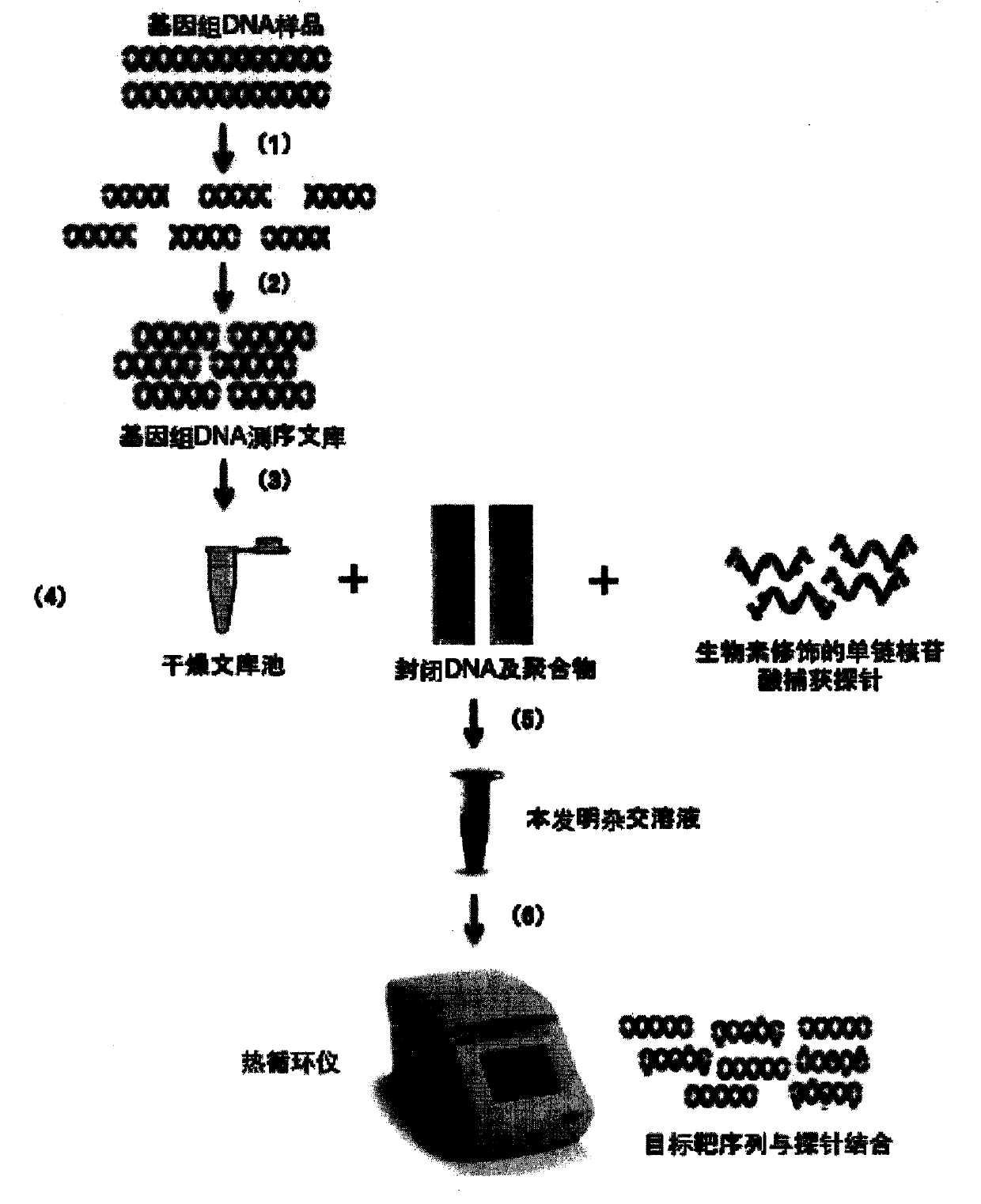
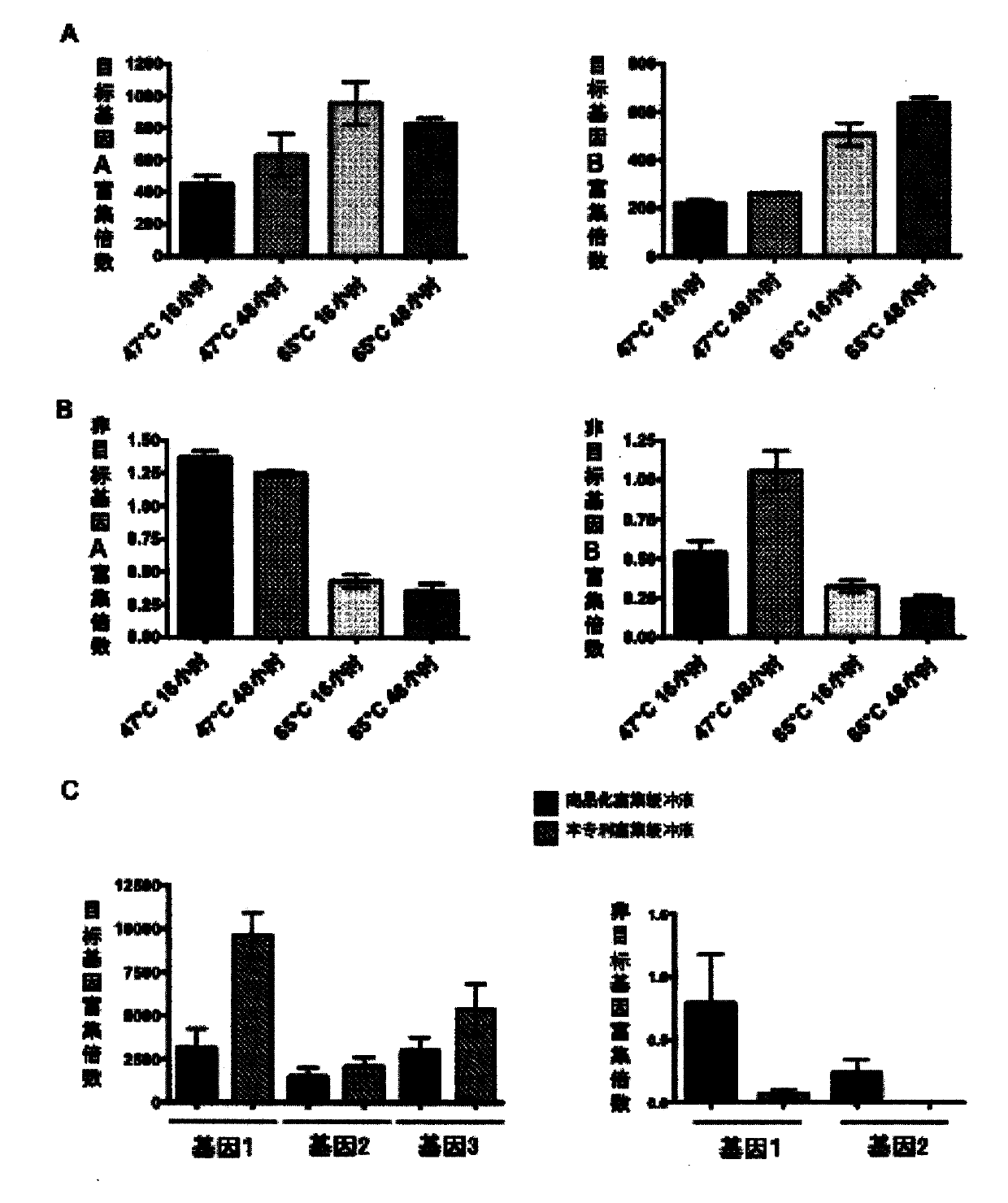
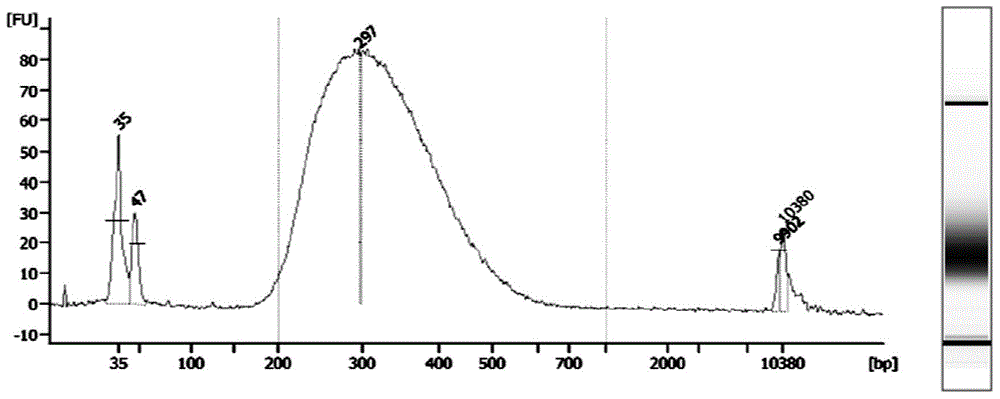
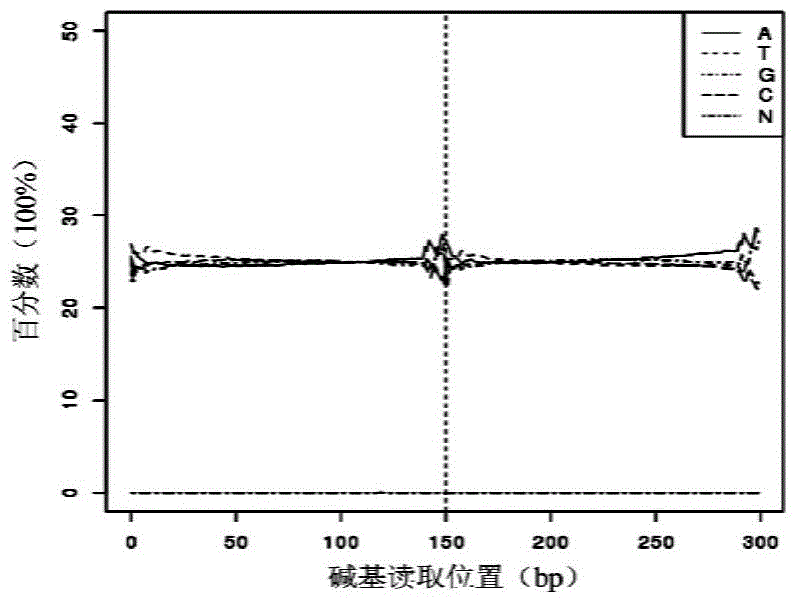
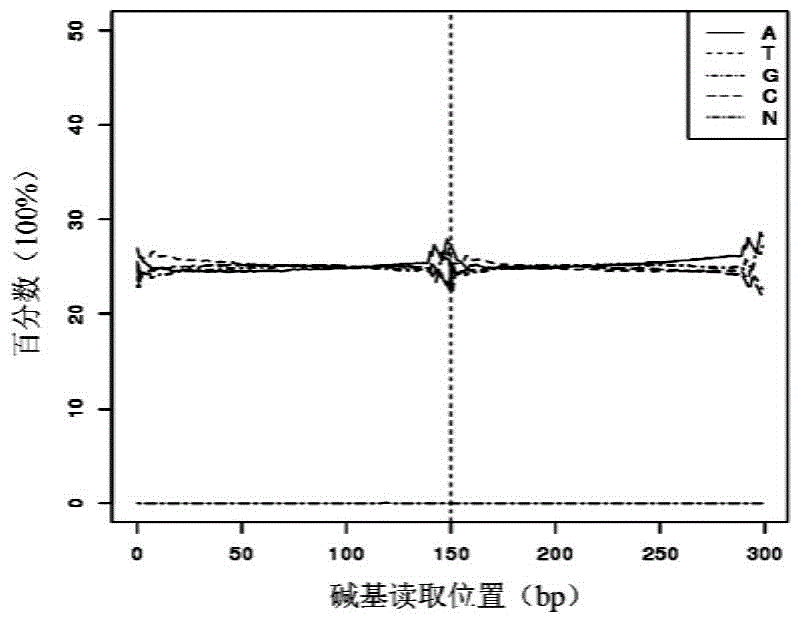
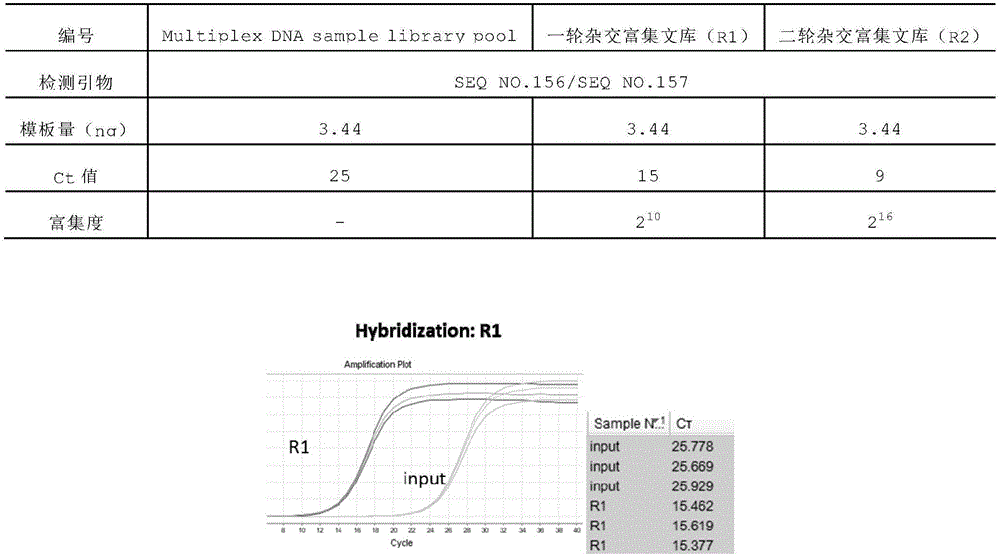
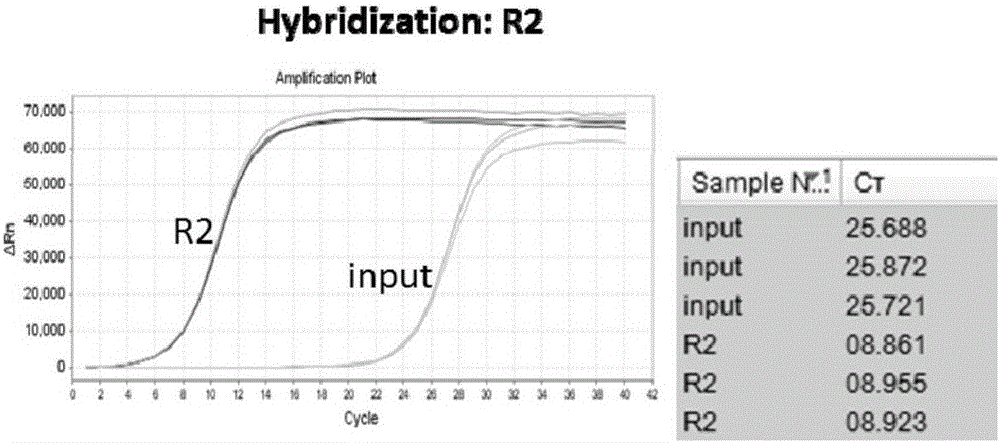
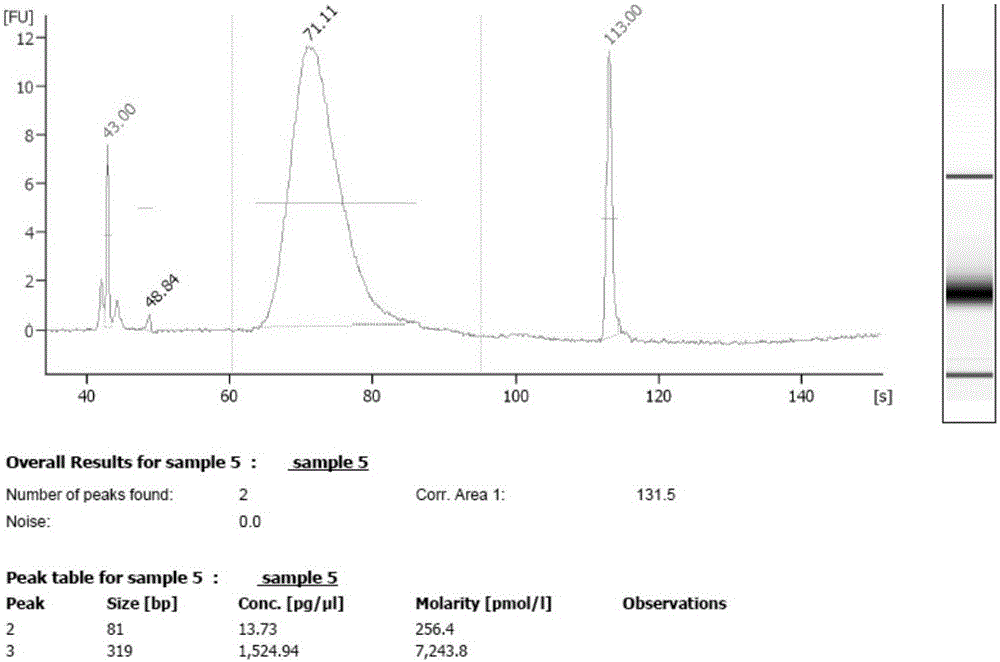
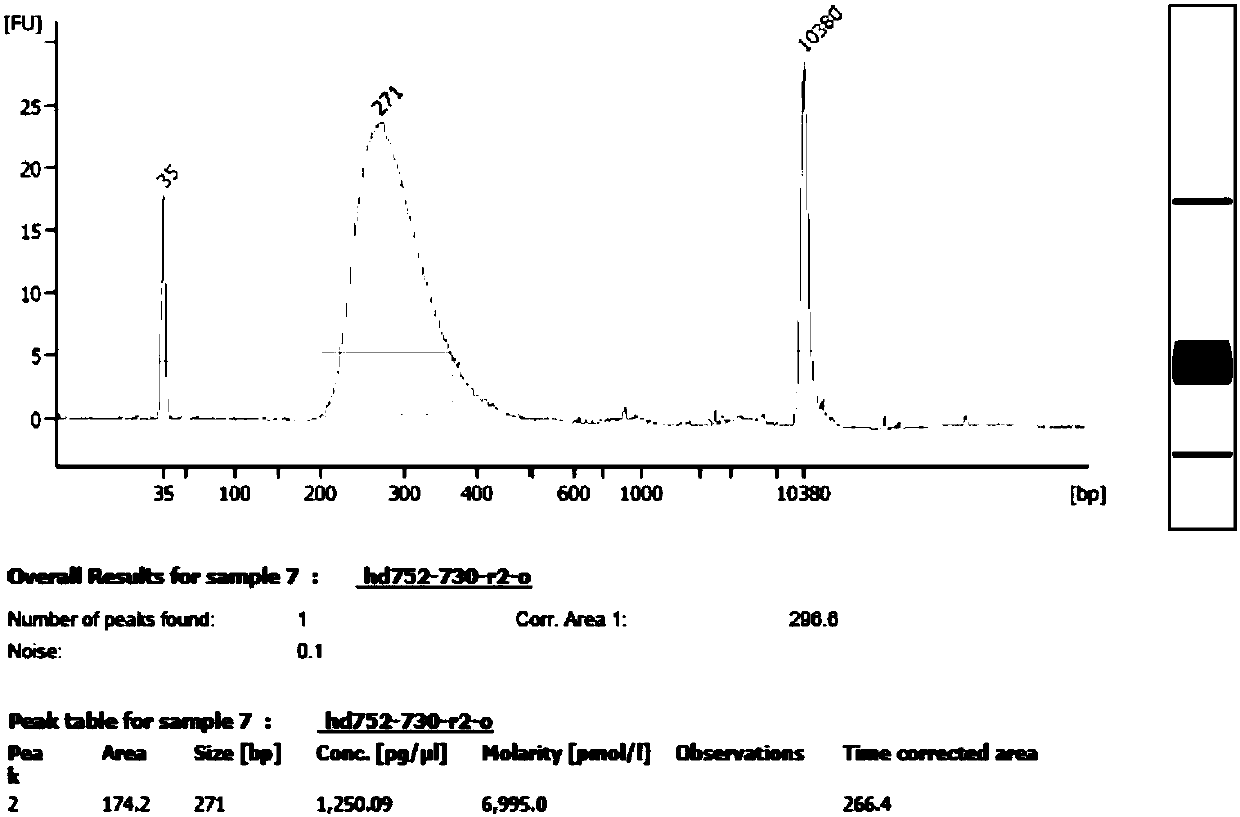
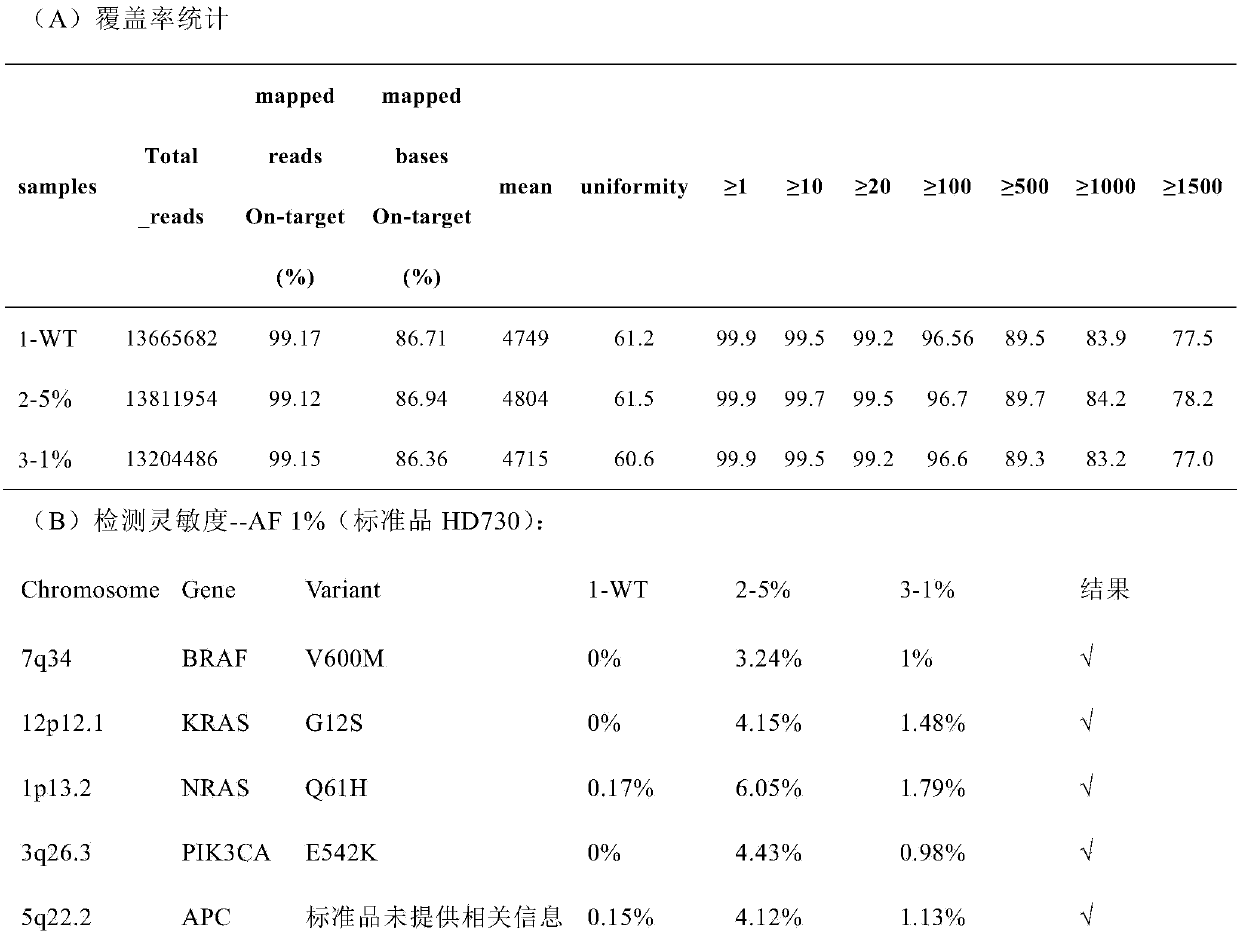
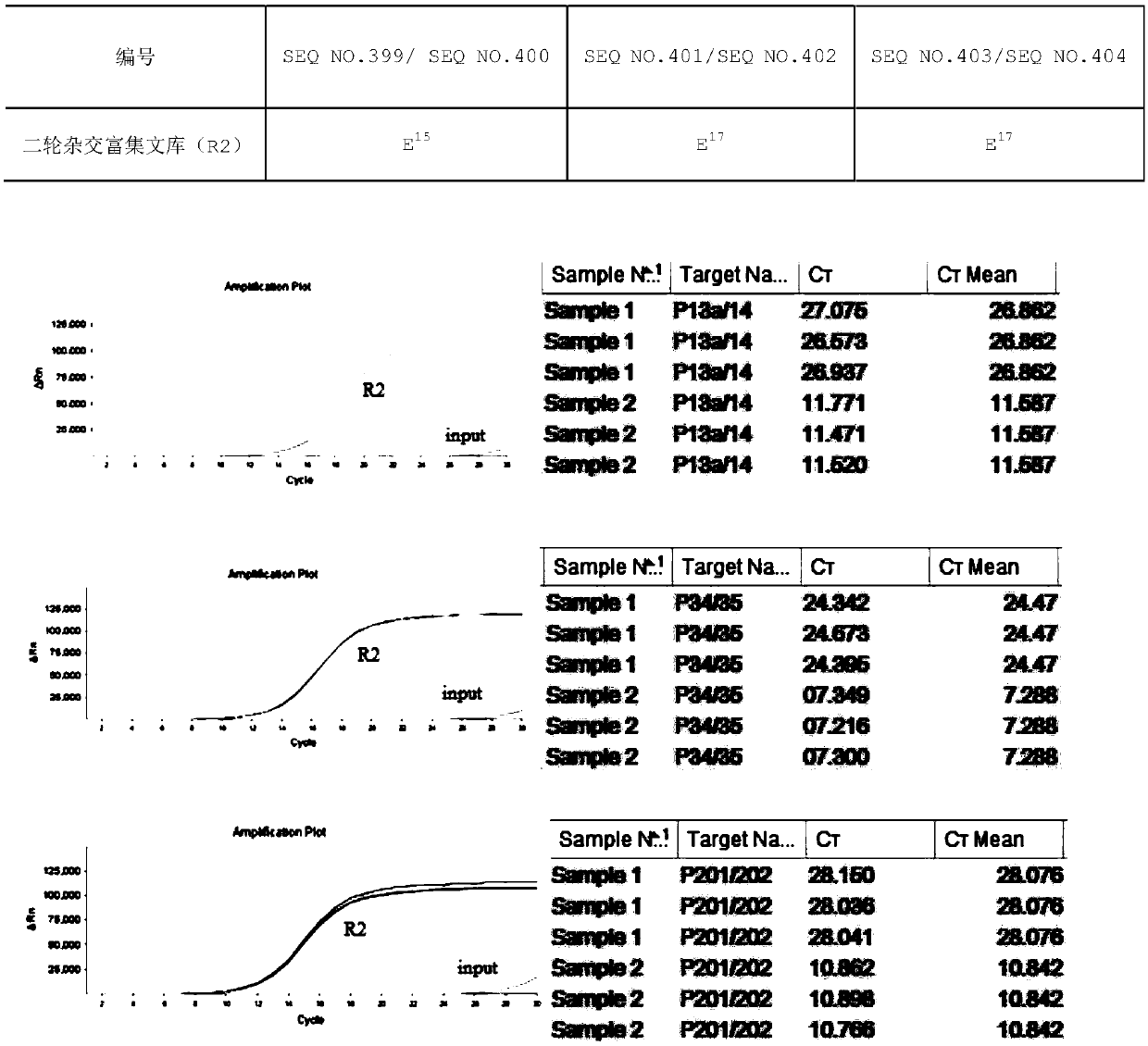
Patents
Literature
62 results about "Hybrid capture" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Hybrid capture based human papillomavirus detection is a test that can detect different types of human papillomavirus, which is used to detect high-grade cervical lesions.
