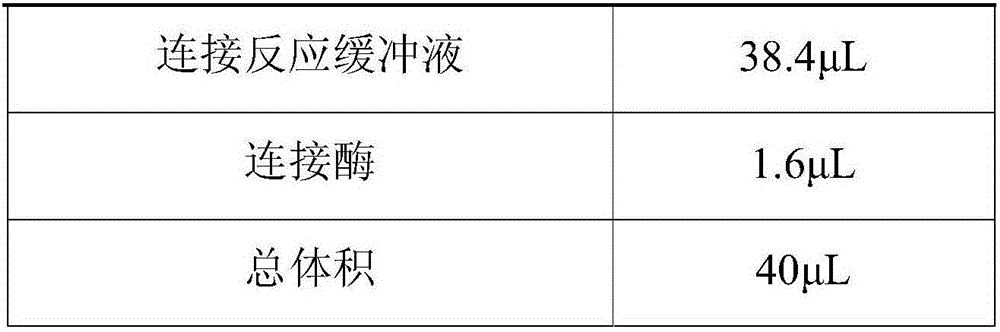
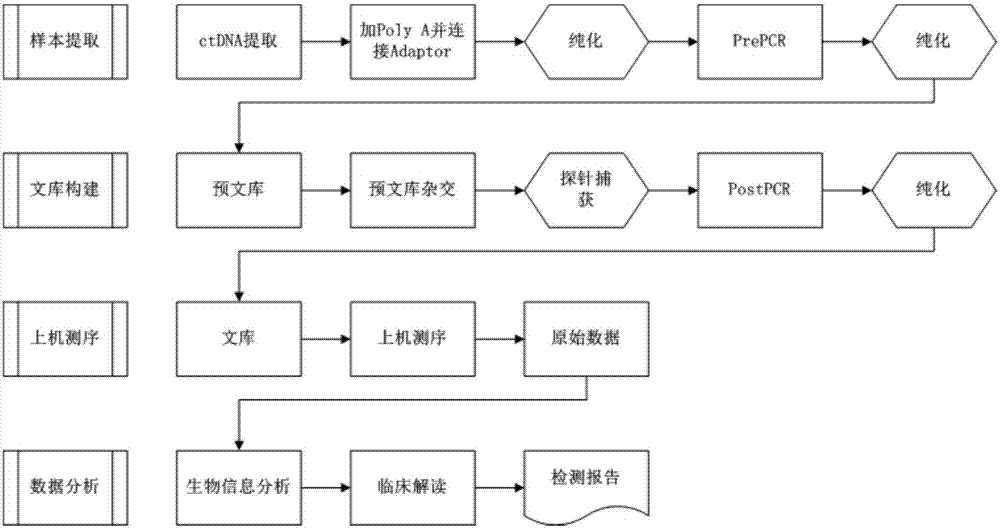
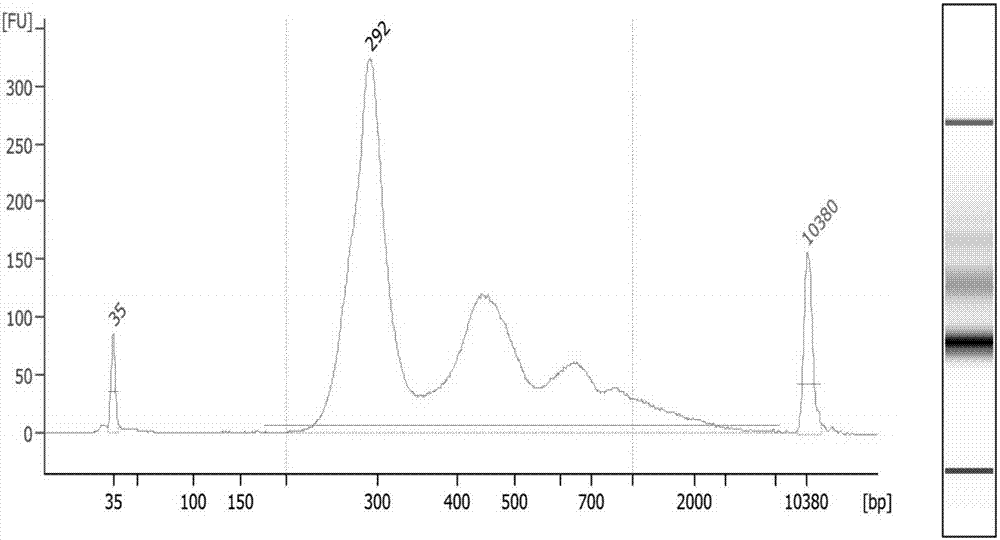
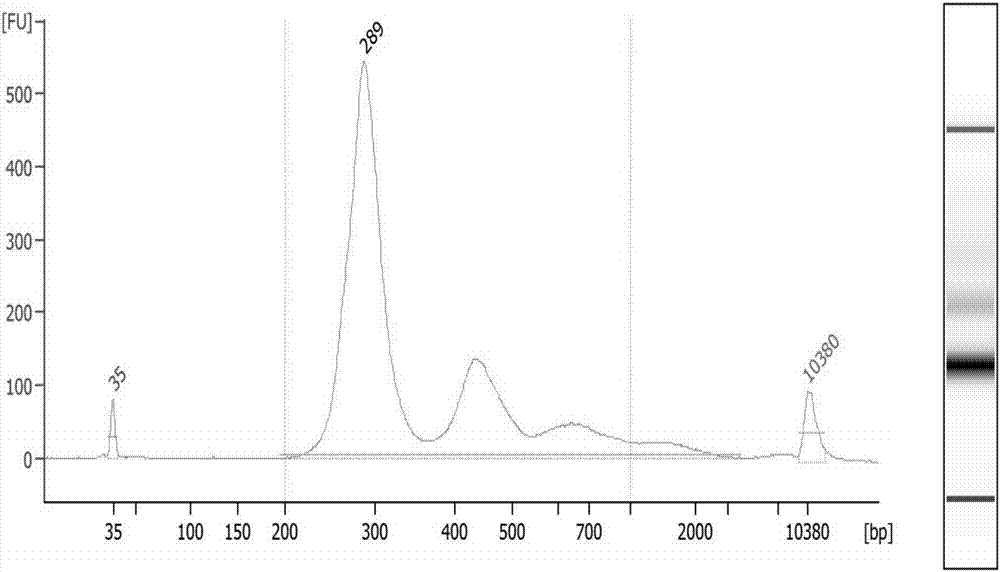
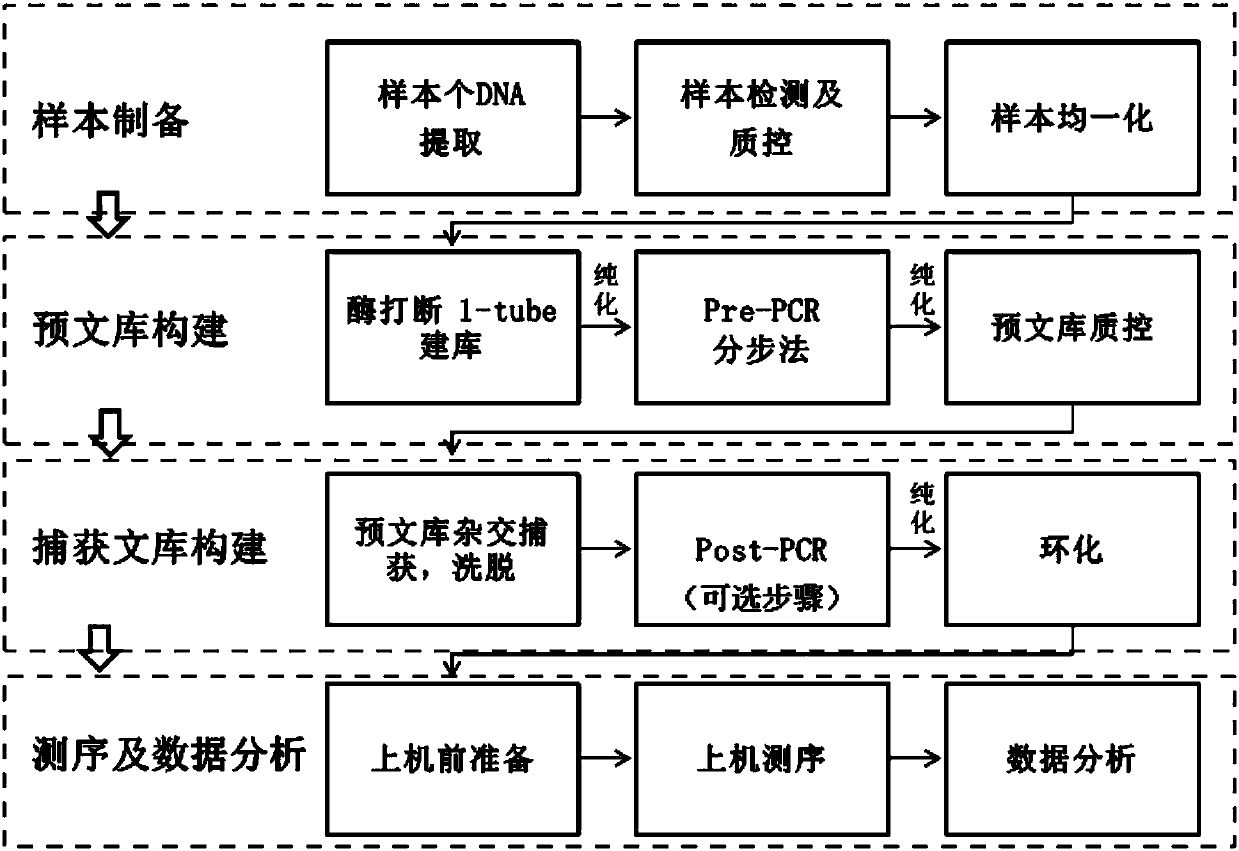
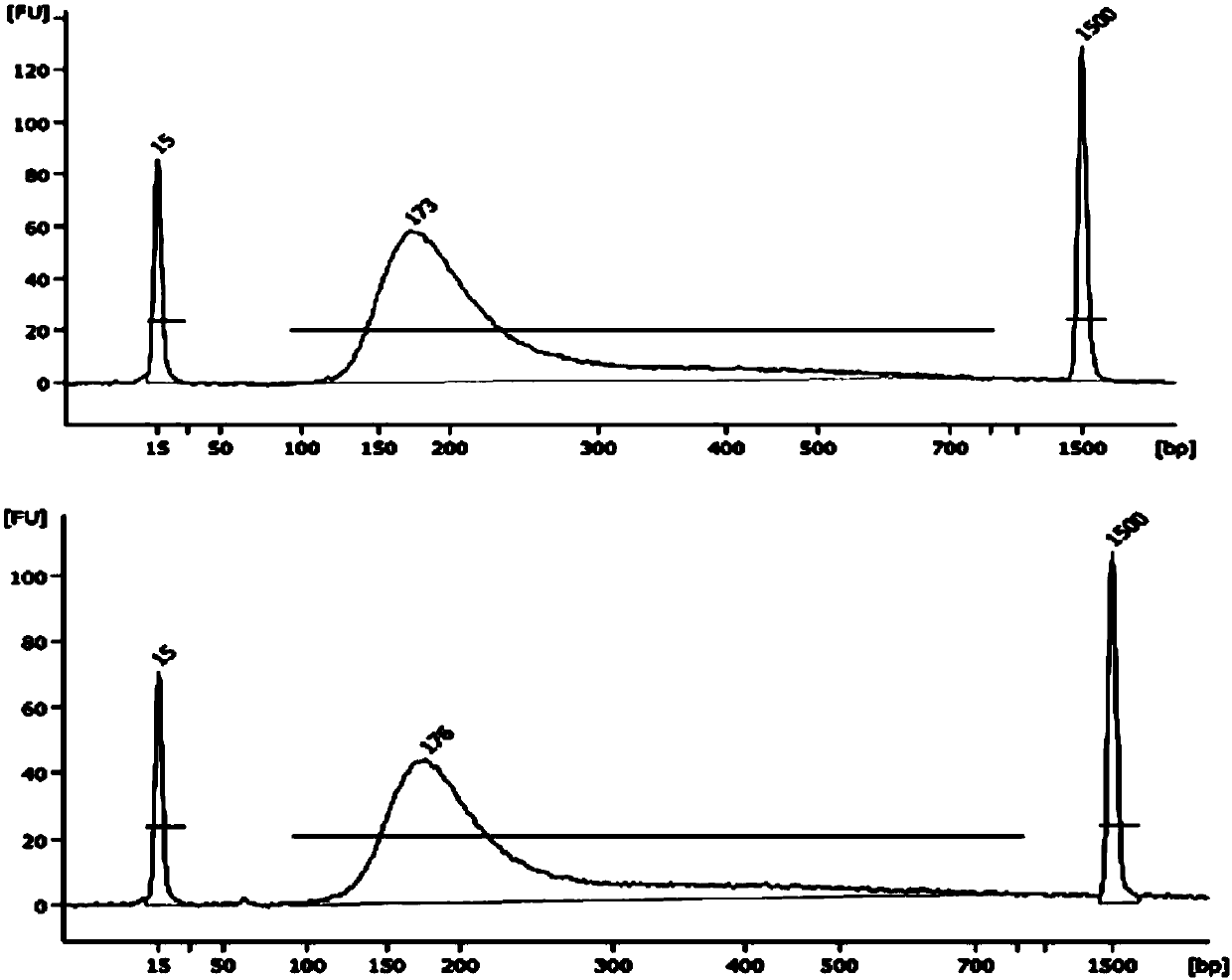
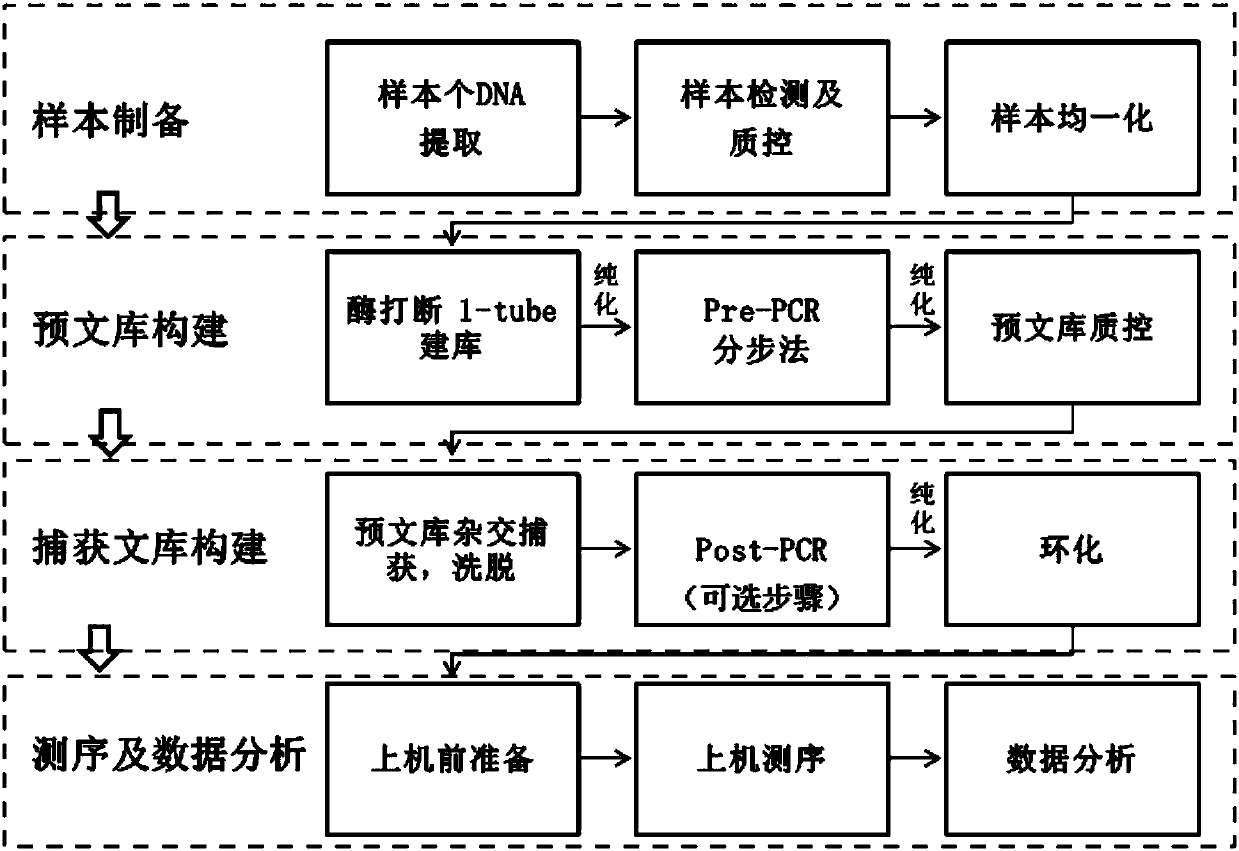
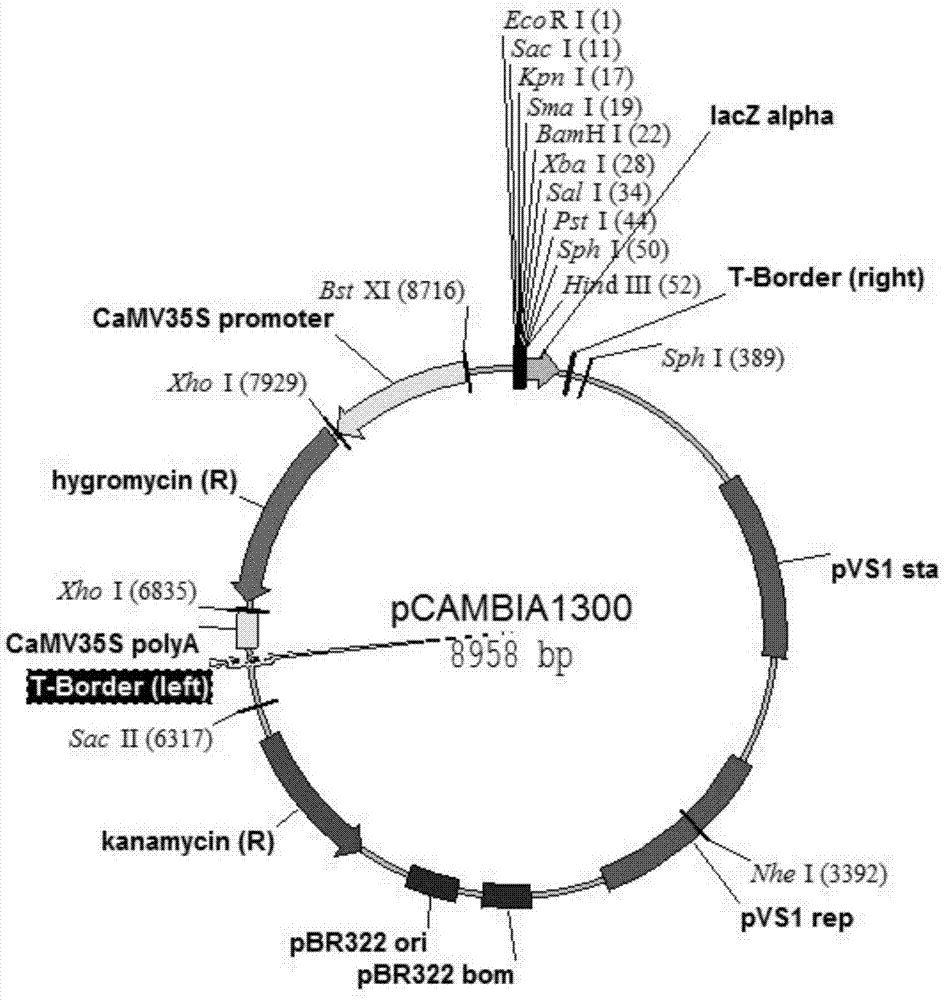
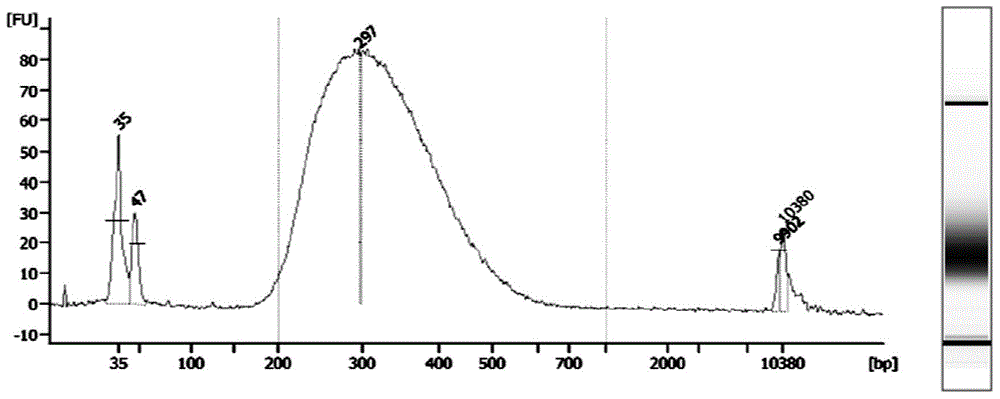
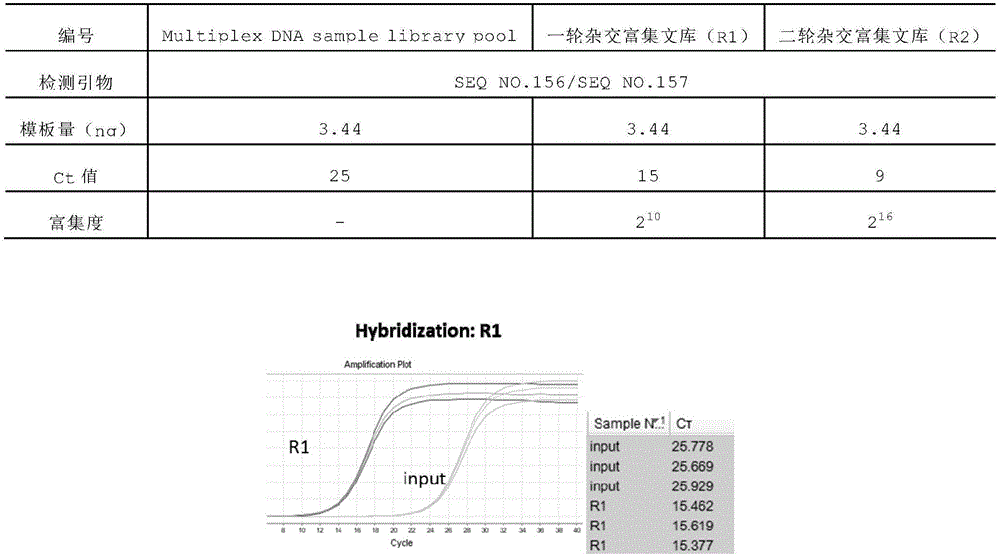
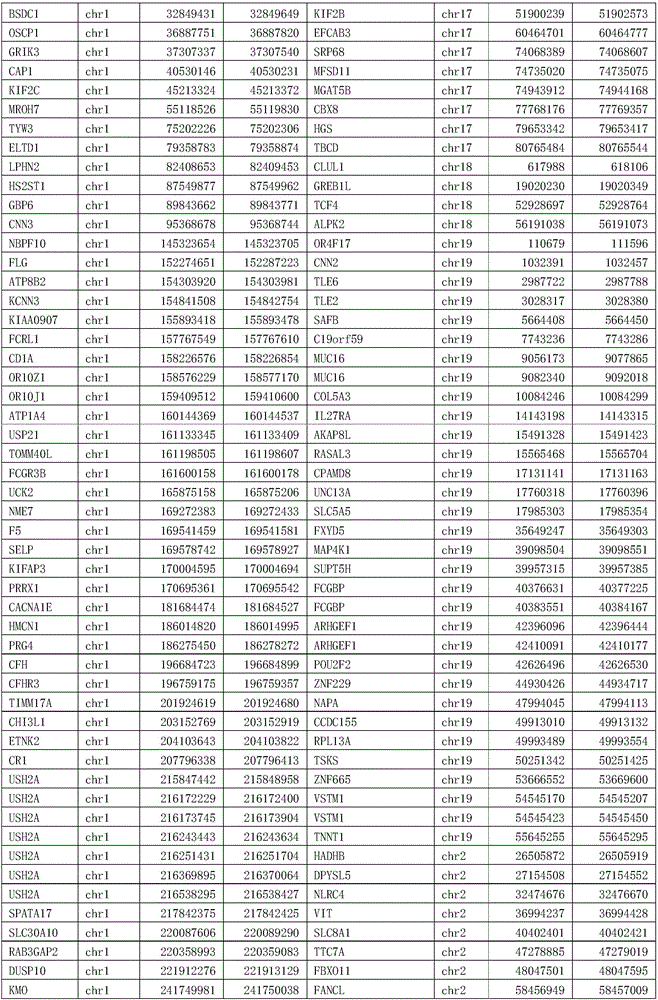
