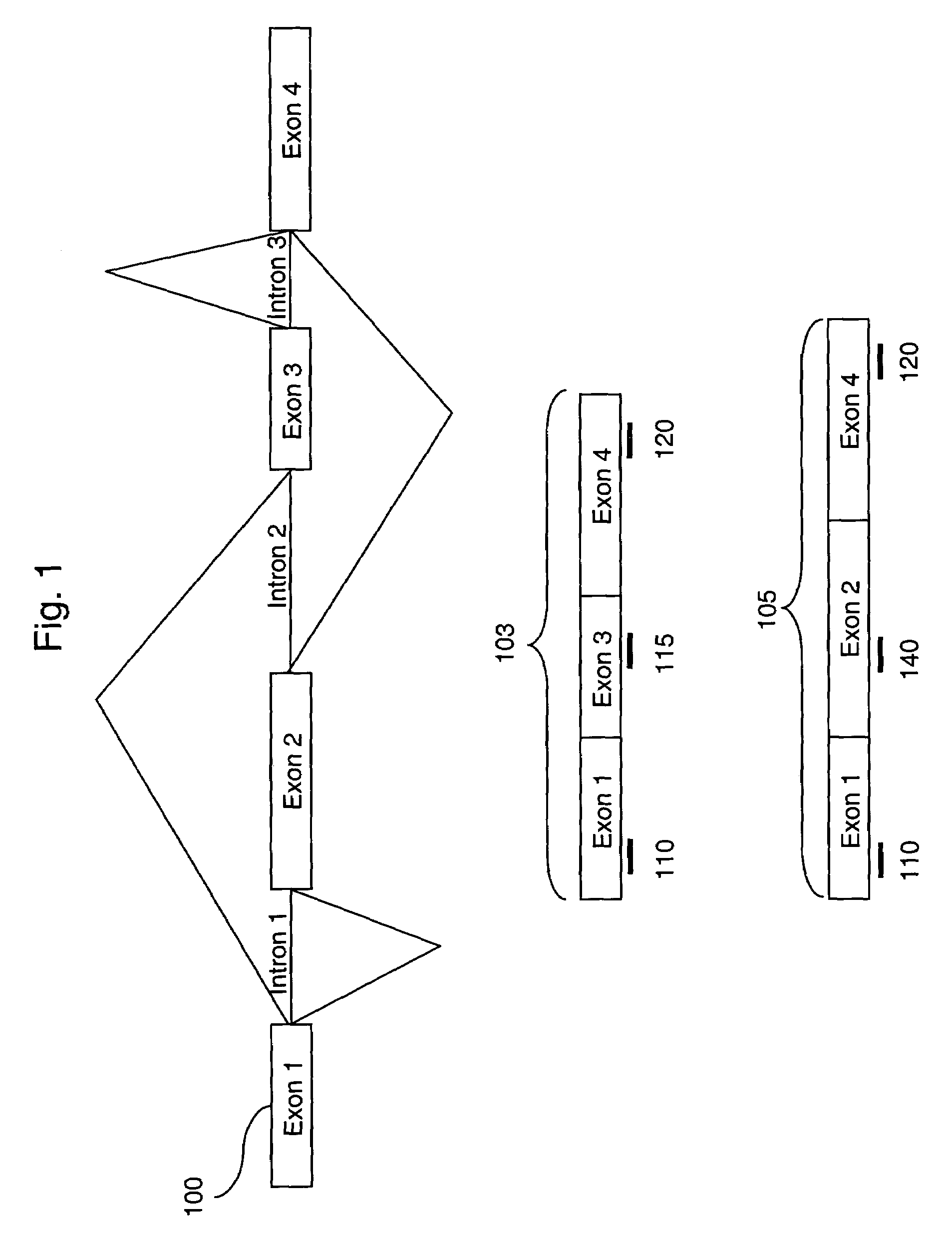
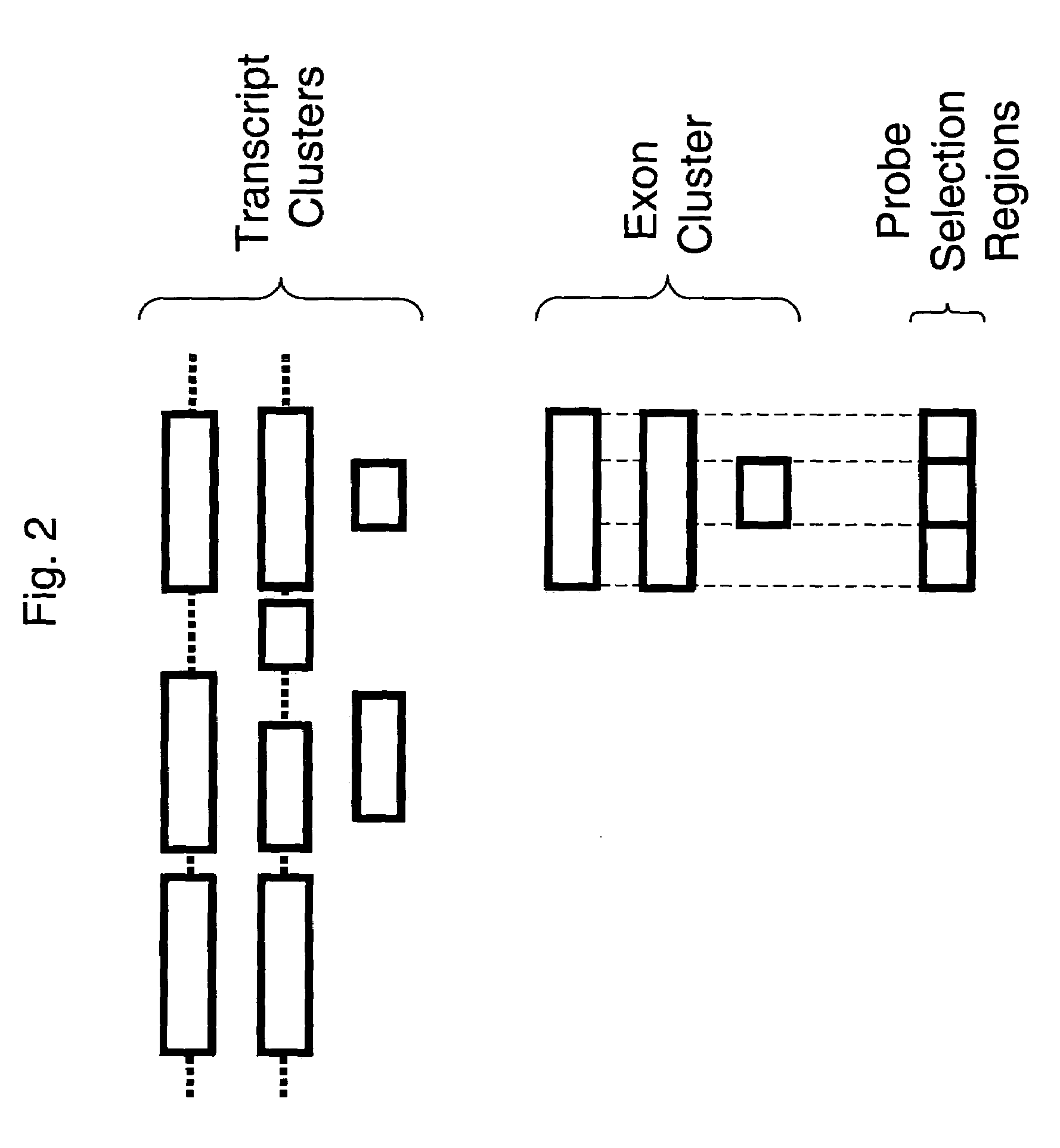
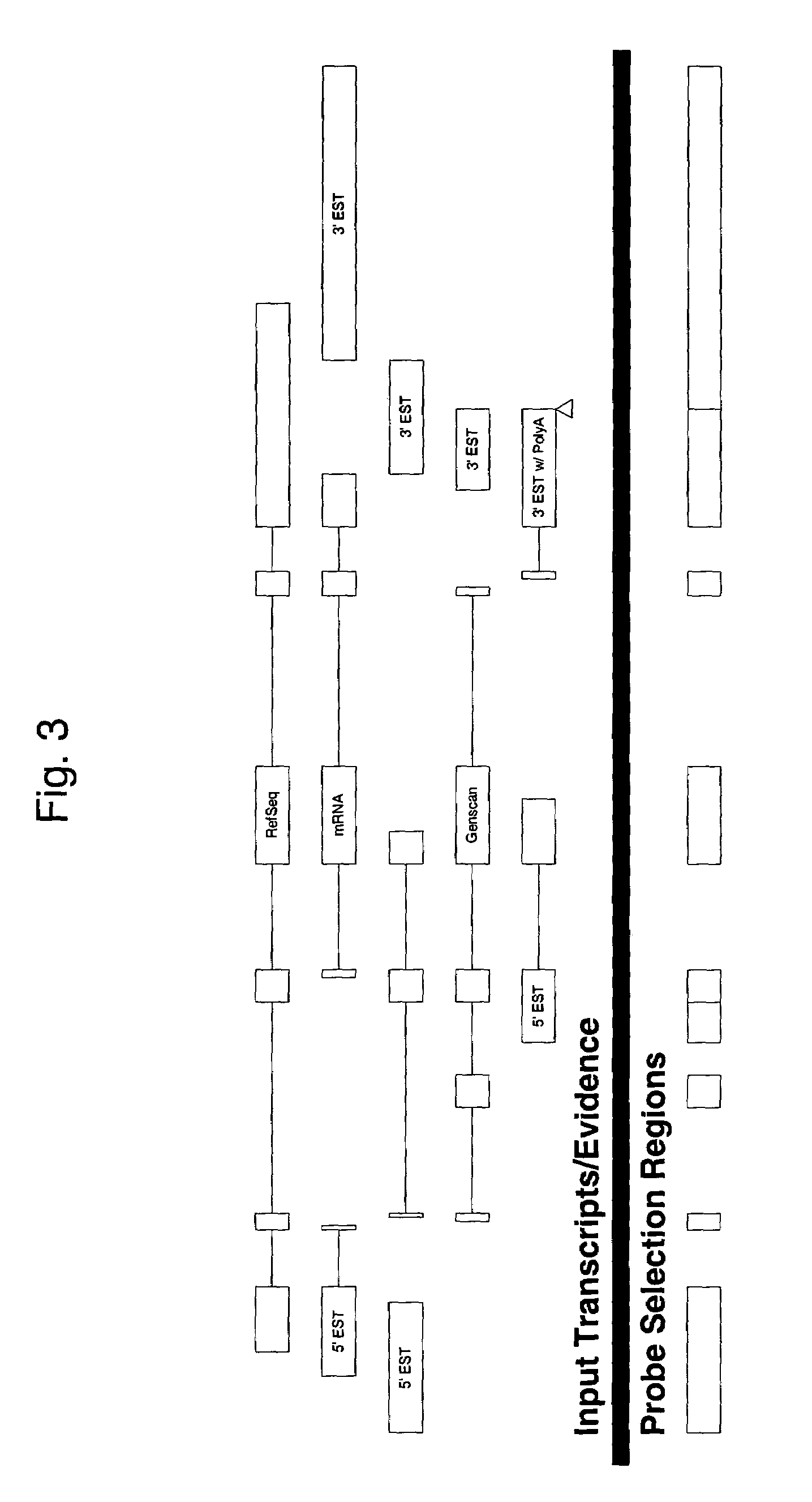
The invention provides nucleic acid sequences which are complementary, in one embodiment, to a wide variety of human exons. The invention provides the sequences in such a way as to make them available for a variety of analyses including analysis of alternative splicing events. In one embodiment the nucleic acid sequences provided are present as an array of probes that may be used to measure gene expression of at least 5,000 alternatively spliced human genes. As such, the invention relates to diverse fields impacted by the nature of molecular interaction, including chemistry, biology, medicine, pharmacology and medical diagnostics.
Owner:AFFYMETRIX INC
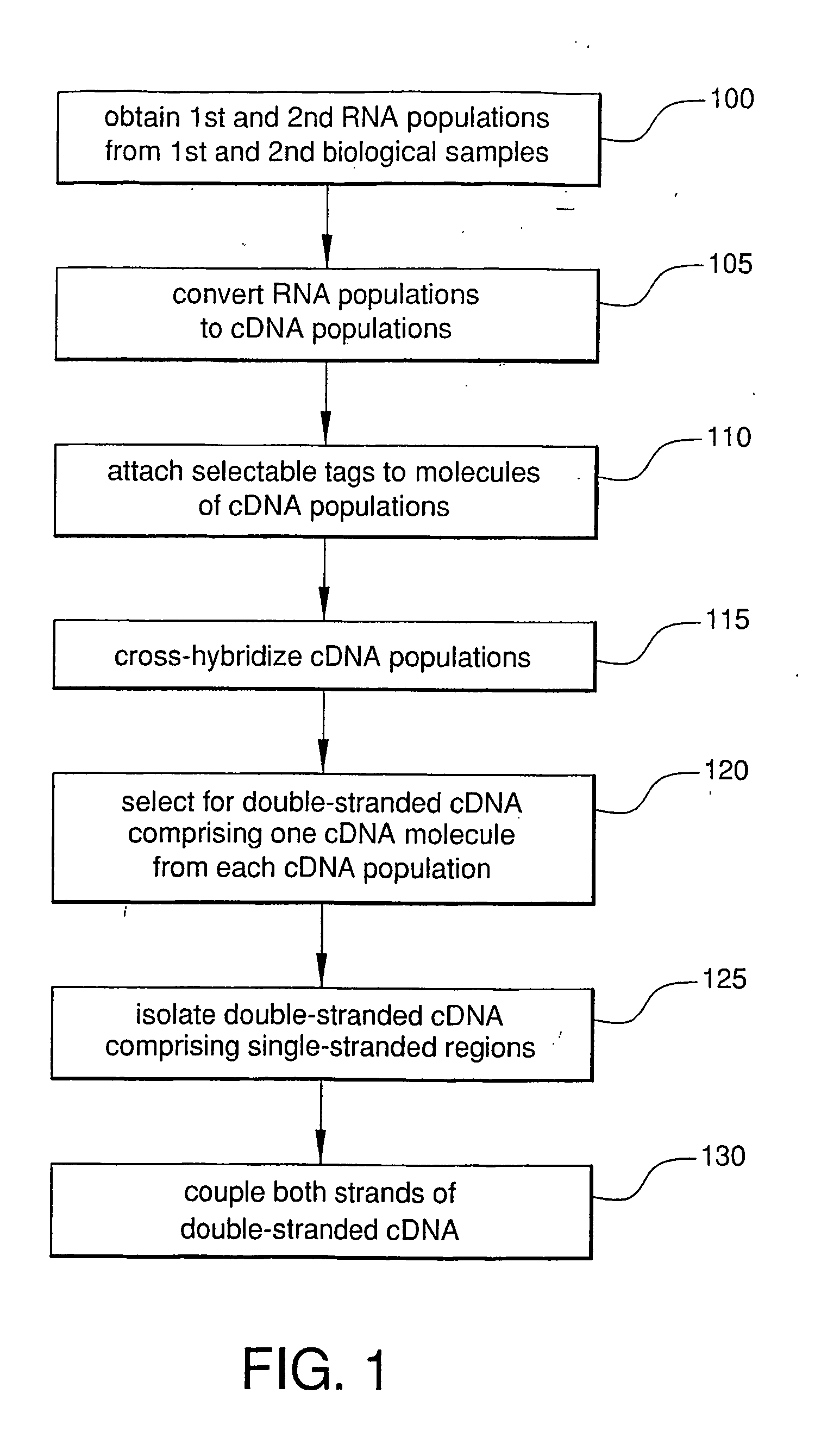
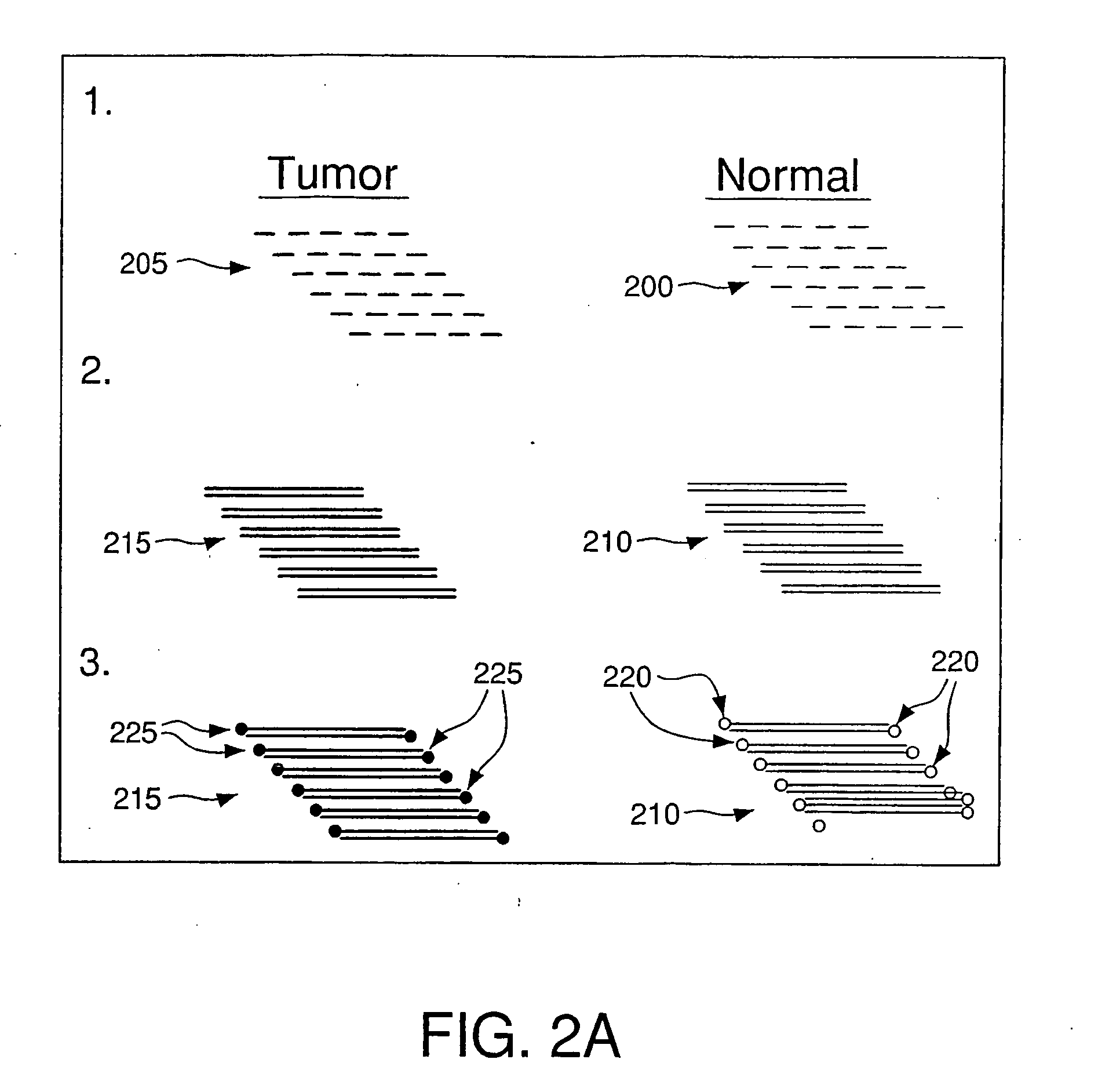
Method For Rapid Identification of Alternative Splicing
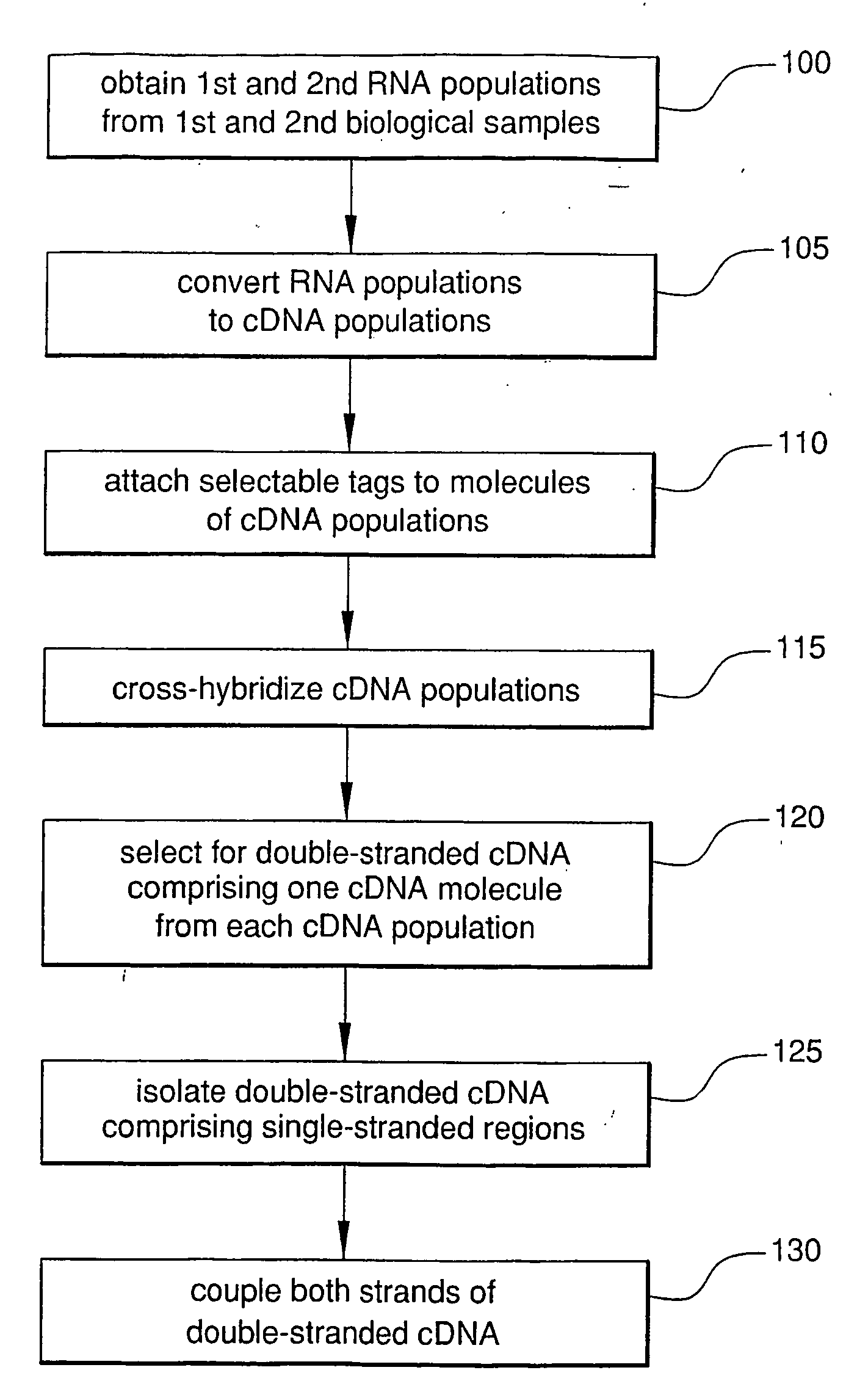
Alternatively spliced RNA, along with their normally-spliced counterparts, can be rapidly identified by hybridizing cDNA from normal tissue to cDNA from an abnormal or test tissue. The two cDNA populations are separately tagged prior to hybridization, which allows isolation of double-stranded cDNA containing both normal and alternatively spliced molecules. Within this population, pairing of cDNA molecules representing an alternatively spliced mRNA with cDNA molecules representing the counterpart normally spliced mRNA will form double-stranded cDNA with single-stranded mismatched regions. The mismatched double-stranded cDNA are isolated with reagents that bind single-stranded nucleic acids. The strands of each mismatched double-stranded cDNA are then coupled and analyzed, simultaneously identifying both normal and alternatively spliced molecules.
