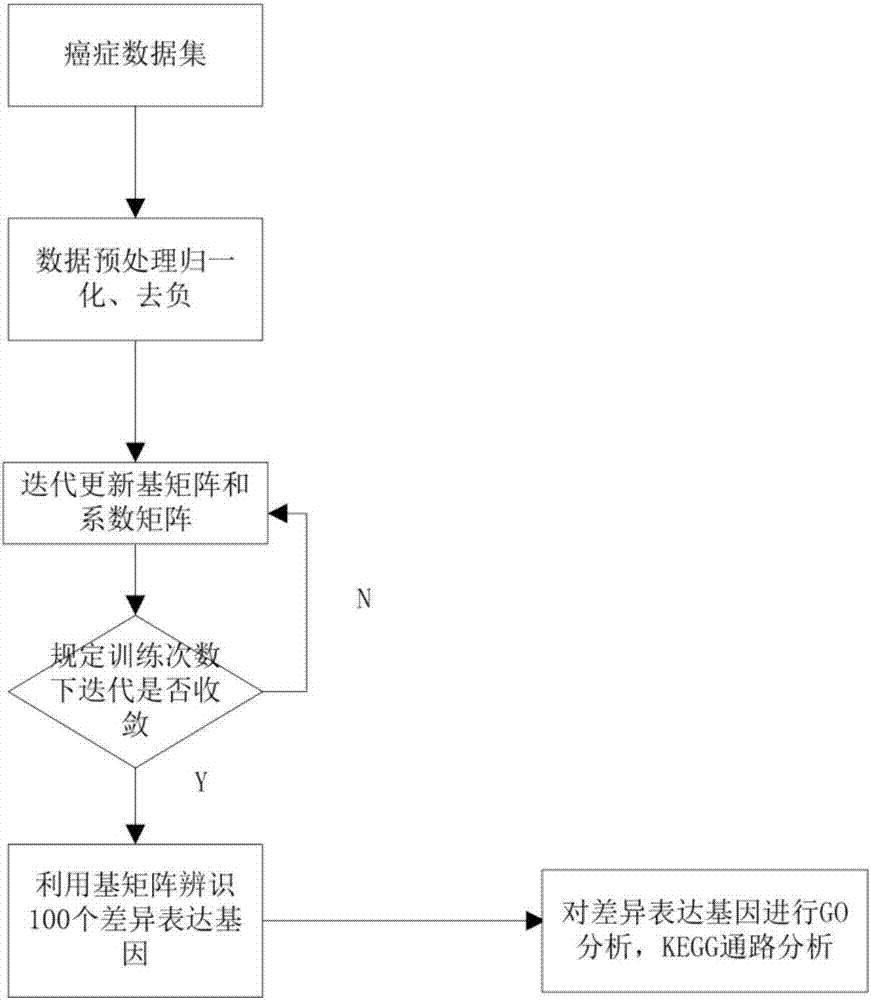
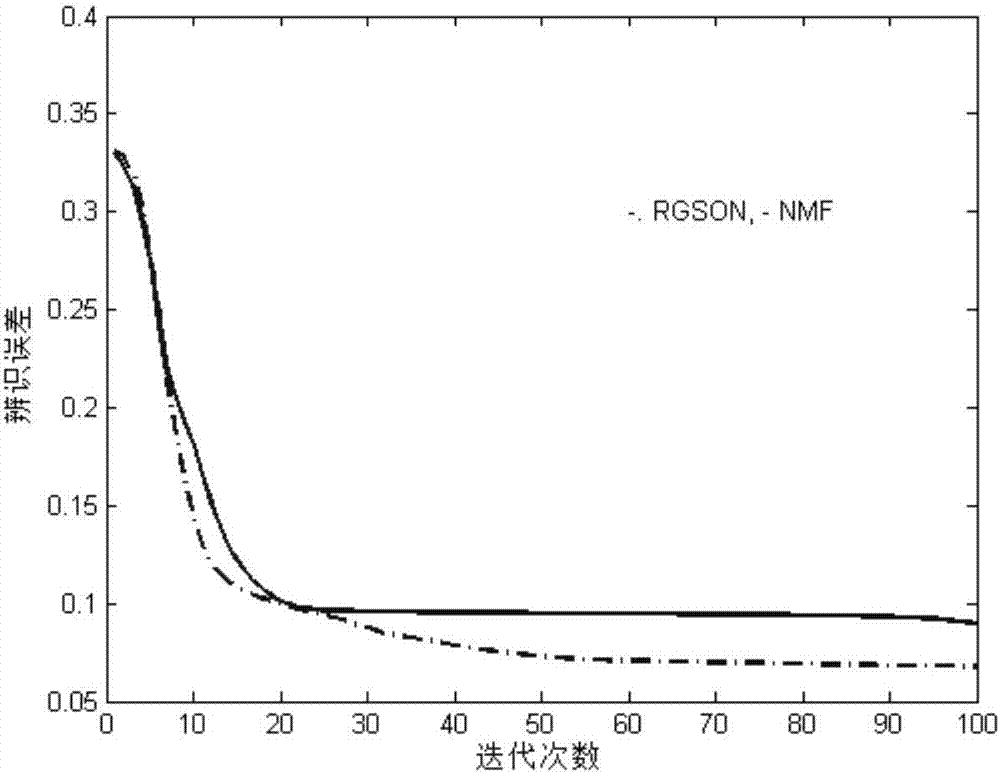
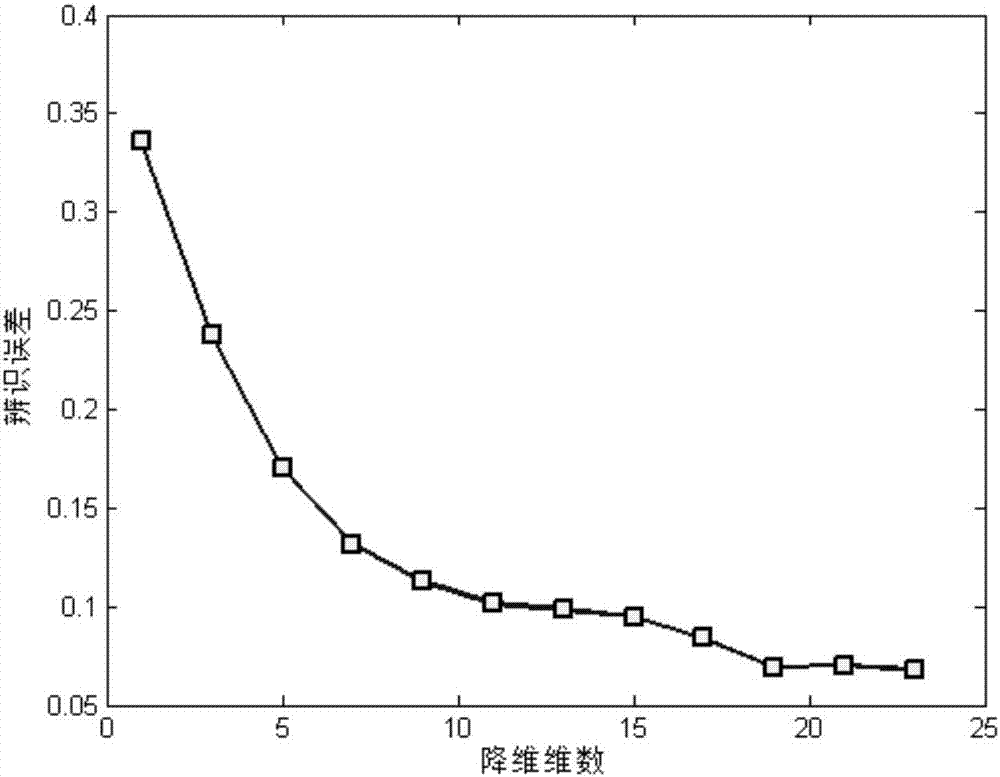
Differentially expressed gene identification method based on combined constraint non-negative matrix factorization
A technology of differentially expressed genes and non-negative matrix decomposition, applied in the field of pattern recognition, can solve the problems of no advantage in differential expression feature selection, lack of sparsity in non-negative matrix decomposition, etc., and achieve the effect of improving robustness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] With the rapid development of deep sequencing technology and gene chip technology, a large amount of gene expression profile data has emerged. Therefore, finding a suitable data analysis method to process huge gene expression profile data has become a research hotspot in bioinformatics. Due to the limitation of experimental conditions, there are usually only a few dozen experimental samples, and gene sequencing technology can monitor tens of thousands of genes at the same time. Therefore, the analysis of gene expression profile data is a typical singular value problem in statistics - high-dimensional small sample problem . Usually, the dimensionality reduction method can be used to reduce the complexity of the data and improve the accuracy of the analysis results. Many dimension processing techniques, such as principal component analysis PCA, singular value decomposition SVD and other algorithms have been widely used. But they still have some shortcomings, the principa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com