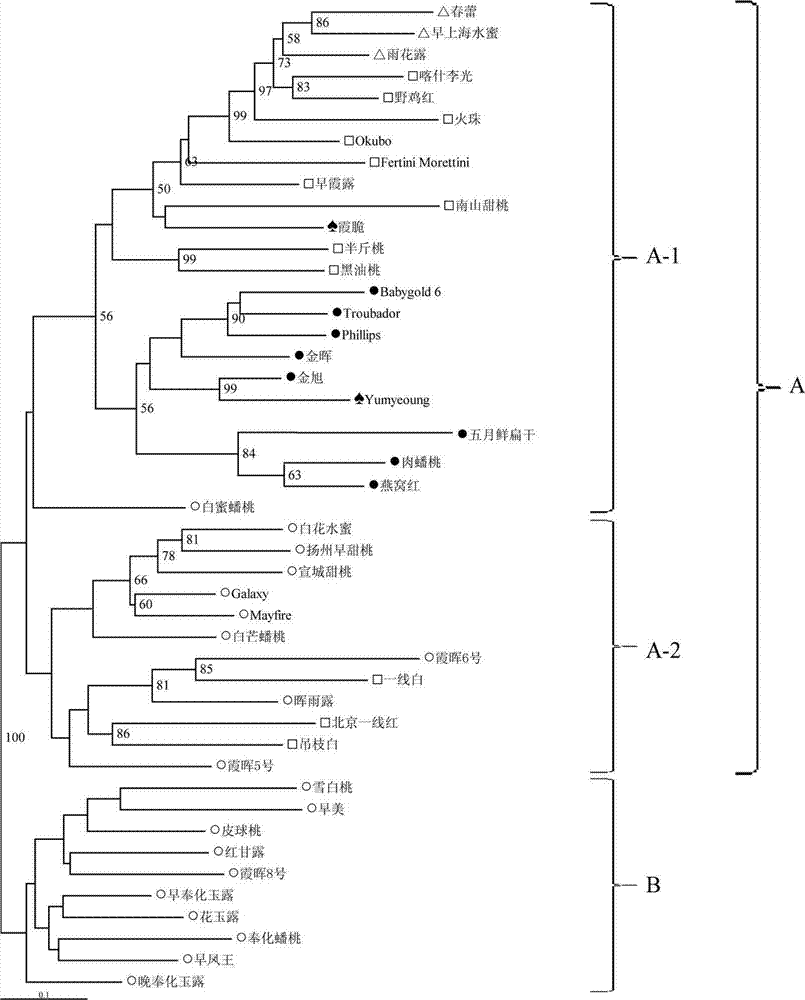
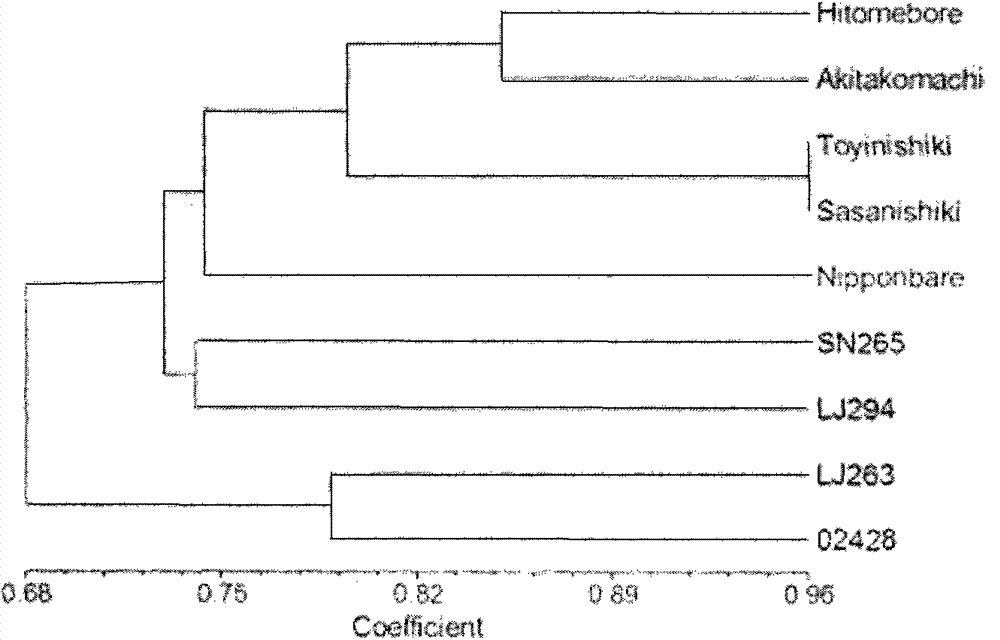
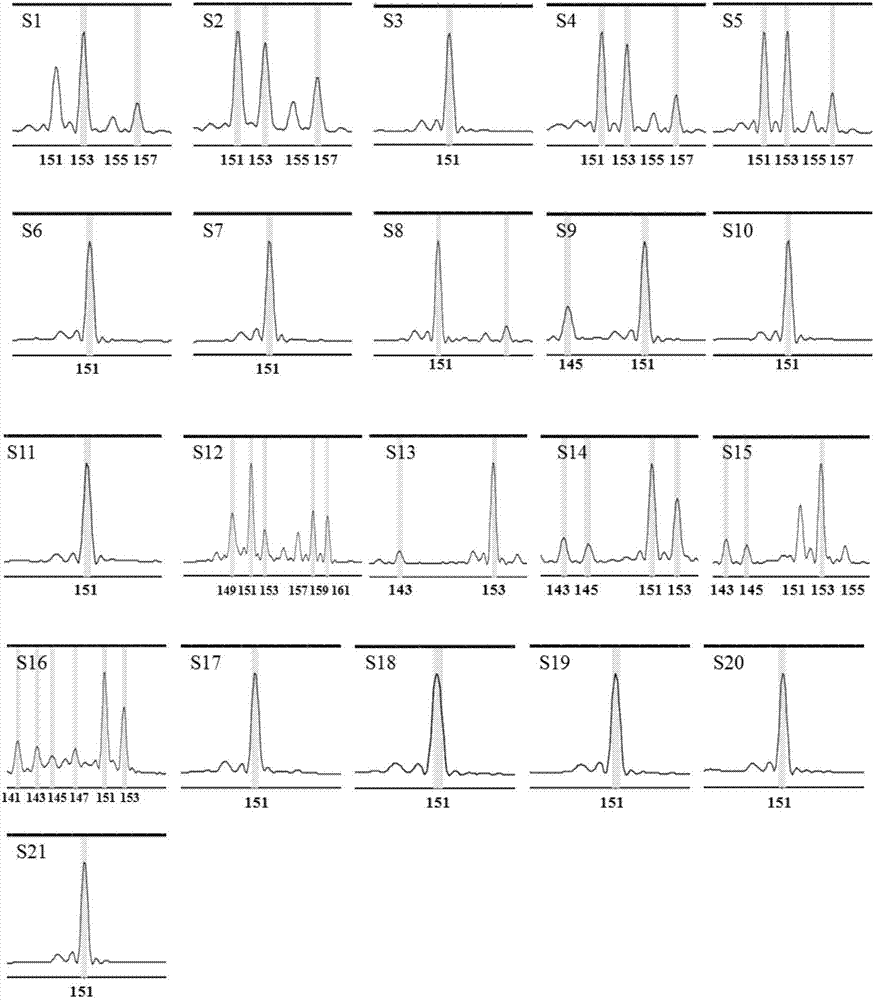
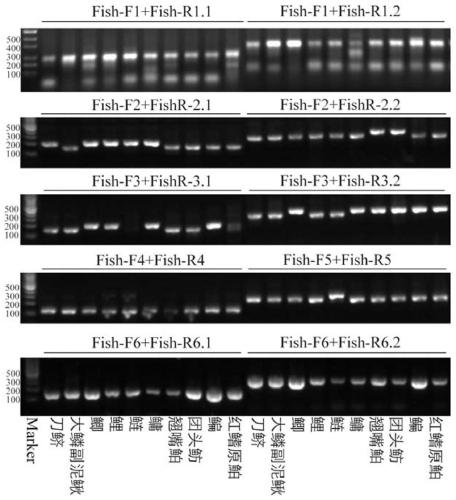
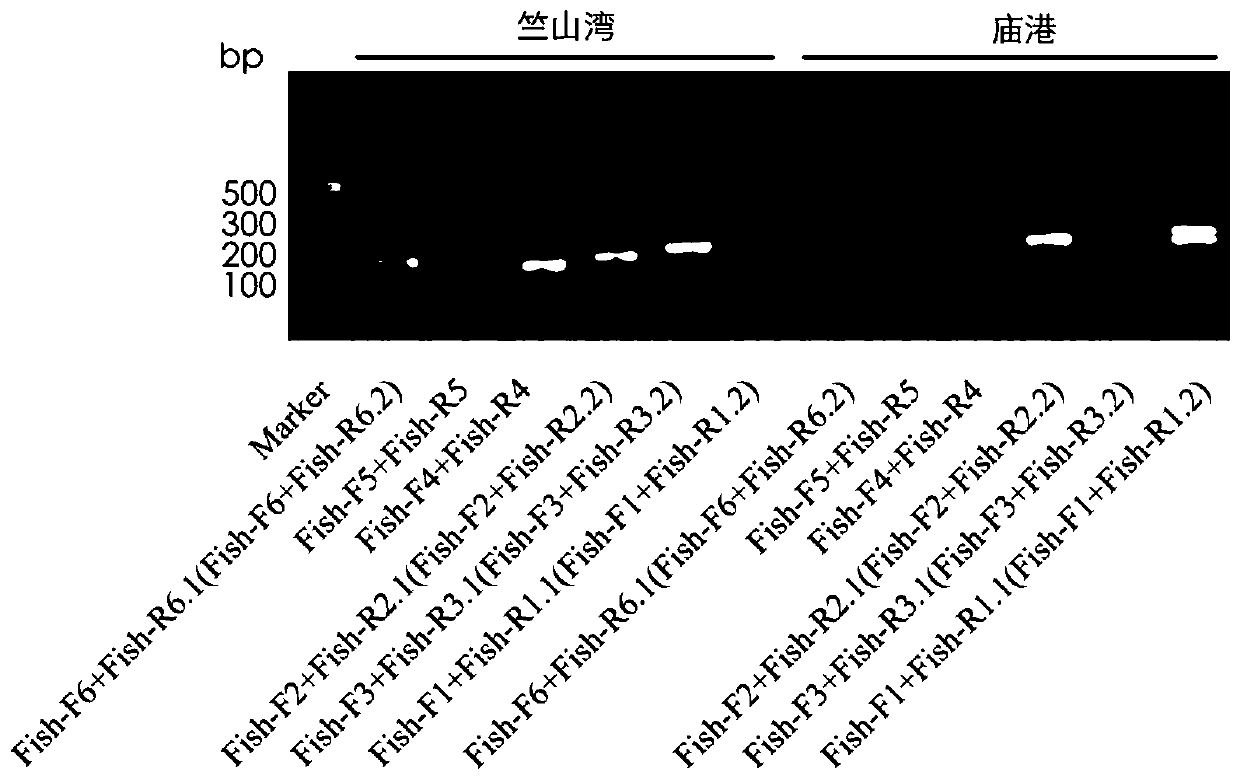
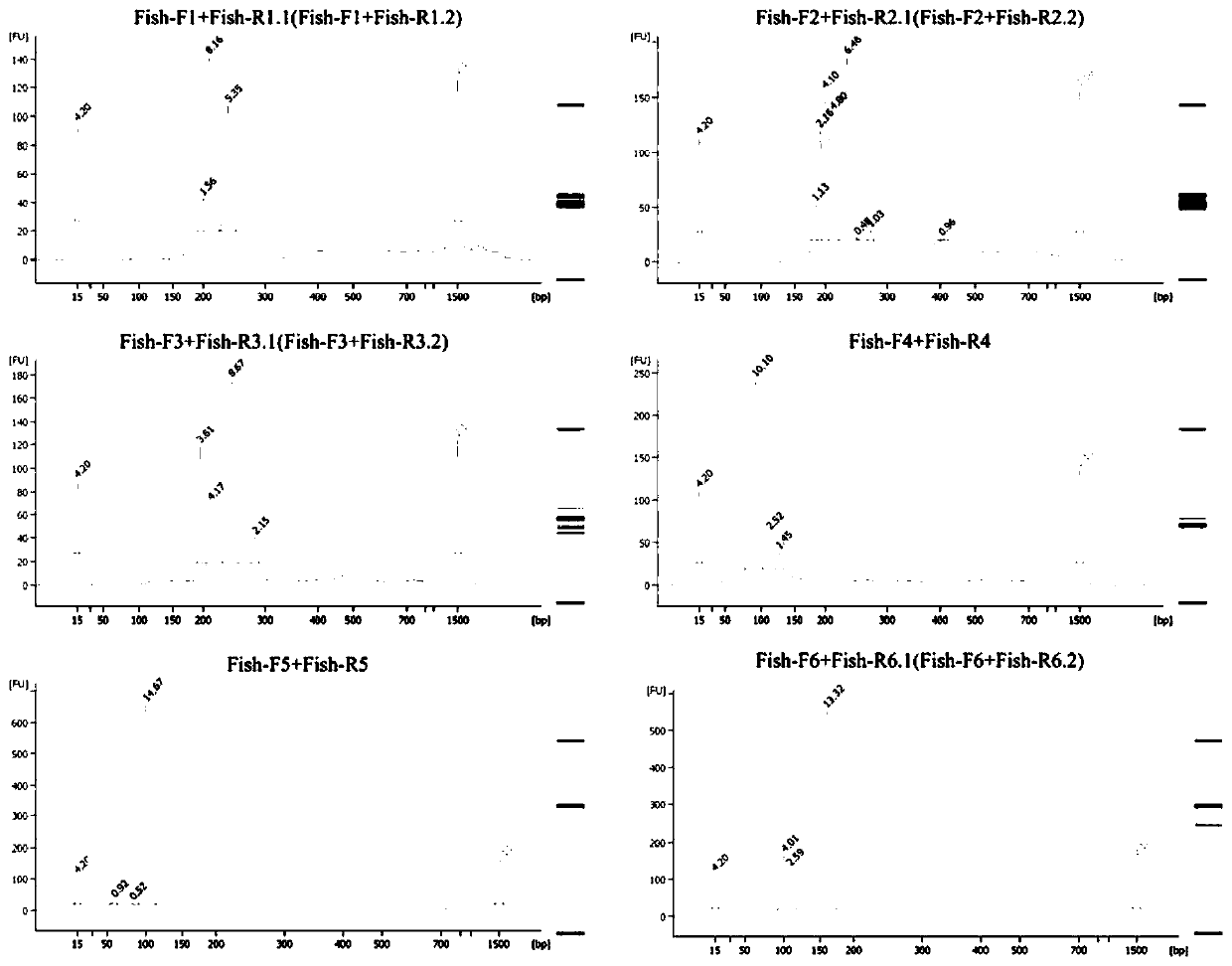
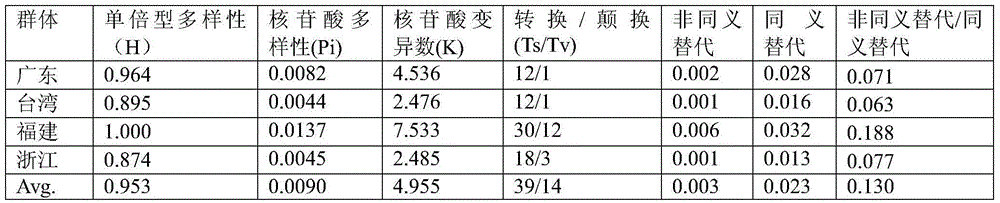
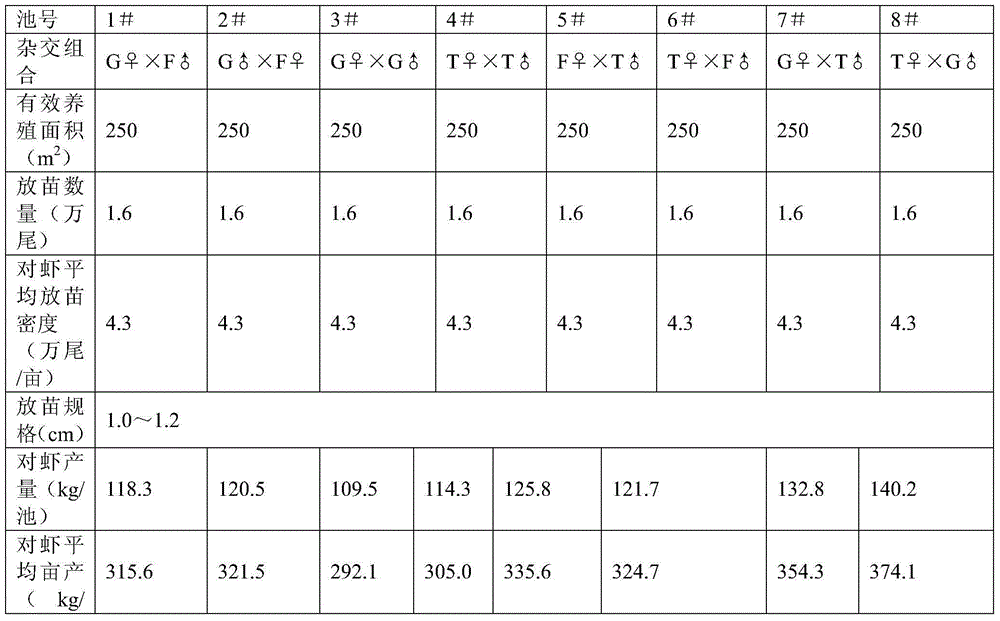
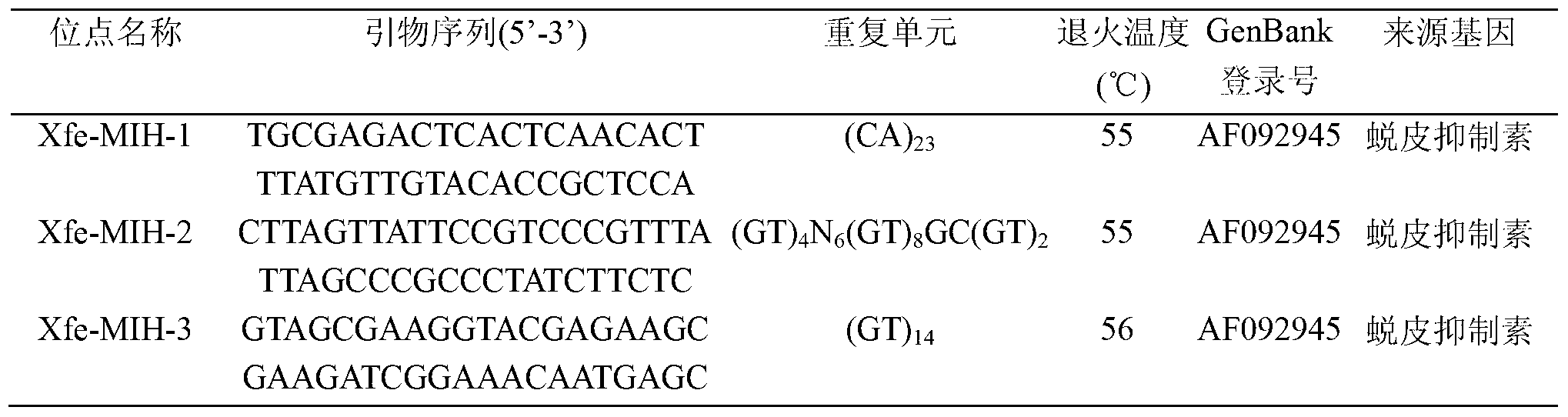
Patents
Literature
168 results about "Diversity analysis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