Owner:WONG ALBERT
Probe biochips and methods for use thereof
InactiveUS20060199183A1Bioreactor/fermenter combinationsBiological substance pretreatmentsPolynucleotideAlternative splicing
The invention relates to fields of use of unlabelled polynucleotide probes able to form hairpins, the biochips comprising such probes and methods allowing use thereof. The present invention also concerns methods for designing such probes and biochips. More particularly, the invention concerns the use of such unlabelled probes and biochips for manipulating and analysing polynucleotide sequences and optionally molecules which are associated therewith. This invention further concerns methods for preparing and use such probes and biochips for analysing mutations, sequencing, detection of alternative splicing variants, gene expression analysis, analysis of allelic imbalances and loss of heterozygosity and the detection of any nucleic acid present in organisms or residues from said organisms.
Owner:GENEWAVE SAS
Proton-based transcriptome sequencing data comparison and analysis method and system
InactiveCN104657628AImprove accuracyImprove reliabilitySpecial data processing applicationsDifferentially expressed genesSingle-nucleotide polymorphism
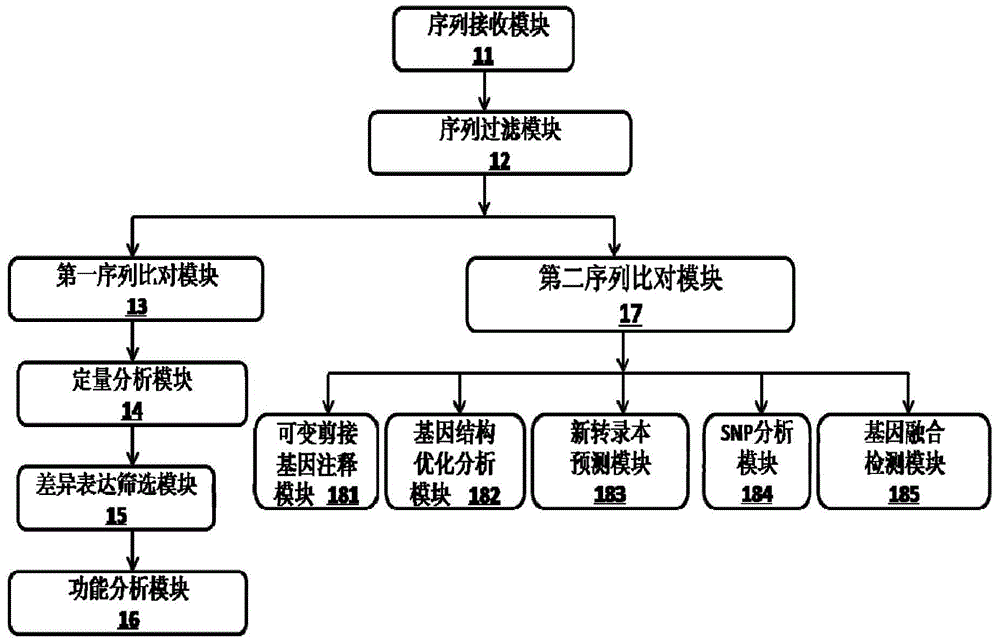
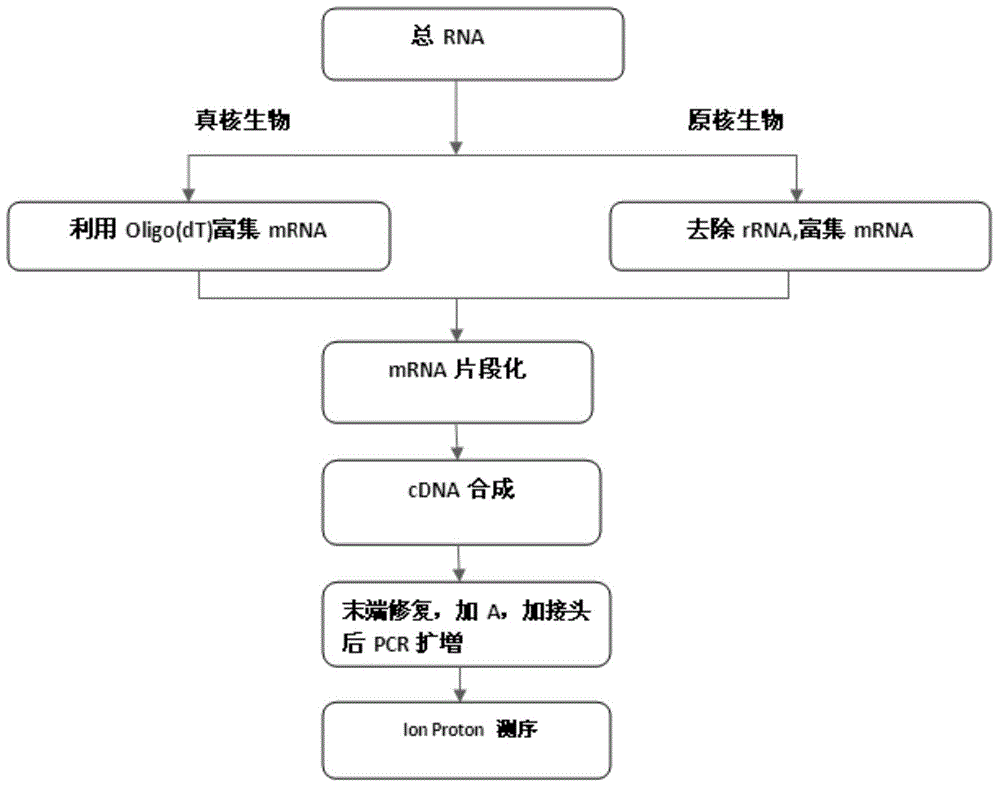
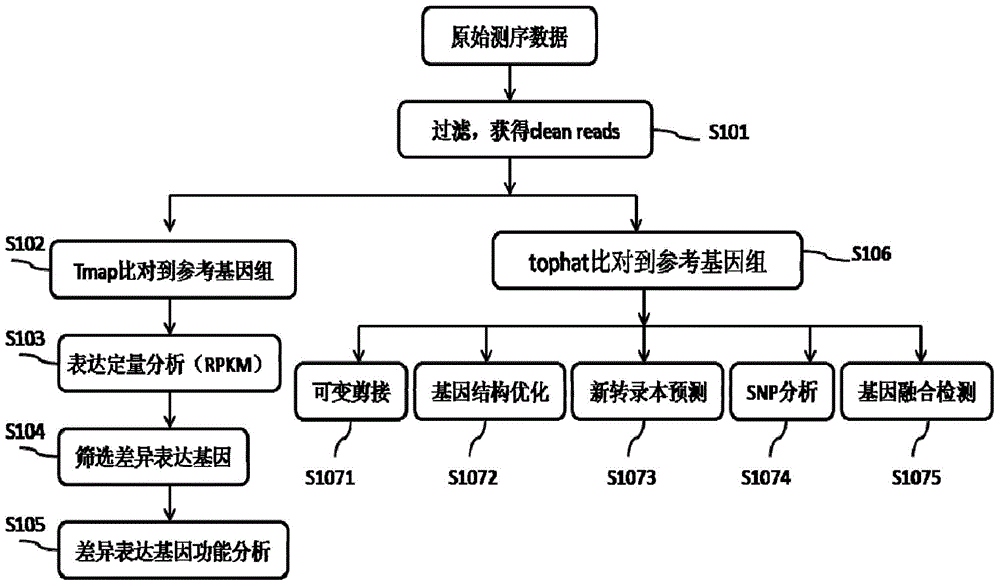
The invention provides a Proton-based transcriptome sequencing data comparison and analysis method and system. The method comprises the following steps: acquiring original sequencing data of at least two transcriptomes of a certain species by virtue of a Proton sequencing platform; filtering unqualified data to obtain clean reads; performing first-step analysis and second-step analysis, wherein the first-step analysis comprises the steps of comparing the clean reads with a reference genome of the species respectively, performing transcript quantitative analysis, screening significantly differently expressed genes and performing significantly differently expressed gene function analysis; the second-step analysis comprises the steps of comparing the clean reads to the reference genome of the species respectively, performing alternative splicing analysis, performing gene structure optimization analysis, performing new transcript prediction, performing SNP (Single Nucleotide Polymorphism) analysis and performing gene fusion detection. According to the method and the system, the transcriptome sequencing data comparison and analysis accuracy and reliability can be improved.
Owner:BGI TECH SOLUTIONS
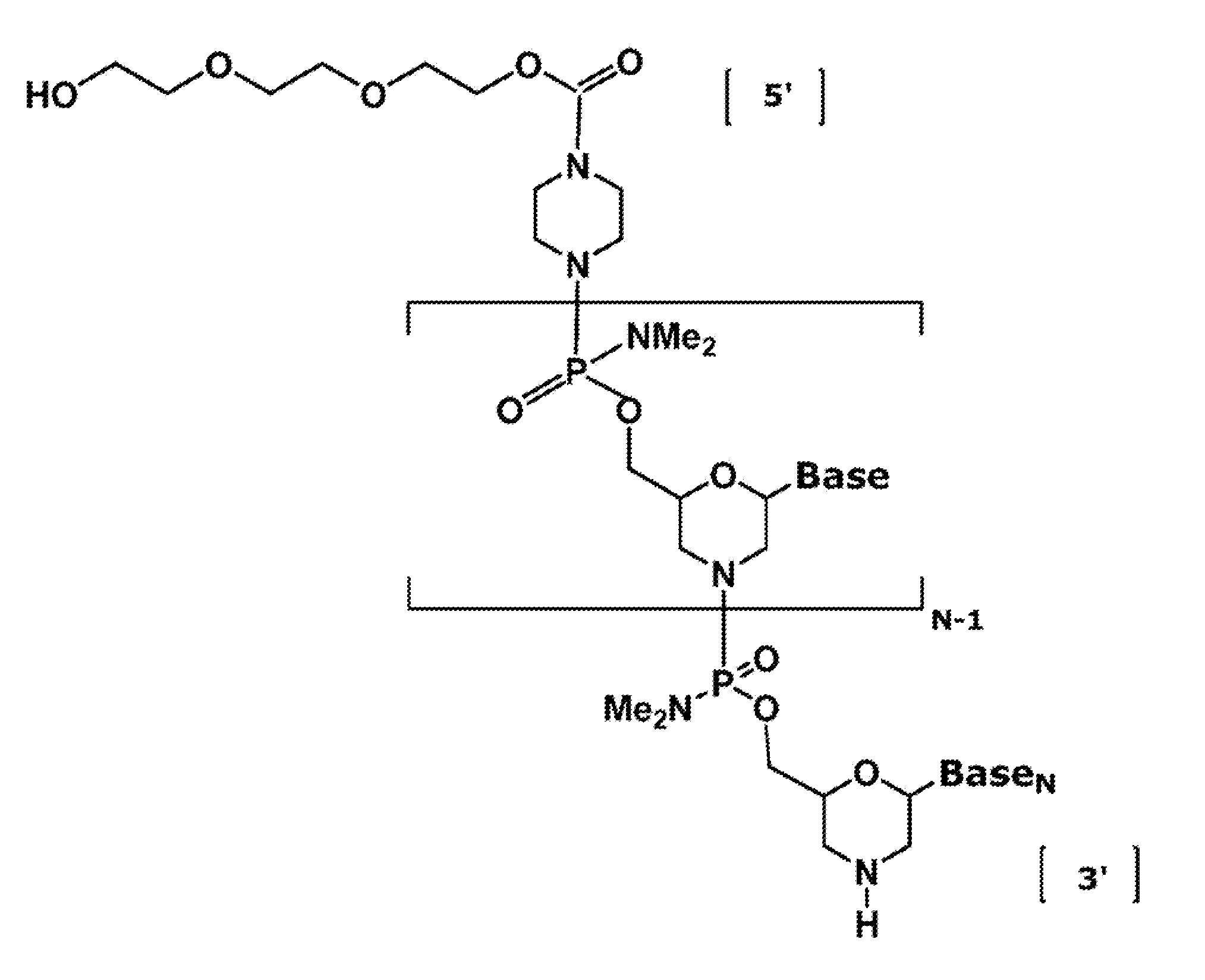
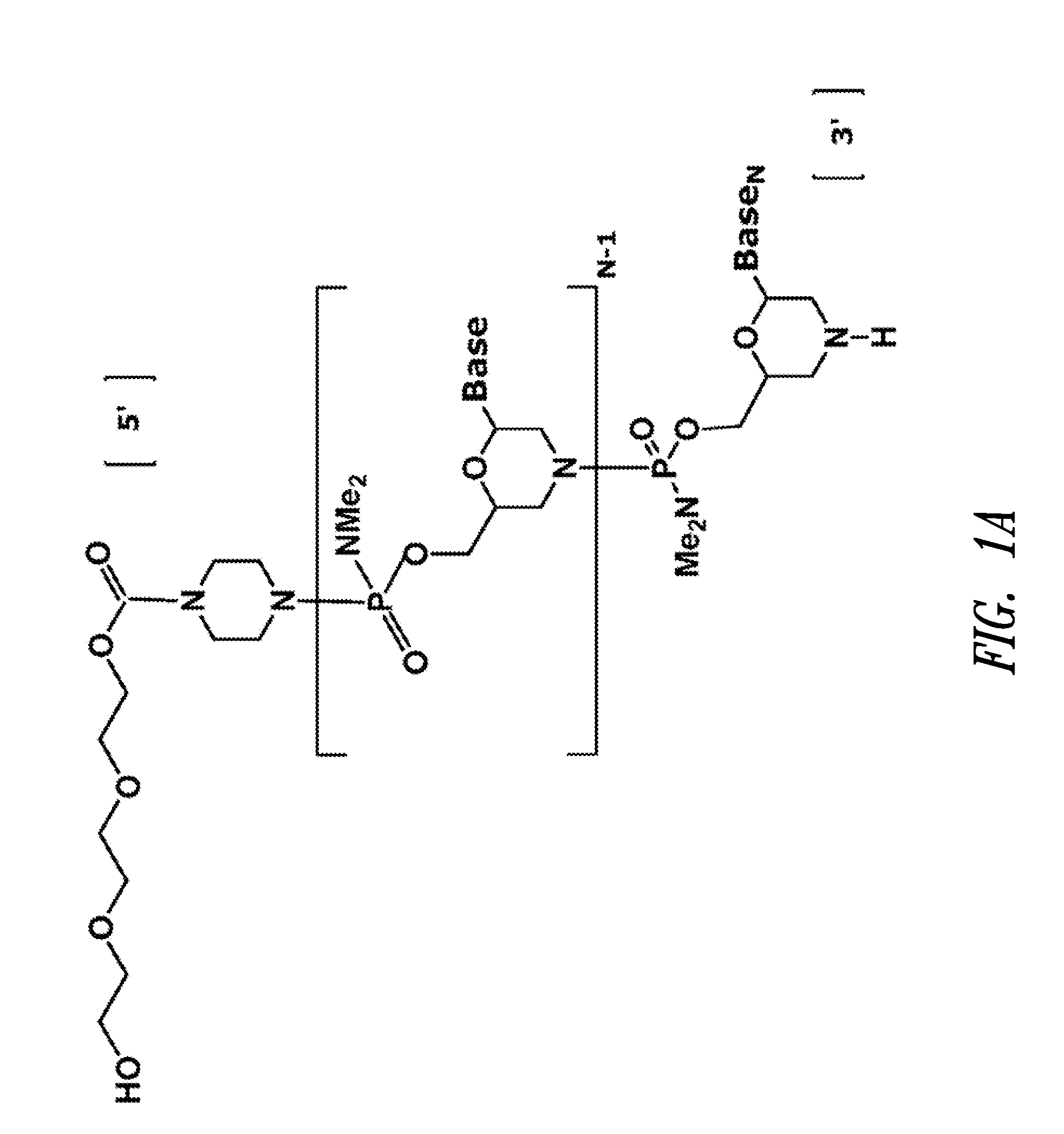
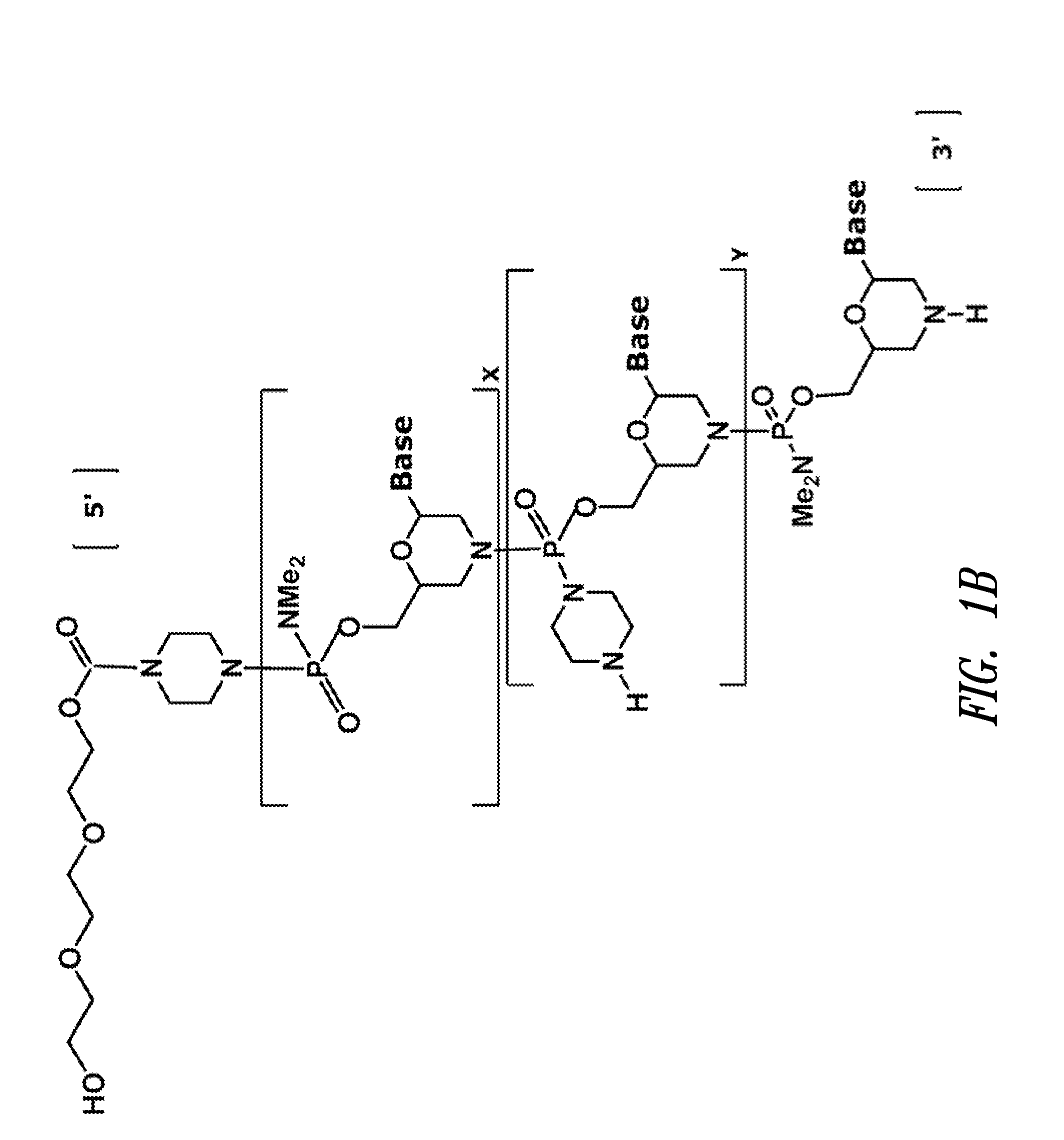
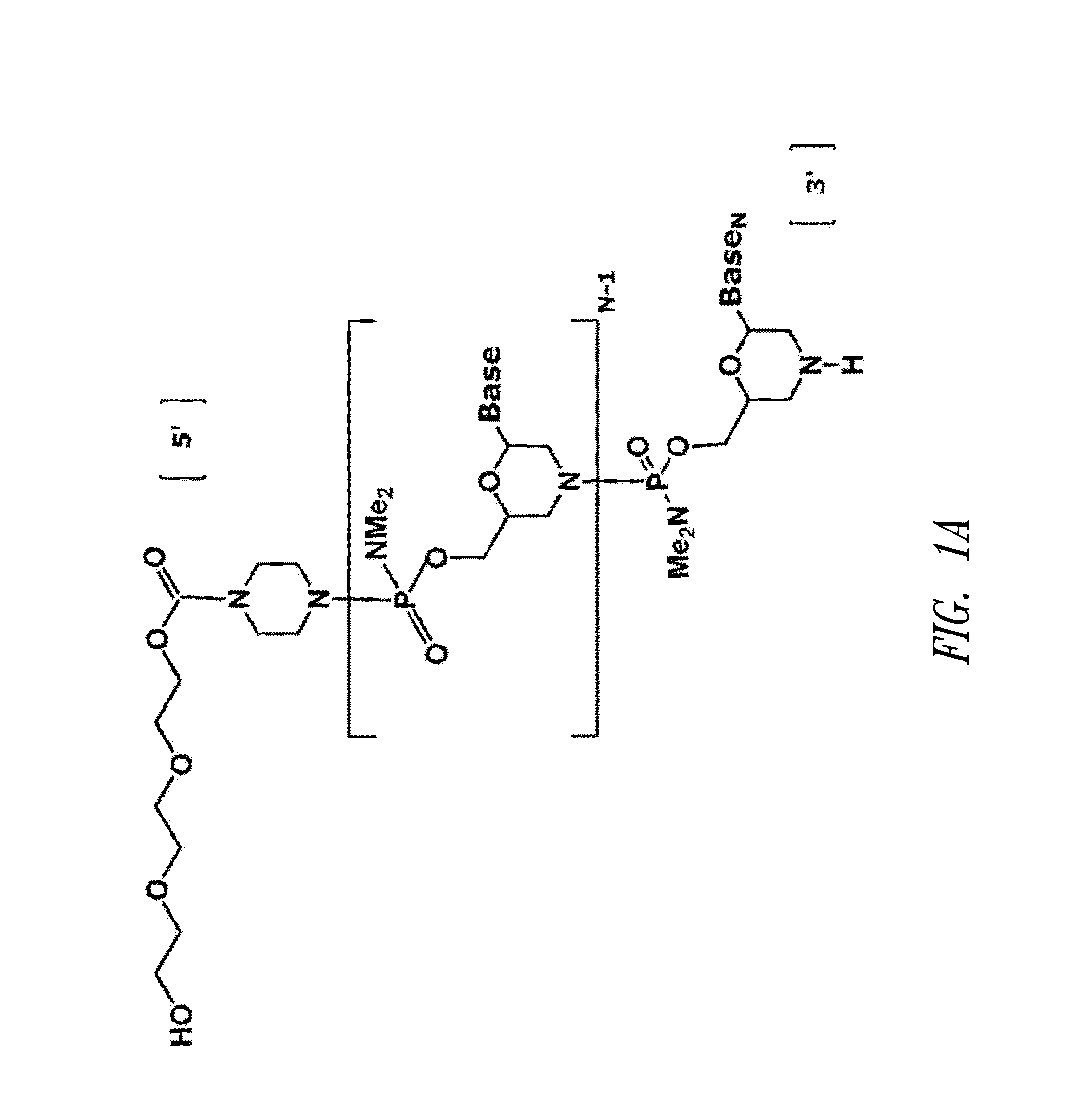
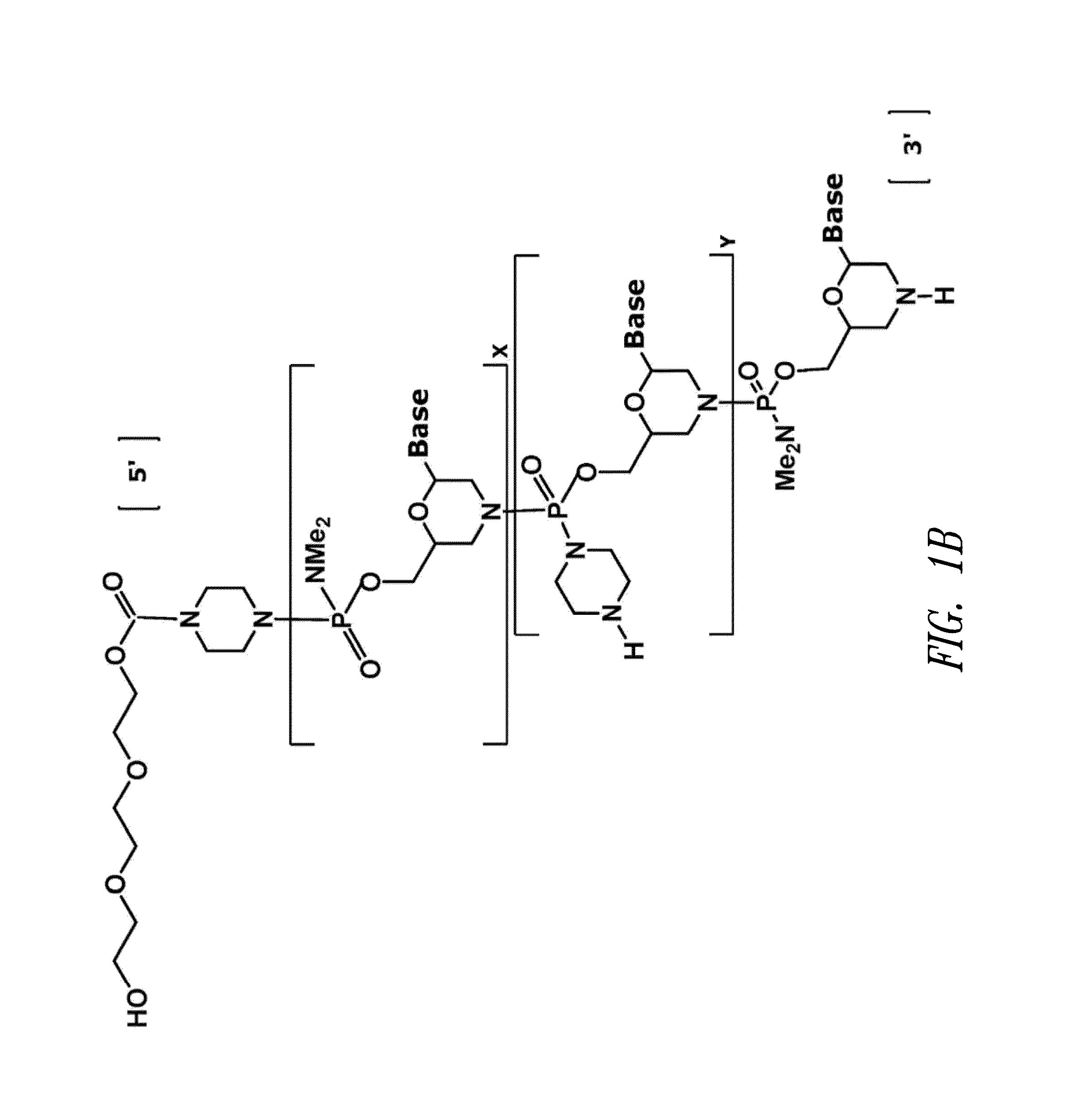
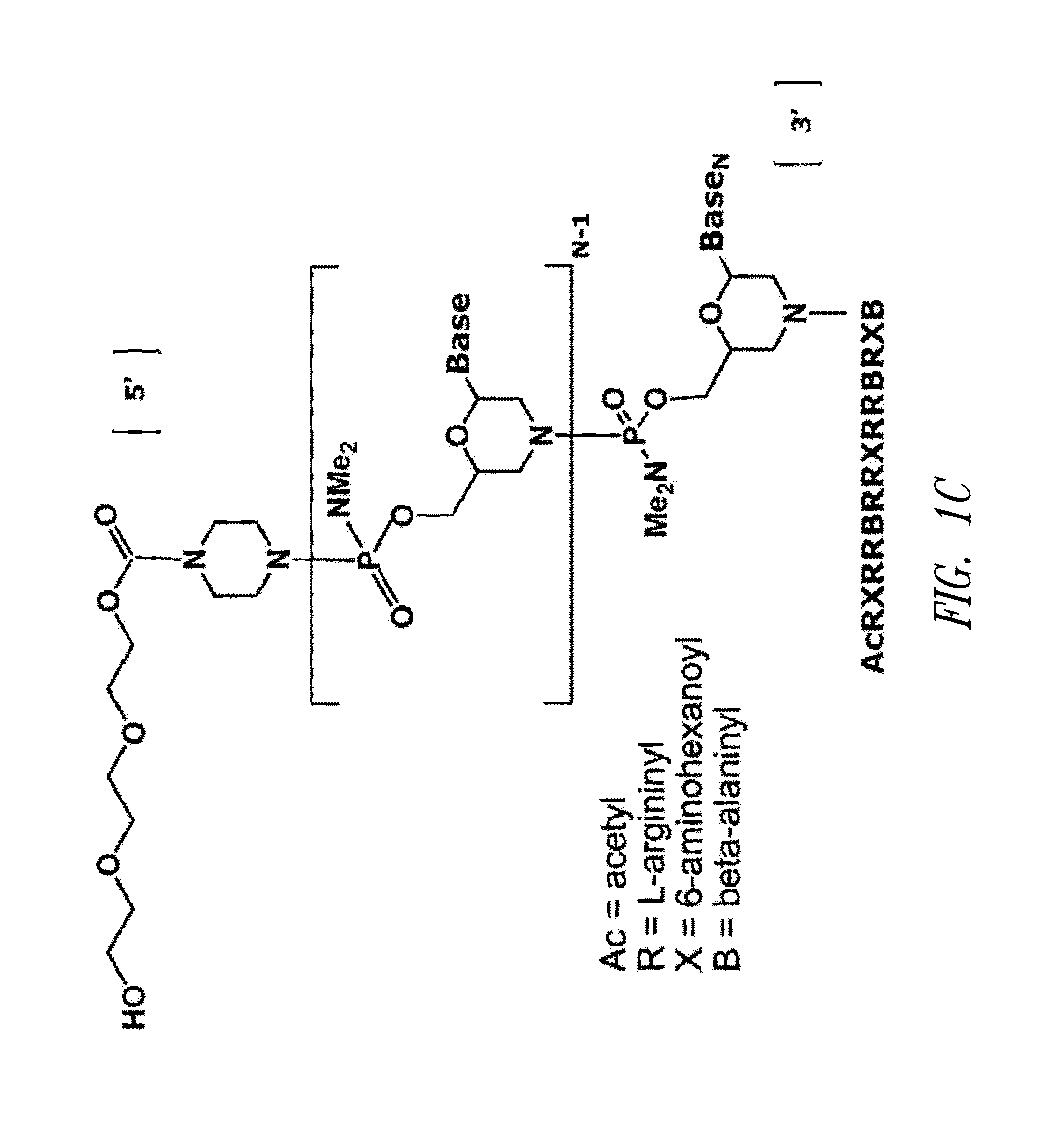
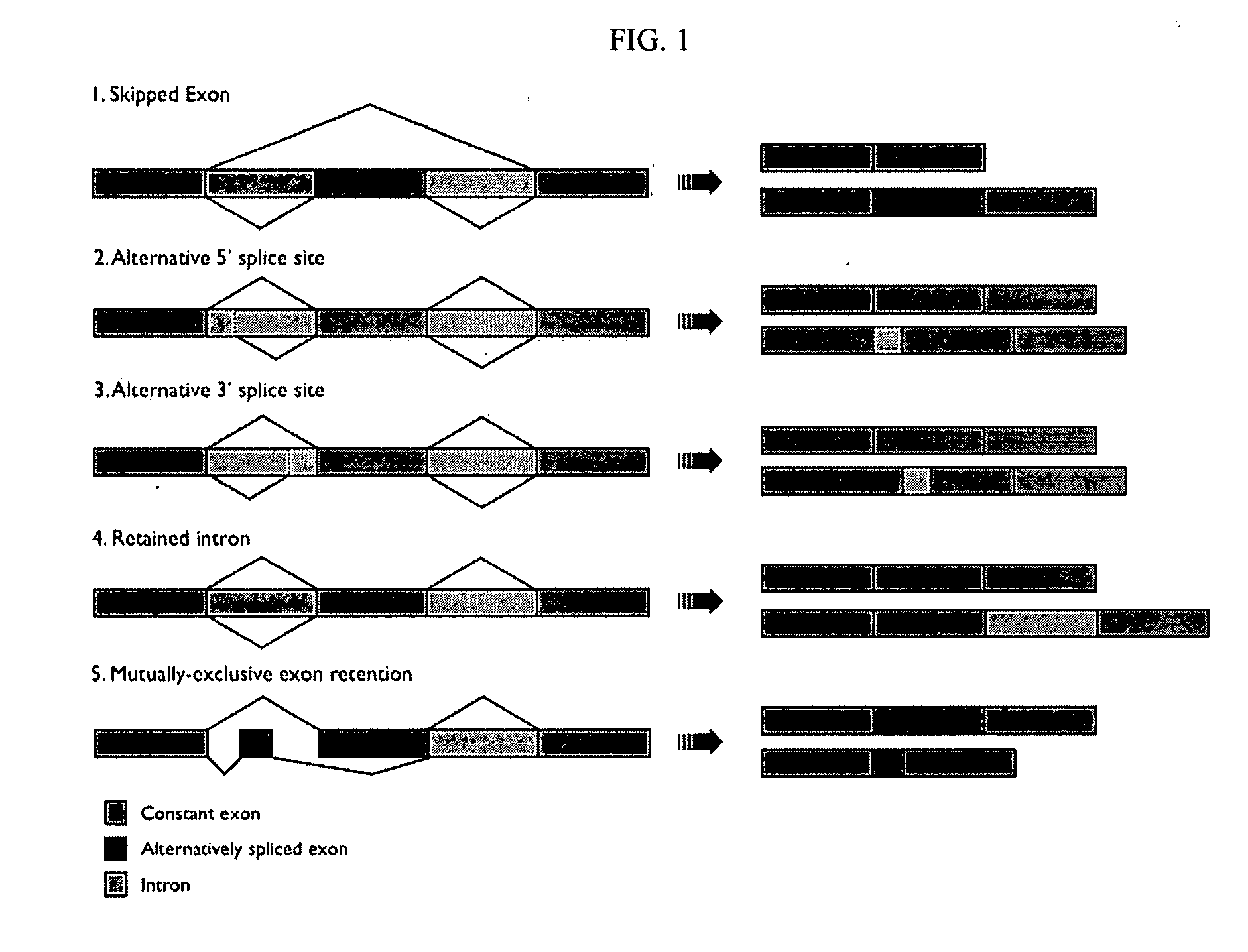
Antisense antiviral compound and method for treating influenza viral infection
ActiveUS20110118334A1Improve throughputLimit deliveryAntibacterial agentsSugar derivativesInfluenzavirus CStart codon
The present invention relates to antisense antiviral compounds and methods of their use and production in inhibition of growth of viruses of the Orthomyxoviridae family and in the treatment of a viral infection. The compounds are particularly useful in the treatment of influenza virus infection in a mammal. Exemplary antisense antiviral compounds are substantially uncharged, or partially positively charged, morpholino oligonucleotides having 1) a nuclease resistant backbone, 2) 12-40 nucleotide bases, and 3) a targeting sequence of at least 12 bases in length that hybridizes to a target region selected from the following: a) the 5′ or 3′ terminal 25 bases of the negative sense viral RNA segment of Influenzavirus A, Influenzavirus B and Influenzavirus C; b) the terminal 30 bases of the 5′ or 3′ terminus of the positive sense vcRNA; c) the 45 bases surrounding the AUG start codon of an influenza viral mRNA and; d) 50 bases surrounding the splice donor or acceptor sites of influenza mRNAs subject to alternative splicing.
