Patents
Literature
159 results about "Cancer gene" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Compositions and methods for treating and diagnosing cancer
InactiveUS20070099209A1Reduce the amount requiredBioreactor/fermenter combinationsBiological substance pretreatmentsCancer geneSolid tumor
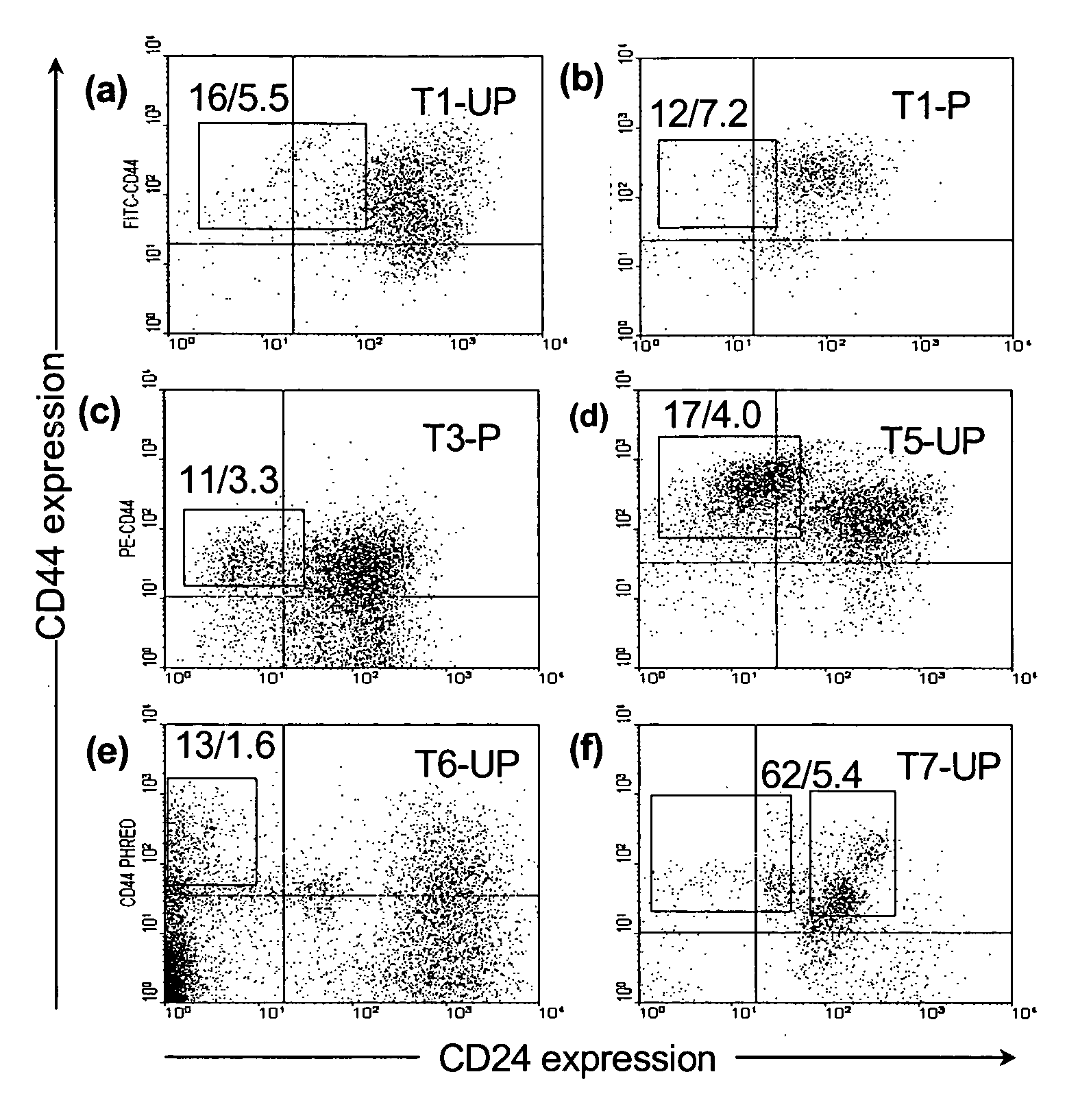
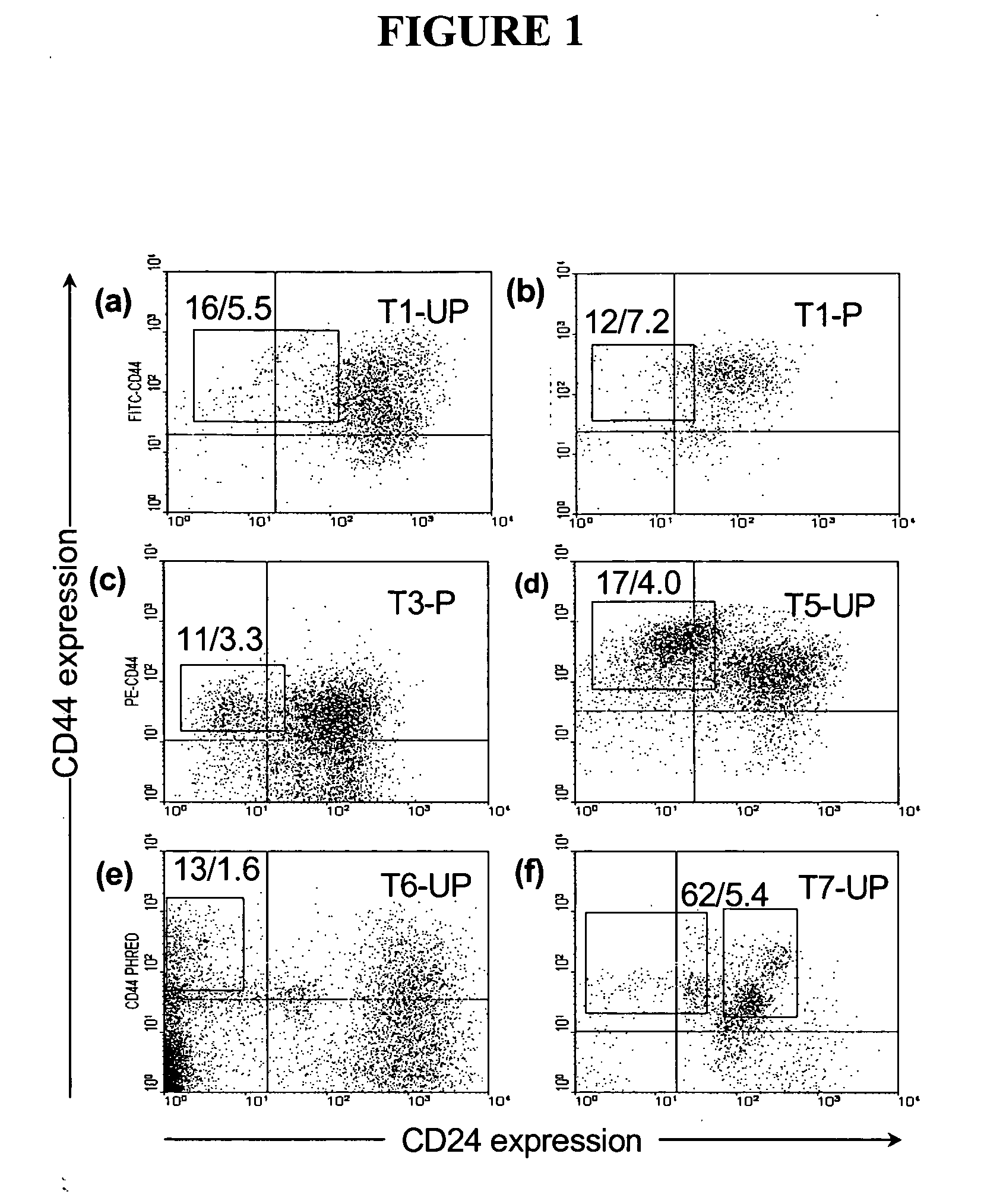
The present invention relates to compositions and methods for treating, characterizing, and diagnosing cancer. In particular, the present invention provides gene expression profiles associated with solid tumor stem cells, as well as novel stem cell cancer gene signatures useful for the diagnosis, characterization, prognosis and treatment of solid tumor stem cells.
Owner:RGT UNIV OF MICHIGAN
Anaerobic bacterium as a drug for cancer gene therapy
InactiveUS20050025745A1Efficient deliveryEffective gene therapyBiocideBacteriaAbnormal tissue growthBifidobacterium
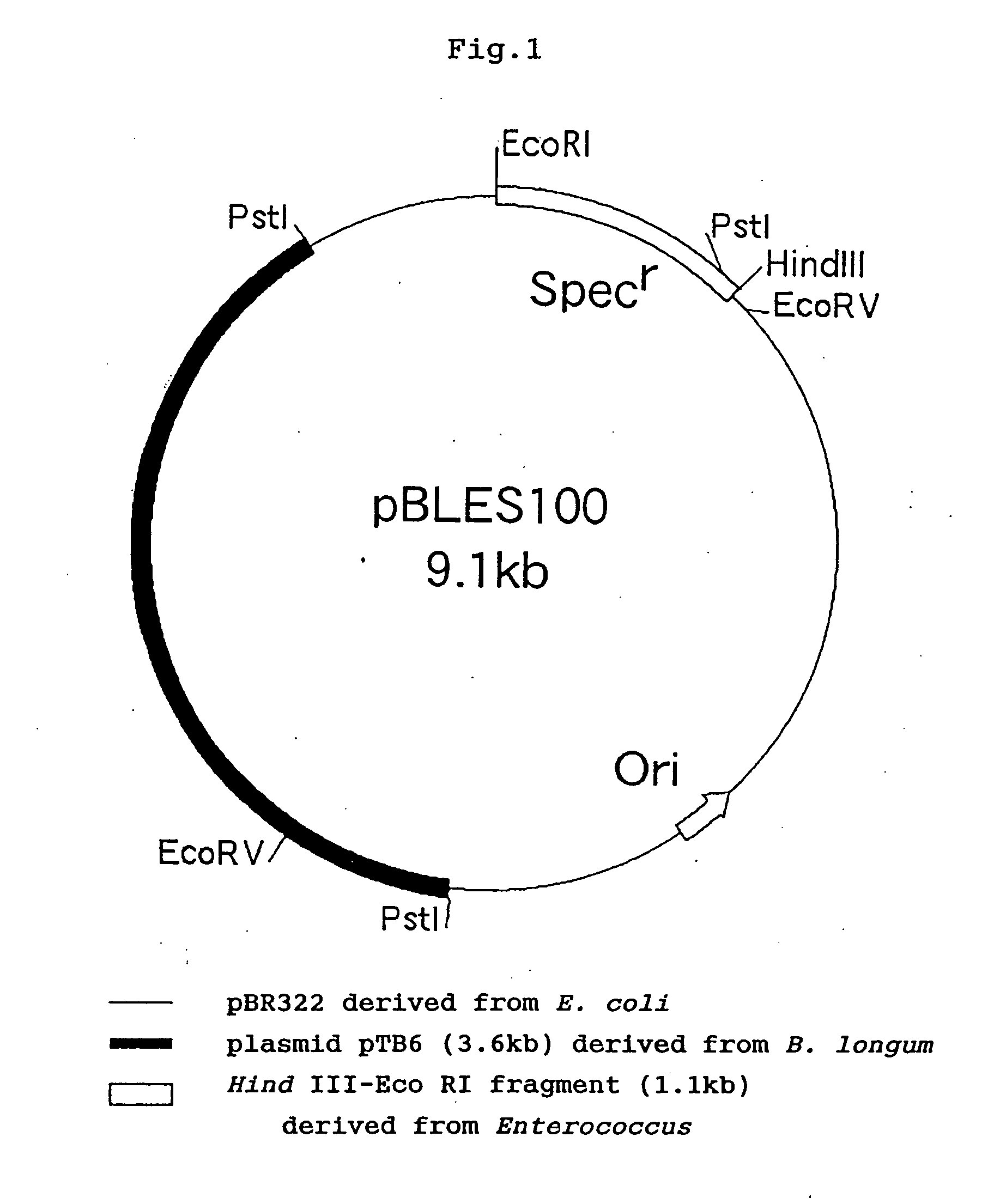
The present invention provides a bacterium belonging to the genus Bifidobacterium, by which DNA coding for a protein having an antitumor activity or DNA coding for a protein having the activity of converting a precursor of an antitumor substance into the antitumor substance is delivered to tumor tissues specifically under anaerobic conditions thereby expressing the protein encoded by the DNA, as well as a pharmaceutical composition comprising said anaerobic bacterium.
Owner:ANAEROPHARMA SCI
CRISPR/Cas9 enrichment sequencing method applied in large-scale screening of cancer genes
InactiveCN105400773AHigh infection efficiencyMeet the packaging ratioMicrobiological testing/measurementLibrary creationInfected cellCancer cell
The invention relates to a CRISPR / Cas9 enrichment sequencing method applied in large-scale screening of cancer genes. Firstly, an sgRNA library is established; then the sgRNA library is packaged by lentiviruses, and viruses are collected; the sgRNA library is screened in a cancer cell line, the obtained cells are extracted and screened, and genome DNAs of precancerous cells are screened; finally, enrichment of sgRNAs in the genome DNAs is carried out. When the provided method is compared with the prior art, the screening process of CRISPR / Cas9 cells is improved, the virus infection efficiency is determined and the virus MOI value is determined through a simple method and by utilization of puromycin resistance of infected cells, importantly, the restriction enzyme cutting method and a high flux method are combined, correct chromatin fragments can be obtained effectively, false positive caused by non-specific PCR amplification can be lowered, a DNA template of sgRNAs can be amplified efficiently, and a library establishment efficiency is raised.
Owner:TONGJI UNIV
Method for applying functionalized poly(amidoamine) dendrimer and nanometer compound thereof in gene transfection
InactiveCN103435815AImprove transfection abilityGood gene transfection effectGenetic material ingredientsPharmaceutical non-active ingredientsCompound sBiology
The invention relates to a method for applying a functionalized poly(amidoamine) dendrimer and a nanometer compound thereof in gene transfection. The method comprises the following steps: preparation of the functionalized poly(amidoamine) dendrimer and the nanometer compound thereof, surface functional modification and characterization; preparation of functionalized poly(amidoamine) dendrimer-nanometer compound / pDNA; and research on gene transfection efficiency of the functionalized poly(amidoamine) dendrimer-nanometer compound / pDNA. The advantages of easy operation, simple transfection conditions, high transfection efficiency, strong specificity and the like are obtained in application of the functionalized poly(amidoamine) dendrimer and the nanometer compound thereof in gene transfection, and the functionalized poly(amidoamine) dendrimer and the nanometer compound thereof have good application prospects in aspects like gene therapy of cancers.
