Patents
Literature
38 results about "Representative sequences" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
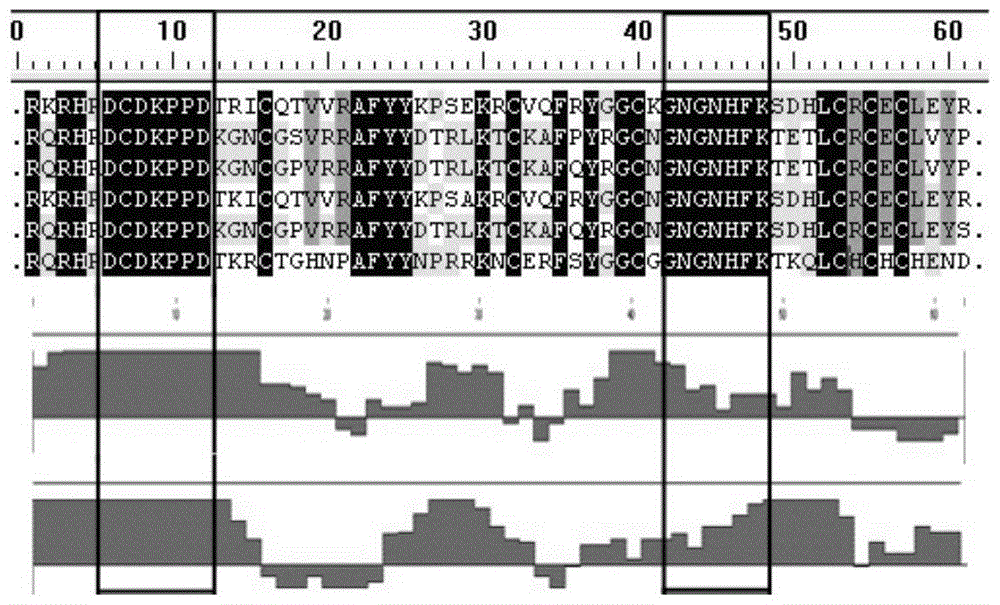
Representative sequences are short regions within protein sequences that can be used to approximate the evolutionary relationships of those proteins, or the organisms from which they come. Representative sequences are contiguous subsequences (typically 300 residues) from ubiquitous, conserved proteins, such that each orthologous family of representative sequences taken alone gives a distance matrix in close agreement with the consensus matrix.
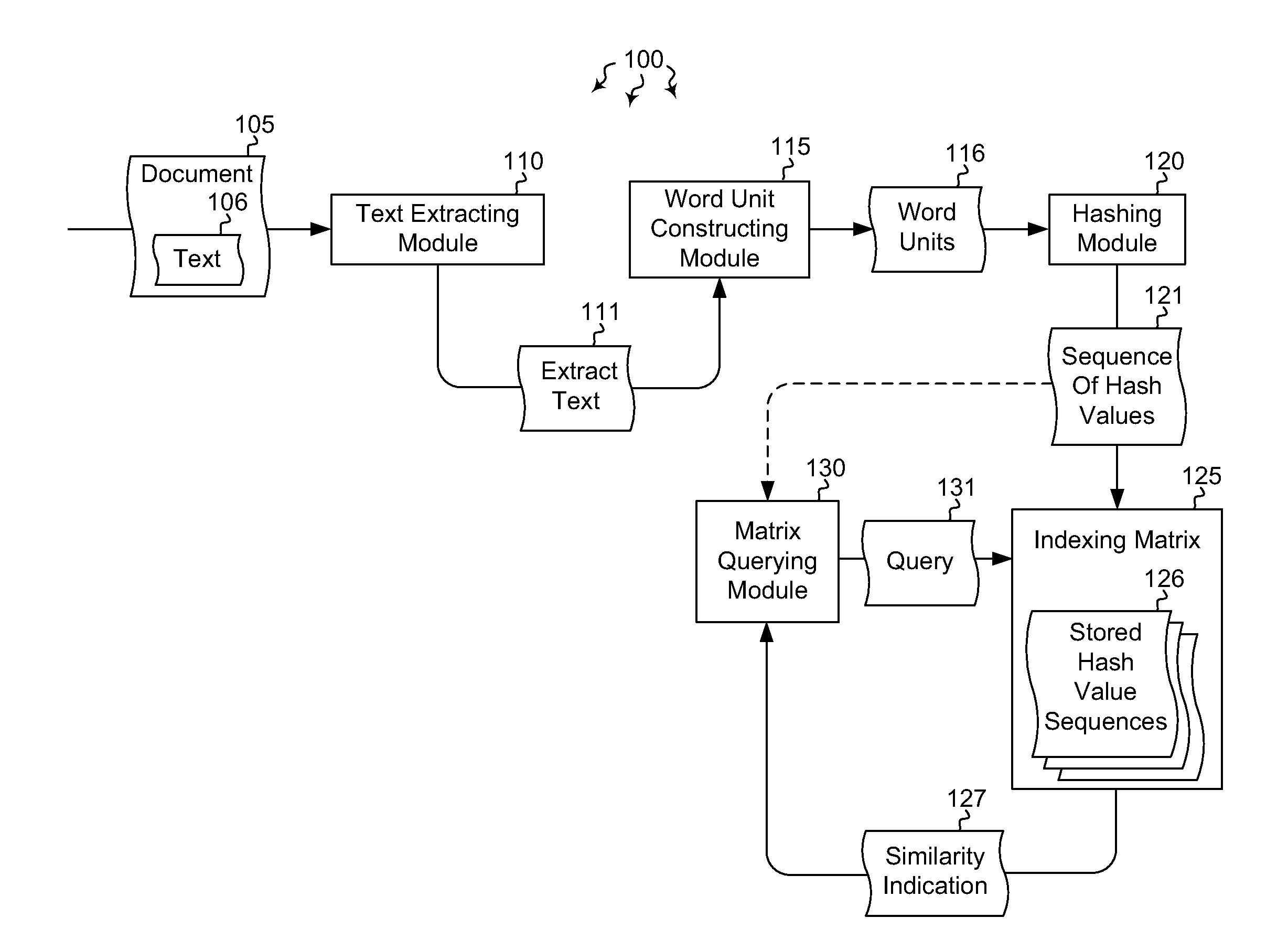
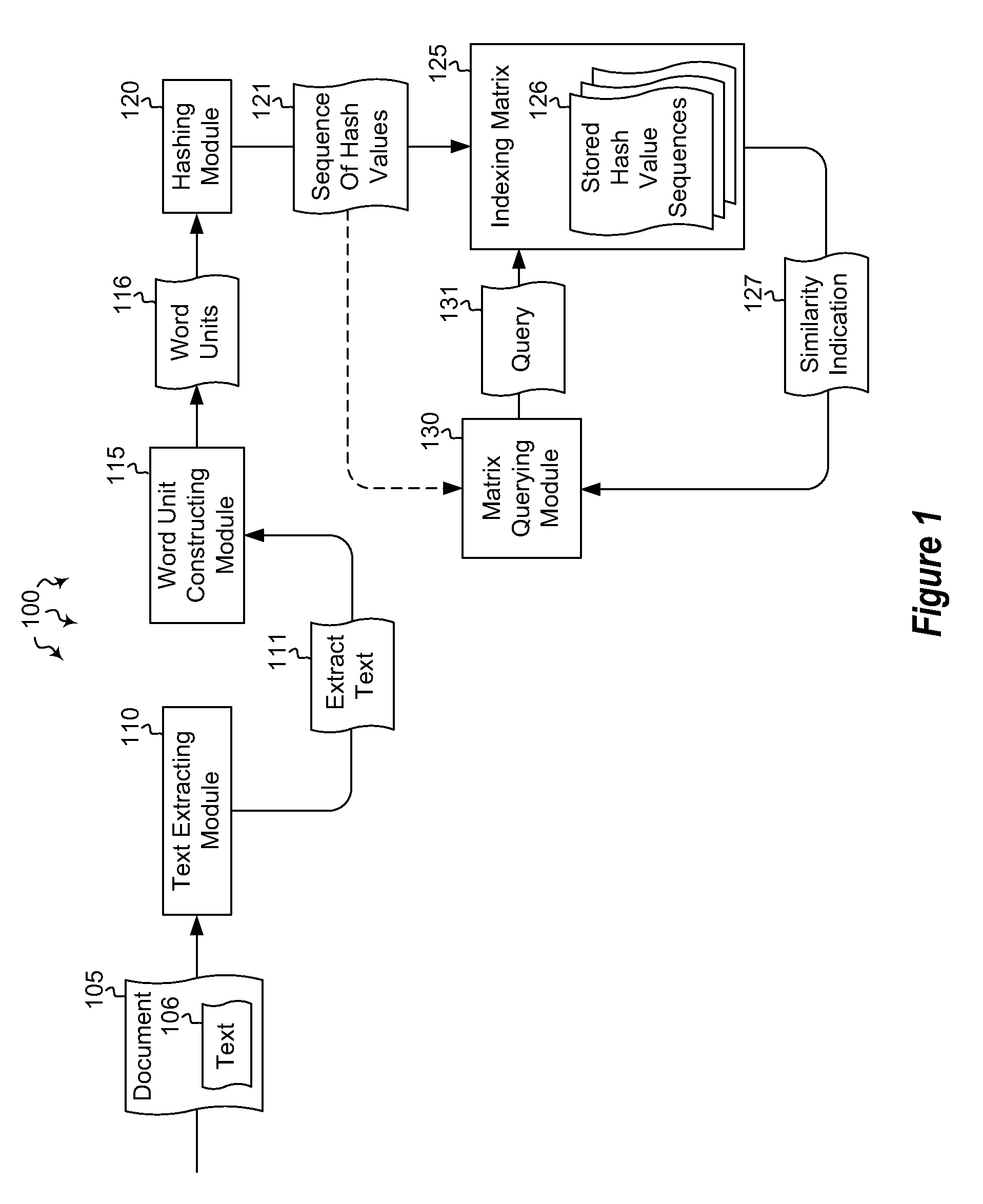
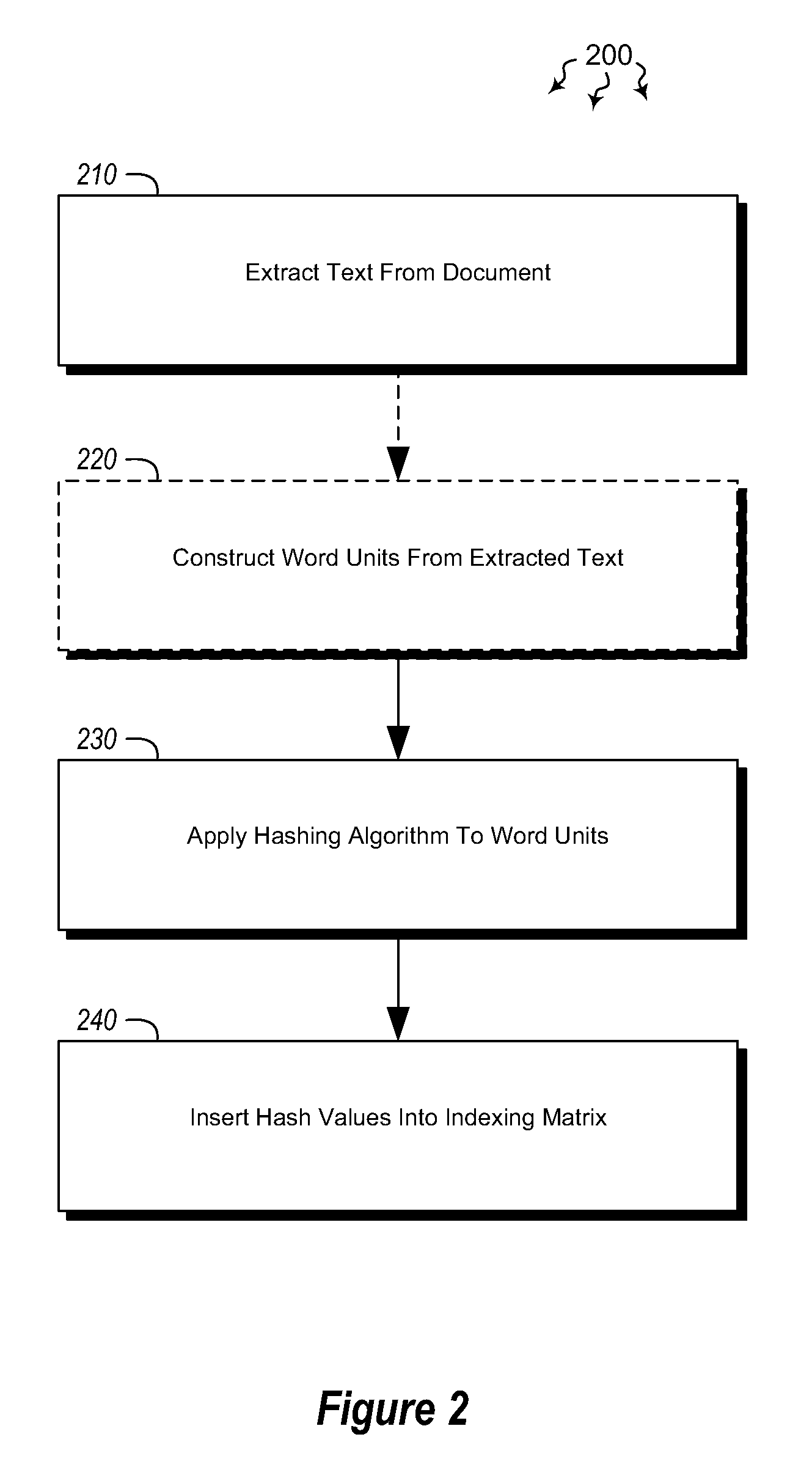
Indexing and querying hash sequence matrices
ActiveUS20120117080A1Well formedDigital data processing detailsText database indexingAlgorithmComputerized system
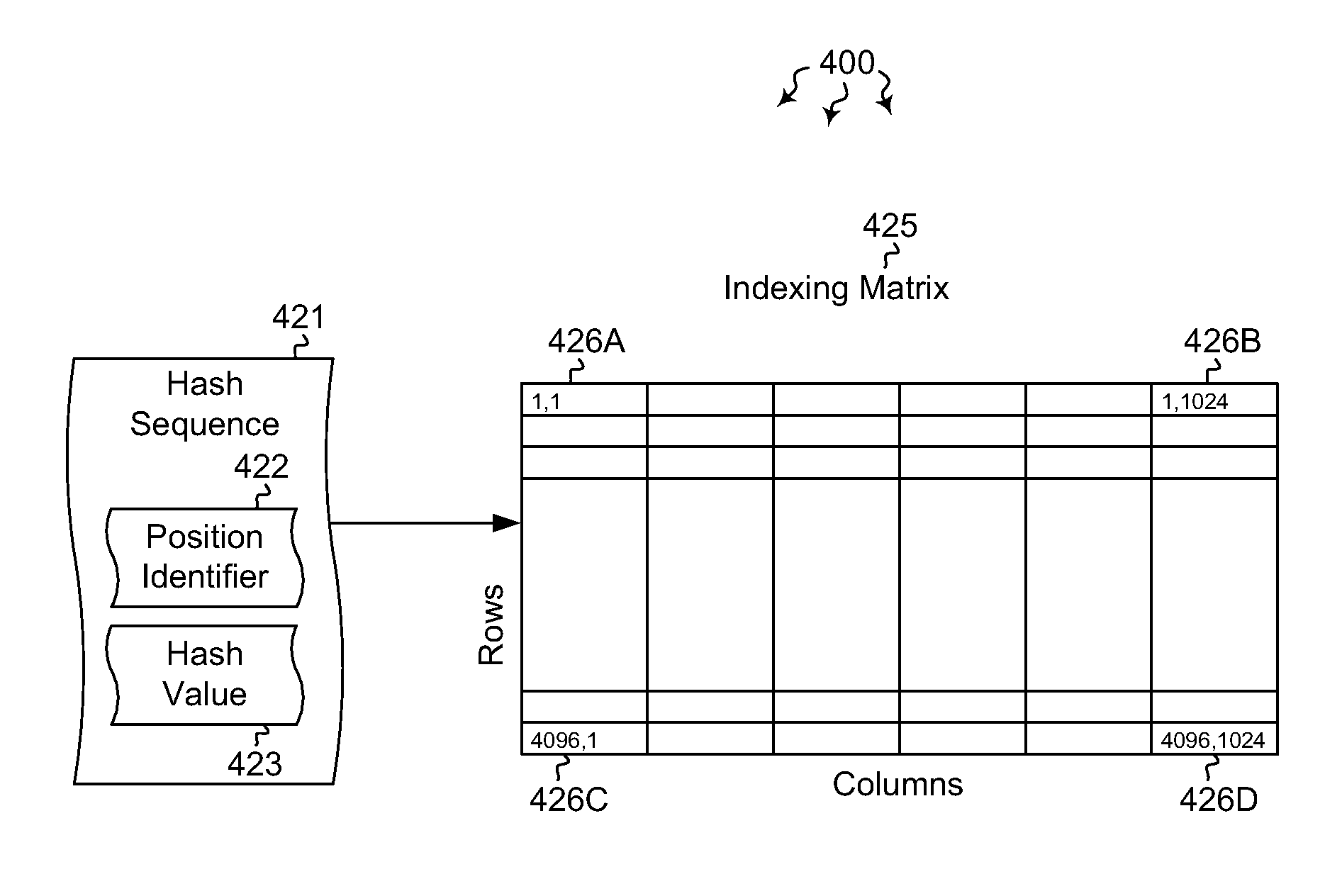
Embodiments are directed to indexing and querying a sequence of hash values in an indexing matrix. A computer system accesses a document to extract a portion of text from the document. The computer system applies a hashing algorithm to the extracted text. The hash values of the extracted text form a representative sequence of hash values. The computer system inserts each hash value of the sequence of hash values into an indexing matrix, which is configured to store multiple different hash value sequences. The computer system also queries the indexing matrix to determine how similar the plurality of hash value sequences are to the selected hash value sequence based on how many hash values of the selected hash value sequence overlap with the hash values of the plurality of stored hash value sequences.
Owner:MICROSOFT TECH LICENSING LLC
Method for detecting diversity of fish species on basis of environmental DNA technology
PendingCN109825563ACause some damagesSimple samplingMicrobiological testing/measurementData informationRepresentative sequences
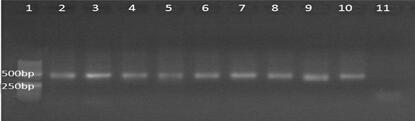
The invention discloses a method for detecting diversity of fish species on the basis of an environmental DNA technology. The method comprises the following steps of (1) conducting sampling; (2) extracting eDNA; (3) performing PCR amplification; (4) recovering and purifying target fragments and constructing a target fragment library; (5) conducting OTU clustering; (6) creating a fish comparison database; (7) conducting comparison analysis on representative sequences of OTUs with data information of the created fish database to determine the species diversity composition of fishes in a to-be-detected water area. According to the method, there is no need to catch the fishes, sampling is simple, and the fish bodies cannot be damaged; detection is convenient, and there is no need to conduct fish appearance identification; detection is simple and fast, the precision is high, and a detection result is accurate.
Owner:ZHEJIANG INST OF FRESH WATER FISHERIES
Lathyrus quinquenervius EST-SSR primer group developed based on transcriptome sequencing, method and application
InactiveCN107345256ALow costMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceSequence database
The invention provides a lathyrus quinquenervius EST-SSR primer group developed based on transcriptome sequencing, a method and application. The method comprises the steps of obtaining a set of all root, stem and leaf transcripts of two lathyrus quinquenervius varieties of different flower colors at a seedling stage to form an original sequence database; removing low-quality sequencing data with a joint and forming a high-quality filtered sequence database; splicing high-quality filtered sequences, reducing the transcripts and carrying out BLAST comparison on the transcripts and a reference protein database NR, reserving the optimal comparison result and determining the maximal sequence as a Unigene representative sequence; carrying out SSR locus scanning in Unigene by using MISA 1.0; and carrying out SSR primer design by adopting primer 3.0, screening SSR primers and carrying out fingerprint map construction on 43 parts of lathyrus quinquenervius materials. The lathyrus quinquenervius EST-SSR primer group is convenient, fast, accurate, low in cost, and a new thought is provided for the development of lathyrus quinquenervius EST-SSR primers and germplasm resources identification.