Owner:SAREPTA THERAPEUTICS INC
Antisense antiviral compound and method for treating influenza viral infection
ActiveUS8697858B2Improve throughputLimit deliveryAntibacterial agentsSugar derivativesInfluenzavirus CBase J
The present invention relates to antisense antiviral compounds and methods of their use and production in inhibition of growth of viruses of the Orthomyxoviridae family and in the treatment of a viral infection. The compounds are particularly useful in the treatment of influenza virus infection in a mammal. Exemplary antisense antiviral compounds are substantially uncharged, or partially positively charged, morpholino oligonucleotides having 1) a nuclease resistant backbone, 2) 12-40 nucleotide bases, and 3) a targeting sequence of at least 12 bases in length that hybridizes to a target region selected from the following: a) the 5′ or 3′ terminal 25 bases of the negative sense viral RNA segment of Influenzavirus A, Influenzavirus B and Influenzavirus C; b) the terminal 30 bases of the 5′ or 3′ terminus of the positive sense vcRNA; c) the 45 bases surrounding the AUG start codon of an influenza viral mRNA and; d) 50 bases surrounding the splice donor or acceptor sites of influenza mRNAs subject to alternative splicing.
Owner:SAREPTA THERAPEUTICS INC
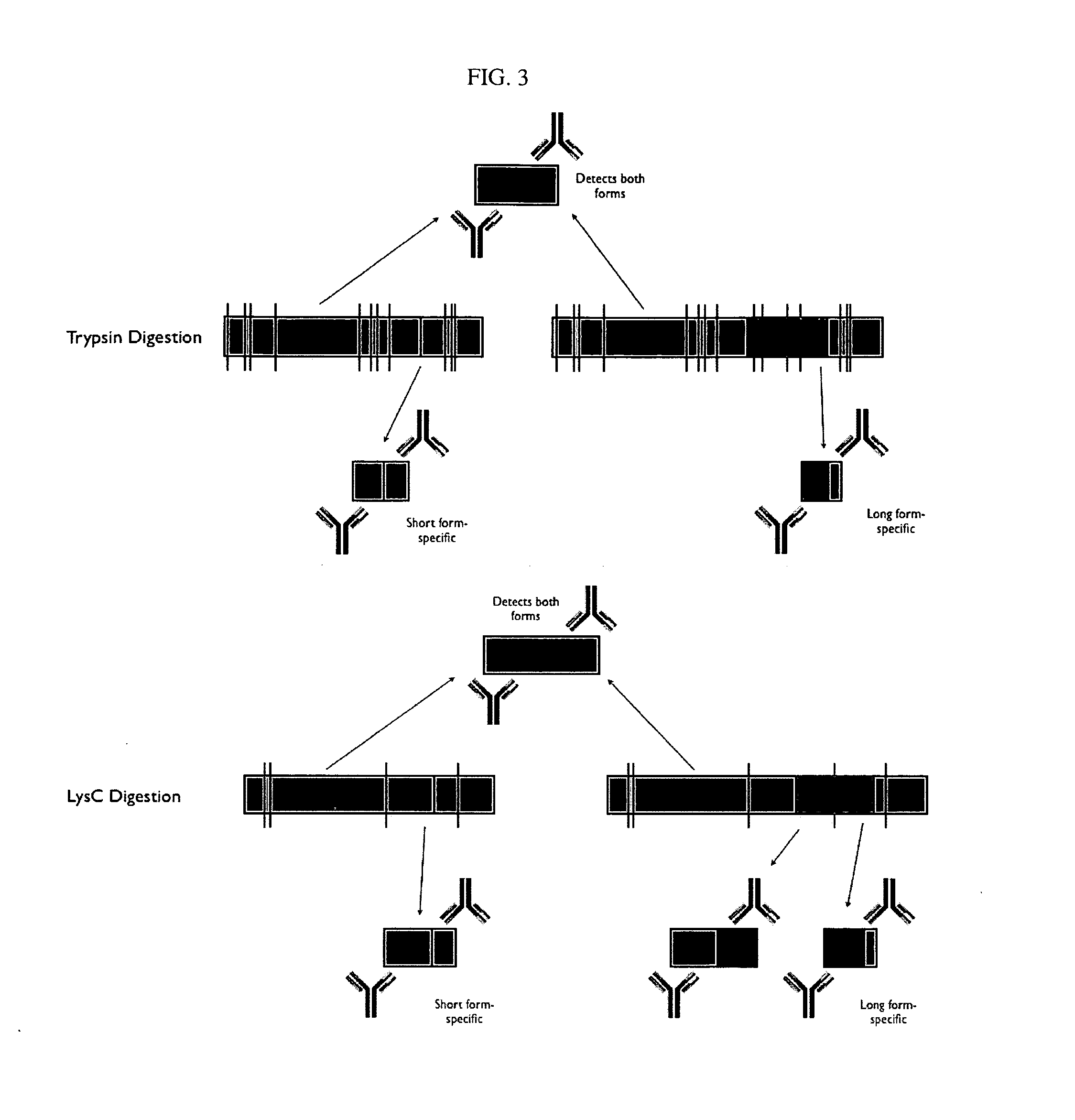
Protein isoform discrimination and quantitative measurements thereof
InactiveUS20070224628A1Reliably retrievedMicrobiological testing/measurementBiological testingAlternative splicingBiology
The invention relates to methods, reagents and apparatus for detecting protein isoforms (e.g., those due to alternative splicing, or different disease protein isoforms or degradation products) in a sample, including using combinations of capture agents to identify the isoforms to be detected / measured.
Owner:MILLIPORE CORP
Method for identification and quantification of nucleic acid expression, splice variant, translocation, copy number, or methylation changes
Owner:CORNELL UNIVERSITY
Methods of analysis of alternative splicing in mouse
InactiveUS7341835B2Bioreactor/fermenter combinationsBiological substance pretreatmentsRNA IsoformsNucleic acid sequencing
The invention provides nucleic acid sequences which are complementary, in one embodiment, to a wide variety of mouse genes. The invention provides the sequences in such a way as to make them available for a variety of analyses. In one embodiment the nucleic acid sequences provided are present as an array of probes that may be used to measure gene expression of different mature RNA isoforms from at least 5,000 alternatively spliced mouse genes. As such, the invention relates to diverse fields impacted by the nature of molecular interaction, including chemistry, biology, medicine, pharmacology and medical diagnostics.
Owner:AFFYMETRIX INC
Method and system for predicting splice variant from DNA chip expression data
InactiveUS20020029113A1Microbiological testing/measurementBiological testingEukaryotic geneMessenger RNA
A system and method predict alternative splicing transcripts using DNA chip expression data as a primary data source. The system and method may perform prediction of alternative splicing of pre-messenger RNA that may be used, for example, for regulating eukaryotic gene expression.