Owner:DONGHUA UNIV
Compositions and methods for diagnosing and treating cancer
InactiveUS20080318234A1High of deathHigh riskMicrobiological testing/measurementProtein nucleotide librariesCancer geneSolid tumor
Owner:ONCOMED PHARMA
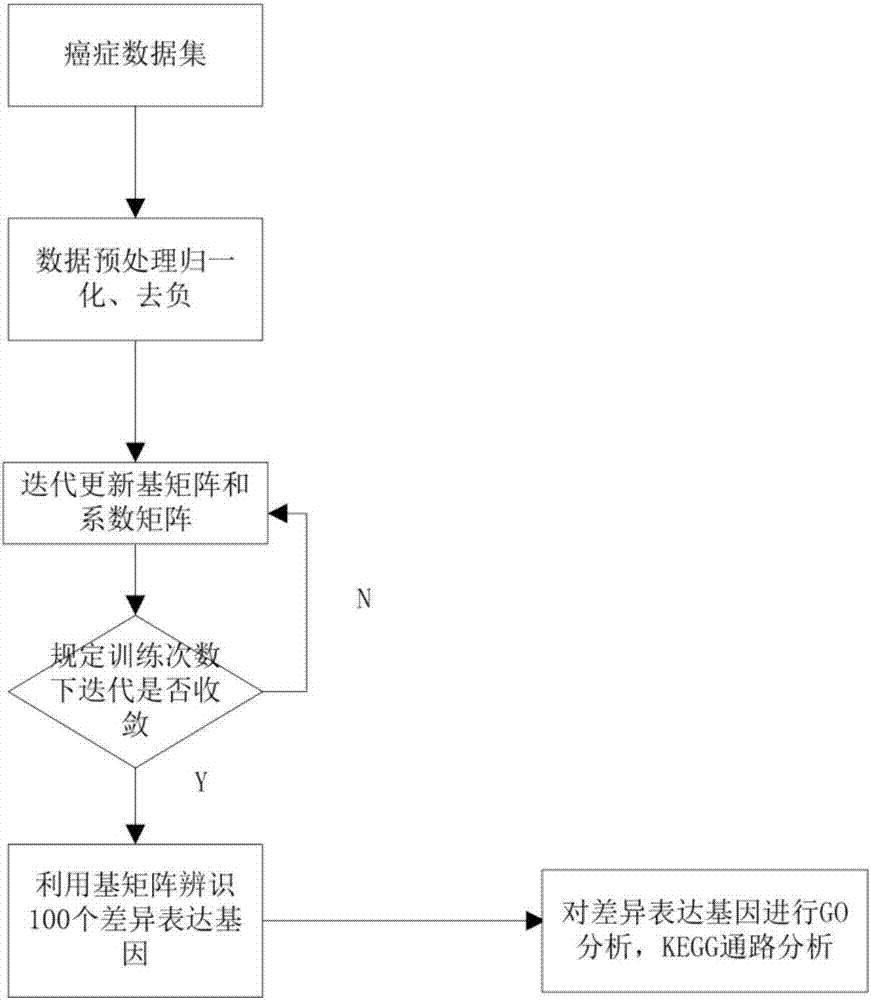
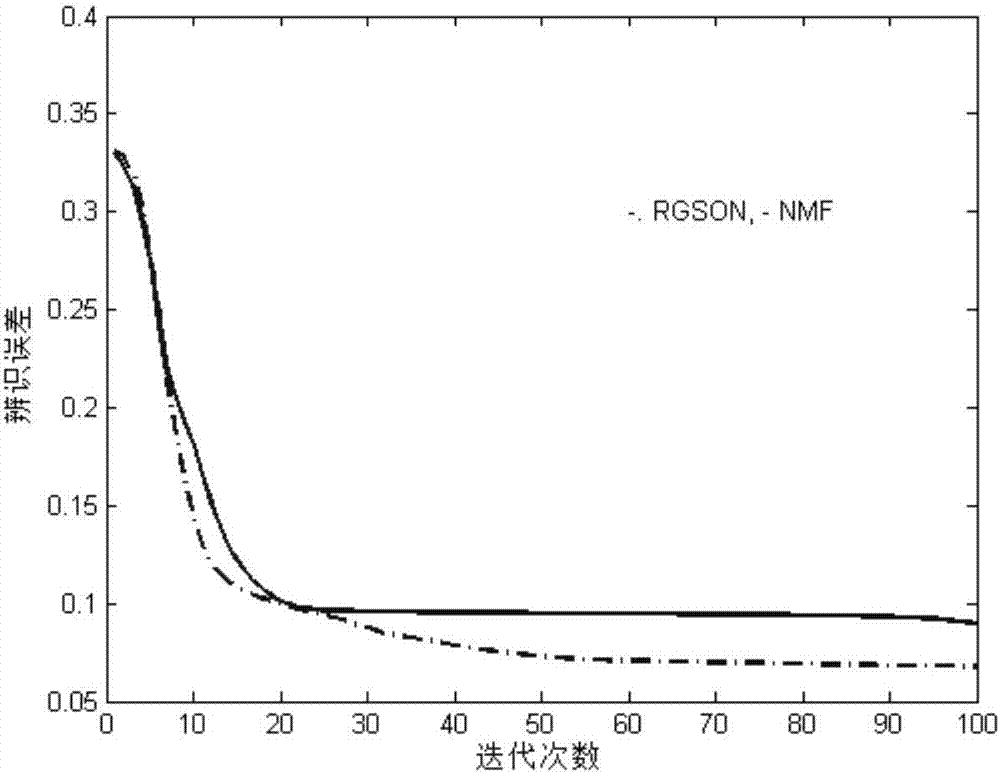
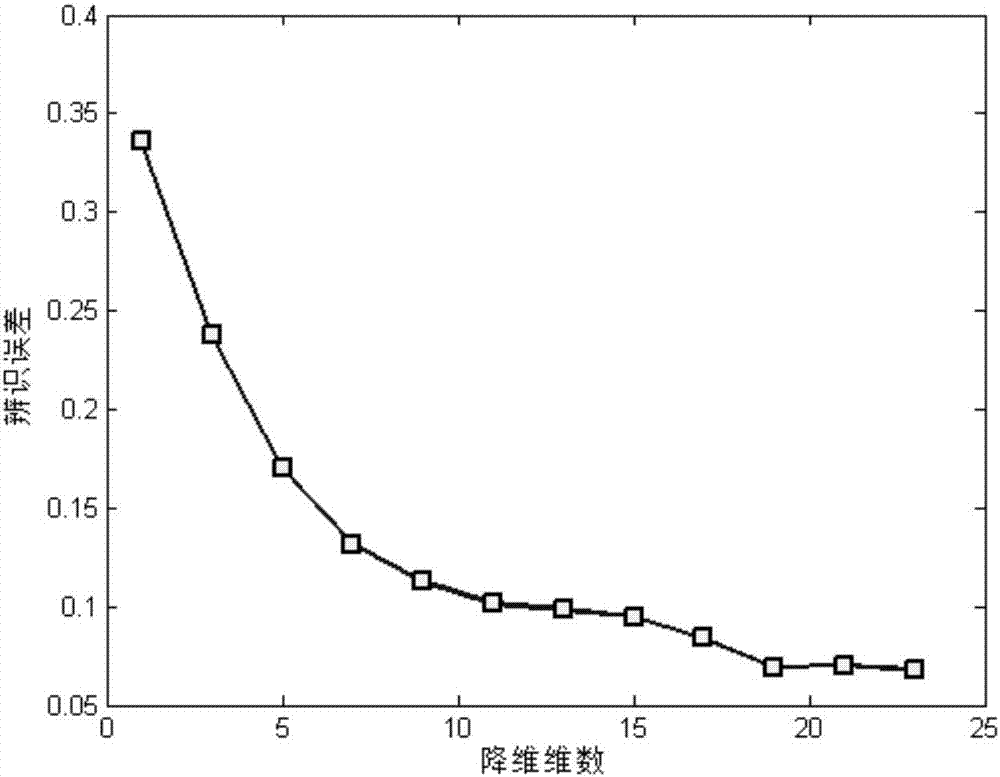
Differentially expressed gene identification method based on combined constraint non-negative matrix factorization
ActiveCN107016261AEffective decomposition resultsEfficient Sparse Decomposition ResultsSpecial data processing applicationsData setAlgorithm
The invention discloses a differentially expressed gene identification method based on combined constraint non-negative matrix factorization. The method comprises the following steps of 1, representing a cancer-gene expression data set with a non-negative matrix X, 2, constructing a diagonal matrix Q and an element-full matrix E, 3, introducing manifold learning in the classical non-negative matrix factorization method, conducting orthogonal-constraint sparseness and constraint on a coefficient matrix G, and obtaining a combined constraint non-negative matrix factorization target function, 4, calculating the target function, and obtaining iterative formulas of a basis matrix F and the coefficient matrix G, 5, conducting semi-supervision non-negative matrix factorization on the non-negative data set X, and obtaining the basis matrix F and the coefficient matrix G after iteration convergence, 6, obtaining an evaluation vector (the formula is shown in the description), sorting elements in the evaluation vector (the formula is shown in the description) from large to small according to the basis matrix F, and obtaining differentially expressed genes, 7, testing and analyzing the identified differentially expressed genes through a GO tool. The identification method can effectively extract the differentially expressed genes where cancer data is concentrated, and be applied in discovering differential features in a human disease gene database. The identification method has important clinical significance for early diagnosis and target treatment of diseases.
Owner:HANGZHOU HANGENE BIOTECH CO LTD
Molecule substitution label sequencing parallel detection method-oligomictic nucleic acid coding label molecule library micro-sphere array analysis
InactiveCN101100764AImprove reliabilityGreat advantageMicrobiological testing/measurementLibrary member identificationBiotechnologyMolecular replacement
Molecular displacement label sequence parallel inspection - oligonucleo coded label molecular pool micro-ball array analysis includes: using artificial sequential oligo nucleo-labeled DNA coded molecular pool, coating micro-balls with ligands in correspondent pools to form catching micro-balls, caught ligands combined with labeled ciding molecular pools to form labeled micro-balls array pools. Sample molecules competitively combined with micro-balls catching ligands when sample contains molecules same as those in labeled micro-balls array pools, calculating amount of displaced coded molecular sequence computing kinds and mounts of molecules to be tested. It verifies high flux of inspection with good repeatability, high reliability and sensitivity, so that it is suitable to precisely parallel quantitative analysis concerned with cancer-inhibiting genes and cancer genes pools, cell factor pools, etc.
Owner:北京万达因生物医学技术有限责任公司
Nanoporous silicon-based electrochemical nucleic acid biosensor
InactiveUS20090050492A1Immobilised enzymesBioreactor/fermenter combinationsMicroorganismNanoporous silicon
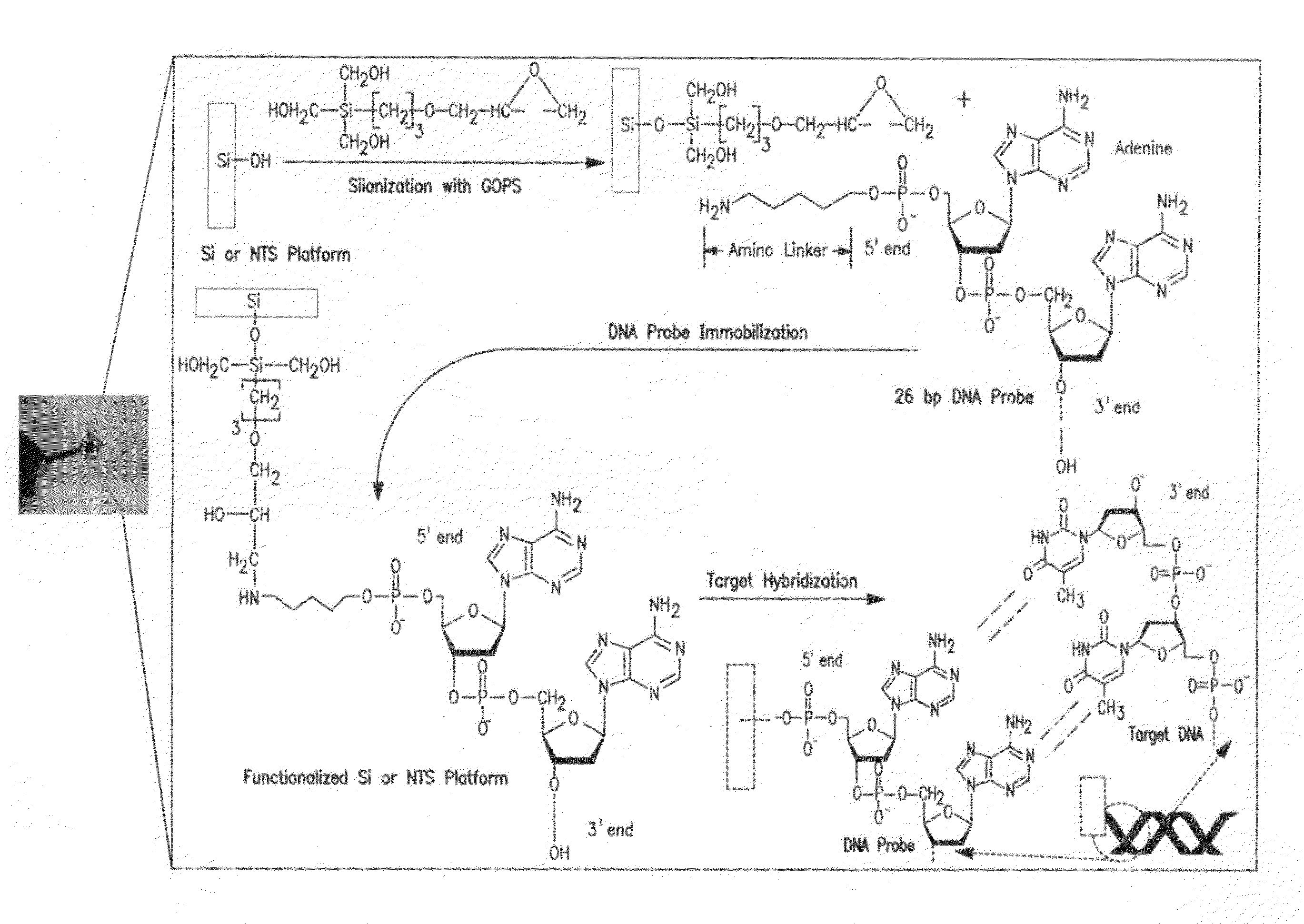
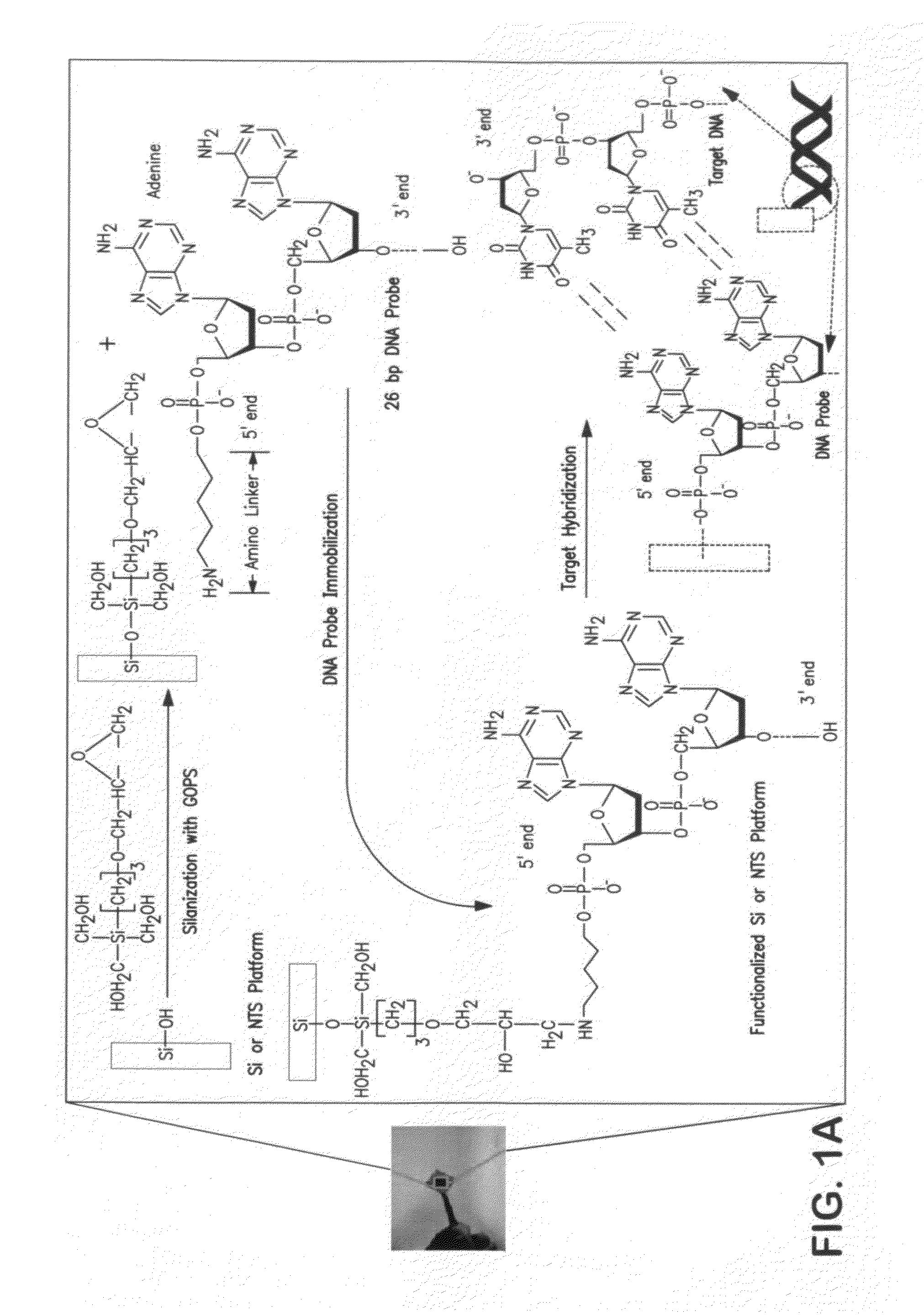
A method and biosensor device for detecting single strand target nucleic acid by cyclic voltammetry is described. A porous silicon chip is linked to bound DNA probe complementary to the target nucleic acid. The device is particularly useful for detecting microorganisms and viruses that may be pathogenic or cancer genes, however any target nucleic acid can be detected by using a specific DNA probe.