Owner:山西省农业科学院农作物品种资源研究所 +1
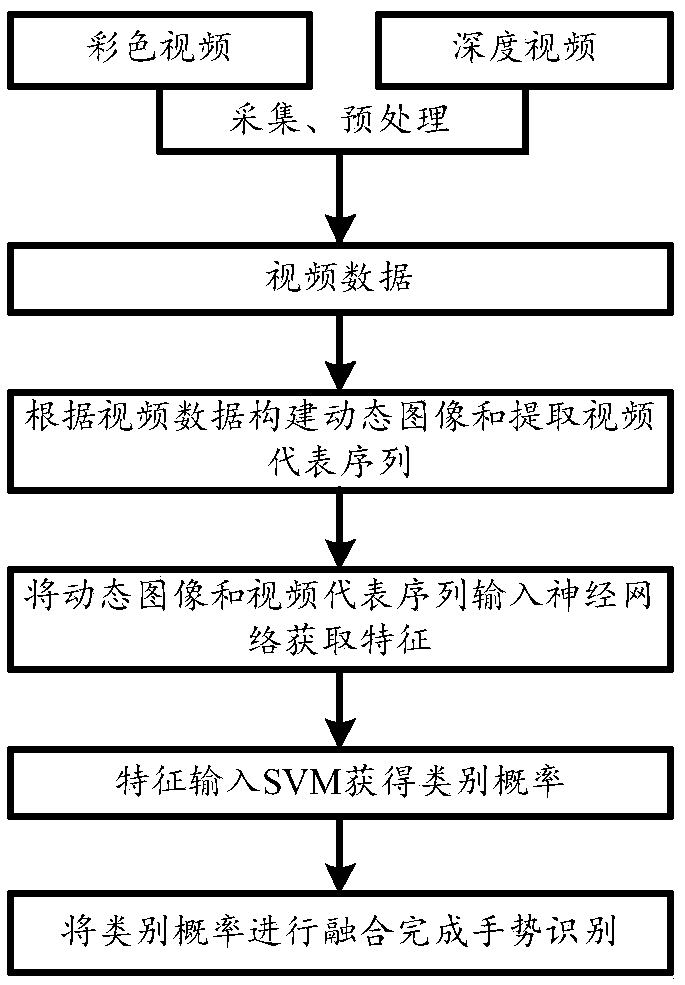
First-person-view gesture recognition method based on dynamic image and video subsequence
InactiveCN109034012AUse the Internet less oftenSimplify the feature extraction processCharacter and pattern recognitionPattern recognitionSvm classifier
The invention discloses a first-person-view gesture recognition method based on a dynamic image and a video subsequence, which relates to the field of first person angle of view gesture recognition. The method comprises the steps of: 1, collecting color video and depth video, and preprocessing the video to obtain video data; 2, constructing a dynamic image and extracting a video representative sequence according to the video data; 3, inputting the representative sequences of the dynamic image and the video into the fine-tuned neural network model to obtain the features, and inputting the feature into the SVM classifier to obtain the category probability; 4, carrying out fusion on that category probability to complete gesture recognition. The present invention solves the problems of complicated gesture recognition feature extraction process based on depth learning and low precision, and achieves the effects of simplifying gesture recognition process and improving recognition precision.
Owner:SICHUAN UNIV
Method for detecting community structure and abundance of ammonia oxidizing bacteria in wastewater system
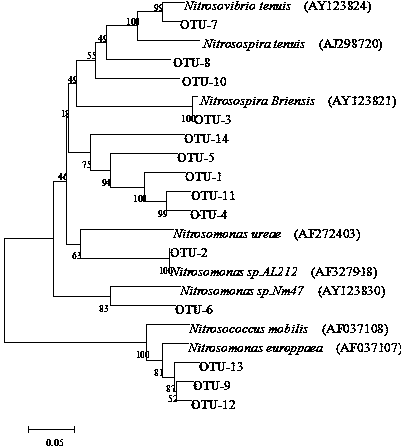
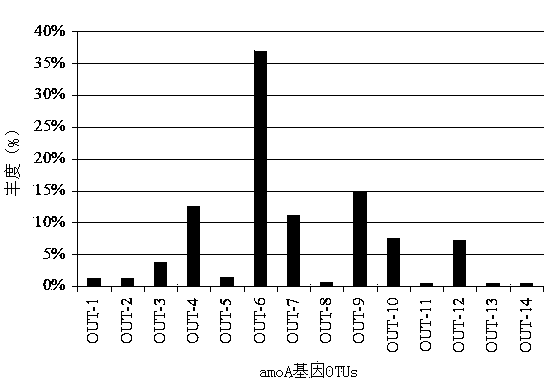
The invention discloses a method for detecting the community structure and abundance of ammonia oxidizing bacteria in a wastewater system. The method comprises the following specific steps: (A) extracting all genome DNA (Deoxyribonucleic Acid) of microorganisms in active sludge of the wastewater system; (B) performing PCR (polymerase chain reaction) amplification by taking all genome DNA as a template and taking amoAlF with a sequence number of SEQ ID No.1 and amoA2R with a sequence number of SEQ ID No.2 as primers; and (C) sequencing an amplification product by adopting a Roche 454 pyrosequencing process, dividing an operational taxonomic unit for a sequencing result by applying a Mothur program, performing online BLASTn comparison on representative sequences with the similarity of 99% in the operational taxonomic unit and sequences disclosed in a database GenBank, and dividing a system evolutionary tree according to a comparison result to obtain the community structure distribution and abundance of the ammonia oxidizing bacteria. The method is simple to perform, low in cost, quick in detection and high in sequence analysis accuracy, can be used for qualitatively analyzing different community structures of the ammonia oxidizing bacteria and quantitatively analyzing the abundance of different species of the ammonia oxidizing bacteria, can be used for simultaneously analyzing a plurality of samples, and is relatively high in practicality and easy to popularize and apply.
Owner:HIGH TECH RES INST NANJING UNIV LIANYUNGANG +1
Correlation-associated wind power output scene construction method
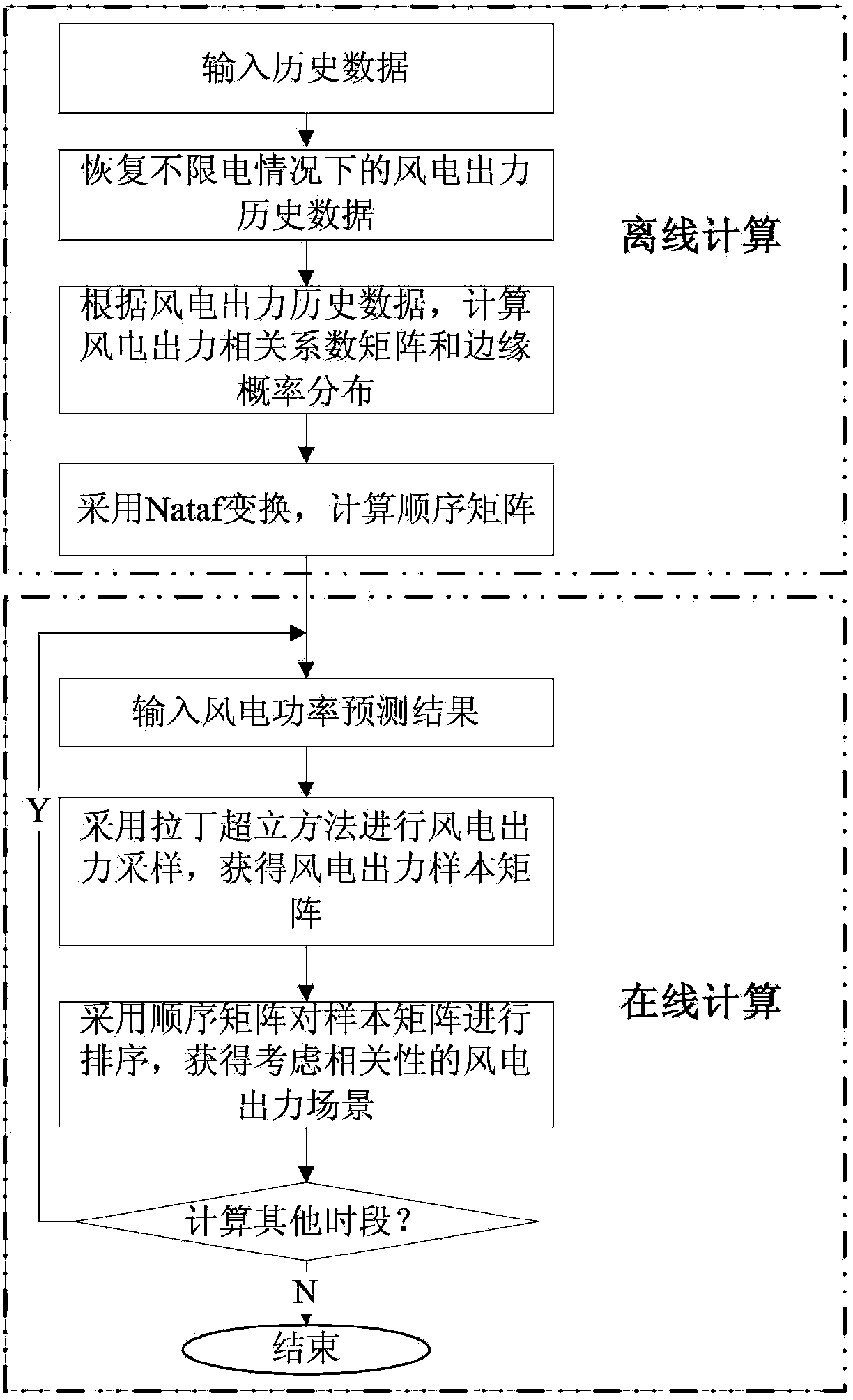
InactiveCN104182910AReduce scene construction timeShorten construction timeData processing applicationsSystems intergating technologiesElectricityEngineering
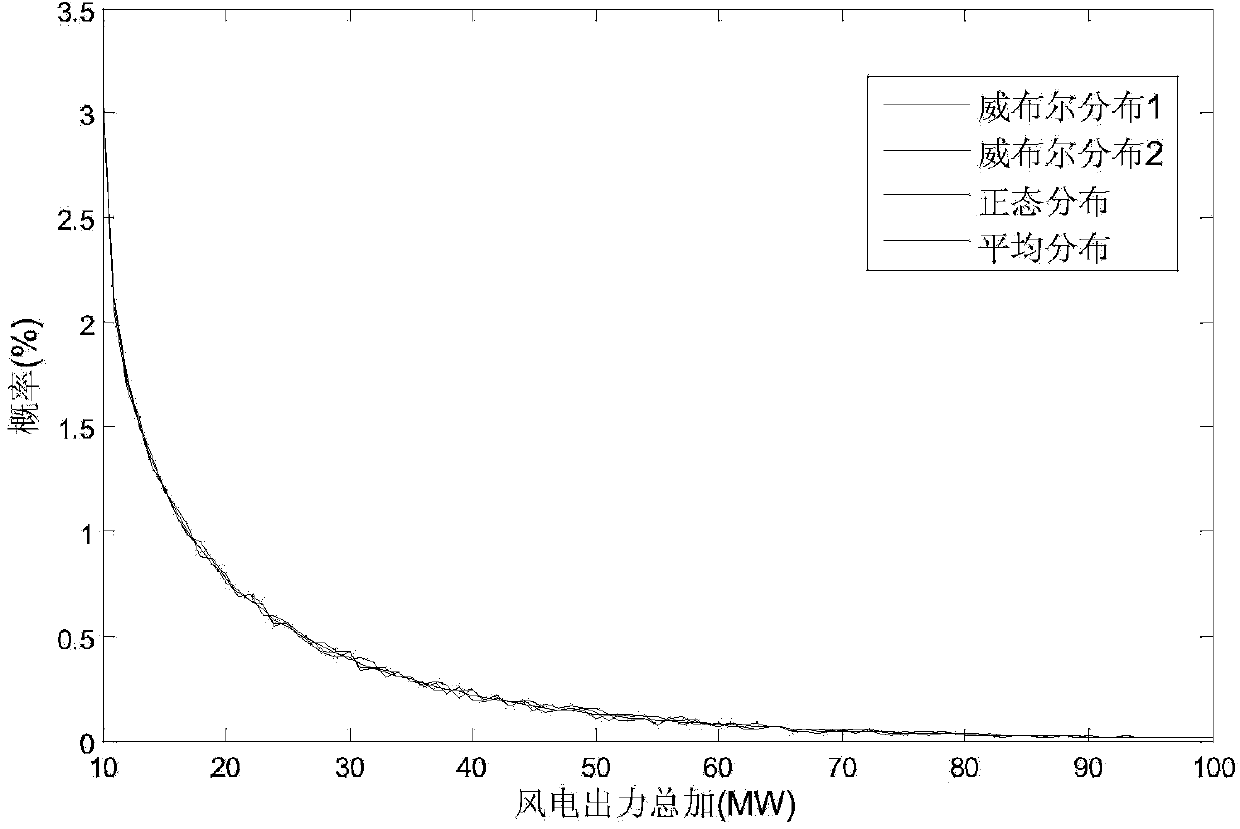
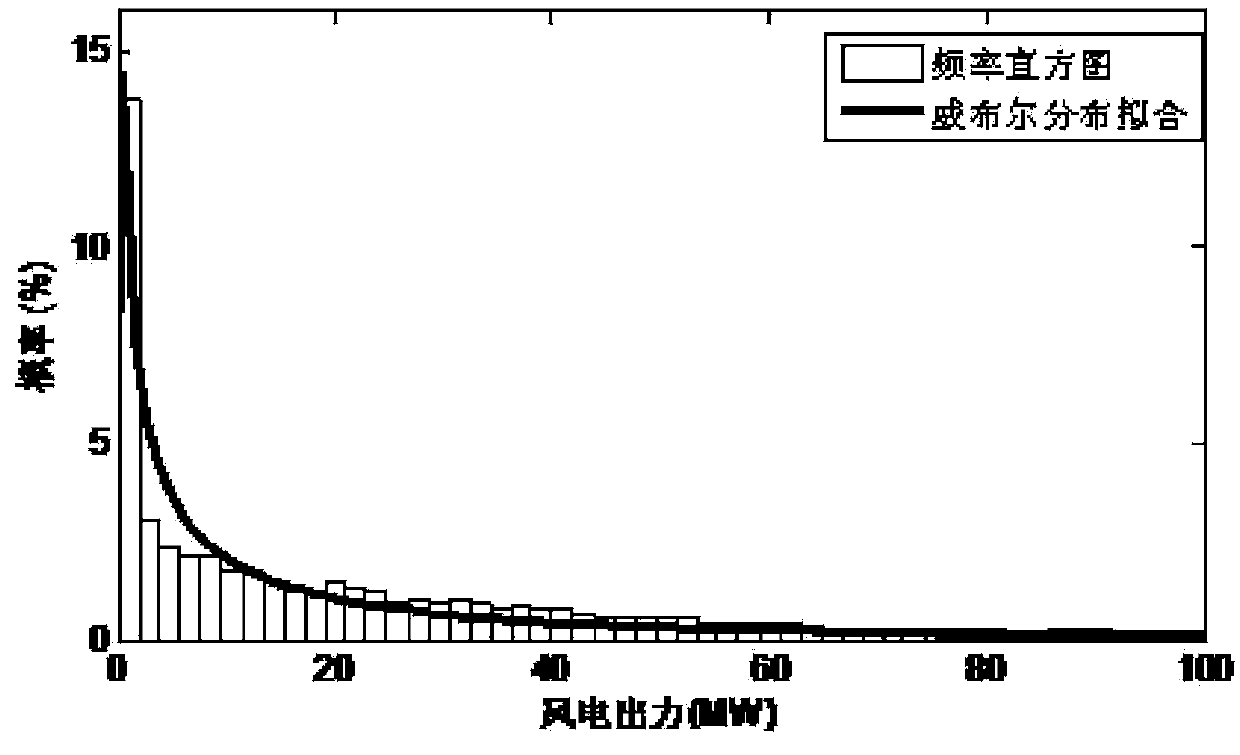
The invention provides a correlation-associated wind power output scene construction method. The method comprises the following steps: firstly, according to a historical output data statics result of a wind power field, extracting a representative wind power output edge probability distribution function and an output correlation index matrix among the wind power fields; calculating a representative sequence matrix in advance by adopting an off-line mode; and when a wind power output scene needs to be constructed, Latin hypercube sampling is carried out according to a wind power probability prediction result, a sampling matrix is sorted by directly adopting the sequence matrix which is calculated in advance to quickly construct the wind power output scene with correlation, so that the on-line calculation time of scene construction is greatly shortened to meet the practical requirements of engineering.
Owner:STATE GRID CORP OF CHINA +3
Microbial community functional gene analysis method based on protein sequence similarity
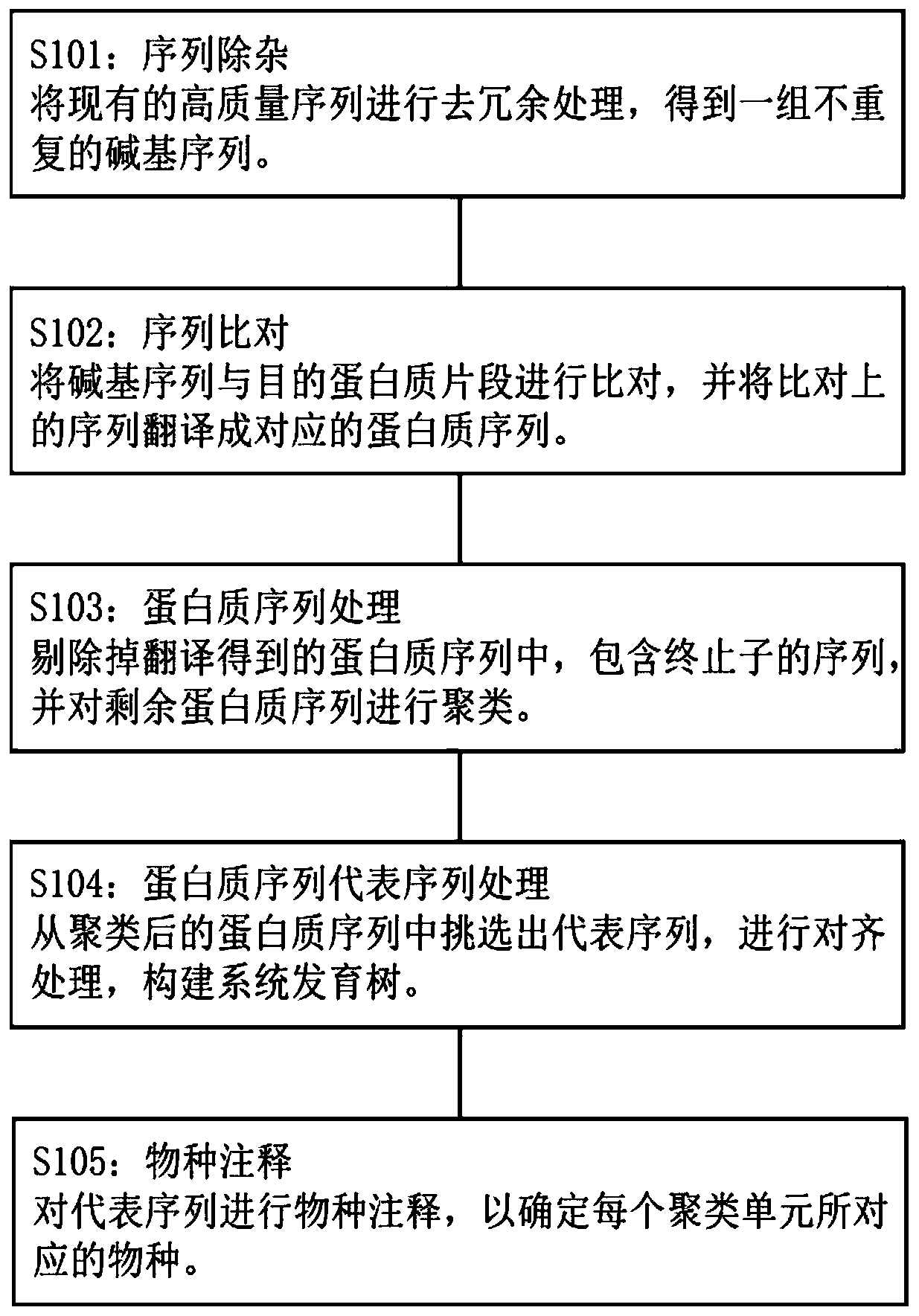
The invention discloses a microbial community functional gene analysis method based on protein sequence similarity. The microbial community functional gene analysis method comprises the following steps: sequence impurity removal step; sequence comparison step; protein sequence processing step; protein sequence representative sequence processing step; and species annotation step. According to the method, compared with the OUT method, the data analysis is more simplified, and merged protein classification units are more centralized. An amino acid sequence rather than a nucleic acid sequence is taken as a basis for sequence merging. Factors such as degeneracy and termination codons are fully considered. For a target fragment of a specific functional gene, the method has enough good directivity, and interference sequences except the target fragment can be effectively eliminated.