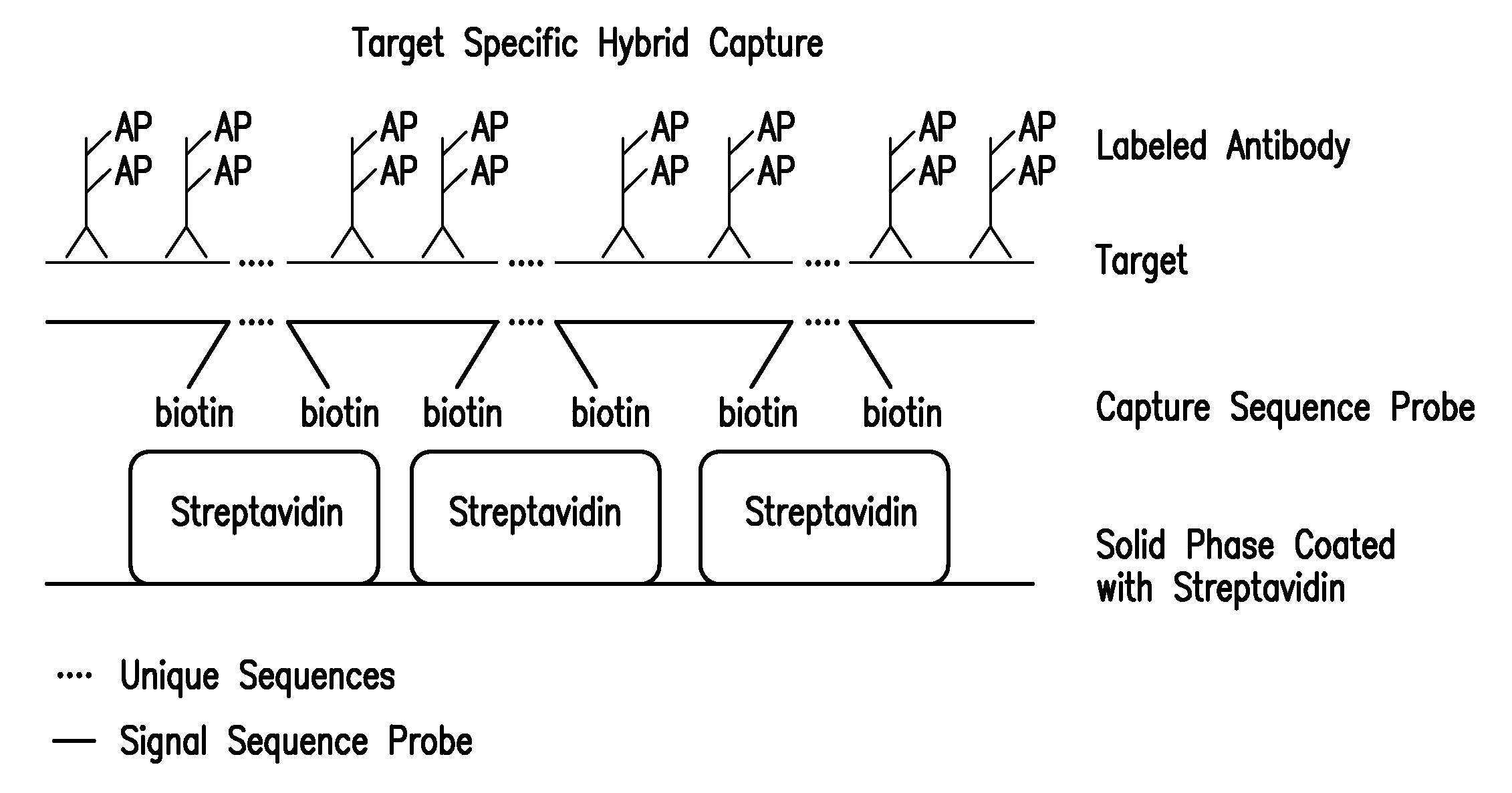
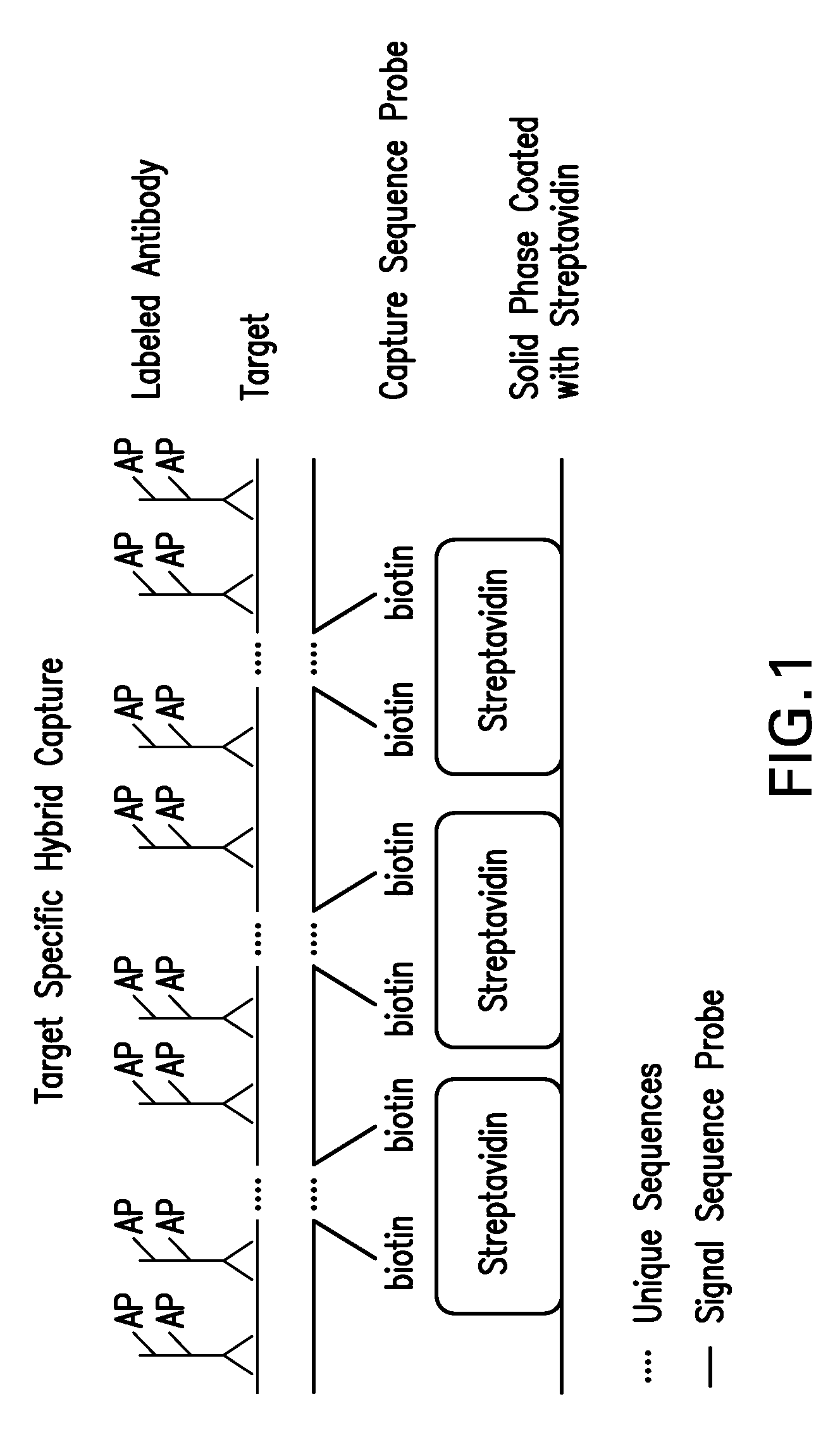
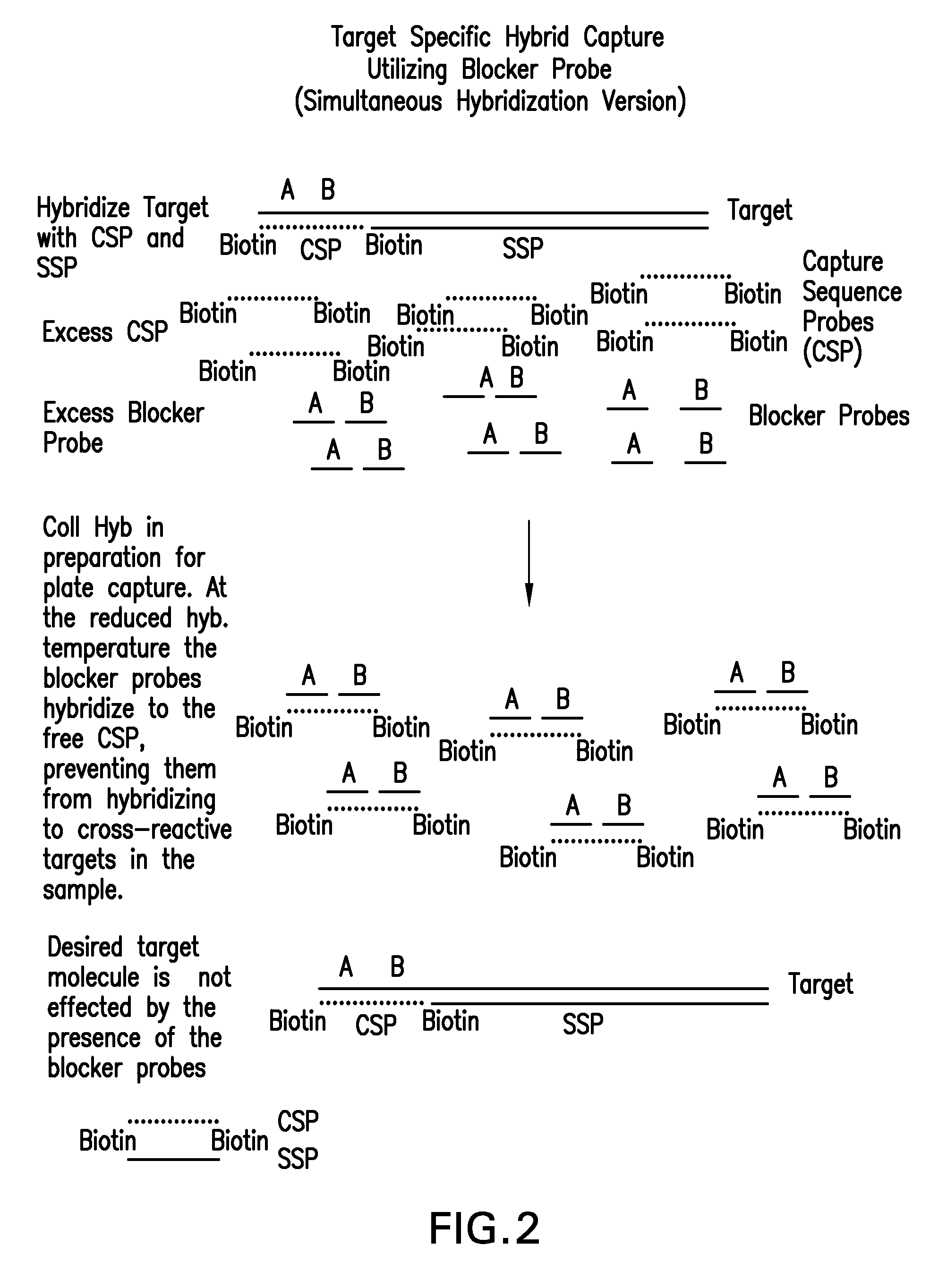
Detection of nucleic acids by target-specific hybrid capture method
InactiveUS20060051809A1Rapid and sensitive and accurateFunction increaseSugar derivativesMicrobiological testing/measurementNucleic acid detectionDNA
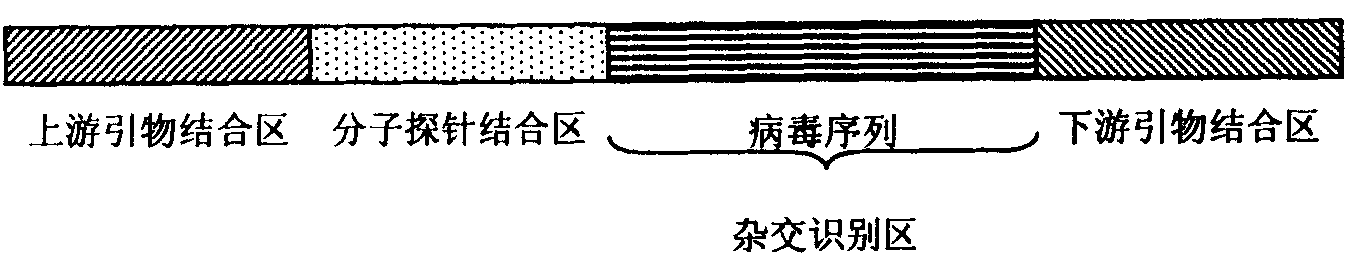
Target-specific hybrid capture (TSHC) provides a nucleic acid detection method that is not only rapid and sensitive, but is also highly specific and capable of discriminating highly homologous nucleic acid target sequences. The method produces DNA:RNA hybrids which can be detected by a variety of methods.