Owner:WARNER-LAMBERT CO
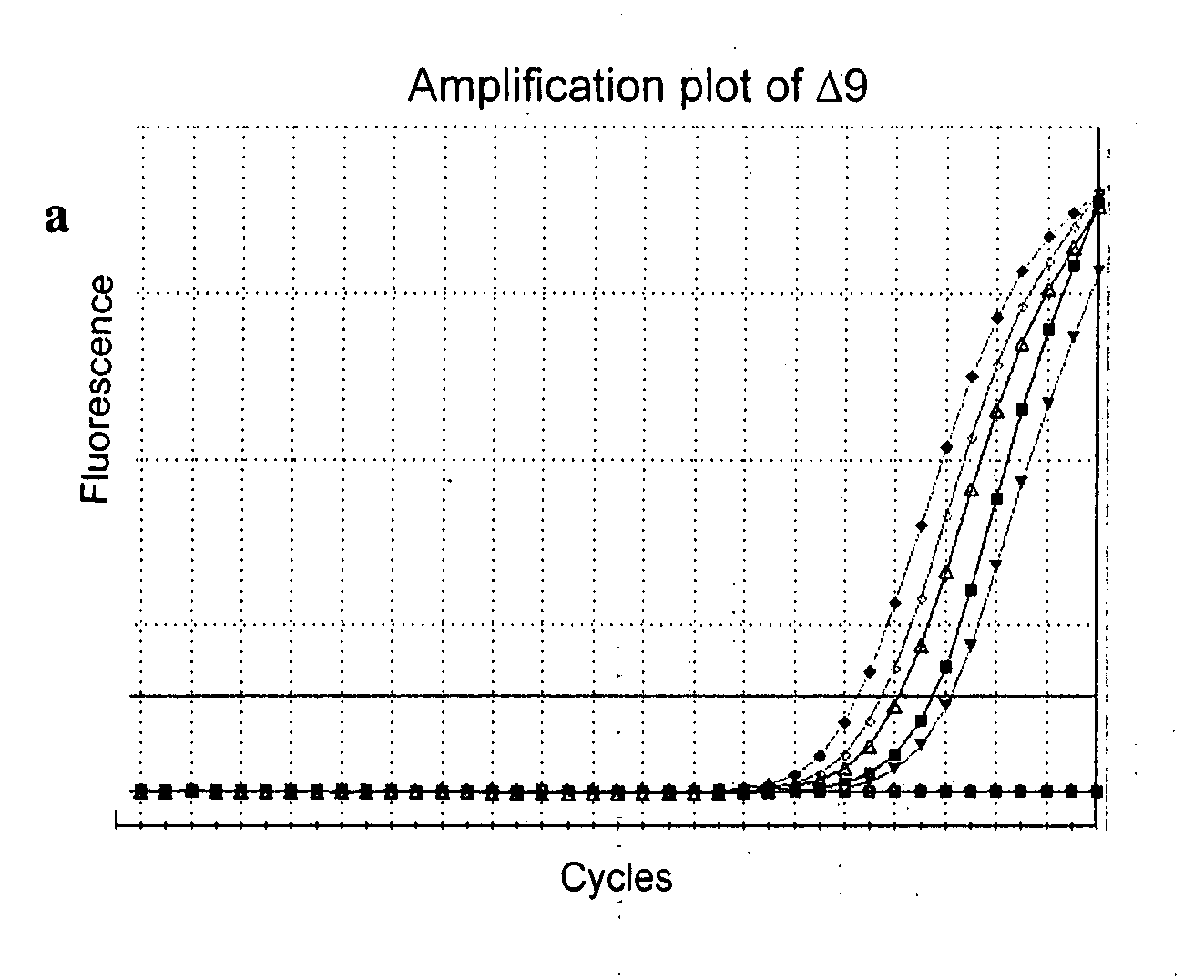
Splice variants of human IL-23 receptor (IL-23R) mRNA and use of a delta 9 isoform in predicting inflammatory bowel diseases
ActiveUS20090311694A1Increased riskDecreased in levelSugar derivativesMicrobiological testing/measurementCervical tissueCrohn's disease
There is disclosed the cloning and identification of human IL-23R splice variants caused by alternative splicing of the IL-23R mRNA in human. Alternative mRNA forms occur through skipping one, multiple full exons or partial exons, within the IL-23R gene. A total of twenty-five (25) different IL-23R transcripts were identified. A novel exon deletion (exon 9) isoform in the interleukin 23 receptor is disclosed, denoted as Δ9. The present application also describes a quantitative assay to measure different IL-23R isoform. Detection of Δ9 isoform of IL-23R is predominantly present in colon and cervical tissues. A decrease in Δ9 is observed in inflamed colon tissues in Crohn's patients. There is disclosed a method of predicting Crohn's disease by measuring Δ9 isoform of IL-23R.
Owner:MEDICAL DIAGNOSTIC LAB
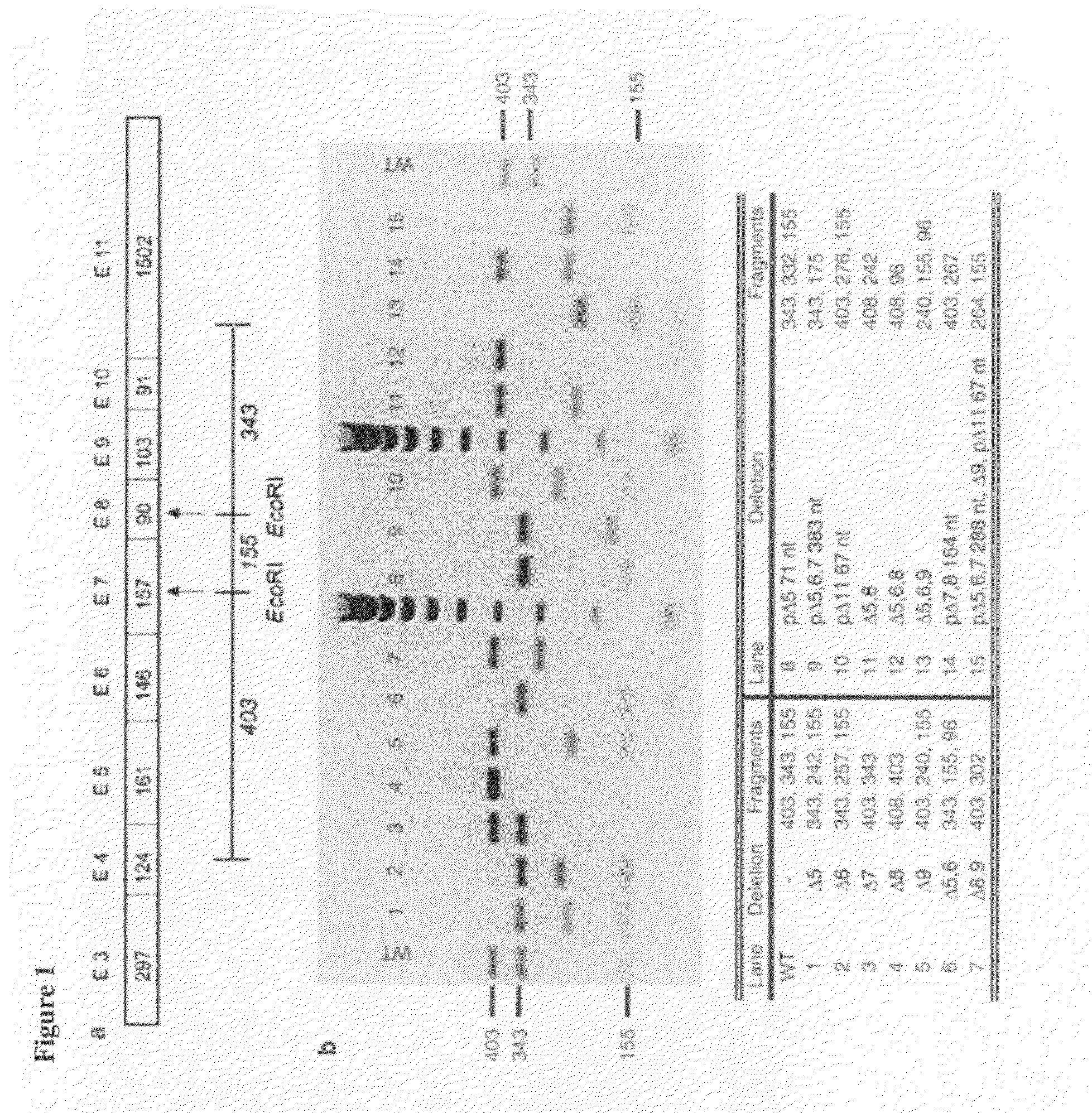
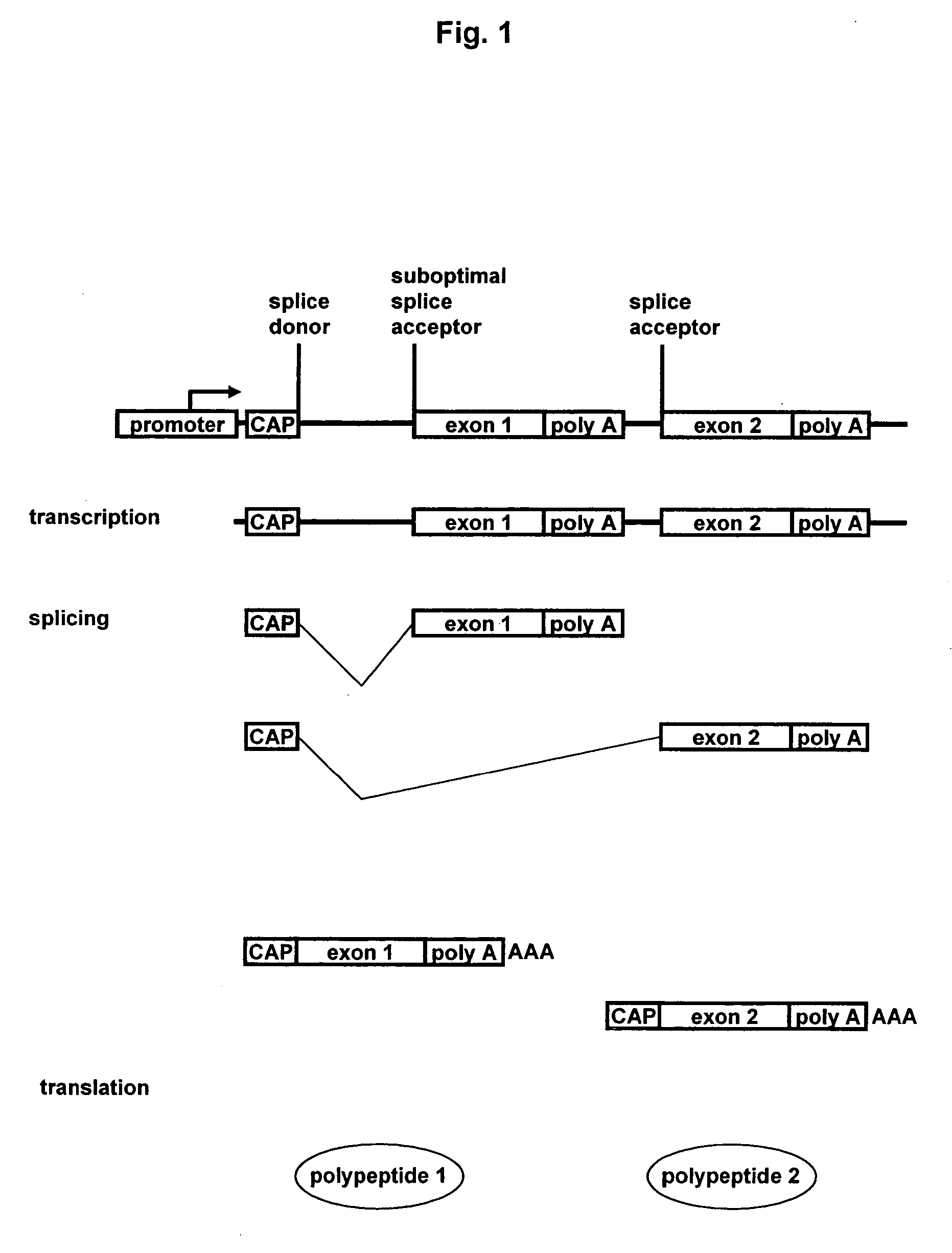
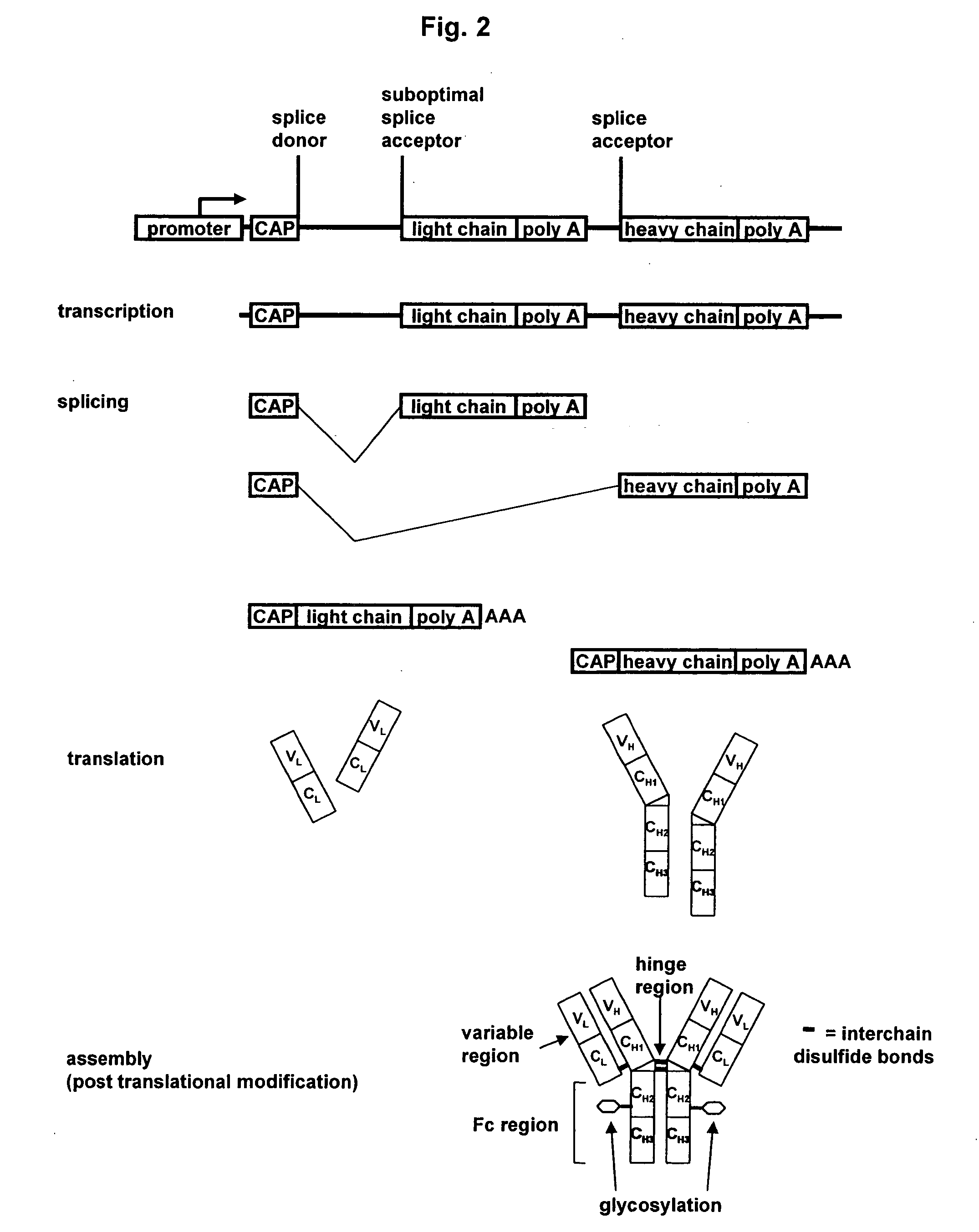
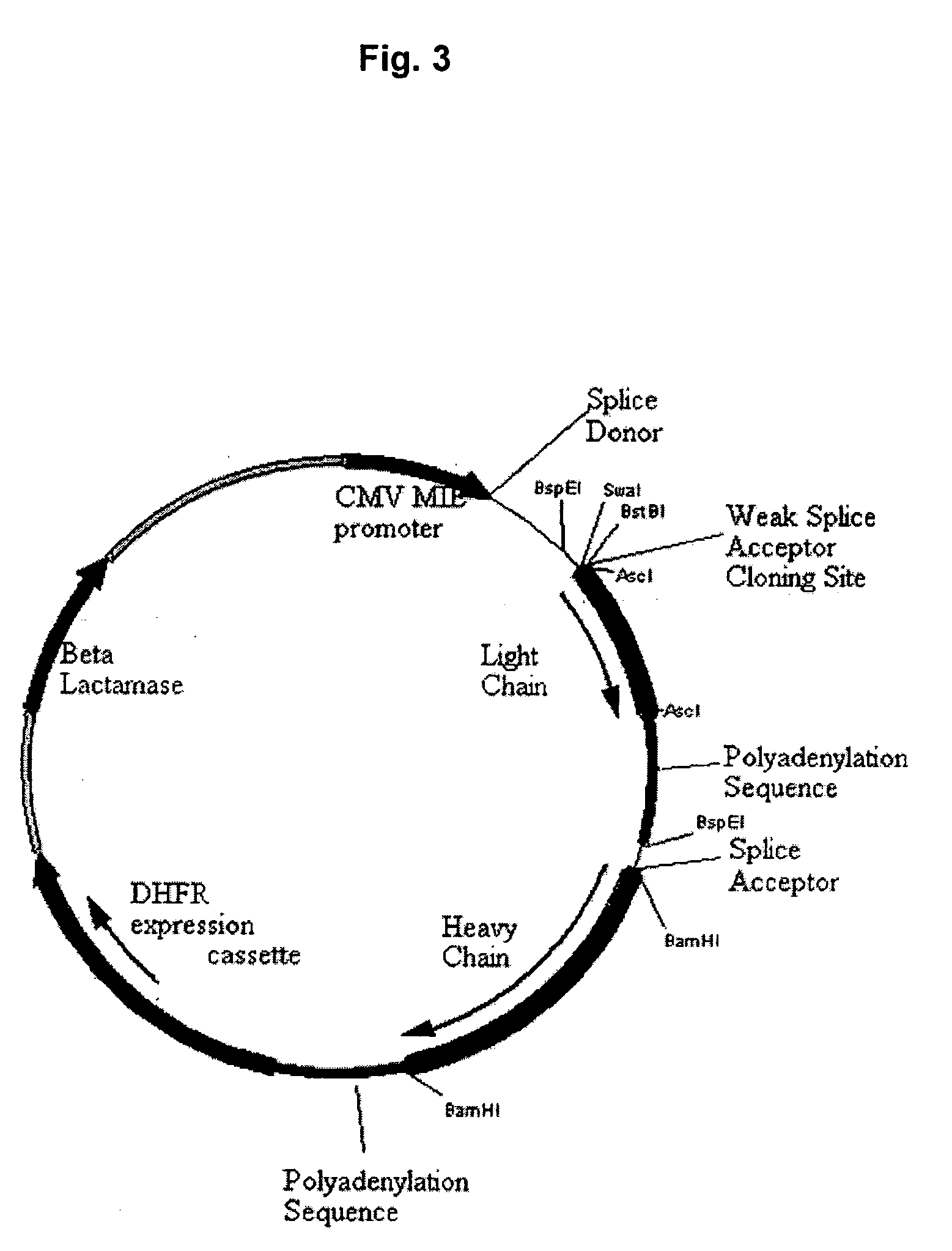
Methods and constructs for expressing polypeptide multimers in eukaryotic cells using alternative splicing
ActiveUS20050221430A1Efficient productionAntibacterial agentsPeptide/protein ingredientsAntibody fragmentsGene product
The invention provides a method of producing multiple polypeptides, such as antibodies or antibody fragments, in a eukaryotic cell using a single expression vector. The expression vector is engineered to comprise two or more expression cassettes under the control of a single promoter wherein the expression cassettes have splice sites which allow for their alternative splicing and expression as two or more independent gene products at a desired ratio. Use of the vector for the efficient expression of recombinant antibodies in eukaryotic host cells is disclosed as well as the use of such antibodies in diagnostic and therapeutic applications.