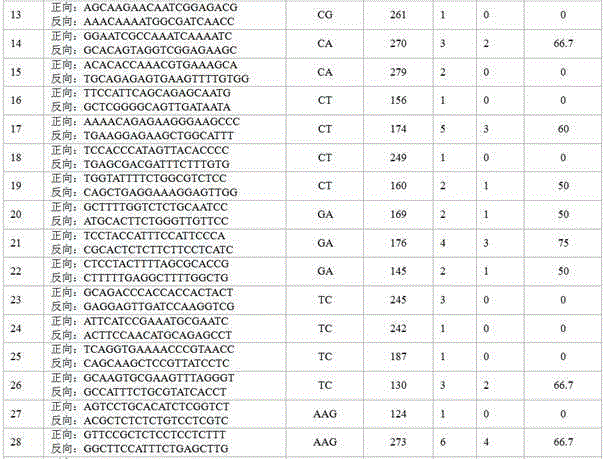
Application Year
Inventor
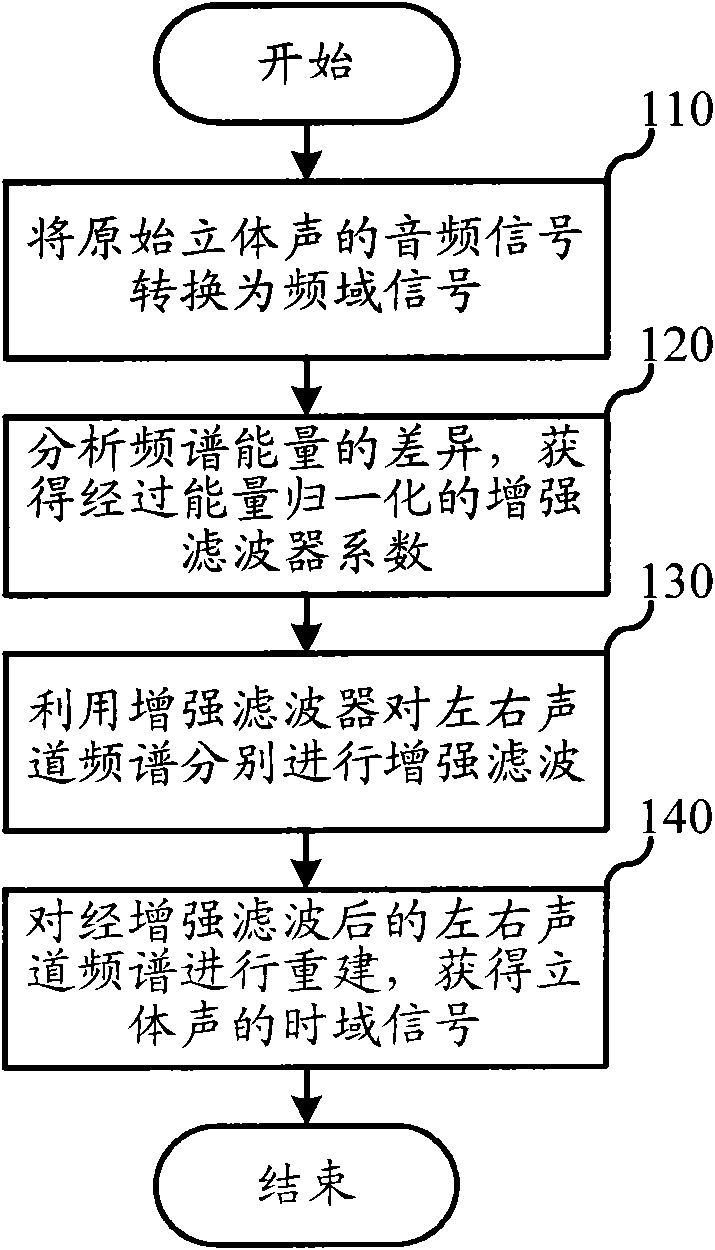
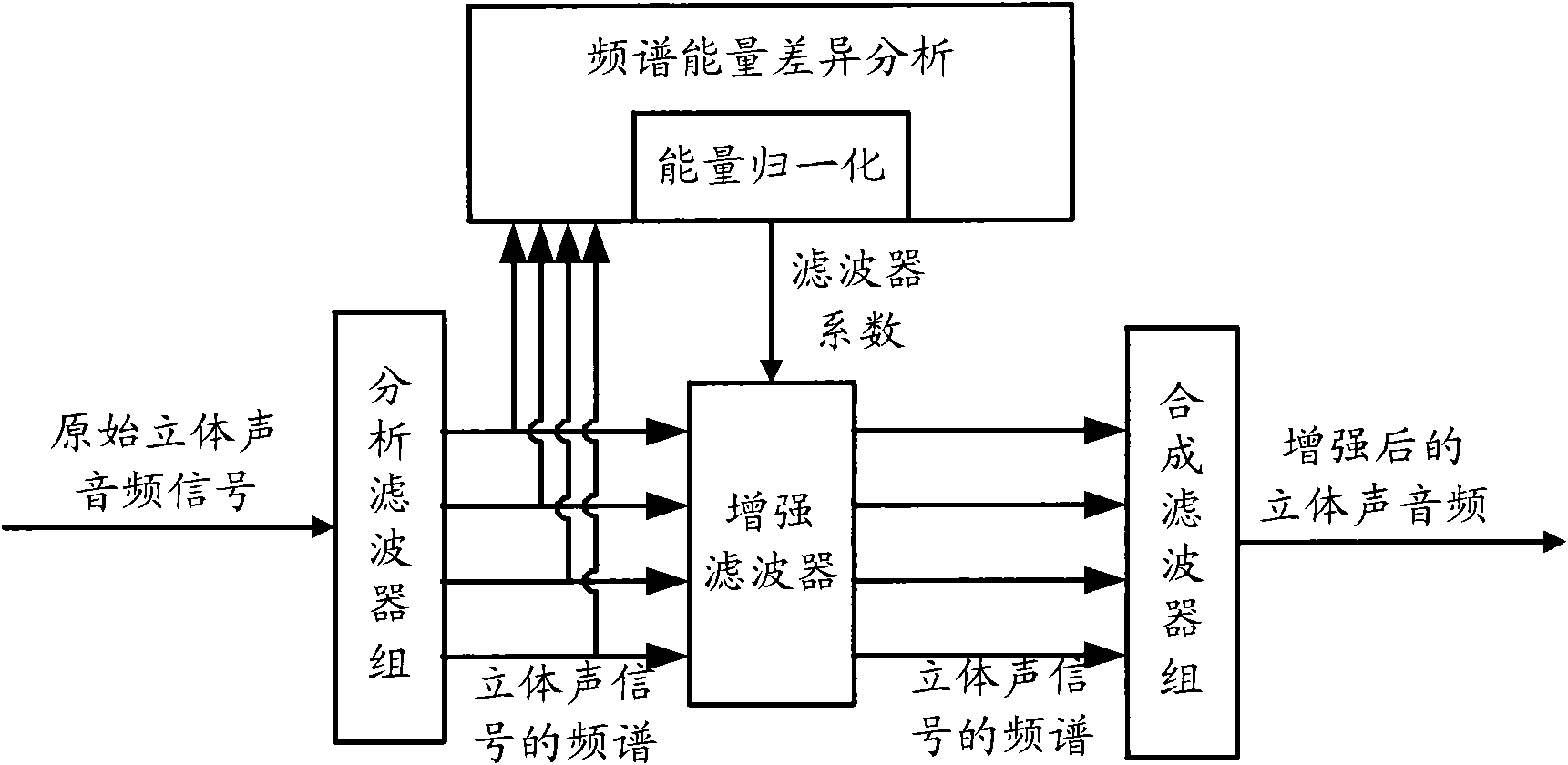
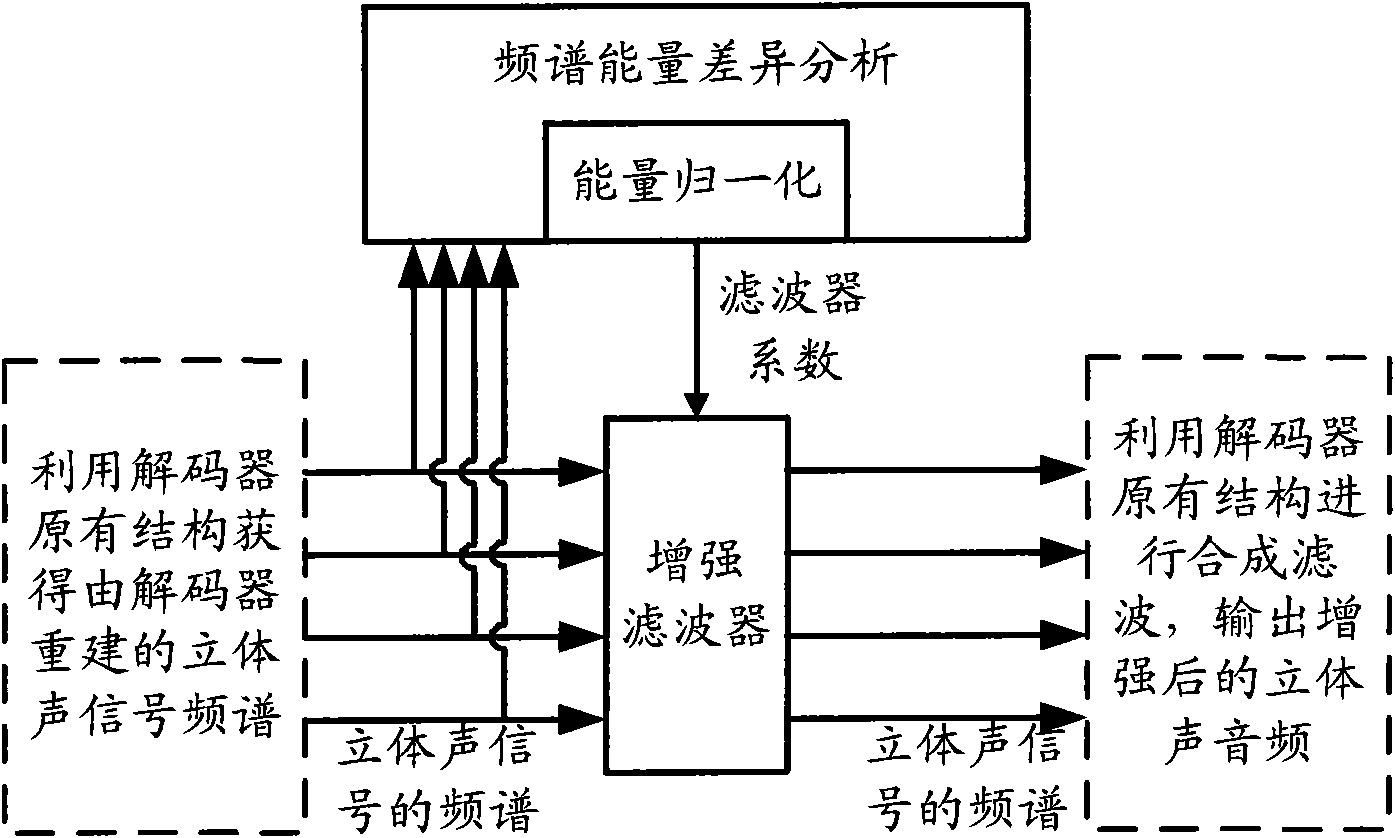
Audio processing method and device thereof
ActiveCN101894559ASimple calculationThe implementation mode can be flexible and simpleSpeech analysisStereophonic systemsFrequency spectrumFiltration
