Second-generation sequencing method for dimolecular self-checking library preparation and hybrid capture used for trace DNA ultralow frequency mutation detection
A DNA molecule and DNA library technology, applied in the biological field, can solve problems such as inherent sequencing errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
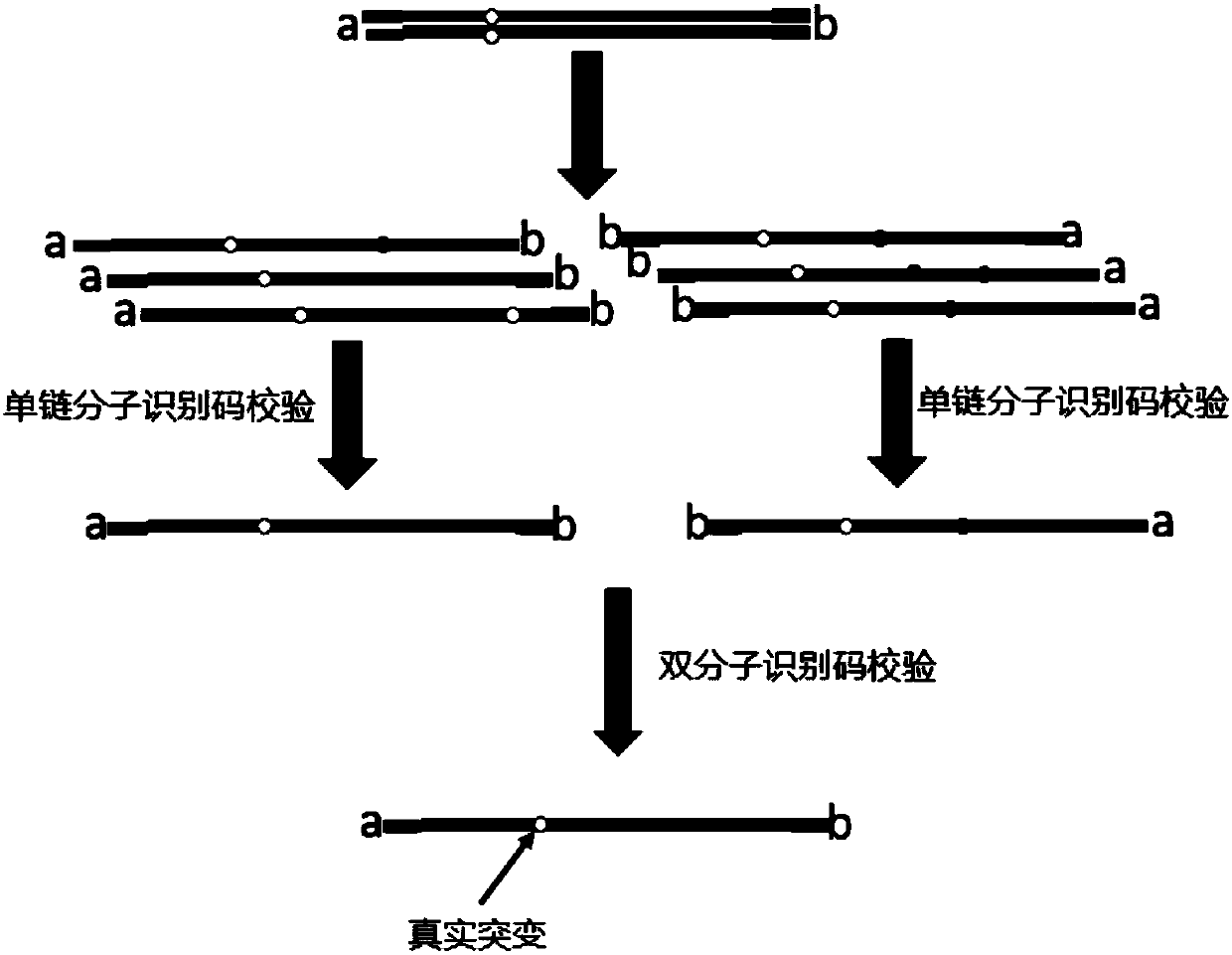
[0148] Example 1. Preparation and purification of self-verifying bimolecular identification code hairpin adapter
[0149] The self-verifying bimolecular identification code hairpin connector of the present invention is as follows: figure 1 shown.
[0150] 1. Synthesis of linker primers
[0151] Design and synthesize the following four single-stranded DNA molecules: single-stranded DNA molecule A, single-stranded DNA molecule B, single-stranded DNA molecule C and single-stranded DNA molecule D, which are named DYMB-6a, DYMB-6b, and DYMB- 6c and DYMB-6d. The sequence is as follows:
[0152] DYMB-6a:
[0153]
[0154] DYMB-6b:
[0155]
[0156] DYMB-6c:
[0157]
[0158] DYMB-6d:
[0159]
[0160] Each single-stranded DNA molecule includes, from the 5' end to the 3' end, the protection sequence of the enzyme recognition site, the enzyme recognition site, the fixed spacer sequence, the random molecular tag sequence, the stem A (Illumina standard sequencing prim...