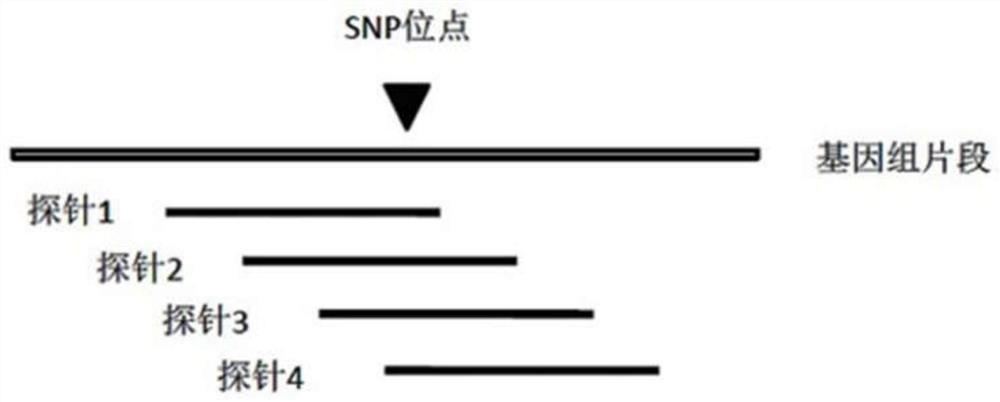
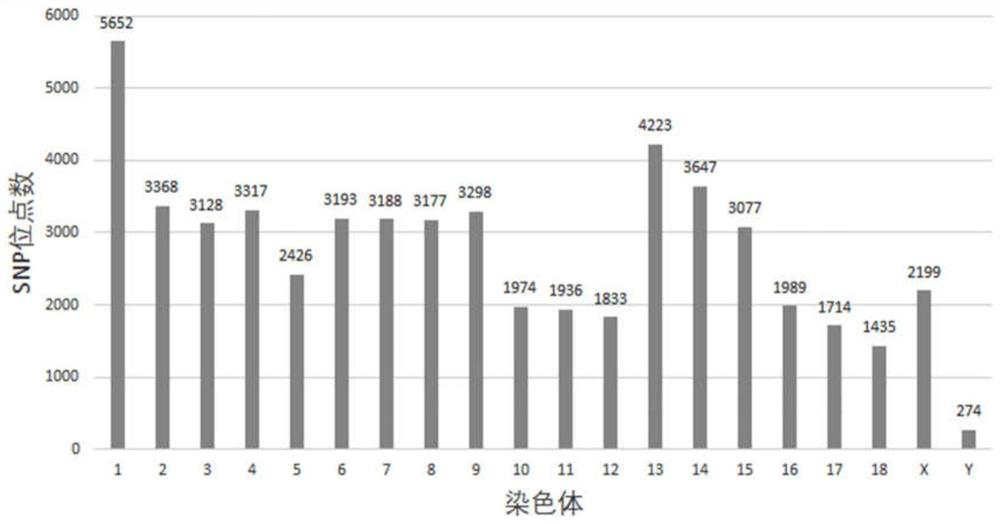
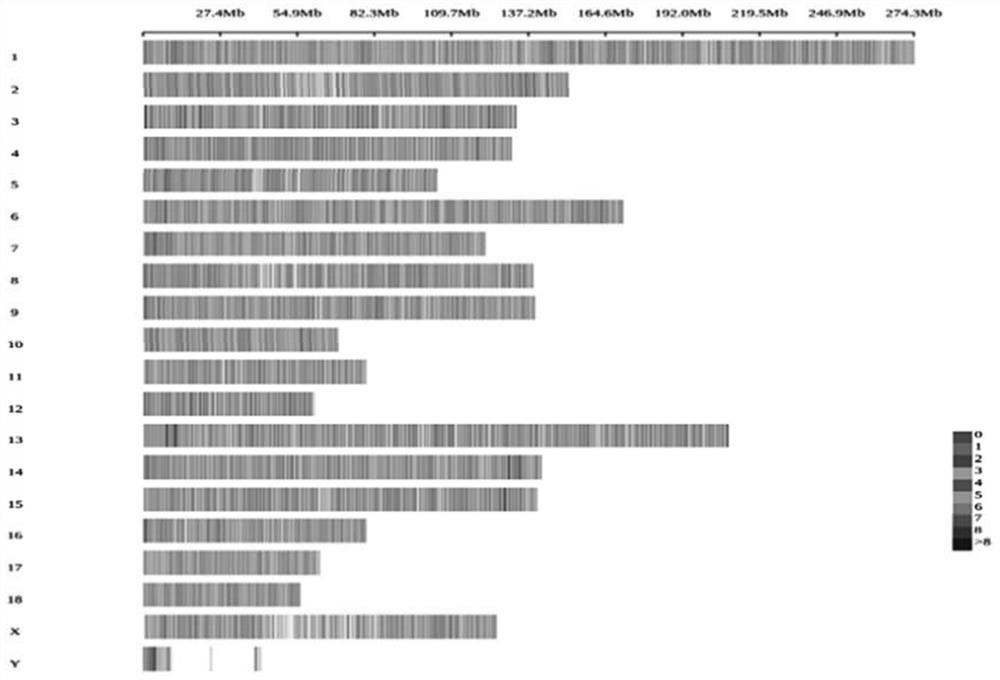
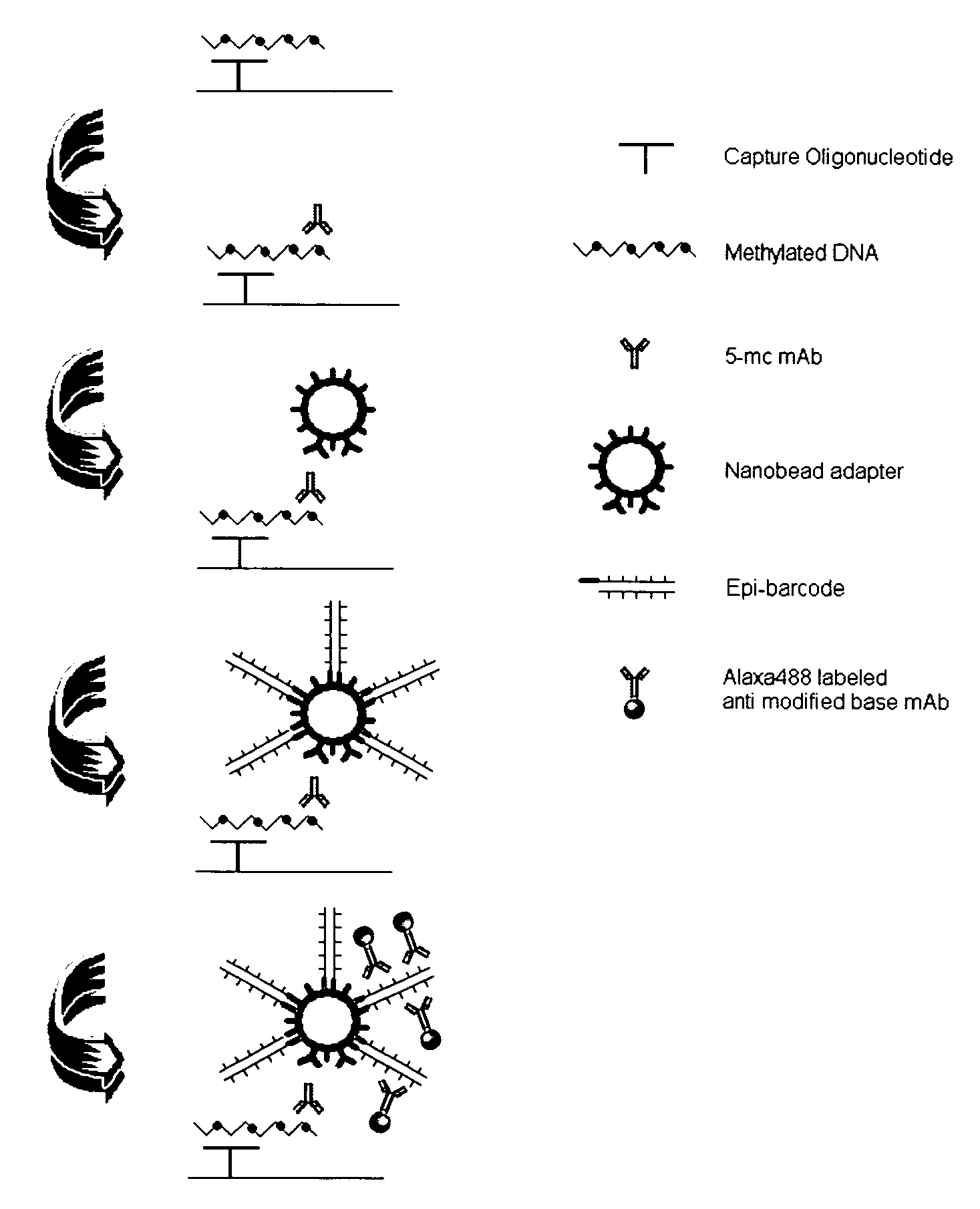
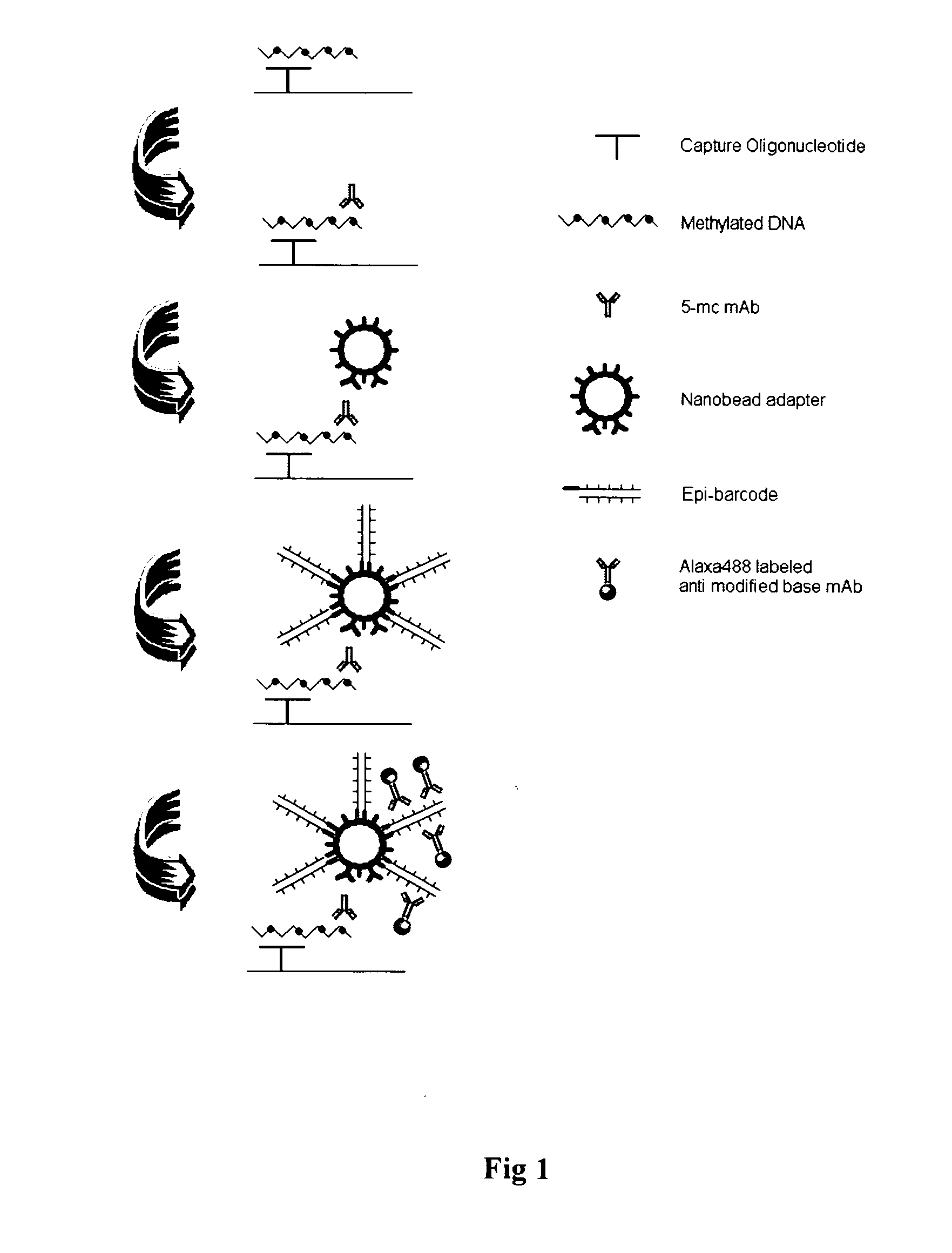
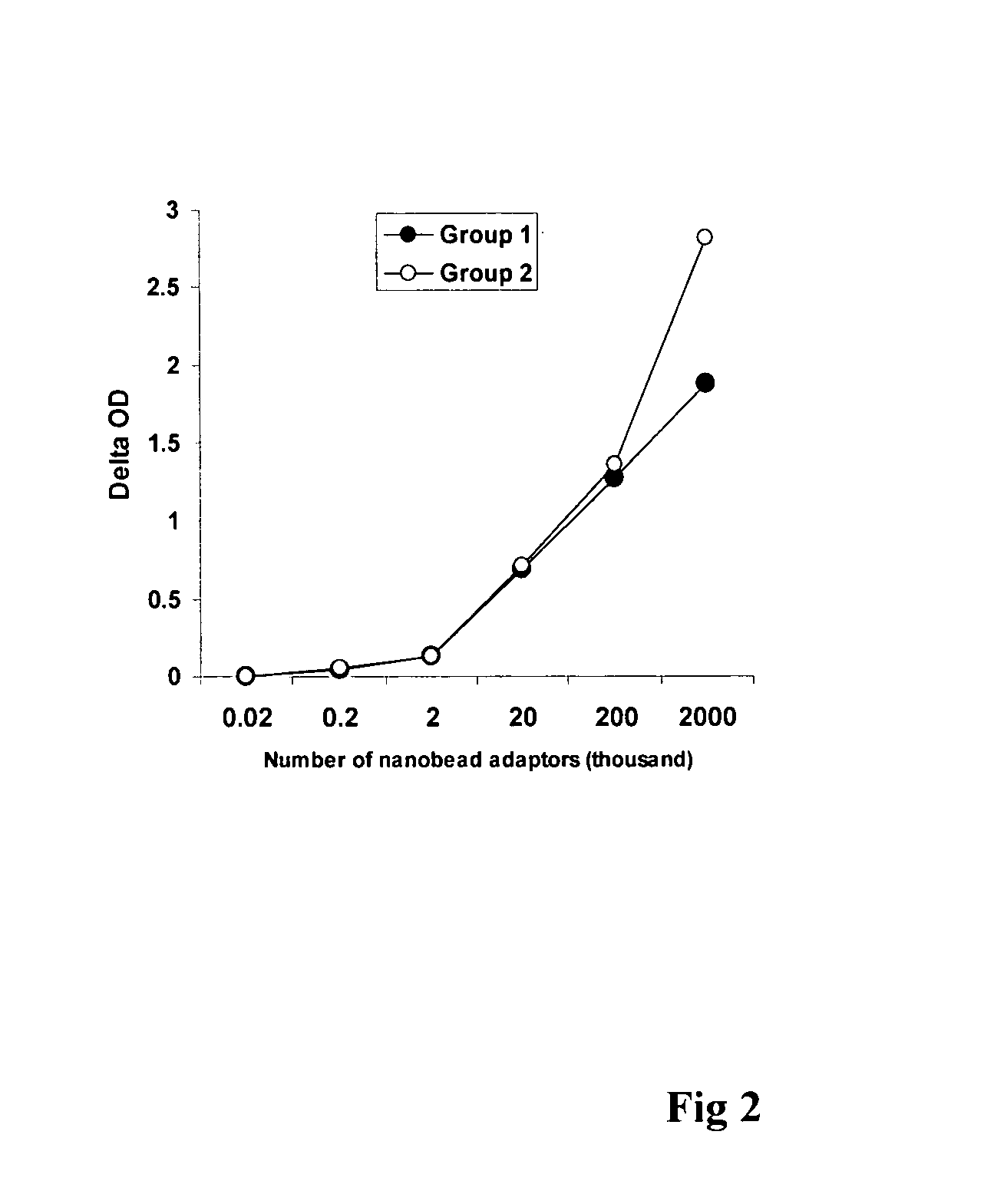
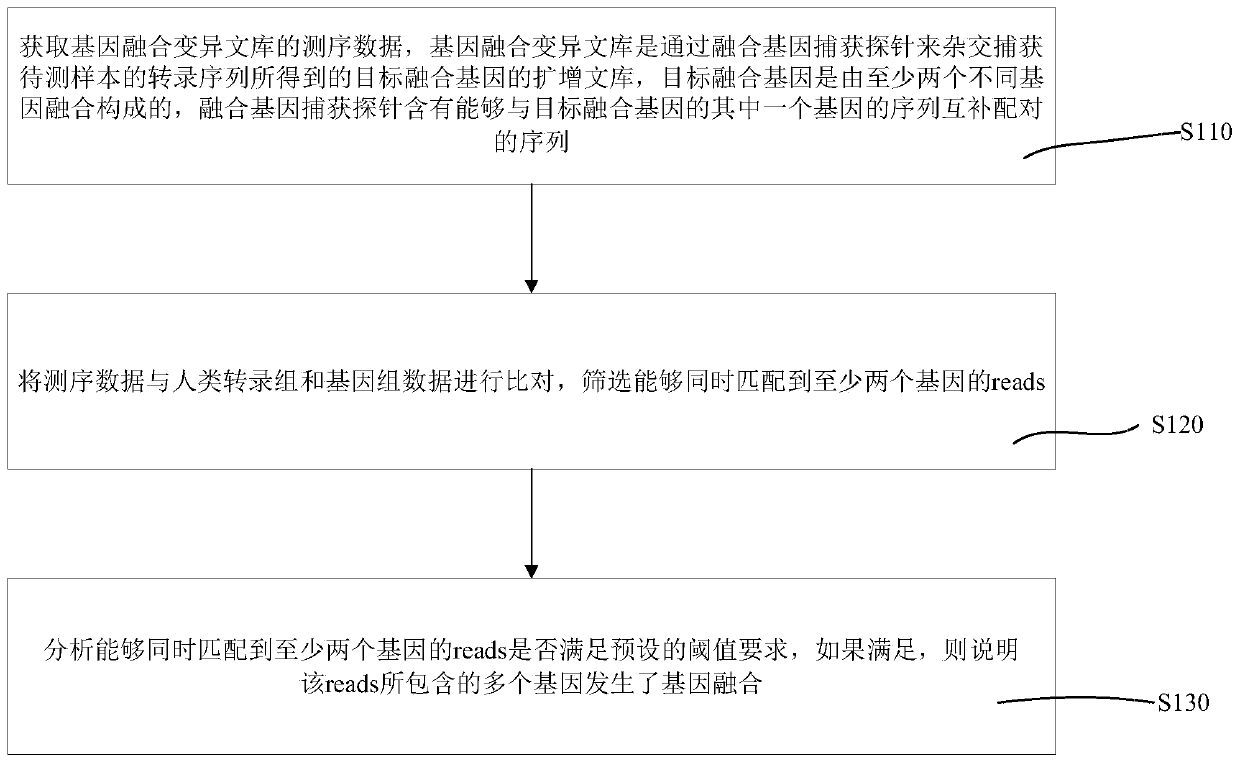
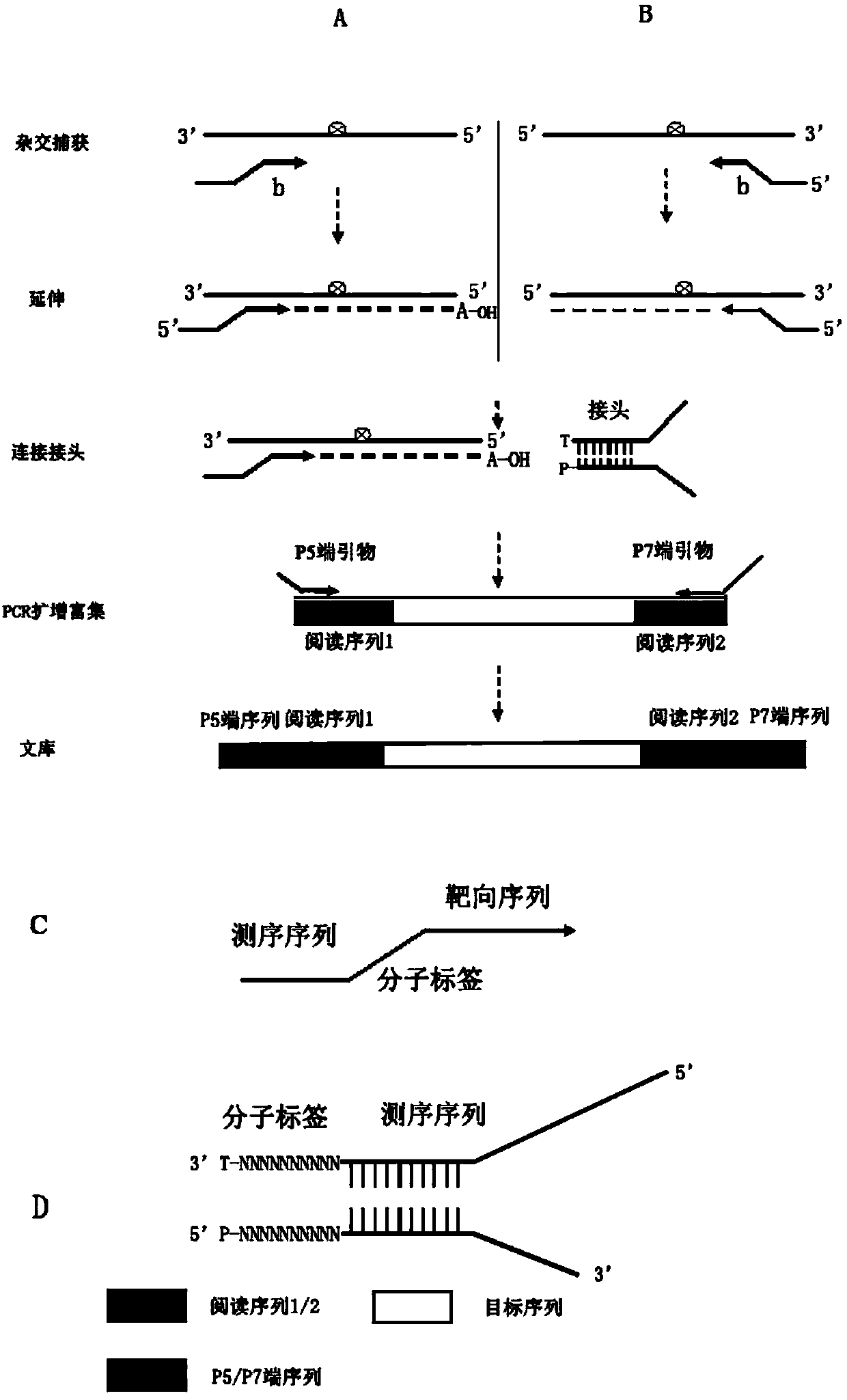
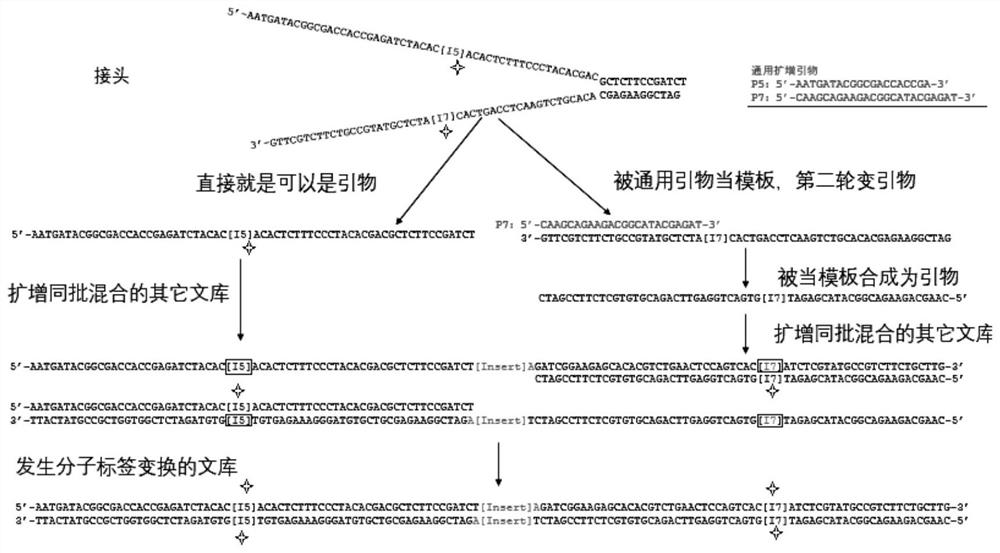
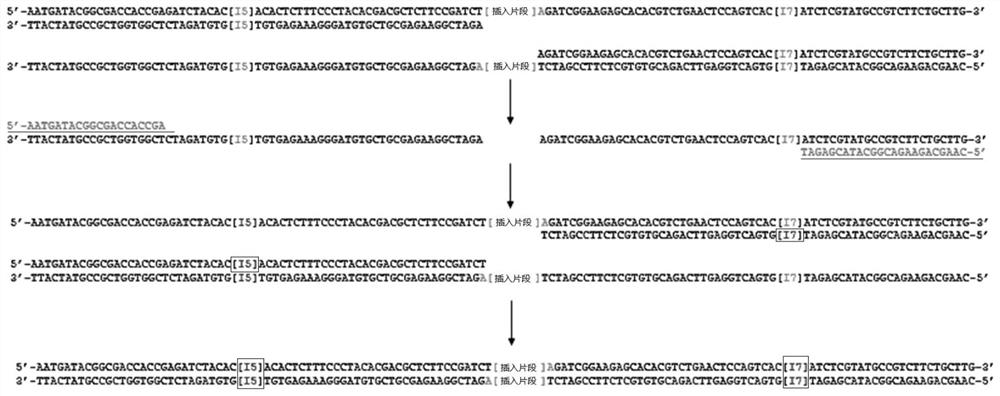