Owner:BOARD OF TRUSTEES OPERATING MICHIGAN STATE UNIV
Application of gastric cancer genes
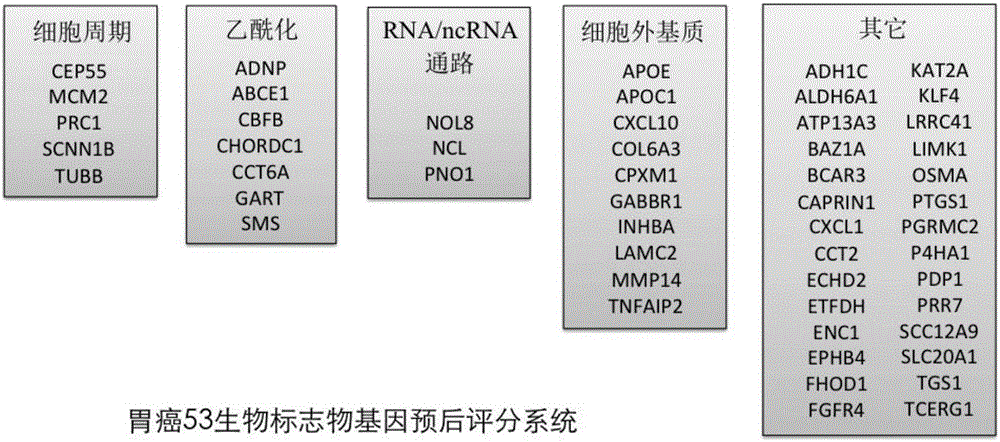
ActiveCN106834462AAvoid overdosingReduce medical costsMicrobiological testing/measurementDNA/RNA fragmentationOncologyBiology
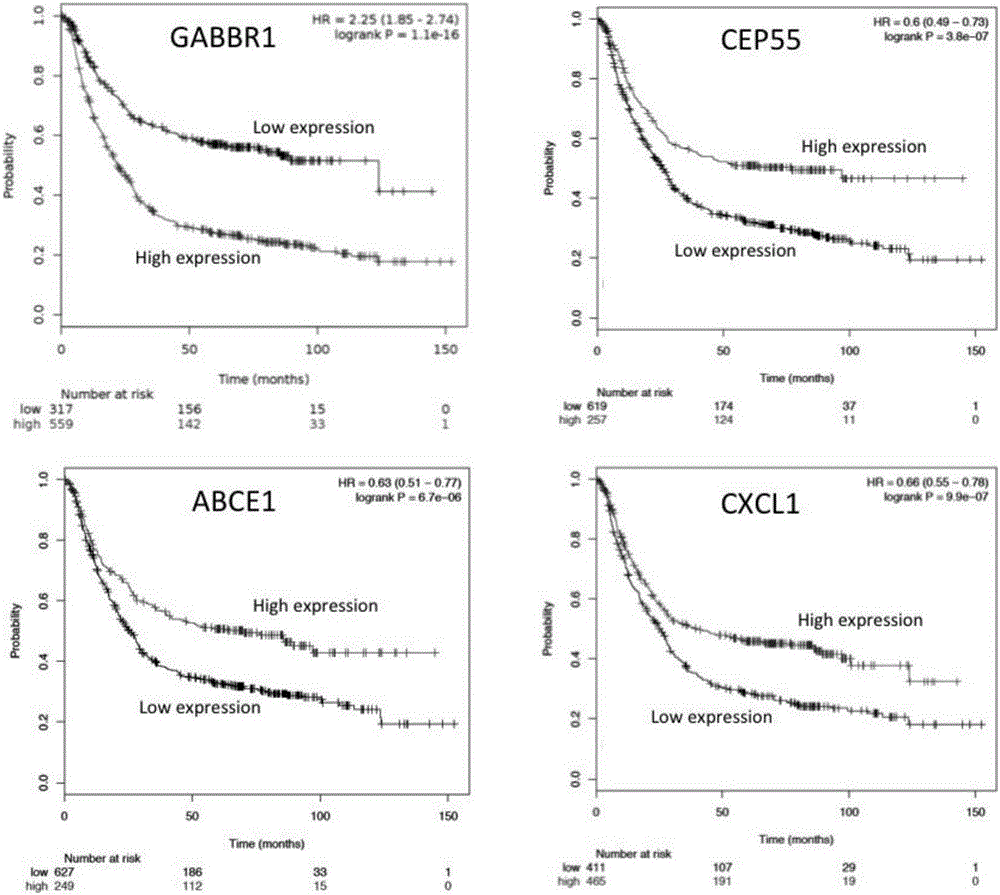
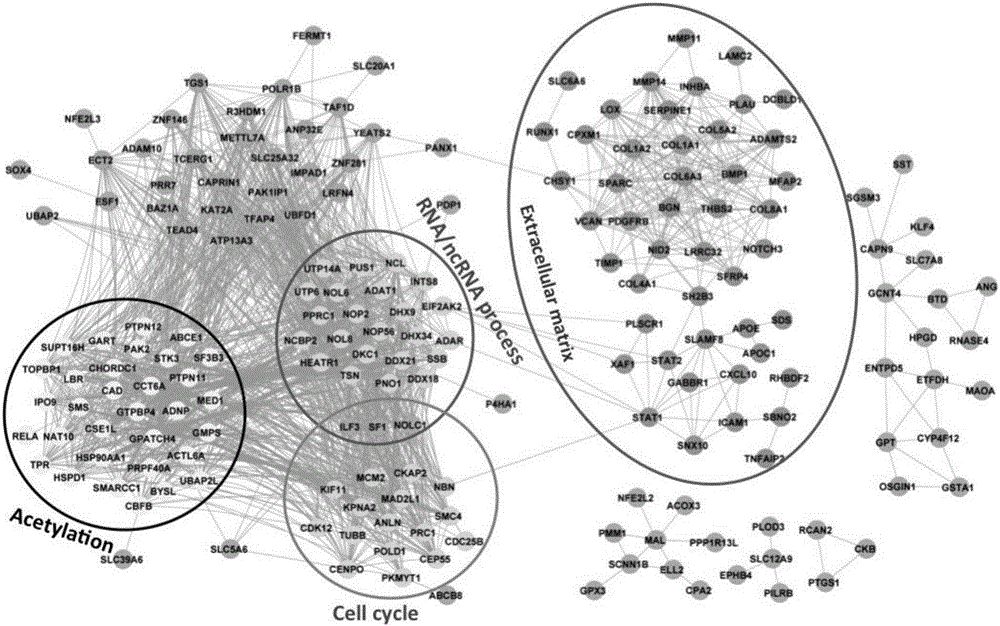
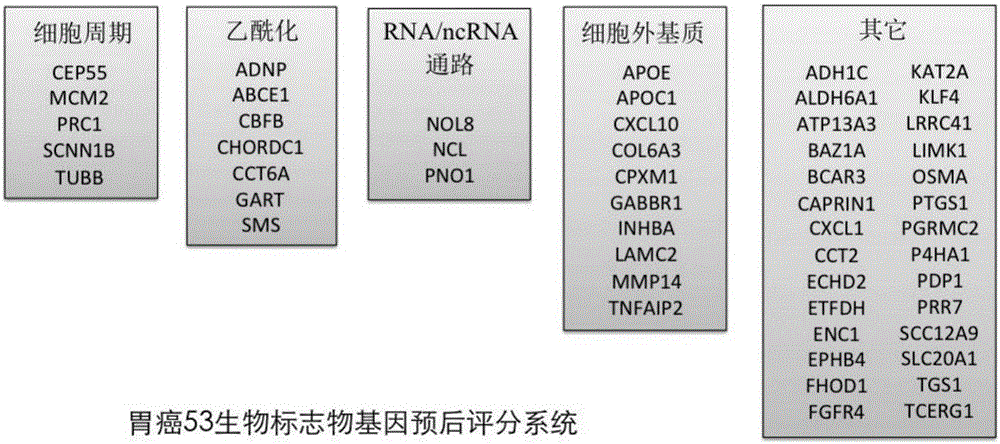
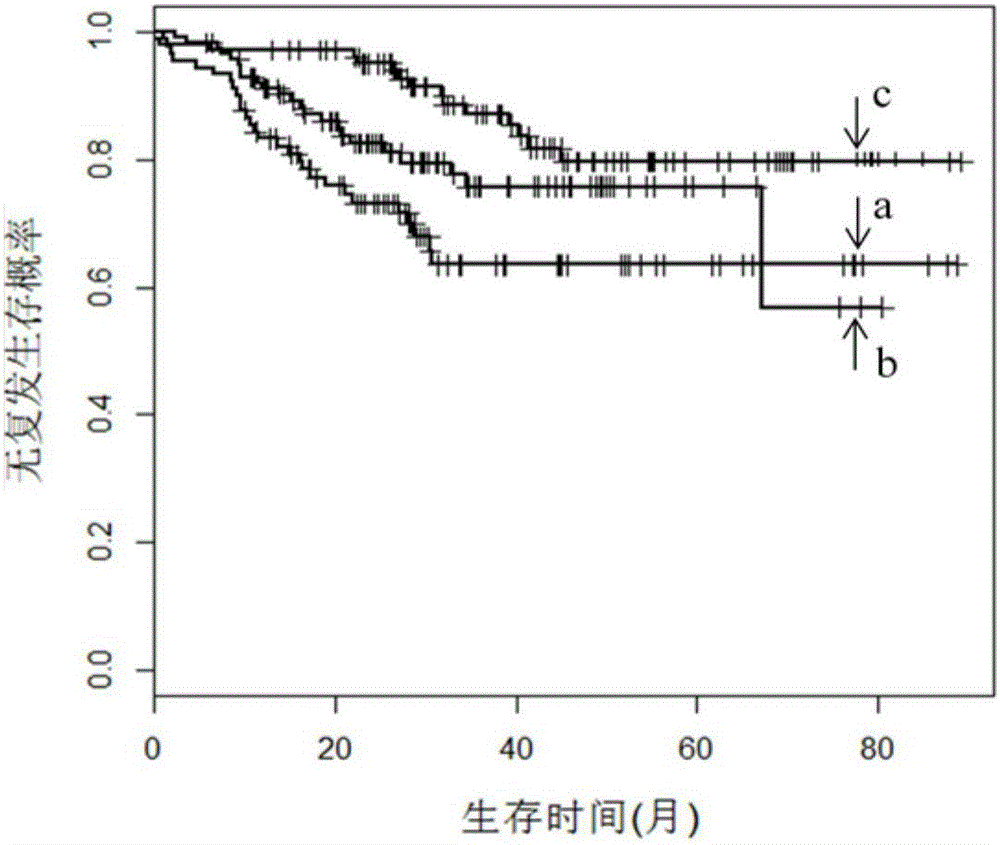
The invention discloses an application of a group of gastric cancer related genes. According to the invention, prediction scores are calculated to assess clinical prognosis of gastric cancers and related applications of the clinical prognosis of the gastric cancers based on a group of 53 prognostic related genes in the gastric cancers and a detection result of the expression level of the related genes in a clinical sample. The system can be used for helping gastric cancer patients in therapeutic choices of and prediction of responses to therapeutic intervention to judge whether the patients benefit from chemical and targeted therapies or not, so as to avoid excessive use of drugs and lower medical costs, and finally reach the purposes of precision or personalized medicine. According to the system and different detection technology platforms, corresponding kits for measuring expression of the 53 genes are designed and developed.
Owner:NANJING KDRB BIOTECH INC LTD
Method for identifying cancer molecular subtype based on spectral clustering algorithm of sparse similar matrix
InactiveCN106529165AEfficient removalImprove forecast accuracyMedical data miningBiostatisticsSpectral clustering algorithmAlgorithm
The invention discloses a method for identifying a cancer molecular subtype based on a spectral clustering algorithm of a sparse similar matrix. The method is characterized in that based on the spectral clustering algorithm of the sparse similar matrix, a cancer molecular subtype prediction model is built by utilizing cancer gene expression profile data as a training set sample; and the prediction model is used for predicting a cancer modular subtype of an independent test set sample, and a cancer sample set is divided into multiple types of molecular subtypes. According to the method, various patients with different prognosis effects are effectively distinguished for high heterogeneity of cancer molecular expression level, and different individual treatment schemes can be made for various cancer patients respectively.
