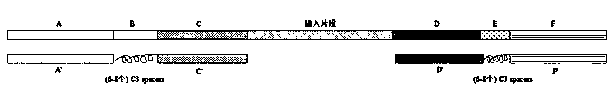
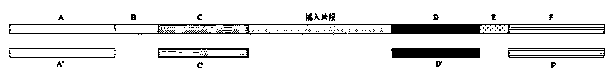
Application of bridge type oligonucleotides to library target region capturing
A technology of sequence capture and target sequence, which is applied in chemical library, microbiological determination/testing, combinatorial chemistry, etc. It can solve the problems of affecting capture rate, poor tag sequence blocking effect, and high cost of synthesizing hypoxanthine, reaching the scope of application wide effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Example 1-Illumina sequencing platform library construction process:
[0066] 1. Library structure and closed oligonucleotide structure
[0067] 1) Illumina sequencing platform prePCR library structure: For clarity, two chains are shown, the structure of the two sequences is as follows:
[0068] a (5' to 3'):AATGATACGGCGACCACCGAGATCTACAC (SEQ ID NO.1) [tag sequence] ACACTCTTTCCCTACACGACGCTCTTCCGATCT (SEQ ID NO.2) [pairing sequence with target sequence] AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC (SEQ ID NO.3) [tag sequence]CTATCTTGTATGTG SEQ ID NO. 4)
[0069] b (3' to 5'): TTACTATGCCGCTGGTGGCTCTAGATGTG (SEQ ID NO.5) [tag sequence]TGTGAGAAAGGGATGTGCTGCGAGAAGGCTAGA (SEQ ID NO.6) [pairing sequence with target sequence] TCTAGCCTTCTCGTGTGCAGACTTGAGGTCAGTG (SEQ ID NO.7) [tag sequence]TAGAGACCATACGG SEQ ID NO. 8)
[0070] The length of the paired sequence with the target sequence is 80-120 bp, preferably 100 bp.
[0071] 2) Blocked oligonucleotide sequence of Illumina sequencing platform:
[00...
Embodiment 2
[0195] Example 2-NEB sequencing platform library construction process
[0196] 1. Library structure and closed oligonucleotide structure
[0197] 1) The structure of the prePCR library of NEB sequencing platform: For clarity, the structure of the front and back strands and the double-stranded two sequences are shown as follows:
[0198] a(5’ to 3’):
[0199] CAAGCAGAAGACGGCATACGAGAT(SEQ ID NO.9)[Tag sequence]GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT(SEQ ID NO.10)[pairing sequence with target sequence]AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTATCATT(SEQ ID NO.14)
[0200] b(3’ to 5’):
[0201] GTTCGTCTTCTGCCGTATGCTCTA(SEQ ID NO.15)[Tag sequence]CACTGACCTCAAGTCTGCACACGAGAAGGCTAGA(SEQ ID NO.16)[pairing sequence with target sequence]TCTAGCCTTCTCGCAGCACATCCCTTTCTCACATCTAGAGCCACCAGCGGCATAGTAA (SEQ ID NO.17)
[0202] The length of the paired sequence with the target sequence is 80-120 bp, preferably 100 bp.
[0203] 2) NEB sequencing platform blocking oligonucleotide sequence:
[0204] a-1: AGA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com