Owner:SHANGHAI PASSION BIOTECHNOLOGY CO LTD
Detection method and system for flora markers and terminal
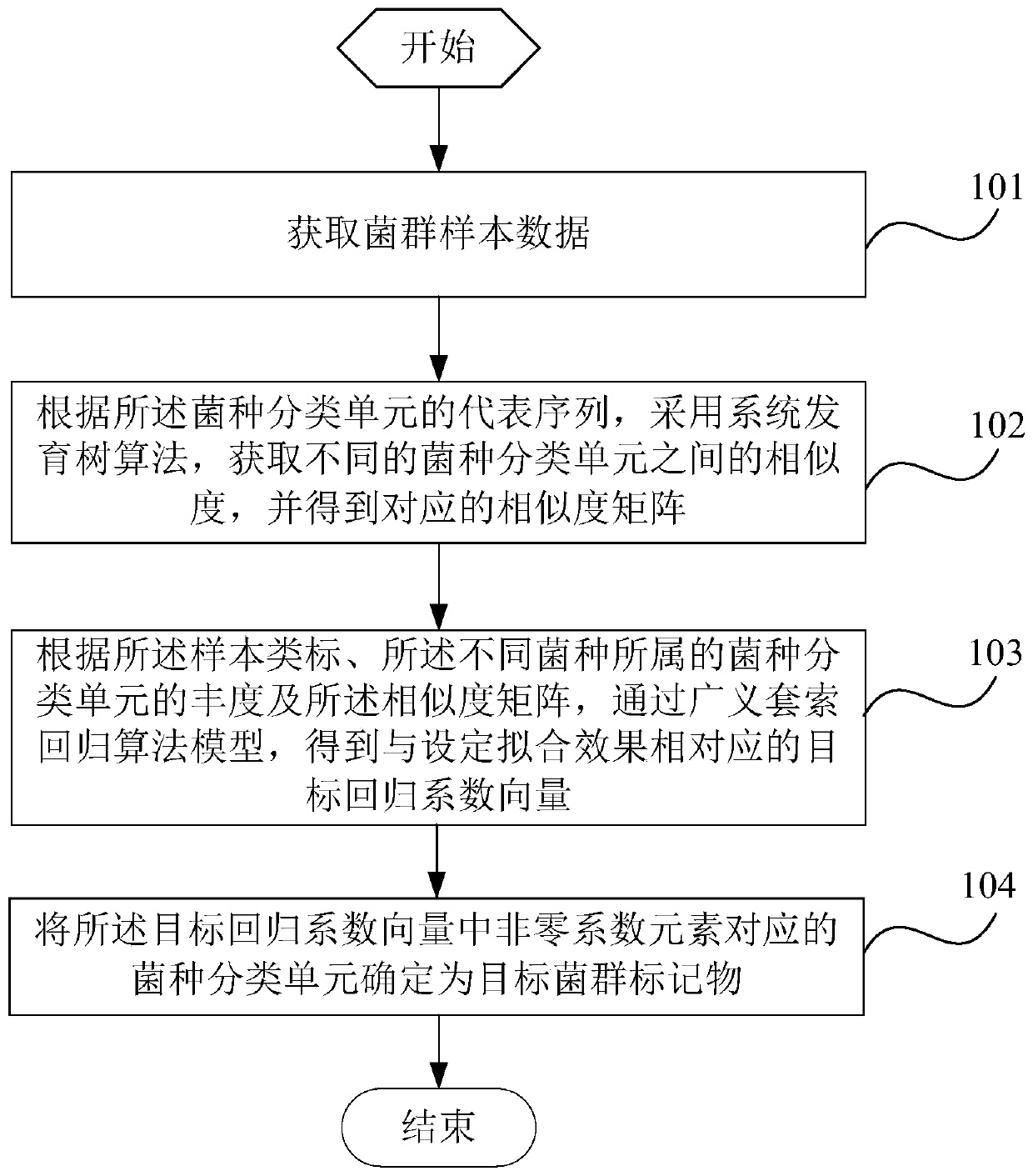
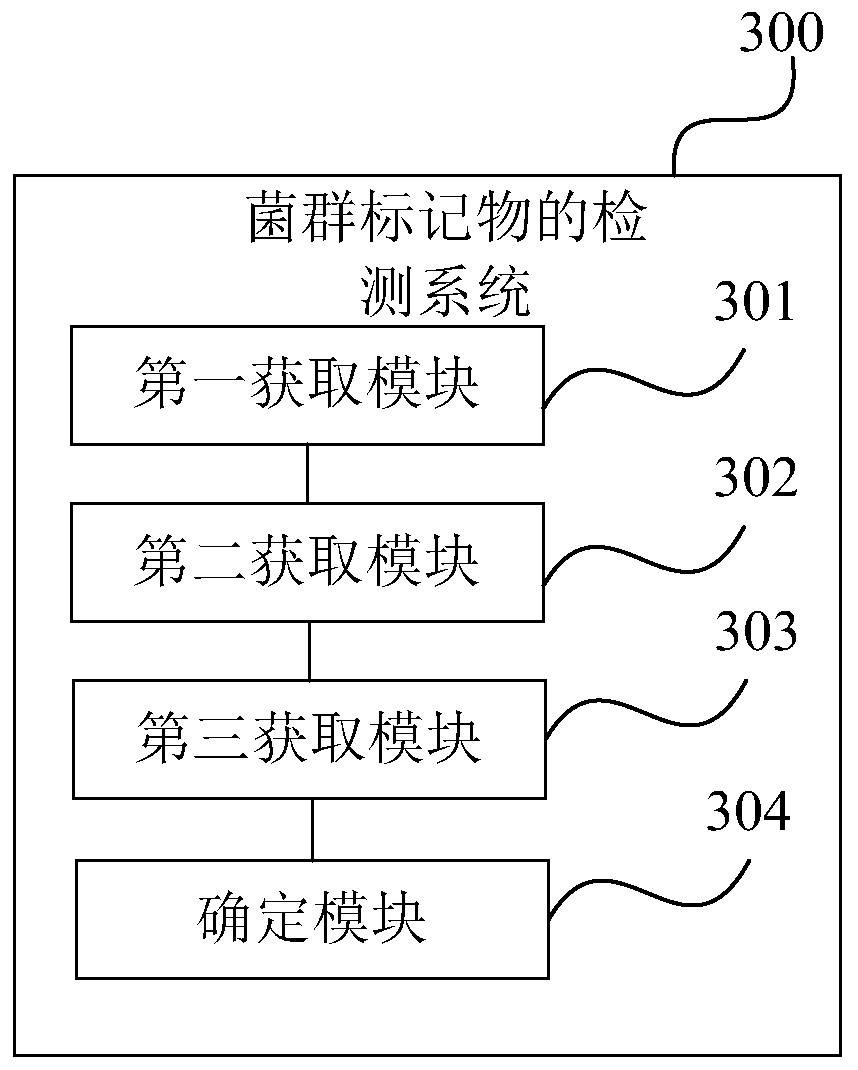
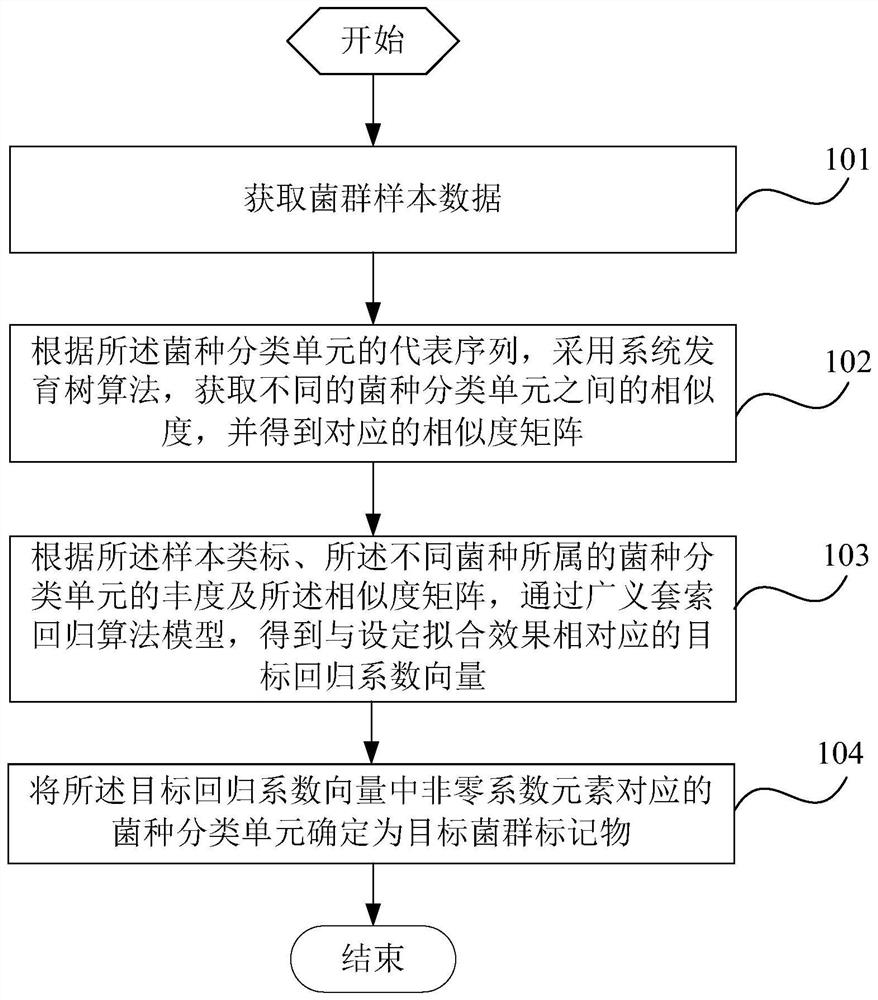
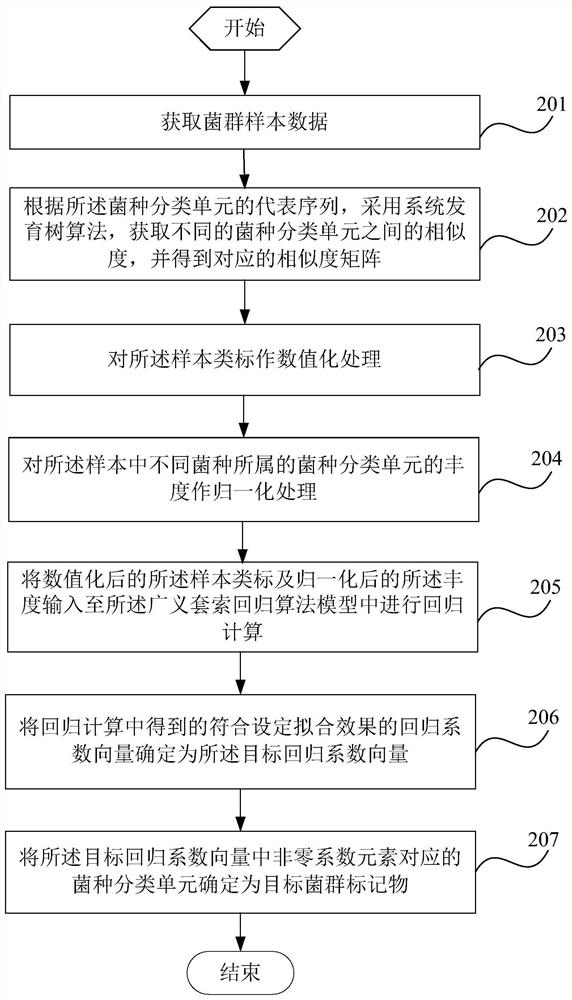
ActiveCN110444254AComprehensive reflection of complex relationshipsReflect complex relationshipsSequence analysisInstrumentsNonzero coefficientsSystem development
The invention is suitable for the technical field of biology and provides a detection method and system for flora markers and a terminal. The method comprises the steps that flora sample data is acquired; according to representative sequences of strain classifications units, by adopting a system development tree algorithm, the similarity of different strain classifications units is acquired, and acorresponding similarity matrix is obtained; according to sample type markers, the abundance of the strain classifications units to which different strains belong, and the similarity matrix, througha generalized lasso regression algorithm model, a target regression coefficient vector corresponding to a set fitting effect is obtained; the strain classifications units, corresponding to nonzero coefficient elements, in the target regression coefficient vector are determined as target flora markers, and the effectiveness of the screened flora markers is improved.
Owner:SHENZHEN INST OF ADVANCED TECH +1
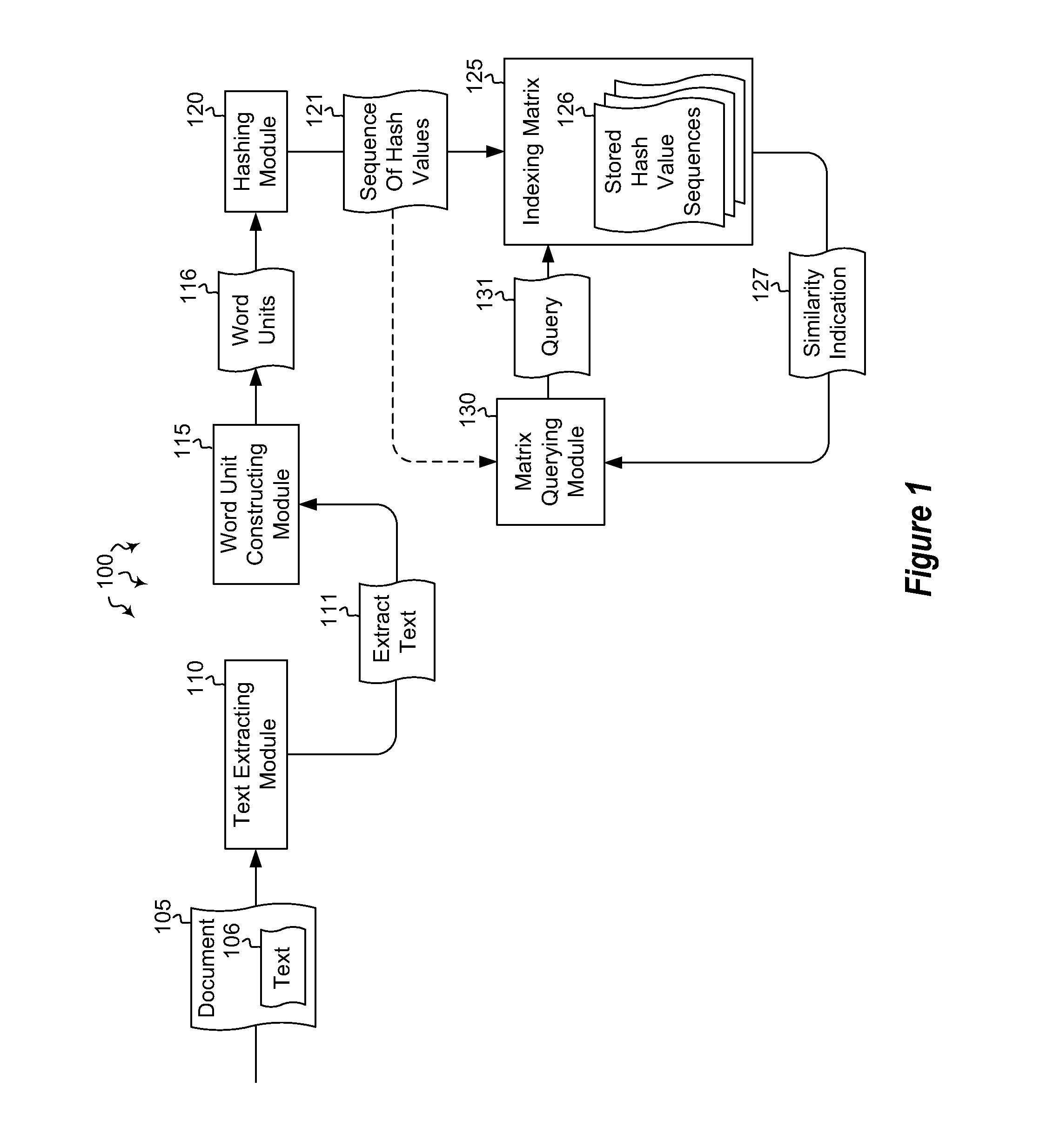
Indexing and querying hash sequence matrices
InactiveUS9129007B2Digital data processing detailsText database indexingAlgorithmComputerized system
Embodiments are directed to indexing and querying a sequence of hash values in an indexing matrix. A computer system accesses a document to extract a portion of text from the document. The computer system applies a hashing algorithm to the extracted text. The hash values of the extracted text form a representative sequence of hash values. The computer system inserts each hash value of the sequence of hash values into an indexing matrix, which is configured to store multiple different hash value sequences. The computer system also queries the indexing matrix to determine how similar the plurality of hash value sequences are to the selected hash value sequence based on how many hash values of the selected hash value sequence overlap with the hash values of the plurality of stored hash value sequences.
Owner:MICROSOFT TECH LICENSING LLC
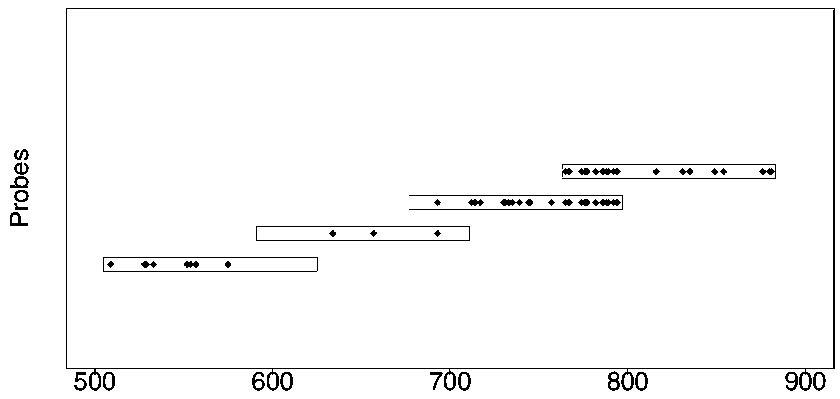
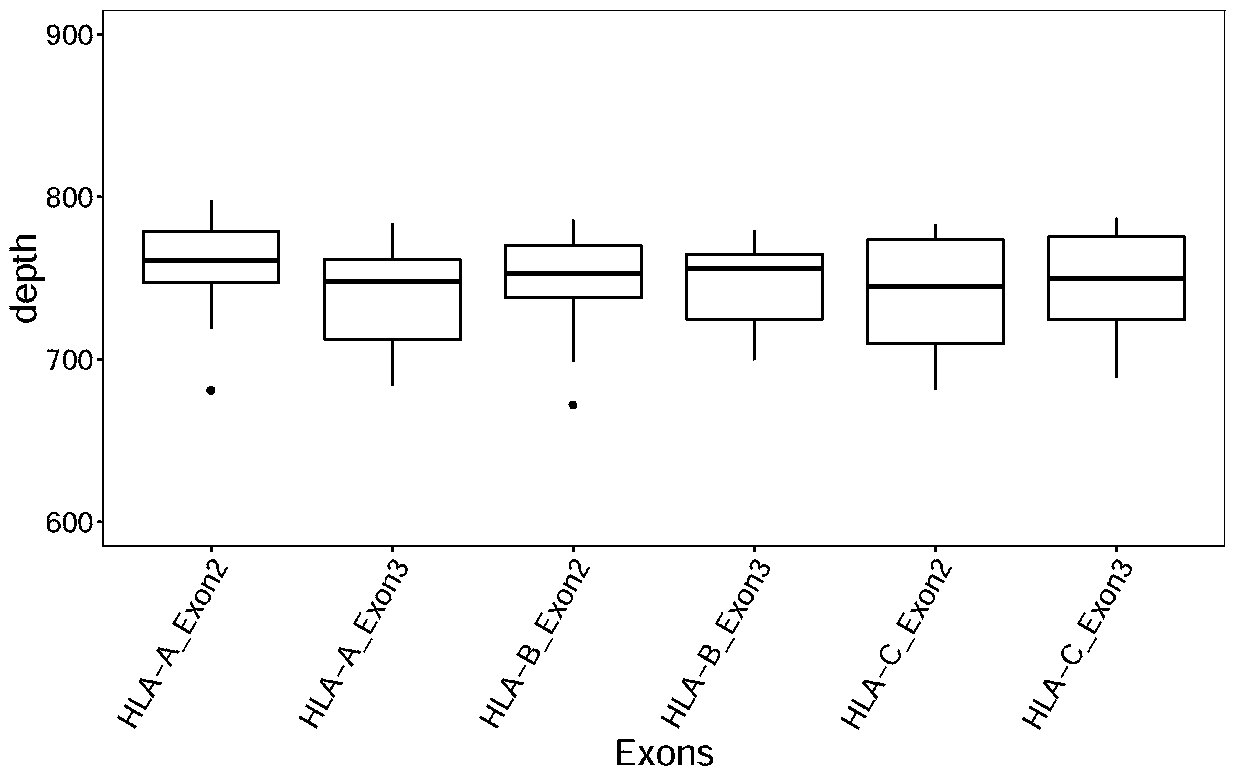
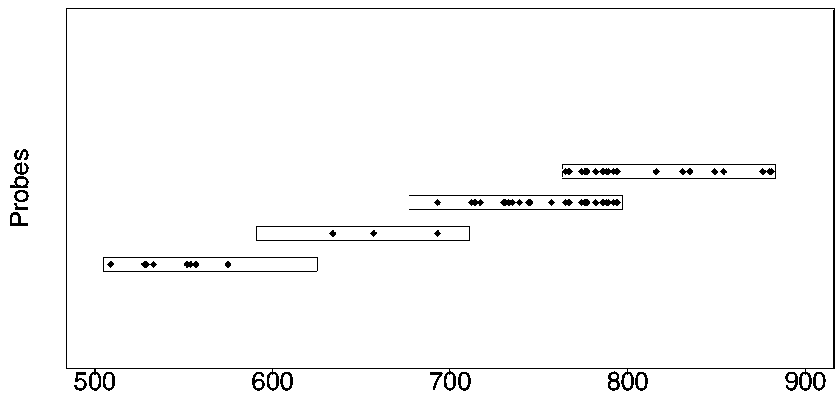
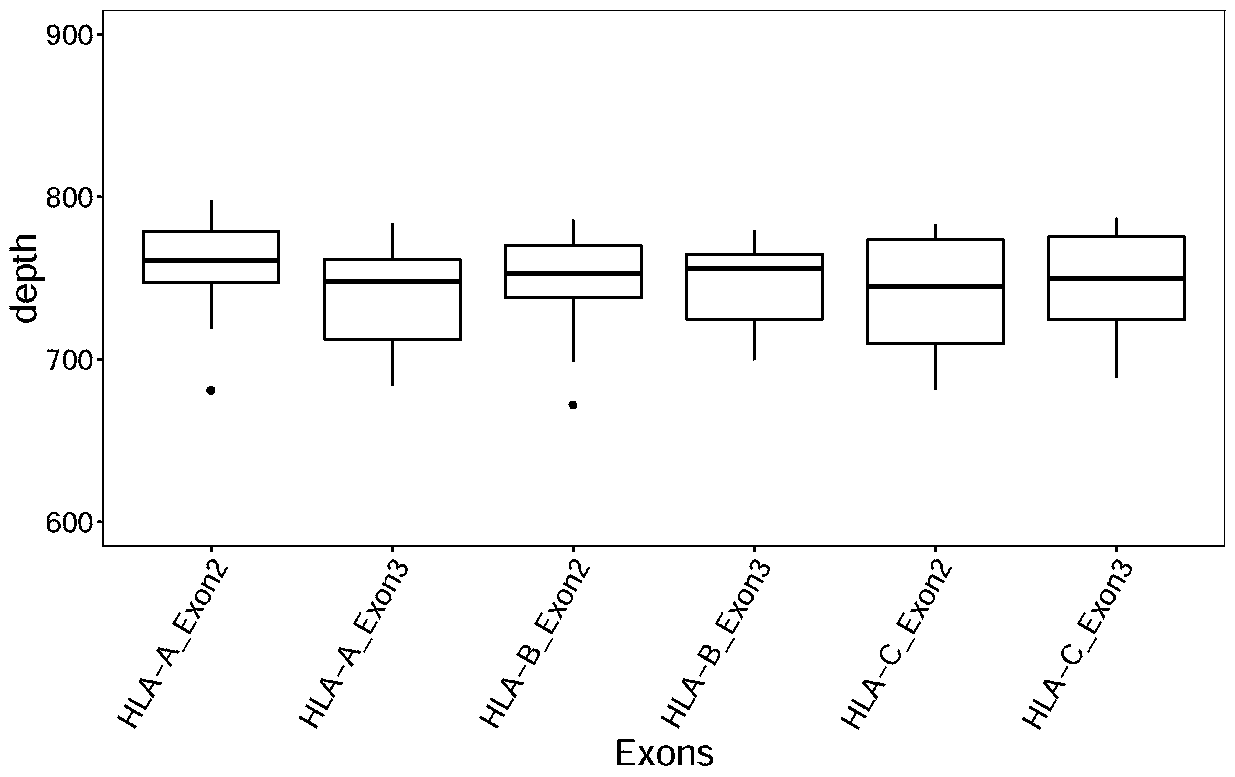
Design method of nucleic acid capture probe for HLA typing
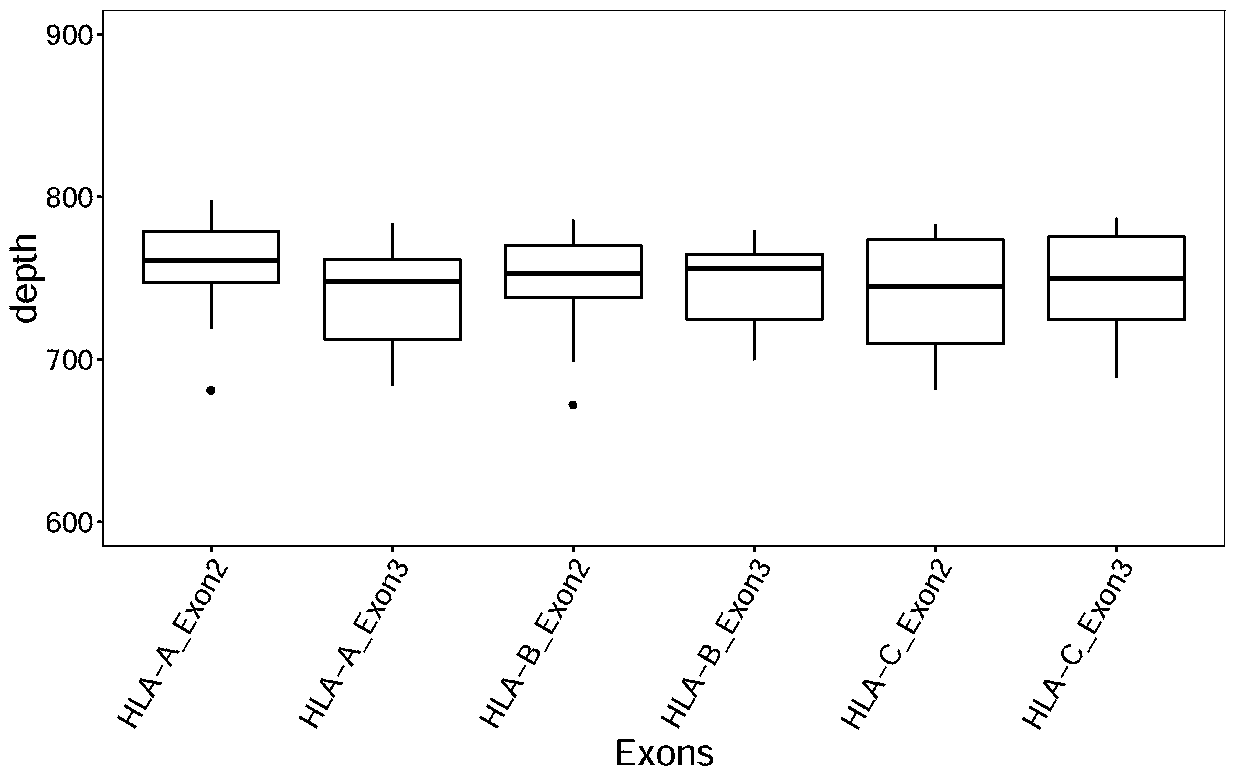
ActiveCN110853708AEfficient captureImprove the effect of hybrid captureSequence analysisHybridisationRepresentative sequencesGene probe
The invention discloses a design method of a nucleic acid capture probe for HLA typing. The design method comprises the following steps: 1) constructing HLA-A, HLA-B and HLA-C sequence libraries; 2) performing multiple sequence alignment, and searching SNP sites on each gene Exon2 and Exon3 and upstream and downstream SNP sites; 3) selecting an area covering a set number of SNP sites as a probe design candidate area by using a sliding window separation algorithm; 4) performing clustering analysis to obtain a representative sequence of each probe design candidate region as a candidate probe; 5)repeating all the candidate probes to obtain capture probes of the three genes. The invention also discloses the probe designed by the method, and the sequence of the probe is shown as SEQ ID NO: 9-88. The nucleic acid capture probe is designed for all polymorphic sites of Exon2 and Exon3 regions with the GC proportion of 60% or above in the HLA gene, and the hybrid capture effect of the HLA geneprobe is improved.