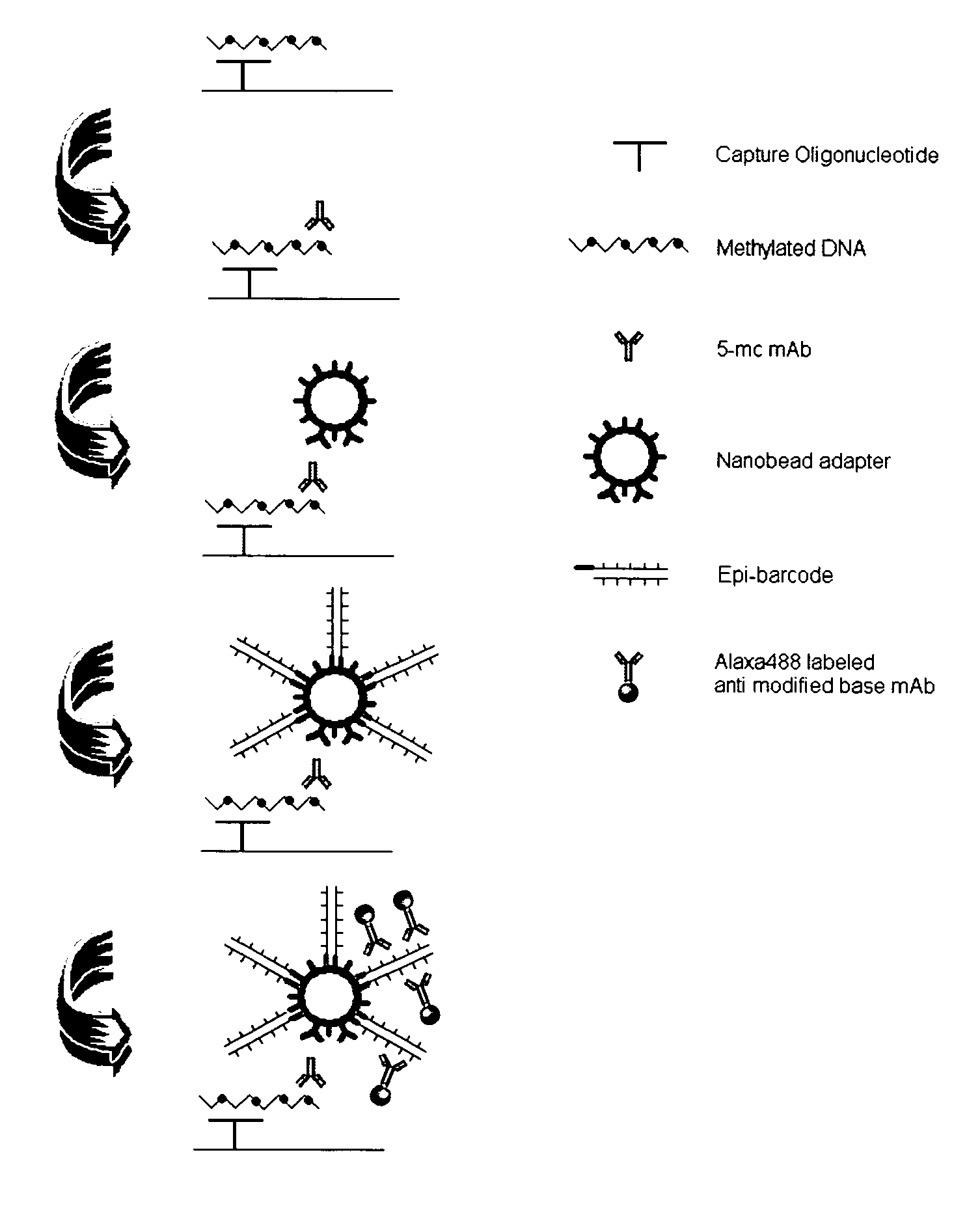
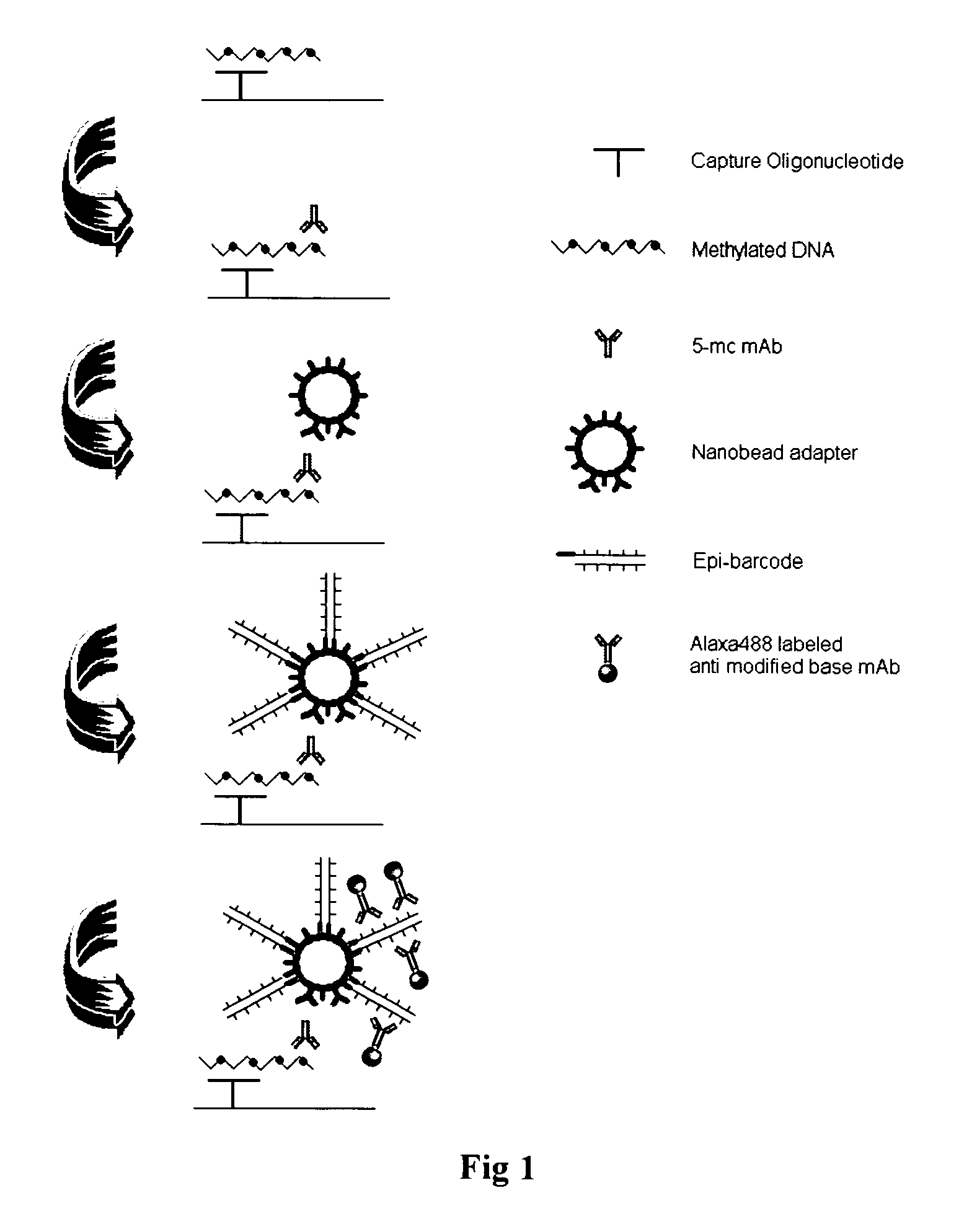
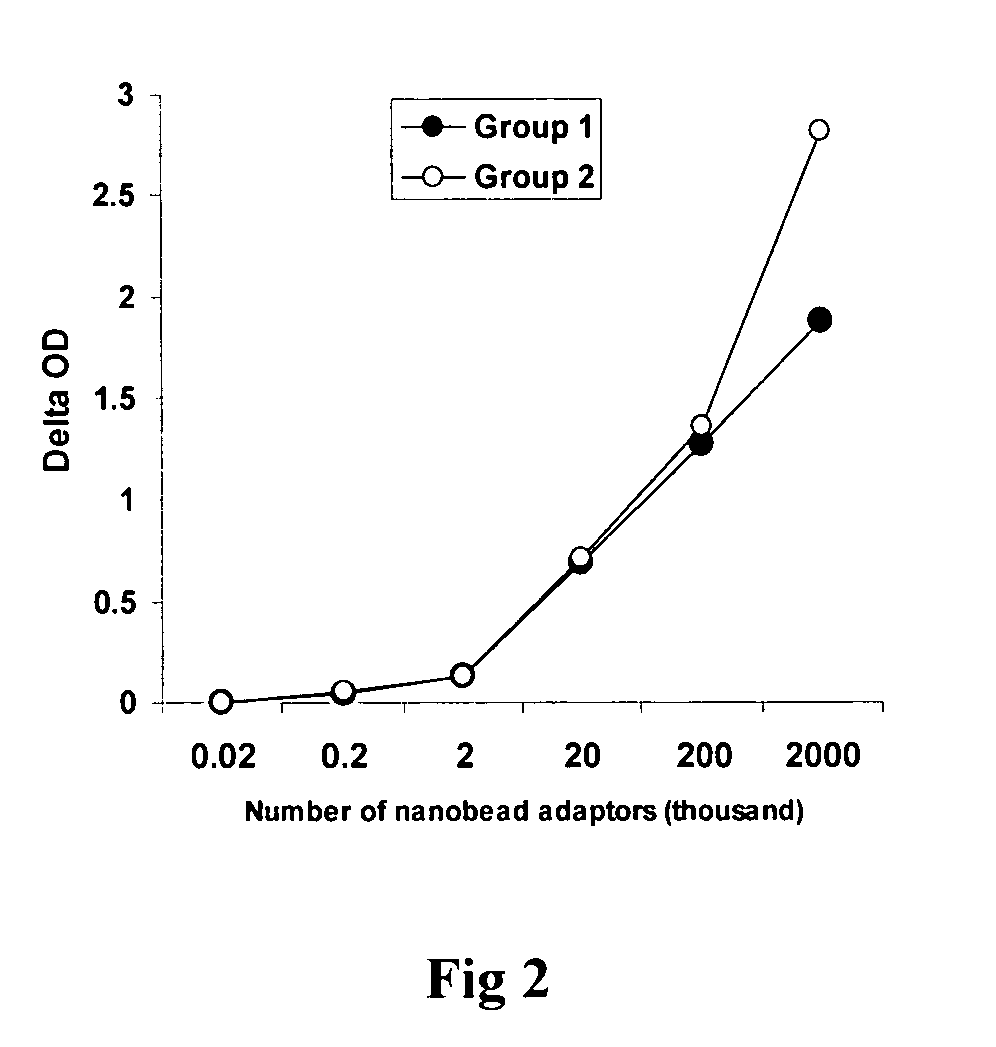
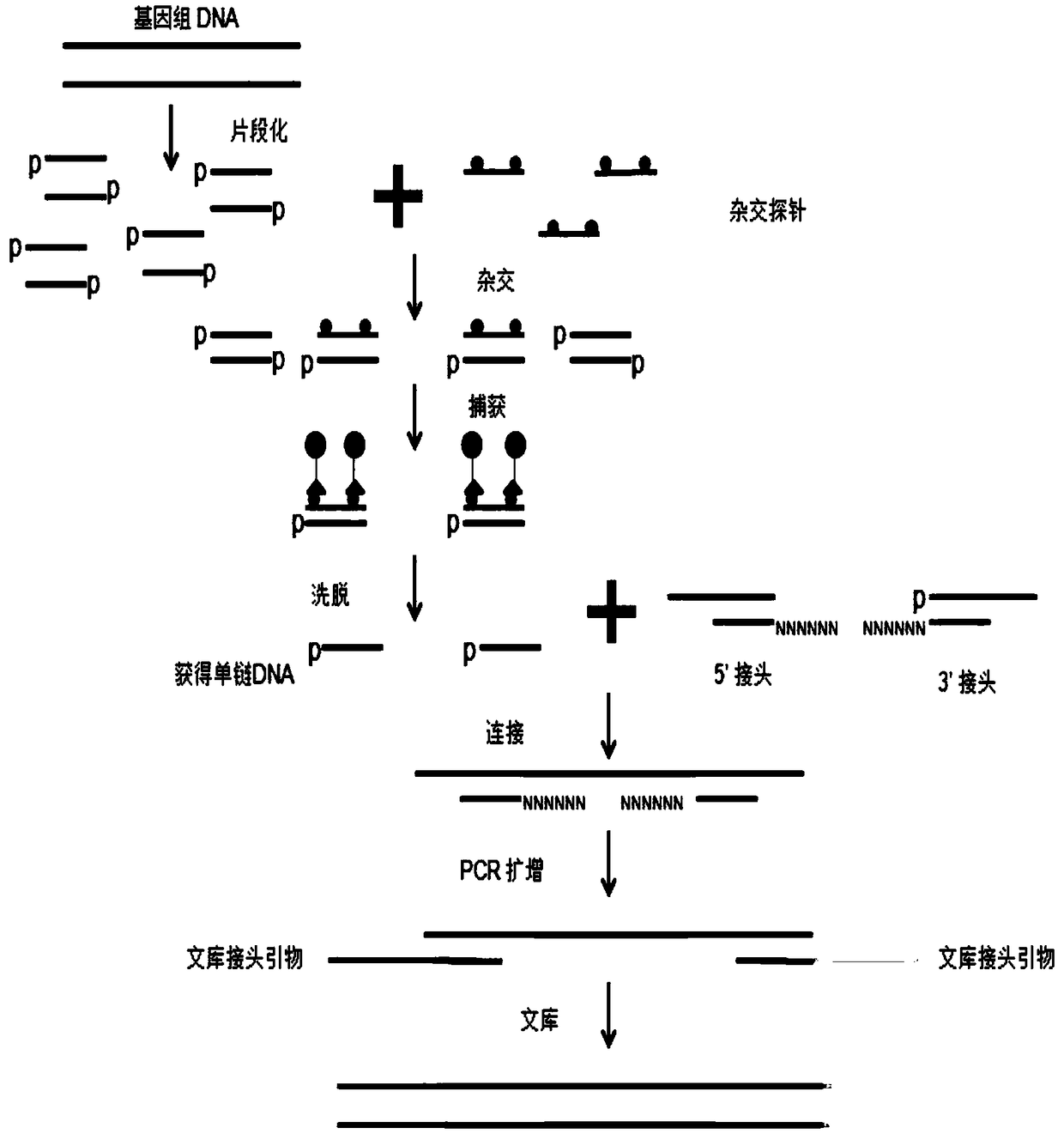
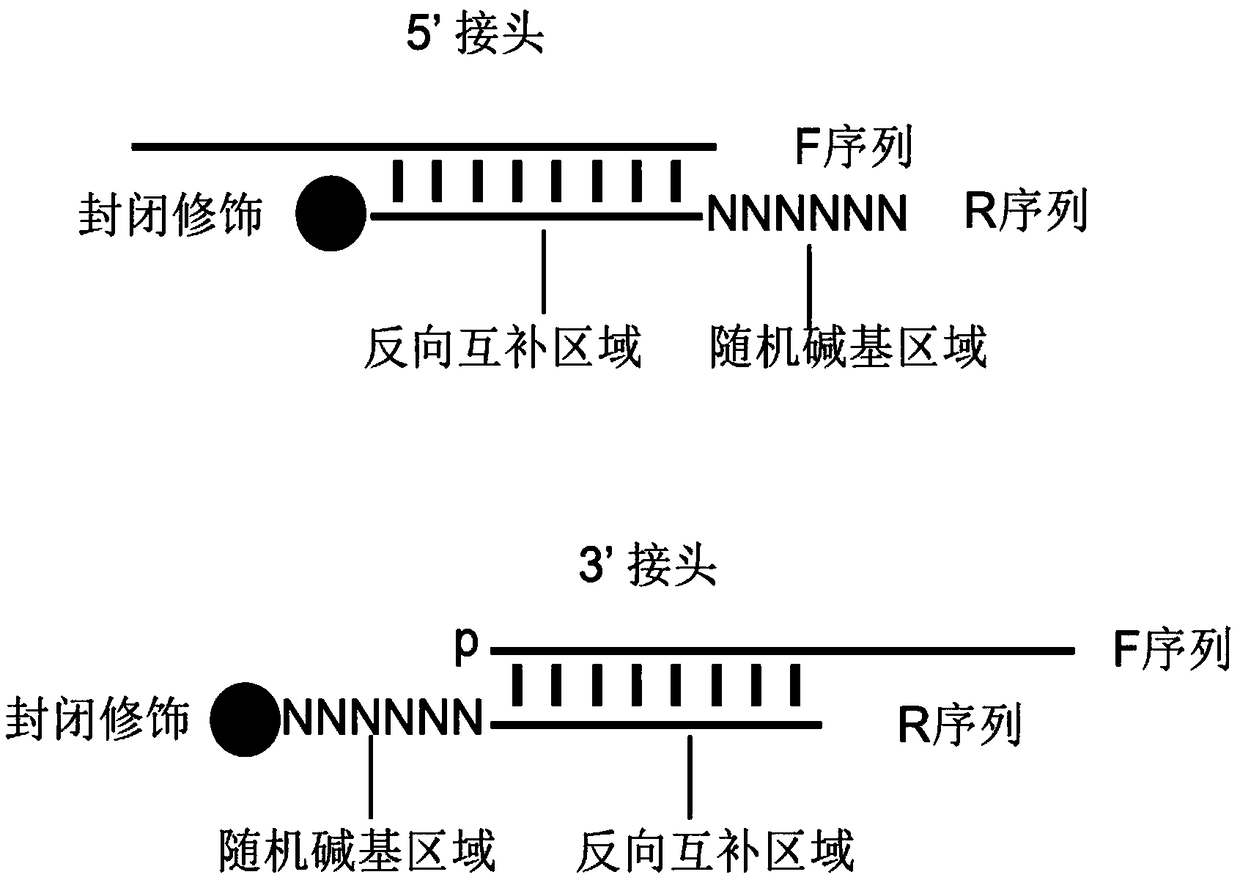
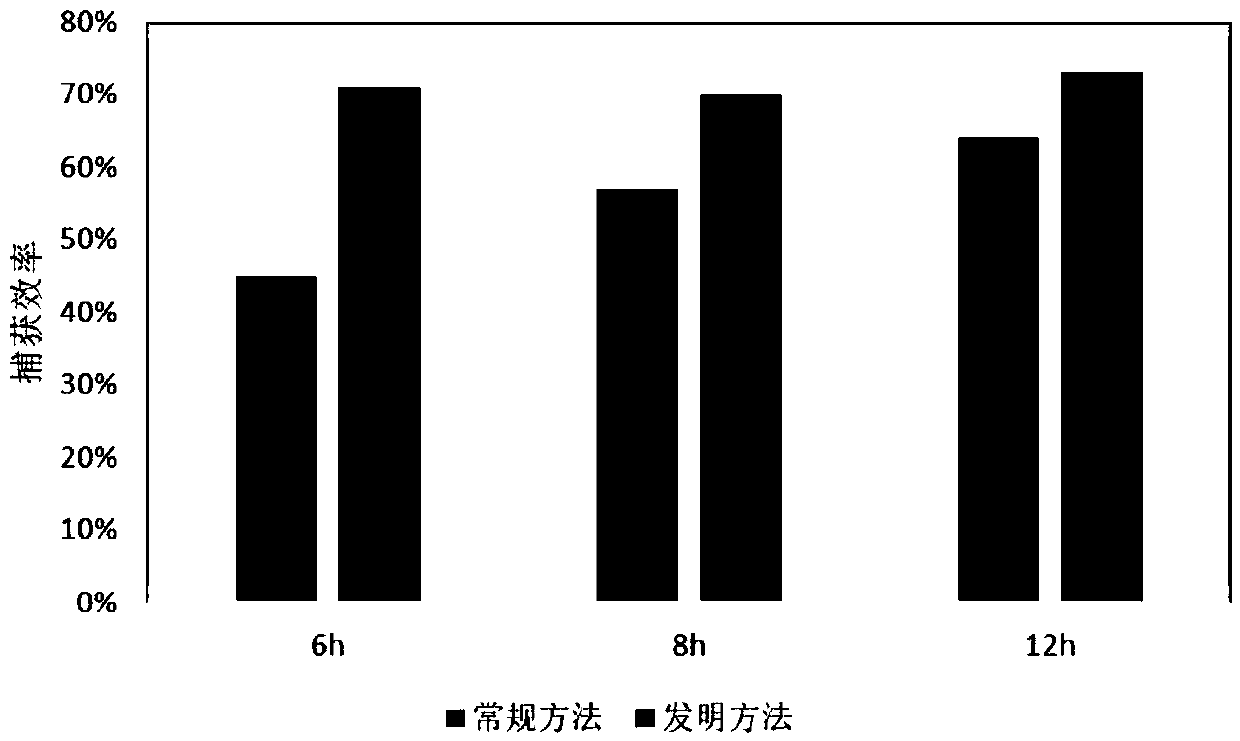
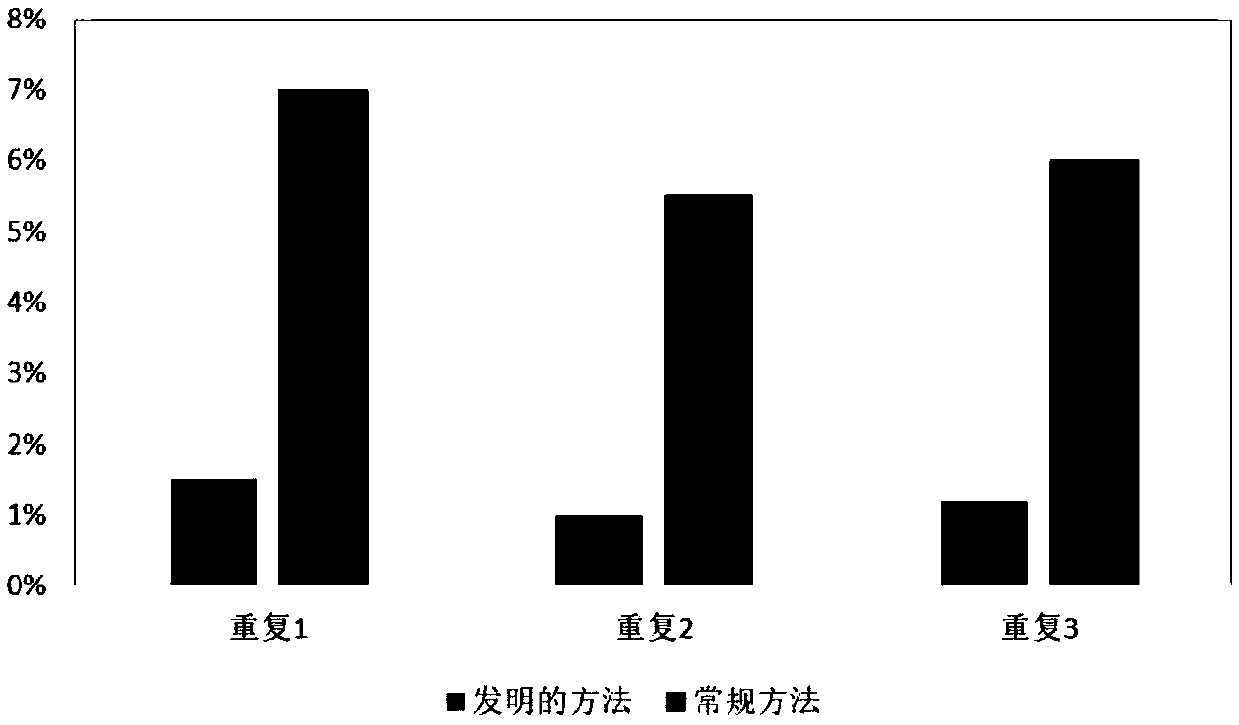
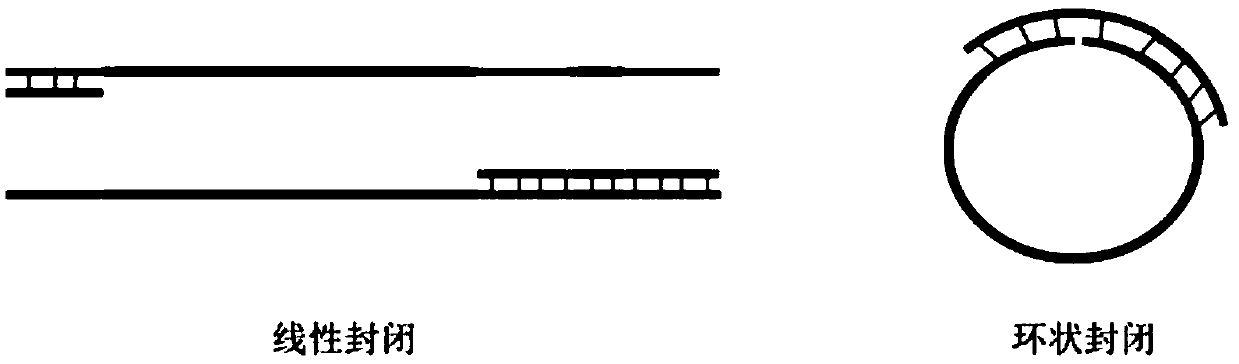
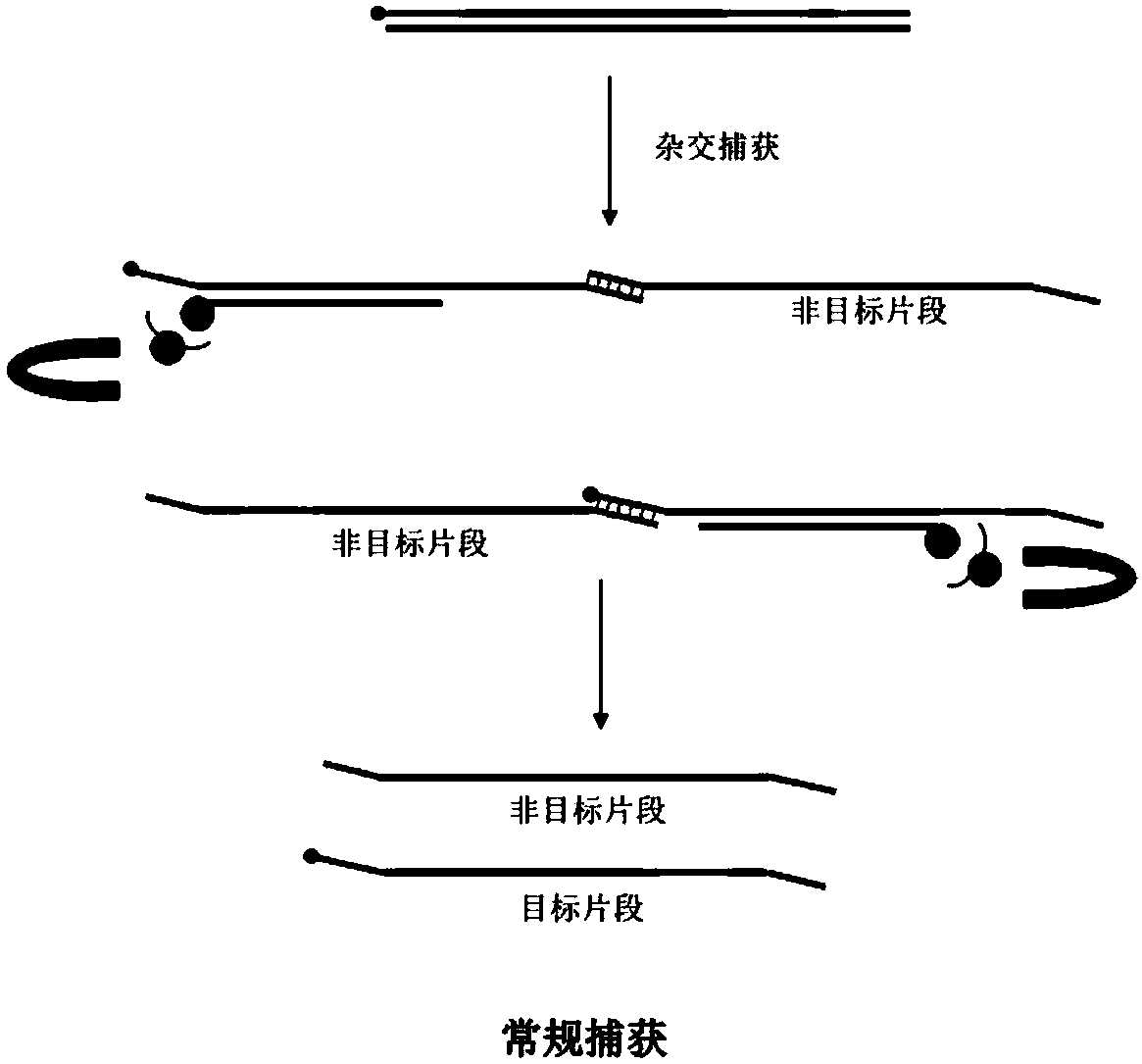
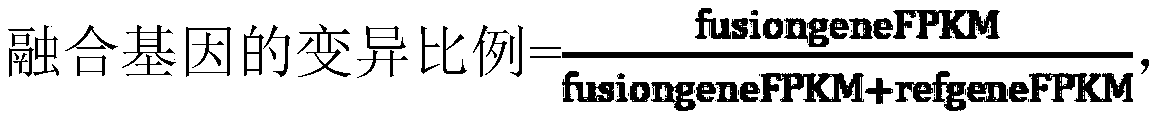Patents
Literature