Owner:QIAGEN GAITHERSBURG
Construction method and application of high-throughout sequencing library
InactiveCN103806111AEfficient constructionEfficiently obtainedBioreactor/fermenter combinationsBiological substance pretreatmentsCytosineDNA fragmentation
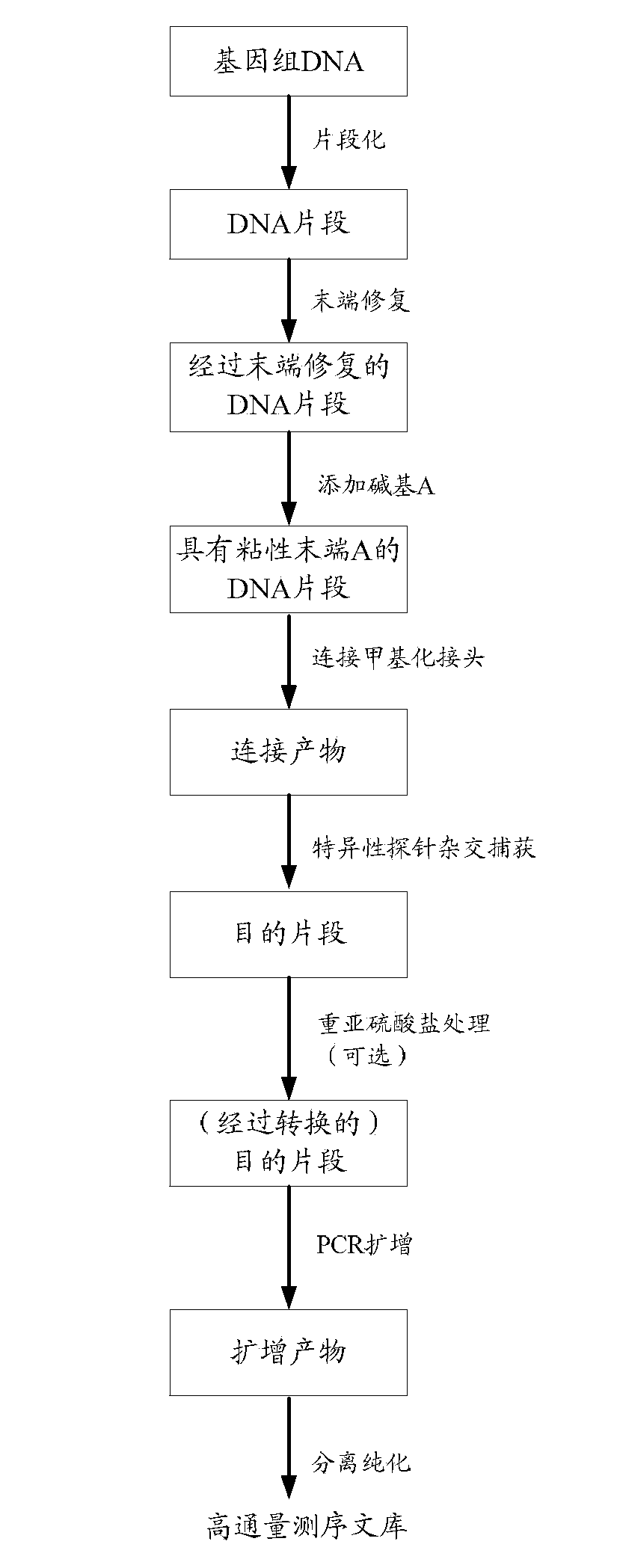
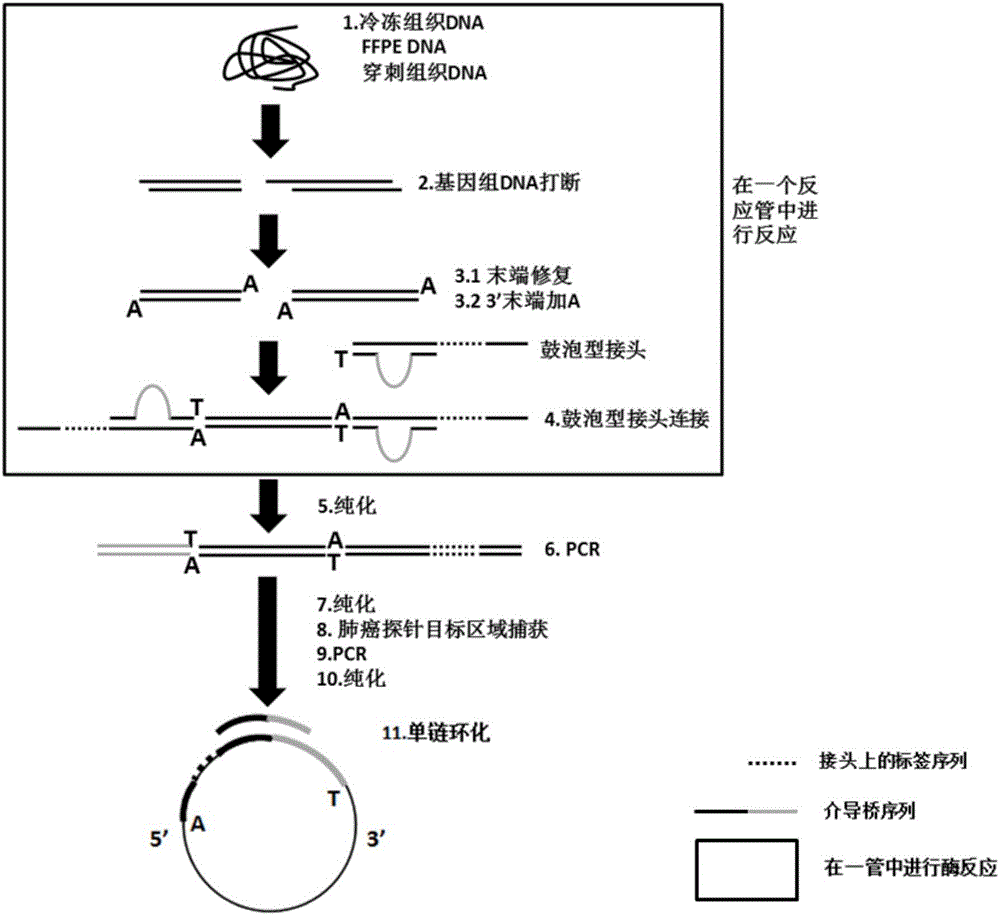
The invention discloses a construction method and an application of a high-throughout sequencing library. The construction method of the high-throughout sequencing library comprises the following steps: fragmenting genome DNA (deoxyribonucleic acid); repairing terminals of DNA fragments; adding a base A to 3' terminals of the DNA fragments with the repaired terminals; connecting the DNA fragments with sticky ends A to methylation linkers; carrying out hybrid capture on connecting products by using a specificity probe so as to obtain target fragments; carrying out bisulfite treatment on the target fragments so as to convert non-methylated cytosine in the target fragments to uracil; carrying out PCR (polymerase chain reaction) amplification on the converted target fragments; separating and purifying amplification products which form the high-throughout sequencing library. According to the method and application of the high-throughout sequencing library, the high-throughout sequencing library of a genome specific zone of a sample can be conveniently and efficiently constructed.
Owner:BGI SHENZHEN CO LTD +1
Building method of library for detecting non-small cell lung cancer gene mutation and kit
PendingCN106497920AImprove recycling efficiencyImprove utilization efficiencyMicrobiological testing/measurementLibrary creationGene targetingGene mutation
The invention discloses a building method of a library for detecting non-small cell lung cancer gene mutation and a kit. The method includes: using tubular reaction to complete genome DNA breaking and connector connection, performing hybrid capture on connection products after amplification and non-small cell lung cancer related gene target area probes, and performing BGISEQ-500 / 1000 platform sequencing and data analysis to obtain mutation conditions. The method has the advantages that the experiment flow is optimized greatly by the tubular reaction, operation complexity and time are reduced, and the requirements on clinical sample initial amount are lowered; multiple genes and multiple sites can be detected in one step, point mutation, insertion and deletion, structural variation and copy number variation are covered, the detecting result is accurate and overcomes the defect that a PCR capture method cannot detect the structural variation in one step, and the effectiveness of the high-throughput sequencing applied to the detection of the non-small cell lung cancer gene mutation; the method is wide in coverage, high in cost performance, capable of providing a reference basis for the diagnosing, treatment and drug use performed by doctors, and the method is suitable for being popularized and used in a large-scale manner.
Owner:BGI BIOTECH WUHAN CO LTD
Detection of nucleic acids by target-specific hybrid capture method
InactiveUS7601497B2Rapid and sensitive and accurateFunction increaseSugar derivativesMicrobiological testing/measurementNucleic acid detectionDNA
Target-specific hybrid capture (TSHC) provides a nucleic acid detection method that is not only rapid and sensitive, but is also highly specific and capable of discriminating highly homologous nucleic acid target sequences. The method produces DNA:RNA hybrids which can be detected by a variety of methods.
Owner:QIAGEN GAITHERSBURG
Gene group and kit for diagnosing lung caner, and diagnosis method thereof
InactiveCN107475370AExcellent detection depthExcellent detection accuracyMicrobiological testing/measurementLibrary creationA-DNABlood plasma
The invention relates to the field of genetic engineering and biotechnologic detection, and concretely relates to a gene group and a kit for diagnosing lung cancer, and a diagnosis method thereof. The gene group for diagnosing the lung cancer includes ABCB1, AKT1, ALK, APC, ATIC and other 63 genes. The method using the gene group to diagnose the lung cancer comprises the following steps: (1) extracting free DNA and genome DNA from the plasma of a sample to be detected; and (2) breaking the genome DNA into fragments with the length of 150-250 bp, carrying out hybrid capture on the broken genome DNA and the free DNA to construct a DNA library, carrying out online sequencing, and analyzing the obtained sequencing result. Exon and partial intron regions of the 68 genes of the free DNA are enriched at one time by a probe capture technology to realize multi-gene and multi-target parallel deep high-throughput sequencing with high accuracy, optimize the detection flow and improve the detection precision, so liquid biopsy, low-frequency detection and tumor diagnosis become possible.
Owner:天津脉络医学检验有限公司
Process for microsatellite instability detection
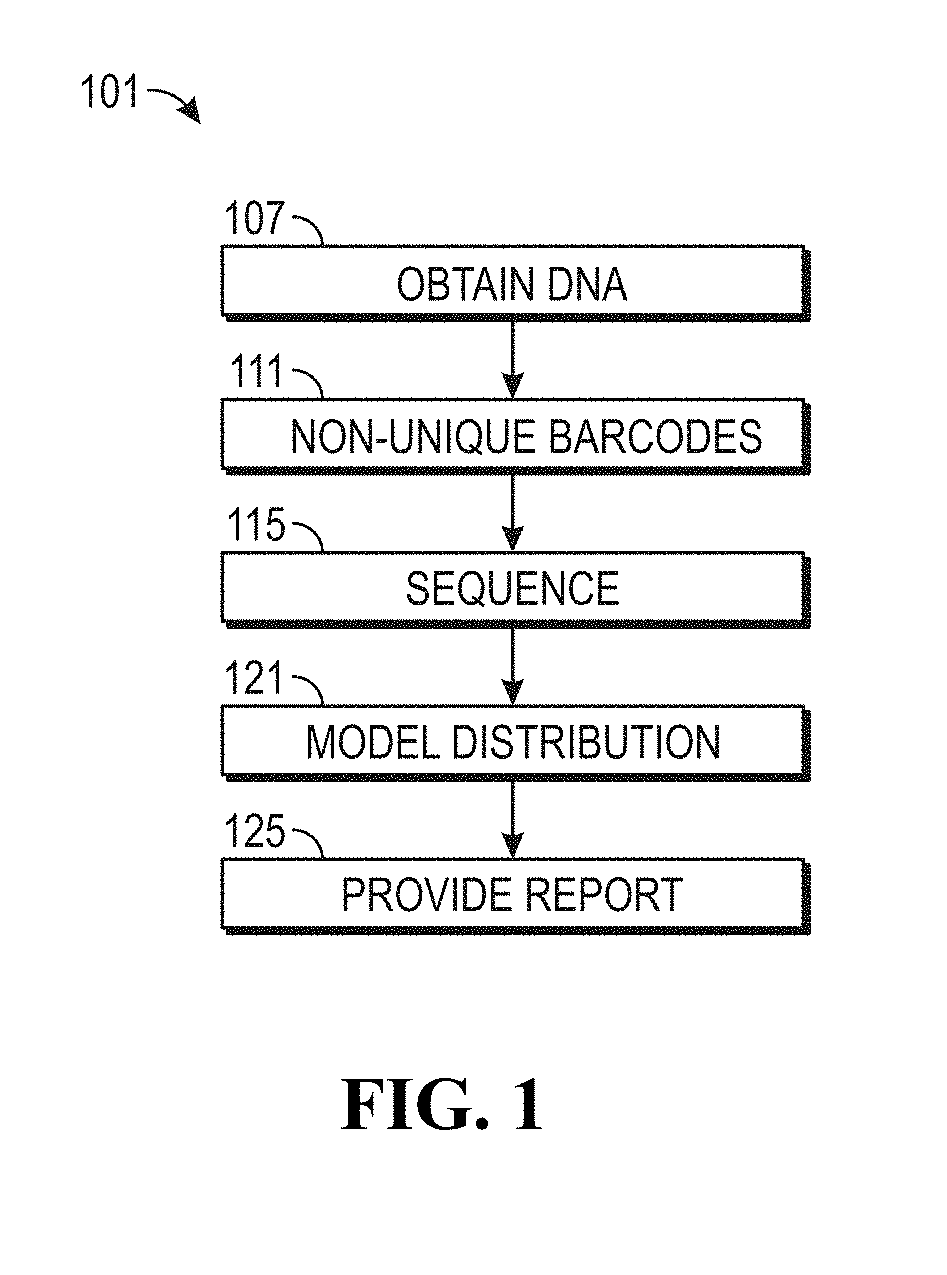
ActiveUS20190169685A1Useful in detectionIncrease the burdenTherapiesHealth-index calculationCell freeNucleotide
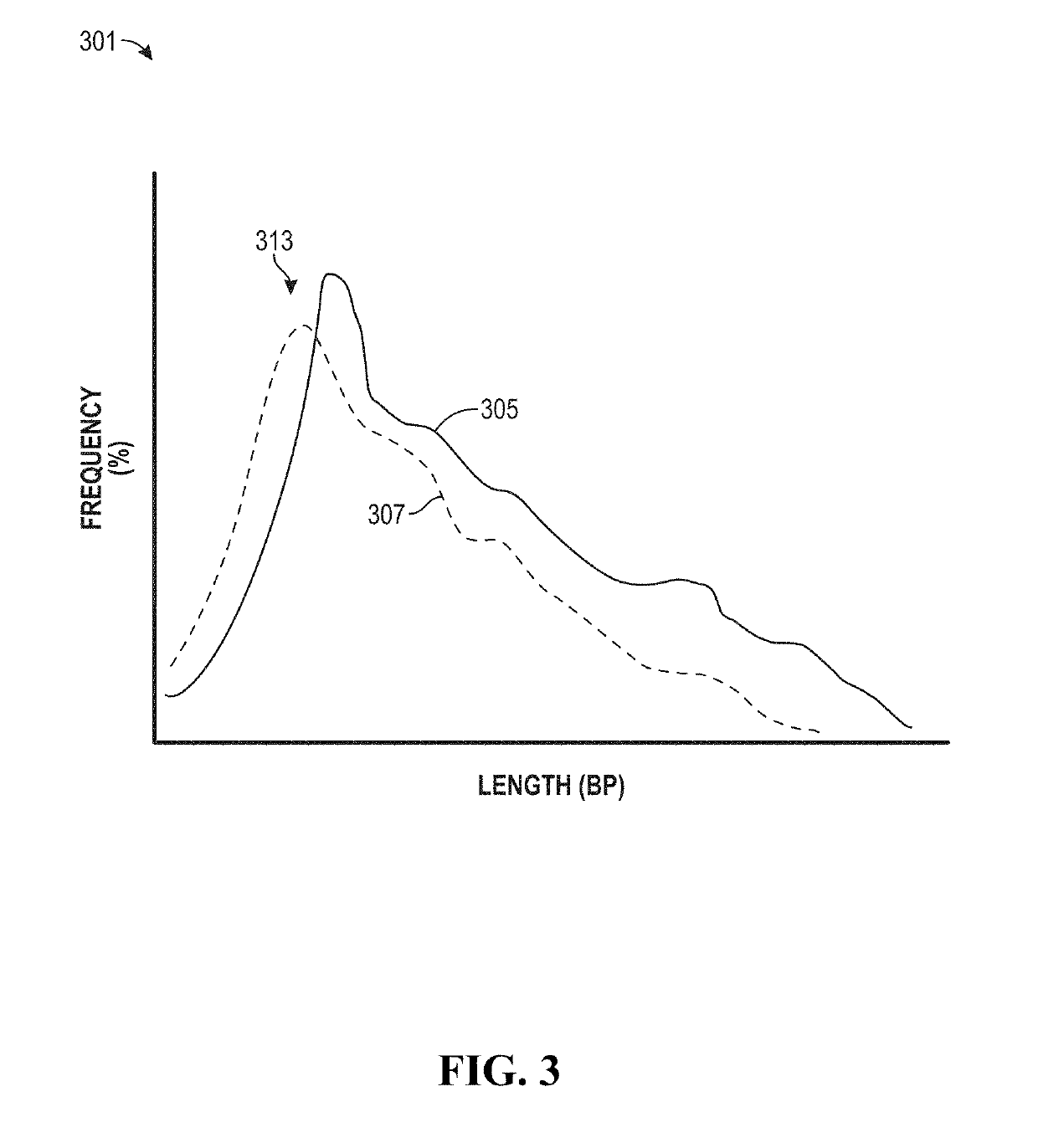
The invention provides methods for determining the MSI status of a patient by liquid biopsy with sample preparation using hybrid capture and non-unique barcodes. In certain aspects, the invention provides a method of detecting microsatellite instability (MSI). The method includes obtaining cell-free DNA (cfDNA) from a sample of blood or plasma from a patient and sequencing portions of the cfDNA to obtain sequences of a plurality of tracts of nucleotide repeats in the cfDNA. A report is provided describing an MSI status in the patient when a distribution of lengths of the plurality of tracts has peaks that deviate significantly from peaks in a reference distribution.
Owner:PERSONAL GENOME DIAGNOSTICS INC
Nucleic acid detection method combining RNA amplification with hybrid capture method
InactiveCN103525942AEnable high-throughput detectionNot easy to inactivateMicrobiological testing/measurementBiotin-streptavidin complexStreptolydigin
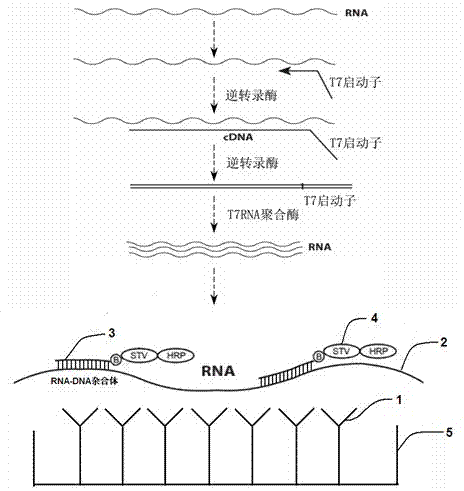
The invention relates to a nucleic acid detection method combining RNA amplification with a hybrid capture method. The nucleic acid detection method includes the following steps of extracting nucleic acid from a sample or splitting the sample to release the nucleic acid to be detected, conducting transcription and amplification on the nucleic acid to be detected to obtain an RNA product, adding the obtained RNA product together with a DNA probe marked with biotins to a solid phase so that hybridization can be conducted, forming a DNA / RNA heterozygote through hybridization, coating the solid phase with specific antibodies for recognizing the heterozygote or avidin or streptavidin, capturing the heterozygote through the solid phase, and adding avidin or streptavidin or antibodies for recognizing the DNA / RNA heterozygote to the solid phase to achieve obtaining of a nucleic acid signal, wherein the avidin or the streptavidin or the antibodies for recognizing the DNA / RNA heterozygote are marked with enzymes with the secondary amplification capacity. According to the nucleic acid detection method, the inherent pollution problem of the PCR is avoided, RNA molecules can be directly detected, inverse transcription and pre-dissociation of RNA do not need to be conducted, and the operation steps are greatly simplified.