Owner:上海仁东医学检验所有限公司
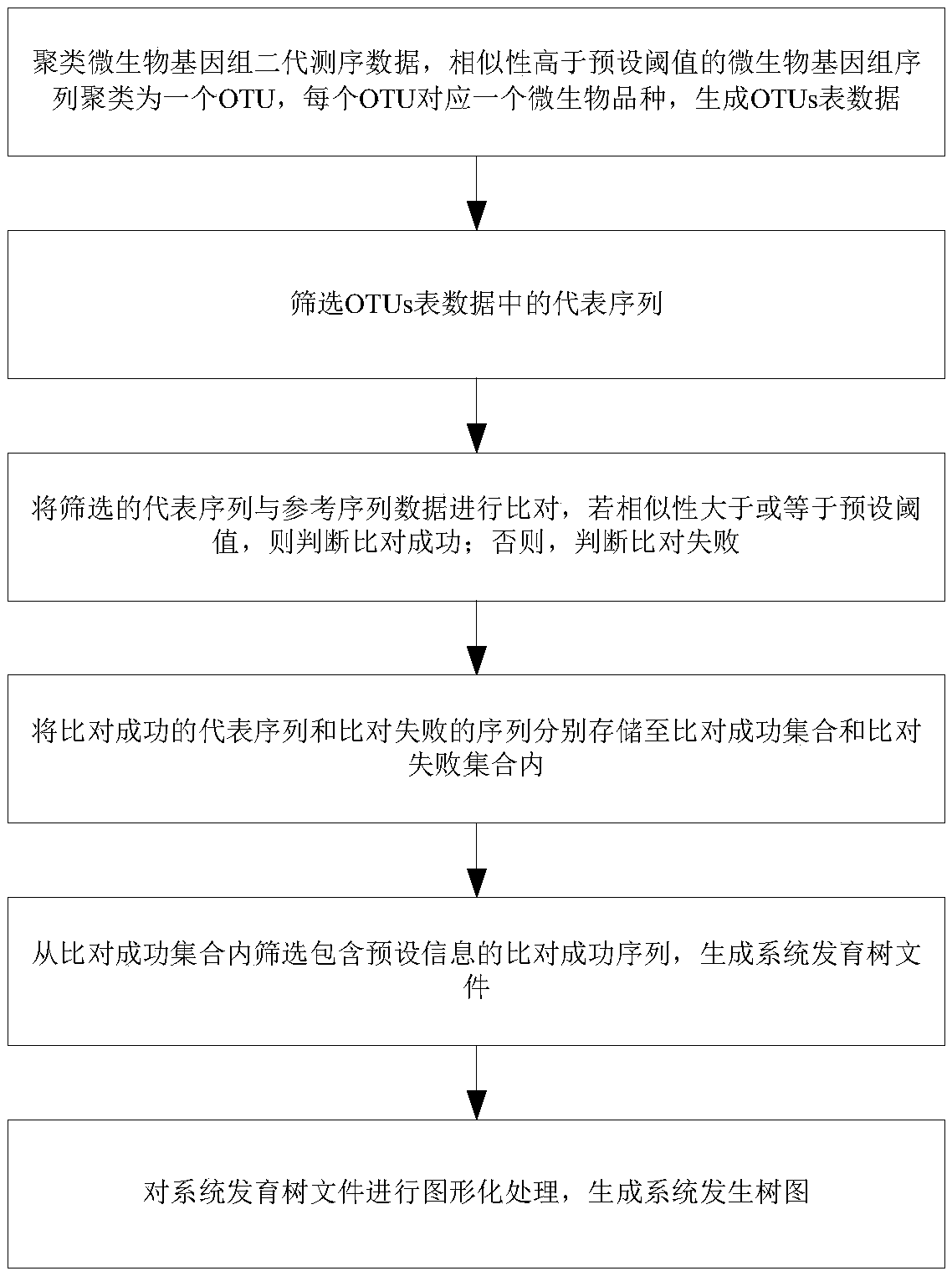
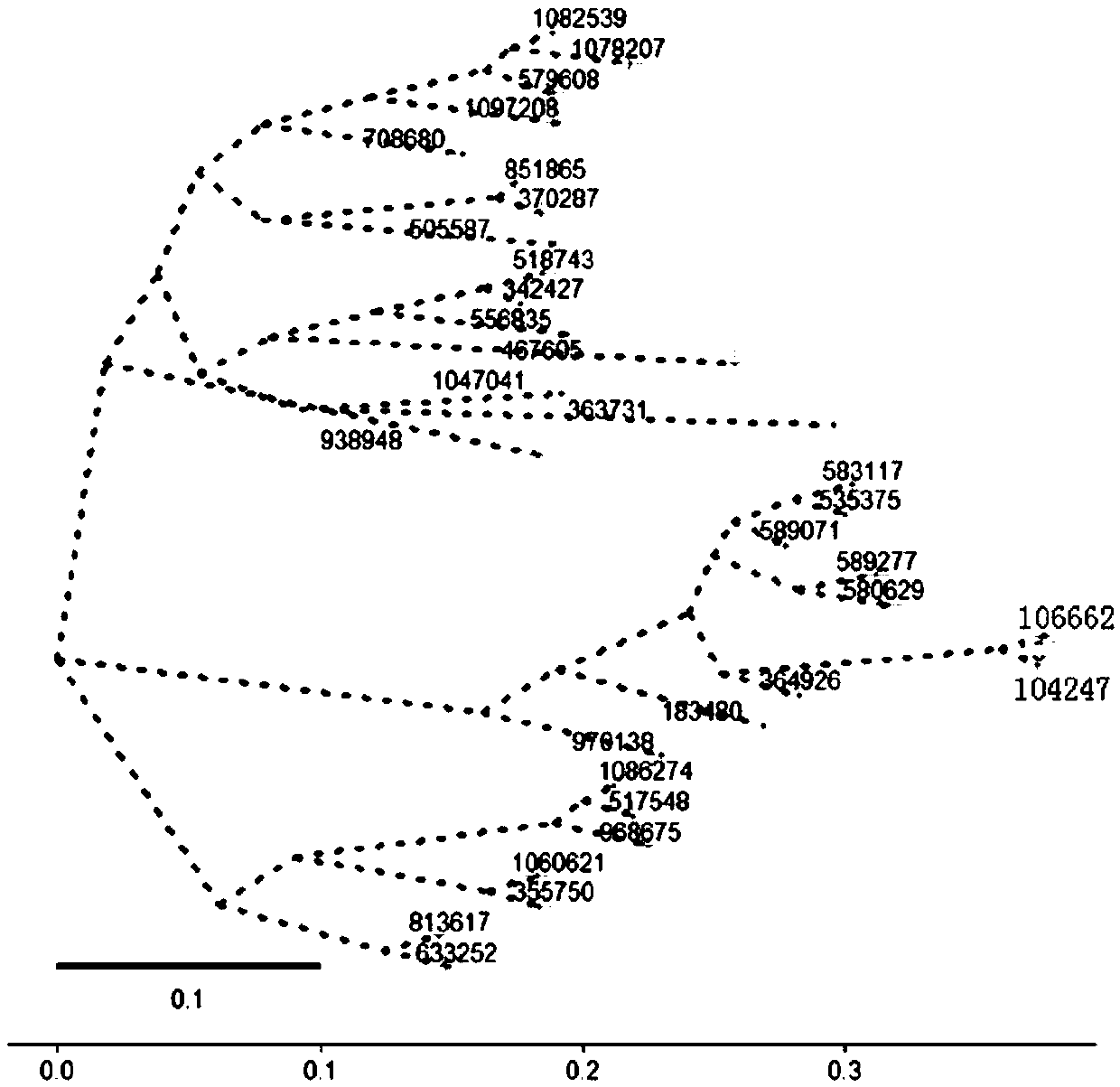
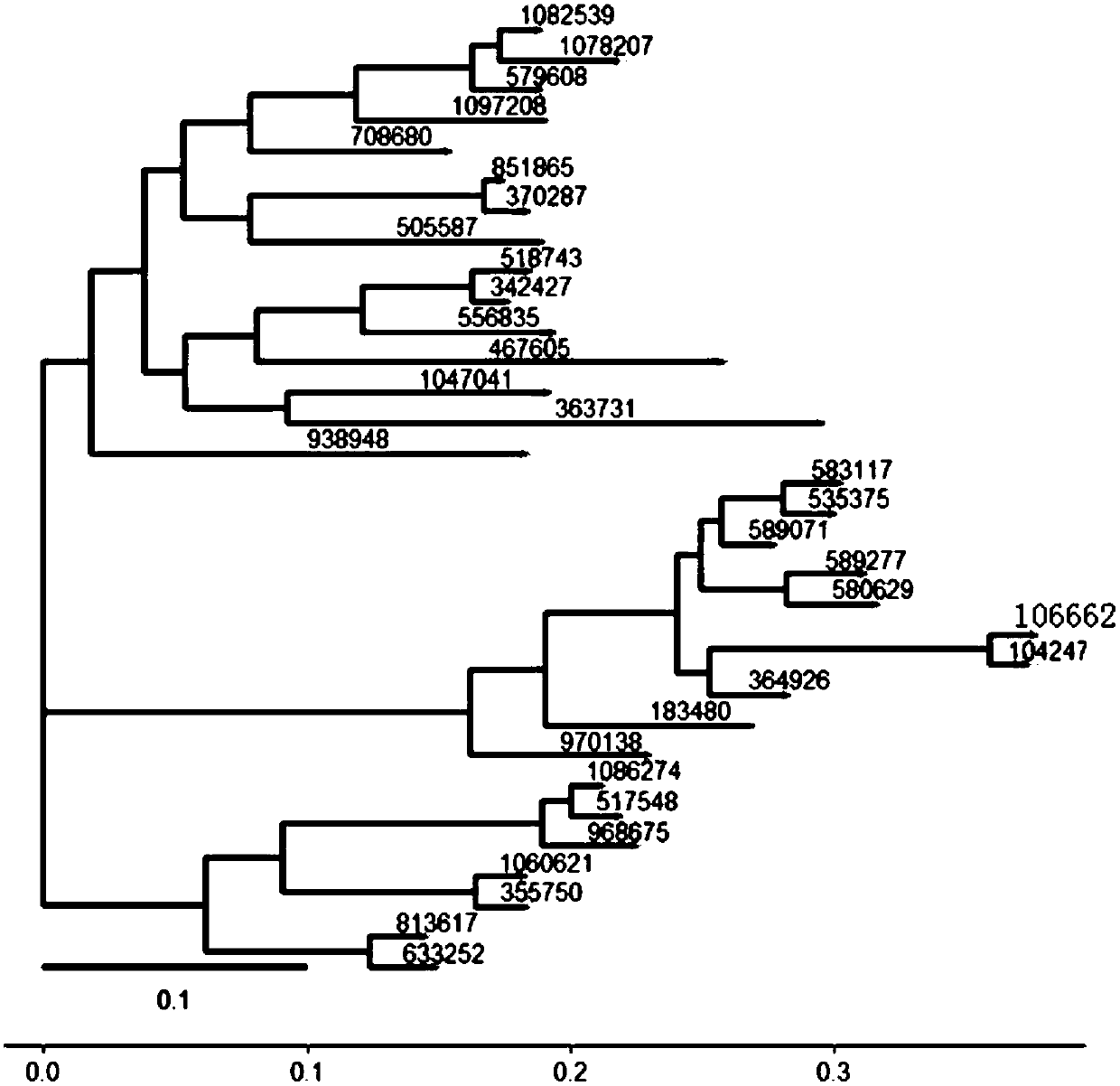
System generation tree diagram manufacturing method and system thereof
PendingCN109686406AConvenient, fast and accurate generationSimplify the drawing processBiostatisticsSequence analysisPhylogenetic treeRepresentative sequences
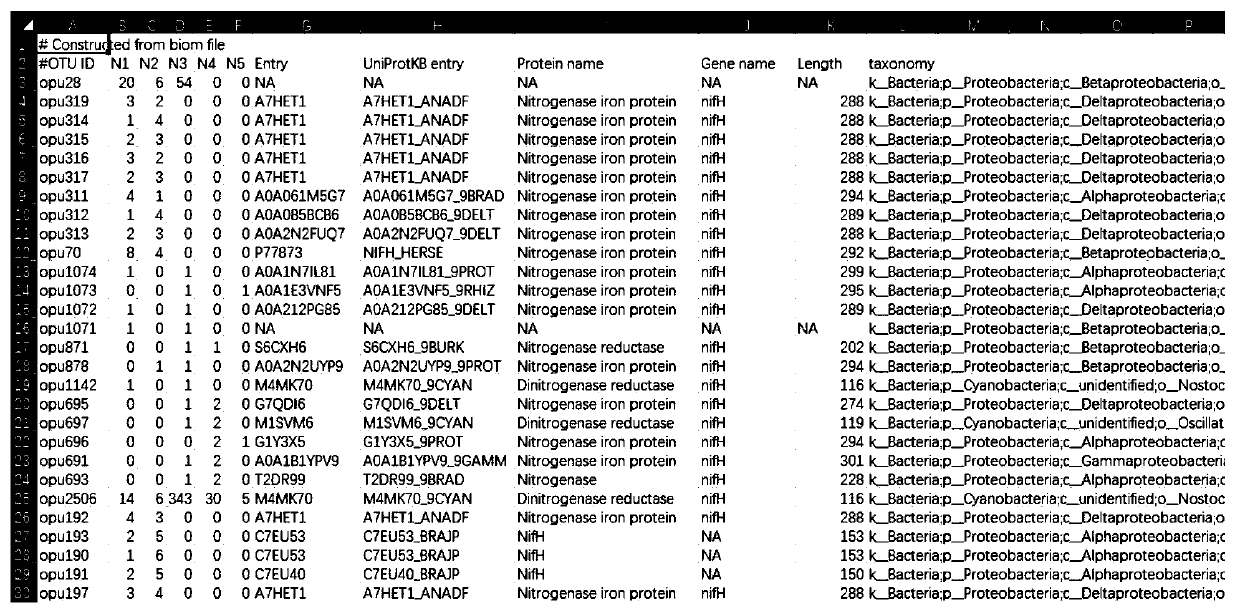
The invention provides a system generation tree diagram manufacturing method and a system thereof, wherein the system generation tree diagram manufacturing method comprises the steps of clustering microbe genome second-generation sequencing data, clustering the microbe genome sequence with similarity which is higher than a preset threshold for forming one OTU, wherein each OUT corresponds with onemicrobe kind, and generating OTUs table data; screening a representative sequence in the OTUs table data; comparing the screened representative sequence with reference sequence data, if similarity ishigher than or equal with a preset threshold, determining successful comparison; and otherwise, determining failed comparison; respectively storing the representative sequence after successful comparison and the sequence after failed comparison into a successful comparison set and a failed comparison set; screening the successful comparison sequence which contains preset information from the successful comparison set, and generating a phylogenetic tree file; and performing patterning processing on the phylogenetic tree file and generating a phylogenetic tree diagram.