Owner:HEFEI UNIV OF TECH
Eukaryotic expression vector for expressing shRNA (short hairpin Ribonucleic Acid) in manner of targeting in cancer cells
InactiveCN101993892ASolve the problem of non-specific interferenceGenetic material ingredientsMicroorganism based processesCancer cellReverse transcriptase
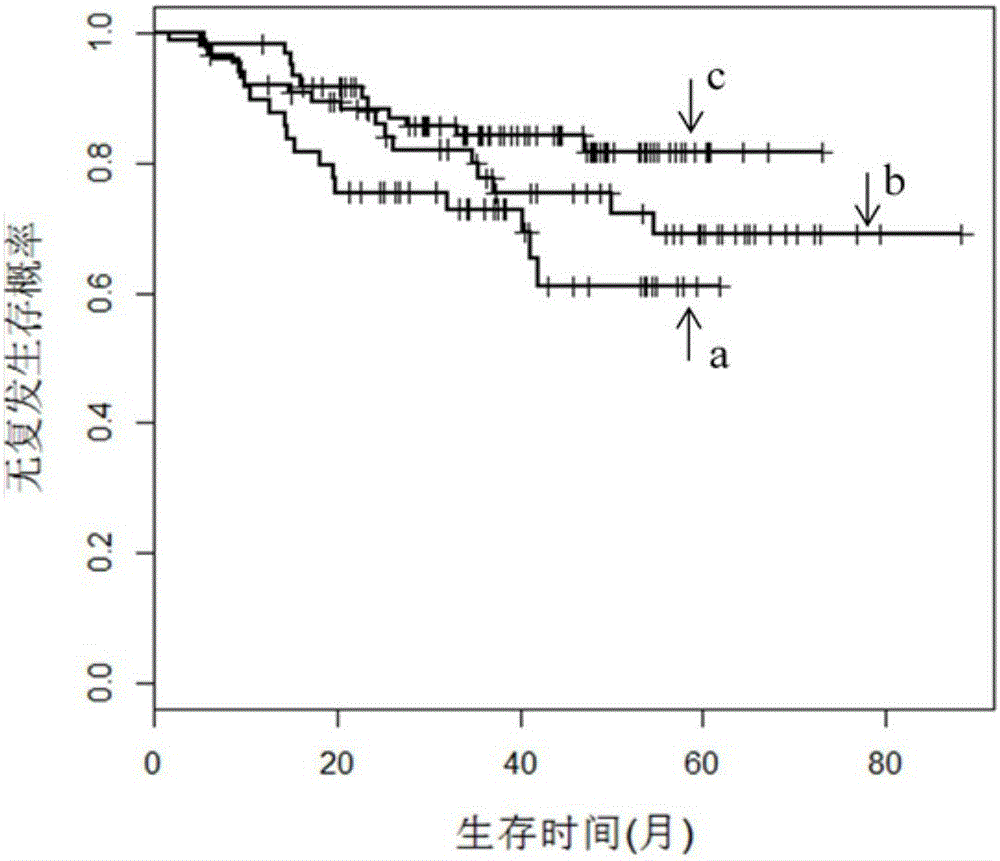
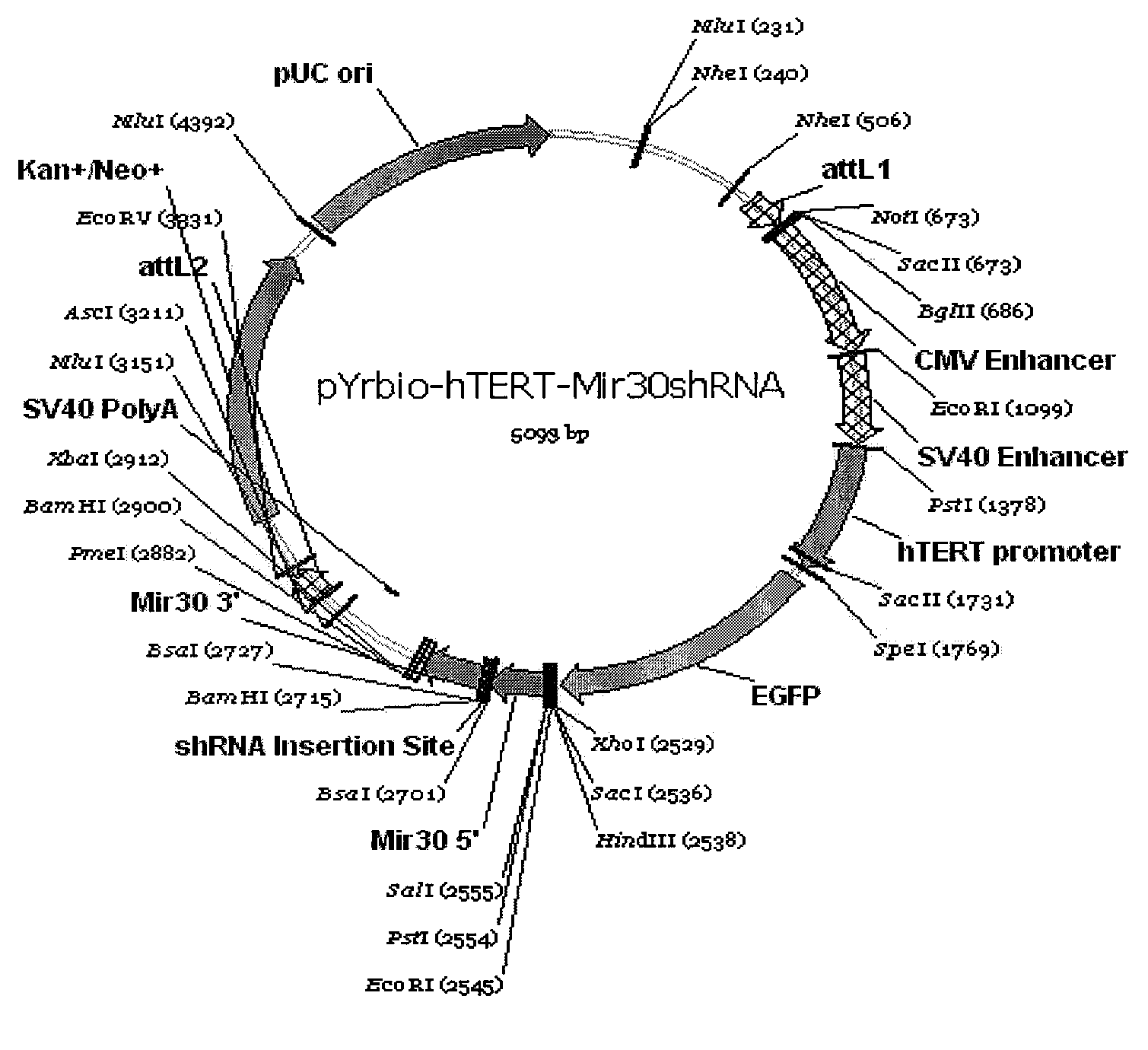
The invention provides a eukaryotic expression vector for expressing shRNA (short hairpin Ribonucleic Acid) in manner of targeting in cancer cells, comprising the structure as follows: (1) an expression cassette structure which is driven by a polII-type promoter and connects a fluorescent protein gene and a mirshRNA structure in series together; (2) a hTERT (human telomerase reverse transcriptase) promoter enhanced by a CMV (cytomegalovirus) enhancer and a SV40 (simian virus 40) enhancer; (3) mirshRNA structures based on mir30: mir30 left arm-shRNA-mir30 right arm, and multiple mirshRNA structures can be connected in series; (4) a Kan or Amp resistance selection marker; and (5) LR homologous recombination arms. By utilizing the method to construct the shRNA eukaryotic expression vector, one or more than one shRNA can be specifically expressed in the cancer cells in manner of targeting; normal cells are not influenced while the RNA interference and the gene therapy are carried out on the cancer cells, thereby solving the problem of non-specific interference during carrying out the gene therapy by utilizing the RNA interference and being beneficial to the research and the application of the RNA interference in the cancer gene therapy aspect.
Owner:HUNAN NENGRUN MEDICAL DIAGNOSIS TECH
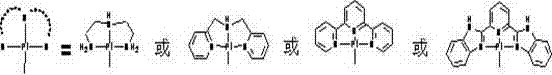
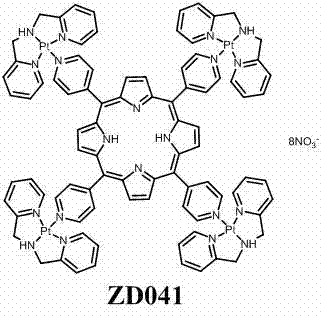
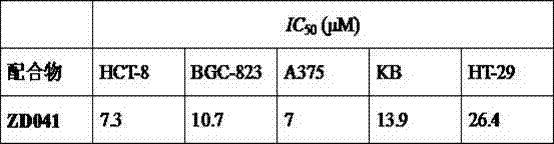
Tetrapyridylporphine bridged crossed tetra-palladium complexes, and preparation method and antitumor activity thereof
InactiveCN102408452AGood antitumor activityOrganic active ingredientsPlatinum organic compoundsTelomeraseTumor target
The invention relates to a preparation method of tetra-palladium complexes in a crossed structure which synthesize tetrapyridylporphine bridge by using supramolecular self-assembly. The tetra-palladium complexes are possibly used for treating human cancers. The MTT experiment data indicates that the compounds have high antitumor activity. A single-teeth platinum ligand, of which the chloride ion is substituted by nitrate, and a bridging ligand tetrapyridylporphine are subjected to self-assembly, thereby obtaining new potential anticancer drugs with excellent anticancer activity and tumor targeting property. The complexes have excellent tumor cell targeting property, and induce the G-rich sequence of the cancer gene to form a G-quadruplex structure, thereby inhibiting the activity of the telomere enzyme and further having high antitumor activity. In addition, since the preparation method is simple and easy to implement, has lower cost, and can be complemented in a common chemical laboratory, and does not have environment pollution in the production process. Therefore, the tetrapyridylporphine bridged tetra-palladium complexes in a crossed structure can be used as novel potential antineoplastic drugs.
Owner:SUN YAT SEN UNIV
Anaerobic bacterium as a drug for cancer gene therapy
The present invention provides a bacterium belonging to the genus Bifidobacterium, by which DNA coding for a protein having an antitumor activity or DNA coding for a protein having the activity of converting a precursor of an antitumor substance into the antitumor substance is delivered to tumor tissues specifically under anaerobic conditions thereby expressing the protein encoded by the DNA, as well as a pharmaceutical composition comprising said anaerobic bacterium.
Owner:ANAEROPHARMA SCI
Induced malignant stem cells
The present invention addresses the problem of providing malignant stem cells that can be grown in vitro and are useful in cancer therapy research and drug discovery research for cancer therapy, a method for producing same, cancer cells induced from the cells, and use for the cells. Provided are induced malignant stem cells that can be grown in vitro, the induced malignant stem cells being characterized in (1) having at least one type of abnormality selected from among (a) abnormal methylation (hypermethylation or hypomethylation) of a cancer suppressor gene or cancer-related gene region in the endogenous genome DNA, (b) somatic mutation of a cancer suppressor gene in the endogenous genome DNA or somatic mutation of an endogenous cancer-related gene, (c) abnormal expression (increased expression or decreased / lost expression); of an endogenous cancer gene or endogenous cancer suppressor gene, (d) abnormal expression (increased expression or decreased / lost expression) of non-coding RNA such as endogenous cancer-related micro RNA, (e) abnormal expression (increased expression or decreased / lost expression) of an endogenous cancer-related protein, (f) endogenous cancer-related metabolism abnormality (hypermetabolism or hypometabolism), or (g) abnormal endogenous cancer-related carbohydrate; and (2) expressing the POU5F1 gene, NANOG gene, SOX2 gene, and ZFP42 gene.
Owner:NAT CANCER CENT +1
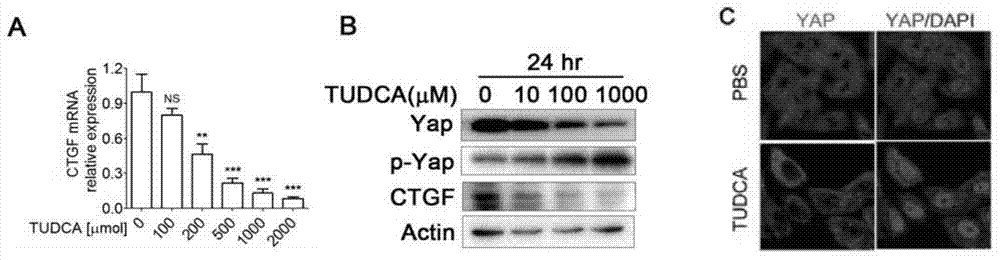
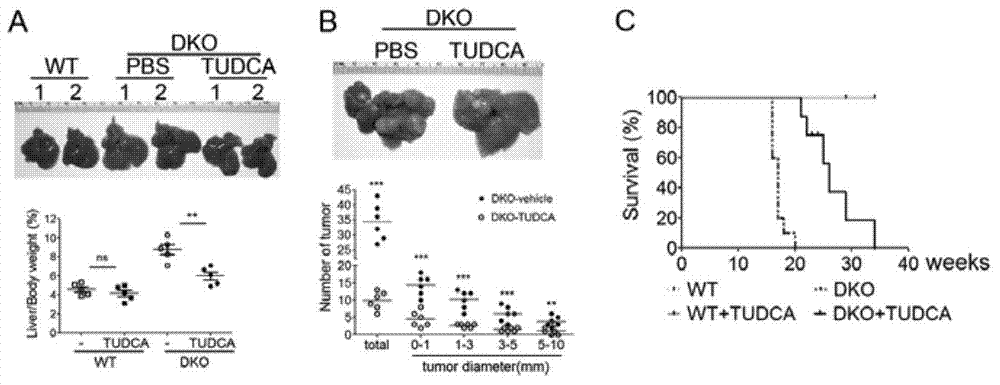
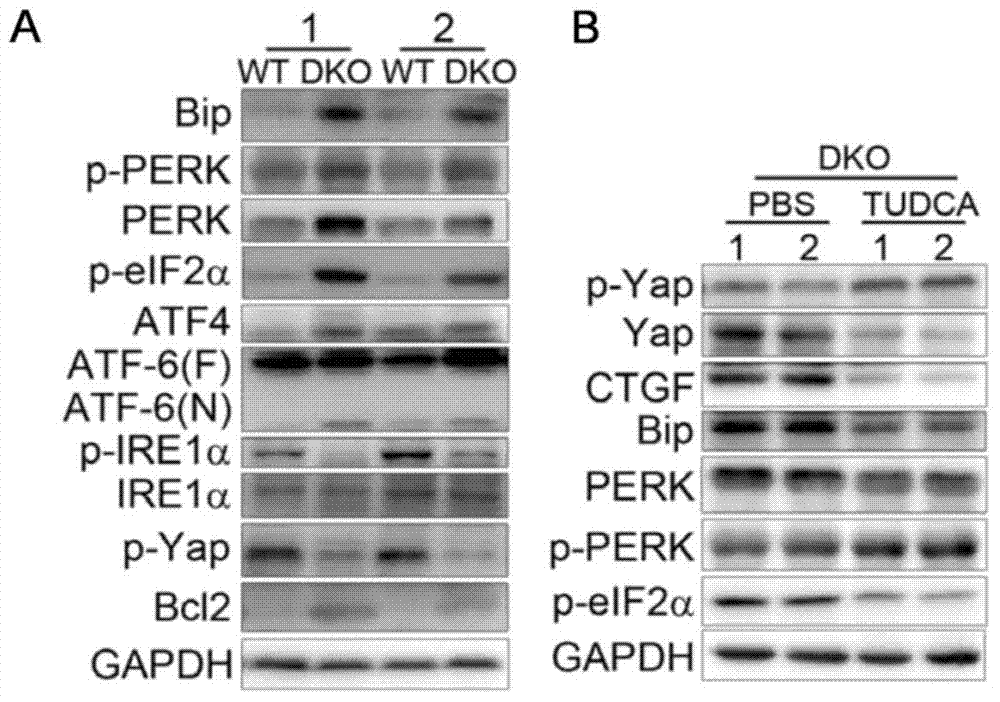
Pharmaceutical application of tauroursodeoxycholic acid and acceptable salts thereof
InactiveCN103919787AGood inhibitory effectWide variety of sourcesOrganic active ingredientsAntineoplastic agentsActivation functionMedicine
The invention discloses pharmaceutical application of tauroursodeoxycholic acid and acceptable salts thereof. The tauroursodeoxycholic acid can reduce the expression and activity of cancer genes Yap and inhibit the activity of a UPR (unfolded protein response) signal channel component; and as verified in two liver cancer models with Yap activation and UPR signal channel activation functions, the tauroursodeoxycholic acid has an effect of preventing and treating a liver cancer, and has a certain application prospect in preparation of a medicament for preventing and treating the liver cancer, particularly in preparation of a medicament for preventing and treating the liver cancer caused by Yap activation or UPR signal channel activation.