Owner:武汉中帜生物科技股份有限公司
Methods and compositions for sequence-specific purification and multiplex analysis of nucleic acids
InactiveUS20120045756A1Microbiological testing/measurementAgainst vector-borne diseasesMultiplexNucleic acid detection
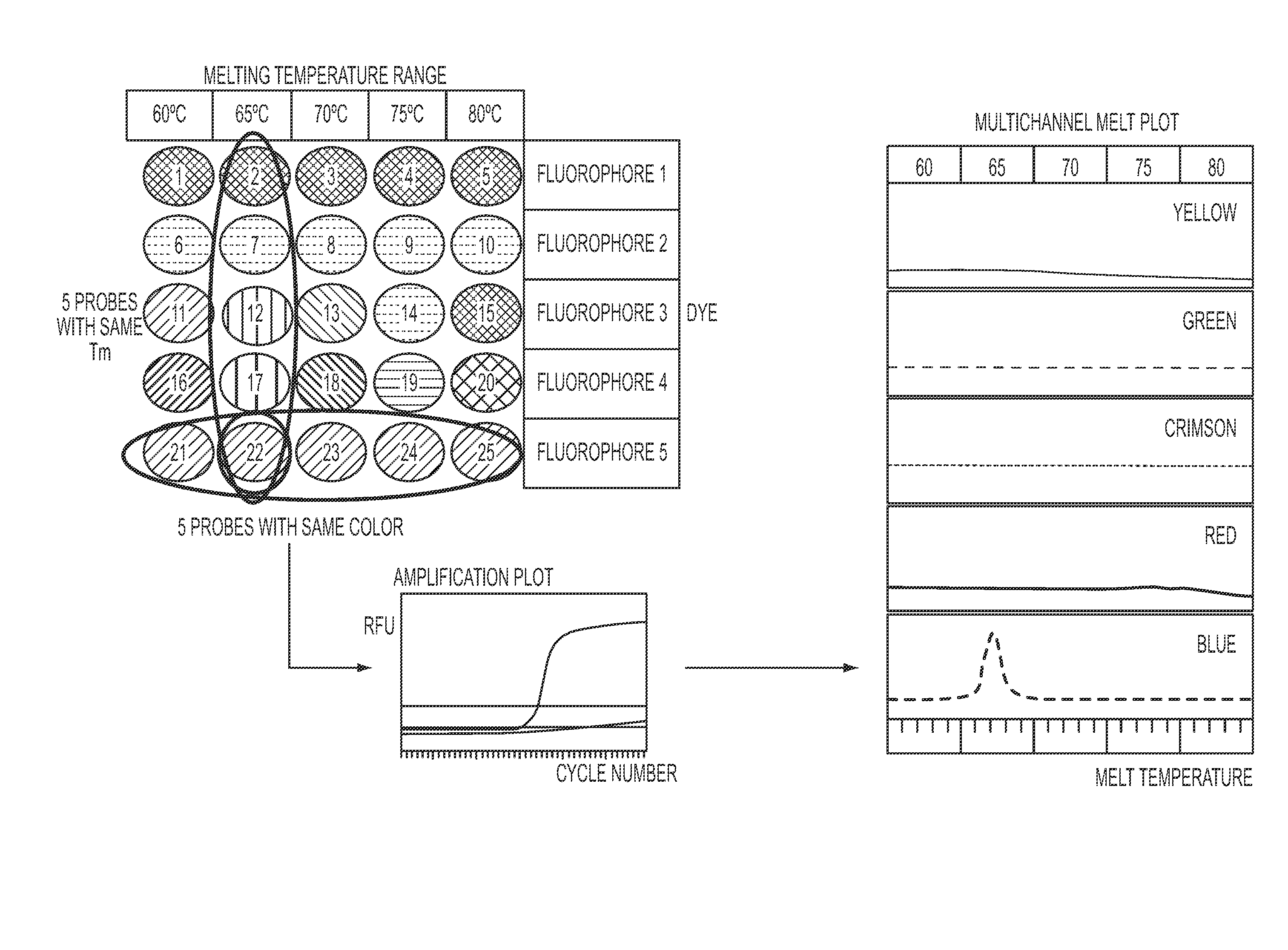
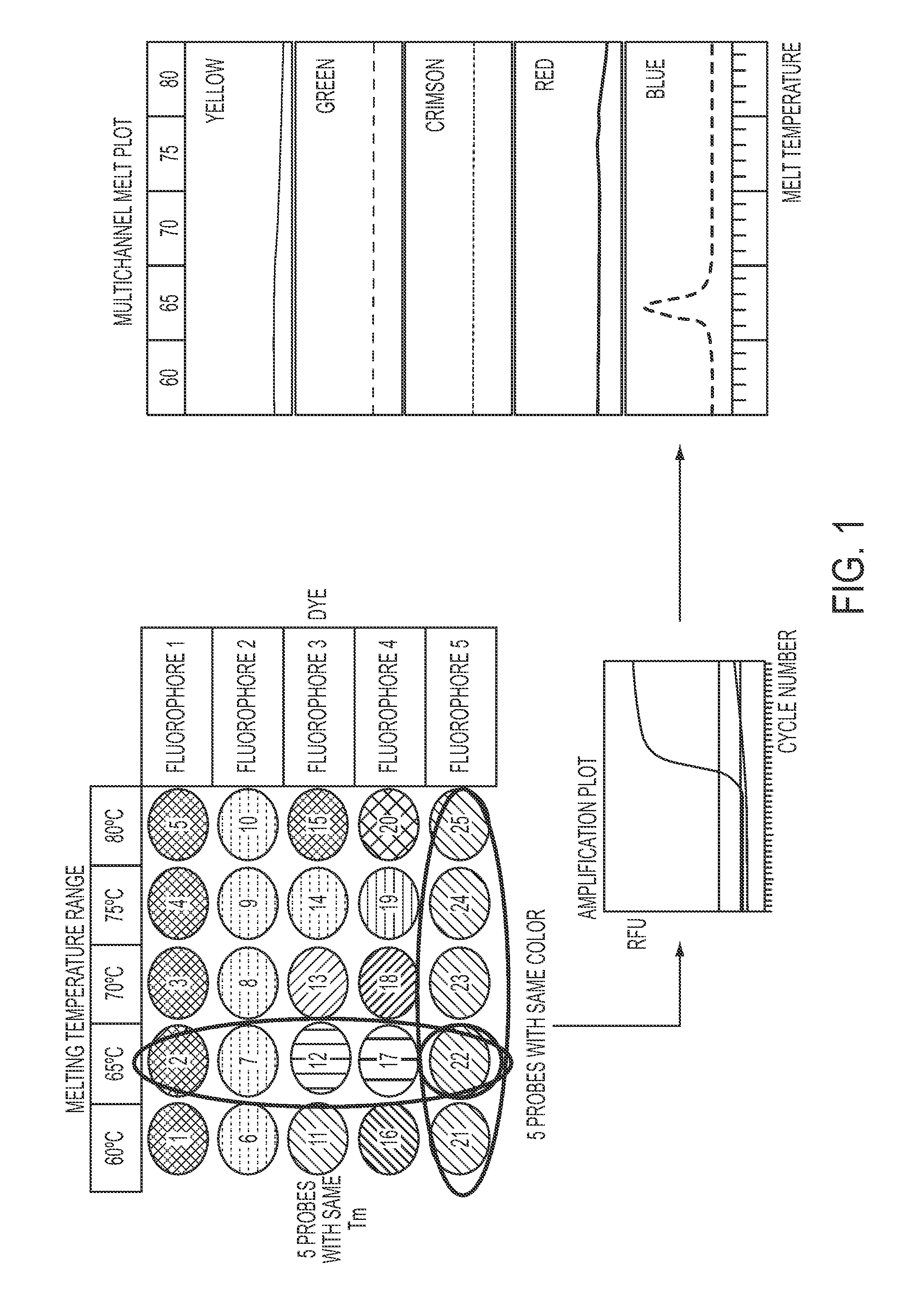
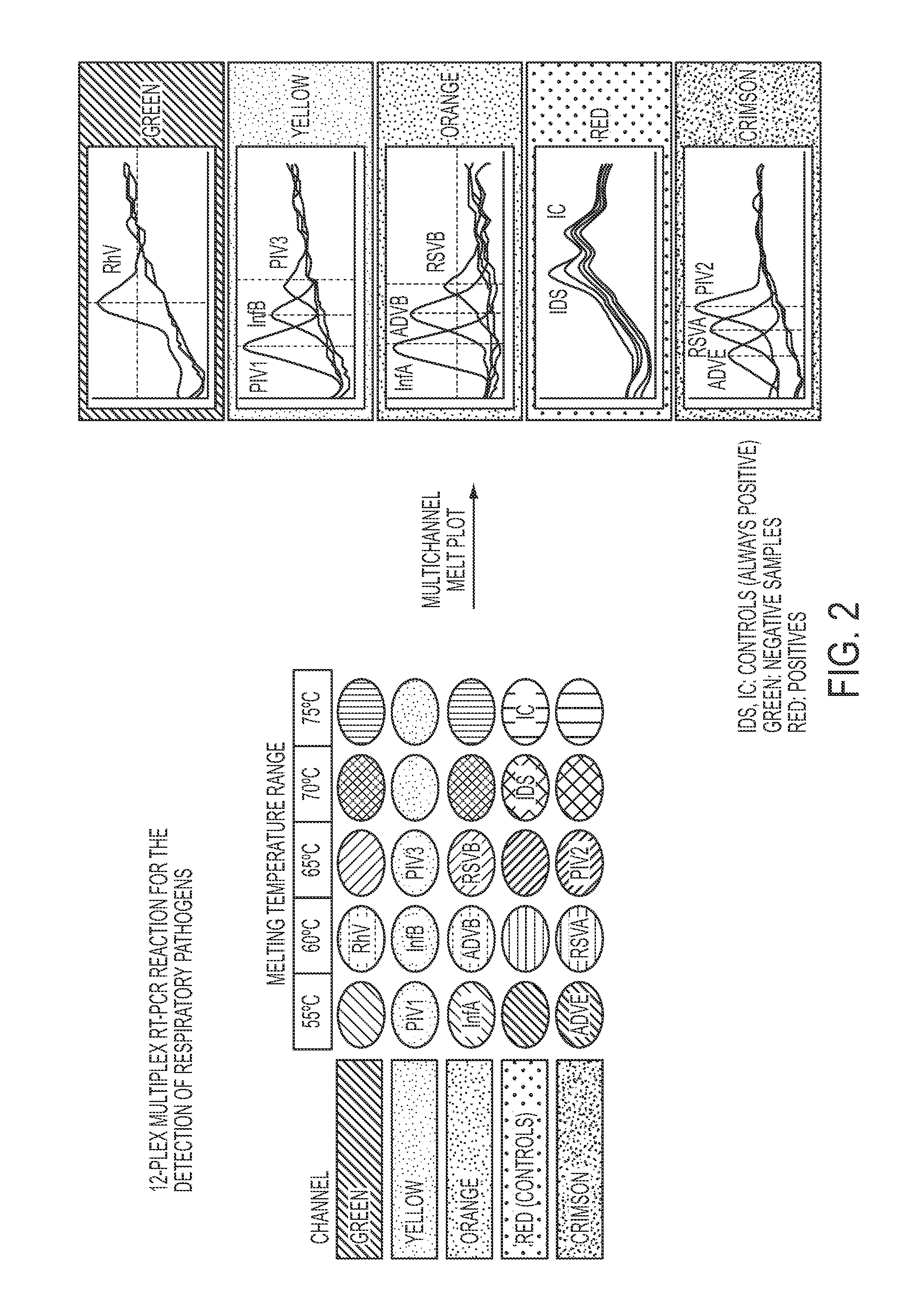
Methods and materials for determining the presence of at least one nucleic acid in a sample are provided, said methods comprising (1) a purification step using sequence specific hybrid capture; (2) an amplification step; and (3) a detection step comprising contacting the target nucleic acid with a plurality of detectably labeled nucleic acid detection probes, wherein each (a) bears a different detectable label from the other detection probes, and / or (b) has a different melting temperature from probes bearing the same detectable label. Also disclosed are compositions and kits for use in such a method.
Owner:QIAGEN GAITHERSBURG
Liquid-phase hybrid capture enriching liquor for genome DNA sequencing library and hybrid method adopting liquid-phase hybrid capture enriching liquor
InactiveCN104178478AThe method is simpleSimple designMicrobiological testing/measurementDNA preparationEthylene diaminePhosphate
The invention discloses a liquid-phase hybrid capture enriching liquor for a genome DNA sequencing library and a hybrid method adopting the liquid-phase hybrid capture enriching liquor. The hybrid enriching liquor comprises phosphate buffer, sodium citrate buffer (SSC), SDS (lauryl sodium sulfate), EDTA (ethylene diamine tetraacetic acid) and Denhardt liquor. The hybrid liquor and the corresponding hybrid method are low in cost, simple and convenient to implement; the hybrid time can be effectively shortened, the needed DNA fragment can be precisely extracted, influences of a background value on a signal value can be reduced, and the hybrid target enriching effects can be optimized.
Owner:GENESEEQ TECH INC
Method for fragmenting trace DNA sample and method for establishing DNA library by utilizing trace DNA sample
ActiveCN104946629AAvoid damagePlay a protective effectMicrobiological testing/measurementLibrary creationCDNA libraryFragment processing
The invention discloses a method for fragmenting a trace DNA sample and a method for establishing a DNA library by utilizing the trace DNA sample. The method for fragmenting the trace DNA sample comprises the steps that 1, Carrier RNA is added into the trace DNA sample to obtain a sample to be fragmented; 2, fragmenting processing is conducted on the sample to be fragmented. An inventor discovers that after the exogenetic Carrier RNA is added into the target DNA, that is to say, the target DNA is mixed with the Carrier RNA and then fragmenting is conducted on the mixture, the protective action to the target DNA can be achieved, the destructive action of a fragmenting instrument to the trace DNA is reduced to the maximum extent, and full gene establishing library sequencing and even hybrid capture sequencing can be conducted on DNA of a trace FFPE sample.
Owner:天津诺禾医学检验所有限公司
Hybrid capture kit and method for detecting mutation of breast cancer susceptibility genes BRCA1 and BRCA2
ActiveCN106676169AImplement mutation detectionLow costMicrobiological testing/measurementBreast cancer susceptibility genesMutation detection
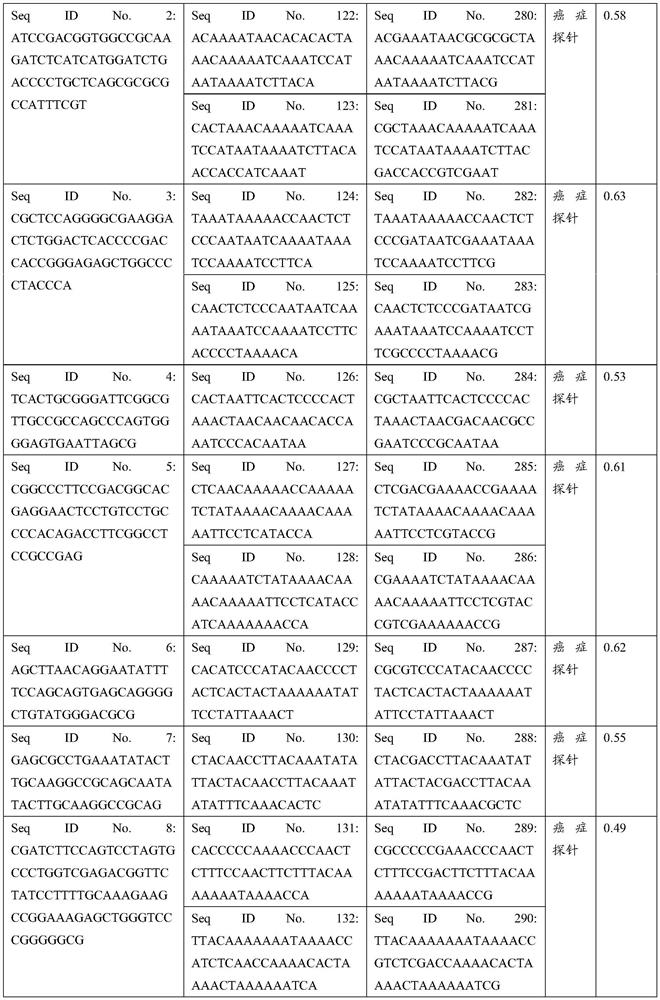
The invention discloses a hybrid capture kit for detecting mutation of breast cancer susceptibility genes BRCA1 and BRCA2. The hybrid capture kit comprises a hybridization reagent, a PCR amplification reagent and an enrichment degree detection reagent, wherein the hybridization reagent comprises probe mixtures from SEQ NO.1 to SEQ NO. 173, and the mole ratio is SEQ NO.1: SEQ NO. (2-172): SEQ NO. 173=1: 1: 1. During detection, the use amount of each of the probe mixtures with the concentration of 2nM is 1 [mu]l. The kit disclosed by the invention can be used for high-sensitivity, high-flux, low-cost and easy-operation full-explicit mutation detection for the genes BRCA1 and BRCA2, and can assist a clinical doctor in screening out individuals with the breast cancer susceptibility risk to fulfill the aim of preventing occurrence of breast cancers.
Owner:SHANGHAI PERSONAL BIOTECH
Methods and compositions for sequence-specific purification and multiplex analysis of nucleic acids
InactiveUS9422593B2Sugar derivativesMicrobiological testing/measurementNucleic acid detectionBiology
Methods and materials for determining the presence of at least one nucleic acid in a sample are provided, said methods comprising (1) a purification step using sequence specific hybrid capture; (2) an amplification step; and (3) a detection step comprising contacting the target nucleic acid with a plurality of detectably labeled nucleic acid detection probes, wherein each (a) bears a different detectable label from the other detection probes, and / or (b) has a different melting temperature from probes bearing the same detectable label. Also disclosed are compositions and kits for use in such a method.
Owner:QIAGEN GAITHERSBURG
Non-amplification nucleic acid hybrid capture system and application thereof in nucleic acid test
InactiveCN107523611AFast detection methodLow costMicrobiological testing/measurementNucleic acid chemistryNucleic acid detection
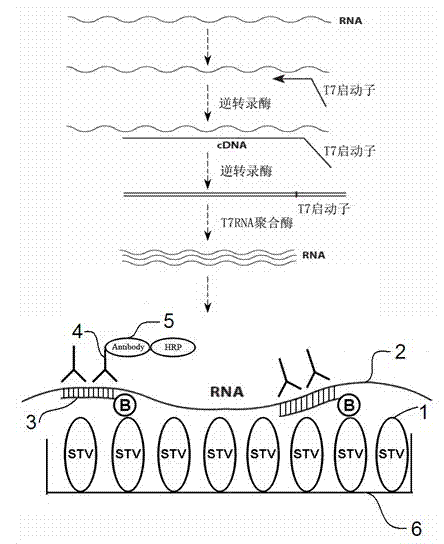
The invention belongs to the technical field of molecular biology, immunology and nucleic acid chemistry and in particular relates to a non-amplification nucleic acid hybrid capture system and application thereof. The kit is an immunoassay method for capturing hybrid nucleic acids based on a non-amplification anti-nucleic antibody; inverse transcription, PCR (Polymerase Chain Reaction) and other amplification technologies are not needed in the detection process, while a probe complementary to a to-be-detected nucleic acid is subjected to annealing hybridization with the to-be-detected nucleic acid so as to form a structure which can be recognized by the anti-nucleic antibody, and the detection signal is amplified by utilizing enzyme linked immunosorbent assay. The detection method has the advantages of being rapid, low in cost, high in sensitivity, high in specificity, free in complicated amplification and detection instruments and capable of detecting initial viral load.
Owner:NANJING UNIV
Application of bridge type oligonucleotides to library target region capturing
ActiveCN108676846AImprove bindingOmit controllabilityMicrobiological testing/measurementLibrary creationBridge typeDesign strategy
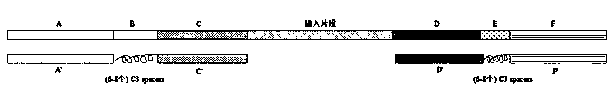
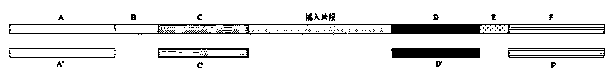
The invention discloses an improved sequence capturing method, which comprises the steps of 1, extracting DNA; breaking the DNA; 2, connecting the two ends of the broken DNA with a connector, whereinthe remote end of a connector sequence at one end of the broken DNA includes a tag sequence with the length being 6 to 8nt; 3, performing Pre-PCR on the DNA of the connecting connector; 4, mixing a probe and a sealing reagent; capturing the Pre-PCR, wherein the sealing reagent includes sealing sequences complementary with sequences except the broken DNA, and the tag sequences are sealed by C3 spacer arms with the same length; 5, after the hybrid capture, performing PCR amplification. A bridge type sealed design strategy is used; corresponding sealing sequences are respectively designed for connector sequences at two ends of inserting fragments; the tag sequences at the middle part are subjected to bridge type connection by the C3 spacer arms; the cost is reduced; the complexity in the library building process is simplified.