124 results about "Hybridization capture" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
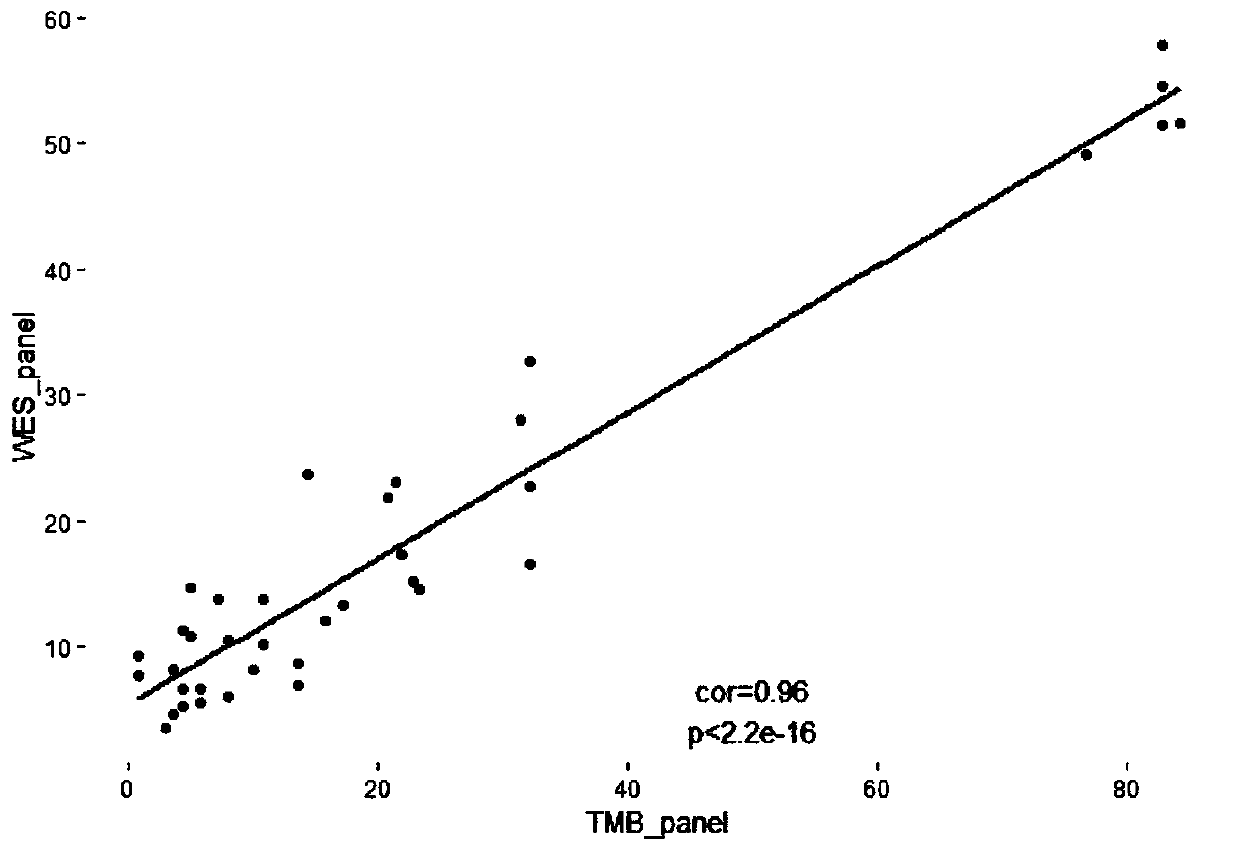
DNA capture via hybridization allows the efficient exploitation of current high-throughput sequencing for population genetic analyses using aDNA samples. Specifically, hybridization capture allows larger data sets to be generated for multiple target loci as well as for multiple samples in parallel.