Owner:XIAMEN UNIV
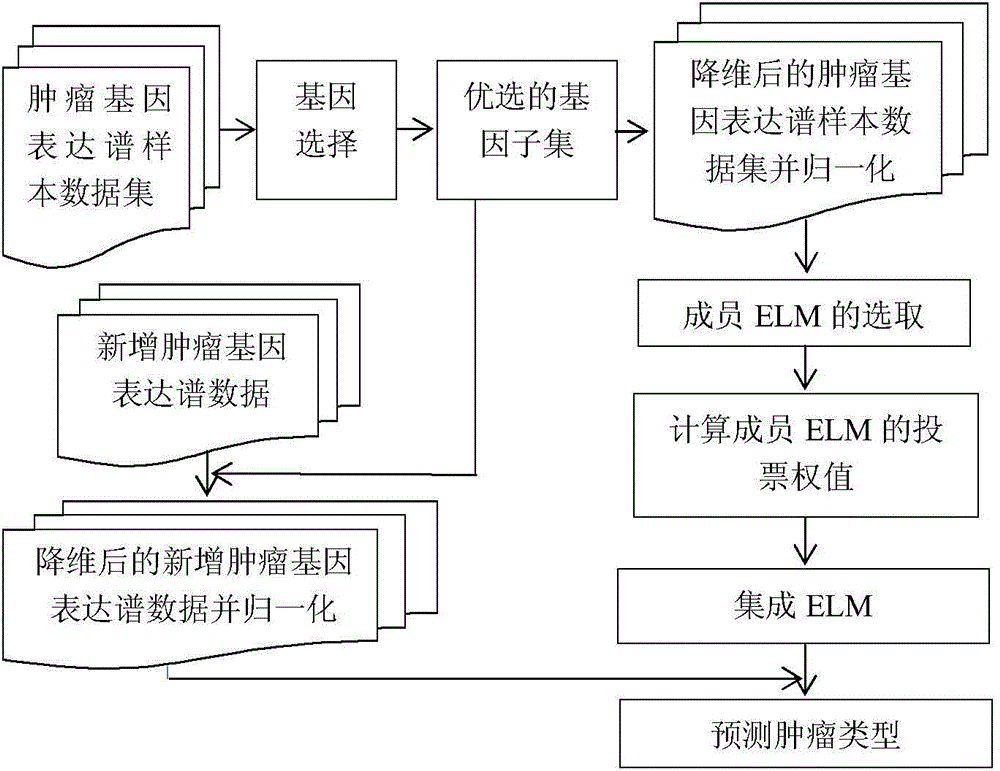
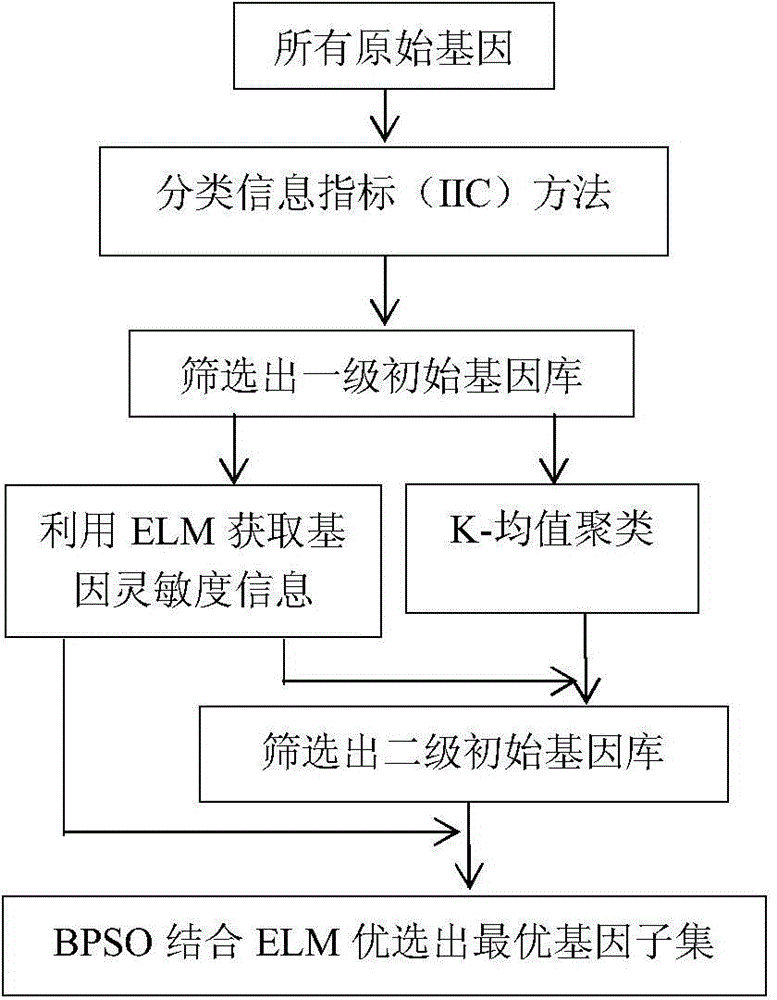
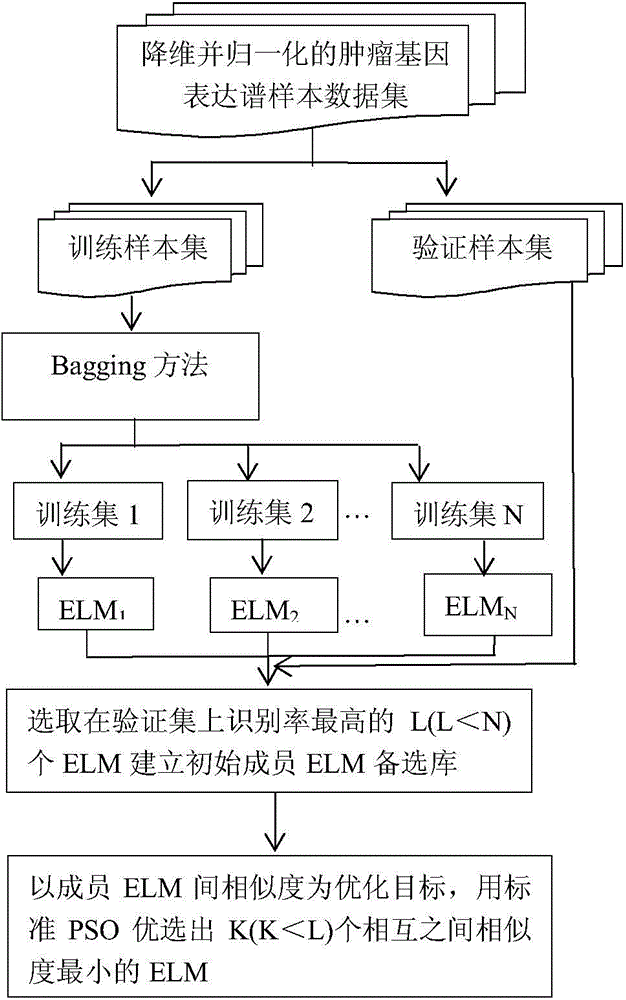
Cancer gene expression profile data identification method based on integration of extreme learning machines
InactiveCN104463251AImprove the accuracy of tumor recognitionReduce learning costsBiostatisticsCharacter and pattern recognitionData setGene selection
The invention discloses a cancer gene expression profile data identification method based on integration of extreme learning machines. The method includes the steps of selection and integration of member extreme learning machines (ELMs). The method concretely includes the steps that preprocessing is carried out on a cancer gene expression profile data set, wherein the preprocessing includes gene selection and normalization of expression profile data; N sample sets are generated through a Bagging method, and each sample set is divided into a training set and a verification set according to a certain proportion; N ELMs are generated on the N training sets in a learning mode, and L ELMs (L&1t; N) with the highest recognition rate on the corresponding verification sets are selected to form an alternative member ELM base; K member ELMs (K&1t; L) forming an integrated system are selected from L ELMs based on the particle swarm optimization algorithm; the integrated vote weight of K member ELMs is worked out by utilizing the minimum-norm least square method; an integrated ELM system is obtained, and the integrated ELM system is used for performing tumor recognition on a newly increased cancer gene expression profile sample. Through the method, the cancer gene expression profile data can be quickly and accurately recognized.
Owner:JIANGSU UNIV OF SCI & TECH
Method for preparing chorionic mesenchymal stem cells
ActiveCN104560869AReduced Chances of ContaminationReduce pollutionSkeletal/connective tissue cellsEmbryonic cellsTelomeraseSurface marker
The invention provides a method for preparing chorionic mesenchymal stem cells. The method comprises the following steps: performing aseptic collection of placentas; separating chorionic tissues; acquiring chorionic mesenchymal stem cells; culturing the chorionic mesenchymal stem cells; performing frozen preservation on the chorionic mesenchymal stem cells; and reviving the chorionic mesenchymal stem cells. The method provided by the invention has the following technical effects: by adopting a pollution prevention method to reduce the probability of pollution from sources, the probability of pollution is effectively reduced by flushing a surface for multiple times; the pollution of hybrid cells is reduced; a single enzyme is adopted for digestion, thereby simplifying the process; the use of animal source components is reduced by using a serum-free culture medium for culture, the cell performance is stable, the in vitro long-term cultivation of the chorionic mesenchymal stem cells can be maintained, the form, proliferation ability, MSC surface marker expression and differentiation ability of the cells are maintained, and the expression of telomerase, the stable expression of cancer genes and the stable karyotype are maintained; and the enzyme digestion, frozen preservation and reviving cause no damage to the cells.