Owner:BIOGEN MA INC
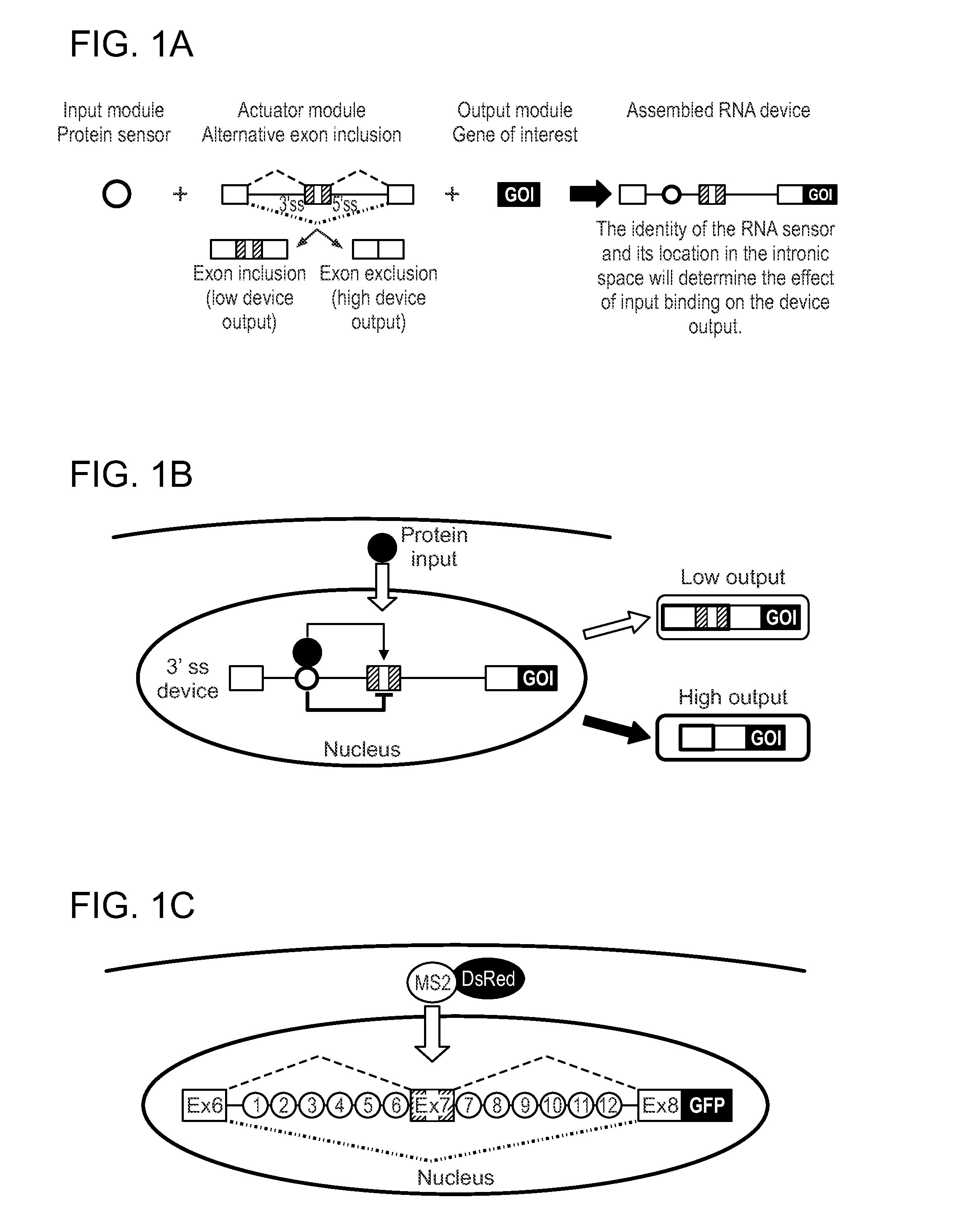
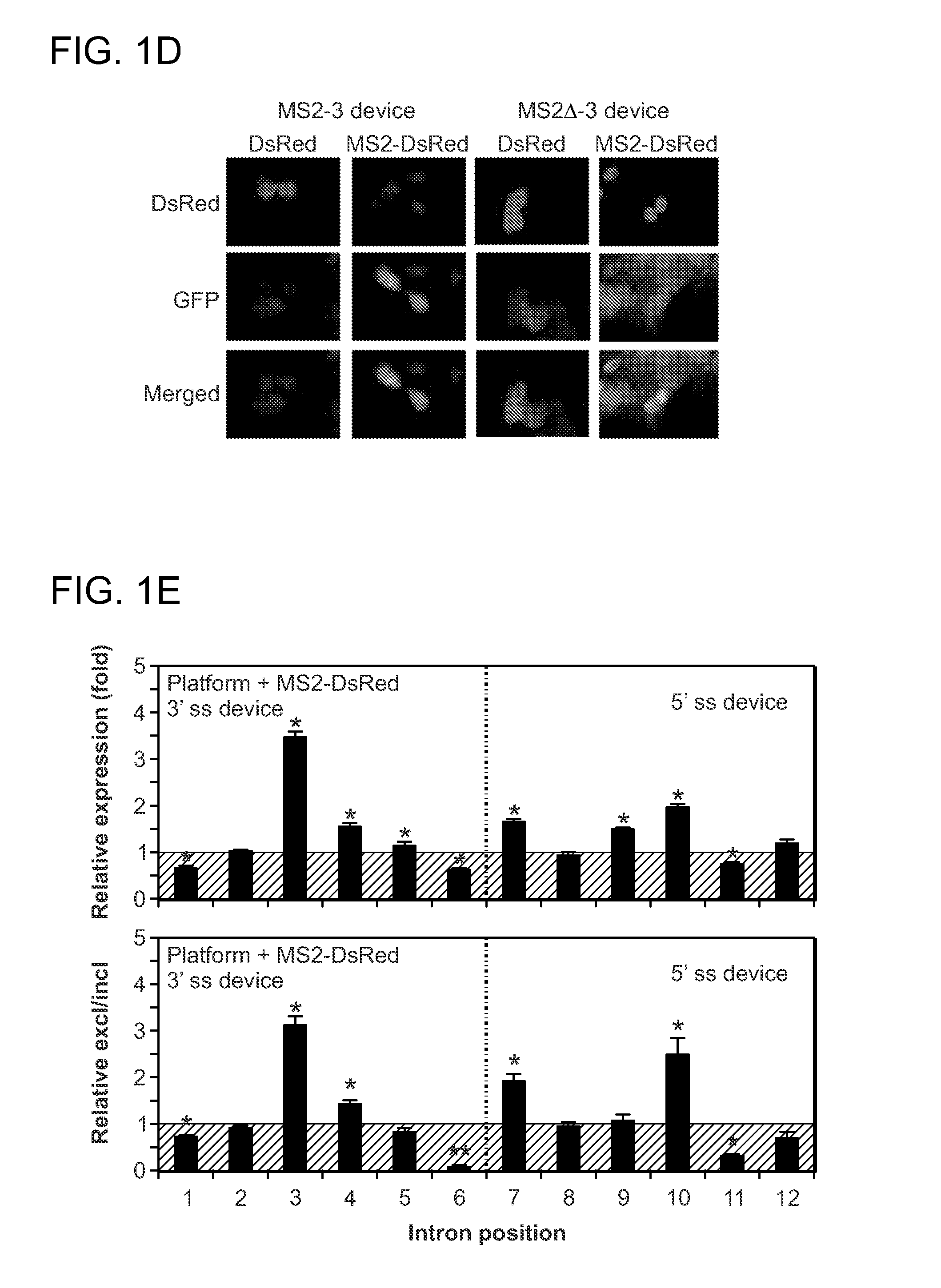
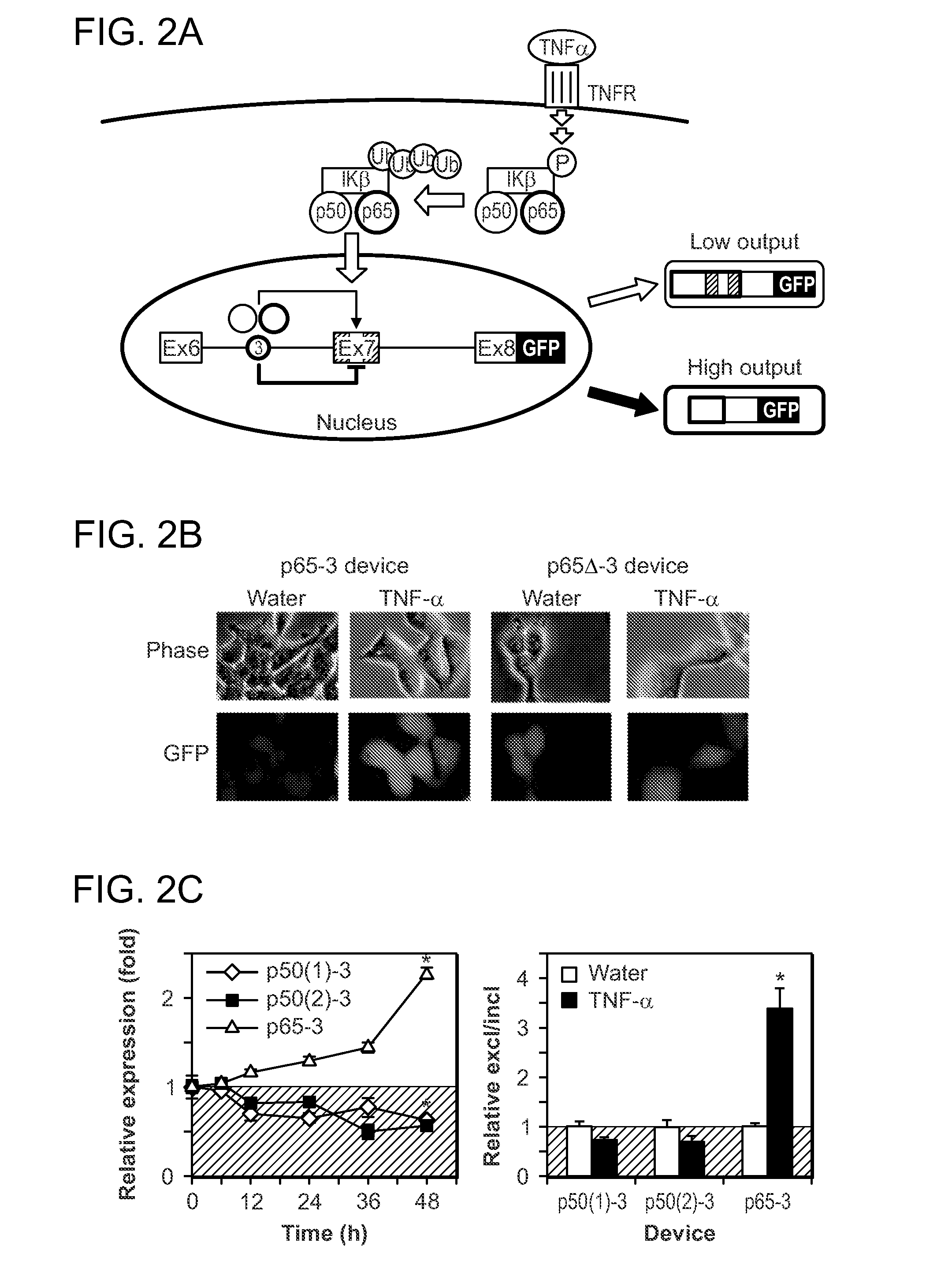
Protein-Responsive RNA Control Devices and Uses Thereof
ActiveUS20110111411A1Modulating the expression of the GOI in the cellAltered expressionSugar derivativesMicrobiological testing/measurementAlternative splicingBiology
The invention described herein relates to an RNA-based control device that senses the presence and / or concentration of at least one protein ligand, preferably through its protein-binding aptamer domain, and regulates a target gene expression through alternative splicing of the target gene in which the RNA-based control device is integrated. The device has uses in therapeutic as well as diagnostic applications.
Owner:CALIFORNIA INST OF TECH
Protein splice variant / isoform discrimination and quantitative measurements thereof
The invention relates to methods, reagents and apparatus for detecting protein isoforms (e.g., those due to alternative splicing, or different disease protein isoforms or degradation products) in a sample, including using combinations of capture agents, each combination being unique to the splicing variant to be detected / measured.
Owner:MILLIPORE CORP
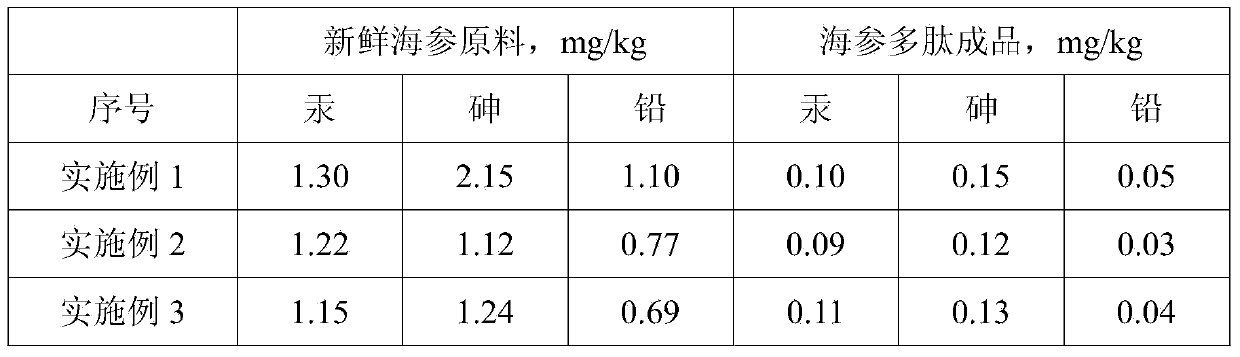
Preparation method of sea cucumber polypeptide
The invention discloses a preparation method of a sea cucumber polypeptide. The method comprises the following steps: screening raw materials; cleaning the raw materials; carrying out low-temperature enzymolysis; carrying out first concentration filtration; carrying out second concentration filtration; and drying. The reaction condition in the whole preparation process disclosed by the invention is mild; and the activity of the sea cucumber polypeptide can be ensured. Low-temperature enzymolysis at 40-60 DEG C is adopted, and enzyme cutting sites of sea cucumber protein is subjected to alternative splicing through papain and collagenase, so that the enzymolysis of the sea cucumber protein is relatively thorough; and the sea cucumber protein with high molecular weight is decomposed into a plurality of micro-molecule polypeptides with smaller molecular weight, so that the absorption requirements of a human body are met, full absorption of the human body on the sea cucumber polypeptide is facilitated, and the absorptivity can reach over 80%. The sea cucumber polypeptide prepared by the method disclosed by the invention is very low in harmful heavy metal ion content, and safe and non-toxic. The preparation method disclosed by the invention is low in wastewater discharge in the preparation process and beneficial to reduction of energy consumption, is capable of utilizing the value of the sea cucumber to the maximal extent, and has important social and economic significance.
Owner:冯群力
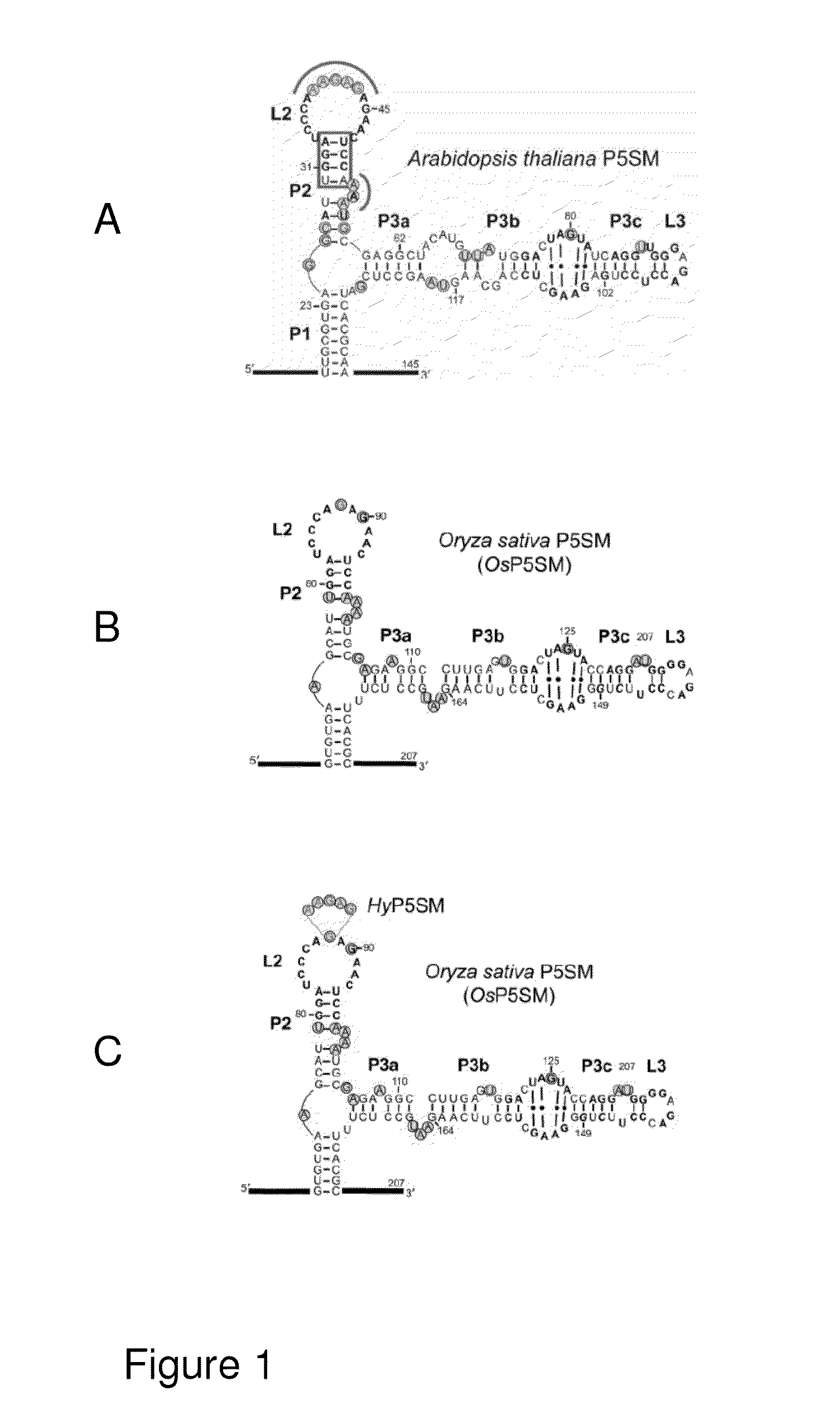
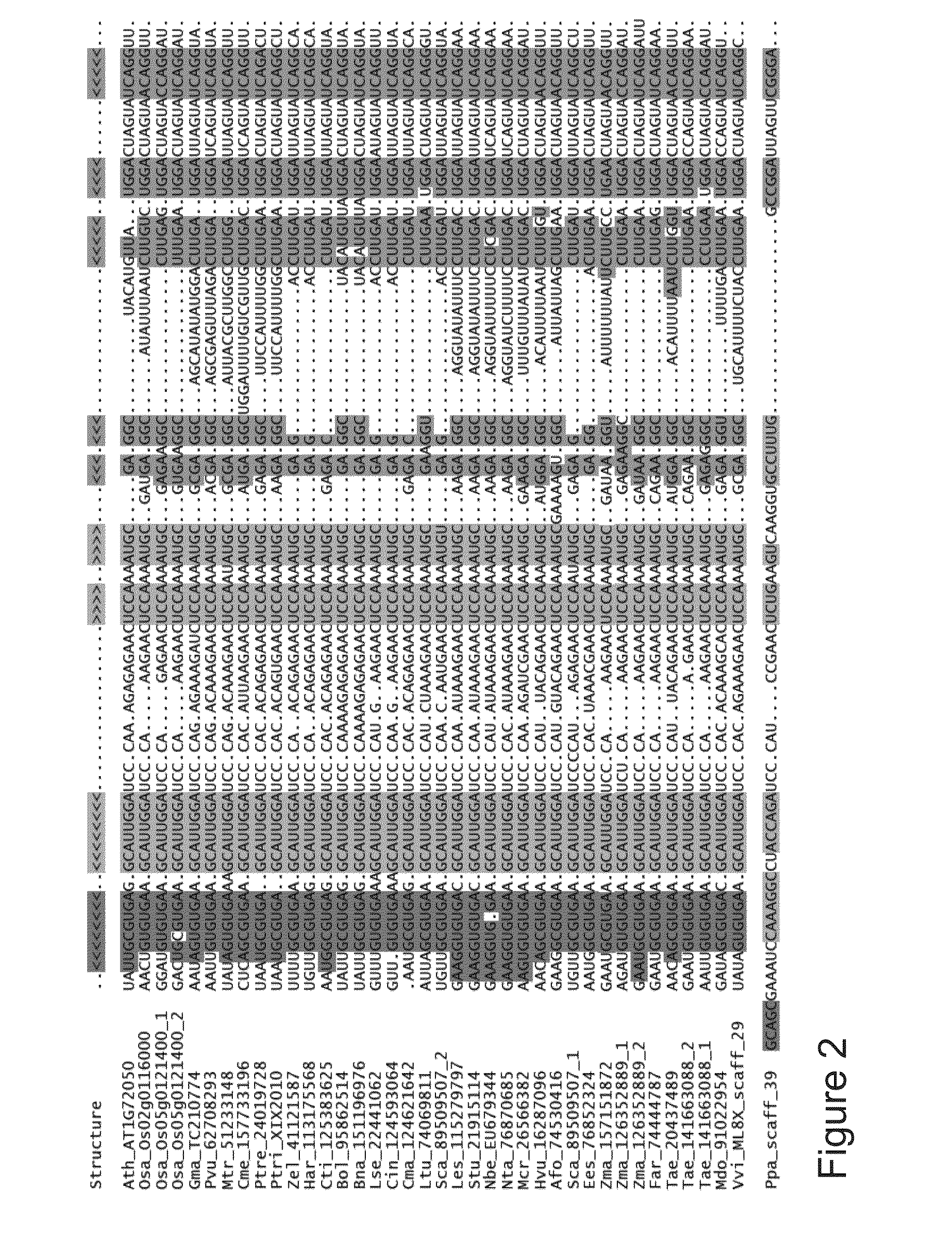
P5sm suicide exon for regulating gene expression
ActiveUS20130291226A1Robust gene activationMaintain fidelitySugar derivativesFermentationRegulator geneAlternative splicing
Owner:RGT UNIV OF CALIFORNIA
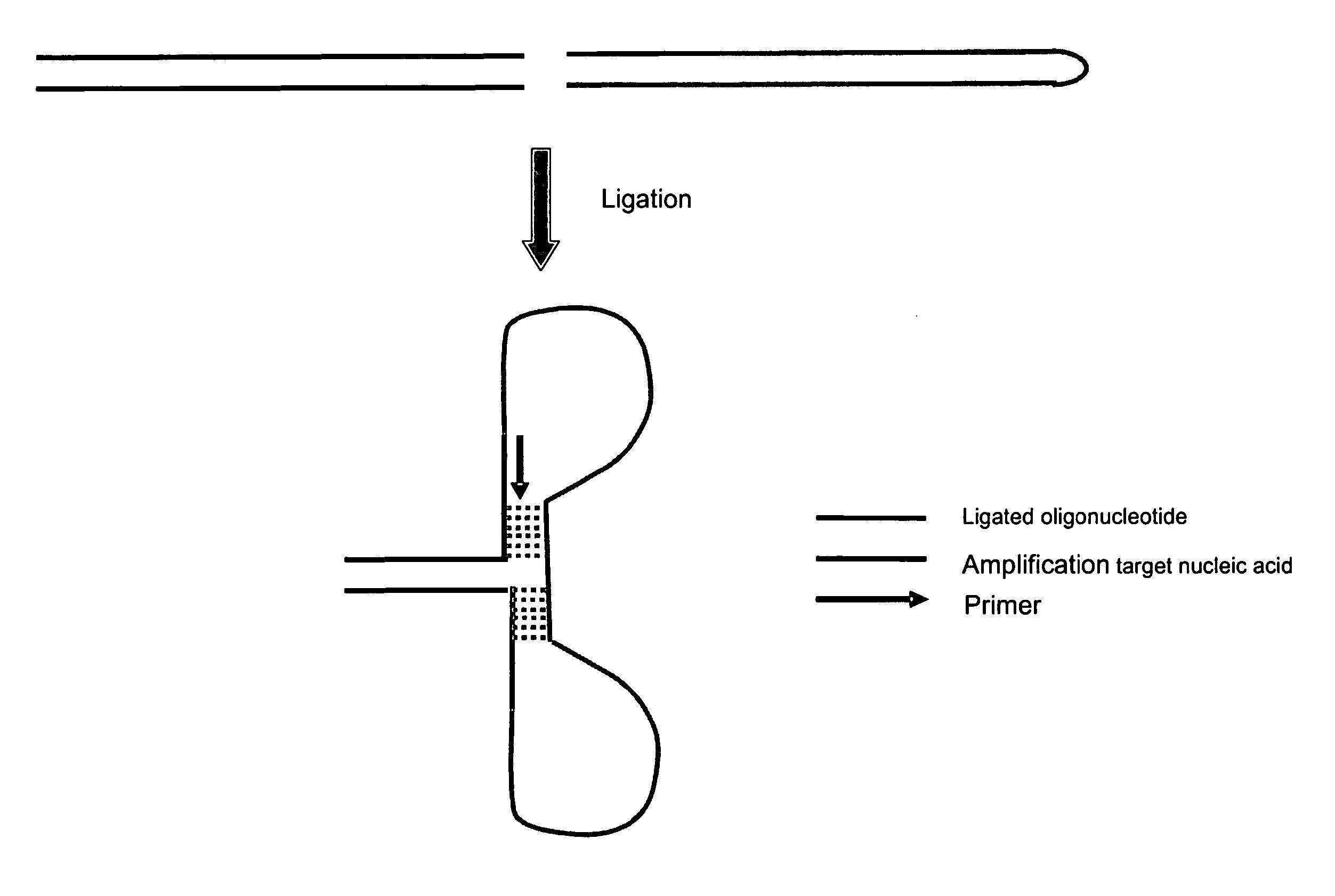
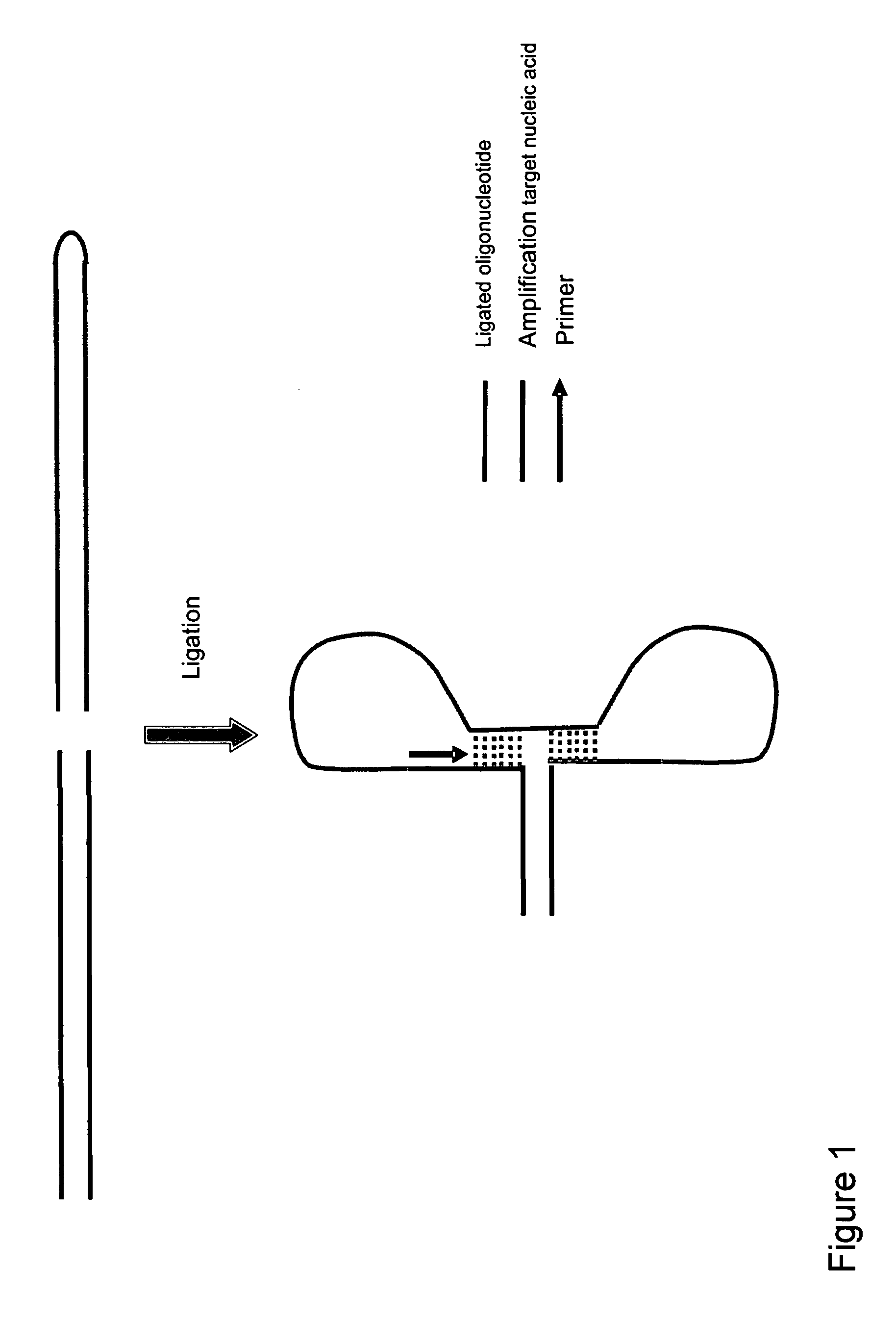
Method for amplifying nucleic acids
InactiveUS20060110745A1Increase ratingsEliminate amplificationMicrobiological testing/measurementRecombinant DNA-technologyNucleic acid sequencingA-DNA
The invention provides a sequence specific method for amplifying nucleic acids. More particularly, the invention provides a method for amplifying nucleic acid sequences which enables such sequences to be detected with high precision, rapidity and high specificity as compared to conventional methods. The present invention also provides a simple method for cloning nucleic acids, particularly, a rapid and simple method for amplifying alternative splicing forms synthesized by an alternative splicing which is performed in a process of preparing a matured mRNA from a DNA.