The invention relates to the multimedia technology, and discloses an audio processing method and a device thereof. The method comprises the following steps of: acquiring a normalized energy diversity ratio of spectrums of left and right sound channels by performing diversity analysis on the spectrums of the left and right sound channels, obtaining an enhanced filter coefficient according to the normalized energy diversity ratio and a sequence for shaping, and performing enhanced filtration on the spectrum of a stereo signal according to the obtained enhanced filter coefficient. Because the enhanced filtration is finished in a frequency domain by using the enhanced filter before a frequency domain signal is converted into au audio signal through a synthesis filter, the audio processing effect can be greatly improved so that the finally output audio has better stereo enhancing effect or voice removing effect.
Owner:SPREADTRUM COMM (SHANGHAI) CO LTD
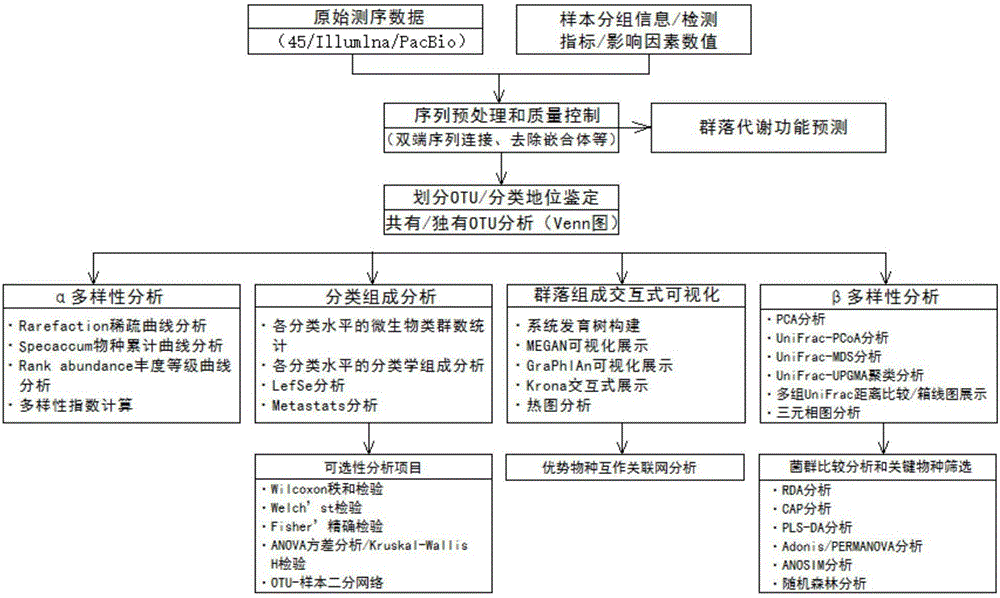
Automatic method for analyzing constitution and diversity of bacterial community of 16SrRNA gene
ActiveCN106815492AVarious methods of data mining analysisIn-depth analysis of data mining methodsSequence analysisSpecial data processing applicationsTechnical standardModularity
The invention discloses an automatic method for analyzing constitution and diversity of a bacterial community of a 16SrRNA gene. The automatic method comprises the following steps: in 16SrRNA sequencing data analysis process, by taking original sequencing sequence data as input, calling a standard analysis tool (such as Mothur and QIIME) of the industry, and performing visualization on data finally, thereby obtaining analysis results which are easy to analyze. The automatic method includes popular main analysis items at present, meanwhile analysis contents can be modularized, relatively rich and deep data mining analysis methods can be available, different analysis module contents can be combined according to different demands, and process procedures can be relatively reasonably arranged; and in addition, analysis errors caused by different sequencing depths can be eliminated, and the analysis results can be relatively comprehensive, accurate and reliable.
