Method for identifying cancer molecular subtype based on spectral clustering algorithm of sparse similar matrix
A spectral clustering algorithm and molecular subtyping technology, which is applied in the field of identifying cancer molecular subtypes based on sparse similarity matrix spectral clustering algorithm, can solve the problems of different prognosis, poor prognosis and unpredictability of patients. Achieve the effects of avoiding singularity problems, improving prediction accuracy, and reducing computational complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
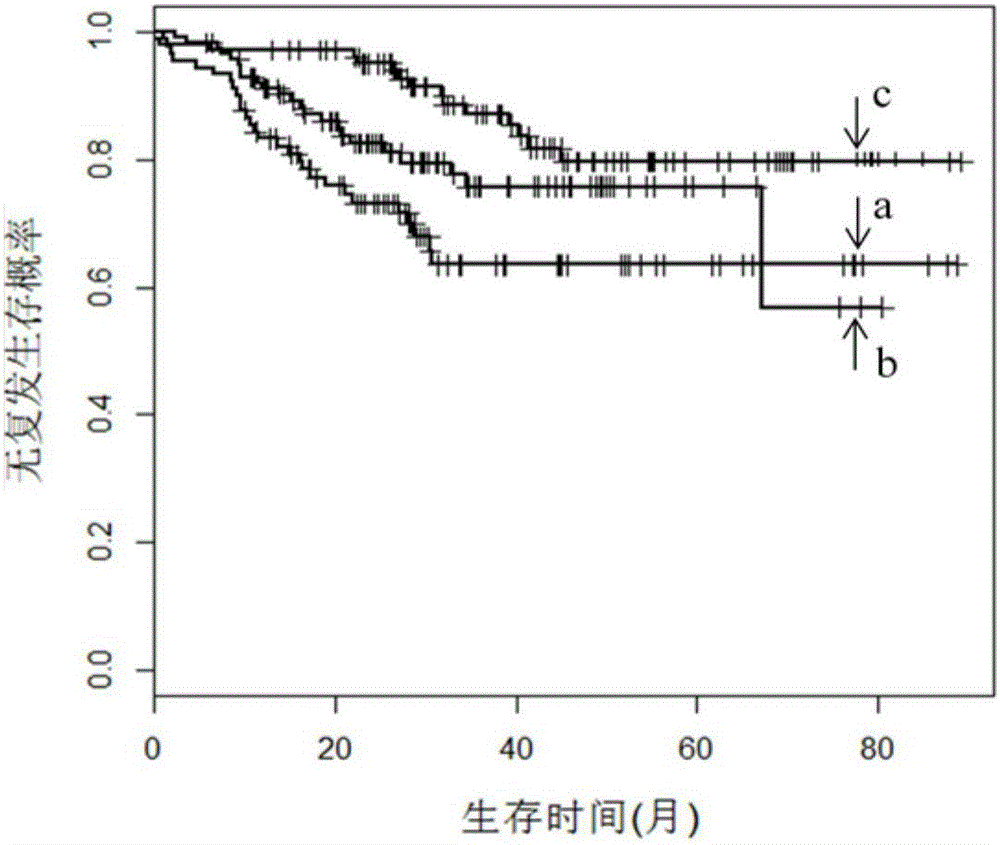
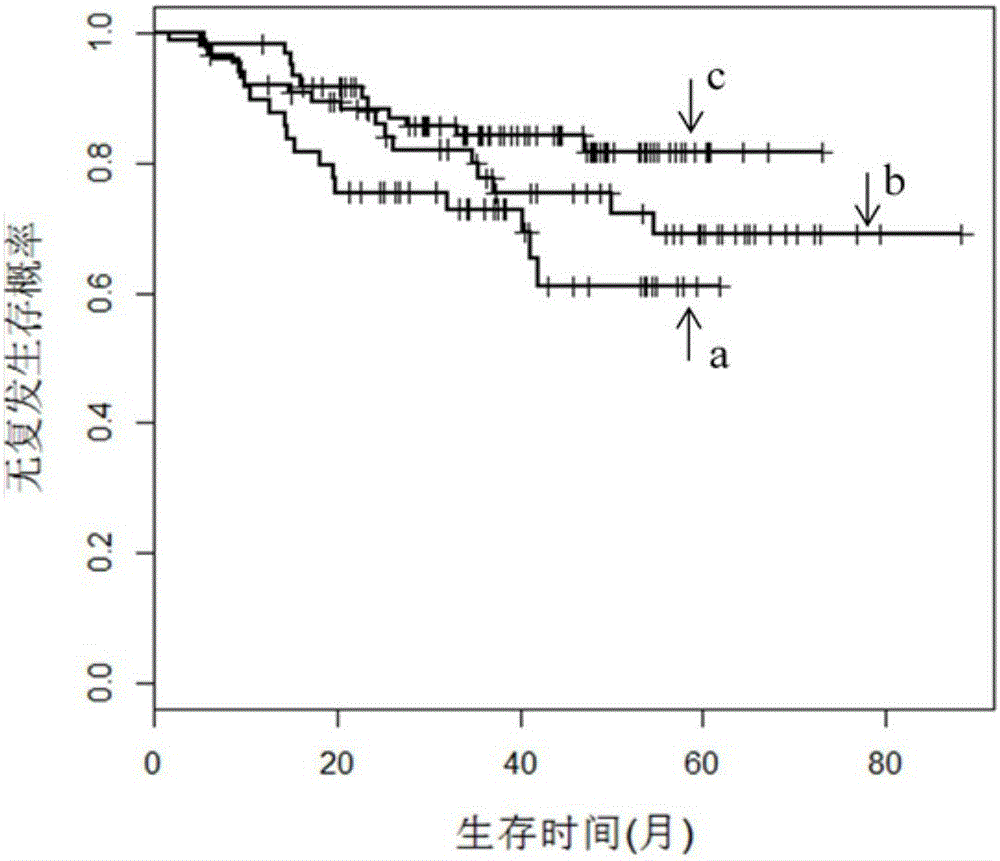
[0027] In this embodiment, a spectral clustering algorithm based on a sparse similarity matrix uses cancer gene expression profile data as a training set sample to construct a cancer molecular subtype prediction model; the prediction model is used to predict the cancer molecular subtype of an independent test set sample, Thus, the independent test set samples are divided into multiple classes of molecular subtypes.
[0028] Specifically, proceed as follows:
[0029] Step 1. Calculate the similarity matrix SL(n×n) between any two cancer samples in the cancer gene expression profile data as the training set samples.
[0030] A cancer sample refers to a vector whose columns are gene expression profile data; the similarity value s between two cancer samples is calculated according to a Gaussian function ij , with similarity value s ij Construct similarity matrix SL(n×n); where x i and x j is a cancer sample, 1≤i≤n, 1≤j≤n, n is the number of samples in the cancer gene express...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com