Owner:DNAFORM +1
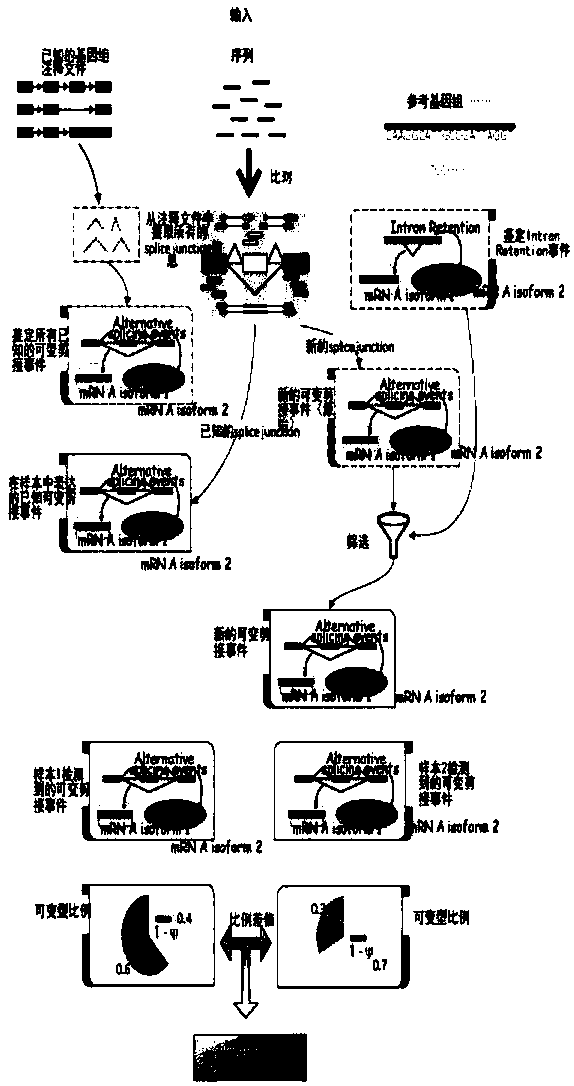
Eucaryon alternative splicing analysis method and system based on RNA-seq data
InactiveCN107766696AAccurate identificationHybridisationSpecial data processing applicationsAnalysis dataData filtering
The invention provides a eucaryon alternative splicing analysis method and system based on RNA-seq data. The method comprises the steps that transcriptome original sequencing data of one or more samples of a certain eucaryon with a reference genome and annotation is acquired through an illumina next-generation sequencing platform; unqualified data is filtered out, and the remaining data serves asto-be-analyzed data; next, basic analysis is performed, wherein the to-be-analyzed data of all the transcriptome samples is compared to the reference genome of the species, and a unique comparison result is screened out; expression quantities of all sample genes are calculated; the genes with significant difference expression are screened out; functional annotation and analysis are performed on the differential genes; then alternative splicing analysis is performed, wherein a known alternative splicing event is identified; a new alternative splicing event is identified; the difference betweenalternative splicing events of samples (sample groups) is analyzed; the correlation between alternative splicing and gene expression is analyzed; the alternative splicing analysis result is subjectedto statistical analysis, and a report is generated; and an alternative splicing visual graph is generated.
Owner:武汉生命之美科技有限公司
Treatment of amyotrophic lateral sclerosis
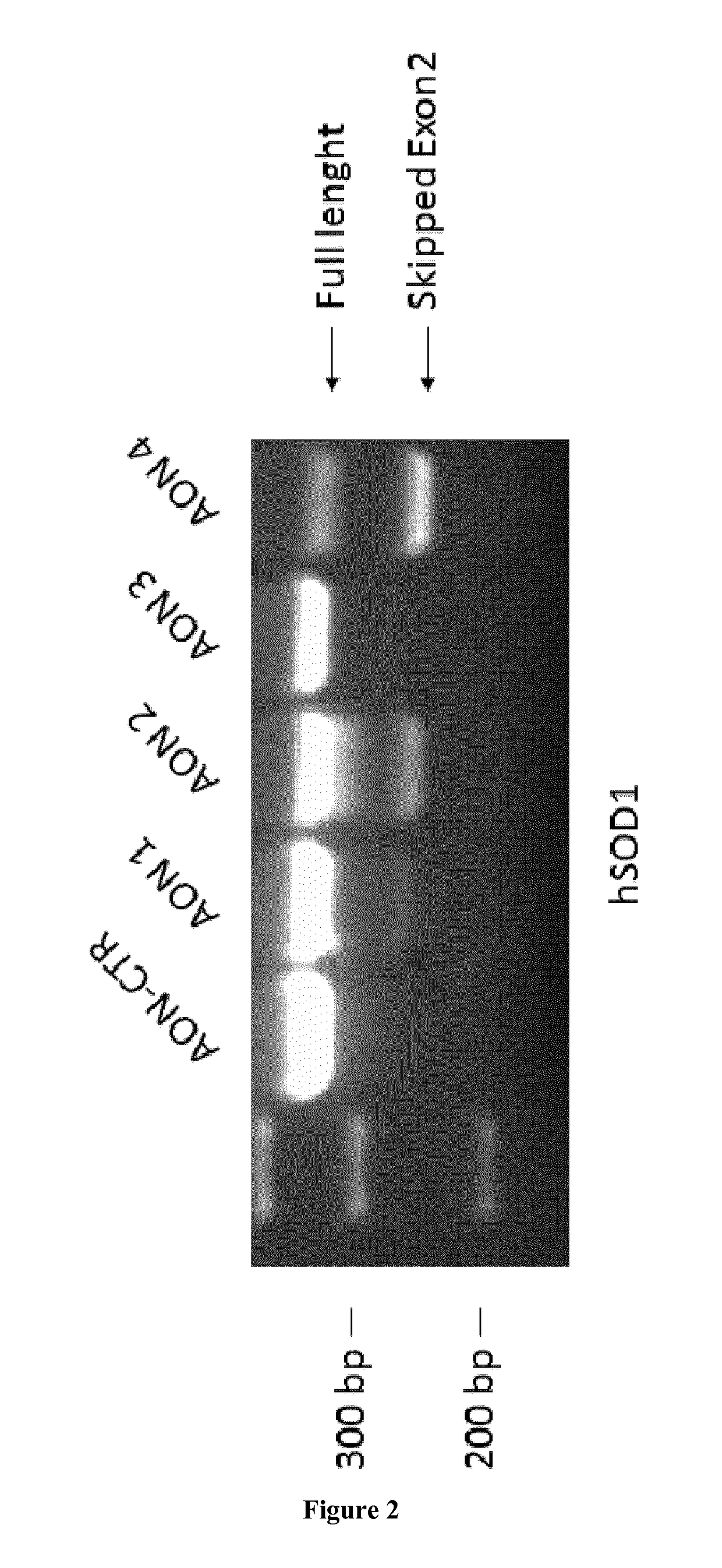
ActiveUS20170152517A1Improve survivalLower Level RequirementsOrganic active ingredientsSplicing alterationSOD1Alternative splicing
The invention relates to a method for the treatment of amyotrophic lateral sclerosis (ALS). Specifically, the invention implements the use of an antisense sequence adapted to affect alternative splicing in a human SOD1 pre-mRNA, thereby leading to the destruction of the skipped m RNA by the cell machinery.
Owner:AZOTIC TECH LTD +3
Protein isoform discrimination and quantitative measurements thereof
InactiveUS7645586B2Reliably retrievedMicrobiological testing/measurementBiological testingDiseaseAlternative splicing
The invention relates to methods, reagents and apparatus for detecting protein isoforms (e.g., those due to alternative splicing, or different disease protein isoforms or degradation products) in a sample, including using combinations of capture agents to identify the isoforms to be detected / measured.
Owner:MILLIPORE CORP
Gastrin compositions and formulations, and methods of use and preparation
Owner:WARATAH PHARMA INC
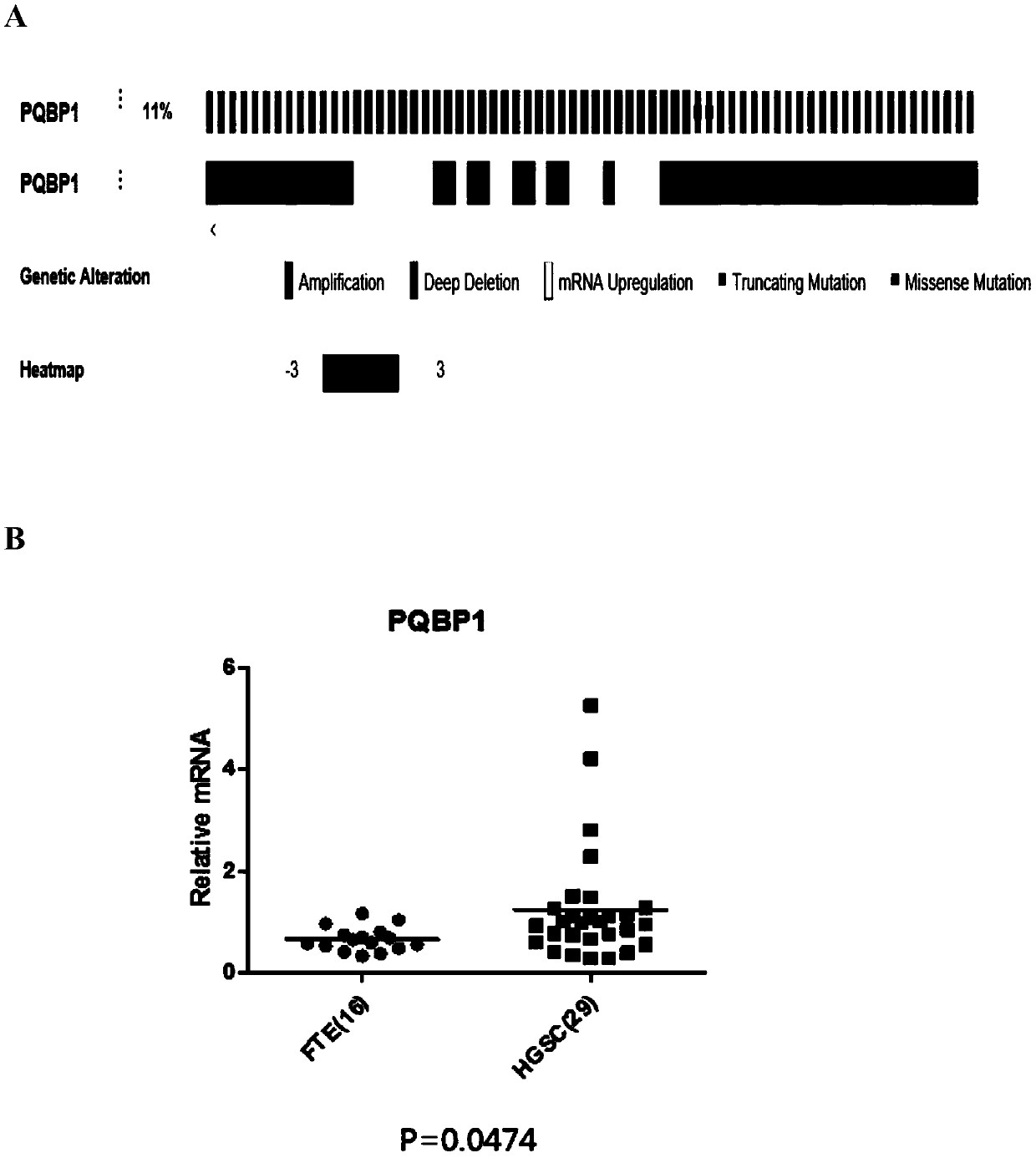
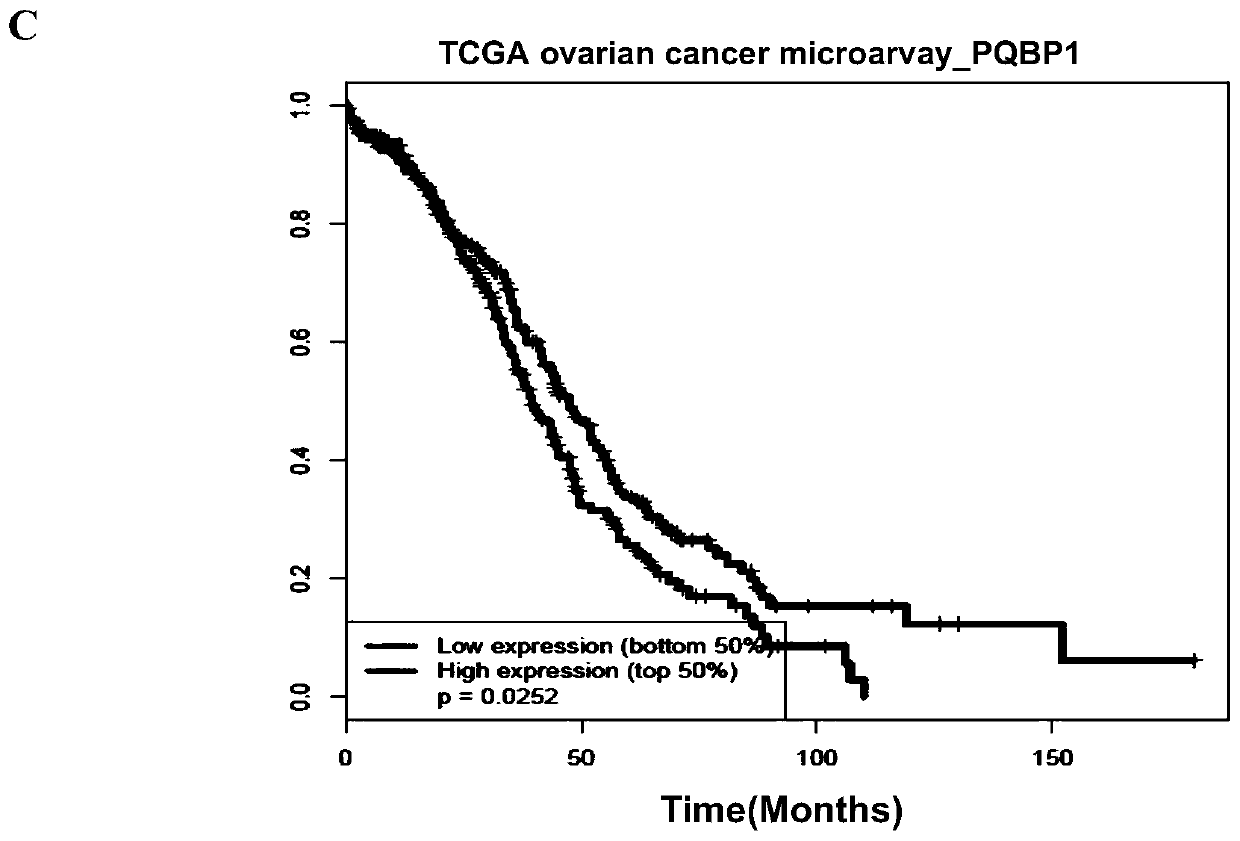
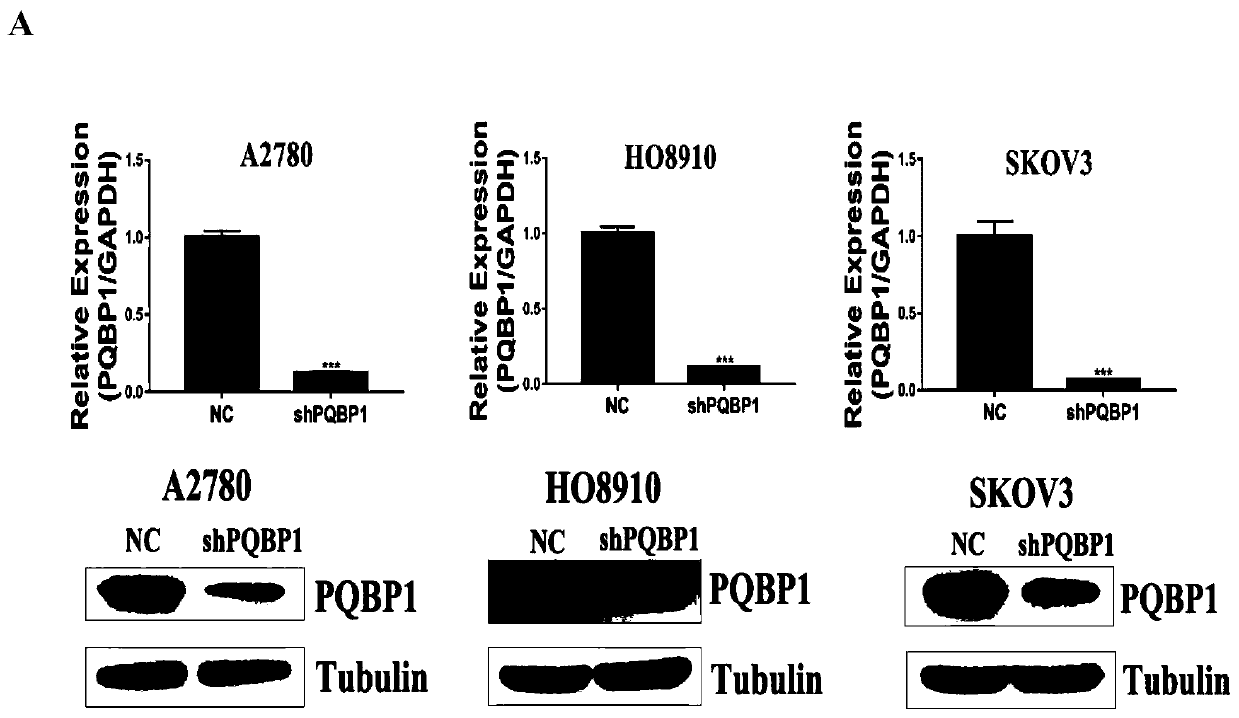
Application of PQBP1 in ovarian cancer diagnosis and treatment
ActiveCN110129447ATreatment reachedPromote proliferationOrganic active ingredientsMicrobiological testing/measurementTreatment targetsApoptosis
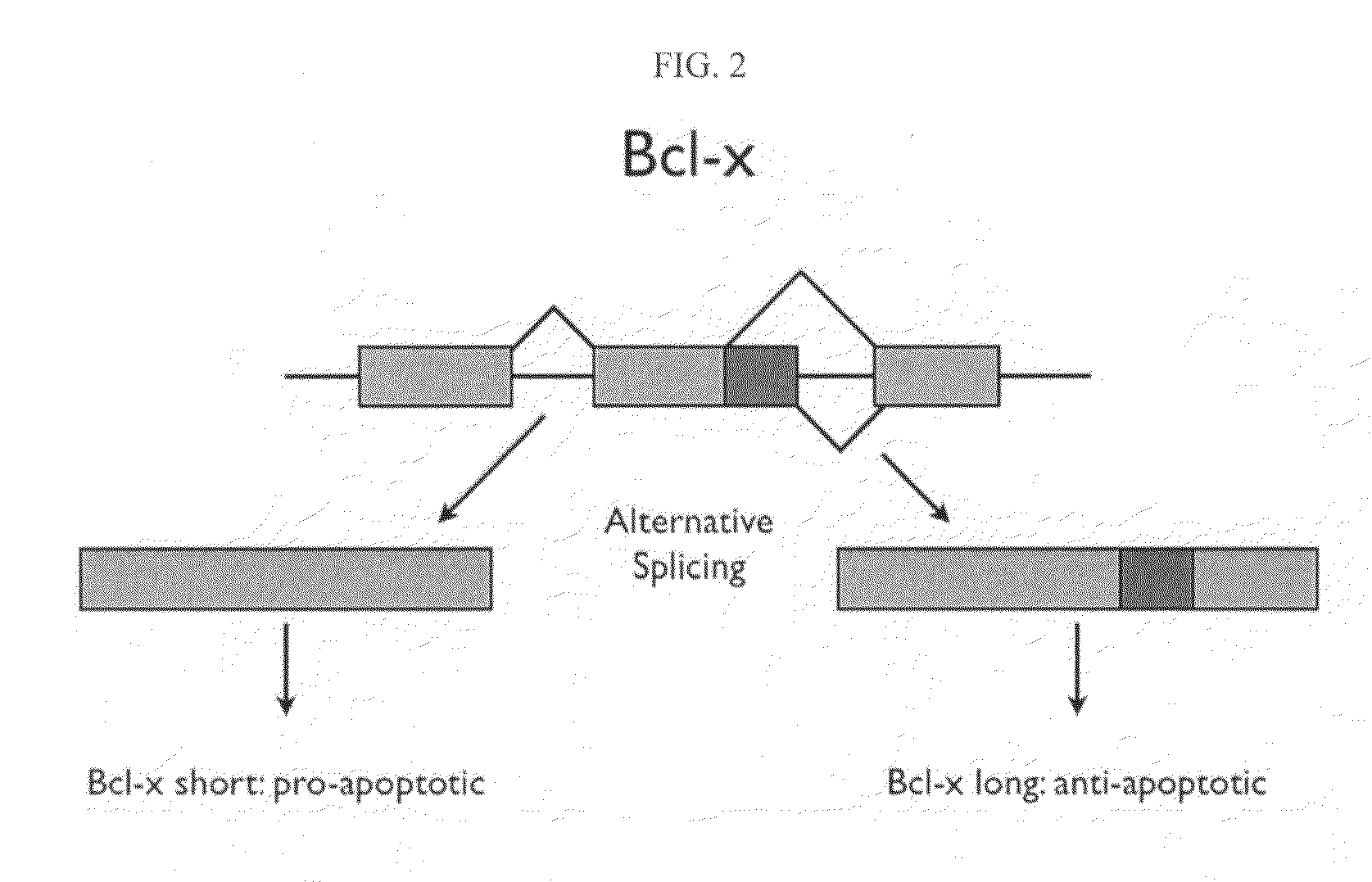
The invention provides application of PQBP1 in ovarian cancer diagnosis and treatment. The researches of the invention discovers that PQBP1 is high in expression in ovarian cancer patients, and in-vivo and in-vitro functional tests show that the PQBP1 can promote the proliferation and invasive migration of ovarian cancer cells. In addition, the researches discover that the PQBP1 can affect the selective splicing of endogenous Bcl-x, to be more specific, the PQBP1 can promote the expression of apoptosis inhibiting transcript Bcl-xL and inhibit the expression of apoptosis promoting transcript Bcl-xS to finally inhibit the apoptosis of the ovarian cancer cells. The researches show that the PQBP1 is a splicing factor participating in ovarian cancer occurrence and development, can be used as the biological marker and treatment target of ovarian cancer and has a good actual application value.