Owner:IGENETECH BIOTECH (BEIJING) CO LTD
Preparation method of modified DNA (deoxyribonucleic acid) hybridization probe for targeted hybrid capture
ActiveCN105647907AImprove bindingLow environmental requirementsMicrobiological testing/measurementDNA preparationHybridization probeEnzyme digestion
Owner:HANGZHOU LC BIOTECH
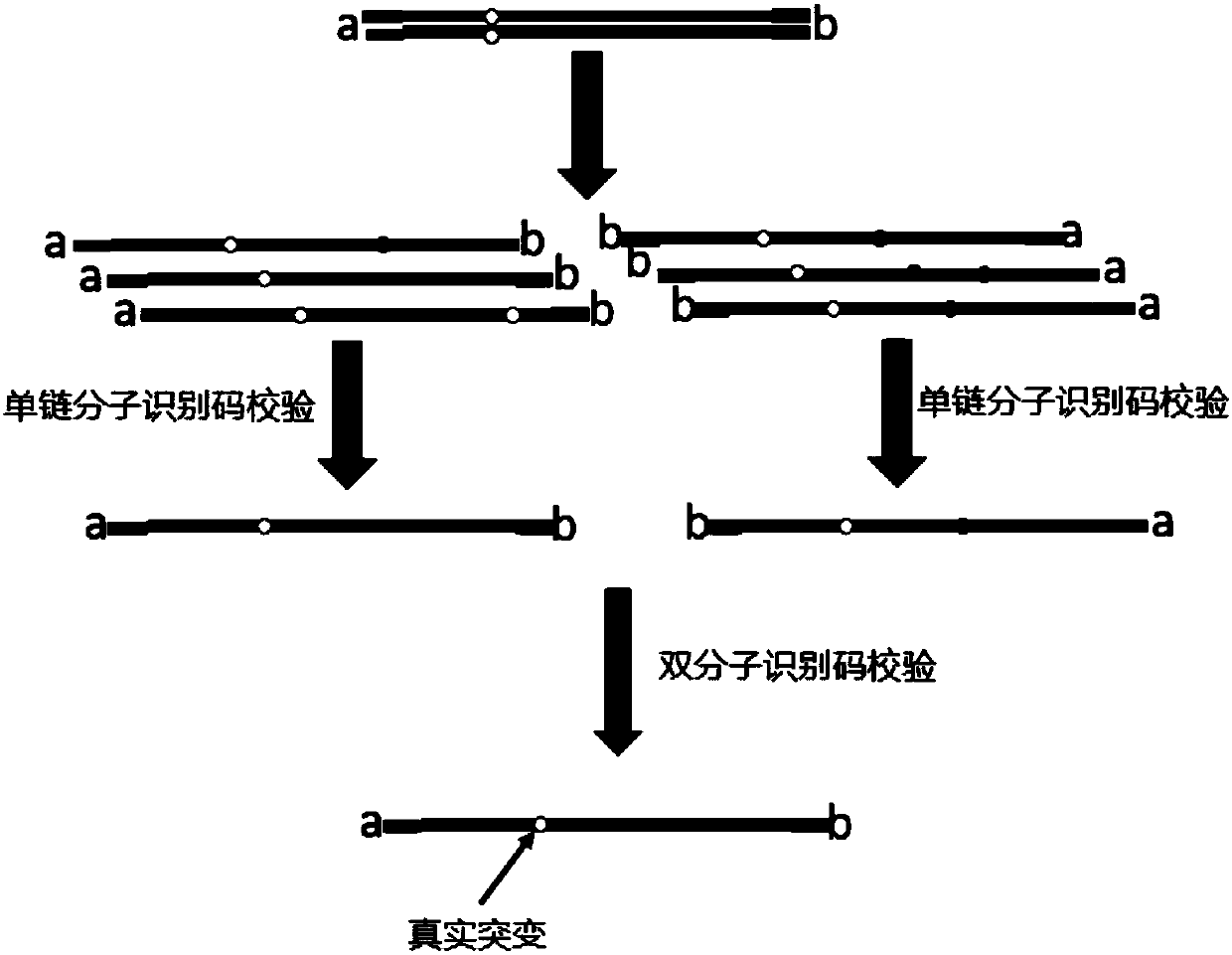
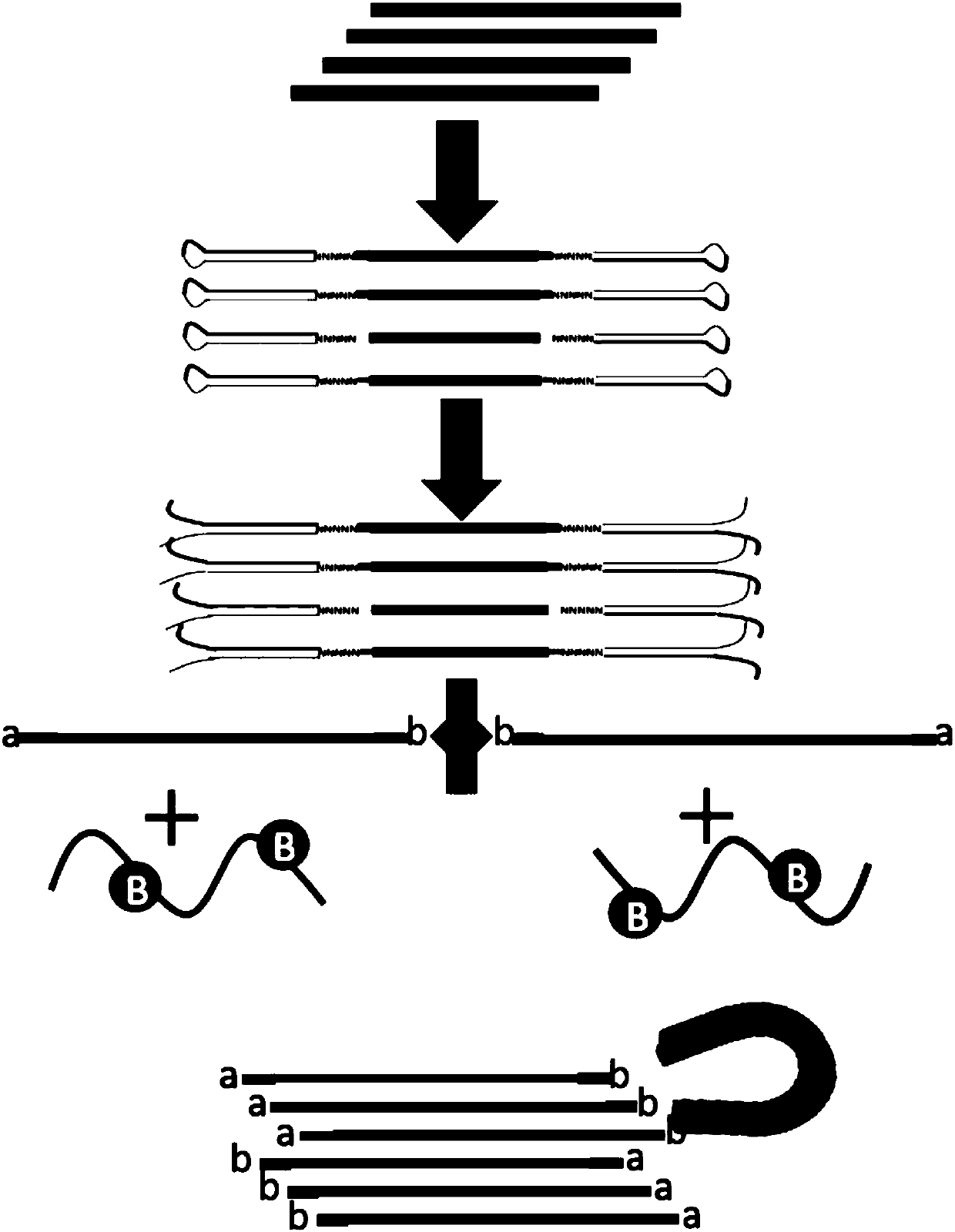
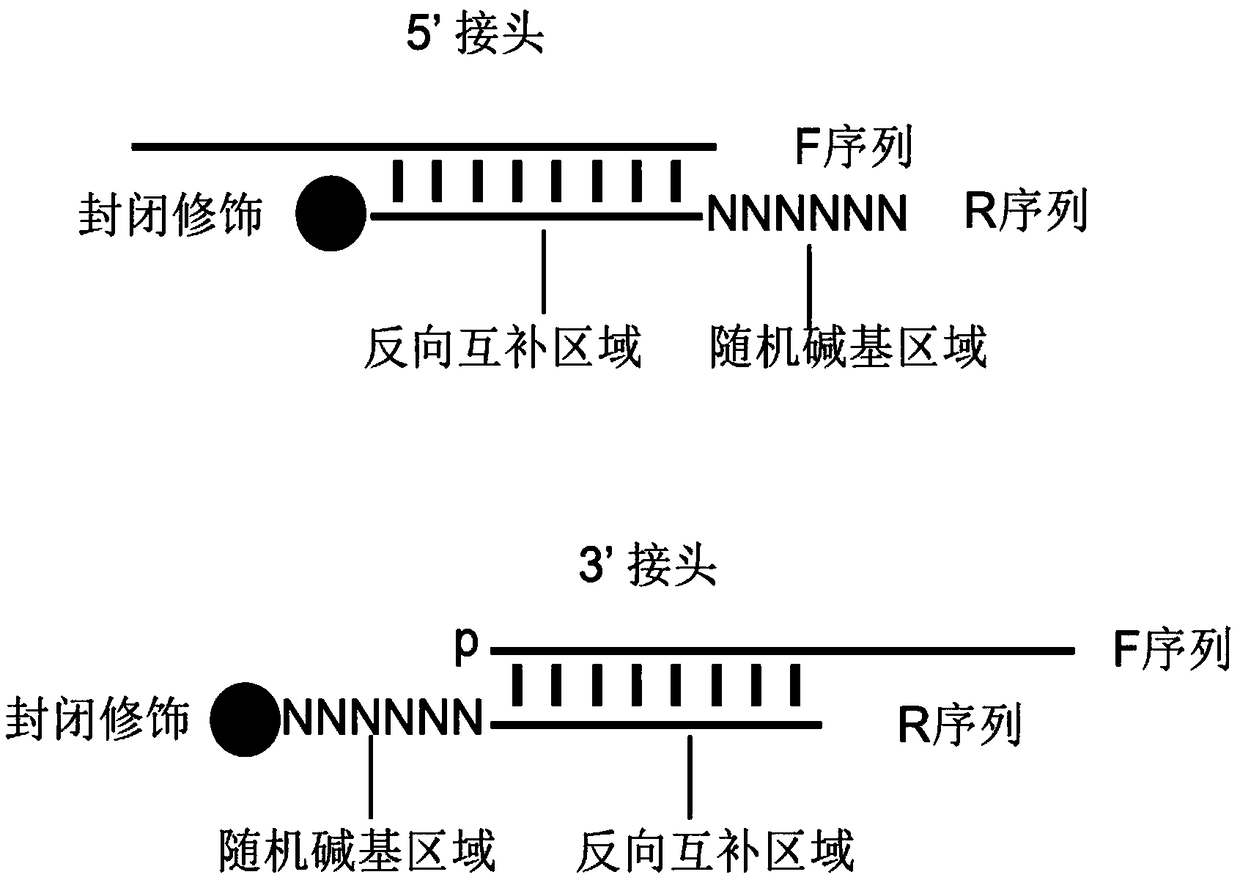
Second-generation sequencing method for dimolecular self-checking library preparation and hybrid capture used for trace DNA ultralow frequency mutation detection
ActiveCN107604046AIncrease profitHigh yieldNucleotide librariesMicrobiological testing/measurementLibrary preparationData error
The invention discloses a second-generation sequencing method for dimolecular self-checking library preparation and hybrid capture used for trace DNA ultralow frequency mutation detection. The methodcomprises the following steps: extracting plasma free DNA, performing DNA chemical error repair, preparing a self-checking dimolecular identification code hairpin type joint, repairing plasma free DNA, connecting DNA with the joint, and performing Pre-PCR amplification, excessive hybrid capture, Post-PCR amplification, computer sequencing, data error correction, and mutation analysis and annotation. The method disclosed by the invention can efficiently realize low frequency mutation detection of plasma free DNA. DNA error repair and dual redundancy checking technology can enable the method tohave ultralow false positive rate and high sensitity while detecting trace samples, thus avoiding defects of existing plasma circulating free DNA detection methods, not only realizing cancer mutationdetection and targeted medication guidance, but also realizing early screening of genetic and birth defects of fetus.
Owner:SHANGHAI DYNASTYGENE CO
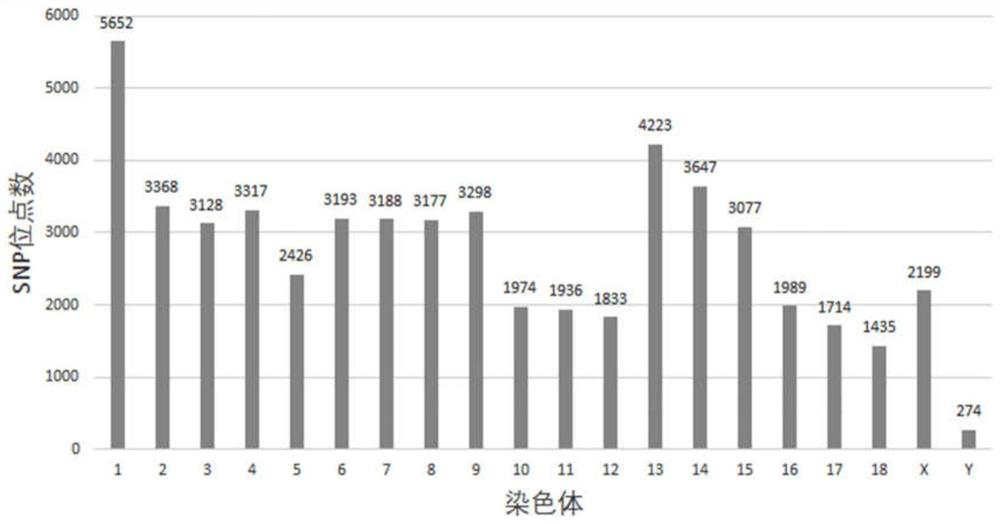
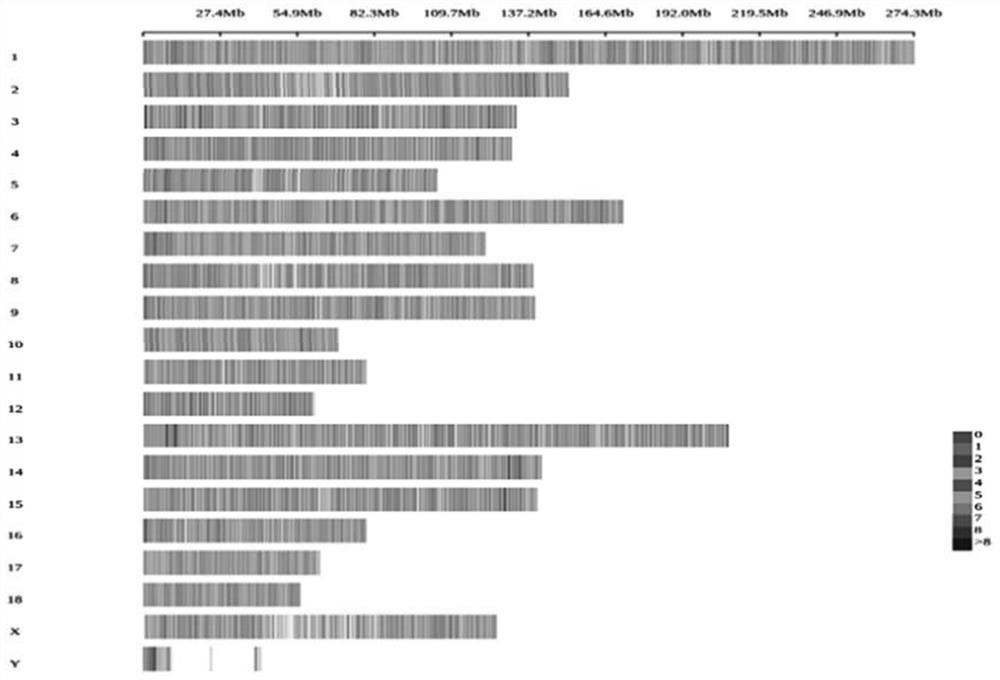
Pig 50K liquid phase chip based on targeted capture sequencing, and application thereof
ActiveCN112921076AAvoid uniformityGuaranteed specific capture rateNucleotide librariesMicrobiological testing/measurementPig farmsNucleotide
The invention relates to a pig 50K liquid phase chip based on targeted capture sequencing, and application thereof. The pig 50K liquid phase chip is composed of pig 50K probe mixed liquor and a hybridization capture reagent which are independently packaged; pig 50K probes are DNA double-stranded probes and are nucleotide sequences designed and synthesized according to screened SNP sites; and the SNP sites are screened by comparing the sequencing results of the whole genome of the pig to a pig reference genome. The pig 50K liquid phase chip prepared by the invention is mixed with a pig DNA high-throughput sequencing library; DNA fragments containing target sites in the pig DNA high-throughput sequencing library are captured for amplification and purification; and, after a product is subjected to high-throughput sequencing, a sequencing result is replied to a pig reference genome for comparison, so that the genotyping of the genome of a to-be-detected pig is obtained. When the 50K liquid phase chip prepared by the invention is used for genotyping pigs; and the problems of high cost, inflexibility and incapability of large-scale use in pig farms in China in the prior art can be solved.