Owner:INST OF BASIC MEDICINE OF SAMS
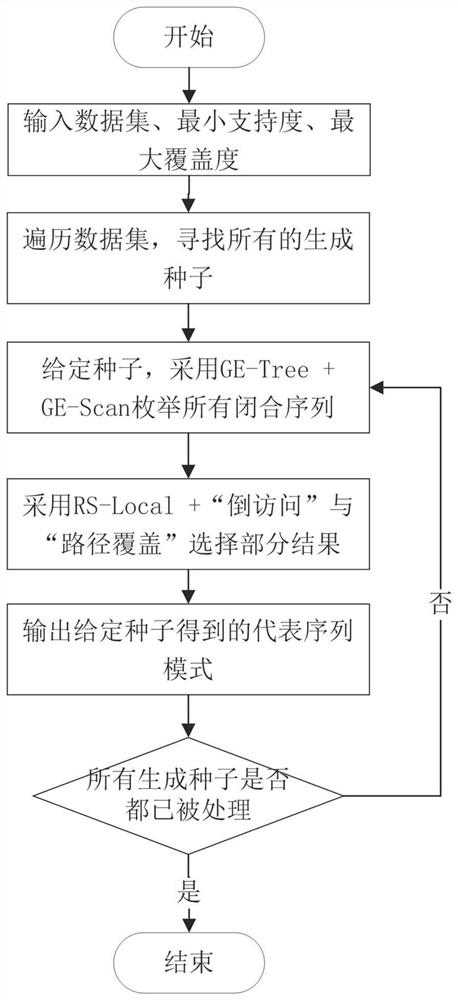
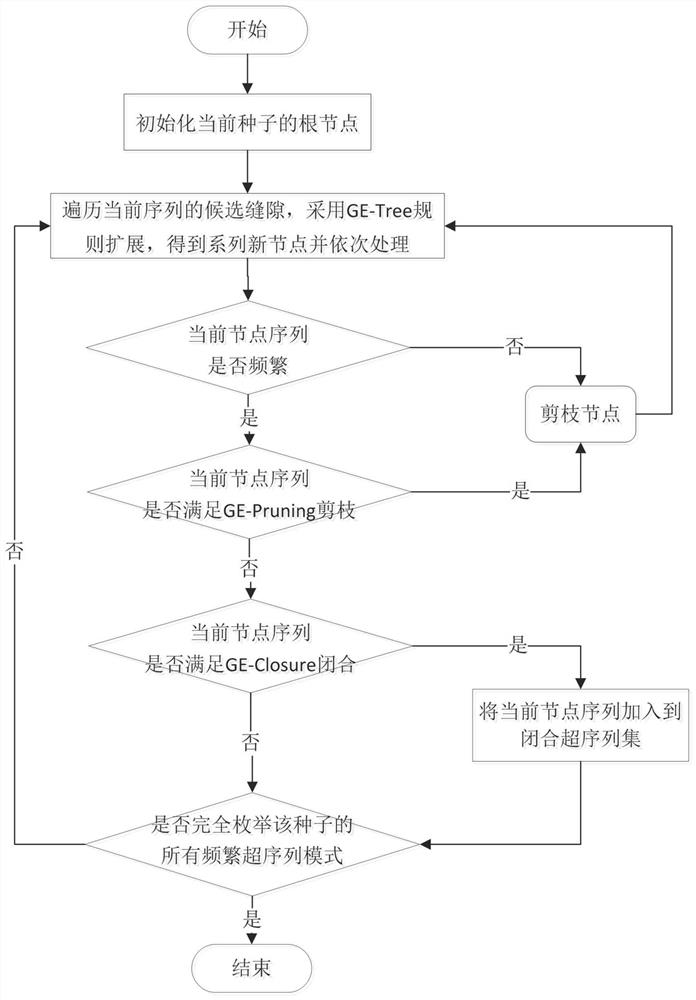
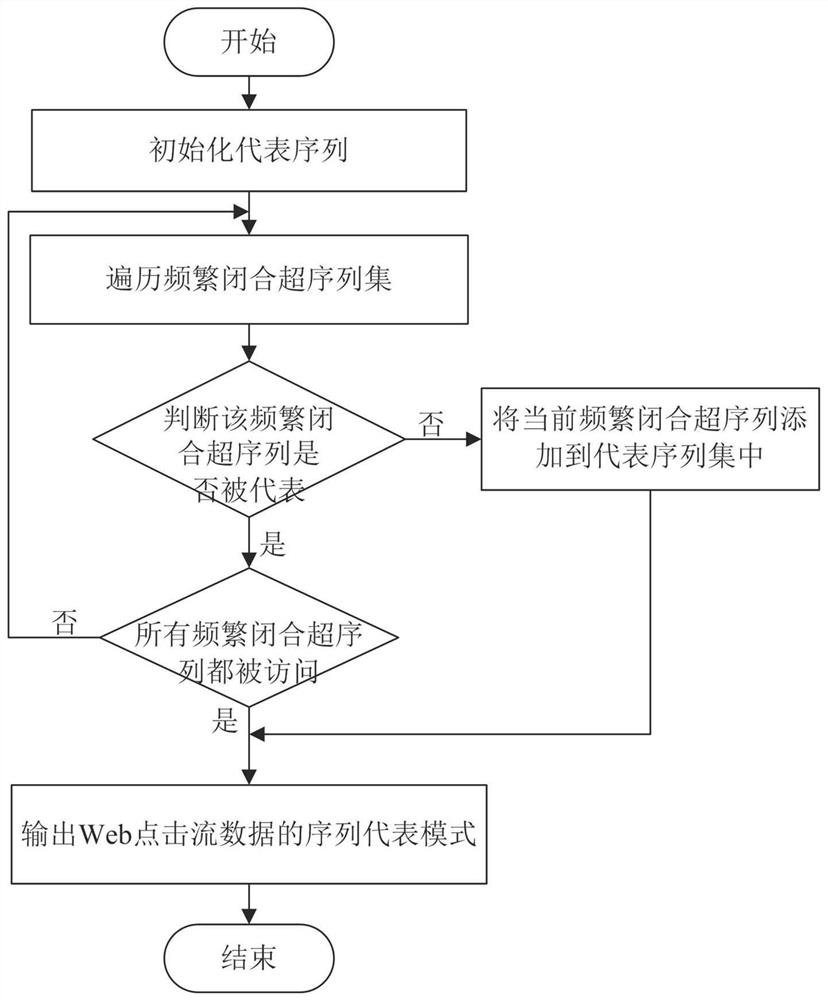
Method for mining representative sequence pattern from Web click stream data
ActiveCN112765469AAvoid efficiency lossImprove usabilityData miningOther databases indexingStreaming dataData set
The invention provides a method for mining a representative sequence pattern from Web click stream data, and relates to the technical field of sequence pattern mining. The method comprises the following steps: firstly, inputting a Web click stream sequence data set, a minimum support degree and a maximum coverage degree, traversing the data set once, and reserving all frequent sites which are not less than the minimum support degree as sequence generation seeds; adopting a gap expansion enumeration tree for each sequence generation seed, and combining a gap scanning pruning strategy and closure check to obtain all frequently closed super sequences of the seed; further selecting all representative sequences of the seed by adopting a local representative sequence screening technology; traversing all the sequences to generate seeds, and outputting a representative sequence of each seed to obtain all representative sequence modes of the Web click stream data. The method has the advantages that the representative sequence pattern can be used to effectively overcome the contradiction that the frequent sequence pattern is large in number and low in availability, and the availability of the result can be enhanced; and reference is provided for applications of online user behavior analysis, information recommendation, engine optimization and the like of the Web click stream.
Owner:NORTHEASTERN UNIV LIAONING
Method for monitoring diversity of freshwater benthic animal communities based on environmental DNA technology
PendingCN112662783ASimple samplingEasy to operateMicrobiological testing/measurementSequence analysisFisheryHigh throughput sequence
Owner:PEKING UNIV
Bungatotoxin antigen epitope gene and application of bungatotoxin antigen epitope gene to preparation of gene vaccine and antigen
InactiveCN103865934ANon-immunogenicEasy to storeGenetic material ingredientsImmunoglobulins against animals/humansProtein targetNeutralizing antibody
The invention relates to a bungatotoxin antigen epitope gene and application of the bungatotoxin antigen epitope gene to the preparation of a gene vaccine and an antigen. Two representative sequences are extracted from each of a long chain of alpha-BGT, chains A and B of beta-BGT and kappa-BGT of a cDNA sequence of bungatotoxin, the signal peptide and propeptide parts of the sequences are removed, mature peptide parts are collected, 10 antigen sites are obtained through analysis by a bioinformatics method, and a gene sequence containing the 10 sites is manually synthesized, and is connected to an eukaryotic expression vector pIRESneo to immunize an animal as the gene vaccine to obtain an anti-bungatotoxin neutralizing antibody; the sequence can also be connected to a prokaryotic expression vector PET-28a and then transformed to expression bacterium BL21 to induce the expression of the protein, and the target protein is separated to immunize the animal as an immunogen to obtain the anti-bungatotoxin neutralizing antibody.
Owner:中国人民解放军成都军区疾病预防控制中心
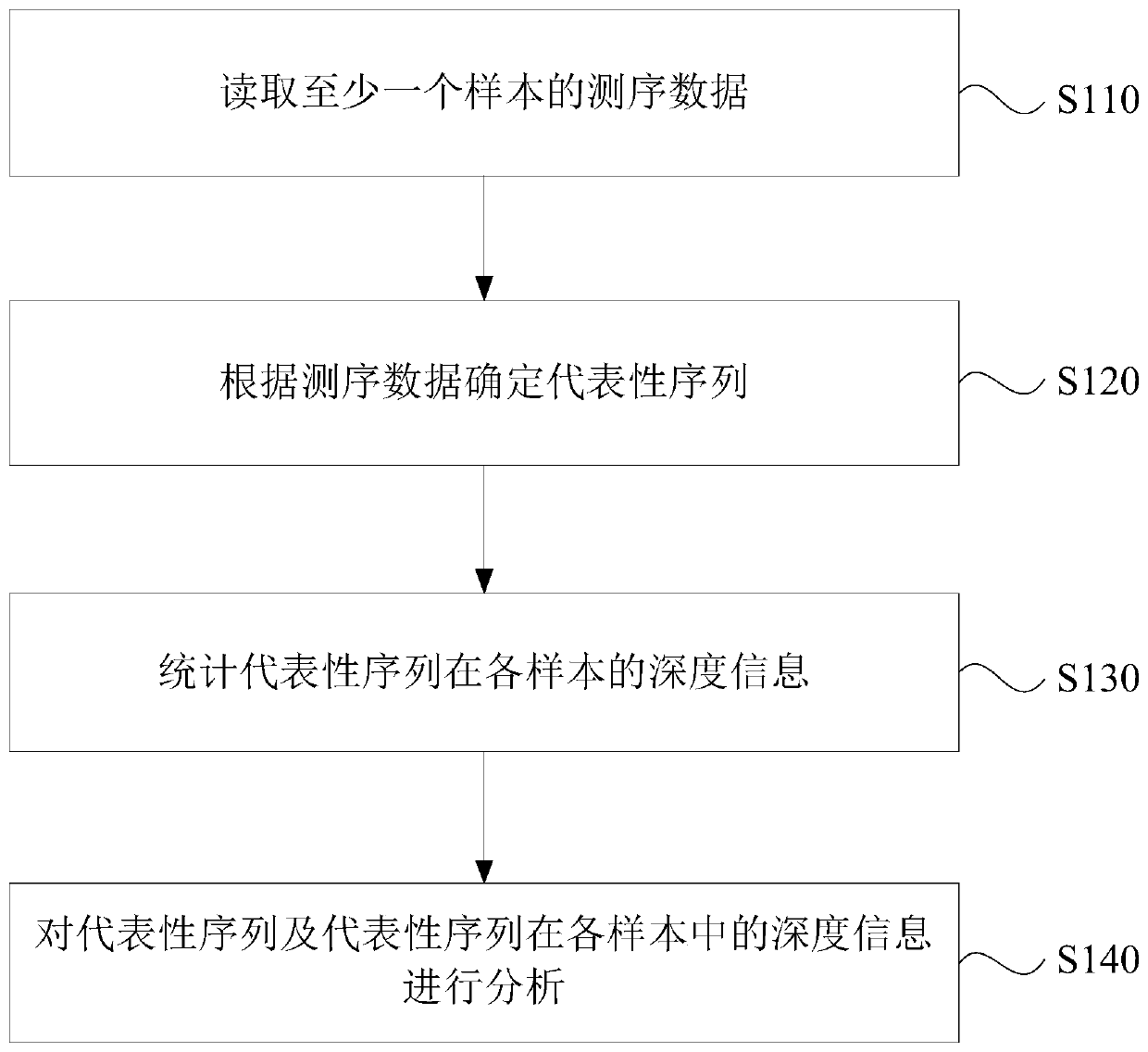
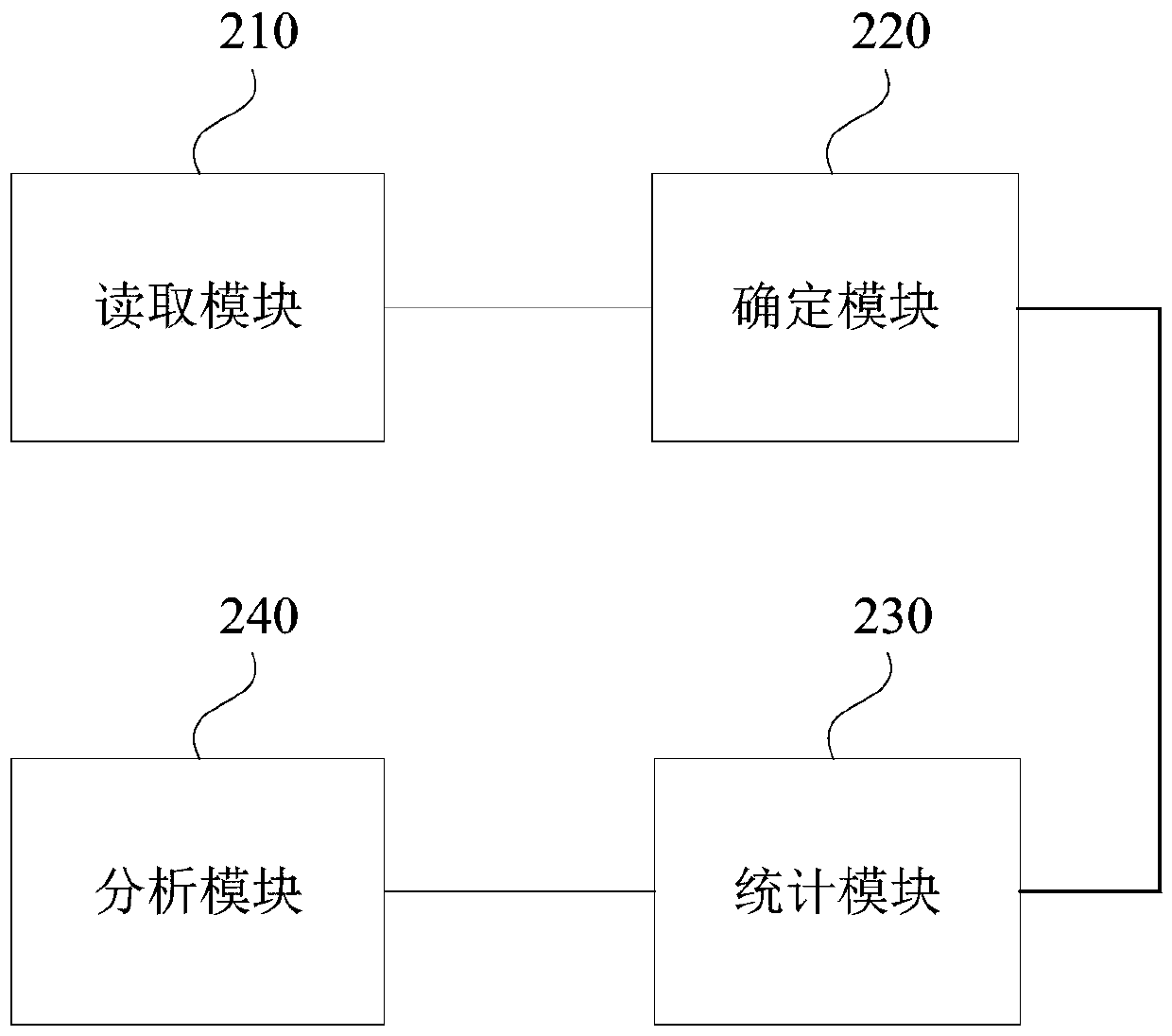
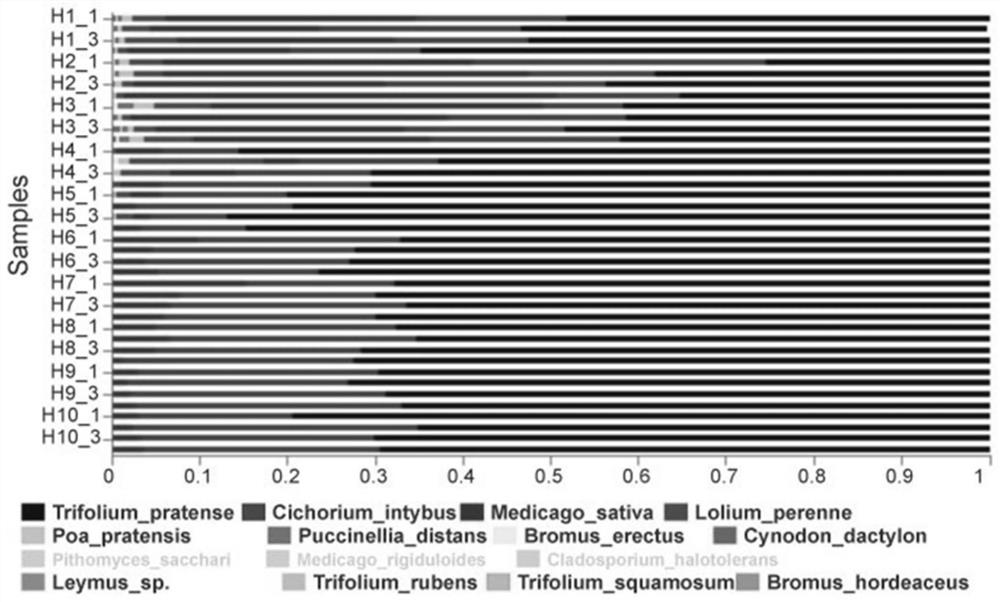
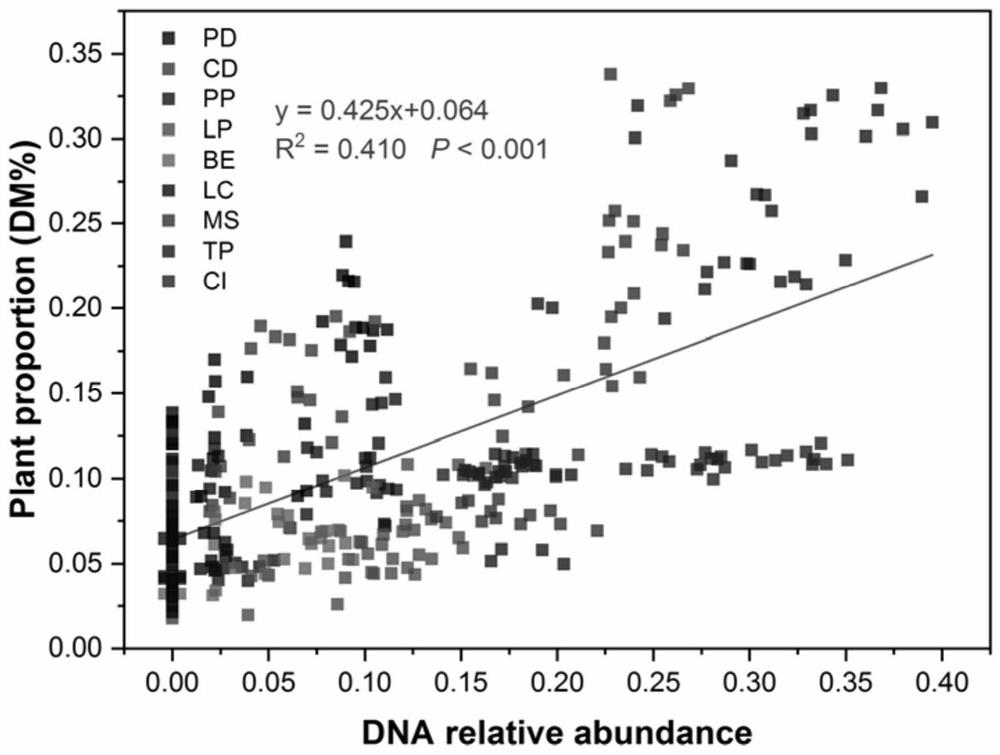
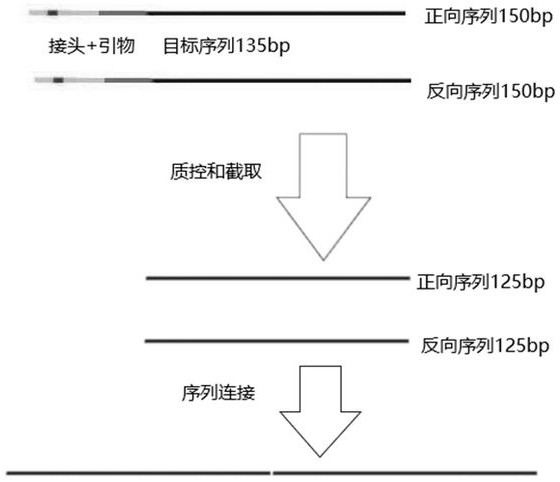
Data analysis method, device and equipment and storage medium
The invention discloses a data analysis method, device and equipment and a storage medium. The method comprises the following steps: reading sequencing data of at least one sample; determining a representative sequence according to the sequencing data; counting depth information of the representative sequence in each sample; and analyzing the representative sequence and the depth information of the representative sequence in each sample. Through the technical scheme of the invention, the components can be detected more completely and accurately, and the detection sensitivity and integrity areexpanded.
Owner:GENEWIZ INC SZ
Construction method of livestock ingestion composition correction model based on macro bar code
PendingCN112599200AImprove accuracyData processing applicationsSequence analysisBiotechnologyLinear regression
The invention provides a construction method of a livestock ingestion composition correction model based on a macro bar code, which is characterized by comprising the following steps: (1) selecting different forage grasses to prepare a simulated ration as a correction sample, extracting DNA, carrying out ITS2 bar code PCR amplification, and carrying out amplicon Illumina MiSeq double-end sequencing to obtain original sequencing data; (2) carrying out filtering and quality control processing on the original sequencing data, and performing OTU clustering on effective sequences; (3) comparing theOTU representative sequence with an NCBI database Blastn to complete species annotation; (4) measuring the forage grass gene sequence of the sample by using a macro bar code technology to obtain a relative proportion measured value of the forage grass DNA sequence, and analyzing to obtain the components of the simulated ration; and (5) correlating the measured value of each component of the simulated ration and the true value of the dry matter proportion of each component by using a linear regression method, and establishing a correction model of the ration composition.