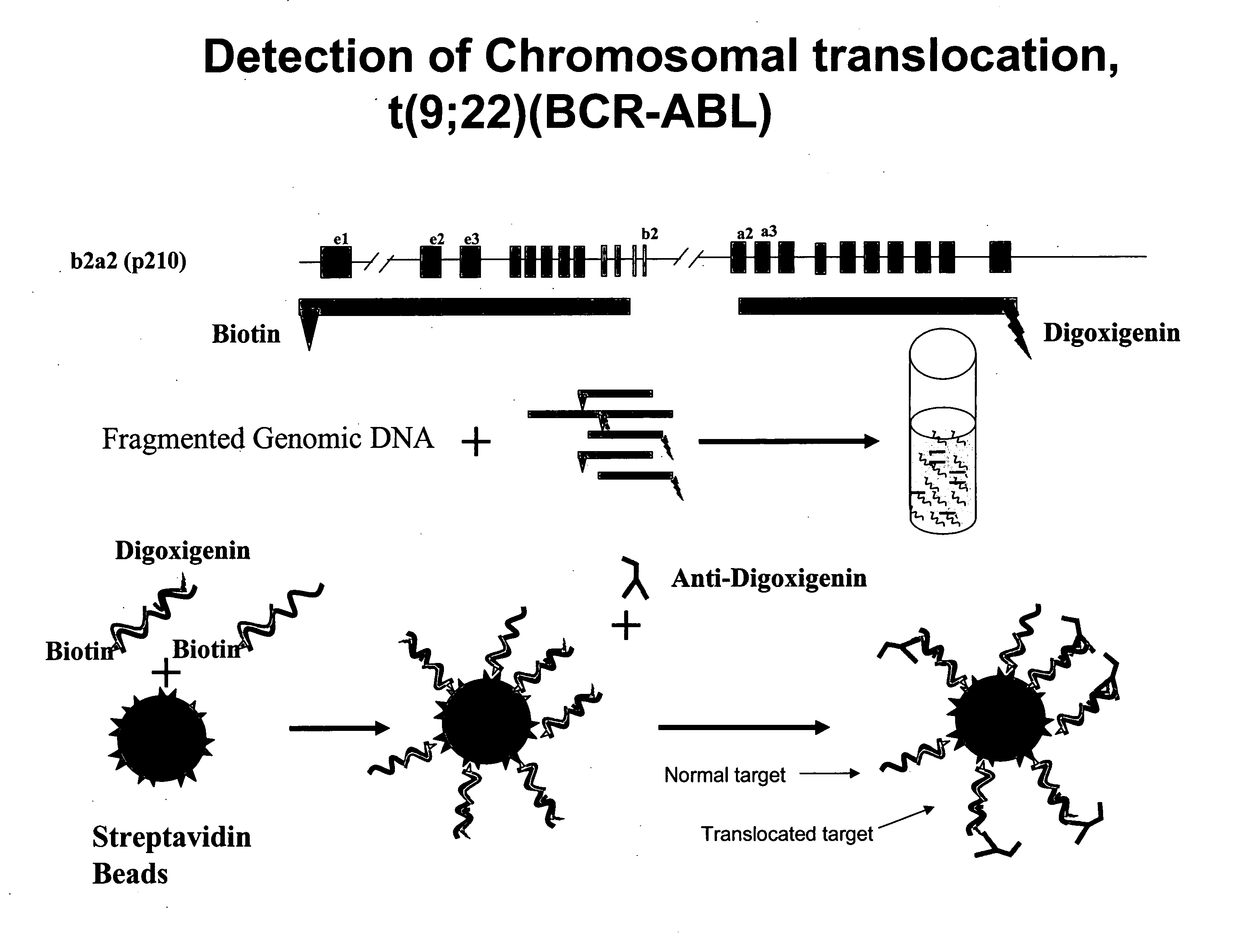
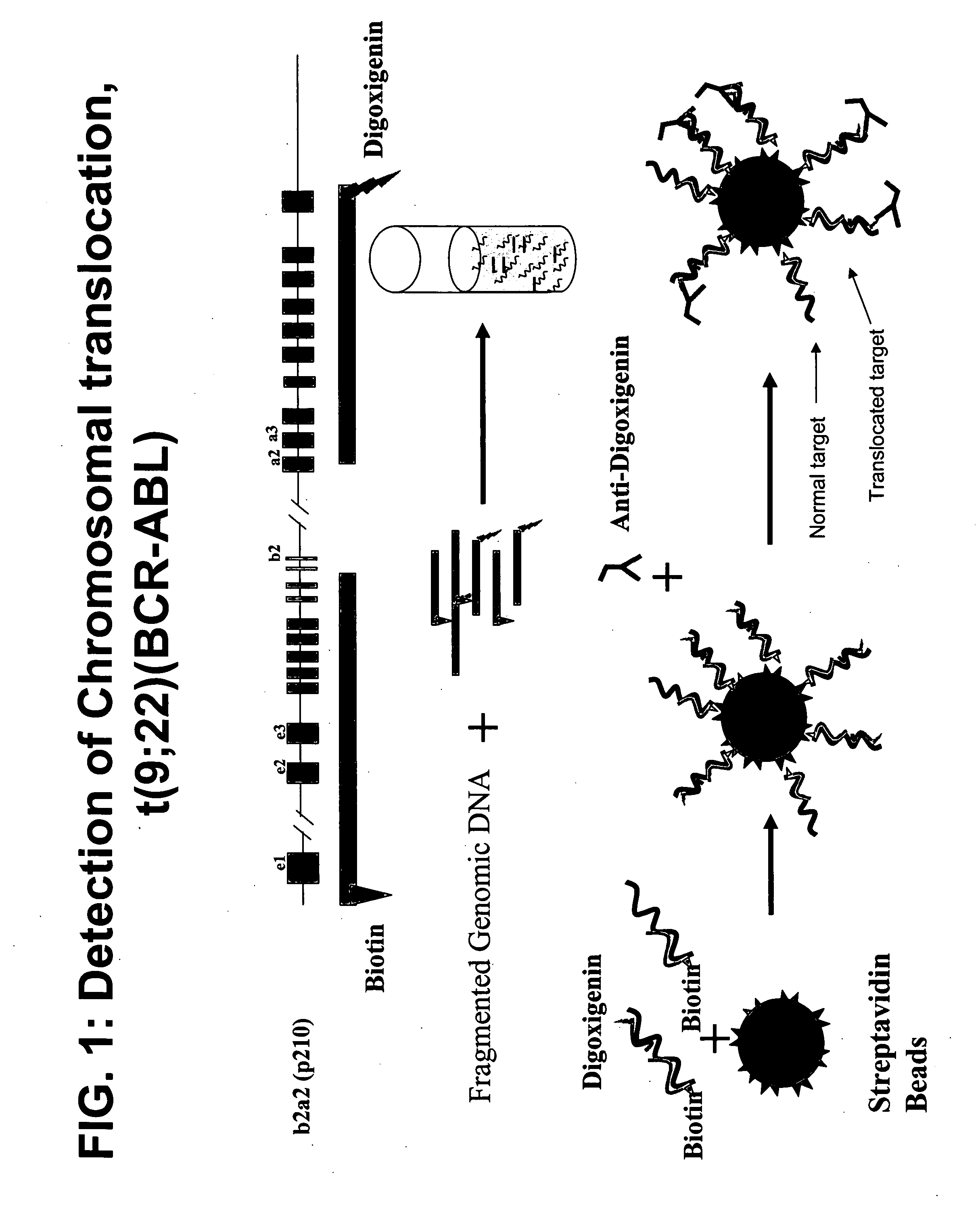
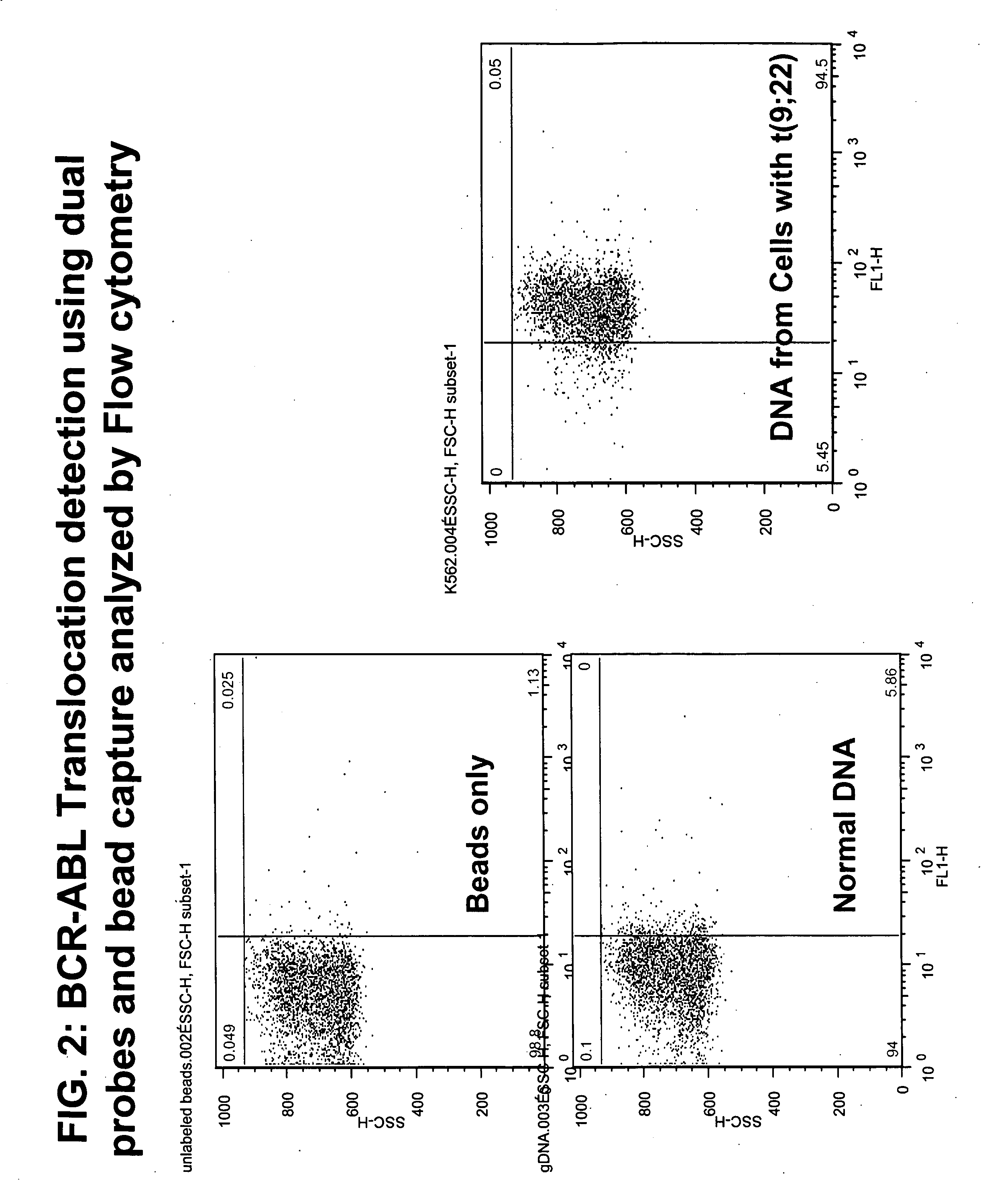
Non-in situ hybridization method for detecting chromosomal abnormalities
The present invention provides methods of detecting chromosomal or genetic abnormalities associated with various diseases or with predisposition to various diseases. In particular, the present invention provides advanced methods of performing DNA hybridization, capture, and detection on solid support. Invention methods are useful for the detection, diagnosis, predicting response to therapy, detecting minimal residual disease, prognosis, or monitoring of disease treatment or progression of particular disease conditions such as cell proliferative disorders
Owner:QUEST DIAGNOSTICS INVESTMENTS INC
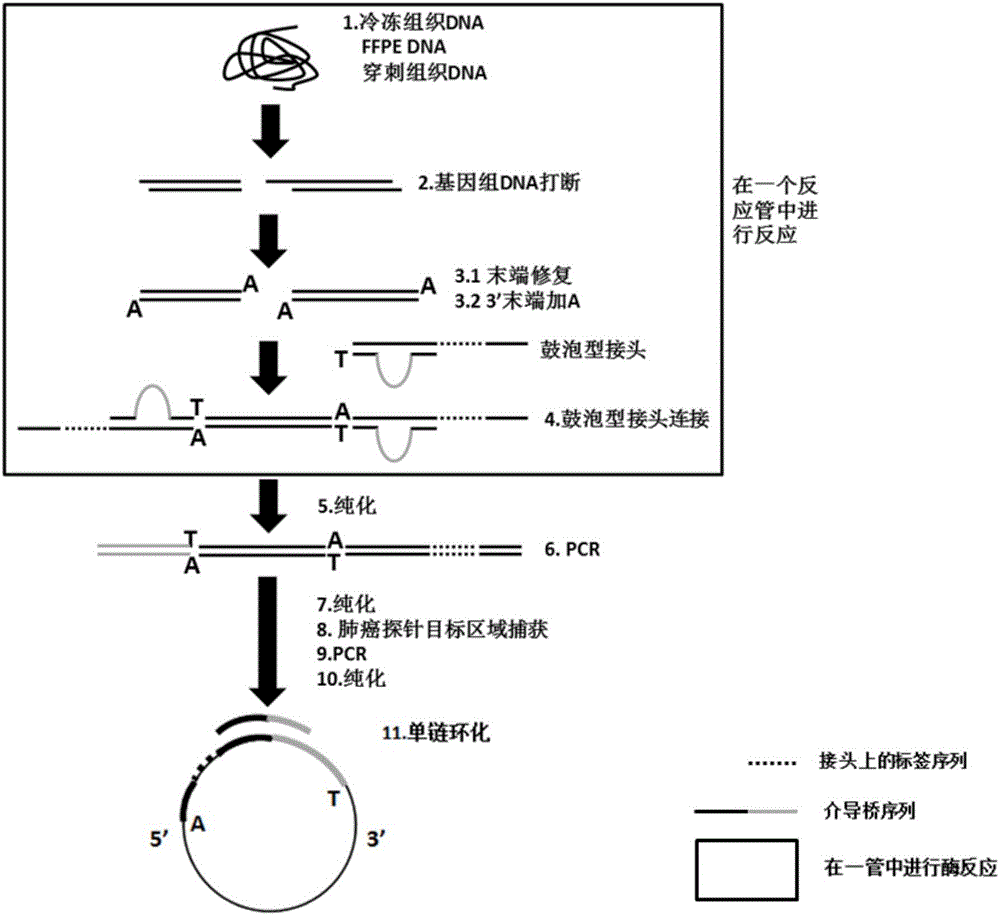
Building method of library for detecting non-small cell lung cancer gene mutation and kit
PendingCN106497920AImprove recycling efficiencyImprove utilization efficiencyMicrobiological testing/measurementLibrary creationGene targetingGene mutation
The invention discloses a building method of a library for detecting non-small cell lung cancer gene mutation and a kit. The method includes: using tubular reaction to complete genome DNA breaking and connector connection, performing hybrid capture on connection products after amplification and non-small cell lung cancer related gene target area probes, and performing BGISEQ-500 / 1000 platform sequencing and data analysis to obtain mutation conditions. The method has the advantages that the experiment flow is optimized greatly by the tubular reaction, operation complexity and time are reduced, and the requirements on clinical sample initial amount are lowered; multiple genes and multiple sites can be detected in one step, point mutation, insertion and deletion, structural variation and copy number variation are covered, the detecting result is accurate and overcomes the defect that a PCR capture method cannot detect the structural variation in one step, and the effectiveness of the high-throughput sequencing applied to the detection of the non-small cell lung cancer gene mutation; the method is wide in coverage, high in cost performance, capable of providing a reference basis for the diagnosing, treatment and drug use performed by doctors, and the method is suitable for being popularized and used in a large-scale manner.
Owner:BGI BIOTECH WUHAN CO LTD
Gene group and kit for diagnosing lung caner, and diagnosis method thereof
InactiveCN107475370AExcellent detection depthExcellent detection accuracyMicrobiological testing/measurementLibrary creationA-DNABlood plasma
The invention relates to the field of genetic engineering and biotechnologic detection, and concretely relates to a gene group and a kit for diagnosing lung cancer, and a diagnosis method thereof. The gene group for diagnosing the lung cancer includes ABCB1, AKT1, ALK, APC, ATIC and other 63 genes. The method using the gene group to diagnose the lung cancer comprises the following steps: (1) extracting free DNA and genome DNA from the plasma of a sample to be detected; and (2) breaking the genome DNA into fragments with the length of 150-250 bp, carrying out hybrid capture on the broken genome DNA and the free DNA to construct a DNA library, carrying out online sequencing, and analyzing the obtained sequencing result. Exon and partial intron regions of the 68 genes of the free DNA are enriched at one time by a probe capture technology to realize multi-gene and multi-target parallel deep high-throughput sequencing with high accuracy, optimize the detection flow and improve the detection precision, so liquid biopsy, low-frequency detection and tumor diagnosis become possible.
Owner:天津脉络医学检验有限公司
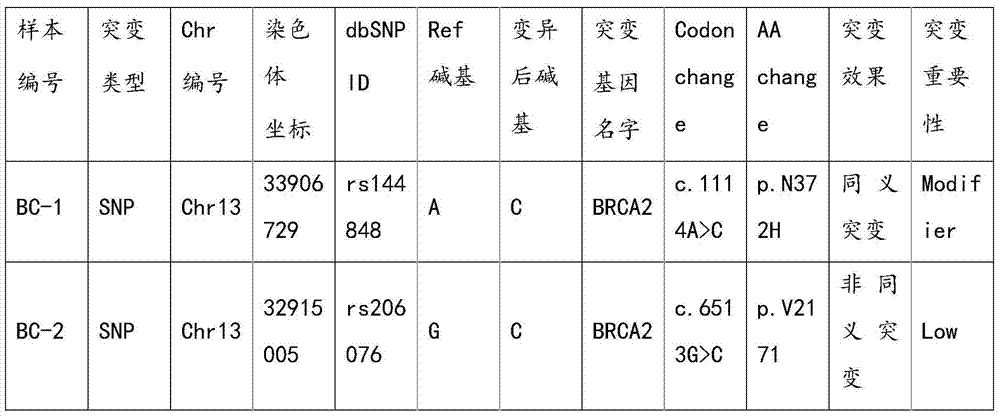
Method and reagent kit for constructing BRCA 1/2 (breast cancer susceptible genes 1/2) detection libraries
InactiveCN106987905AReduce purification operationsReduce workloadMicrobiological testing/measurementLibrary creationSingle strandDNA polymerase
The invention discloses a method and a reagent kit for constructing BRCA 1 / 2 (breast cancer susceptible genes 1 / 2) detection libraries. The method includes carrying out fragmentation and tail-end repair on homogenized DNA (deoxyribonucleic acid) samples in DNA fragmentation and tail-end repair systems under the effects of interruption enzymes and tail-end repair enzymes; linking products obtained at the previous step with linker sequences under the effects of DNA ligase; utilizing linker linking products obtained at the previous step as templates and carrying out PCR (polymerase chain reaction) amplification by the aid of primers of linker sequences under the effects of DNA polymerases; hybridizing probe sequences with BRCA 1 / 2 and products obtained at the previous step to trap DNA fragments with the BRCA 1 / 2 and carrying out elution to obtain hybridization trapped products; carrying out cyclization on single strands of the hybridization trapped products under the effects of ligase to obtain the BRCA 1 / 2 detection libraries. The method and the reagent kit have the advantages that steps for constructing the libraries are simple, convenient and speedy, the cost can be effectively reduced, workload can be relieved, and variation types are comprehensive and accurate and are high in flux.
Owner:BGI GENOMICS CO LTD +3
Method for efficiently and rapidly separating T-DNA insertion site flanking sequence, and uses thereof
InactiveCN107190003AQuick analysisEasy to analyzeMicrobiological testing/measurementDNA preparationMagnetic beadSeparation technology
The invention provides a method for efficiently and rapidly separating a T-DNA insertion site flanking sequence. The method comprises: extracting the genomic DNA of an Agrobacterium-mediated transgenic plant; fragmenting the genomic DNA, carrying out DNA repairing, adding A, linking, carrying out fragment selection by using magnetic beads, carrying out library construction, carrying out hybridization capture on the constructed library and a T-DNA boundary sequence, and carrying out PCR enrichment; and carrying out high-throughput sequencing on the library, and carrying out bioinformatics analysis on the sequenced data so as to obtain the T-DNA insertion site. According to the present invention, the capture is performed with the target DNA, and the low-throughput sequencing is specially performed on the T-DNA boundary sequence; the disadvantages of low throughput, low efficiency and low success rate of the existing flanking sequence separation technology are overcome with the method of the present invention; and with the method of the present invention, the DNA capture and the second-generation sequencing technology are combined, the T-DNA boundary sequence is specially sequenced and the simple analysis is performed to obtain the T-DNA insertion site flanking sequence, and the method has characteristics of high efficiency, economy, and simple analysis.