Owner:江苏省北科生物科技有限公司 +1
Hla-restricted epitopes encoded by somatically mutated genes
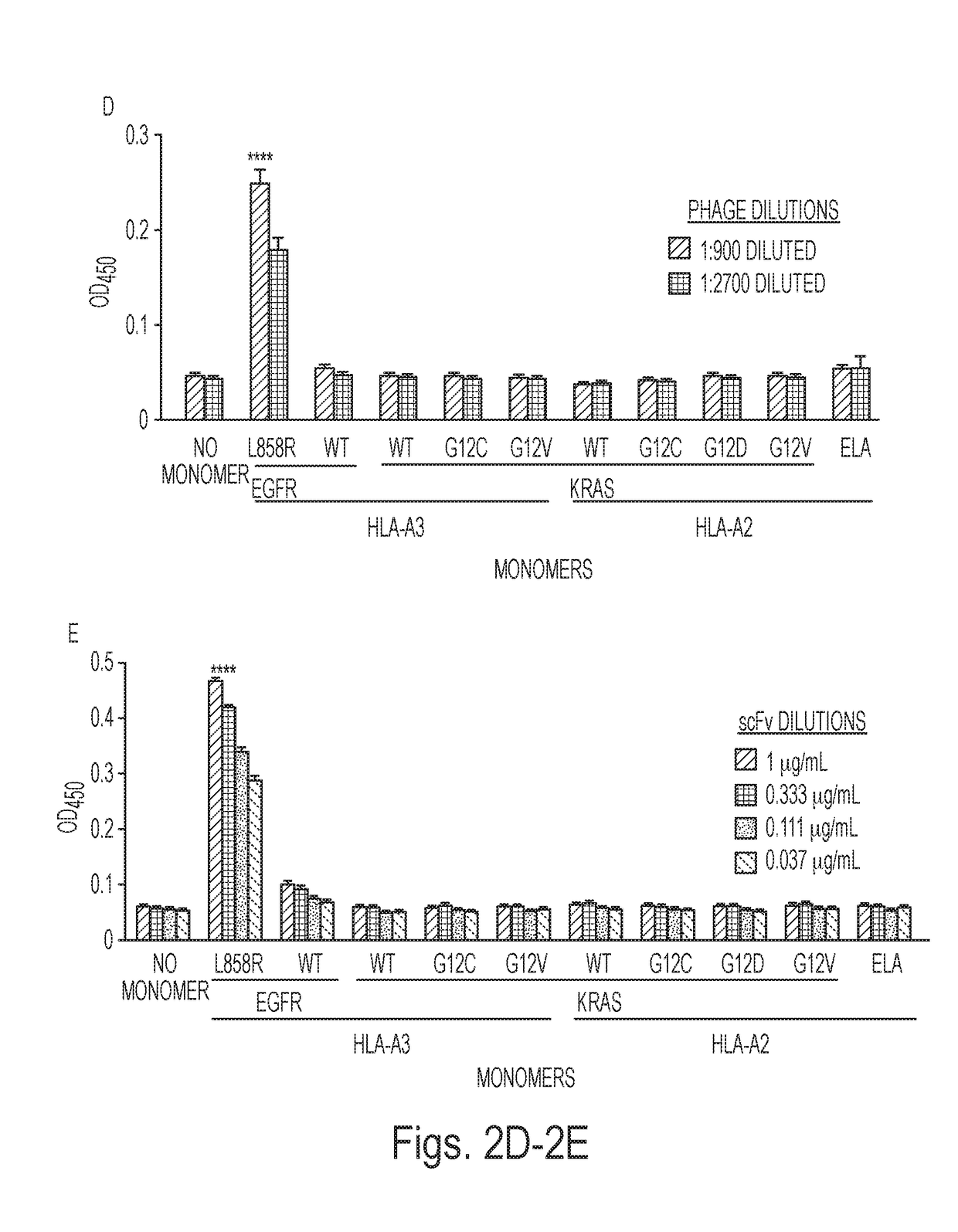
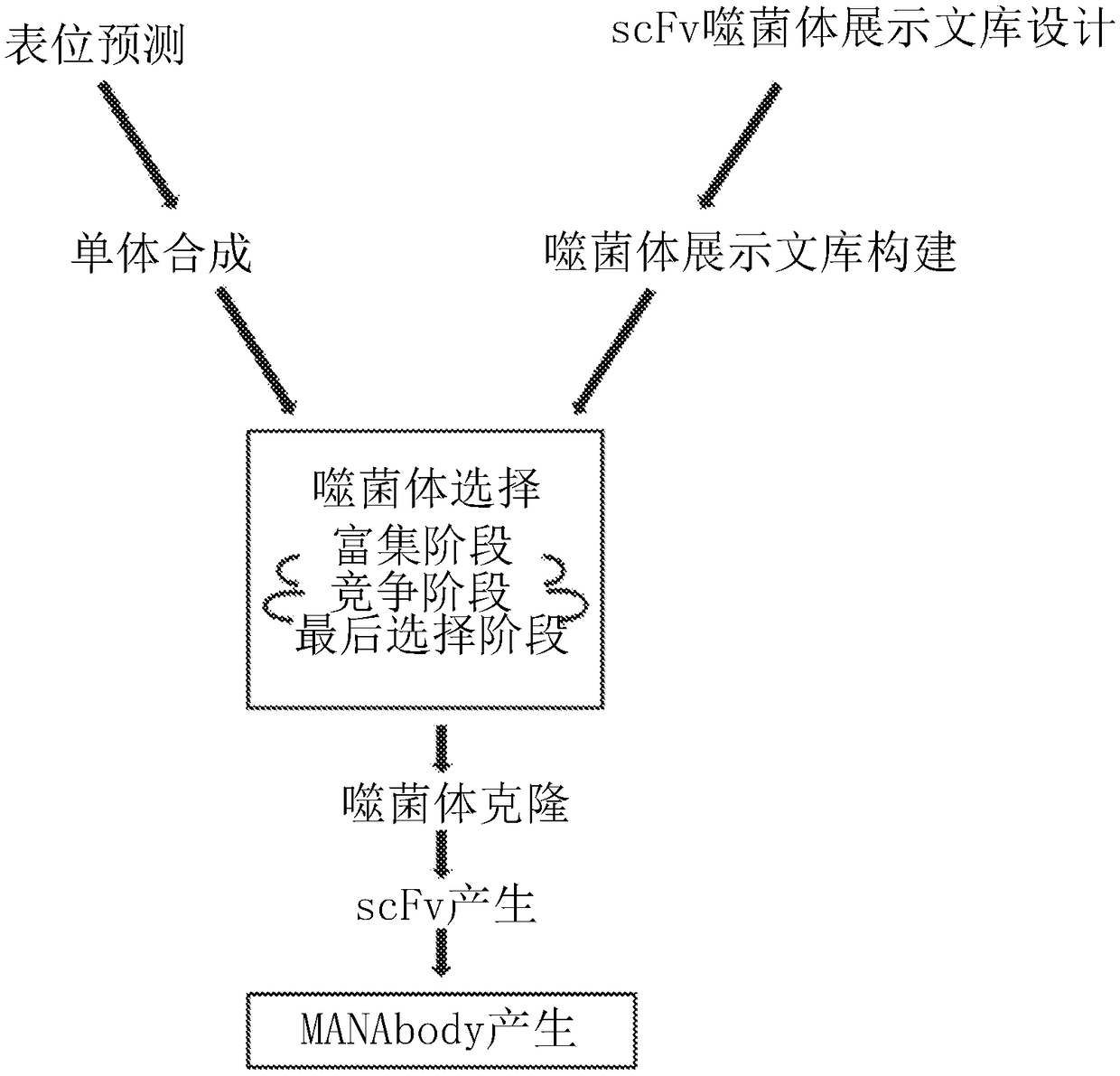
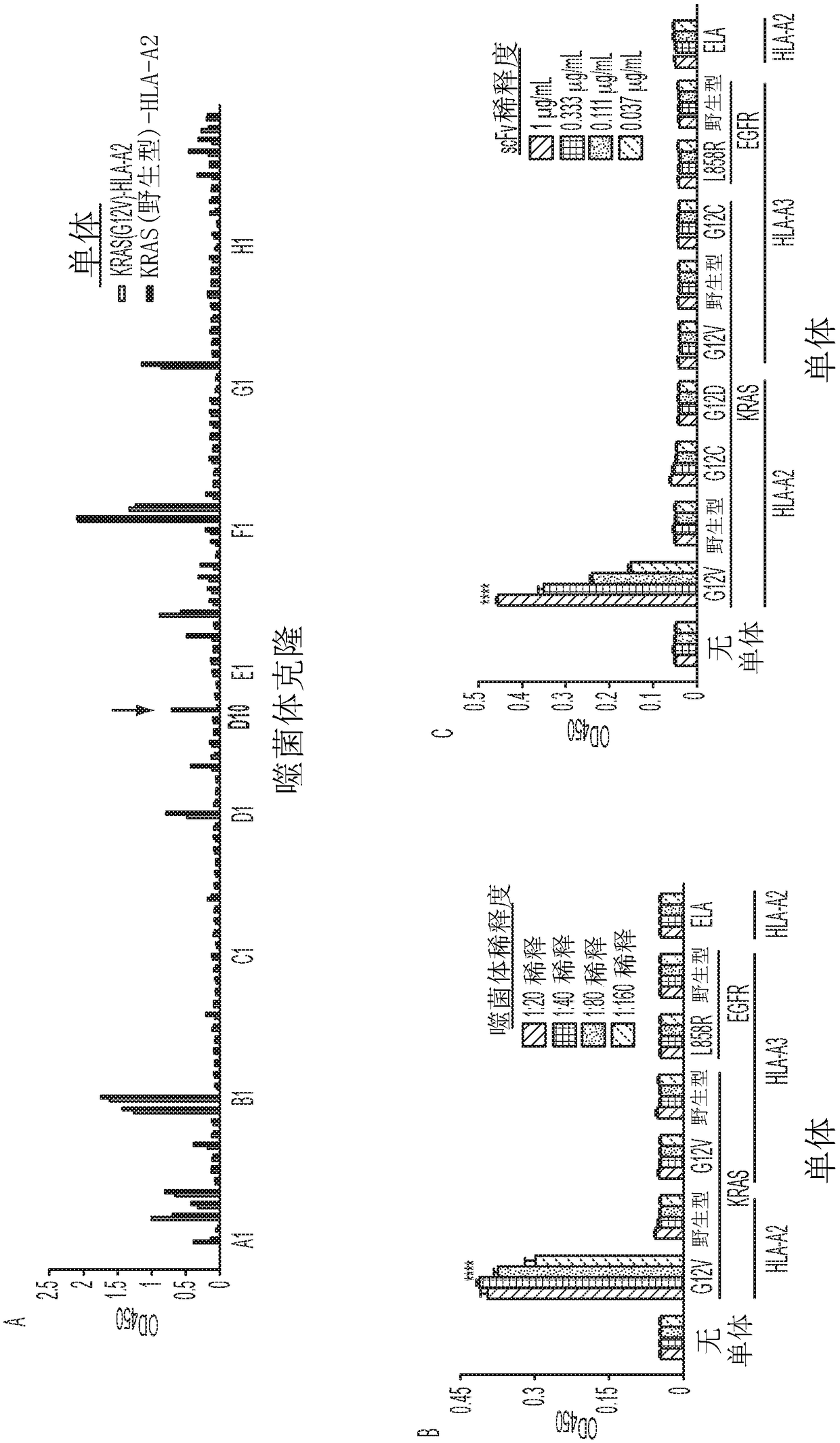
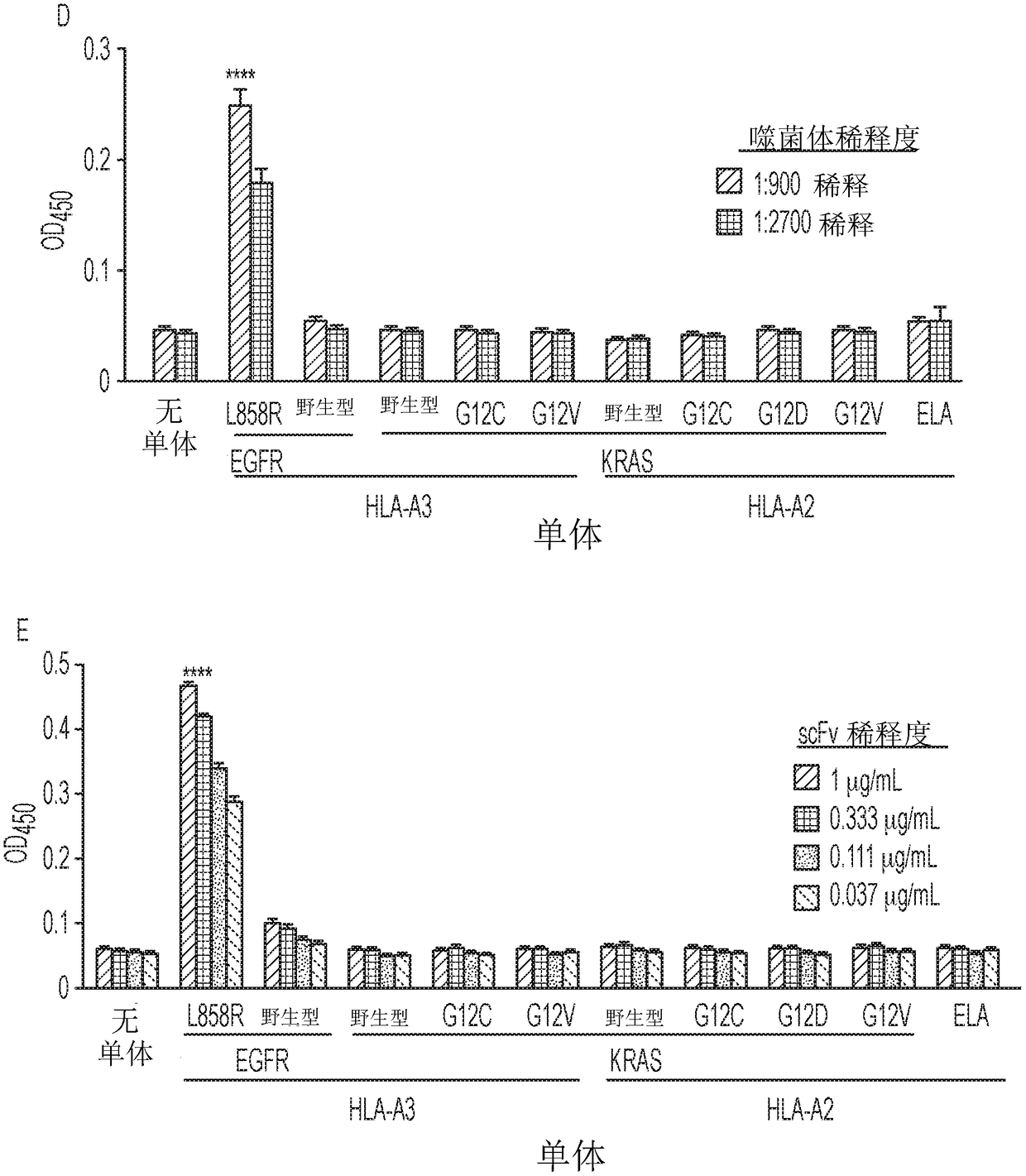
InactiveUS20180086832A1Raise the ratioPolypeptide with localisation/targeting motifAntibody mimetics/scaffoldsWild typeCancer research
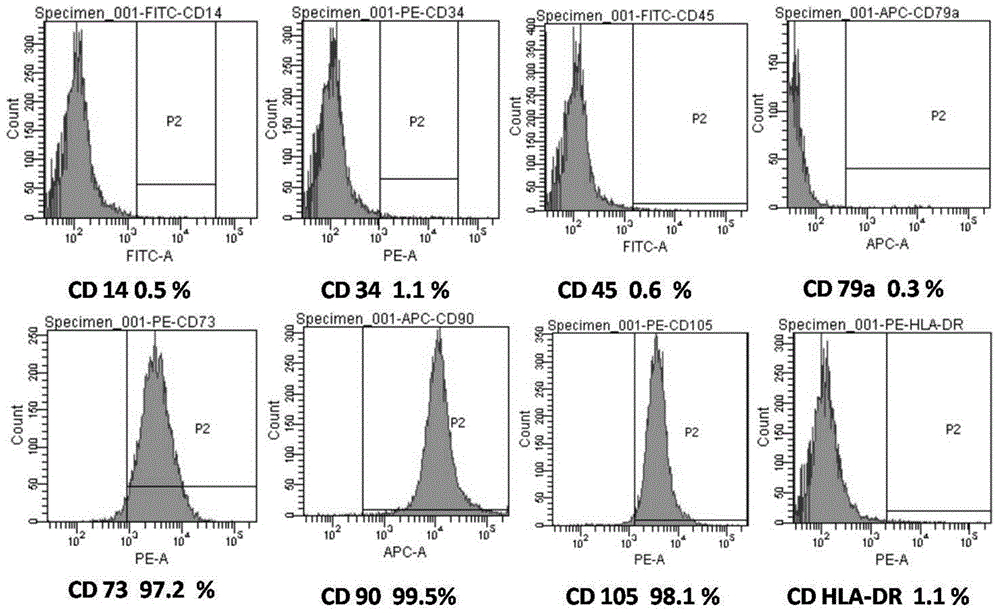
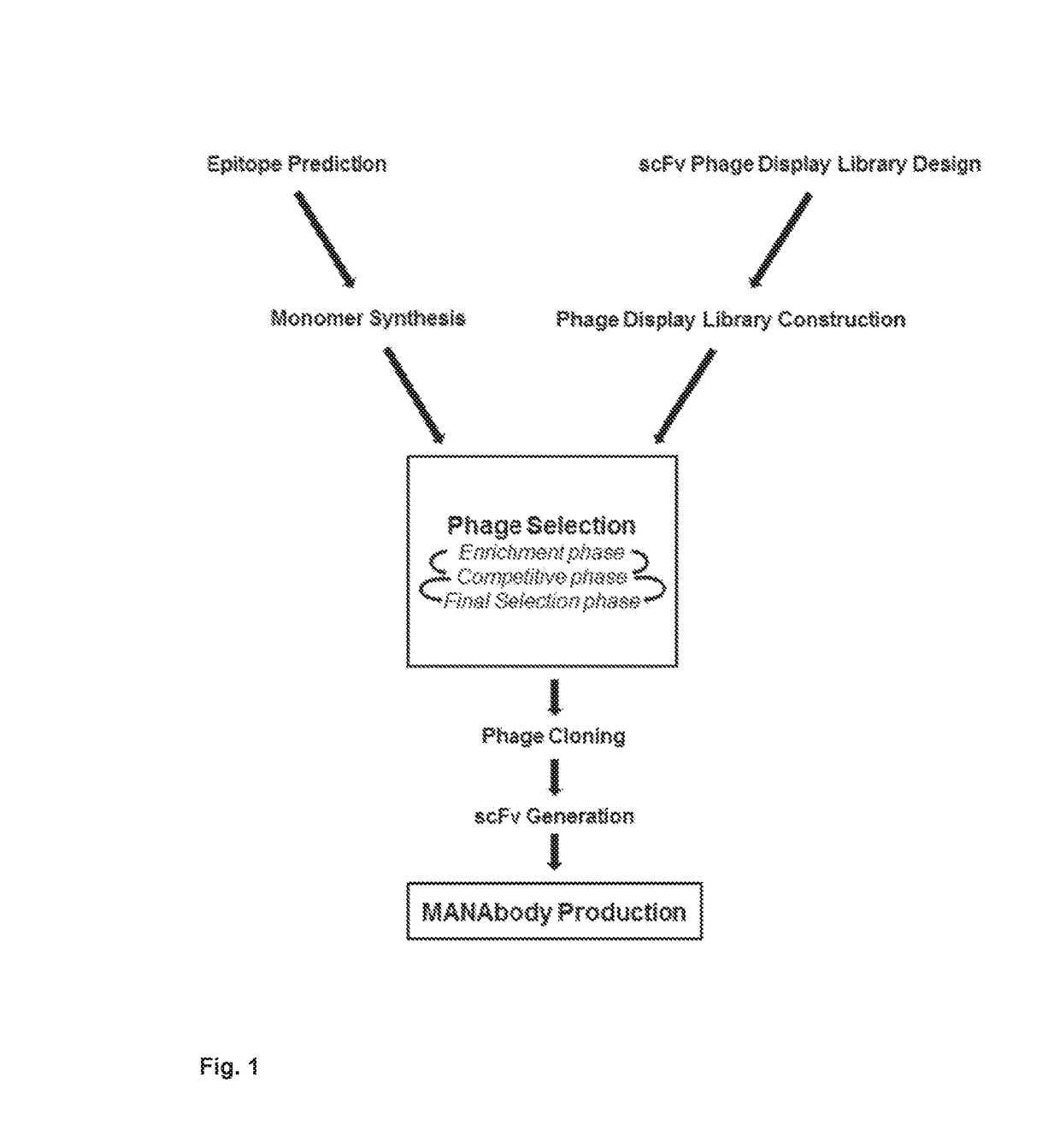
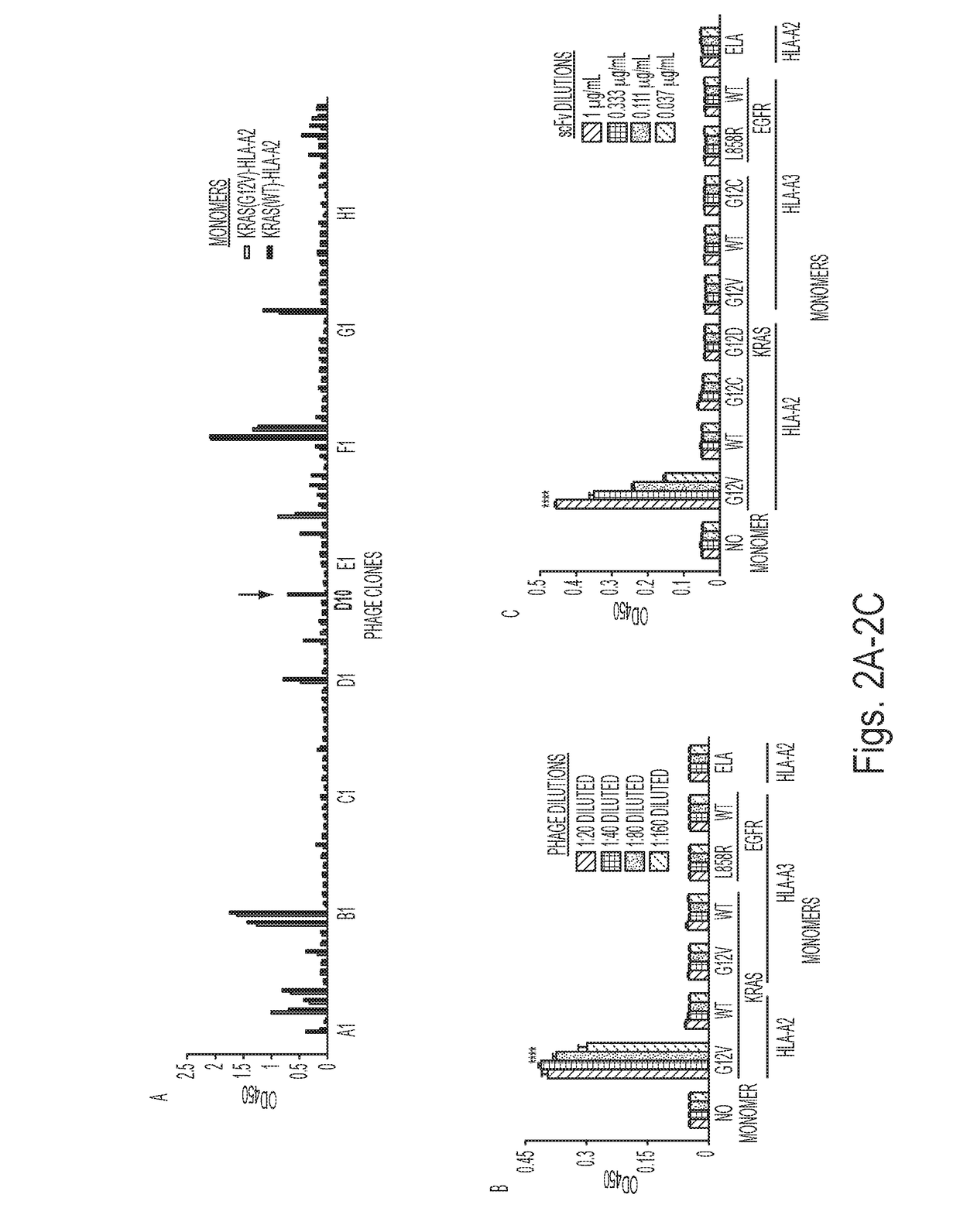
Mutant epitopes encoded by cancer genes are virtually always located in the interior of cells, making them invisible to conventional antibodies. We generated single chain variable fragments (scFvs) specific for mutant peptides presented on the cell surface by human leukocyte antigen (HLA) molecules. These scFvs can be converted to full-length antibodies, termed MANAbodies, targeting “Mutation Associated Neo-Antigens” bound to HLA. A phage display library representing a highly diverse array of single-chain variable fragment sequences was first designed and constructed. A competitive selection protocol was then used to identify clones specific for peptides bound to pre-defined HLA types. In this way, we obtained scFvs, including one specific for a peptide encoded by a common KRAS mutant and another by a common EGFR mutant. Molecules targeting MANA can be developed that specifically react with mutant peptide-HLA complexes even when these peptides differ by only one amino acid from the normal, wild-type form.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
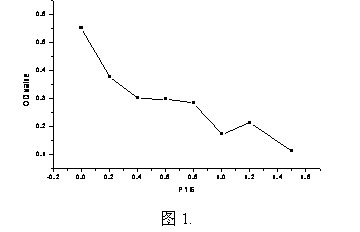
Amino acid sequence for detecting tumor marker P16 antigenic epitope and application of amino acid sequence
The invention discloses an amino acid sequence for detecting a tumor marker P16 antigenic epitope and application of the amino acid sequence, and belongs to the technical field of immunology. The invention provides an antigenic amino acid sequence of a tumor anti-cancer gene P16. The P16 polypeptide antigen is used for detecting corresponding specific antigenic epitope in the blood of a patient with lung cancer and esophagus cancer; the antigenic epitope can be used as a tumor marker for estimating the degree of risk for occurring the lung cancer and the esophagus cancer. And the antigenic polypeptide and an antibody thereof can be used for preparing tumor early-diagnosis reagent and developing a targeted drug for treating the tumors.