Owner:CHINA AGRI UNIV
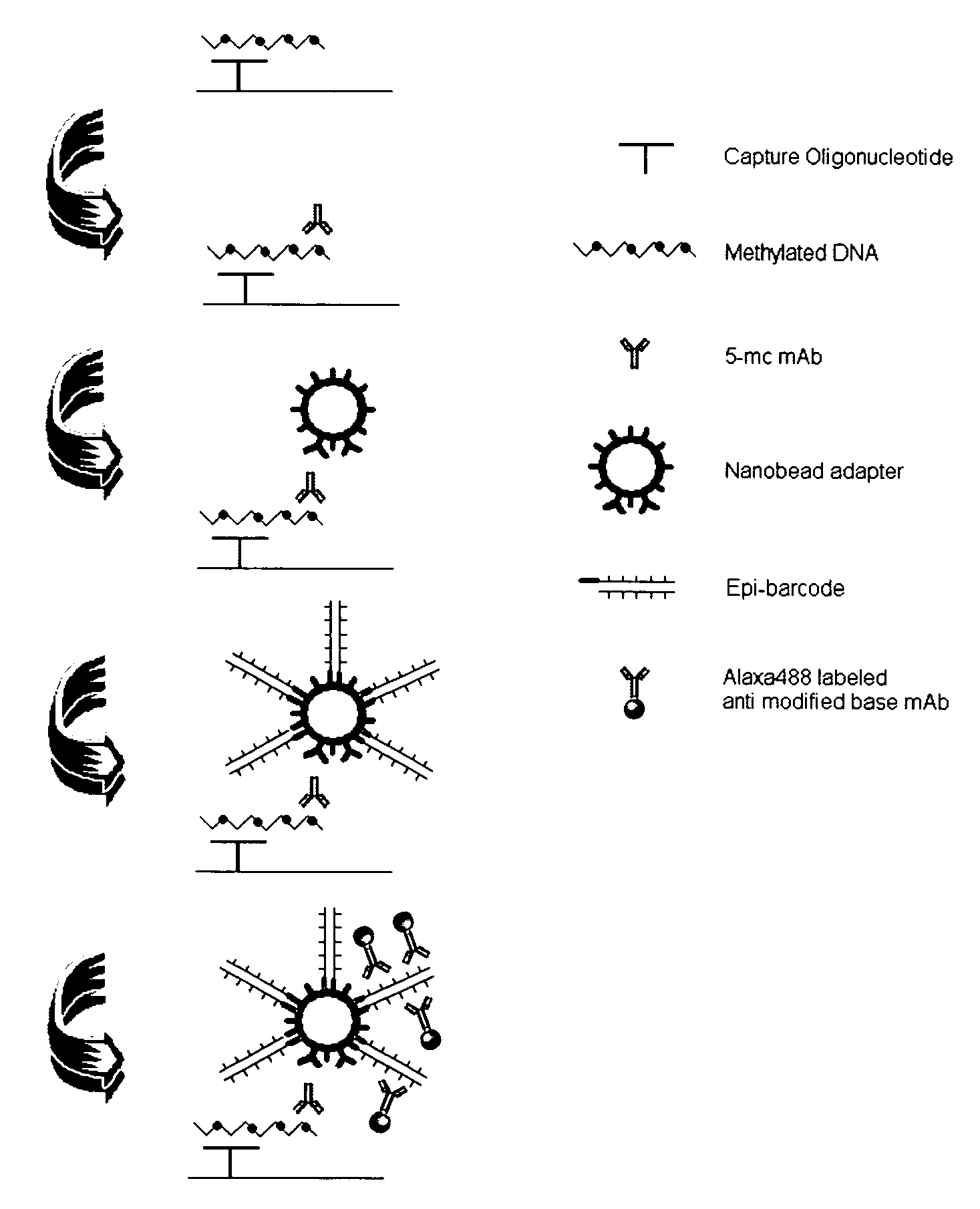
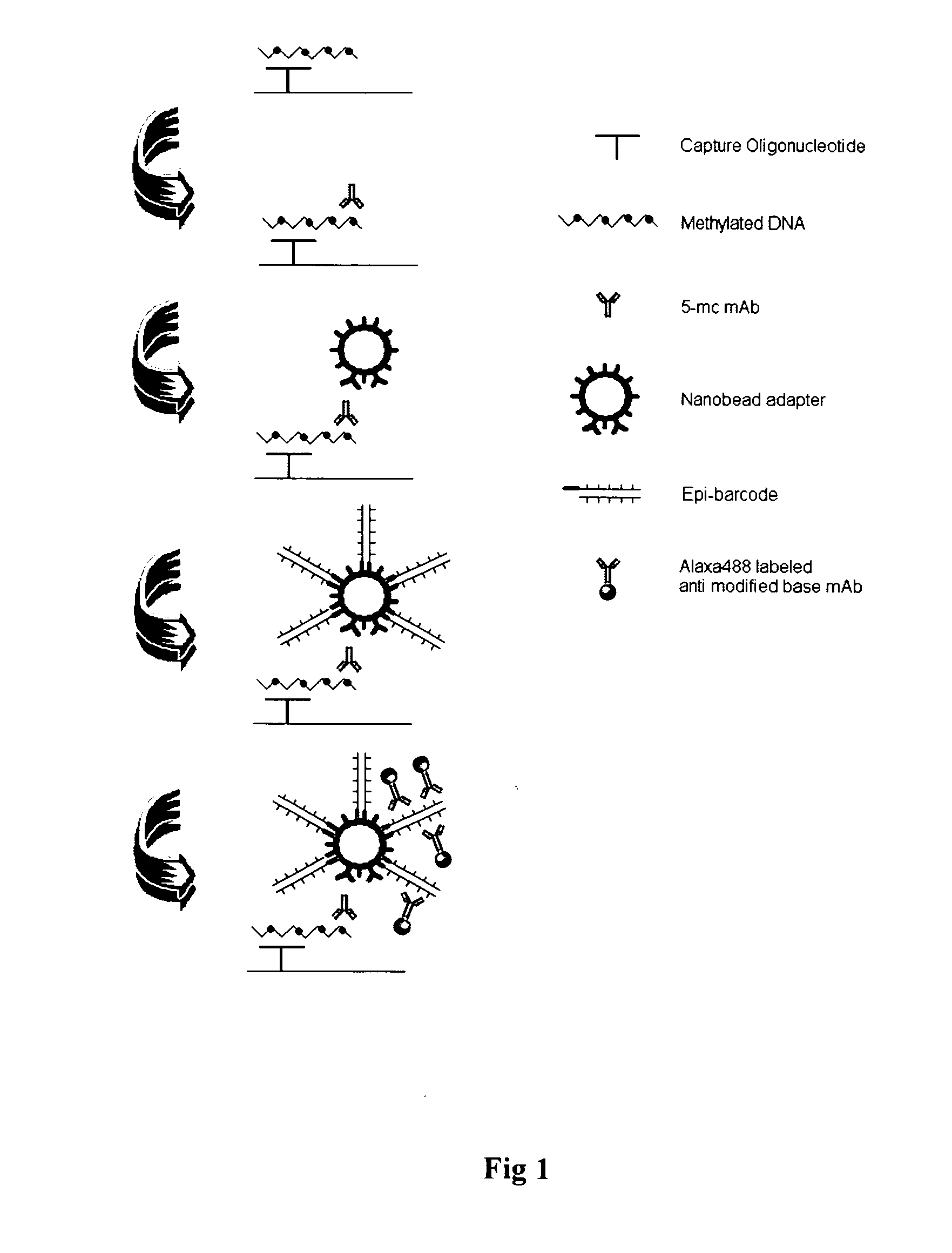
DNA methylation specific signal amplification
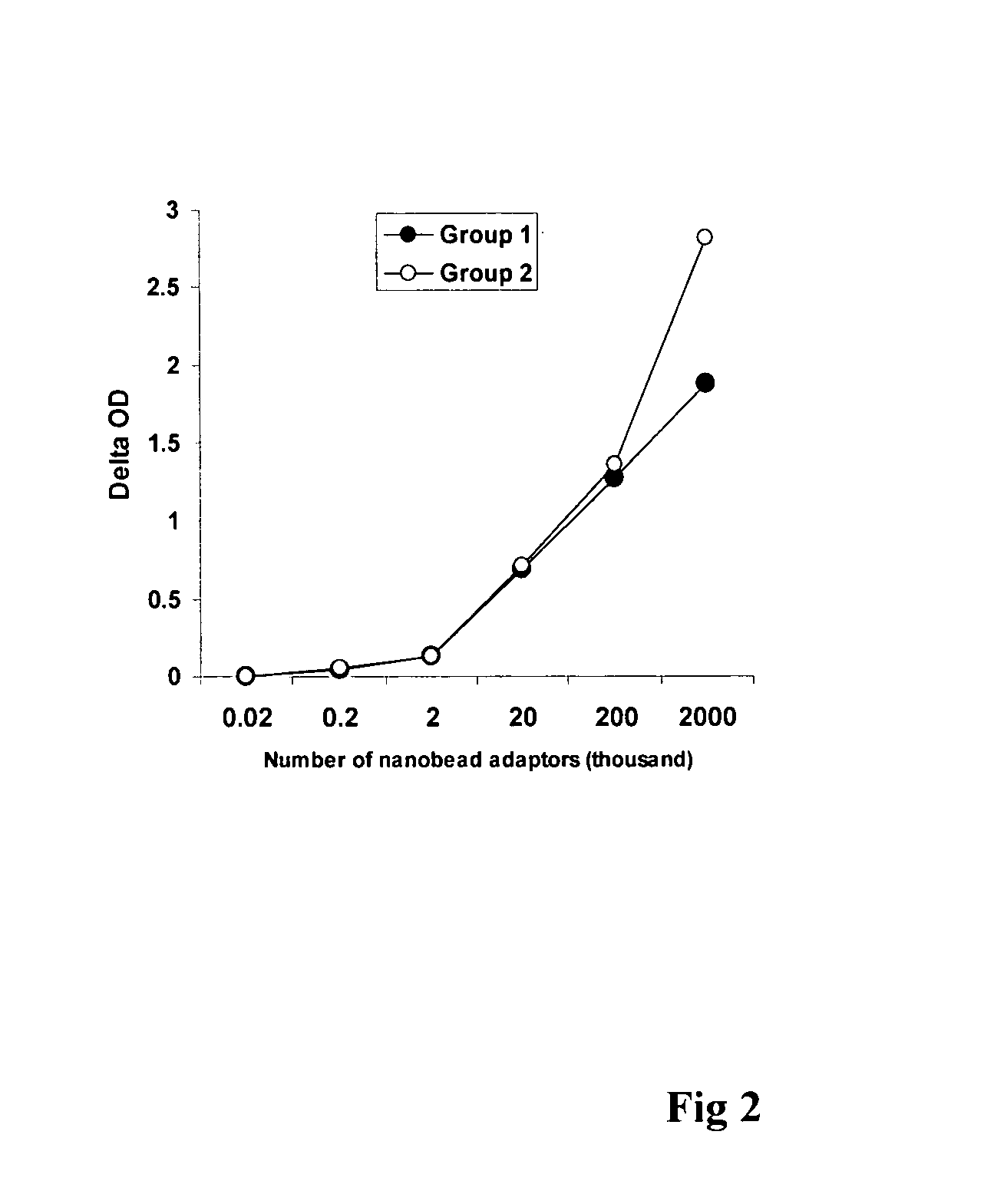
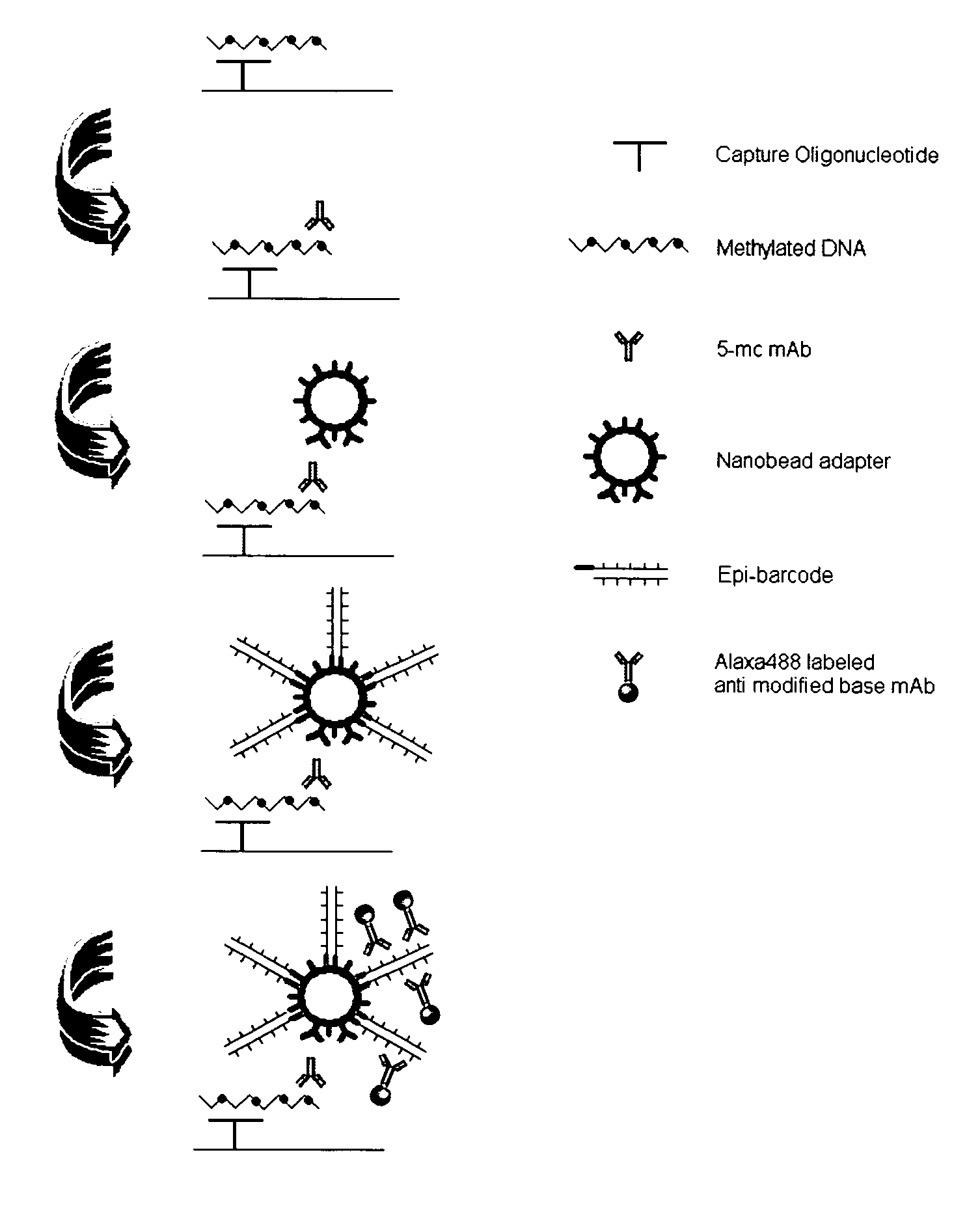
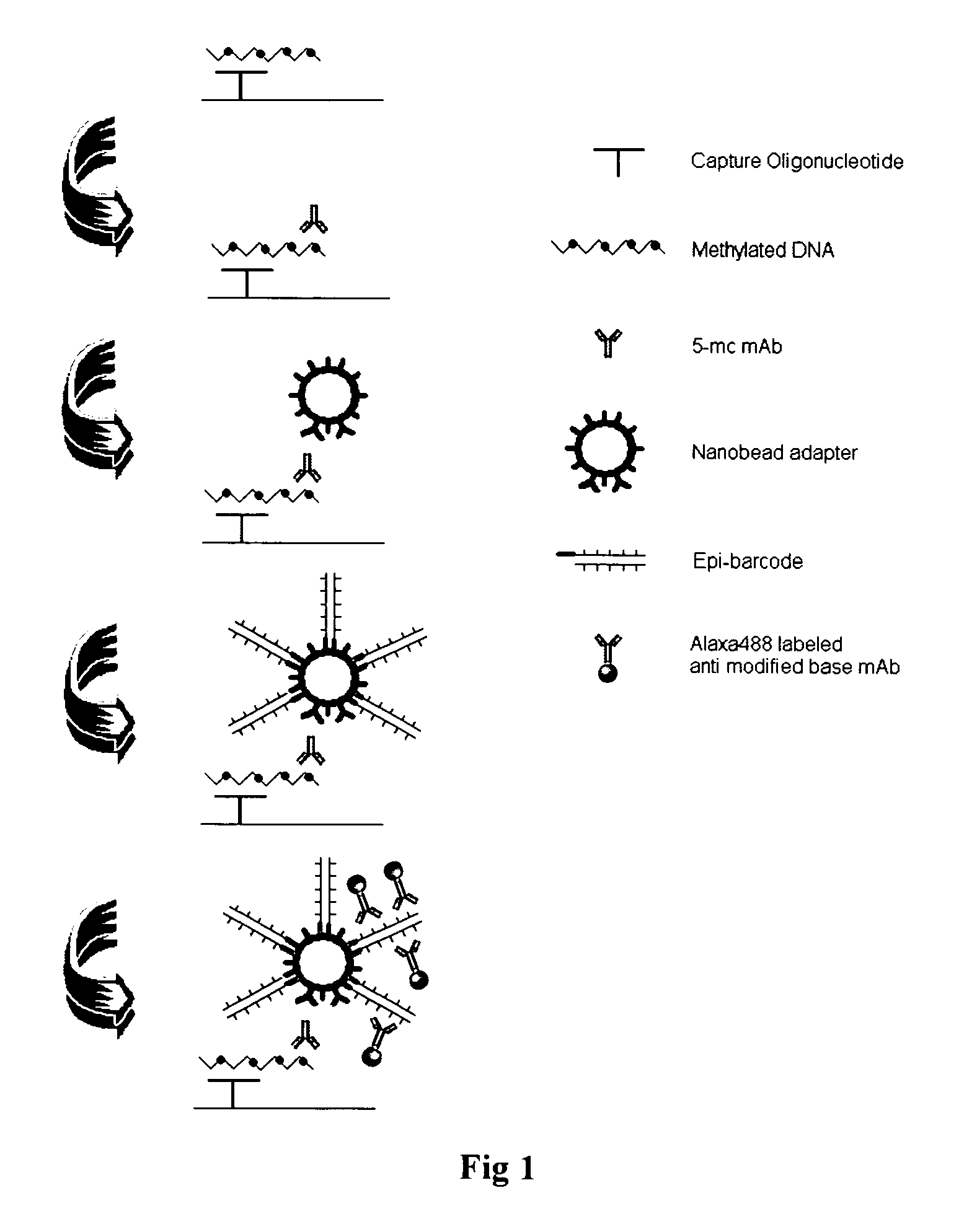
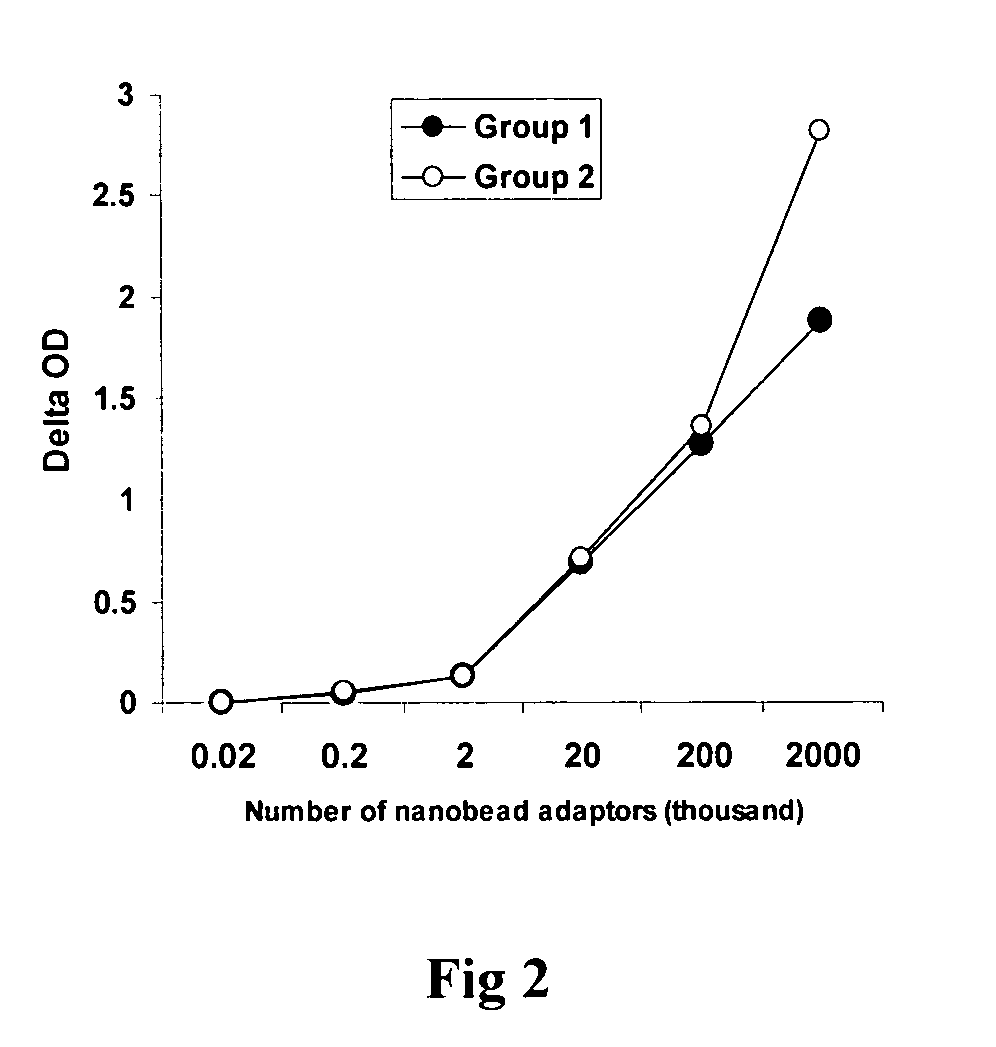
InactiveUS20090181373A1Quick checkAvoid contaminationMicrobiological testing/measurementDNA methylationBarcode
Owner:LI WEIWEI +1
DNA methylation specific signal amplification
InactiveUS7608402B2Quick checkAvoid contaminationMicrobiological testing/measurementDNA methylationBarcode
Owner:LI WEIWEI +1
Detection of nucleic acids by target-specific hybrid capture method
InactiveUS20100036104A1Rapid and sensitive and accurateFunction increaseSugar derivativesMicrobiological testing/measurementNucleic acid detectionDNA
Target-specific hybrid capture (TSHC) provides a nucleic acid detection method that is not only rapid and sensitive, but is also highly specific and capable of discriminating highly homologous nucleic acid target sequences. The method produces DNA:RNA hybrids which can be detected by a variety of methods.
Owner:QIAGEN GAITHERSBURG INC
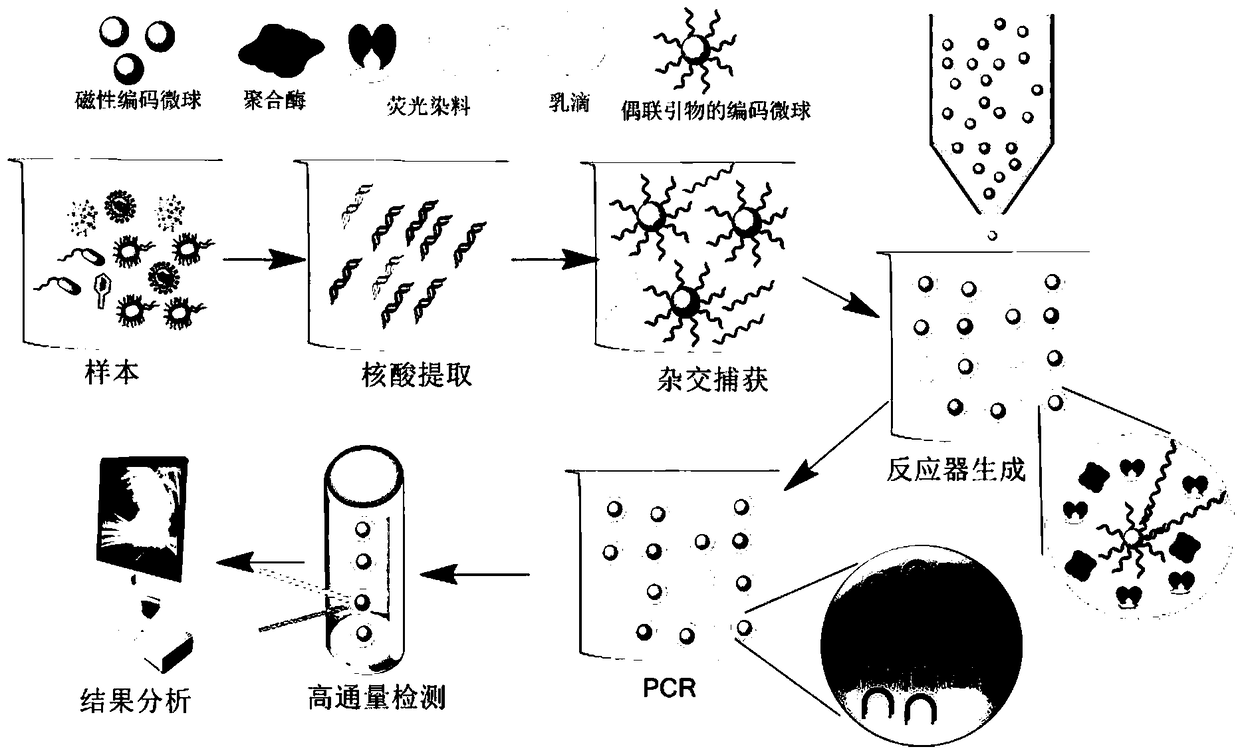
Digital multiple-nucleic-acid detection method on basis of encoding micro-sphere reactors
InactiveCN108660191AAchieve scaleReduce non-specific hybridization reactionsMicrobiological testing/measurementNucleic acid detectionComputer science
The invention discloses a digital multiple-nucleic-acid detection method on the basis of encoding micro-sphere reactors. The digital multiple-nucleic-acid detection method includes steps of extractingsample nucleic acid; designing primers; labeling the primers; carrying out hybrid capture; generating micro-drop; carrying out amplification reaction; carrying out scanning detection and the like. The digital multiple-nucleic-acid detection method has the advantages that multiple-gene detection merits of hybridization technologies and high-sensitivity quantitative detection merits of digital PCR(polymerase chain reaction) are combined with one another, accordingly, gap in the technical field of multiple-gene quantitative detection can be filled, and the digital multiple-nucleic-acid detection method has profound influence.