Owner:武汉天问生物科技有限公司
Nucleic acid detection method combining RNA amplification with hybrid capture method
InactiveCN103525942AEnable high-throughput detectionNot easy to inactivateMicrobiological testing/measurementBiotin-streptavidin complexStreptolydigin
The invention relates to a nucleic acid detection method combining RNA amplification with a hybrid capture method. The nucleic acid detection method includes the following steps of extracting nucleic acid from a sample or splitting the sample to release the nucleic acid to be detected, conducting transcription and amplification on the nucleic acid to be detected to obtain an RNA product, adding the obtained RNA product together with a DNA probe marked with biotins to a solid phase so that hybridization can be conducted, forming a DNA / RNA heterozygote through hybridization, coating the solid phase with specific antibodies for recognizing the heterozygote or avidin or streptavidin, capturing the heterozygote through the solid phase, and adding avidin or streptavidin or antibodies for recognizing the DNA / RNA heterozygote to the solid phase to achieve obtaining of a nucleic acid signal, wherein the avidin or the streptavidin or the antibodies for recognizing the DNA / RNA heterozygote are marked with enzymes with the secondary amplification capacity. According to the nucleic acid detection method, the inherent pollution problem of the PCR is avoided, RNA molecules can be directly detected, inverse transcription and pre-dissociation of RNA do not need to be conducted, and the operation steps are greatly simplified.
Owner:武汉中帜生物科技股份有限公司
Fluorescence detection method for DNA and kit thereof
ActiveCN101240329AHigh selectivityHigh sensitivityMicrobiological testing/measurementFluorescence/phosphorescenceSolubilityDisease
Owner:SHANGHAI INST OF APPLIED PHYSICS - CHINESE ACAD OF SCI
Method for fragmenting trace DNA sample and method for establishing DNA library by utilizing trace DNA sample
ActiveCN104946629AAvoid damagePlay a protective effectMicrobiological testing/measurementLibrary creationCDNA libraryFragment processing
The invention discloses a method for fragmenting a trace DNA sample and a method for establishing a DNA library by utilizing the trace DNA sample. The method for fragmenting the trace DNA sample comprises the steps that 1, Carrier RNA is added into the trace DNA sample to obtain a sample to be fragmented; 2, fragmenting processing is conducted on the sample to be fragmented. An inventor discovers that after the exogenetic Carrier RNA is added into the target DNA, that is to say, the target DNA is mixed with the Carrier RNA and then fragmenting is conducted on the mixture, the protective action to the target DNA can be achieved, the destructive action of a fragmenting instrument to the trace DNA is reduced to the maximum extent, and full gene establishing library sequencing and even hybrid capture sequencing can be conducted on DNA of a trace FFPE sample.
Owner:天津诺禾医学检验所有限公司
Hybrid capture kit and method for detecting mutation of breast cancer susceptibility genes BRCA1 and BRCA2
ActiveCN106676169AImplement mutation detectionLow costMicrobiological testing/measurementBreast cancer susceptibility genesMutation detection
The invention discloses a hybrid capture kit for detecting mutation of breast cancer susceptibility genes BRCA1 and BRCA2. The hybrid capture kit comprises a hybridization reagent, a PCR amplification reagent and an enrichment degree detection reagent, wherein the hybridization reagent comprises probe mixtures from SEQ NO.1 to SEQ NO. 173, and the mole ratio is SEQ NO.1: SEQ NO. (2-172): SEQ NO. 173=1: 1: 1. During detection, the use amount of each of the probe mixtures with the concentration of 2nM is 1 [mu]l. The kit disclosed by the invention can be used for high-sensitivity, high-flux, low-cost and easy-operation full-explicit mutation detection for the genes BRCA1 and BRCA2, and can assist a clinical doctor in screening out individuals with the breast cancer susceptibility risk to fulfill the aim of preventing occurrence of breast cancers.
Owner:SHANGHAI PERSONAL BIOTECH
Construction method of fusion gene detection library based on Ion Torrent sequencing platform and fusion gene detection method
The invention discloses a construction method of a fusion gene detection library based on an Ion Torrent sequencing platform and a fusion gene detection method. The construction method comprises the following steps that 1, fragmentation is conducted on sample DNA to obtain DNA fragments; 2, tail end repairing and basic group A adding are conducted on the DNA fragments; 3, joints with known sequences are connected; 4, PCR amplification is conducted to obtain a sample nucleic acid library; 5, the sample nucleic acid library captures DNA fragments of fusion genes and partner genes on the basis of probe hybridization of a target area capturing platform; 6, a product obtained in the step 5 is introduced into a sequencing joint sequence through PCR, and the fusion gene detection library is obtained. By means of the technical scheme, the problems that in the prior art, due to the fact that an RNA sample is not qualified, the Ion Torrent sequencing platform cannot detect the fusion genes and cannot detect the fusion types of unknown fusion breakpoints are solved.
Owner:天津诺禾医学检验所有限公司
Targeted trapping and sequencing kit for lung cancer-related 285 genes and application of targeted trapping and sequencing kit
InactiveCN108148910AImprove detection efficiencyDetecting Gene FrontiersMicrobiological testing/measurementSmall sampleTrapping
The invention discloses a targeted trapping and sequencing kit for 285 lung cancer-related genes and application of the targeted trapping and sequencing kit. The kit is based on hybrid trapping of a probe, and can be used for preparing a high-throughput library of the lung cancer-related 285 genes and simultaneously detecting all variation loci on exons of the 285 genes, so that the disadvantagesof high cost, high time consumption, tedious operation and the like of conventional single-gene detection are overcome. Detection of a plurality of genes with a single small sample is implemented in the small sample for aspiration biopsy of lung cancer, and the clinical practice requirements of maximally using the small sample and obtaining a maximum amount of information are met. The kit is basedon an Ion Proton platform, has high accuracy and sensitivity, and has obvious advantages compared with a Sanger sequencing method, and a detection result in a residual DNA (deoxyribonucleic acid) sample of a paraffin sample is stable.
Owner:GUANGDONG GENERAL HOSPITAL
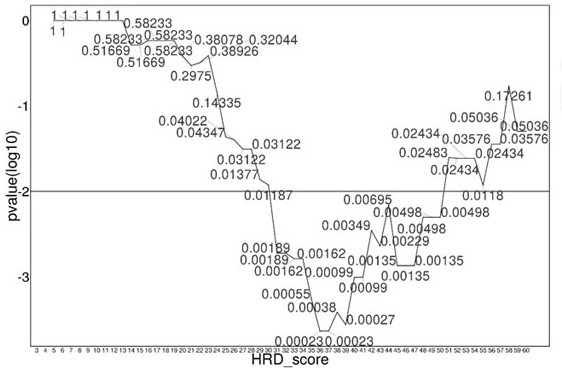
Method and test kit for determining genome instability on basis of next-generation sequencing technology
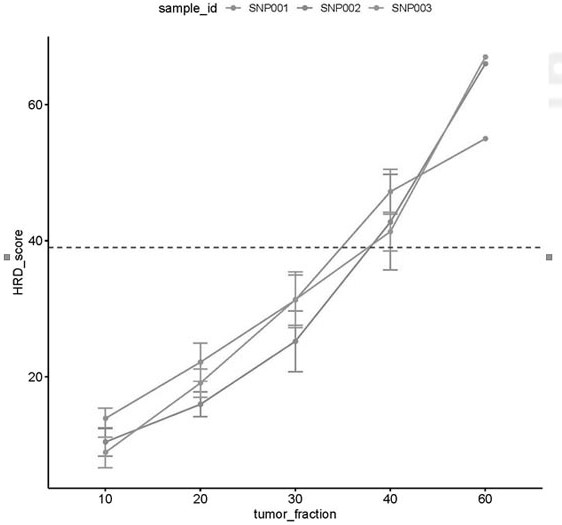
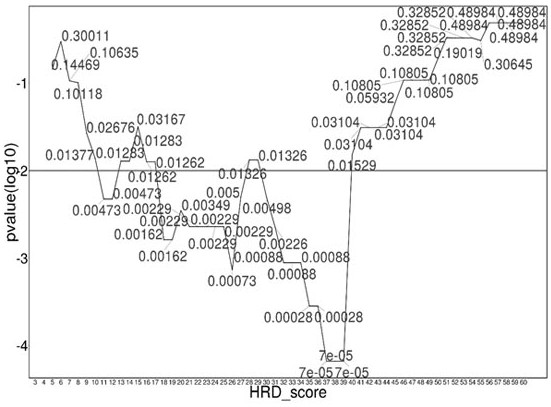
ActiveCN111676277AHigh detection sensitivityImprove capture efficiencyMicrobiological testing/measurementBiostatisticsGenomic StabilityGenome instability
The invention relates to a method and a test kit for determining genome instability on the basis of a next-generation sequencing technology, belonging to the technical field of molecular detection. Inorder to overcome the defect that genome structure variation is not considered during detection of homologous recombination defects in the prior art, the invention provides the method and the test kit for determining genome instability on the basis of the next-generation sequencing technology. The method comprises the following steps: breaking a to-be-detected gene, then adding an A connector fora corresponding PCR reaction, carrying out hybridization capture through a designed probe, then amplifying a captured DNA library, sequencing the library, and carrying out evaluating through a software so as to determine the homologous recombination defect state. Compared with the prior art, the method provided by the invention can comprehensively evaluate the HRD state, is reliable in data results and is applicable to popularization.
Owner:臻和(北京)生物科技有限公司 +1
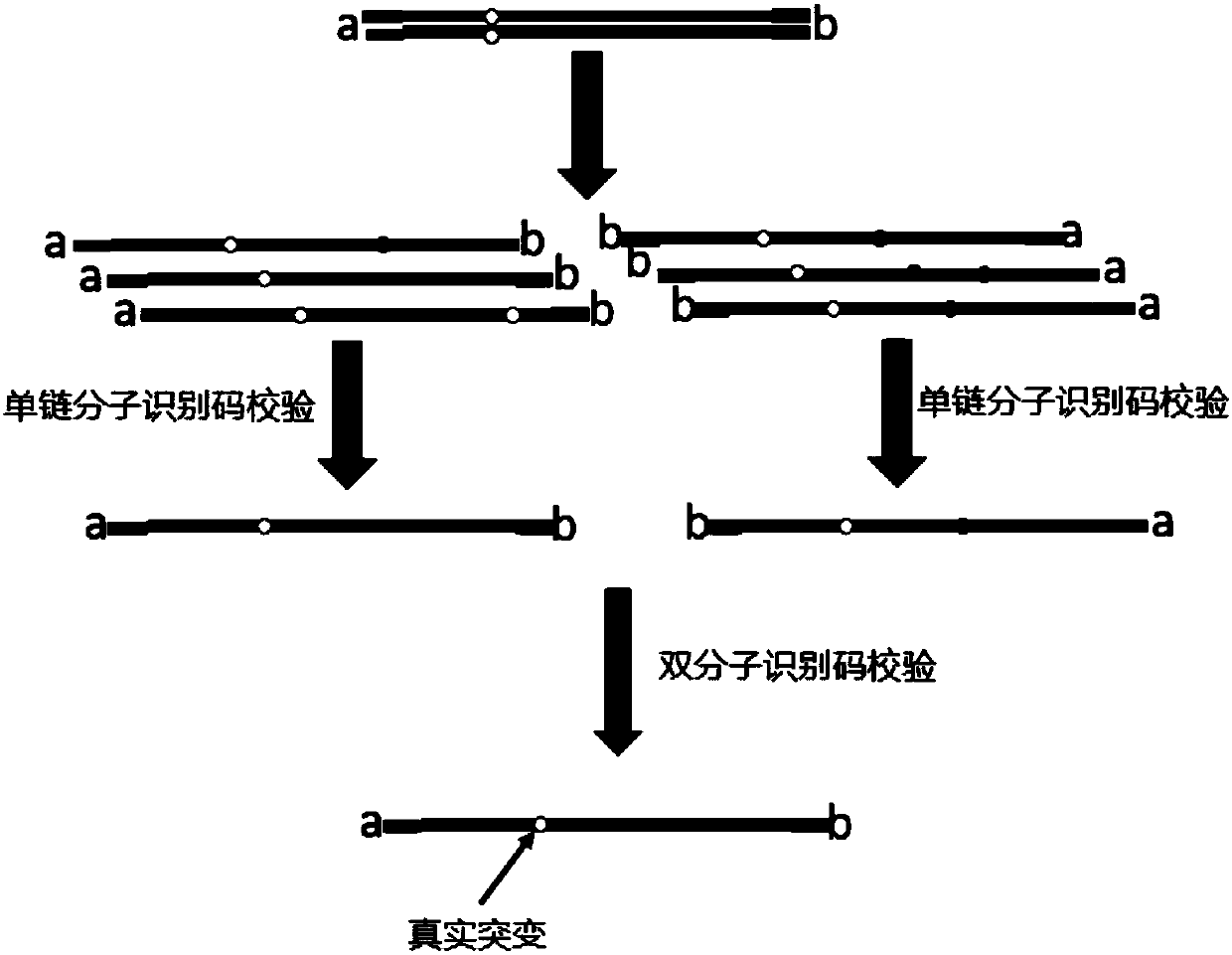
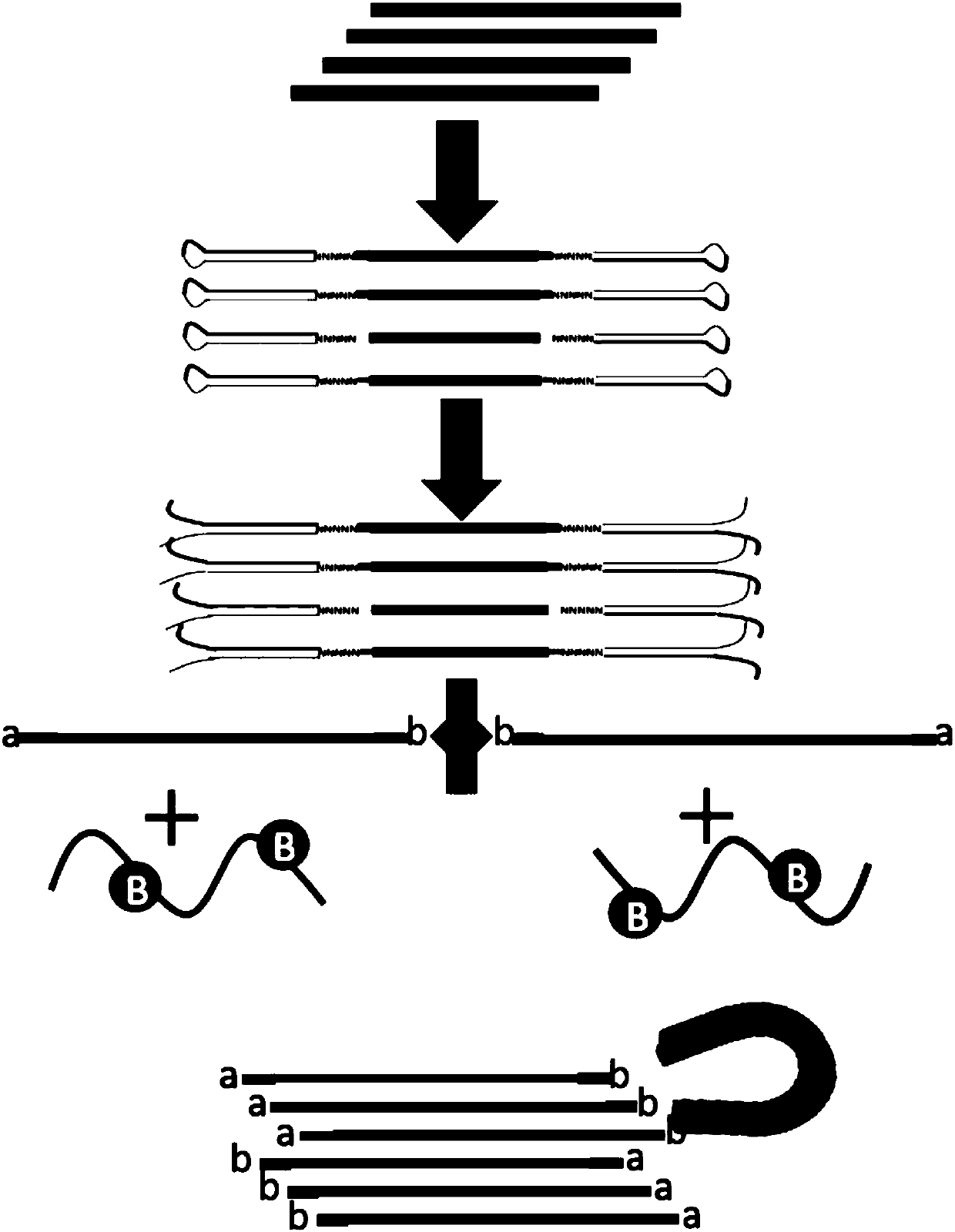
Second-generation sequencing method for dimolecular self-checking library preparation and hybrid capture used for trace DNA ultralow frequency mutation detection
ActiveCN107604046AIncrease profitHigh yieldNucleotide librariesMicrobiological testing/measurementLibrary preparationData error
The invention discloses a second-generation sequencing method for dimolecular self-checking library preparation and hybrid capture used for trace DNA ultralow frequency mutation detection. The methodcomprises the following steps: extracting plasma free DNA, performing DNA chemical error repair, preparing a self-checking dimolecular identification code hairpin type joint, repairing plasma free DNA, connecting DNA with the joint, and performing Pre-PCR amplification, excessive hybrid capture, Post-PCR amplification, computer sequencing, data error correction, and mutation analysis and annotation. The method disclosed by the invention can efficiently realize low frequency mutation detection of plasma free DNA. DNA error repair and dual redundancy checking technology can enable the method tohave ultralow false positive rate and high sensitity while detecting trace samples, thus avoiding defects of existing plasma circulating free DNA detection methods, not only realizing cancer mutationdetection and targeted medication guidance, but also realizing early screening of genetic and birth defects of fetus.