Owner:尉军
siRNA-loading nanoparticle and application thereof
InactiveCN104027821AGood monodispersityExtend cycle timeGenetic material ingredientsMacromolecular non-active ingredientsLysosomal membraneNanoparticle
The invention discloses a siRNA-loading nanoparticle. The siRNA is loaded in a duct of mesoporous carbon dioxide through a nonelectrostatic effect, an electropositive high molecule is wrapped on the surface and high molecular polyethylene glycol with biocompatibility and a polypeptide molecule which modifies a transcellular membrane and an lysosome-breaking membrane are connected, so that the nanoparticle has good biocompatibility and the interference effect of the siRNA is improved. The interference effect of the siRNA is remarkably stronger than that of a commercial lipidosome. The nanoparticle has a wide prospect in preparation of cancer gene therapeutic drugs.
Owner:SHANGHAI JIAO TONG UNIV
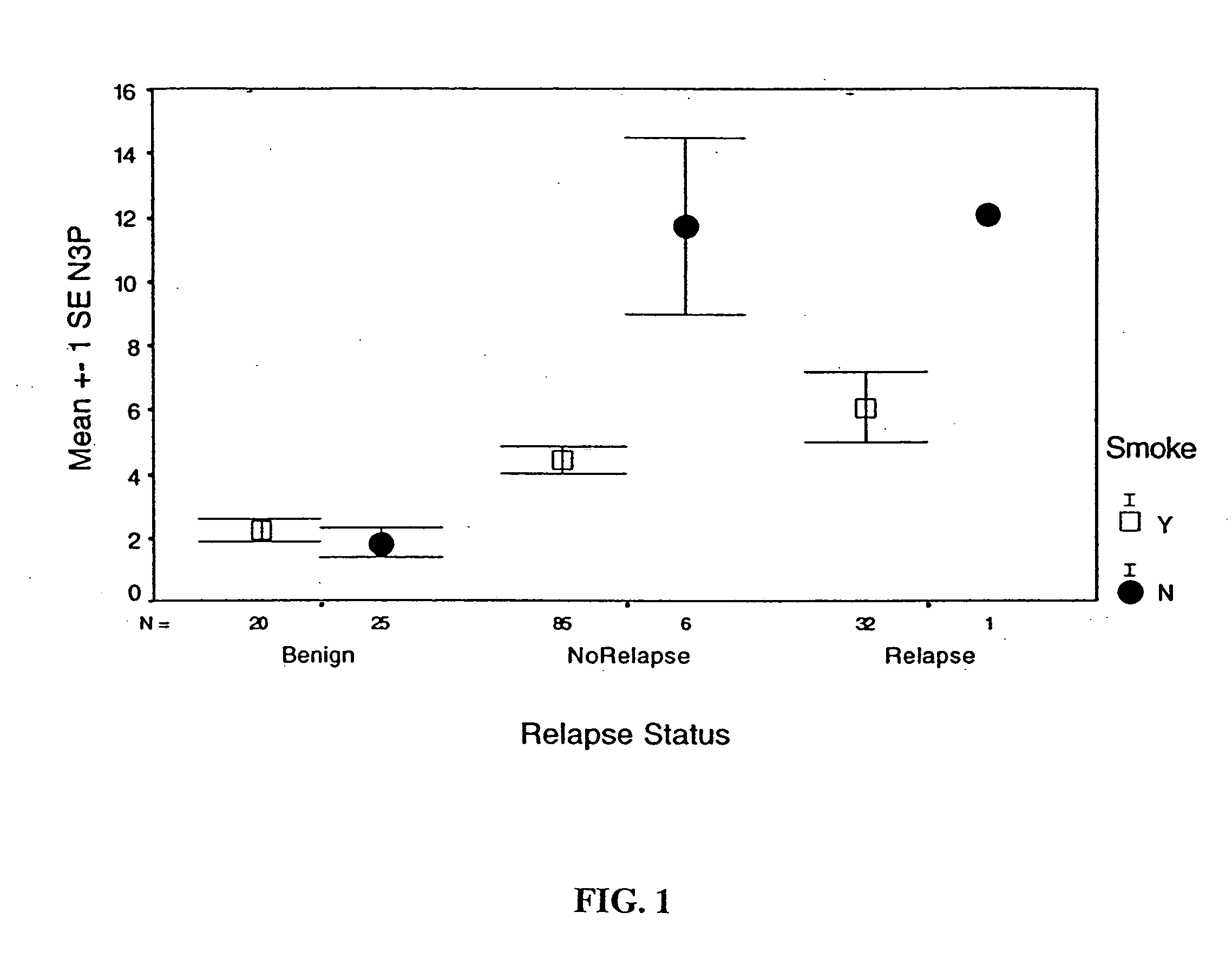
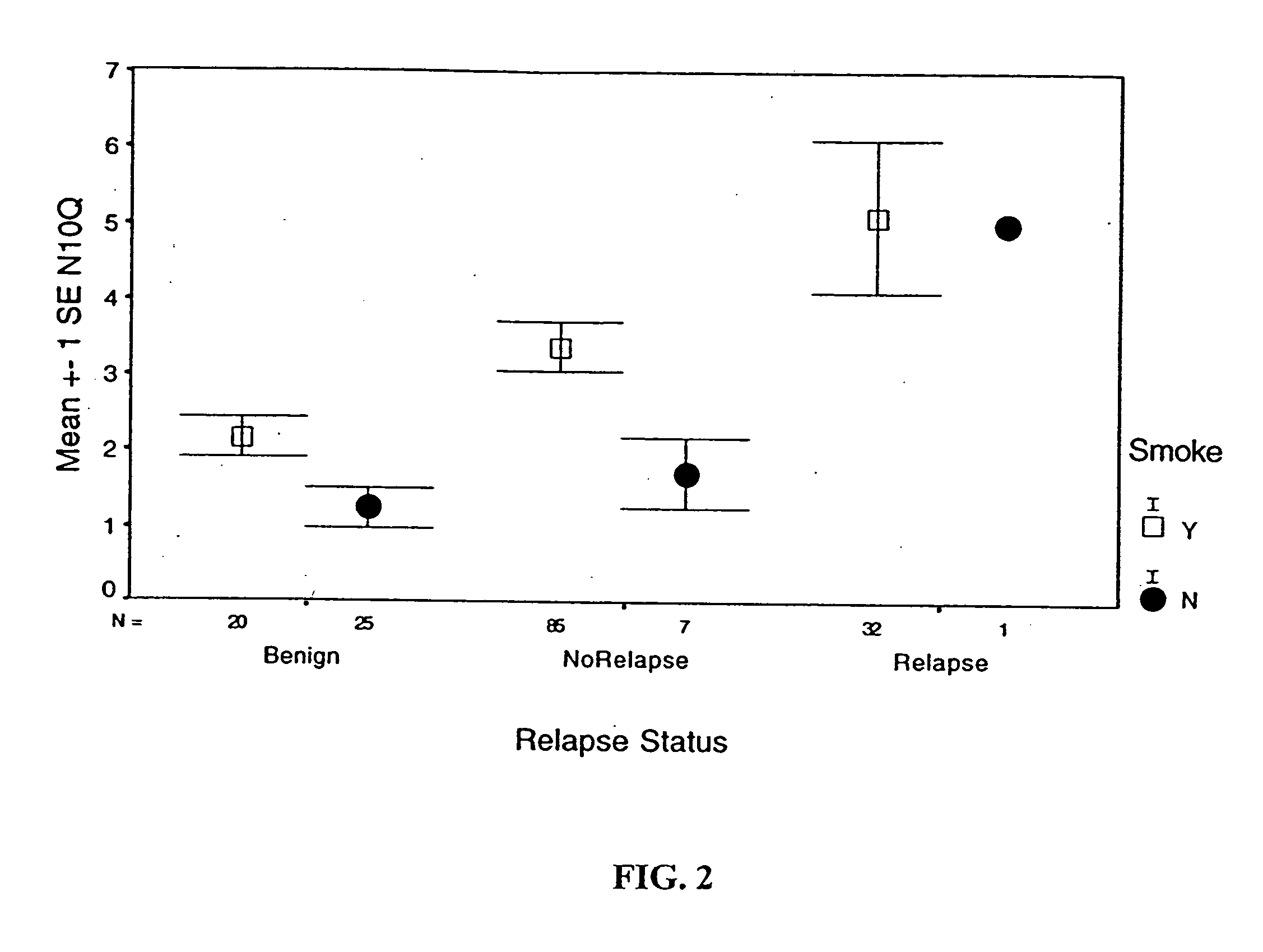
Detection and diagnosis of smoking related cancers
InactiveUS20060078885A1Sugar derivativesMicrobiological testing/measurementCancer genesLow risk group
Gene probes for specific regions of chromosome 3 (3p21.3) and chromosome 10 (10q22) have been found to be tools for the diagnosis and prognosis of smoking related cancers such as non-small cell lung cancer (NSCLC). For example, these probes can be used with fluorescence in situ hybridization (FISH), and used to stratify smokers into high and low risk groups, as well as determine a patients susceptibility to the development of smoking related cancers.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Probe for detecting circulating tumor DNA (Deoxyribonucleic Acid) of human breast cancer and application of probe
InactiveCN105950739AStrong specificityHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationDiseaseIndividualized treatment
The invention discloses a probe for detecting a circulating tumor DNA (Deoxyribonucleic Acid) of a human breast cancer and application of the probe, belonging to the field of diagnosis and prevention of diseases. The invention designs a breast cancer selector which consists of DNA segments in mutation regions of related genes of the breast cancer. The circulating tumor DNA (ctDNA) of a breast cancer patient is subjected to liquid-phase hybridization capture by using the probe designed by the selector; after high-throughput sequencing, specific cancer gene variation of a patient is found, and a tumor load is monitored. Compared with a conventional detection means, the probe and the application thereof disclosed by the invention have the characteristics of no invasiveness, high throughout, good sensitivity and specificity, and the like; the probe and the application thereof can be used for better screening and early-stage diagnosis of the human breast cancer and are applied to individualized treatment of cancer.