Owner:SHANDONG UNIV
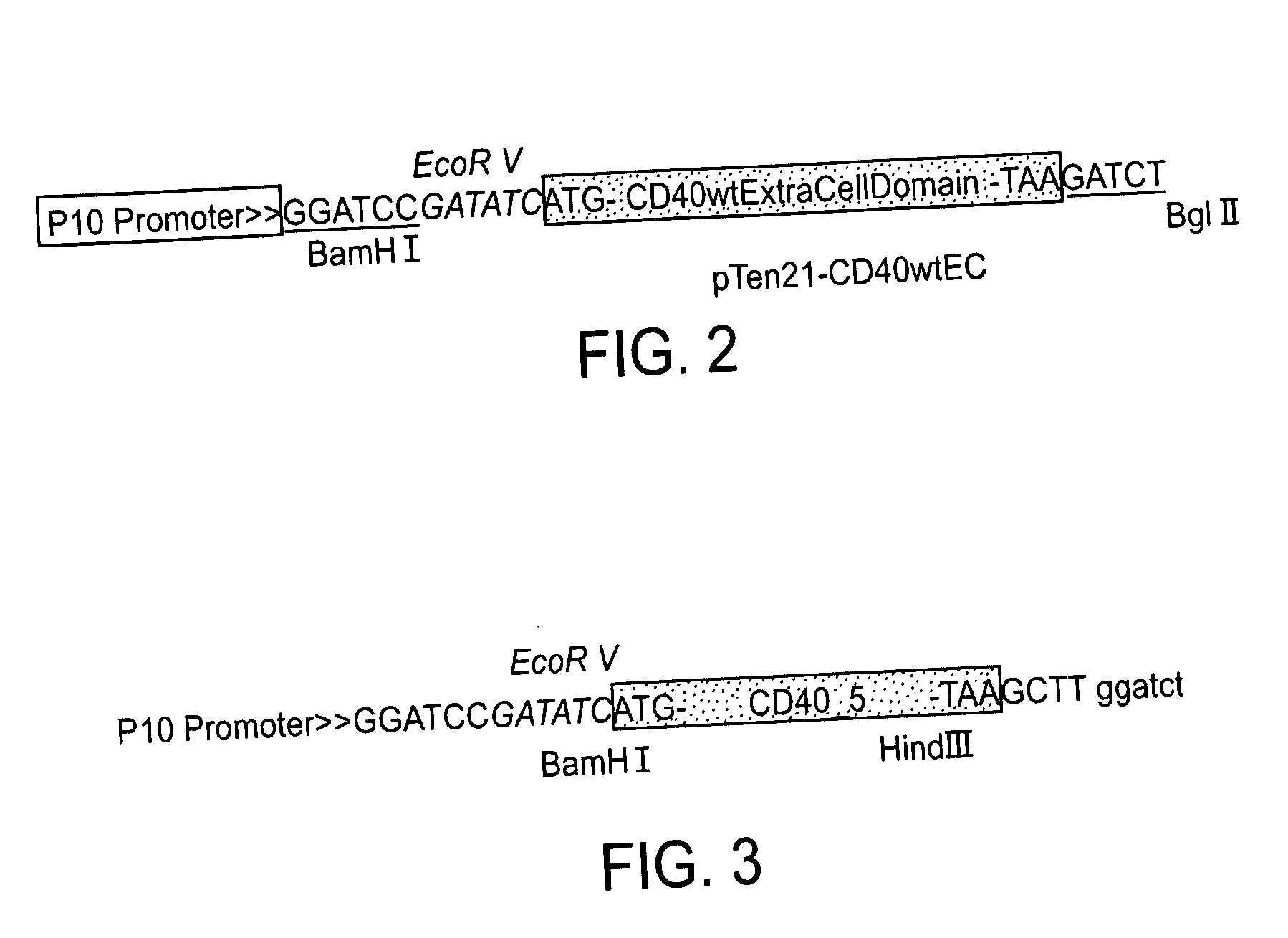
Novel CD40 variants
InactiveUS20060287229A1Unique pharmaceuticalUnique biochemical propertyPeptide/protein ingredientsAntipyreticDiseaseTreatment effect
The invention concerns CD40 skipping 5 nucleic acid sequences and amino acid sequences obtained by alternative splicing of CD40, pharmaceutical compositions comprising said sequences and methods for treatment of a disease, wherein a beneficial therapeutic effect is achieved by the up regulation of the CD40-R-CD40-L interaction. An antibody capable of selectively binding to the amino acid of CD40 skipping 5 and pharmaceutical composition comprising the above antibody and methods for detecting the presence of exon 5 skipping expression in a sample are also within the scope of the invention.
Owner:COMPUGEN
Construction method of gene detection library of familial hypercholesterolemia and gene detection kit
ActiveCN109517884AHigh precisionEasy to operateMicrobiological testing/measurementLibrary creationSubtilisinKexin
The invention discloses a construction method of a gene detection library of familial hypercholesterolemia and a gene detection kit and relates to gene mutation of LDLR (Low-Density Lipoprotein Receptor), APOB (Apolipoprotein B) and PCSK9 (Proprotein Convertase Subtilisin / Kexin type 9). In order to ensure that all target regions (including an entire coding region and an alternative splicing regionof a gene to be detected) are covered and prevent a primer dimer or a short segment from being formed between primers of adjacent amplicons, a PCR (Polymerase Chain Reaction) amplification primer isdivided into two independent primer pools; then primer sequence digestion, sequencing connector connection, library purification and quality detection are carried out; finally, the detection library is constructed. The method has the advantages that library establishment steps are simple and rapid, the cost is effectively reduced, the working amount is reduced, variation types are comprehensive, the detection accuracy reaches 100 percent and the flux is high.
Owner:ANNGEEN BIOTECHNOLOGY CO LTD
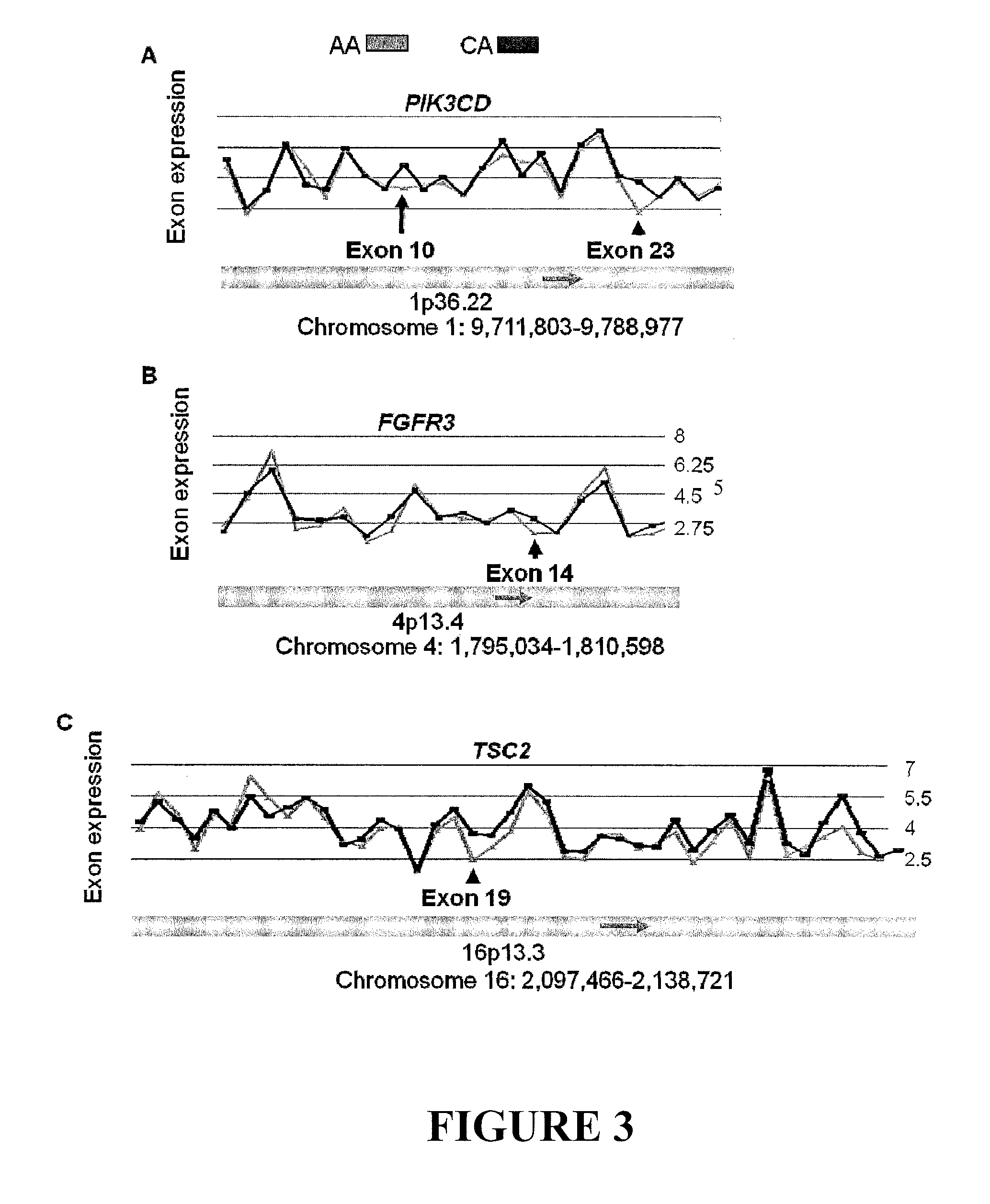
Alternative splicing variants of genes associated with prostate cancer risk and survival
ActiveUS9453261B2High incidence and mortality rateIncreasing the short to long variants ratioTumor rejection antigen precursorsSugar derivativesCvd riskOncology
Disclosed are novel splicing variants of the genes associated with prostate cancer risk and survival, particularly splicing variants of PIK3CD, FGFR3, TSC2, RASGRP2, ITGA4, MET, NF1 and BAK1. The disclosure also relates risk assessment, detection, diagnosis, or prognosis of prostate cancer. More specifically, this disclosure relates to the detection of certain splicing variants of PIK3CD, FGFR3, TSC2, RASGRP2, ITGA4, MET, NF1 and BAK1.
Owner:GEORGE WASHINGTON UNIVERSITY
Alternative splicing factors polynucleotides polypeptides and uses therof
InactiveUS7109390B2Low efficiencyImprove conversion efficiencySugar derivativesClimate change adaptationNucleotideADAMTS Proteins
The invention provides isolated polynucleotides and their encoded proteins that are involved in splicing or modulating splicing activity. The invention further provides recombinant expression cassettes, host cells, transgenic plants, and antibody compositions. The present invention provides methods and compositions relating to altering splicing protein content and / or composition of plants.
Owner:PIONEER HI BRED INT INC
Modulation and detection of a neuronal alternative splicing regulatory network for treatment and diagnosis of neurological disorders
ActiveUS20170360873A1Increase and decreases expression of proteinCompounds screening/testingOrganic active ingredientsNervous systemAutism spectrum disorder
Methods for treatment and diagnosis of neurological disorders such as autism and autism spectrum disorder are disclosed. Also disclosed are modulators of alternative splicing regulators SRRM4 and / or SRRM3 for treating neurological disorders. Further disclosed are agents that modulate the expression of at least one splice variant for treating neurological disorders. Mouse models of neurological disorders having increased or decreased expression of SRRM4 and / or SRRM3 are also disclosed.
Owner:THE GOVERNINIG COUNCIL OF THE UNIV OF TORANTO +1
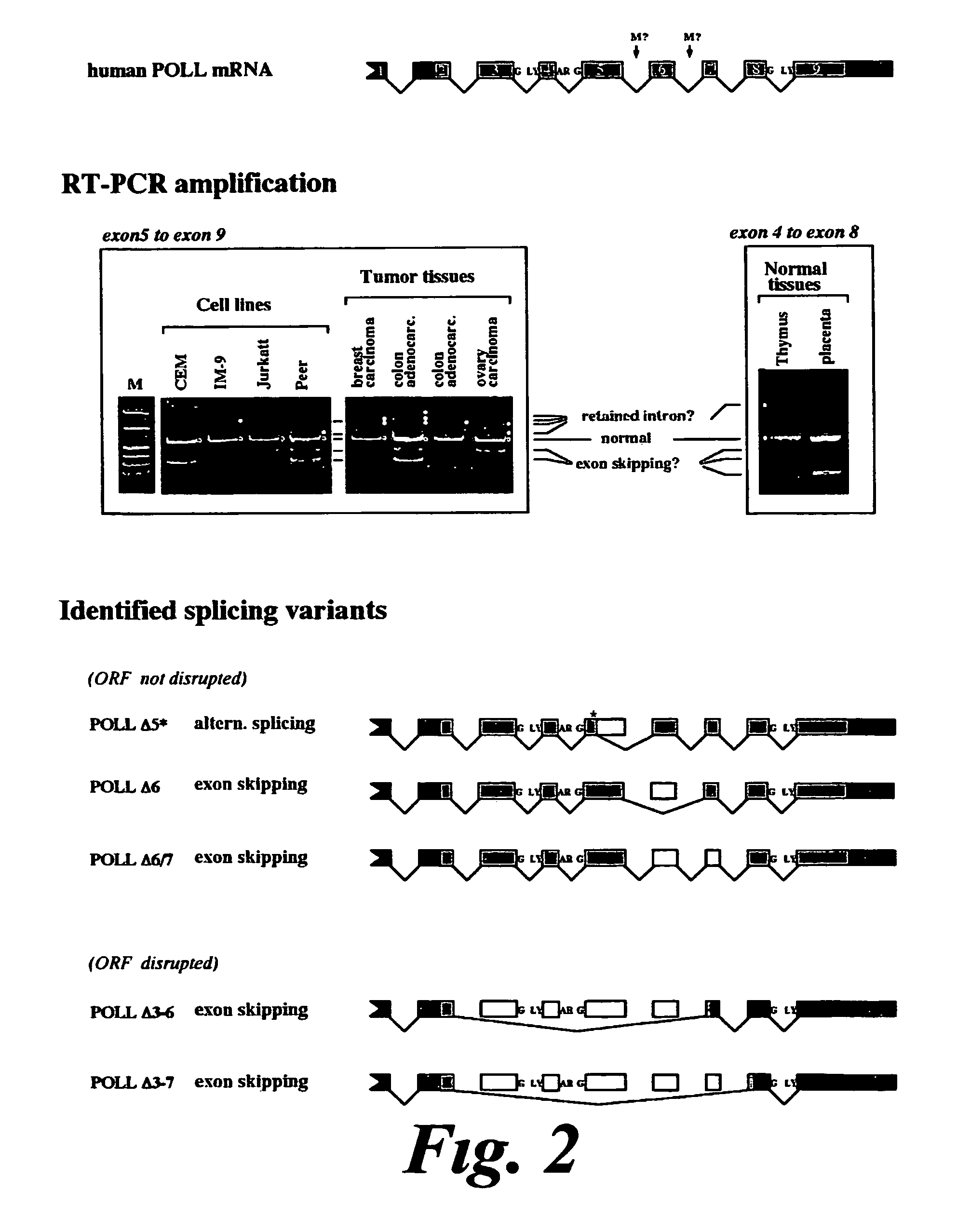
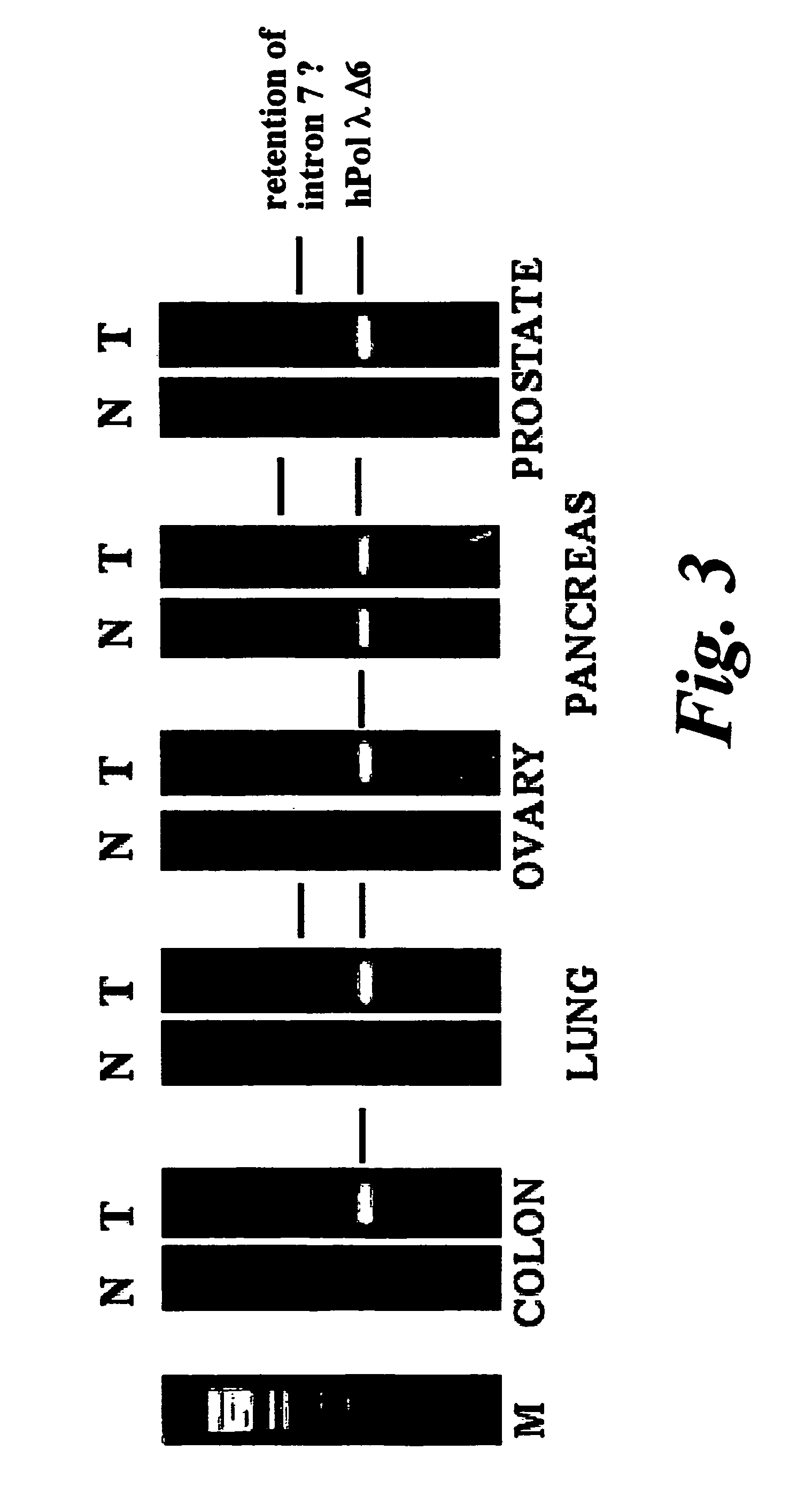
DNA polymerase lambda and uses thereof
InactiveUS7883880B1Increase mutation frequencyLow fidelityFungiNervous disorderNucleotidePolymerase L
The present invention relates to the identification and isolation of a DNA polymerase and uses of this polymerase. In particular, the present invention describes the nucleotide sequence of the human gene for DNA polymerase lambda (Pol λ), the amino acid sequence of Pol λ, and the amino acid sequence of several isoforms derived from alternative splicing of its mRNA. The association of some of these isoforms with tumour samples makes Pol λ a marker for the diagnosis, prognosis and evolution of tumoral processes.