Owner:SHANGHAI DYNASTYGENE CO
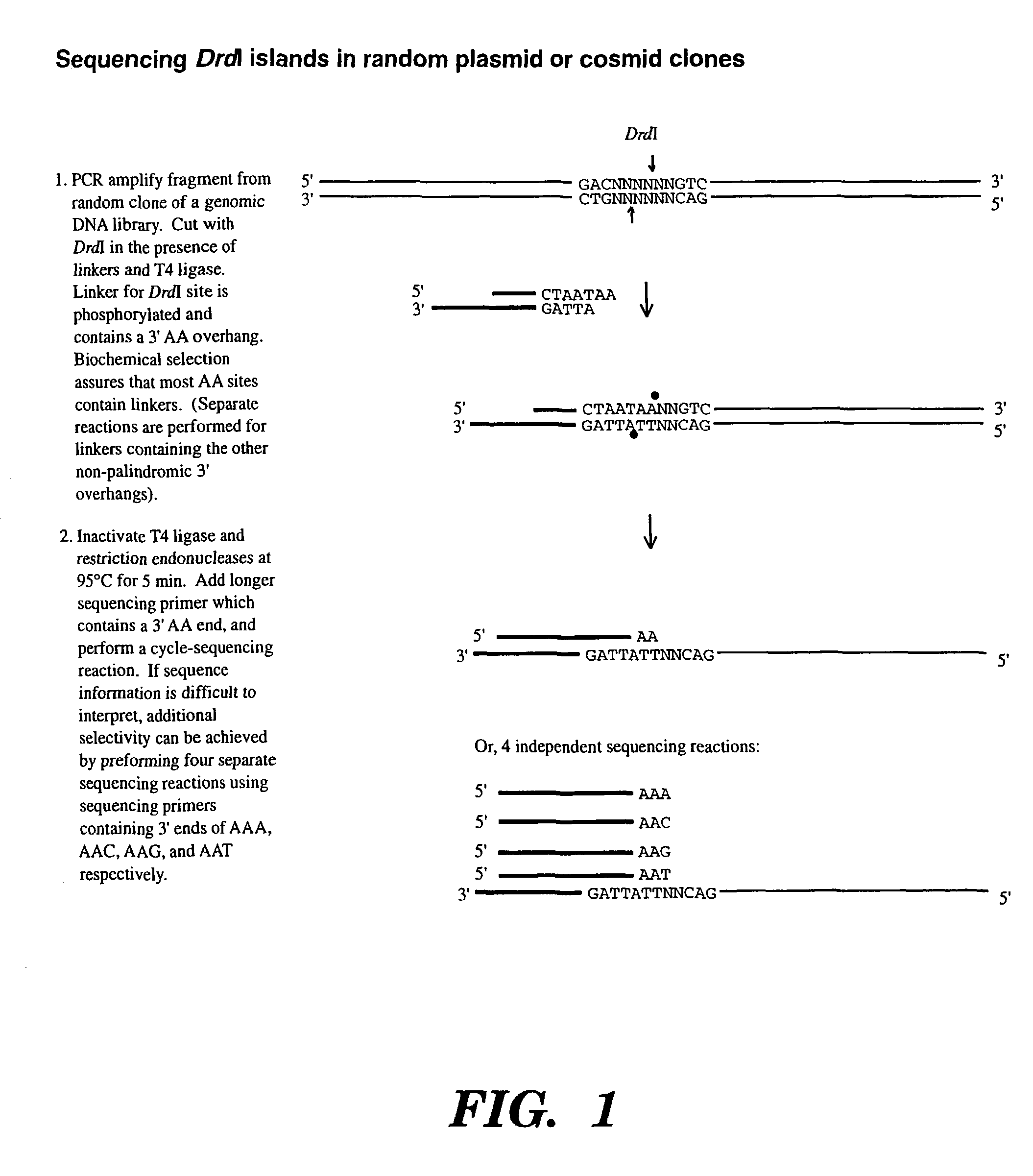
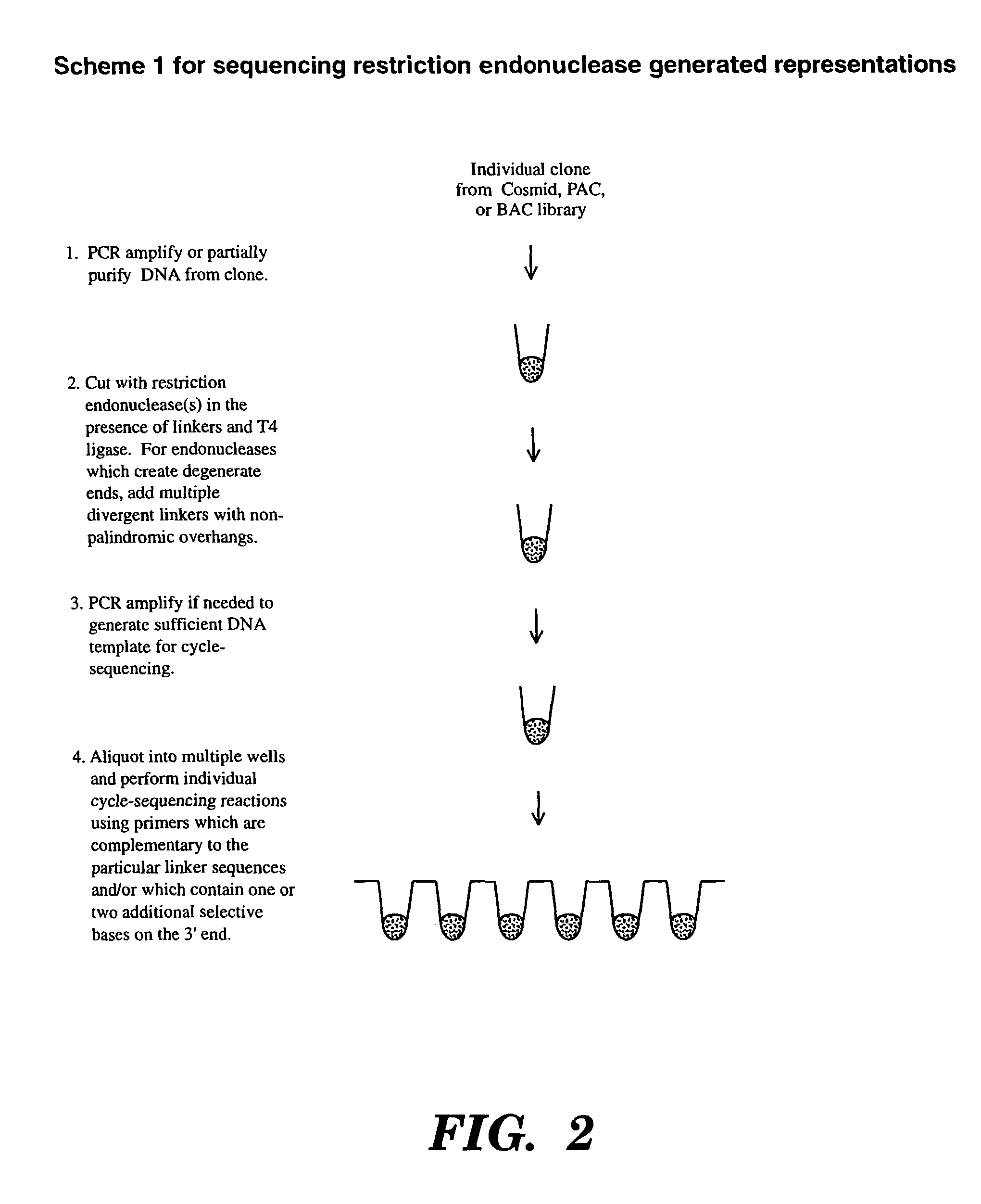
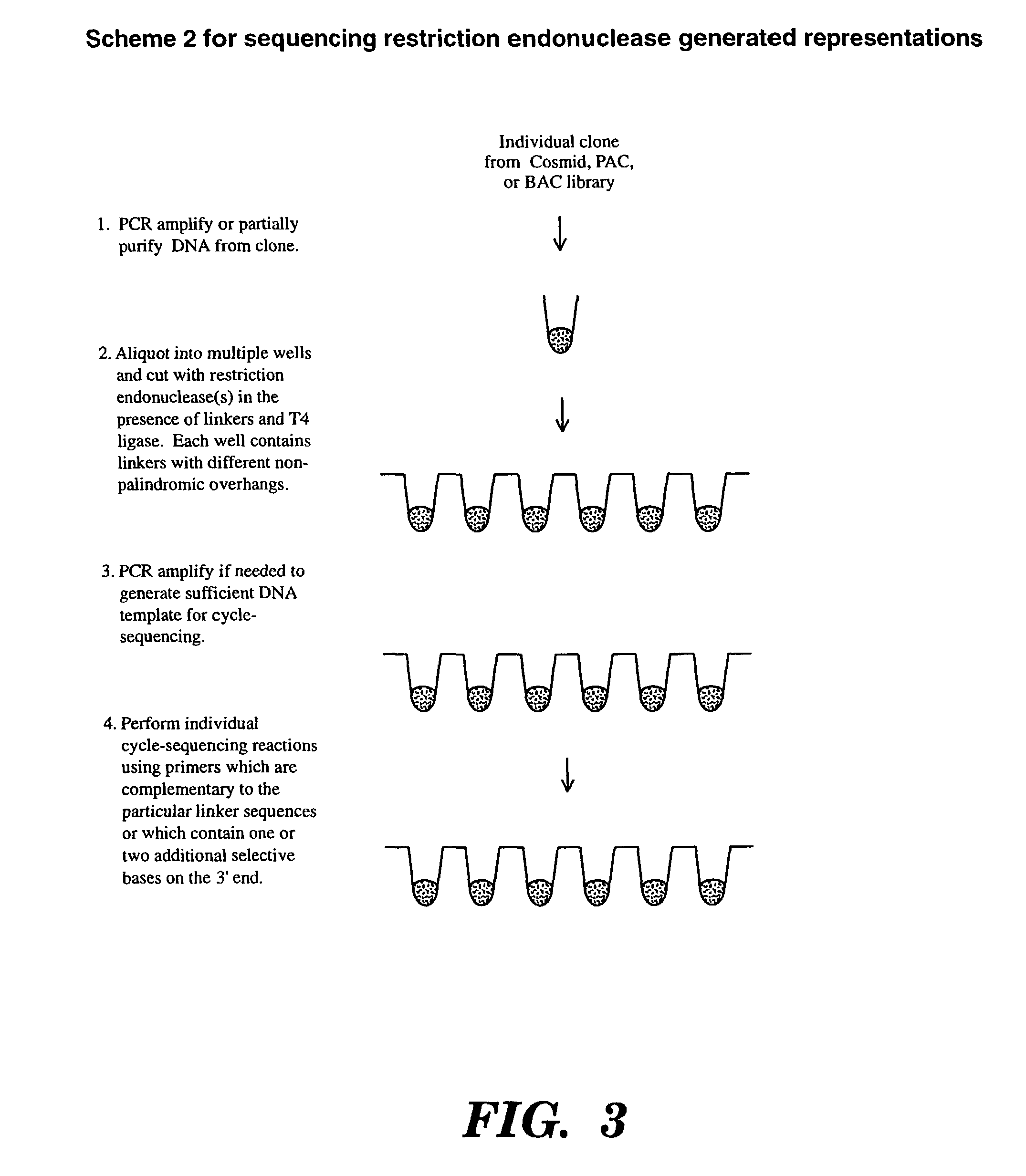
Accelerating identification of single nucleotide polymorphisms and alignment of clones in genomic sequencing
InactiveUS8367322B2Reduce the amount requiredSugar derivativesMicrobiological testing/measurementGenomic SegmentGenomic sequencing
The present invention is directed to a method of assembling genomic maps of an organism's DNA or portions thereof. A library of an organism's DNA is provided where the individual genomic segments or sequences are found on more than one clone in the library. Representations of the genome are created, and nucleic acid sequence information is generated from the representations. The sequence information is analyzed to determine clone overlap from a representation. The clone overlap and sequence information from different representations is combined to assemble a genomic map of the organism. Once the genomic map is obtained, genomic sequence information from multiple individuals can be applied to the map and compared with one another to identify single nucleotide polymorphisms. These single nucleotide polymorphisms can be detected, and alleles quantified, by conducting (1) a global PCR amplification which creates a genome representation, and (2) a ligation detection reaction process whose ligation products are captured by hybridization to a support.
Owner:CORNELL RES FOUNDATION INC +1
Probe for detecting circulating tumor DNA (Deoxyribonucleic Acid) of human breast cancer and application of probe
InactiveCN105950739AStrong specificityHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationDiseaseIndividualized treatment
The invention discloses a probe for detecting a circulating tumor DNA (Deoxyribonucleic Acid) of a human breast cancer and application of the probe, belonging to the field of diagnosis and prevention of diseases. The invention designs a breast cancer selector which consists of DNA segments in mutation regions of related genes of the breast cancer. The circulating tumor DNA (ctDNA) of a breast cancer patient is subjected to liquid-phase hybridization capture by using the probe designed by the selector; after high-throughput sequencing, specific cancer gene variation of a patient is found, and a tumor load is monitored. Compared with a conventional detection means, the probe and the application thereof disclosed by the invention have the characteristics of no invasiveness, high throughout, good sensitivity and specificity, and the like; the probe and the application thereof can be used for better screening and early-stage diagnosis of the human breast cancer and are applied to individualized treatment of cancer.
Owner:HARBIN MEDICAL UNIVERSITY
Pig 50K liquid phase chip based on targeted capture sequencing, and application thereof
ActiveCN112921076AAvoid uniformityGuaranteed specific capture rateNucleotide librariesMicrobiological testing/measurementPig farmsNucleotide
The invention relates to a pig 50K liquid phase chip based on targeted capture sequencing, and application thereof. The pig 50K liquid phase chip is composed of pig 50K probe mixed liquor and a hybridization capture reagent which are independently packaged; pig 50K probes are DNA double-stranded probes and are nucleotide sequences designed and synthesized according to screened SNP sites; and the SNP sites are screened by comparing the sequencing results of the whole genome of the pig to a pig reference genome. The pig 50K liquid phase chip prepared by the invention is mixed with a pig DNA high-throughput sequencing library; DNA fragments containing target sites in the pig DNA high-throughput sequencing library are captured for amplification and purification; and, after a product is subjected to high-throughput sequencing, a sequencing result is replied to a pig reference genome for comparison, so that the genotyping of the genome of a to-be-detected pig is obtained. When the 50K liquid phase chip prepared by the invention is used for genotyping pigs; and the problems of high cost, inflexibility and incapability of large-scale use in pig farms in China in the prior art can be solved.
Owner:CHINA AGRI UNIV
DNA methylation specific signal amplification
InactiveUS20090181373A1Quick checkAvoid contaminationMicrobiological testing/measurementDNA methylationBarcode
Owner:LI WEIWEI +1
Gene fusion mutation library construction method, gene fusion mutation detection method, gene fusion mutation detection device, equipment and storage medium
PendingCN111321202AImprove economyAccurate expression of magnitudeMicrobiological testing/measurementProteomicsA-DNACore gene
The present invention relates to a gene fusion mutation library construction method, a gene fusion mutation detection method, a gene fusion mutation detection device, computer equipment and a computerstorage medium. The above-mentioned gene fusion mutation library construction method and the gene fusion mutation detection method and device are based on a DNA probe hybridization capture multi-geneRNA targeted sequencing technology, through a fusion gene capture probe hybridization capture target fusion gene, the gene fusion mutation library is constructed, the library can be used for high-throughput sequencing and bioinformatics analysis can identify core genes and partner genes there of constitute breakpoints. Furthermore, the present invention also designs a quantitative analysis methodof the fusion gene, a mutation ratio of the fusion gene can be obtained by calculation, accurate expression value of the fusion gene can be further obtained, and the quantitative analysis method of the fusion gene is pioneering and solves a quantitative analysis problem of using a NGS method to detect the fusion gene.
Owner:GUANGZHOU KINGMED DIAGNOSTICS GRP CO LTD
Preparation method for DNA library and kit
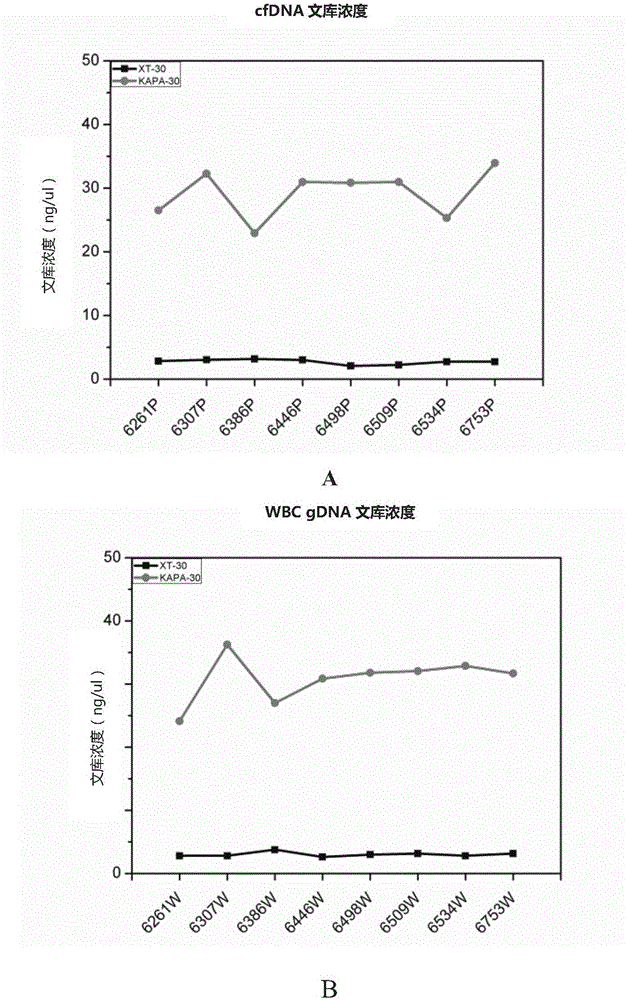
The invention provides a method for preparing a DNA library and specifically capturing and a kit, and the method is suitable for constructing the DNA library of cfDNA, white cell gDNA and DNA from a tissue sample as well as hybridizing and capturing a special target gene area. In the preparation method for the DNA library, terminal repair and 3' end A addition are performed in the same reaction system, and hybridization input is improved, and a blocking preparation in hybridizing and capturing is optimized, so that higher capturing efficiency, higher library variety and more uniform coverage degree are obtained, and therefore, the preparation method is especially suitable for constructing a trace sample library with low-abundance mutation, and is beneficial for realizing saving in technical process. The method provided by the invention can be used for conveniently realizing all variations such as mutation, deletion, insertion, fusion as well as detection on copy number amplification of a target gene, and overcomes the defects that target gene sequences are enriched through a PCR method on DNA level at present, mutation and deletion of the target gene only can be detected, and fusion cannot be detected.
Owner:GUANGZHOU BURNING ROCK DX CO LTD
Capture of breast cancer related genes, and preparation method and application of probes
InactiveCN103757709ARapid early detectionEfficient Early DetectionMicrobiological testing/measurementMicroorganism librariesRelated geneNucleic acid sequence
A capture method of breast cancer related genes is provided. Probes obtained according to a construction method of a breast cancer related gene capture probe database are subjected to hybridization with a to-be-captured target genome region-containing DNA template in a to-be-detected sample. Fragments obtained by capture are subjected to high throughput sequencing; and then based on known information of breast cancer pathogenic genes, data analysis is carried out by utilizing a bioinformatics method, and thus heritable variation conditions of nucleic acid sequences in the to-be-detected sample are obtained. The provided various methods and the obtained database (or clone database) can adjust the relative capture efficiency so as to ensure consistency capture on all the target genes, and the problem that the use efficiency of an NGS sequencing platform is greatly hindered because of heterogeneity and high cost of chip capture reported in current literatures is solved.
Owner:SHANGHAI MAJORBIO BIO PHARM TECH
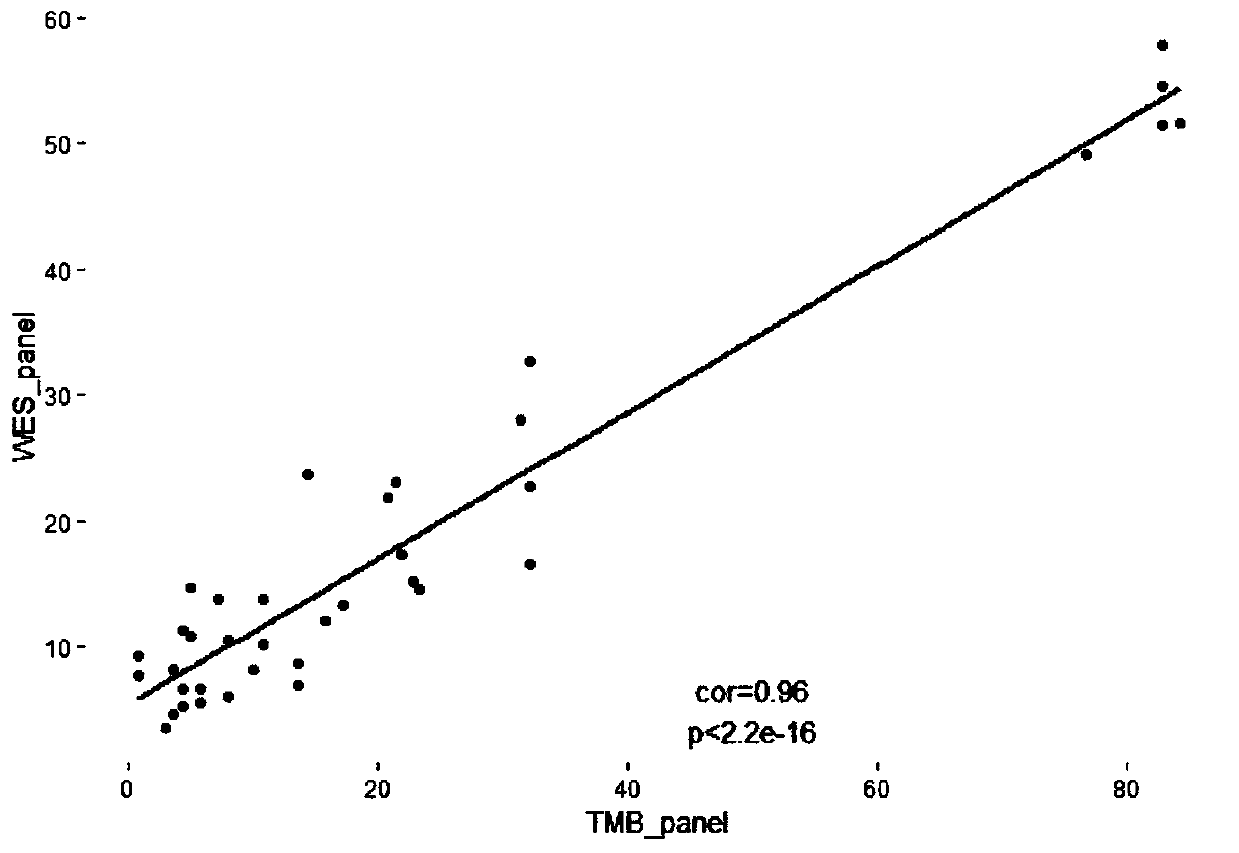
System capable of simultaneously detecting tumor targeting therapy related target spots and immunotherapy related TMB and MSI
PendingCN109988838AFew samplesShorten the timeMicrobiological testing/measurementProteomicsTumor targetTumor targeting
The invention discloses a system capable of simultaneously detecting tumor targeting therapy related target spots and immunotherapy related TMB and MSI. The system comprises a library preparation reagent, a probe, a hybridization capture reagent and a data analysis system, wherein the library preparation reagent, the probe and the hybridization capture reagent can be used for sequencing a library,and the data analysis system can analyze results including data quality control information and statistical information, mutation site information and TMB values.
