Method for detecting diversity of fish species on basis of environmental DNA technology
A technology of technical detection and diversity, applied in the fields of biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems such as the accuracy of the results to be improved, the detection accuracy is not enough, etc., to achieve simple sampling, accurate detection results, and convenient detection. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] A method for detecting fish species diversity based on environmental DNA technology, comprising the steps of:
[0032] 1. Water sample collection and DNA extraction
[0033] 5 L of surface water samples were collected from 9 sampling points in the waters of the Fuchun River, brought back to the laboratory and placed in a 4°C freezer, and the water samples were filtered within 24 hours, using a filter membrane with a pore size of 0.45 μm. The eDNA in the filter membrane was extracted using the kit EZNA waterDNA kit produced by Omega Company. The extracted DNA was dissolved in TE buffer and stored at -20°C for future use.
[0034] 2. PCR amplification and quality inspection
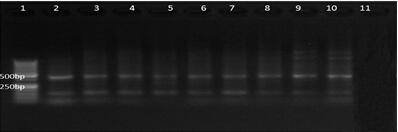
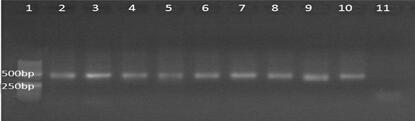
[0035] Cytb and 16S primers were used to amplify water sample DNA, and the primer sequences are shown in Table 1. The 25 μL reaction system included: 2.5 μL 10×buffer, 2 μL dNTP (2.5 mmol·L -1 ), each 1 μL of forward and reverse primers (10 μmol﹐L -1 ), 0.15μL Taq enzyme (5 UoμL -1 ), 1 μL DNA t...
Embodiment 2
[0052] The same sampling point was selected twice in the Qiandao Lake National Aquatic Germplasm Resources Protection Area to collect 5L of surface water samples, and they were brought back to the laboratory and placed in a 4°C freezer. DNA extraction, PCR amplification and quality inspection, target fragment library construction, sequencing and data processing, fish taxonomic annotation methods are the same as in Example 1. The results are shown in Table 4. A total of 9 fish species were annotated with Cytb F / Cytb R primers, and 27 fish species were annotated with 16S F / 16S R primers. Most of the annotated species are species that can be collected in Qiandao Lake by traditional fishing methods. In addition to the common indigenous fishes detected in Qiandao Lake, some exotic species and hybrid species were also detected, such as tilapia, fork-tailed betta, round-tailed betta, spoon-nose sturgeon, Russian sturgeon, hybrid sturgeon, bream×triangle bream.
[0053]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com