Owner:CHINA AGRI UNIV
Nucleic acid capture probe for HLA typing and design method thereof
ActiveCN110600082AEfficient captureImprove the effect of hybrid captureMicrobiological testing/measurementSequence analysisHLA-BRepresentative sequences
The invention discloses a method for designing a nucleic acid capture probe for HLA typing. The method includes the steps: 1) constructing HLA-A, HLA-B, and HLA-C sequence libraries; 2) performing multiple sequence alignment, searching for SNP loci on Exon2 and Exon3 and upstream and downstream of each gene; 3) using a sliding window separation algorithm to select regions that cover a predetermined number of SNP loci as probe design candidate regions; 4) performing cluster analysis to obtain the representative sequences of the respective probe design candidate regions as candidate probes; and5) deduplicating the all candidate probes to obtain the respective capture probes of the three genes. The invention also discloses a probe designed by the above method, and its sequence is shown in SEQIDZNO: 9-88. The method designs the nucleic acid capture probes for all polymorphic loci in the Exon2 and Exon3 regions with a GC ratio of more than 60% in the HLA gene, thereby improving the effectof hybridization capture of the HLA gene probe.
Owner:上海仁东医学检验所有限公司
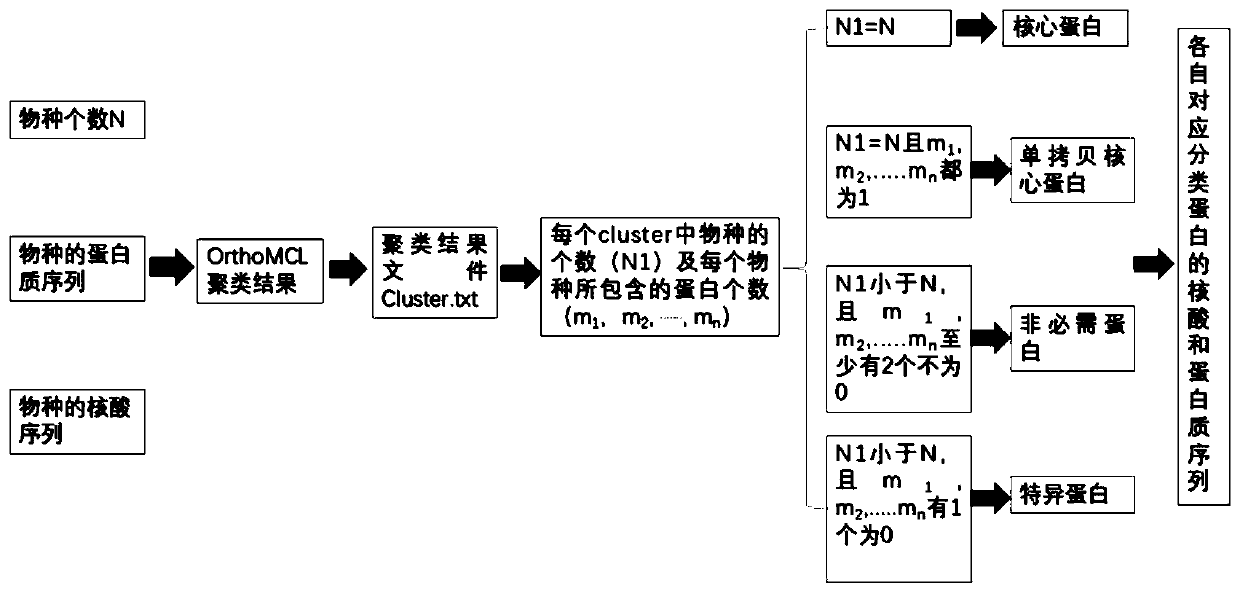
Rapid analysis method based on OrthoMCL clustering result
The invention discloses a rapid analysis method based on an OrthoMCL clustering result, and belongs to the field of comparative genomics and bioinformatics. The method is based on an OrthoMCL clustering result and comprises the steps of: establishing automatic identification of various proteins in generic analysis, including all representative proteins, core proteins, single-copy core proteins andspecies-specific proteins; based on the respective classification of the proteins, counting the number of the classified proteins existing in the respective species, and outputting the result according to the category. The method achieves output of representative sequences of proteins in various classifications and output of representative sequences of various proteins in each species. In addition, according to the method, protein homologous clustering results are output according to the sequence corresponding to each homologous protein, and a foundation is laid for achieving higher-level personalized analysis in generic analysis.
Owner:ANHUI MEDICAL UNIV
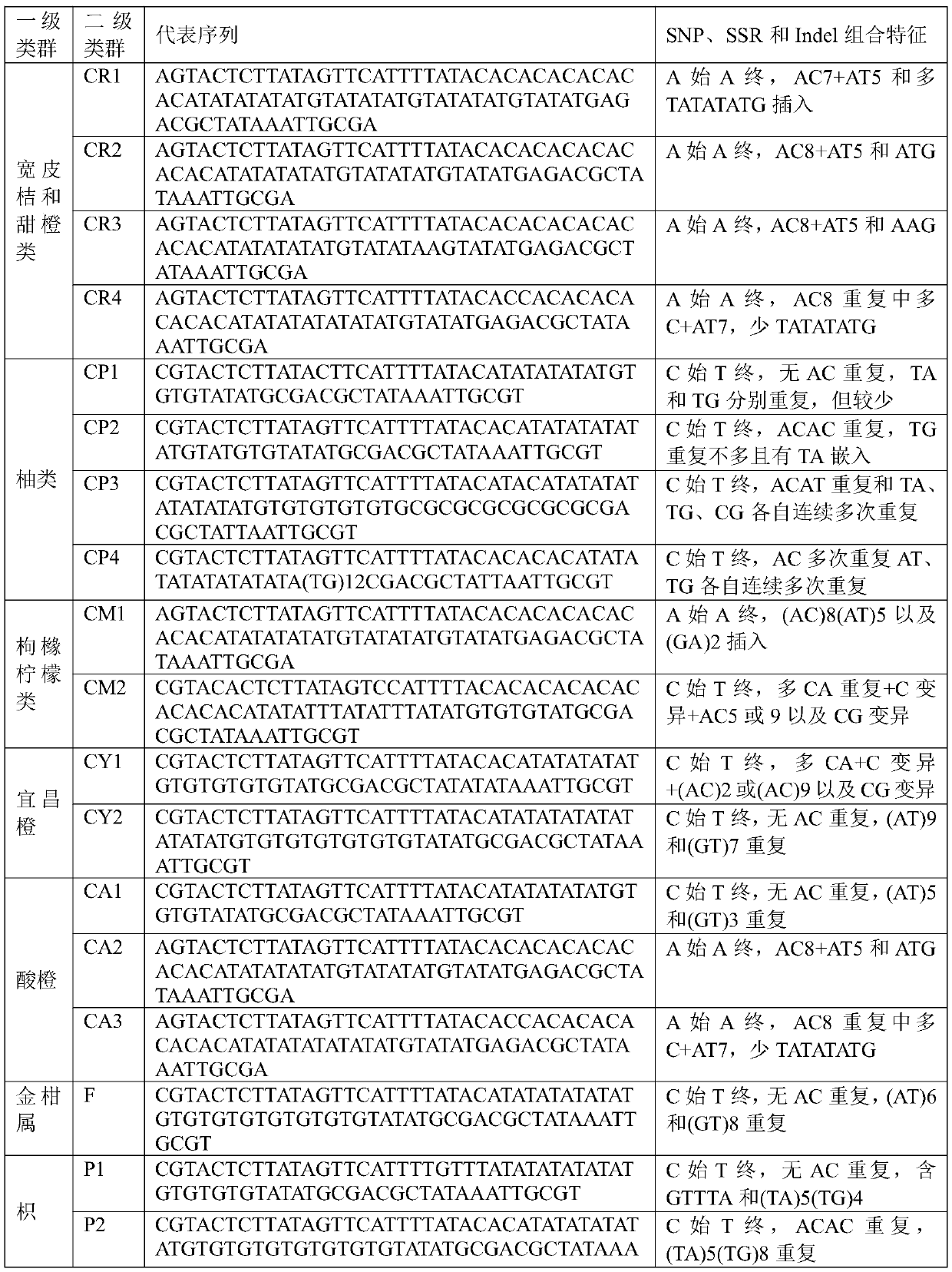
Application of short DNA sequence in identification of citrus groups and citrus seedlings
ActiveCN111593049ATaxa are well differentiatedImprove reliabilityMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyFruit tree
The invention belongs to the technical field of genetic engineering, and discloses an application of a short DNA sequence in the identification of citrus groups and citrus seedlings. Primer sequencesfor identifying the short DNA sequence are SEQ ID NO:1 and SEQ ID NO:2. The invention also provides representative sequences in main citrus groups of the sequence defined by the primers, and a SNP, SSR and Indel combination characteristic table of the representative sequences. The citrus genome is amplified by the given primers, a target short sequence is obtained and the cloning and sequencing analysis are performed, and the heterozygous state, citrus group and citrus seedling of a citrus can be identified according to the fragment SSR, Indel and / or SNP variation characteristics or change combination. The analysis and identification of citrus fruit tree different materials are completed by cloning and sequencing the specific short sequence, and comparing characteristics of the sequencingsequence. The citrus fruit trees are classified into groups through performing genome amplification, cloning and sequencing on sequences and performing comparison and analysis with the combination characteristic table.
Owner:SOUTHWEST UNIVERSITY
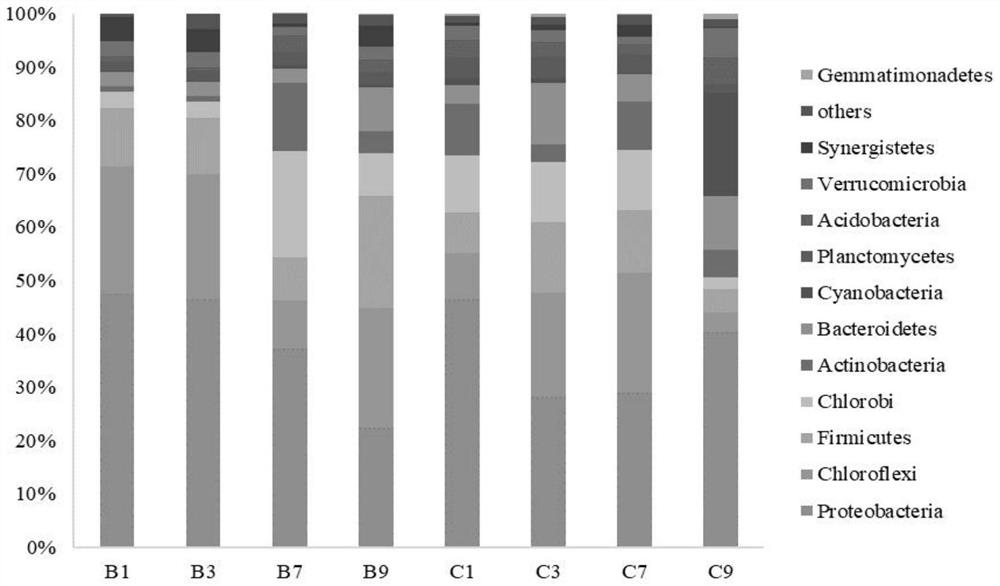
Method for detecting diversity influence of pinkflower clover on constructed wetland substrate microorganisms and application of method
The invention provides a method for detecting the diversity influence of pinkflower grass on constructed wetland matrix microorganisms and application. The method comprises the following steps: (1) preparing and collecting a sample; (2) extracting DNA of a sample, detecting the concentration and purity of the DNA, and then detecting the integrity of the DNA sample; (3) carrying out PCR (Polymerase Chain Reaction) amplification; (4) performing high-throughput sequencing; (5) performing quality control, filtering, splicing and chimera removal; performing taxonomic analysis on the OTU representative sequence, and calculating alpha diversity according to the OTU data to obtain a correlation index; according to the invention, by comparing the community composition difference of a gravel surface matrix biological membrane under the condition of planting or not planting pinkgrass, amplifying and sequencing the bacterium 16S rDNA gene by using a high-throughput sequencing technology, and comparing the diversity of microorganisms in different constructed wetland environments, the flora structure and dominant bacteria of the microorganisms can be researched, and the application of the bacteria 16S rDNA gene to the artificial wetland environment can be realized. And help is provided for finally designing a more scientific constructed wetland scheme.
Owner:WENZHOU MEDICAL UNIV
System and process for data-driven design, synthesis, and application of molecular probes
ActiveCN109071590ASugar derivativesMicrobiological testing/measurementNucleic Acid ProbesMolecular probe
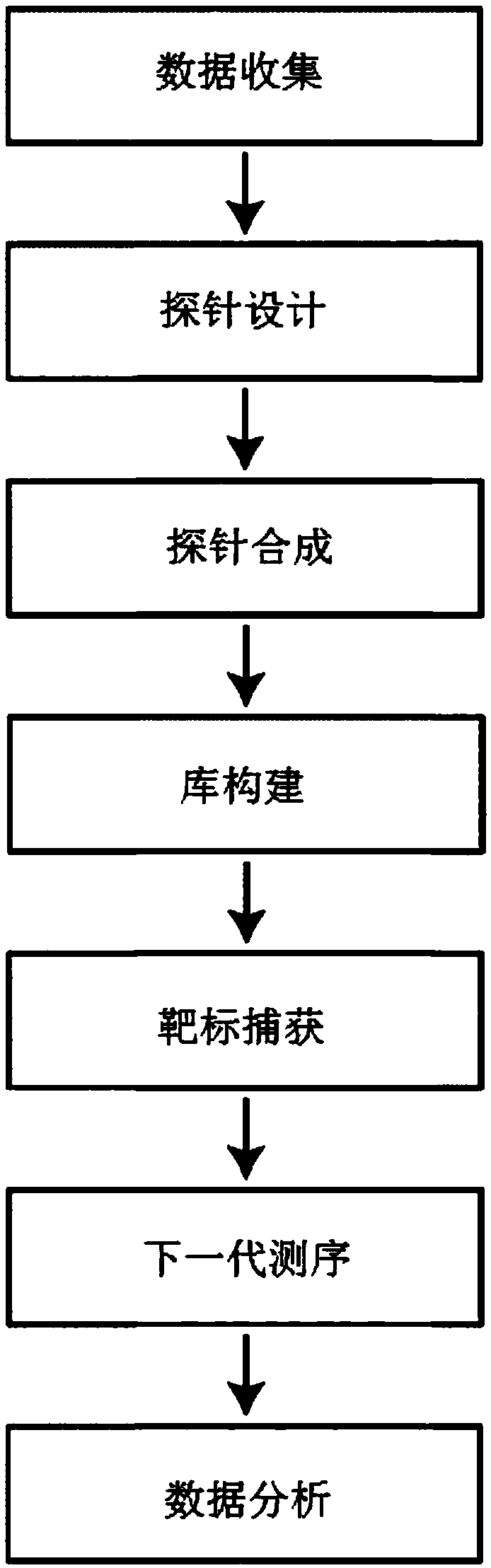
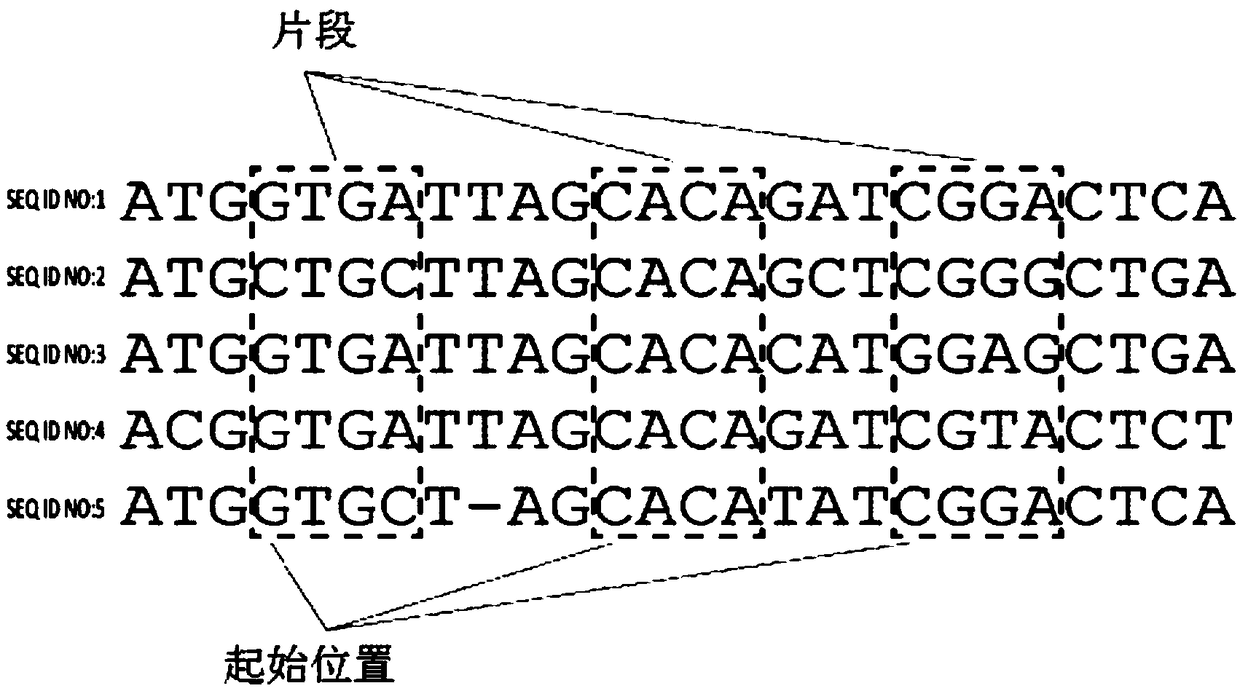
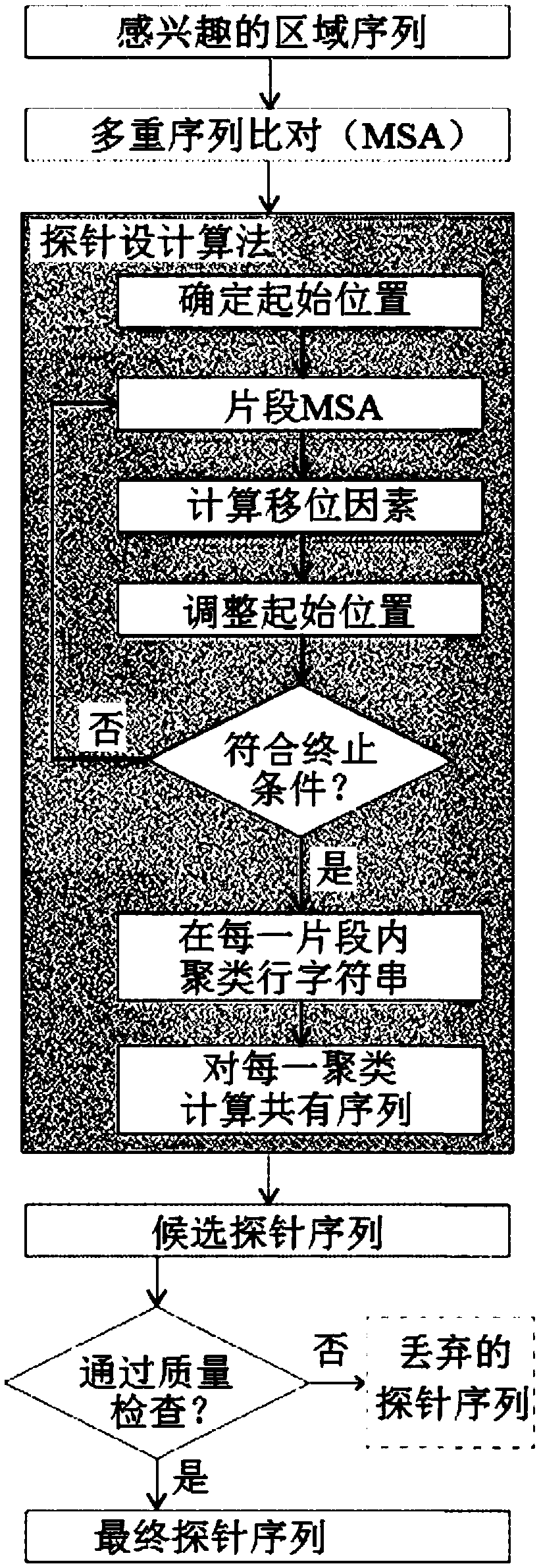
The disclosure provides methods and systems for designing and synthesizing probes to capture a representative sample of genomic variants of a target genome from a sample. The methods include providinga multiple sequence alignment (MSA), designing a plurality of representative subsequences, and optionally synthesizing a nucleic acid probe. The designing step can comprise designating a plurality ofintervals in the MSA, shifting start positions for each MSA subset, clustering the aligned subsequences within each adjusted subset, and determining a representative sequence for each reduced MSA subset. The disclosure also encompasses methods of isolating a plurality of nucleic acid variants of a targeted genomic subregion from a sample using the disclosed probe design, as well as the probe compositions themselves.