Owner:AMOY DIAGNOSTICS CO LTD
Detection method of low requency mutated DNA fragment and library building method
ActiveCN108192955AEasy to useImprove detection limitMicrobiological testing/measurementLibrary creationMutation detectionEngineering
The invention belongs to the technical field of gene engineering, and particularly relates to a detection method of a low frequency mutated DNA fragment and a library building method. The technical problems that in the existing detection technique, the DNA fragment capturing efficiency is low, wrong mutation is introduced in the library building process, the detection limit is low, and the sensitivity is low are solved by adopting the hybrid capturing and molecular tagging technique and meanwhile combining with a special library building mode, the library specificity is improved, the false positive rate is lowered, and the template utilization rate is improved.
Owner:HUNAN YEARTH BIOTECHNOLOGICAL CO LTD
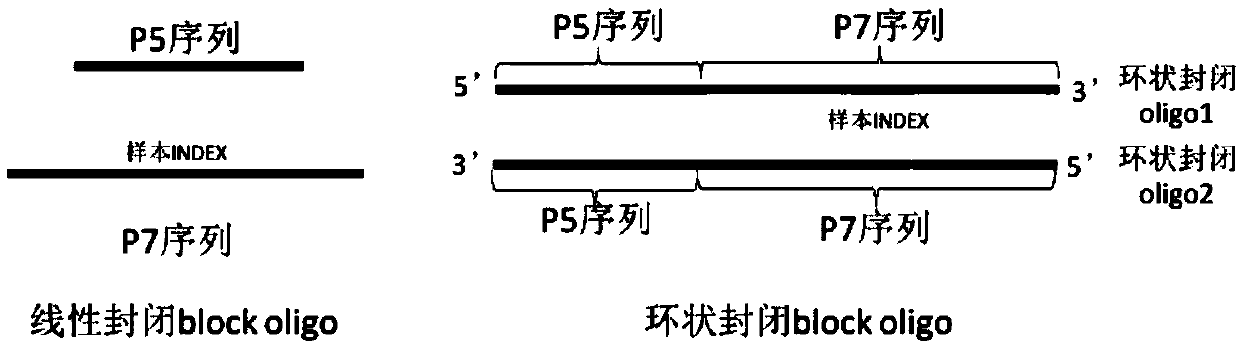
Closed sequence, capture kit, library hybridization capture method and library building method
ActiveCN111748551AImprove bindingReduce or avoid amplificationMicrobiological testing/measurementLibrary creationAlgorithmEngineering
The invention provides a closed sequence, a capture kit, a library hybridization capture method and a library building method. Wherein the closed sequence comprises a first closed segment and a secondclosed segment in the direction from 5'to 3', the second closed segment is arranged at the downstream of the first closed segment, and the 3' ends of the first closed segment and the second closed segment are provided with closed modifications; and part of basic groups on the first closed section and the second closed section are LNA or BNA modified basic groups. LNA or BNA modification is carried out on part of basic groups on the first closed segment and the second closed segment, so that the binding capacity of a to-be-closed sequence can be enhanced, and the closing effect is improved; the 3'ends of the first closed segment and the second closed segment are subjected to closed modification, so that redundant joints in the library cannot be used as primers to amplify the joints of other libraries, and the improvements in the two aspects enable the closed sequence disclosed by the invention to reduce or avoid unnecessary amplification of the redundant joints in the library and sample label skipping phenomena.
Owner:NANODIGMBIO (NANJING) BIOTECHNOLOGY CO LTD
DNA methylation specific signal amplification
InactiveUS7608402B2Quick checkAvoid contaminationMicrobiological testing/measurementDNA methylationBarcode
Owner:LI WEIWEI +1
Method for constructing a hybrid capture sequencing library and application thereof
ActiveCN109234356AImprove capture efficiencyImprove uniformityMicrobiological testing/measurementLibrary creationGenomic DNAPaper document
The invention belongs to the field of gene detection, in particular to a strategy for direct hybridization of genomic DNA and a method for constructing a DNA library and an application thereof in thefield of nucleic acid detection. The method comprises the following steps: genomic DNA is fragmented and phosphorylated; Fragmented genomic DNA hybridized with capture probes; after hybridization, thecaptured single-stranded DNA was eluted. A ligation product is obtained by linking a single strand DNA with a linker; The ligation product was used as template for PCR amplification to obtain the amplified product, and the amplified product was purified to obtain the sequencing library. The invention provides a method for directly hybridizing fragmented genomic DNA, which is universally applicable to a solid-phase chip capture hybridization system and a liquid-phase capture hybridization system, and greatly improves capture efficiency and balance. At that same time, the invention further provide a DNA library construction method, which has the characteristics of simple method, low cost, wide applicability and the like, and can be use for the construction of a high-throughput sequencing document library of DNA or RNA.
Owner:江苏安科华捷生物科技有限公司
Construction method and application of single-stranded sequencing library
ActiveCN109536579AImprove capture efficiencyShort hybridization timeMicrobiological testing/measurementLibrary creationEnzymatic digestionExonuclease I
The invention discloses a construction method and application of a high-throughput sequencing library, wherein the construction method of the high-throughput sequencing library includes the steps: carrying out fragmentation of genomic DNA and adding a base A to the 3' end of an end-repaired DNA fragment, connecting with a connector and amplifying, and digesting the DNA library with exonucleases toobtain a single-stranded DNA library; and carrying out hybrid capturing of the connected product by a specific probe, so as to obtain a target fragment. The DNA hybridization process is improved, theconventional double-stranded DNA template is digested into a single strand by enzymatic digestion, then the introduced connector and tag sequence are completely sealed by cyclic oligonucleotides, andthe single-stranded DNA template is captured with a probe (RNA or DNA); therefore, the time of hybrid capturing can be shortened, the efficiency of capturing the target DNA sequence by the probe is improved, and the preference of GC regional capturing is reduced.
Owner:深圳市艾斯基因科技有限公司
Construction method and application of high-throughput sequencing library
ActiveCN109576346AImprove capture efficiencyReduce the ratioMicrobiological testing/measurementLibrary creationSequence designGenomic DNA
The invention discloses a construction method and application of a high-throughput sequencing library. The method for constructing the high-throughput sequencing library comprises the steps that genomic DNA is fragmented, a basic group A is added to the 3'end of a DNA fragment subjected to end repairing, the DNA fragment is connected with joints, amplification is conducted, then a DNA library is mixed with annular closed oligonucleotides and a specific probe so as to conduct hybrid capturing, and the introduced joints and an introduced tag sequence are closed by the annular closed oligonucleotides, wherein the annular closed oligonucleotides are designed for the joints and the tag sequence, and the annular closed oligonucleotides and the joint sequences at the two ends of the library sequence are subjected to complementary pairing; and a connecting product is subjected to hybrid capturing through the specific probe so as to obtain a target fragment. According to the construction methodand application of the high-throughput sequencing library, a DNA template closing the sequencing joint sequences in an annular mode is captured, and the efficiency of capturing the target DNA sequence by the probe is improved.
Owner:深圳市艾斯基因科技有限公司
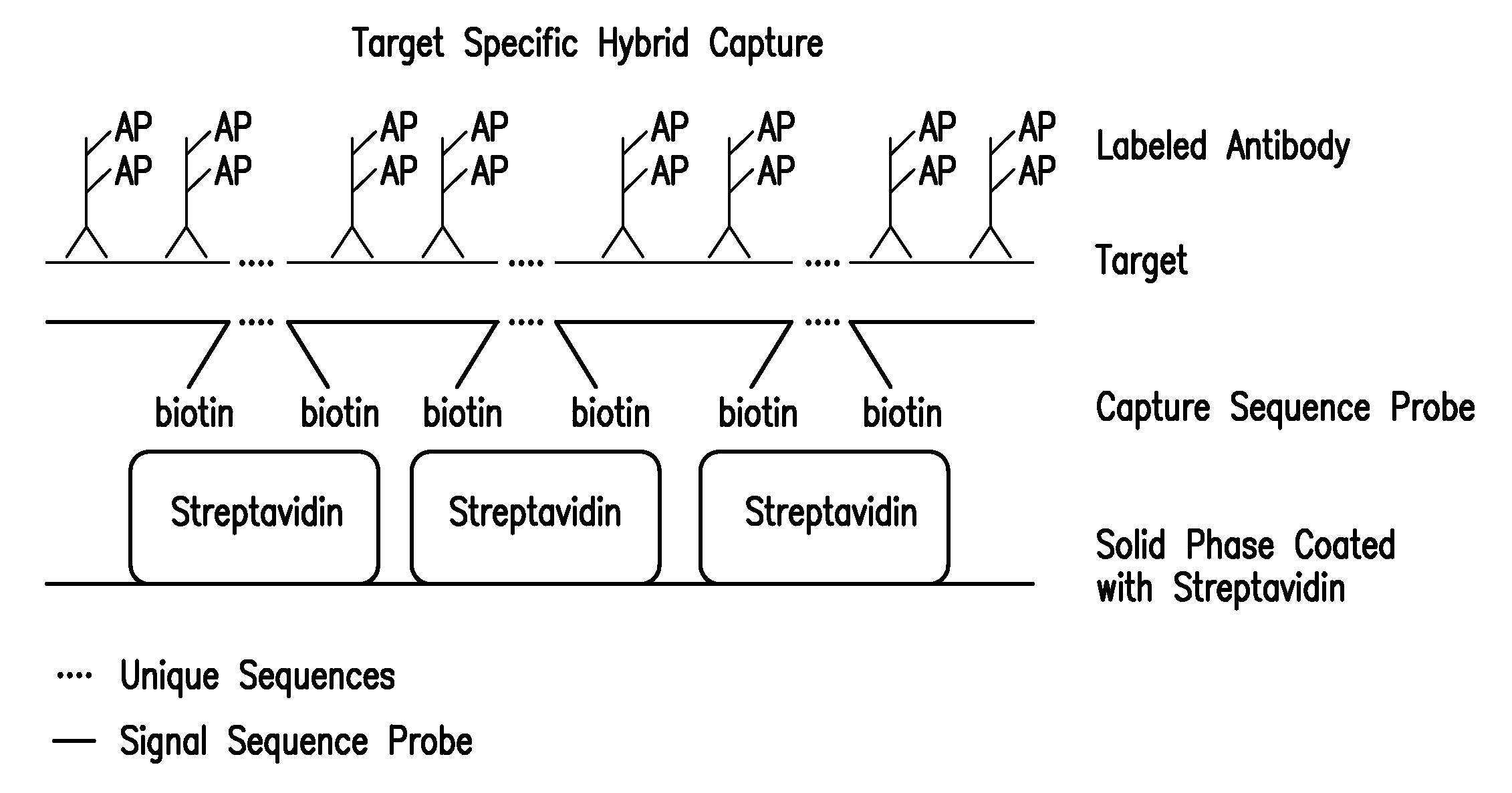
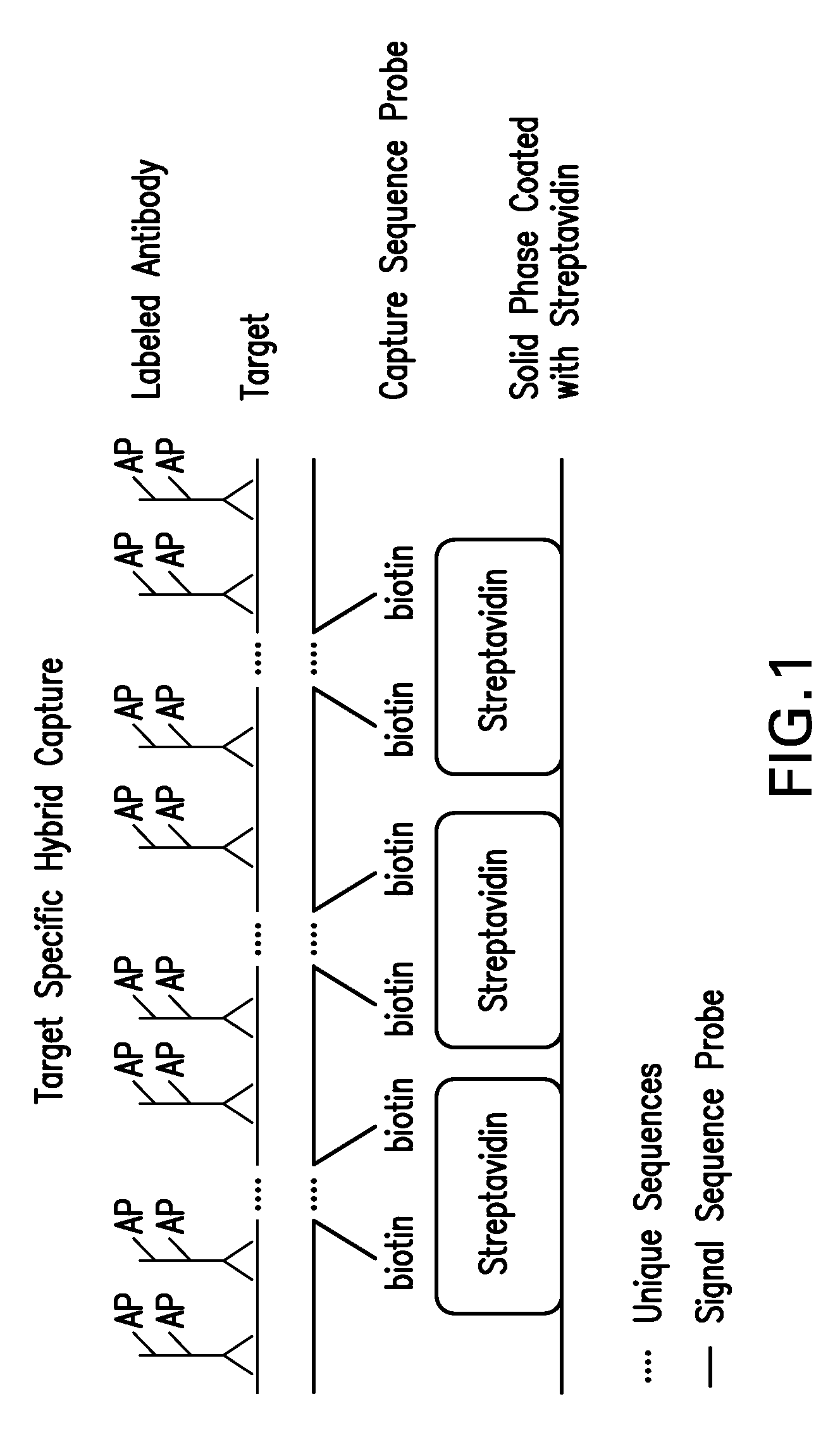
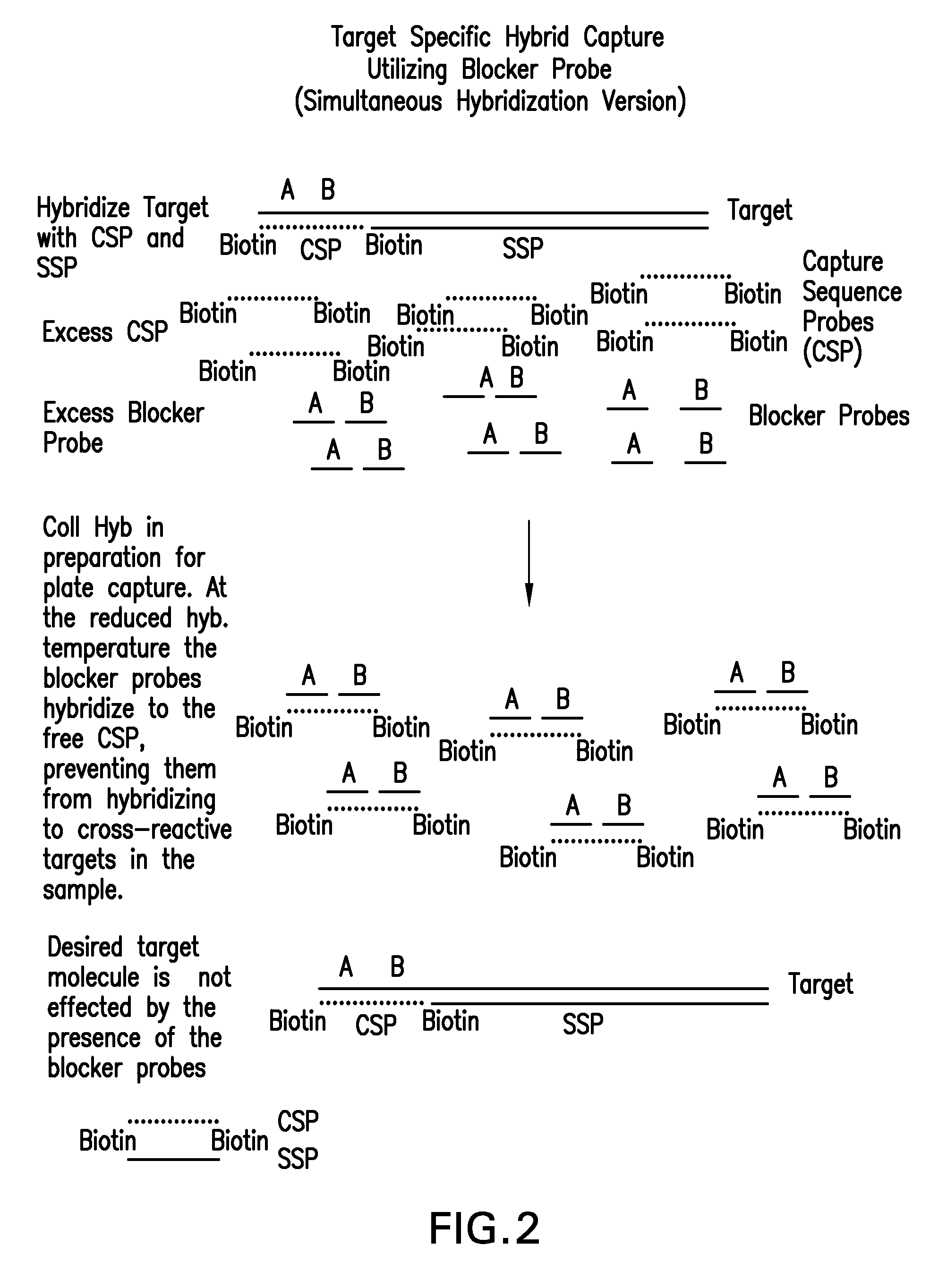
Detection of nucleic acids by type specific hybrid capture method
InactiveUS20090162851A1Reduce background signalHigh detection specificitySugar derivativesMicrobiological testing/measurementNucleic acid detectionType specific
Target-specific hybrid capture (TSHC) provides a nucleic acid detection method that is not only rapid and sensitive, but is also highly specific and capable of discriminating highly homologous nucleic acid target sequences. The method produces DNA-RNA hybrids which can be detected by a variety of methods.
Owner:QIAGEN GAITHERSBURG
Nucleic acid hybridization chemiluminiscence detection method
InactiveCN107058584ARealize detectionImprove stabilityMicrobiological testing/measurementBiotechnologyMicrosphere
The invention belongs to the technical field of biology, and relates to a nucleic acid detection method, in particular to a nucleic acid hybridization chemiluminiscence detection method. The method comprises the steps that specific detection is performed on target nucleic acid through a matrix, a capture nucleotide sequence, a target nucleic acid, a labeled biotin signal nucleotide sequence, labeled streptavidin microballoons and biotin-alkaline phosphatase, a chemiluminescent substrate and the like; the target nucleic acid is hybridized and captured through the capture nucleotide sequence, signal amplification can be achieved through the signal nucleotide sequence, the microballoons, the biotin-avidin and the alkaline phosphatase activity, so that signals of the target nucleic acid can be detected. Compared with the PCR technology that amplification is needed, the target nucleic acid can be detected on the premise that the concentration of a detection object is not increased, and the method has the advantages of being good in stability, low in cost, high in detection speed, low in environmental requirement and the like.
Owner:浙江殷欣生物技术有限公司
Hybridization buffer solution for hybrid capture of DNA sequencing library
InactiveCN107988350AImprove capture efficiencySimple recipeMicrobiological testing/measurementDextranBuffer solution
The invention discloses a hybridization buffer solution for hybrid capture of a DNA sequencing library. The hybridization buffer solution is prepared from the following components in parts by volume:25 parts of 2*SSC, 1 part of 1*Denhardt's, 9 parts of SDS with mass percent concentration of 0.5%, and 14 parts of dextran solfate with mass percent concentration of 10%. The hybridization buffer solution has a simple formula, is easy to prepare, is low-cost, is stable and is easy to store; the hybridization method is simple, is easy to operate and needs no special instruments; the hybridization effect is good, the background value is low, so that the sample capture efficiency is improved; and the method is easy to implement, and uses no extra reagents and operations, so that cost loss causedby over long hybridization time is reduced, and the processing time is shortened.
Owner:北京中源维康基因科技有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com