Owner:SUZHOU INST OF BIOMEDICAL ENG & TECH CHINESE ACADEMY OF SCI
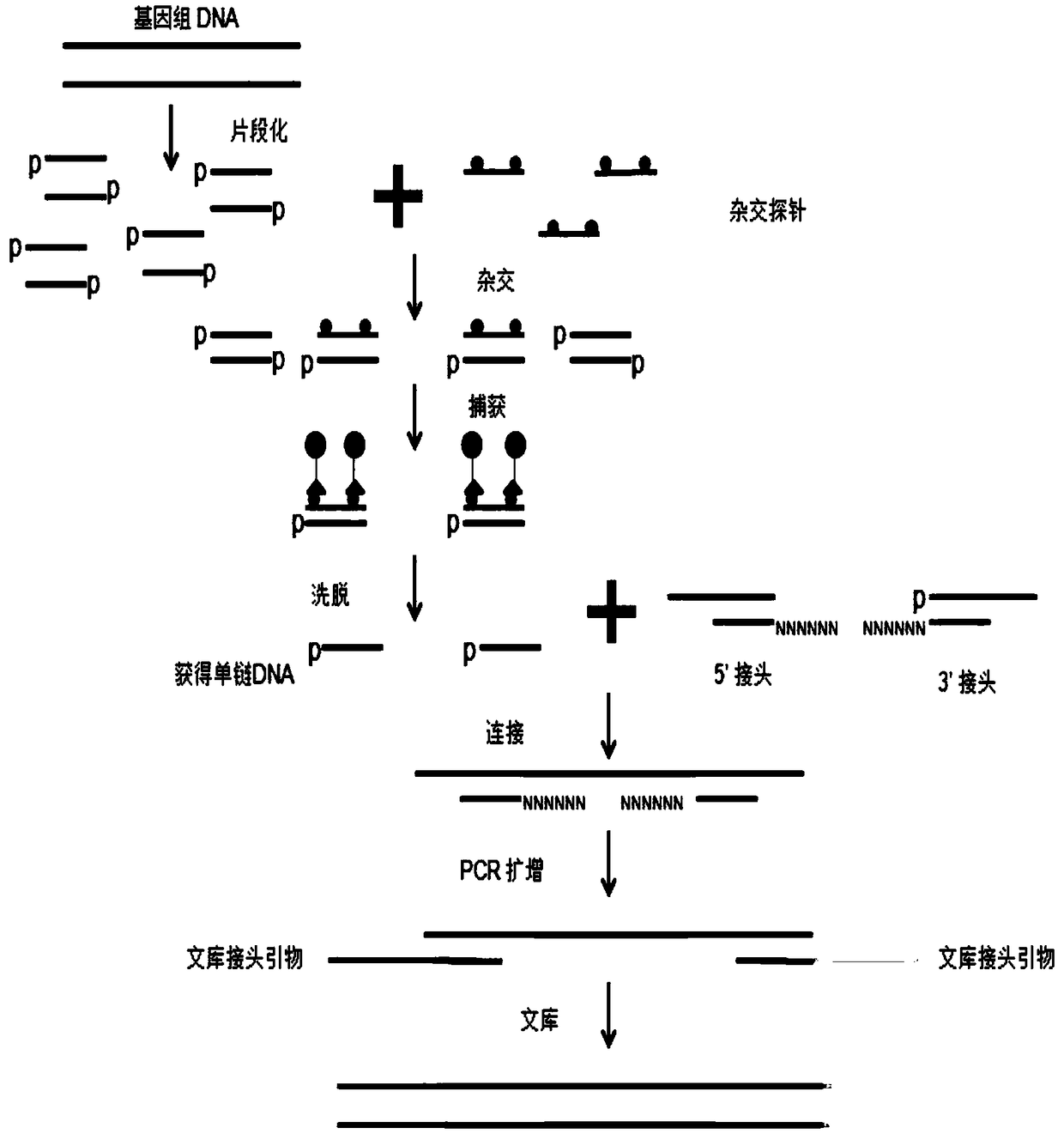
Method for constructing a hybrid capture sequencing library and application thereof
ActiveCN109234356AImprove capture efficiencyImprove uniformityMicrobiological testing/measurementLibrary creationGenomic DNAPaper document
The invention belongs to the field of gene detection, in particular to a strategy for direct hybridization of genomic DNA and a method for constructing a DNA library and an application thereof in thefield of nucleic acid detection. The method comprises the following steps: genomic DNA is fragmented and phosphorylated; Fragmented genomic DNA hybridized with capture probes; after hybridization, thecaptured single-stranded DNA was eluted. A ligation product is obtained by linking a single strand DNA with a linker; The ligation product was used as template for PCR amplification to obtain the amplified product, and the amplified product was purified to obtain the sequencing library. The invention provides a method for directly hybridizing fragmented genomic DNA, which is universally applicable to a solid-phase chip capture hybridization system and a liquid-phase capture hybridization system, and greatly improves capture efficiency and balance. At that same time, the invention further provide a DNA library construction method, which has the characteristics of simple method, low cost, wide applicability and the like, and can be use for the construction of a high-throughput sequencing document library of DNA or RNA.
Owner:江苏安科华捷生物科技有限公司
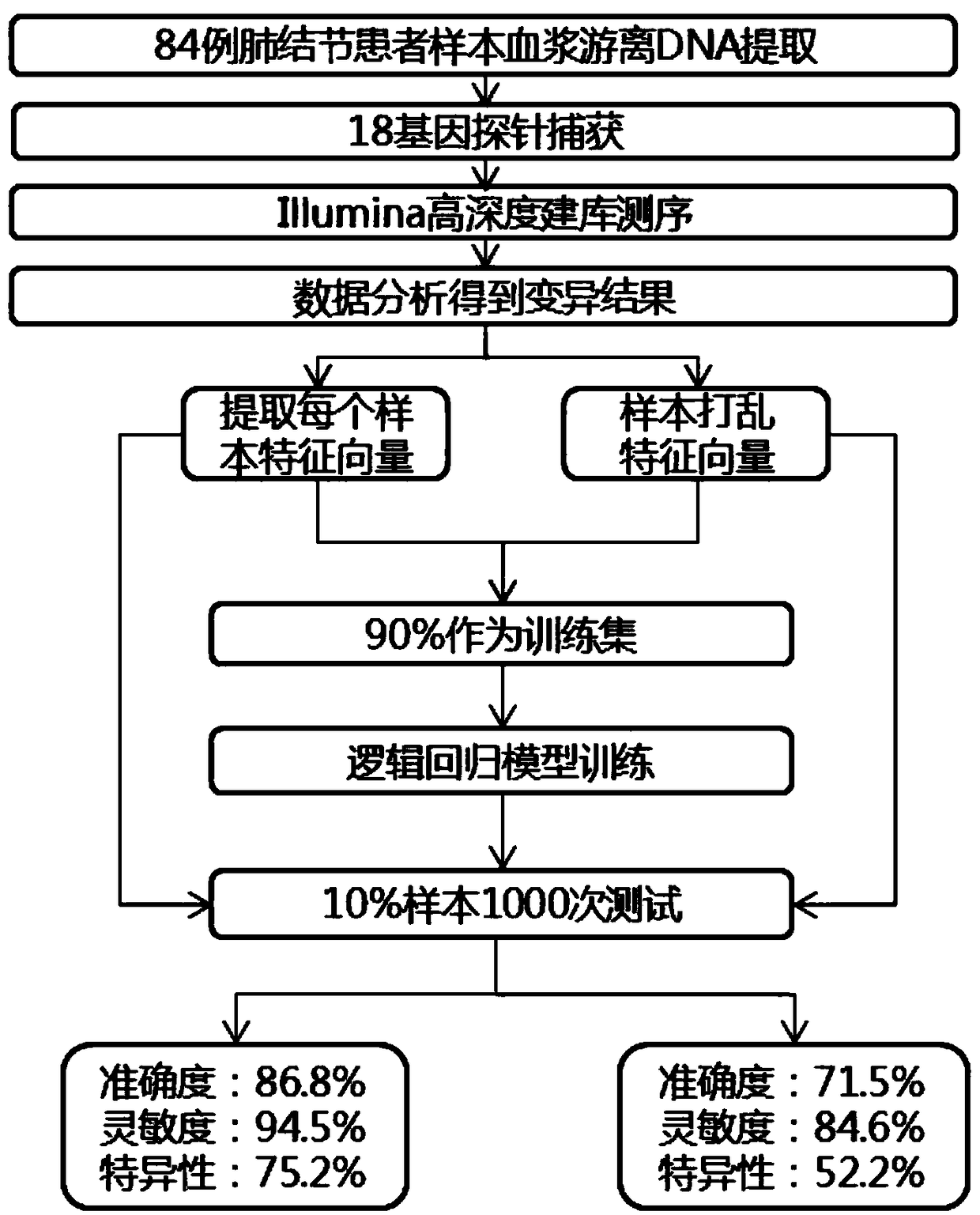
Method for judging benignancy and malignancy of pulmonary nodules by blood sample
InactiveCN108949979AStrong specificityBenign and malignant distinctionMicrobiological testing/measurementPulmonary noduleMutation frequency
The invention discloses a method for judging benignancy and malignancy of pulmonary nodules by a blood sample. Determination is performed through combination of cfDNA polygene variation and machine learning. The method includes the following steps: performing library construction on peripheral blood cfDNA of a pulmonary nodule patient; performing hybrid capture on the cfDNA by utilizing 18 gene capture probes related to a lung cancer; and performing sequencing by utilizing a sequencer, analyzing the cumulative mutation frequency of each of genes, and judging the benignancy and the malignancy of samples by utilizing a logistic regression model. The method can distinguish the benignancy and the malignancy of the pulmonary nodule patient with high sensitivity and high specificity, avoids experience errors of artificial reading CT imaging, and avoids missing detection of a lung cancer and pain of puncture or surgical sampling biopsy of a benign nodule patient.
Owner:SHENZHEN HAPLOX BIOTECH
Detection of nucleic acids by type specific hybrid capture method
InactiveUS20090162851A1Reduce background signalHigh detection specificitySugar derivativesMicrobiological testing/measurementNucleic acid detectionType specific
Target-specific hybrid capture (TSHC) provides a nucleic acid detection method that is not only rapid and sensitive, but is also highly specific and capable of discriminating highly homologous nucleic acid target sequences. The method produces DNA-RNA hybrids which can be detected by a variety of methods.
Owner:QIAGEN GAITHERSBURG
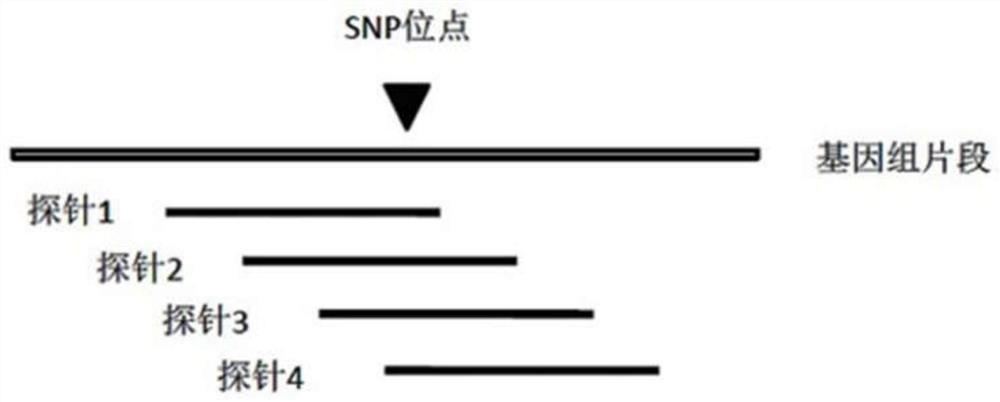
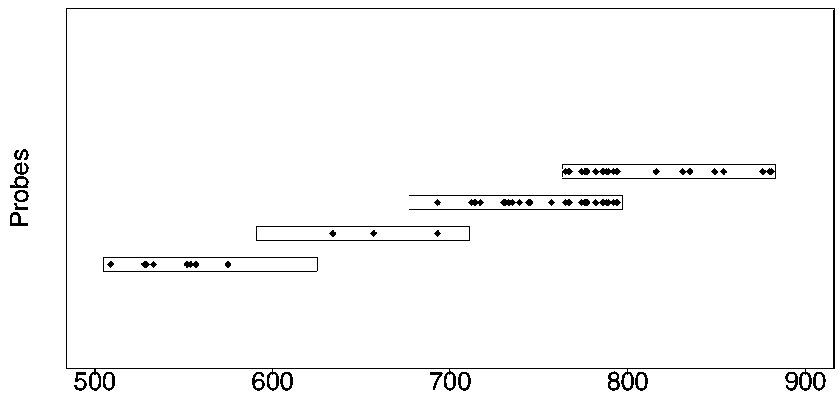
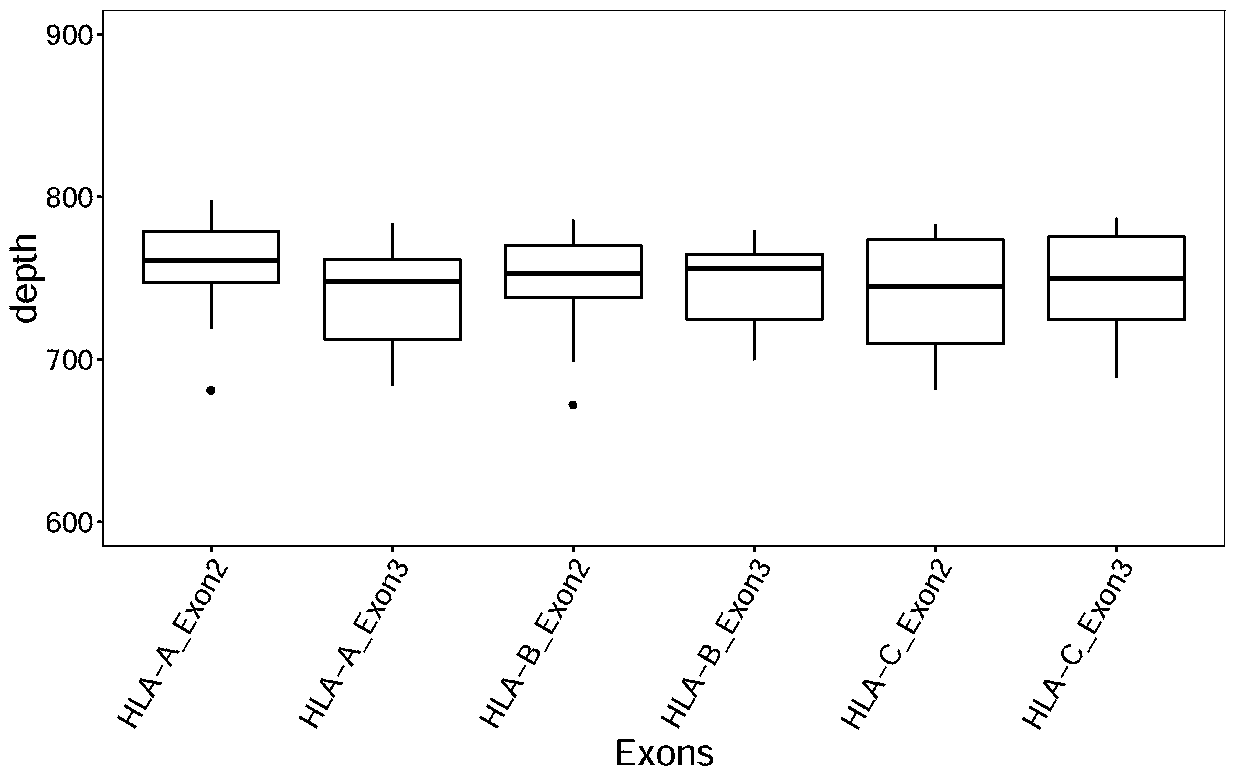
Design method of nucleic acid capture probe for HLA typing
ActiveCN110853708AEfficient captureImprove the effect of hybrid captureSequence analysisHybridisationRepresentative sequencesGene probe
The invention discloses a design method of a nucleic acid capture probe for HLA typing. The design method comprises the following steps: 1) constructing HLA-A, HLA-B and HLA-C sequence libraries; 2) performing multiple sequence alignment, and searching SNP sites on each gene Exon2 and Exon3 and upstream and downstream SNP sites; 3) selecting an area covering a set number of SNP sites as a probe design candidate area by using a sliding window separation algorithm; 4) performing clustering analysis to obtain a representative sequence of each probe design candidate region as a candidate probe; 5)repeating all the candidate probes to obtain capture probes of the three genes. The invention also discloses the probe designed by the method, and the sequence of the probe is shown as SEQ ID NO: 9-88. The nucleic acid capture probe is designed for all polymorphic sites of Exon2 and Exon3 regions with the GC proportion of 60% or above in the HLA gene, and the hybrid capture effect of the HLA geneprobe is improved.
Owner:上海仁东医学检验所有限公司
Hybrid capture kit and method for detecting tumor individually-medicated sixteen gene hot spots
ActiveCN107858426AComprehensive tumor individualized drug selectionComprehensive genetic testingMicrobiological testing/measurementInsertion deletionMutation frequency
The invention discloses a hybrid capture kit for detecting tumor individually-medicated sixteen gene hot spots. The hybrid capture kit comprises a hybridization reagent, a PCR amplification reagent and an enrichment degree detection reagent, wherein the hybridization reagent comprises an SEQ NO.1 to SEQ NO.396 probe mixture, a molar ratio of every two probes is 1:1, and the use volume of the probemixture with the concentration being 10 nM each during detection is 1 [mu]l. The kit is gene detection based on a second-generation sequencing technology, can detect three variation types (mutation,insertion deletion and fusion) one time, and uses a high sequencing depth to cover micro-scale gene variation in order to accurately detect the mutation condition of a patient on the premise of limited samples, and the mutation frequency of 1% can be accurately detected. Polygene and high-sensitivity parallel detection improves the detection rate of medicated gene variation, so the medication opportunity is not missed; and hot spots, rare and even unknown gene variation cane be detected during one-time detection, and relevant drug-resistant mutations (such as KRAS and ALK point mutation) are detected when drug-sensitive variation is found, so the sensitivity and drug resistance information of the medicated patient is accurately reflected.
Owner:SHANGHAI PASSION BIOTECHNOLOGY CO LTD
Kit for detecting integrated viruses of genome in hybrid capture
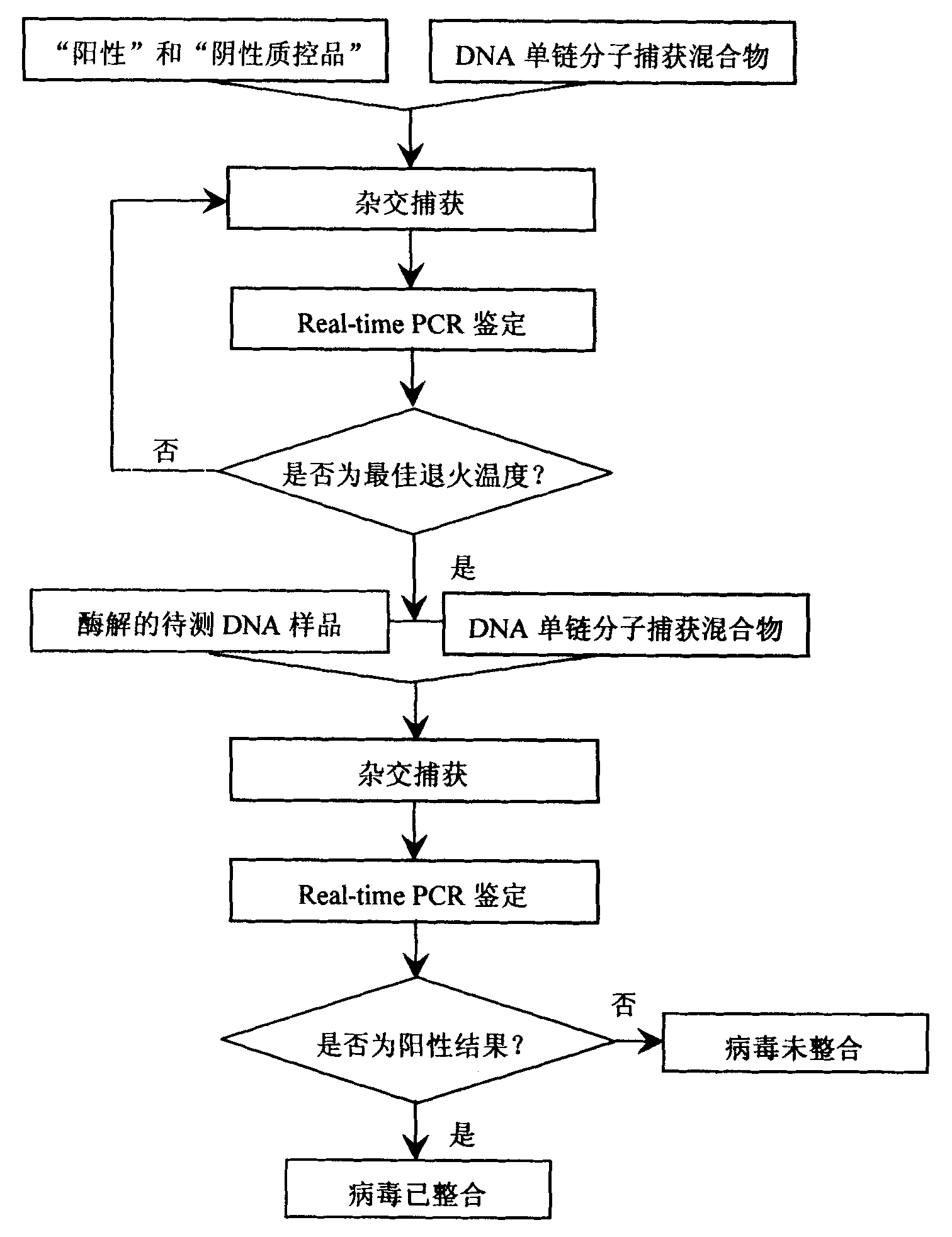
InactiveCN101974652AMicrobiological testing/measurementMicroorganism based processesHuman DNA sequencingBiomedicine
The invention belongs to the field of biomedicine, in particular to a kit for detecting integrated viruses of a genome in hybrid capture. The kit comprises the following main components: (1) a solid phase medium fixed with a capture mixture of human genome DNA (Deoxyribonucleic Acid) single-chain molecules, (2) positive and negative quality control materials for identifying the hybrid capture efficiency and specificity, and (3) a primer pair and a molecular probe for carrying out real-time PCR (Polymerase Chain Reaction) identification to the captured mixture of the target DNA molecules, obtained by capture. By utilizing all or partial components in the invention, a complete integrated genome virus detection kit or an integrated genome virus detection kit with the type specificity can be assembled. The obtained virus detection kit can be used for detecting and identifying the integrated state of the genome in a certain common integrated and oncogenic viruses, such as human hepatitis B viruses, human papilloma viruses, Epstein-Barr viruses and human herpes simplex viruses, in tissue or cytology specimens.