Owner:FUSION GENOMICS CORP
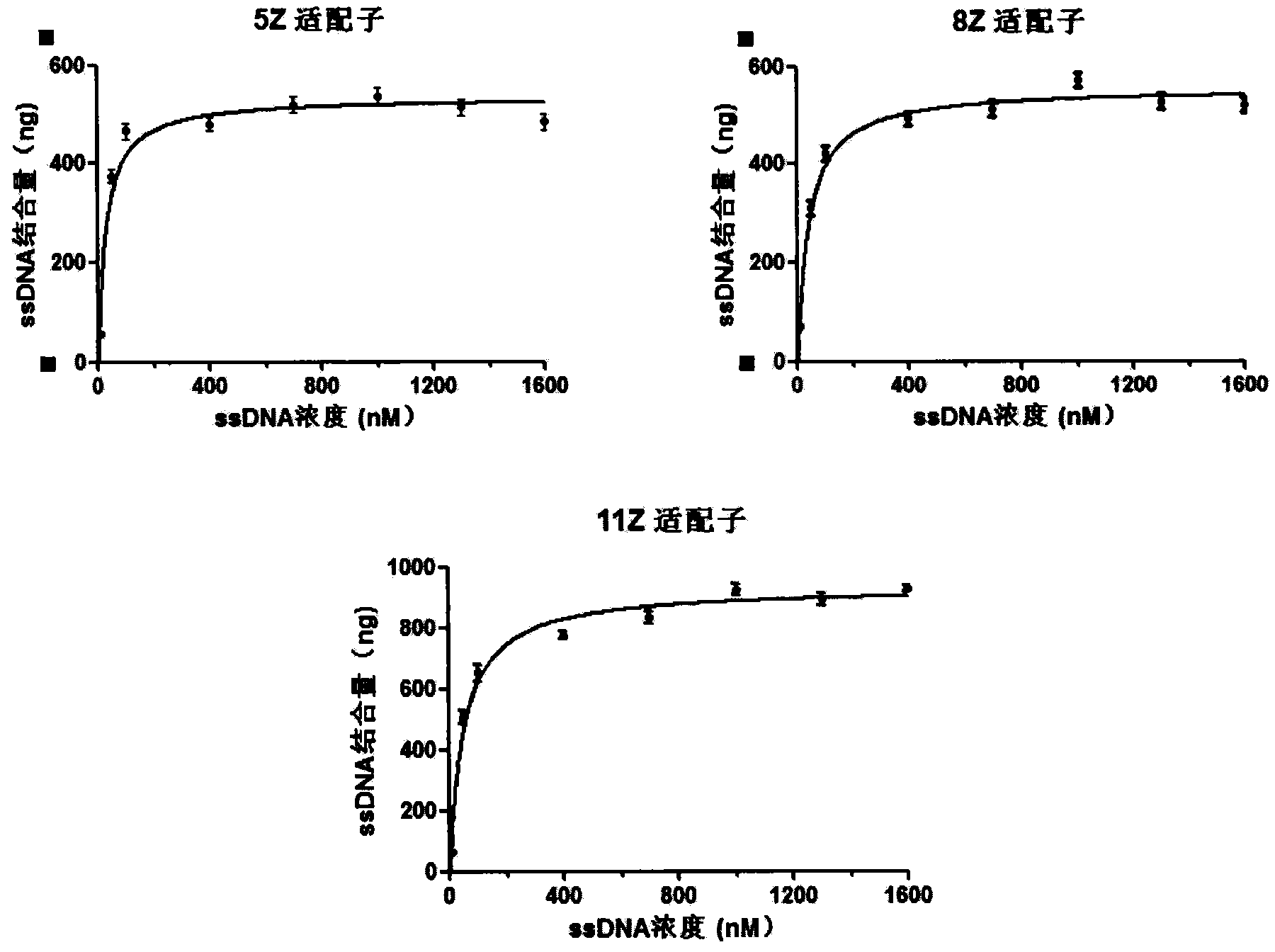
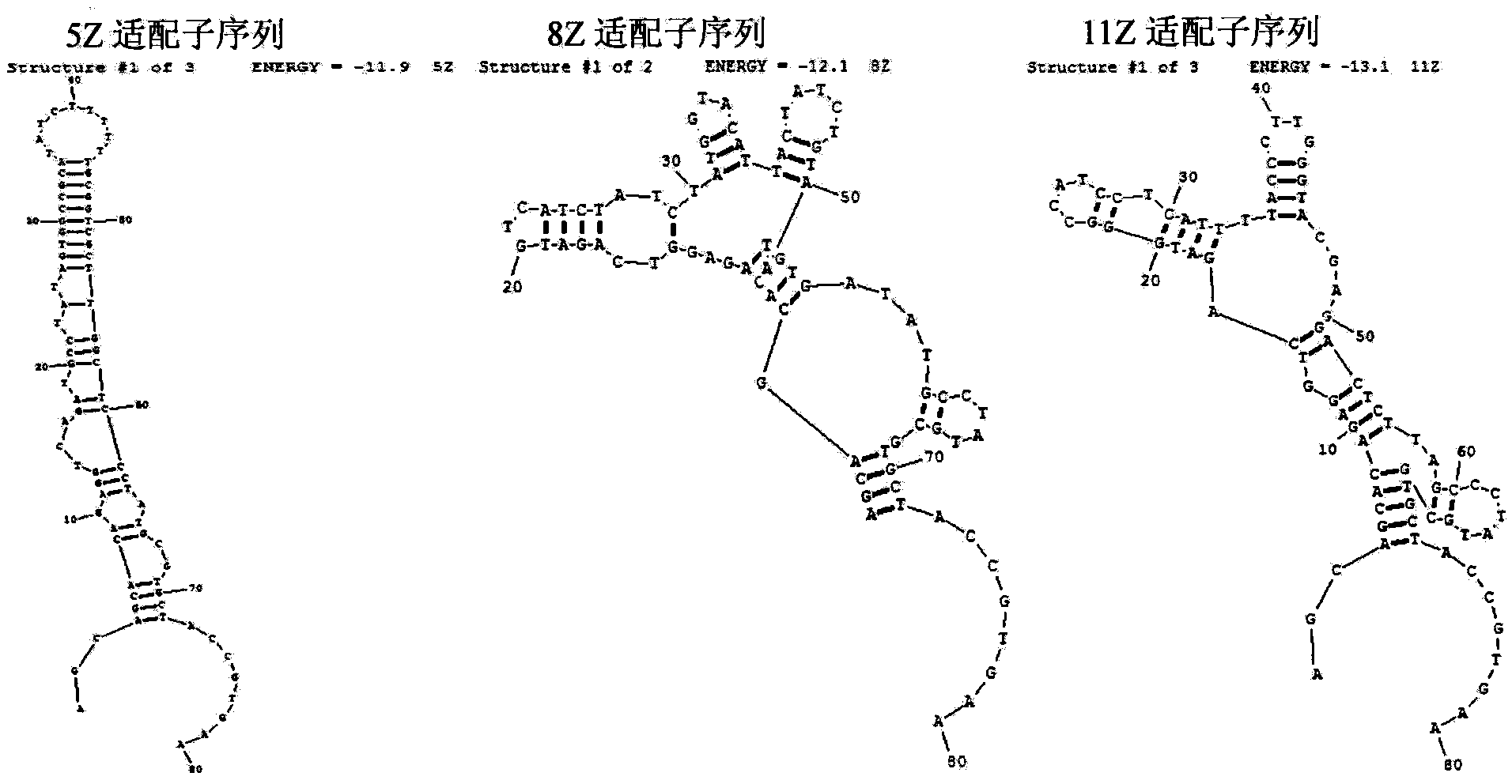
Oligonucleotides aptamer special for distinguishing zearalenone
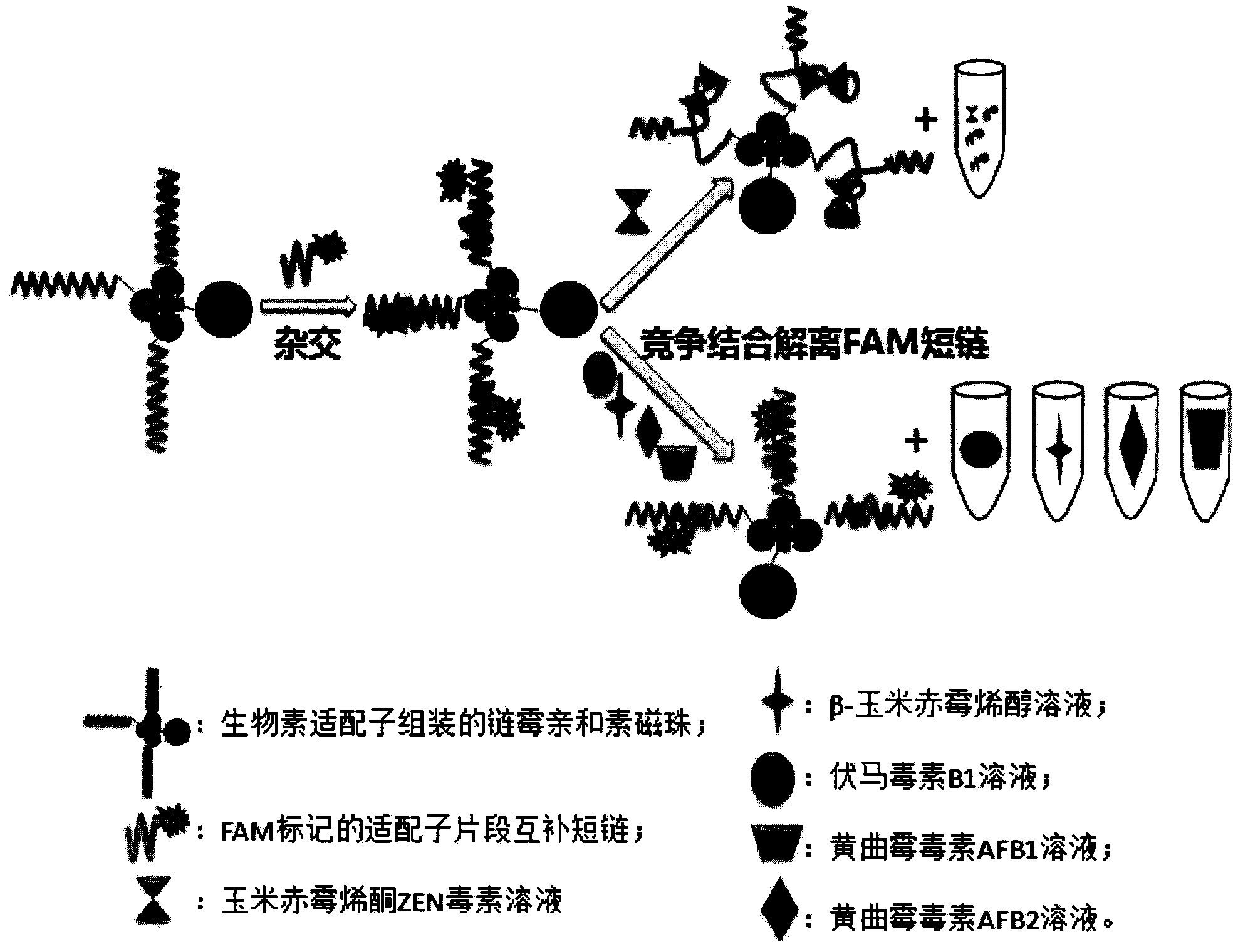
ActiveCN103013998BHigh affinityStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationNucleotideMagnetic bead
Owner:JIANGNAN UNIV
Bungotoxin Antigen Epitope Gene and Its Application in Gene Vaccine and Antigen Preparation
InactiveCN103865934BNon-immunogenicEasy to storeGenetic material ingredientsImmunoglobulins against animals/humansSequence signalProtein target
The invention relates to a bungatotoxin antigen epitope gene and application of the bungatotoxin antigen epitope gene to the preparation of a gene vaccine and an antigen. Two representative sequences are extracted from each of a long chain of alpha-BGT, chains A and B of beta-BGT and kappa-BGT of a cDNA sequence of bungatotoxin, the signal peptide and propeptide parts of the sequences are removed, mature peptide parts are collected, 10 antigen sites are obtained through analysis by a bioinformatics method, and a gene sequence containing the 10 sites is manually synthesized, and is connected to an eukaryotic expression vector pIRESneo to immunize an animal as the gene vaccine to obtain an anti-bungatotoxin neutralizing antibody; the sequence can also be connected to a prokaryotic expression vector PET-28a and then transformed to expression bacterium BL21 to induce the expression of the protein, and the target protein is separated to immunize the animal as an immunogen to obtain the anti-bungatotoxin neutralizing antibody.
Owner:中国人民解放军成都军区疾病预防控制中心
NGS data analysis method of enteric microorganism 16 SrRNA
PendingCN112435713AEasy to implementLow costSequence analysisInstrumentsMicroorganismNaive bayesian classifier
The invention discloses an NGS data analysis method of enteromicroorganism 16 S rRNA, which comprises the following steps: 1), carrying out database preparation: finding out a sequence capable of being expanded by a V4 universal primer from SILVA132 and Duengene13-8-16S databases, cutting off the V4 universal primer from the sequence to obtain a 254bp sequence, respectively intercepting 125bp sequences from positive and negative directions, and connecting the bidirectional 125bp sequences; obtaining a sequence of which the length is 250bp; 2), training a feature classification trainer: training the sequence in the step 1) and the corresponding classification information by using a naive Bayes classifier of the qiime2 to obtain a more accurate feature classifier; 3), evaluating the accuracy, namely deducing the mixed sequencing sample by using a simulated microbial community and DADA2, and comparing the deduced mixed sequencing sample with real annotation information; and 4), performingdiversity analysis: executing multi-sequence alignment on the representative sequence by using a mafft program, and generating a phylogenetic tree based on the filtered alignment result by applying FastTree. The data utilization rate is improved, and the detection effect of longer read-length sequencing can be achieved.
Owner:申友基因组研究院(南京)有限公司
Method for synchronously identifying floating algae and benthic algae in aquatic ecosystem
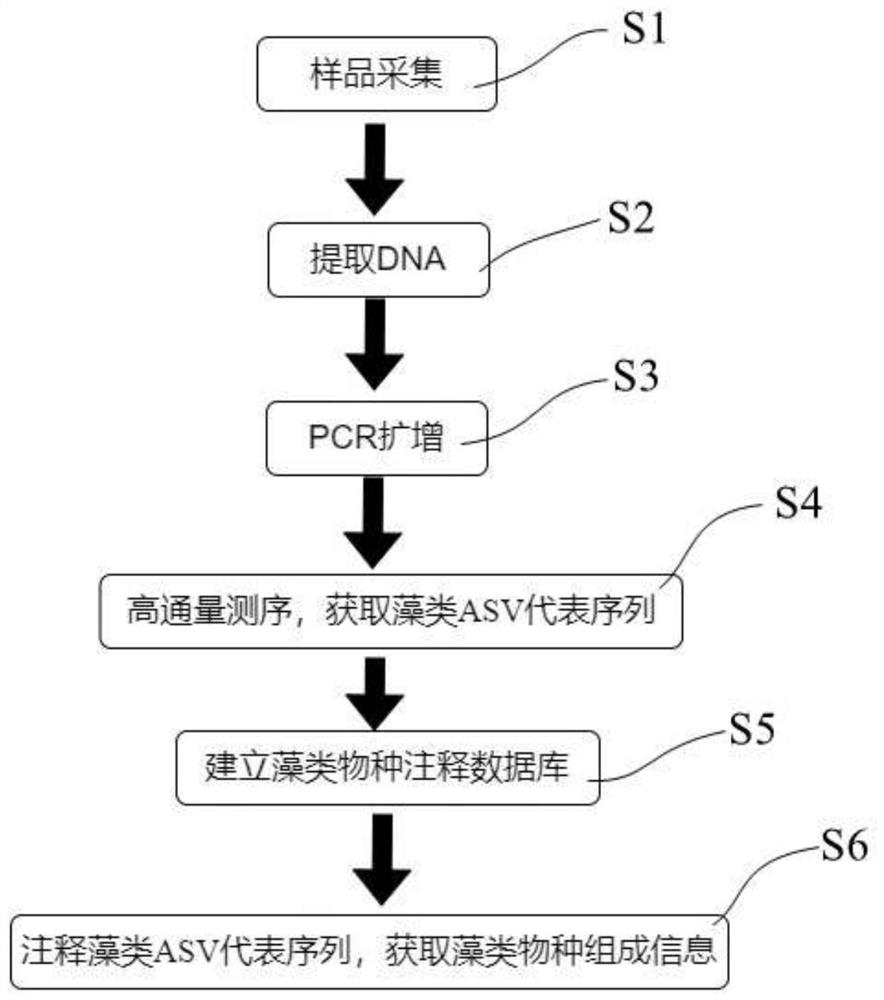
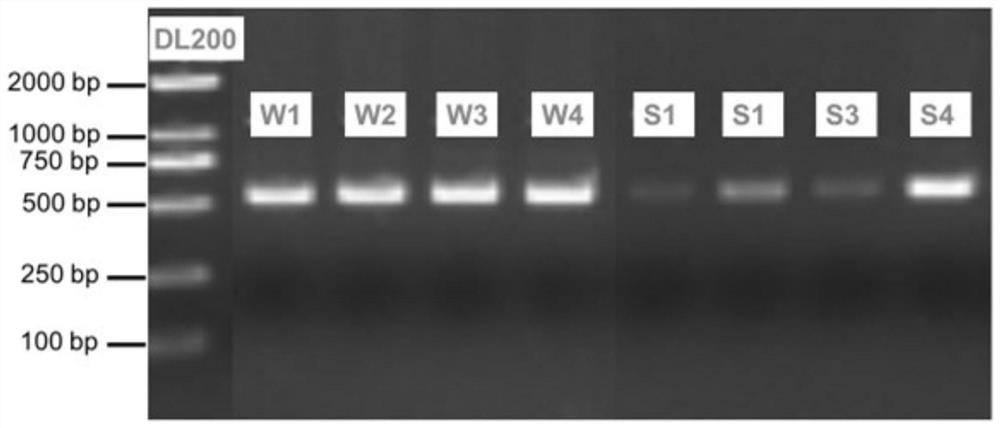
ActiveCN113142034ARapid identificationEfficient identificationClimate change adaptationCultivating equipmentsAquatic ecosystemBenthic algae
The invention provides a method for synchronously identifying floating algae and benthic algae in an aquatic ecosystem. The method comprises the following steps: S1, collecting a water sample and a sediment sample from the aquatic ecosystem; S2, respectively extracting and purifying DNA from the water sample and the sediment sample; S3, carrying out PCR (Polymerase Chain Reaction) amplification by taking the purified DNA as a template; S4, performing high-throughput sequencing on a PCR amplification product to obtain an algae ASV representative sequence; S5, establishing an algae species annotation database; and S6, comparing the algae ASV representative sequence with the sequence data of the algae species annotation database, and carrying out species annotation to obtain algae species composition information. According to the method, the species composition of all eukaryotic and prokaryotic algae in a water body and sediments in the ecological system is efficiently, quickly and synchronously identified through a high-throughput sequencing technology on the basis of the difference of genetic materials of the algae.
Owner:PEKING UNIV
Nucleic acid capture probe for hla typing and its design method
ActiveCN110600082BEfficient captureImprove the effect of hybrid captureMicrobiological testing/measurementSequence analysisRepresentative sequencesNucleic acid
The invention discloses a method for designing a nucleic acid capture probe for HLA typing. The method includes the steps: 1) constructing HLA-A, HLA-B, and HLA-C sequence libraries; 2) performing multiple sequence alignment, searching for SNP loci on Exon2 and Exon3 and upstream and downstream of each gene; 3) using a sliding window separation algorithm to select regions that cover a predetermined number of SNP loci as probe design candidate regions; 4) performing cluster analysis to obtain the representative sequences of the respective probe design candidate regions as candidate probes; and5) deduplicating the all candidate probes to obtain the respective capture probes of the three genes. The invention also discloses a probe designed by the above method, and its sequence is shown in SEQIDZNO: 9-88. The method designs the nucleic acid capture probes for all polymorphic loci in the Exon2 and Exon3 regions with a GC ratio of more than 60% in the HLA gene, thereby improving the effectof hybridization capture of the HLA gene probe.
