NGS data analysis method of enteric microorganism 16 SrRNA
A technology for gut microbes and data analysis, applied in the field of NGS data analysis of gut microbe 16SrRNA, can solve the problems of inaccurate data utilization in data analysis, and achieve the effect of improving data utilization, accurate results and cost saving
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0023] Embodiment: the method comprises the following steps:
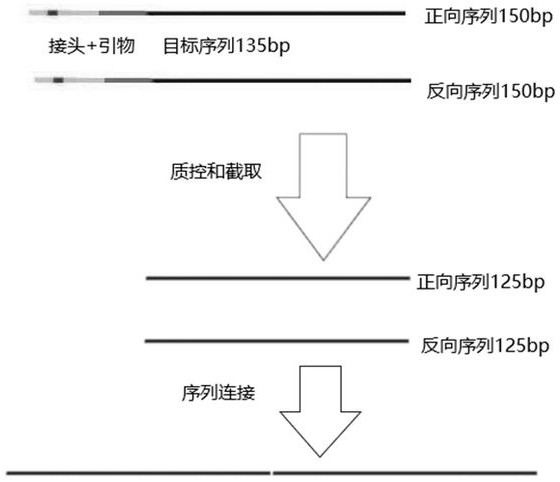
[0024] 1) Database preparation: First, find out the sequence that can be amplified with the V4 universal primer from the SILVA132 and Dreengene13-8-16S databases. Two mismatches are allowed, and the V4 universal primer is cut off from the sequence to obtain a 254bp sequence. The 125bp sequences were intercepted in both directions to adapt to the actual situation of the sequencing sequence, and then the bidirectional 125bp sequences were connected to obtain a sequence with a length of 250bp;
[0025] 2) Feature classification trainer training: By using qiime2's naive Bayesian classifier to train the sequence in 1) and the classification information corresponding to the sequence and obtain a more accurate feature classifier, it can maximize the recognition of each classification group Sensitivity, specificity, precision and negative predictive value for most sequences in
[0026] 3) Accuracy assessment: use a simula...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com