Owner:何以丰
Cancer gene methylation detection system and cancer in-vitro detection method performed in system
PendingCN112662760AProlong lifeExcellent in breadthNucleotide librariesMicrobiological testing/measurementA-DNAHybrid capture
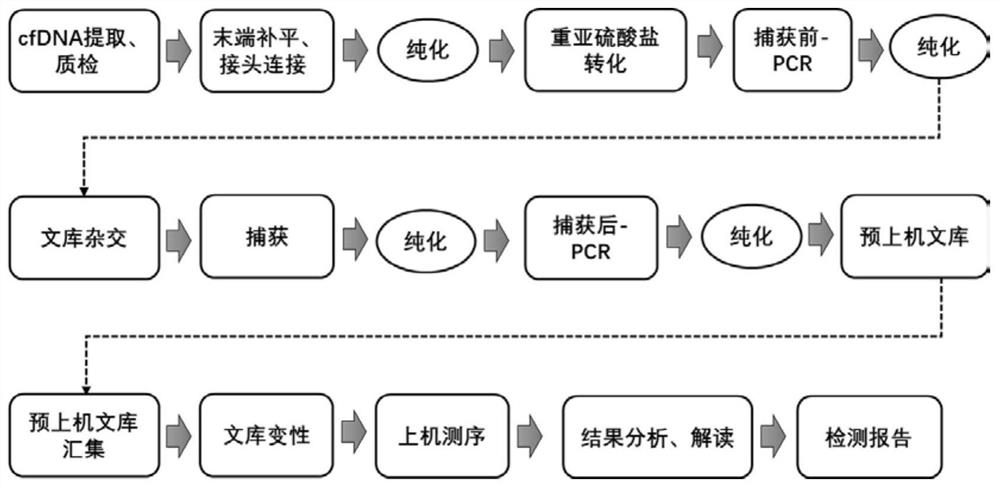
Disclosed herein is a system for detecting cancer methylation. The system comprises a sample collection module for collecting a subject sample, a DNA extraction module used for extracting and purifying DNA in the sample, a library building module used for building a DNA library for sequencing for the purified DNA sample, a conversion module used for converting the constructed DNA library by using a bisulfite, a pre-PCR amplification module used for carrying out pre-PCR amplification on the DNA library subjected to bisulfite conversion, a hybrid capture module used for carrying out hybrid capture on the sample subjected to the pre-PCR amplification by utilizing a probe composition, a post-PCR amplification module used for amplifying a product captured by hybridization by utilizing PCR, a sequencing module used for carrying out high-throughput next-generation sequencing on a product subjected to hybridization capture after PCR amplification, a data analysis module used for analyzing the sequencing data and determining the methylation level of the sample, and an interpretation module used for interpreting the illness condition of the patient based on the methylation level of the sample.
Owner:BIOCHAIN BEIJING SCI & TECH
Lymphangiomyoma joint detection method and application thereof
ActiveCN110885879ARaise positiveImprove positive variant detection rateMicrobiological testing/measurementMultiplexLymphatic vessel
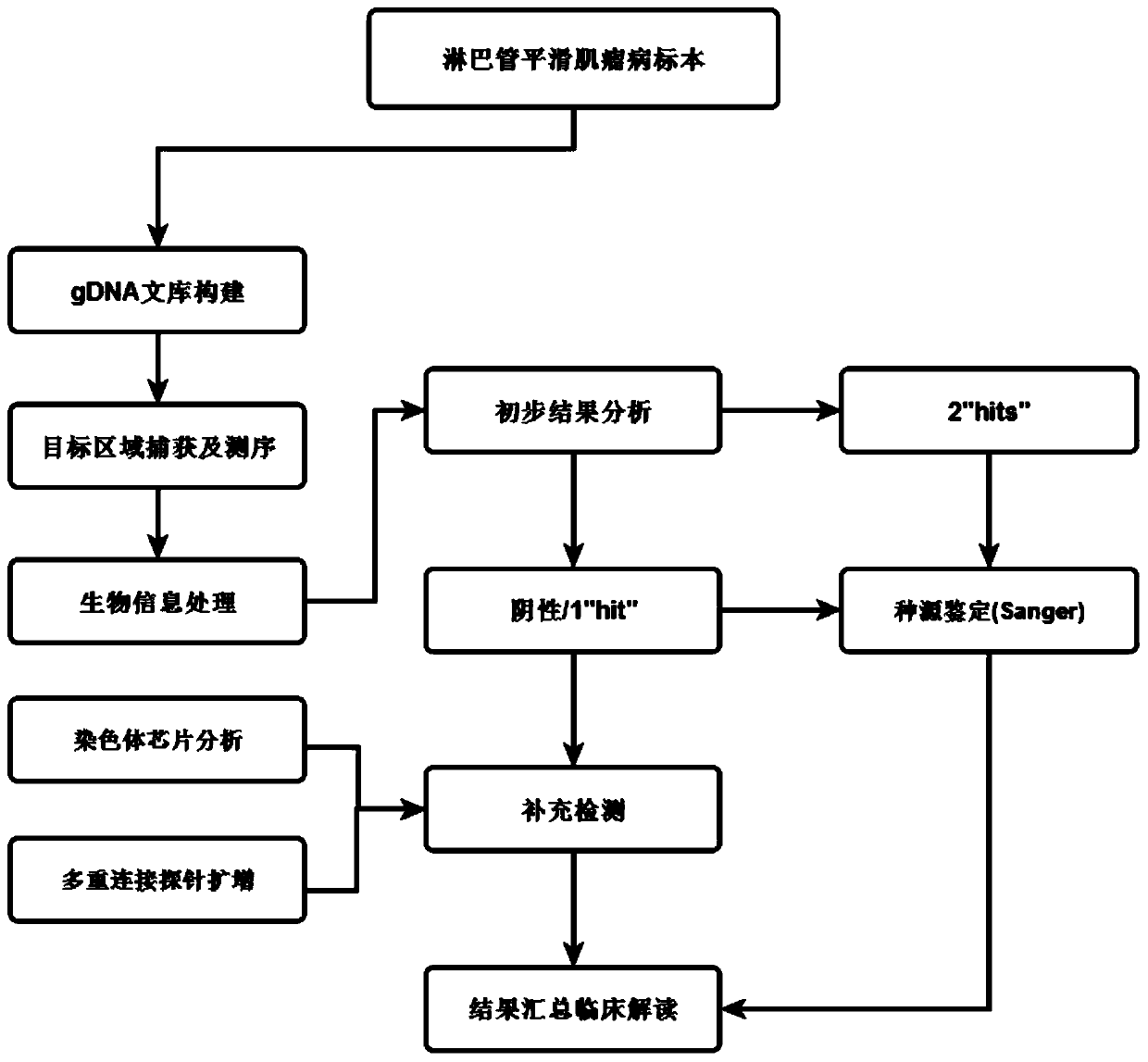
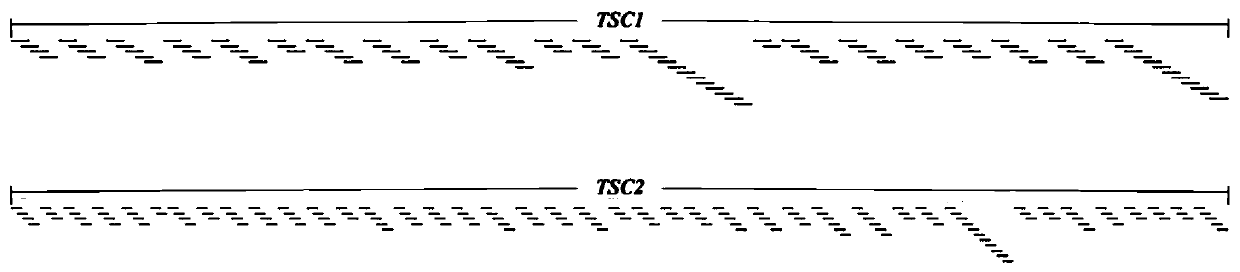
The invention relates to a lymphangiomyoma joint detection method and an application thereof, and belongs to the technical field of gene detection. The method comprises carrying out liquid phase capture targeting sequencing, designing Panel for TSC1 and TSC2 gene whole exon coding regions highly related to LAM and mutant genes closely related to solid tumors, constructing a gDNA library, carryingout hybrid capture, sequencing on a machine, carrying out sorting by analyzing the sequencing data through bioinformatics processing, and if the TSC1 and TSC2 genes are negative, carrying out supplementary detection by adopting a chromosome chip analysis method, if the single strike site is detected, performing supplementary detection by adopting multiplex ligation-dependent probe amplification, and if the sites have no clear sources of the somatic mutation and the embryonic system mutation, carrying out supplementary detection by adopting Sanger detection. The method improves the positive variation detection rate of LAM patients.
Owner:GUANGZHOU KINGMED DIAGNOSTICS GRP CO LTD
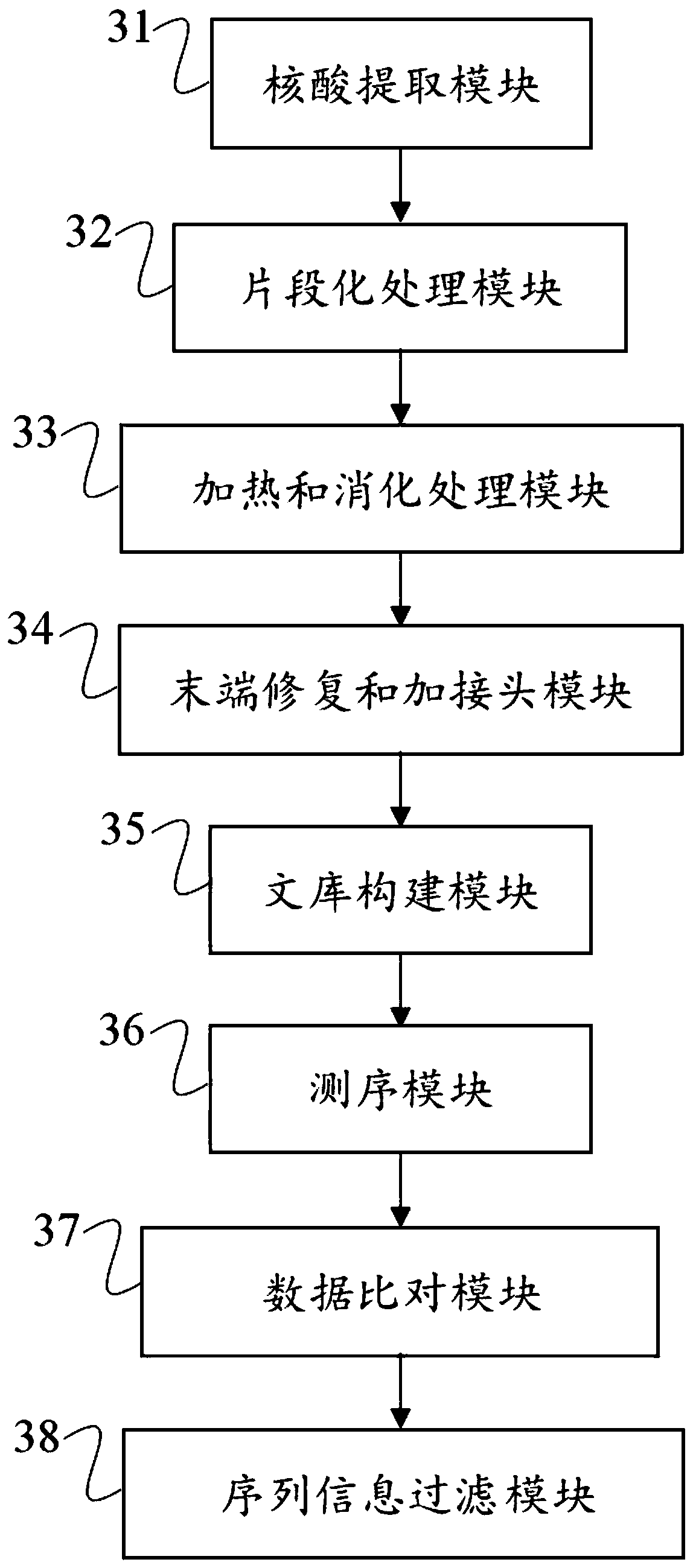
Low-quality FFPE DNA processing method and device and storage medium
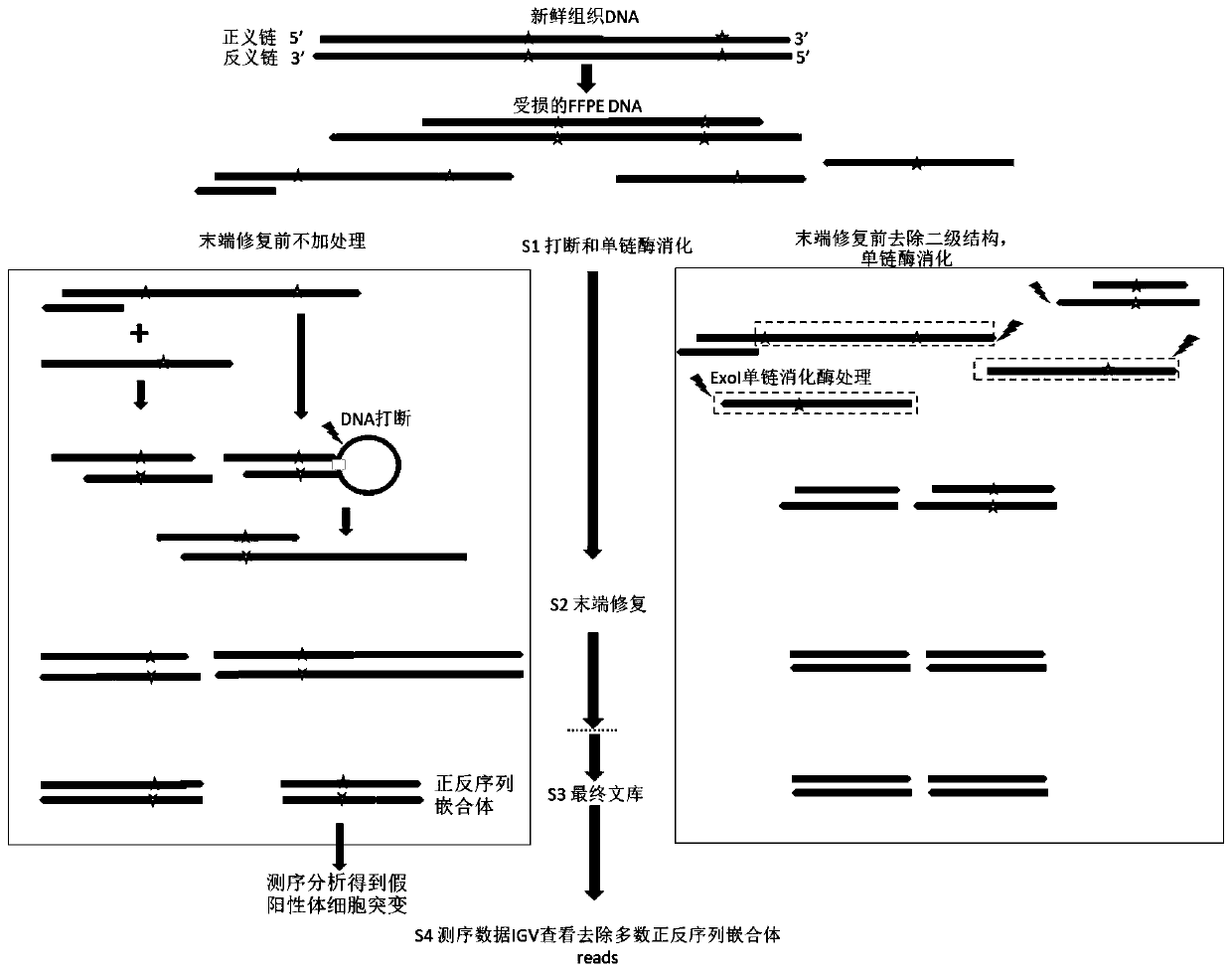
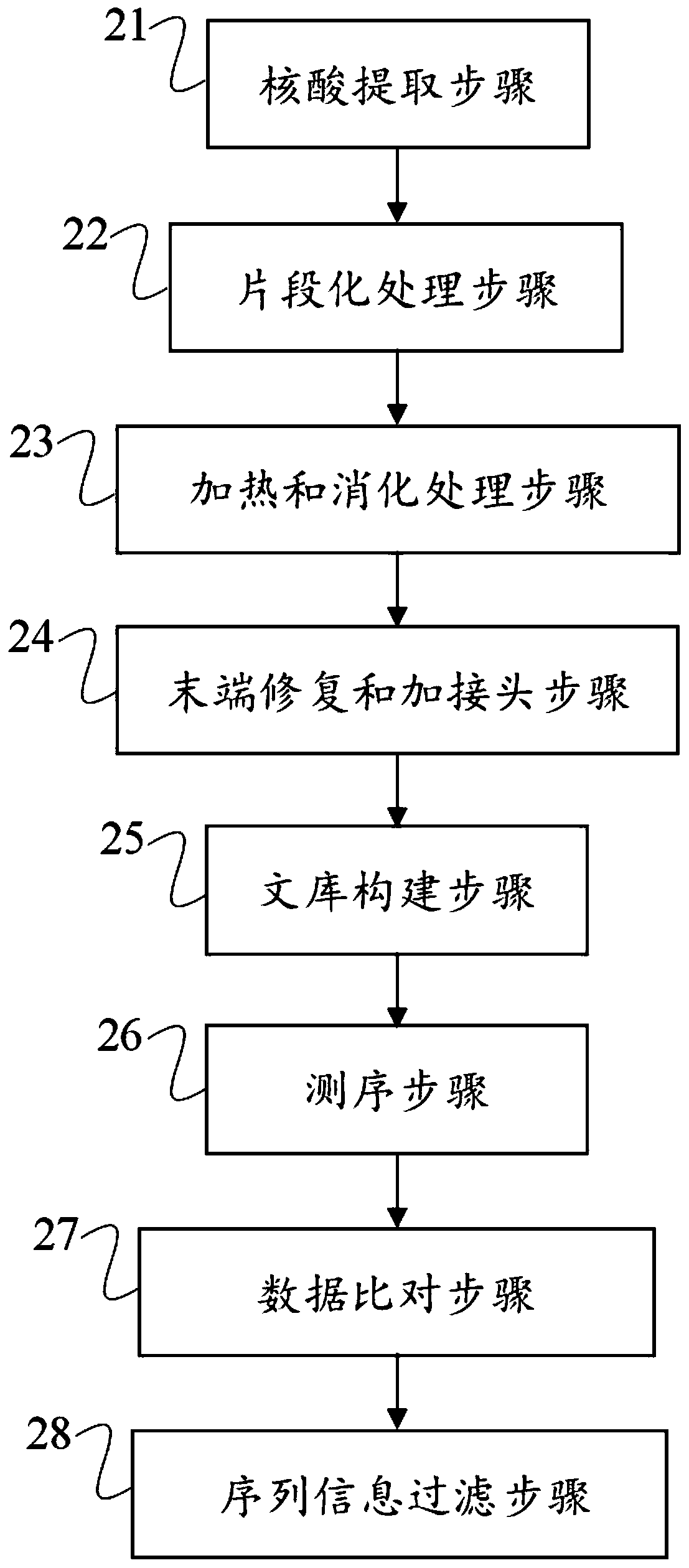
ActiveCN110669830AImprove accuracyImproved detection of point mutationsBioreactor/fermenter combinationsBiological substance pretreatmentsEnzyme digestionDigestion
The invention discloses a low-quality FFPE DNA processing method and device and a storage medium. The processing method comprises the following steps: carrying out fragmentation treatment on FFPE DNA,performing heating to remove a secondary structure, and carrying out enzyme digestion; repairing the tail end after digestion, adding poly A tail, and using an A base connector; then, carrying out PCR amplification, hybrid capture and sequencing to obtain sequencing data; and comparing the sequencing data with a human reference genome, and filtering out sequences which are partially compared to two different places of the same chromosome at the same time and of which the comparison initial position distance is less than 500bp from a comparison result. According to the method disclosed by theinvention, most of single chains are eliminated through the heating and the enzyme digestion, so that the single chains cannot form stable double chains in terminal filling-in after being connected toone another, and false positive of subsequent mutation detection caused by single chain residues is reduced; and through data comparison and filtering, single-chain interference is further removed, and the accuracy of mutation detection results is improved.
Owner:裕策医疗器械江苏有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com