Owner:上海仁东医学检验所有限公司
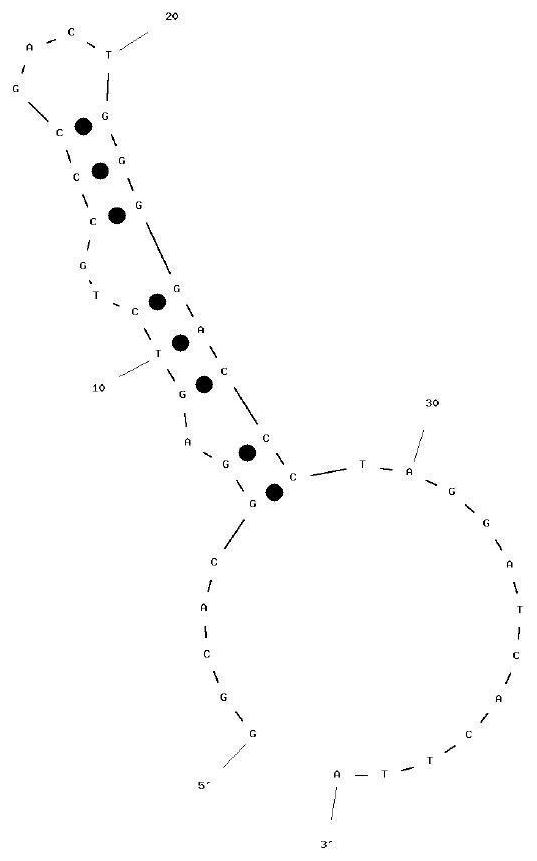
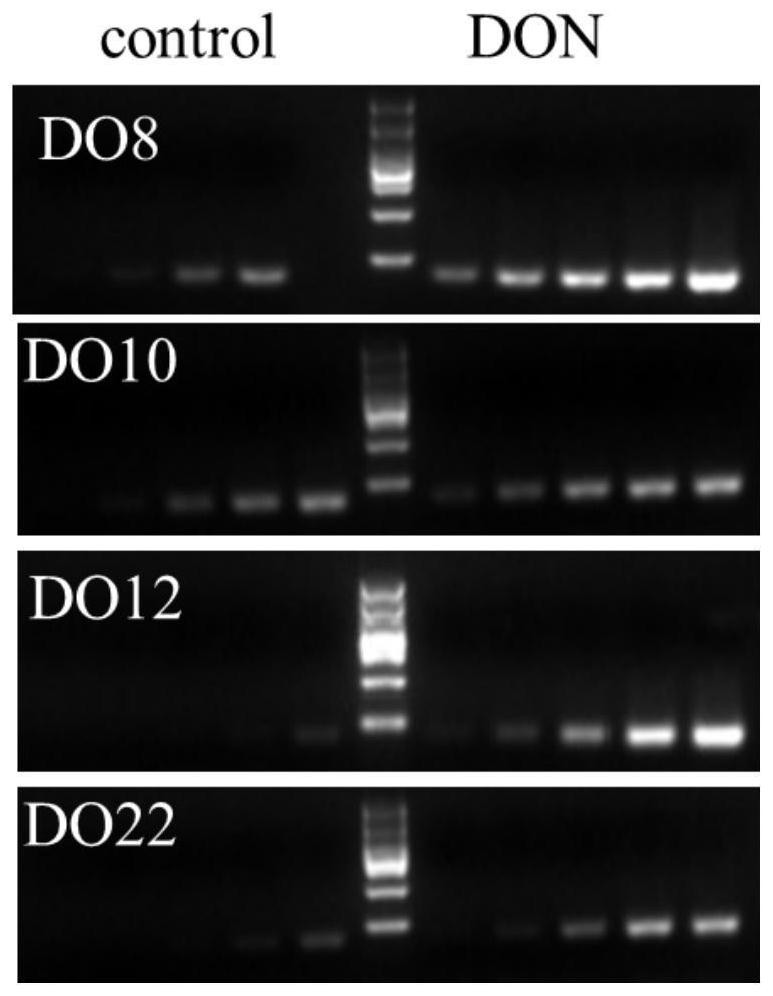
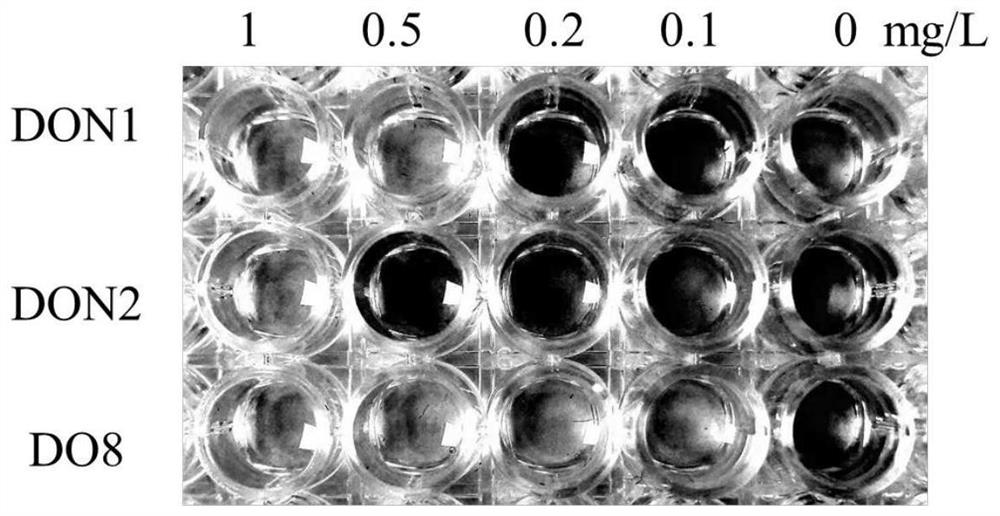
Vomitoxin specific nucleic acid aptamer and application thereof
The invention provides a vomitoxin specific nucleic acid aptamer and application thereof. According to the aptamer, deoxynivalenol (DON) is taken as a target, a competitive SELEX technology is utilized for seven rounds of repeated incubation, cleaning, dissociation, amplification and lambda-exonuclease digestion, and the nucleic acid aptamer capable of being specifically combined with DON is screened from a single-stranded DNA random library. After cloning, sequencing and synthesizing two representative sequences, affinity determination is performed on the representative sequences through SPR and PCR methods, the nucleic acid aptamer DO8 capable of being specifically bound with deoxynivalenol (DON) is obtained, and the sequence is shown as SEQ ID NO: 1. The nucleic acid aptamer provided by the invention has high specificity, can be combined with DON with high affinity, and can be used for rapid detection of polluted DON in agricultural products.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
A detection method, detection system and terminal of bacterial flora markers
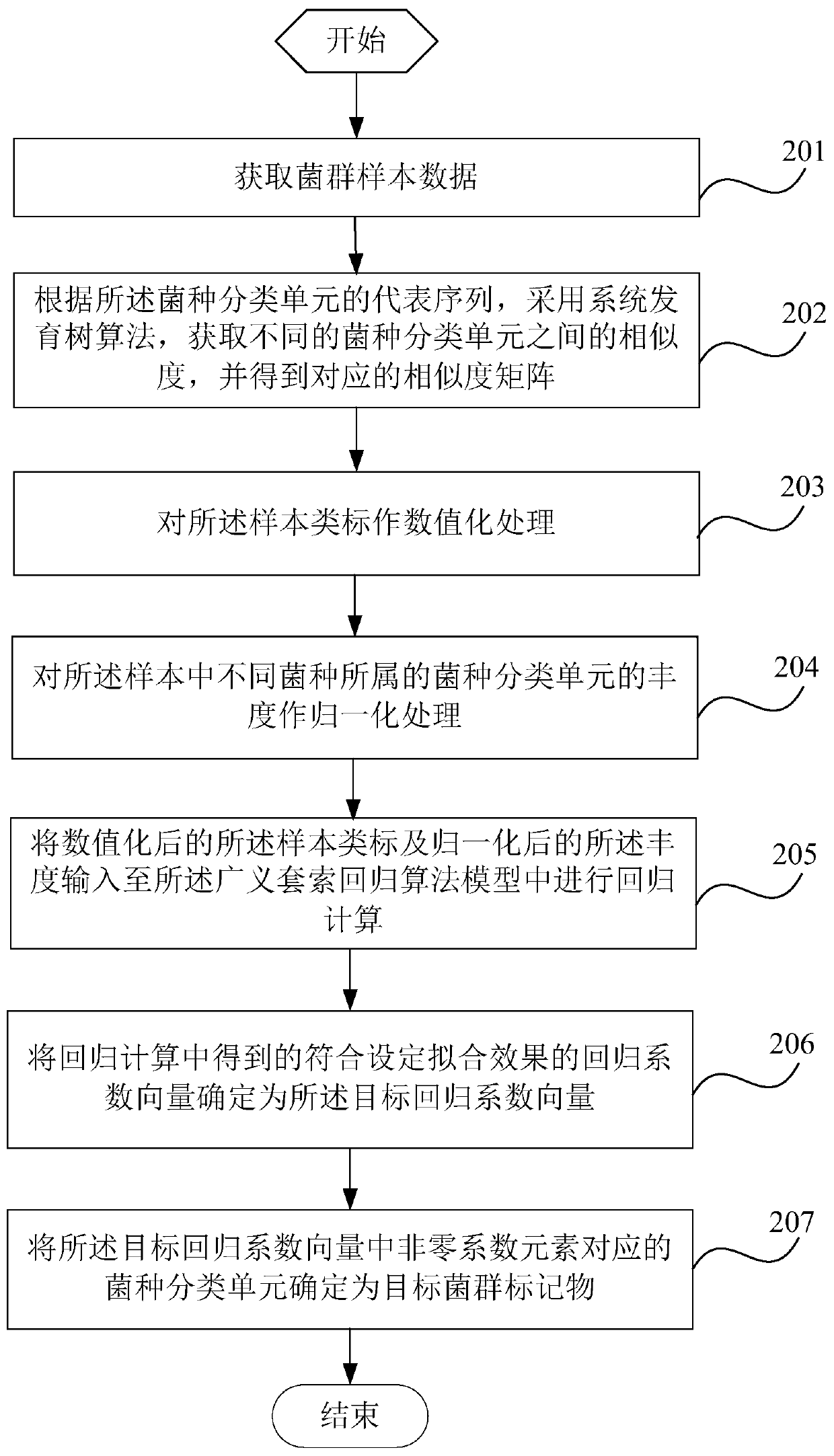
ActiveCN110444254BComprehensive reflection of complex relationshipsReflect complex relationshipsSequence analysisInstrumentsAlgorithmFlora
This application is applicable to the field of biotechnology, and provides a detection method, detection system and terminal of bacterial flora markers, wherein the method includes: obtaining bacterial flora sample data; according to the representative sequence of the bacterial taxon, using the phylogenetic tree algorithm , to obtain the similarity between different bacterial species taxa, and obtain the corresponding similarity matrix; according to the sample class label, the abundance of the bacterial taxa to which the different bacterial species belong and the similarity matrix, Through the generalized lasso regression algorithm model, the target regression coefficient vector corresponding to the set fitting effect is obtained; the bacterial classification unit corresponding to the non-zero coefficient element in the target regression coefficient vector is determined as the target flora marker, and the improvement is achieved. Validity of screened flora markers.
Owner:SHENZHEN INST OF ADVANCED TECH +1
Dynamic generation method for hydrological time series prediction model
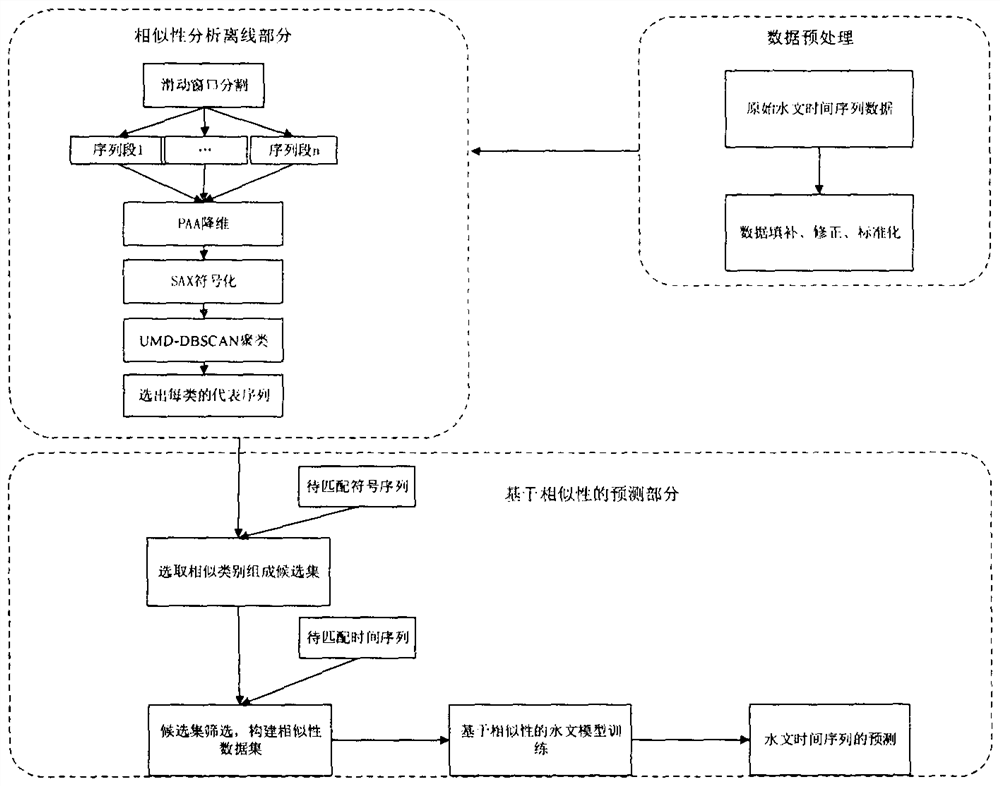
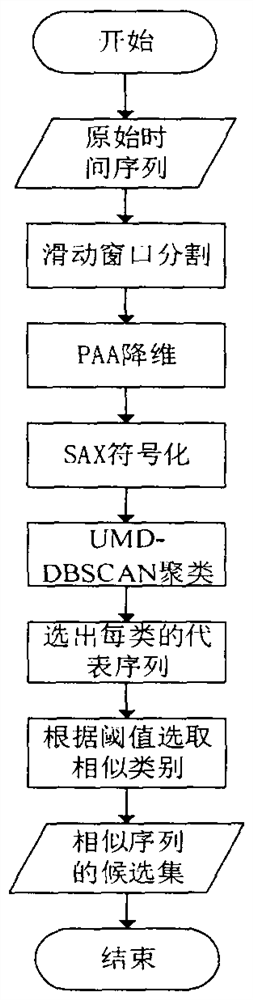
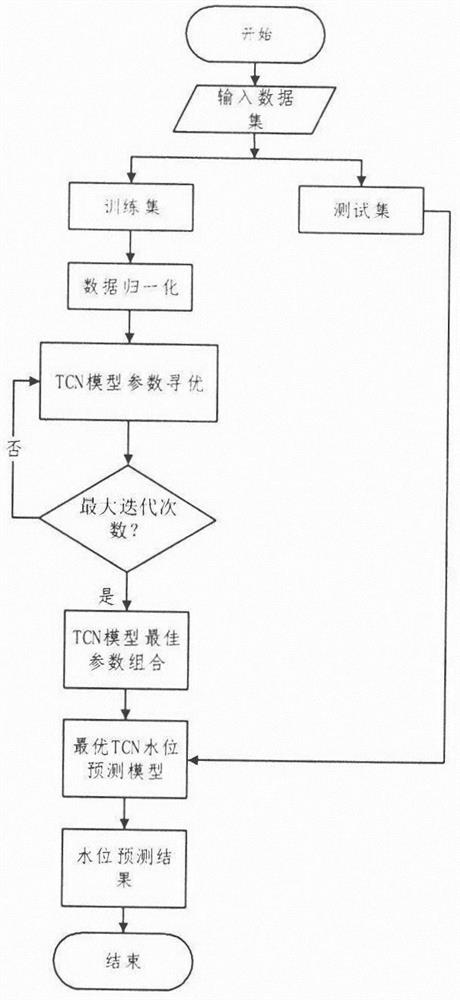
PendingCN114819260AReduce training timeReduce data volumeForecastingCharacter and pattern recognitionHydrometryData set
The invention discloses a hydrological time series prediction model dynamic generation method, which comprises the following steps of: acquiring water level data of a corresponding hydrological station, organizing into a hydrological time series data set and preprocessing; clustering is carried out on the segmented and symbolized sequences in combination with improved symbol distance UMD and DBSCAN clustering; dynamically forming a similar sequence set for each to-be-matched sequence and constructing and training a model, firstly measuring the distance between a representative sequence and a to-be-matched symbolized sequence and selecting similar categories to form candidate sets, secondly screening the similar sequence candidate sets by adopting an improved DTW algorithm, constructing the similar sequence set, and finally optimizing TCN model parameters to obtain a TCN model; training the model by using the similar sequence set to obtain a hydrological time sequence prediction model based on similarity search; the model obtained by the hydrological time series prediction model dynamic generation method is subjected to water level prediction, and the accuracy is higher.
Owner:HOHAI UNIV
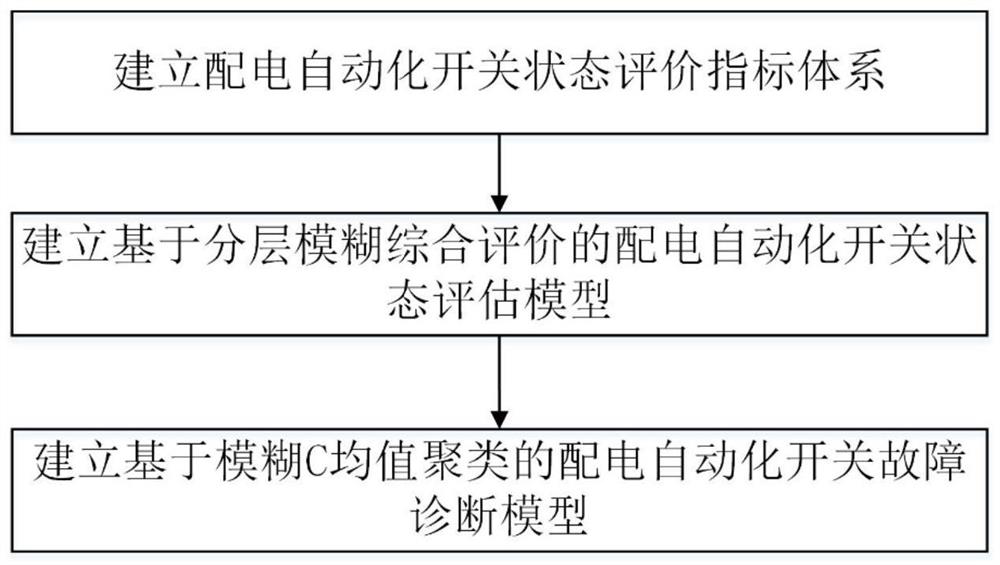
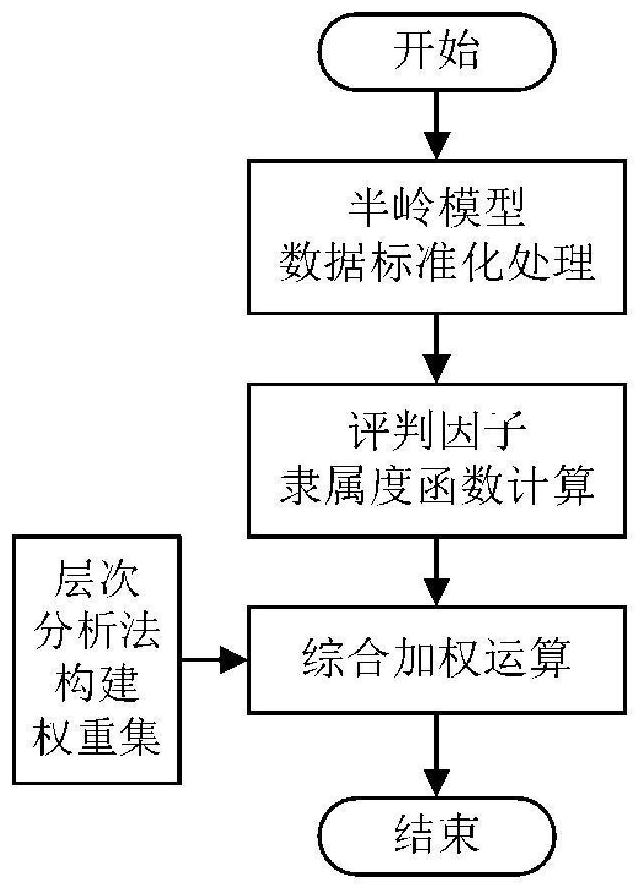
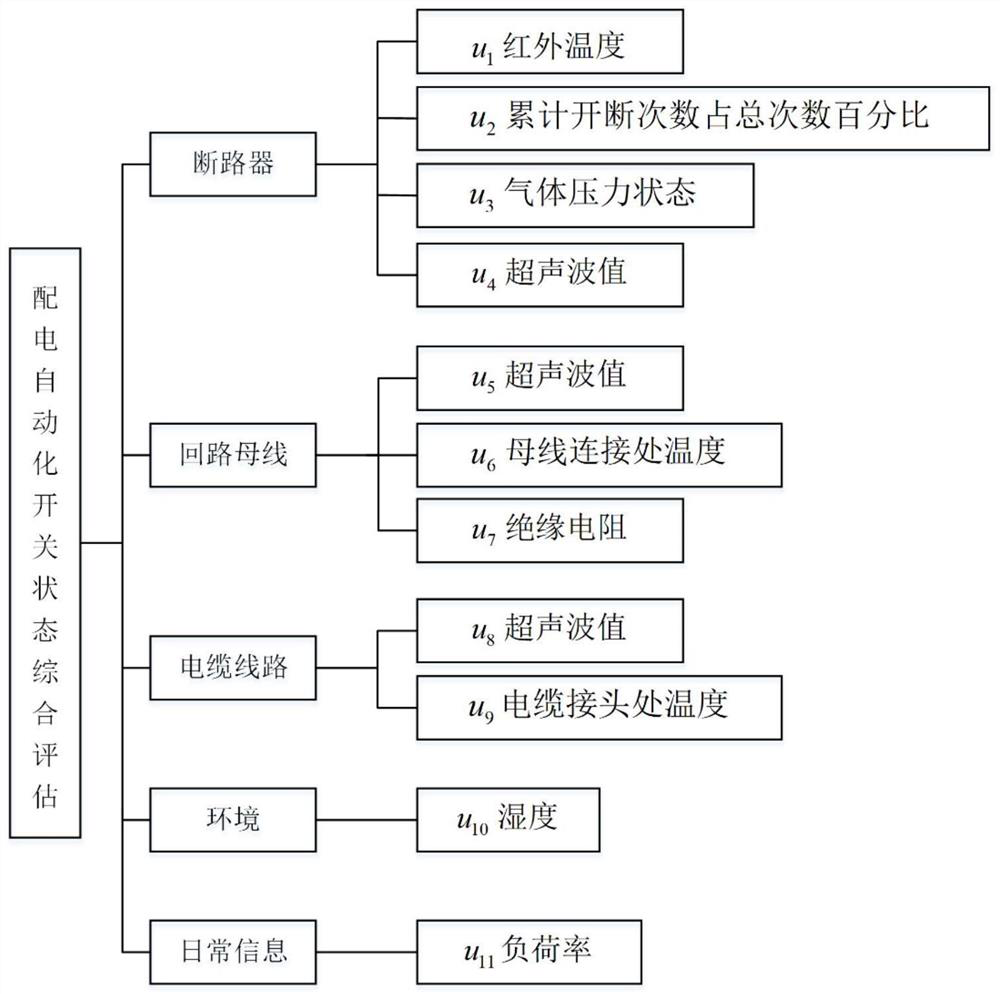
A kind of distribution automation switch fault diagnosis method, equipment and readable storage medium
ActiveCN111638449BImprove accuracyImprove efficiencyCircuit interrupters testingIndex systemComputer science
Owner:STATE GRID BEIJING ELECTRIC POWER +2
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com