Owner:HARBIN MEDICAL UNIVERSITY
Hla-restricted epitopes encoded by somatically mutated genes
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
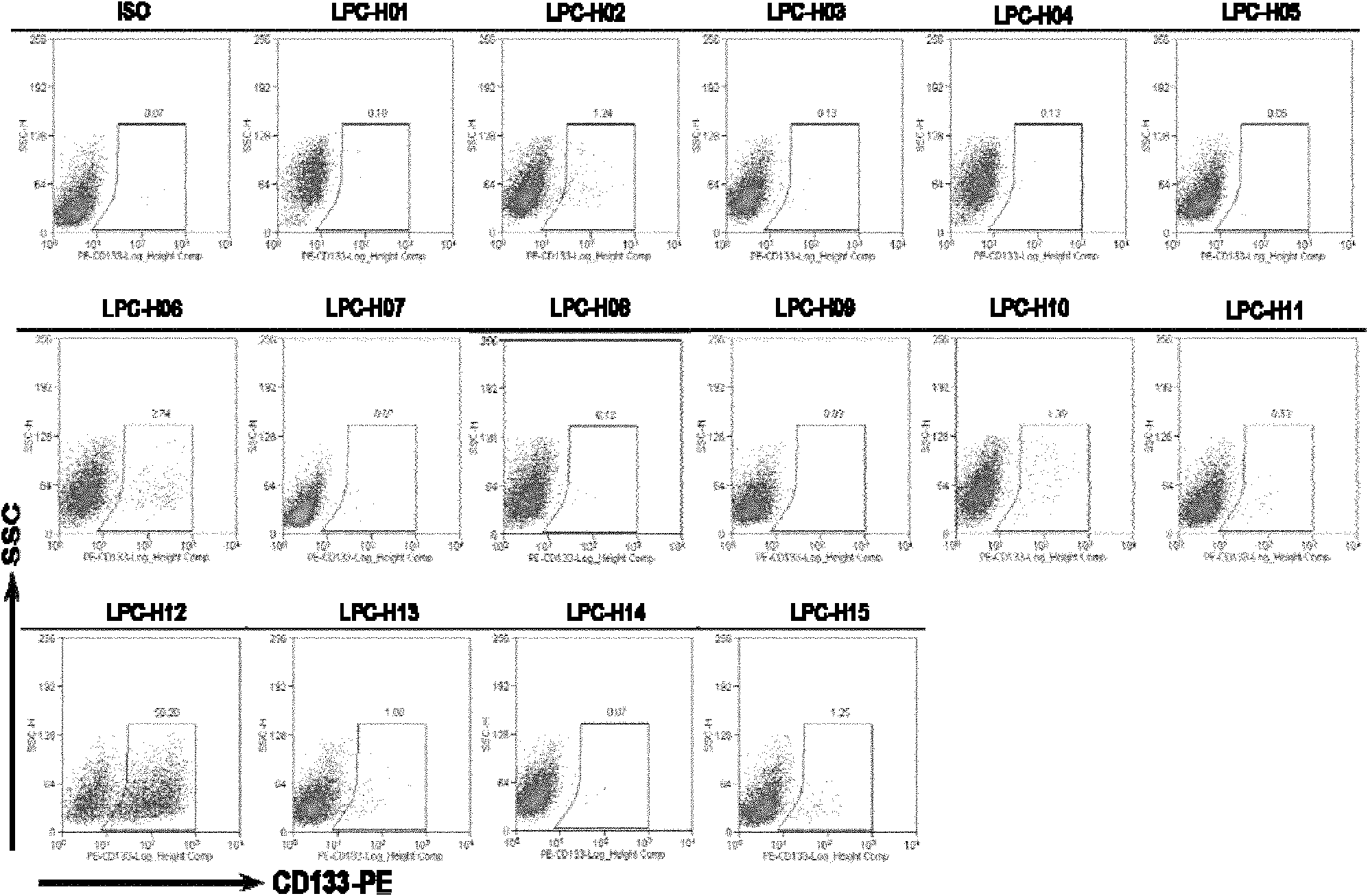
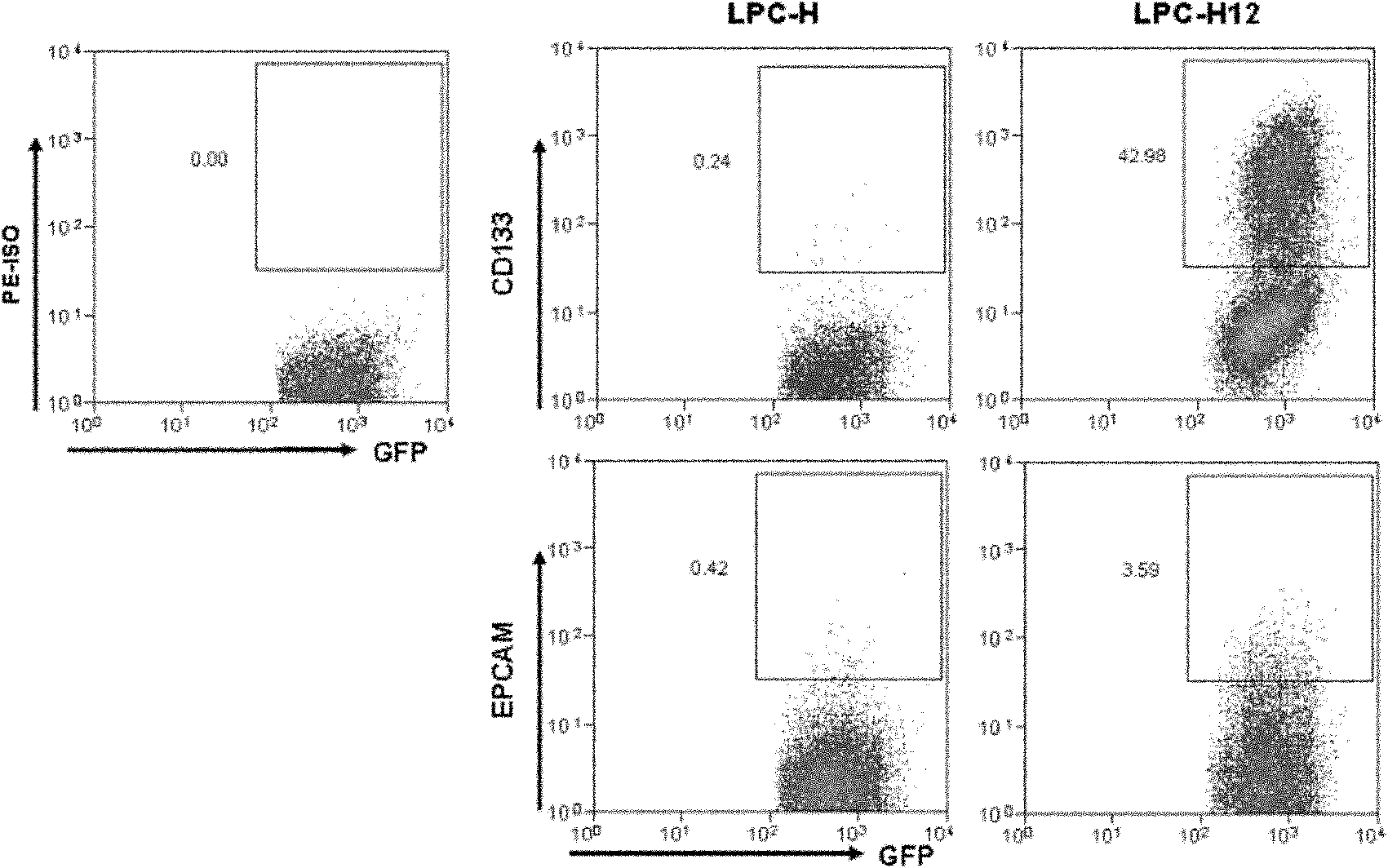
Mouse liver tumor cell line for highly expressing CD133 and preparation method thereof
ActiveCN102181399AHigh expressionHigh in vivo tumorigenicityMicroorganism based processesViruses/bacteriophagesEmbryoCell subpopulations
The invention provides a mouse liver tumor cell line for highly expressing CD133 and a preparation method thereof. The mouse liver tumor cell line with high content of CD133+cell subpopulations is established by the following steps of: importing a cancer gene Hras into p53- / - mouse fetal liver cells, obtaining a single-cell clonal group derived from CD133+cells by utilizing a monoclonal patternmaking technology, and finally screening. The cell line LPC-H12 expresses related genes of various liver stem cells and the stem cells, namely Marker:CD133 and EpCAM, and has high in-vitro balling capacity and in-vitro tumorigenic capacity. The mouse liver tumor cell line provides a powerful tool for researching the action and the mechanism of CD133+liver cancer stem cell subpopulations in the occurrence and development process of liver cancer and screening medicaments used for liver tumor stem cells.
Owner:SHANGHAI INST OF ONCOLOGY
Method for establishing tree-shrew breast cancer model for slow virus by nipple injection
ActiveCN103173496AShort cycleImprove consistencyViruses/bacteriophagesGenetic engineeringOncologyBreast Duct
The invention discloses a method for establishing a tree-shrew breast cancer model for slow virus by nipple injection. Slow virus is introduced to a tree-shrew breast duct by adopting a nipple injection technology, so that a cancer gene is over-expressed or an anti-cancer gene is knocked down after epithelial cells in a mediated duct are infected, and therefore, tree-shrew breast tumors are induced. According to the method for establishing a tree-shrew breast cancer model for slow virus by nipple injection disclosed by the invention, the breast tumors can be touched within two weeks after expressing the cancer gene PyMT; and moreover, the induction success rate is almost 100%, and the method is short in incubation period, strong in specificity, extremely high in induction rate, stable and convenient and simple to operate. The method disclosed by the invention can be used for quickly and efficiently establishing the tree-shrew breast cancer model.
Owner:KUNMING INST OF ZOOLOGY CHINESE ACAD OF SCI
Chinese lung adenocarcinoma cell line with high metastases potentiality of bone, lever and adrenal gland
InactiveCN101955911AGrow fastStrong metabolismMicrobiological testing/measurementMicroorganism based processesTransfer cellLymphatic Spread
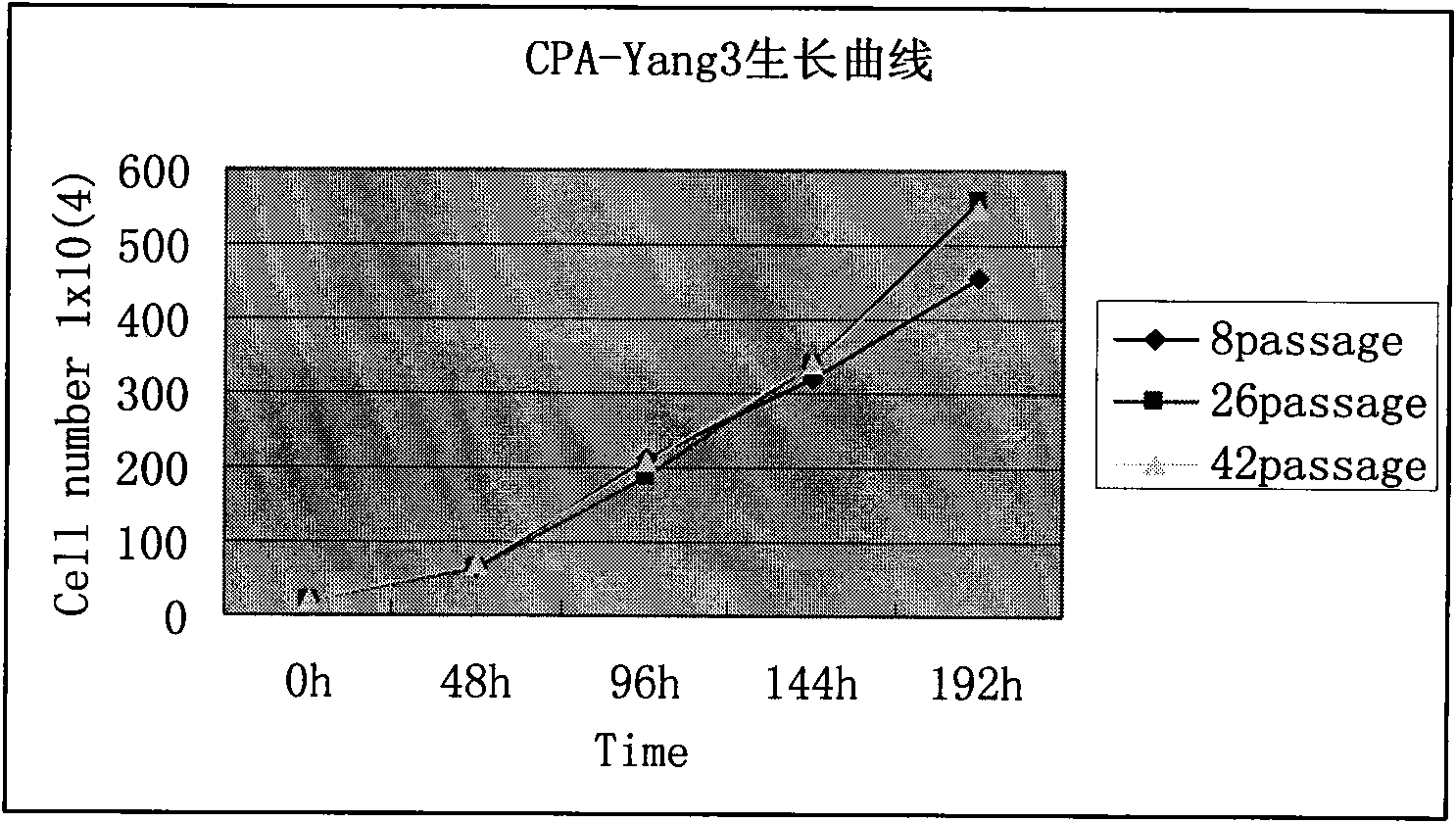
The invention belongs to the filed of microorganism animal cell lines and provides a Chinese lung adenocarcinoma cell line with high metastases potentiality of bone, lever and adrenal gland. According to the invention, hydrothorax of an adenocarcinoma first-diagnosis patient is used as primary culture of cells; the human adenocarcinoma cell has the preservation number of CGMCC No.3137 and classified name as human lung adenocarcinoma cell line CPA-Yang3, can grow adhered to the wall and has the tumorigenesis rate up to 100 percent; the human lung adenocarcinoma cell can grow quickly and metabolize vigorously; the expression levels of cancer genes EMS1, VEGF-C, IL-6, IL-8, SVIL and AR gene are higher than the expression level of SPC-A-1 cell; and the lung adenocarcinoma cancel cell has the biological characteristics of mainly transferring the bone and having high transfer potential of the lever and the adrenal gland, high growth speed of bone transfer cells and complete cell morphology. The invention can be used for providing reference data to early diagnosis for transferring the human lung adenocarcinoma bone, further establishing the relevant gene chip technology and assessing and exploring the curative effects of various medicaments and the like by comprehensively applying the gene chip, quantifying PCR (Polymerase Chain Reaction), western blot in real time and other technologies.
Owner:SHANGHAI CHEST HOSPITAL
Method for establishing pancreatic cancer model
ActiveCN105132463AHigh simulationEfficient methodGenetic engineeringFermentationWilms' tumorTumor suppressor gene
The invention relates to a method for establishing a pancreatic cancer model, and belongs to the technical field of the establishment of an animal model. According to the method, a slow virus for over-expressing one cancer gene and simultaneous knocking down two cancer suppressor genes is constructed according to cancer genes and cancer suppressor genes having the maximum mutation rate in human pancreatic cancer in TCGA database, and the slow virus is applied to the pancreas head of a tree shrew through orthotopic injection so as to induce pancreatic cancer of the tree shrew; and specifically, the method comprises the following steps: (1) construction of a slow virus vector; (2) virus packaging, titre determination and in vitro verification; (3) virus injection; and (4) monitoring and pathological investigation of the tree shrew. The method disclosed by the invention has the advantages that the method is short in inducing cycle, high and stable in morbidity, and simple and convenient in operation, and is capable of rapidly and effectively establishing a tree shrew pancreatic cancer model for simulating the genetic mechanism of human pancreatic cancer to the greatest extent; and the pancreatic cancer model is applicable to the research of the pathogenesis of human pancreatic cancer, the exploration of cancer therapy targets and the development of novel antineoplastic drugs, so as to offer a reference for the treatment of human pancreatic cancer.
Owner:成都豆麦科技有限公司
Adenovirus capable of expressing anti-cancer gene efficiently, regulated by miRNA and capable of specifically proliferating in glioma cells and application thereof
InactiveCN103667347AAvoid damageImprove targetingGenetic material ingredientsFermentationNucleotideNerve cells
The invention provides a recombinant adenovirus vector regulated by miRNA (micro Ribose Nucleic Acid) and capable of specifically proliferating in glioma cells. The recombinant adenovirus vector provided by the invention is characterized in that the target spot nucleotide sequence of at least one miRNA is inserted into the 3-untranslated region of at least one proliferation essential gene of the virus, the expression of miRNA is absent or reduced in glioma cells, but miRNA is normally expressed in nerve cells and glial cells so that the modified recombinant adenovirus specifically proliferate in the adenovirus in which the expression of miRNA is absent or reduced, but do not proliferate in normal cells in which the miRNA is normally expressed; and therefore, the recombinant adenovirus does not generate cytotoxicity to the normal cells.
Owner:刘佳
High throughput screening for cancer genes
InactiveUS20050005311A1Fast generation timeEasy mappingMicrobiological testing/measurementAnimals/human peptidesCancer genesHigh-Throughput Screening Methods
The invention provides high throughput screening systems and in vivo methods for high throughput screening of cancer genes. The invention also is applicable to the discovery of therapeutic agents that block tumor growth and metastasis. The invention further provides kits and compositions to perform such assays.
Owner:US DEPT OF HEALTH & HUMAN SERVICES
Application of group of gastric cancer genes
InactiveCN105986034AAvoid overdosingReduce medical costsMicrobiological testing/measurementClinical prognosisCancer gene
The invention discloses an application of a group of gastric cancer genes. In the invention, based on a group of 53 prognosis related genes in gastric cancer and the detection results of the expression levels thereof in clinical samples, the score is calculated and predicted to evaluate clinical prognosis of gastric cancer as well as related application thereof. The system can help the gastric cancer patients in treatment selection and can predict the reaction to treatment intervention so as to judge whether the patients profit from chemical and targeted therapy, excessive medication is avoided, the medical cost is reduced, and finally, an aim of accurate or individualized medicine is achieved. According to the system and different detection technology platforms, corresponding 53 gene expression measurement kits are designed and developed.
Owner:NANJING KDRB BIOTECH INC LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com