Embodiment 2
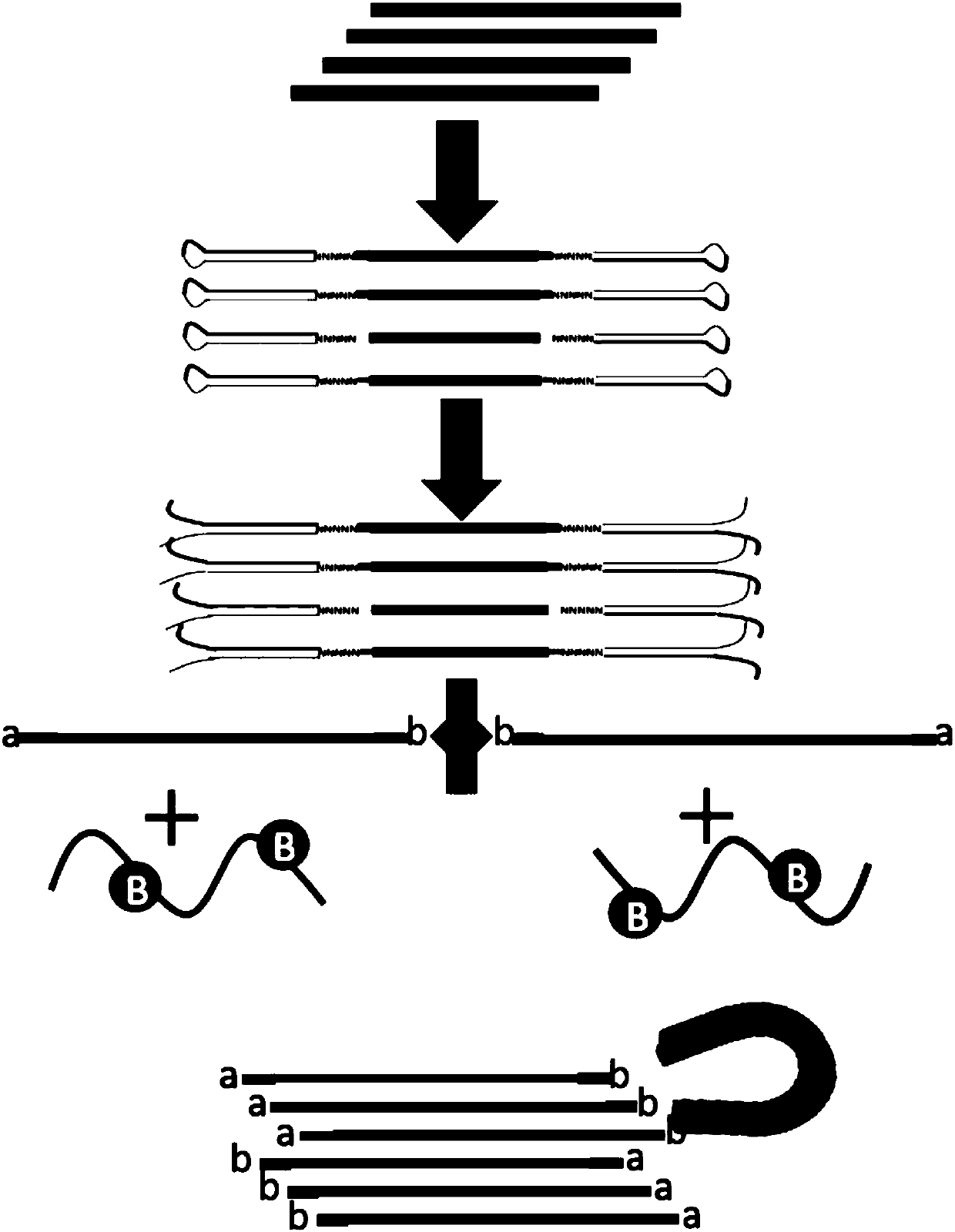
[0198] Embodiment 2, circulating cell-free DNA ultra-low frequency variation detection method
[0199] The experimental flow chart of the circulating cell-free DNA ultra-low frequency variation detection method of the present invention is as follows image 3 shown.
[0200] 1. Construction of library for cfDNA low-frequency mutation detection (DY-Ultra)
[0201] 1. End repair and A-tailing at the 3' end of circulating free DNA (cfDNA) in trace amounts of plasma
[0202] Take qualified cfDNA (cfDNA standard HD779 from Horizon Company, 0.1% mutation frequency, Multiplex I cfDNA Reference Standard, containing 8 known variations: L858R, ΔE746-A750, T790M, V769- D770insASV, G12D of the KRAS gene (Genebank ID 3845), Q61K and A59T of the NRAS gene (Genebank ID 4893), E545K of the PIK3CA (Genebank ID 5290) gene) total 30ng, diluted to 50uL with Low TE, and added 20uL of end repair solution (Wuxi Diying Biotechnology Co., Ltd., D8011A), and incubate at 20 degrees for 30 minutes to o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com