Owner:CONSEJO SUPERIOR DE INVESTIGACIONES CIENTIFICAS (CSIC)
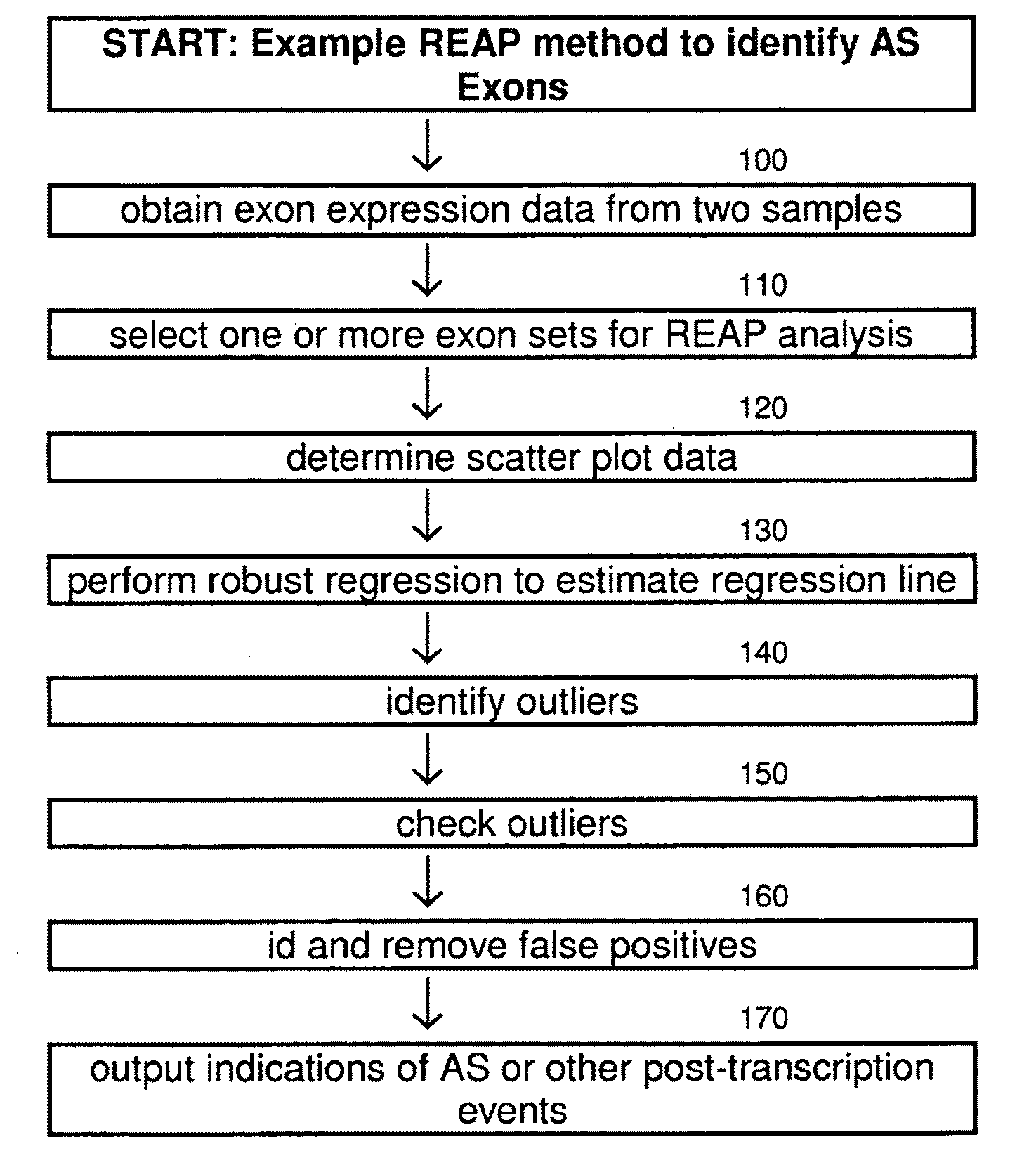
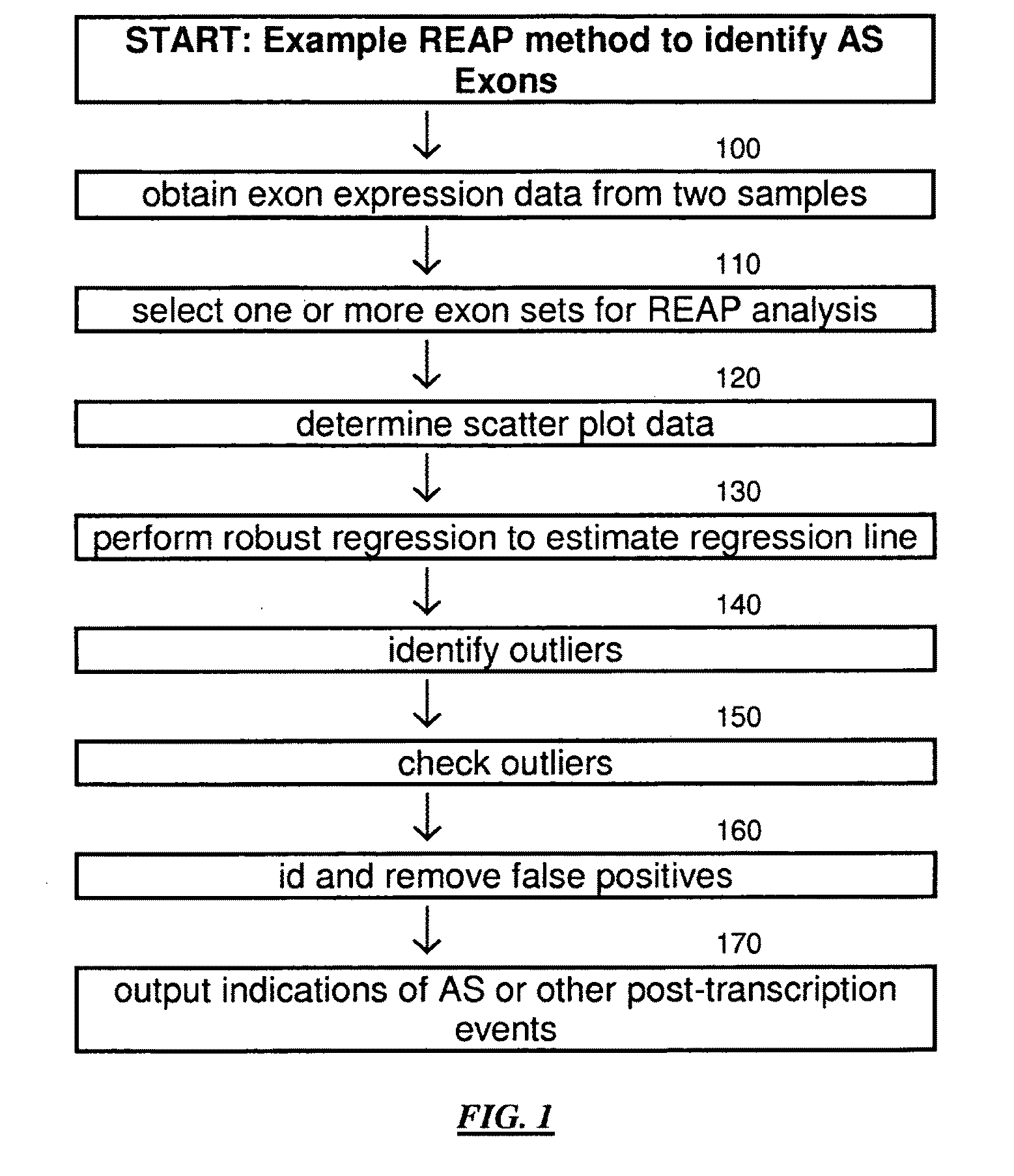
Robust regression based exon array protocol system and applications
InactiveUS20110045996A1Less sensitiveBioreactor/fermenter combinationsBiological substance pretreatmentsData profilingAlternative splicing
An analysis technique for genetic data to detect alternative spliced exons. Exon expression of similar data is analyzed using a robust regression technique to find outliers to the main regression. False outliers are detected and removed. The remaining outliers are identified as potential alternative splicing events.
Owner:SALK INST FOR BIOLOGICAL STUDIES
Mta1s, a steriod hormone receptor corepressor
InactiveUS20060204957A1Improve satisfactionSugar derivativesMicrobiological testing/measurementADAMTS ProteinsHormones regulation
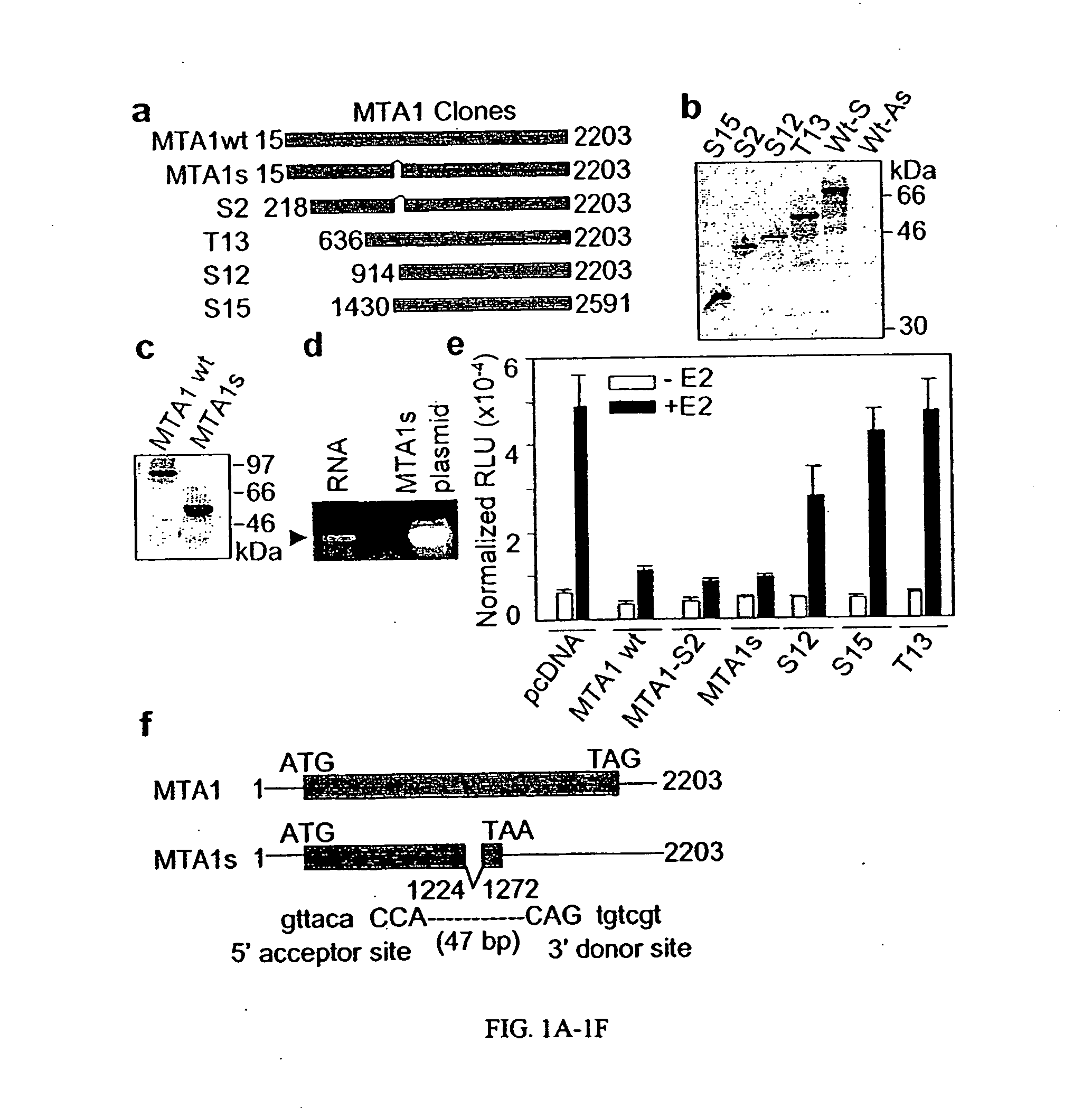
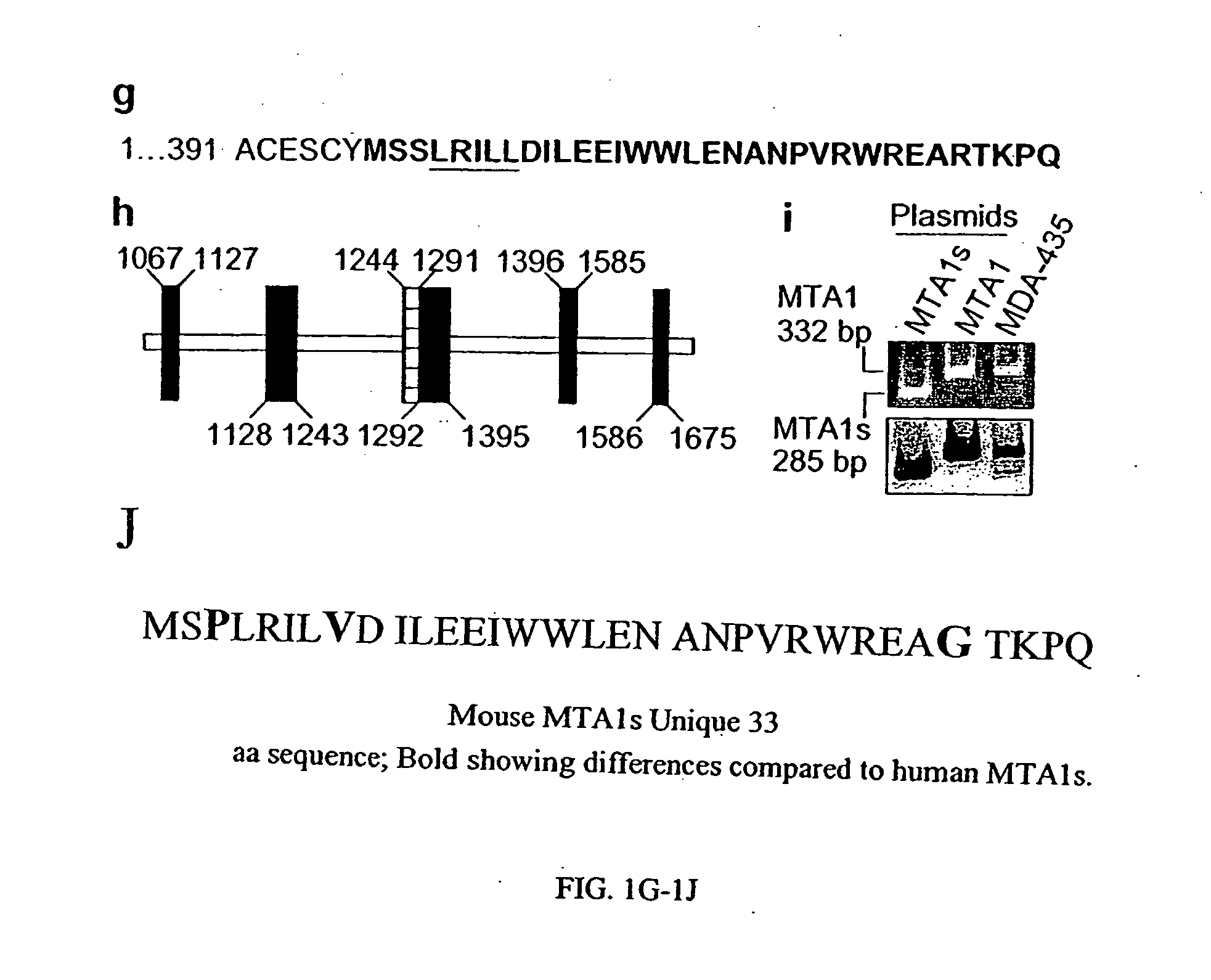
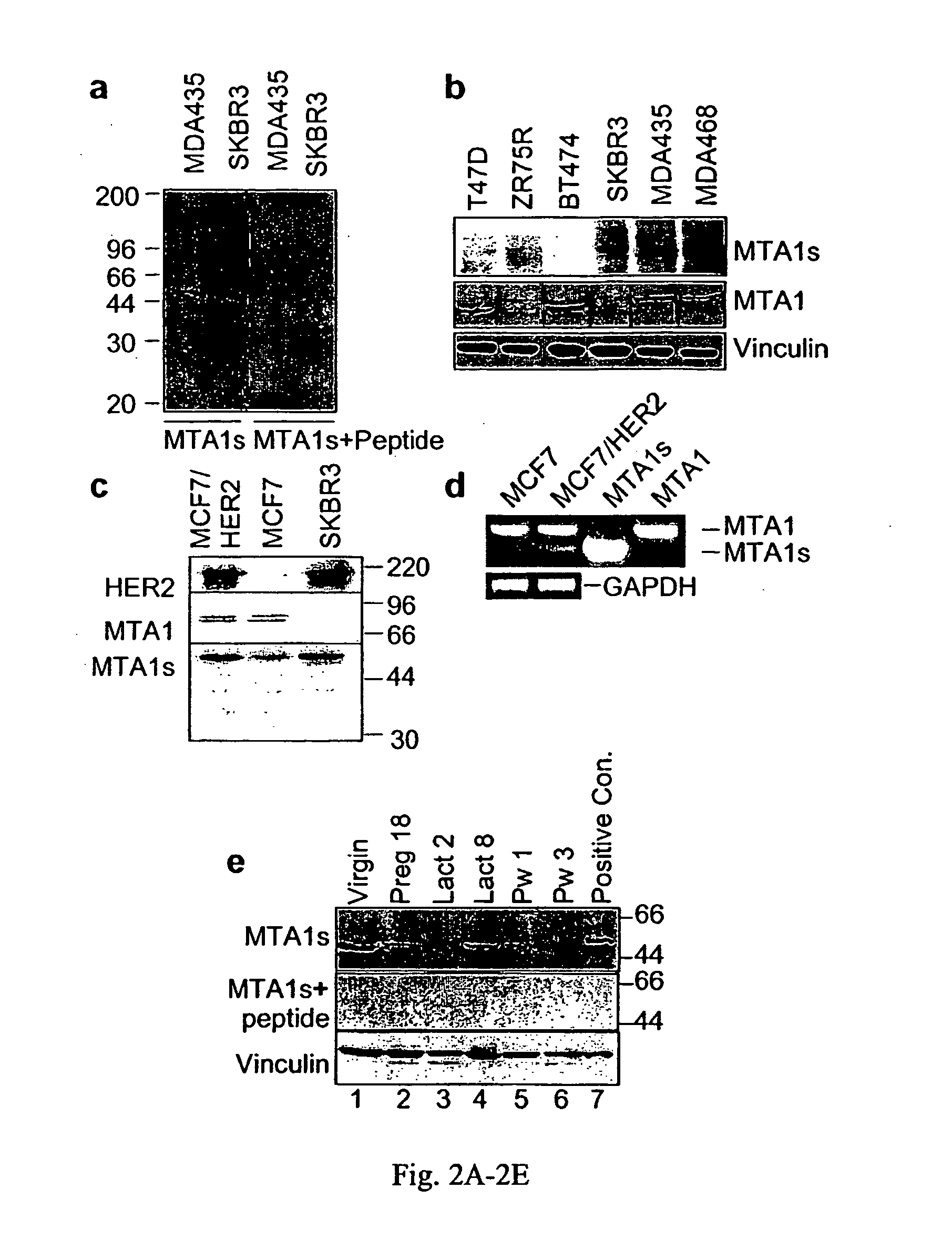
The present invention concerns the use of methods and compositions to diagnose, treat and identify therapeutic molecules for cancer, in particular hormone insensitive cancers. The invention includes polypeptides and nucleic acids encoding polypeptides of MTA1s, a novel protein derived from alternative splicing of the MTA1 gene. Other embodiments include antibody compositions for the diagnosis and prognosis related to the aberrant expression of the MTA1s polypeptide.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com