Owner:SHANGHAI PASSION BIOTECHNOLOGY CO LTD
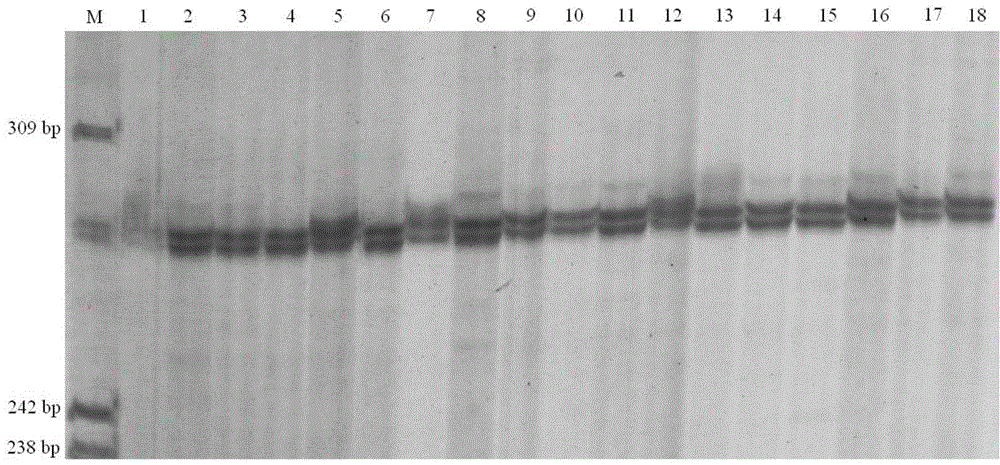
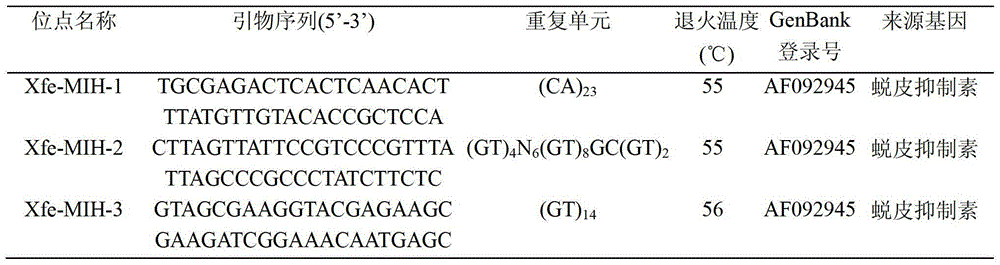
Sextuple PCR (polymerase chain reaction) detection method of portunus trituberculatus miers microsatellite marker
ActiveCN102154500AThe detection method is simpleFast detection methodMicrobiological testing/measurementPcr methodFamily management
The invention provides a sextuple PCR (polymerase chain reaction) detection method of a portunus trituberculatus miers microsatellite marker. The detection method comprises the following steps of: designing and synthesizing a sextuple PCR primer; extracting genome DNA (deoxyribonucleic acid); performing sextuple PCR reaction; and detecting a PCR product, wherein the sextuple PCR reaction comprises a sextuple PCR reaction system and a sextuple PCR reaction procedure. The method builds the sextuple PCR reaction system which performs the portunus trituberculatus miers family and paternity test to simultaneously detect six microsatellite sites in the PCR reaction, so that efficiency is improved by about six times compared with the existing PCR method. The method has the characteristics of being high-efficiency, economical, simple, convenient and the like, thereby being capable of being popularized and applied in the portunus trituberculatus miers genetic diversity analysis, the genetic relationship test, the family management and the improved variety breeding.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
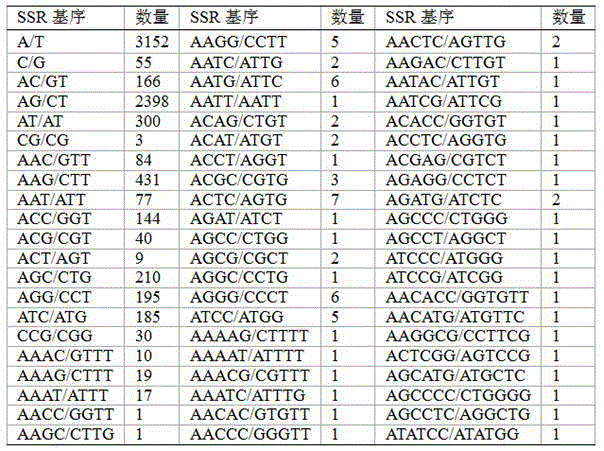
Plum SSR labeled primer pair exploited on basis of transcriptome sequence, and application thereof
ActiveCN105238781AOvercoming complexityOvercome workloadMicrobiological testing/measurementDNA/RNA fragmentationGeneticsGermplasm
The invention belongs to the fields of molecular marker technology exploitation and application, and concretely provides a plum SSR labeled primer pair exploited on the basis of a transcriptome sequence, and an application thereof in plum germplasm resource researches. The primer pair is exploited on the basis of the transcriptome sequence, batch treatment of a large amount of sequence information is carried out on the basis of transcriptome sequencing, and large batch exploitation of SSR labels can be realized through SSR site searching and SSR primer designing, so the exploitation efficiency is greatly improved, and the problems of complex steps, large workload and high exploitation cost of traditional SSR label exploitation methods are solved. The validity of SSR primers is verified through SSR primer screening and diversity analysis. Foundation is laid for plum SSR labeling primer exploitation and researches of the plum germplasm resource diversity, the kind identification and the genetic relationship by using the SSR molecular labels.
Owner:POMOLOGY RES INST FUJIAN ACAD OF AGRI SCI
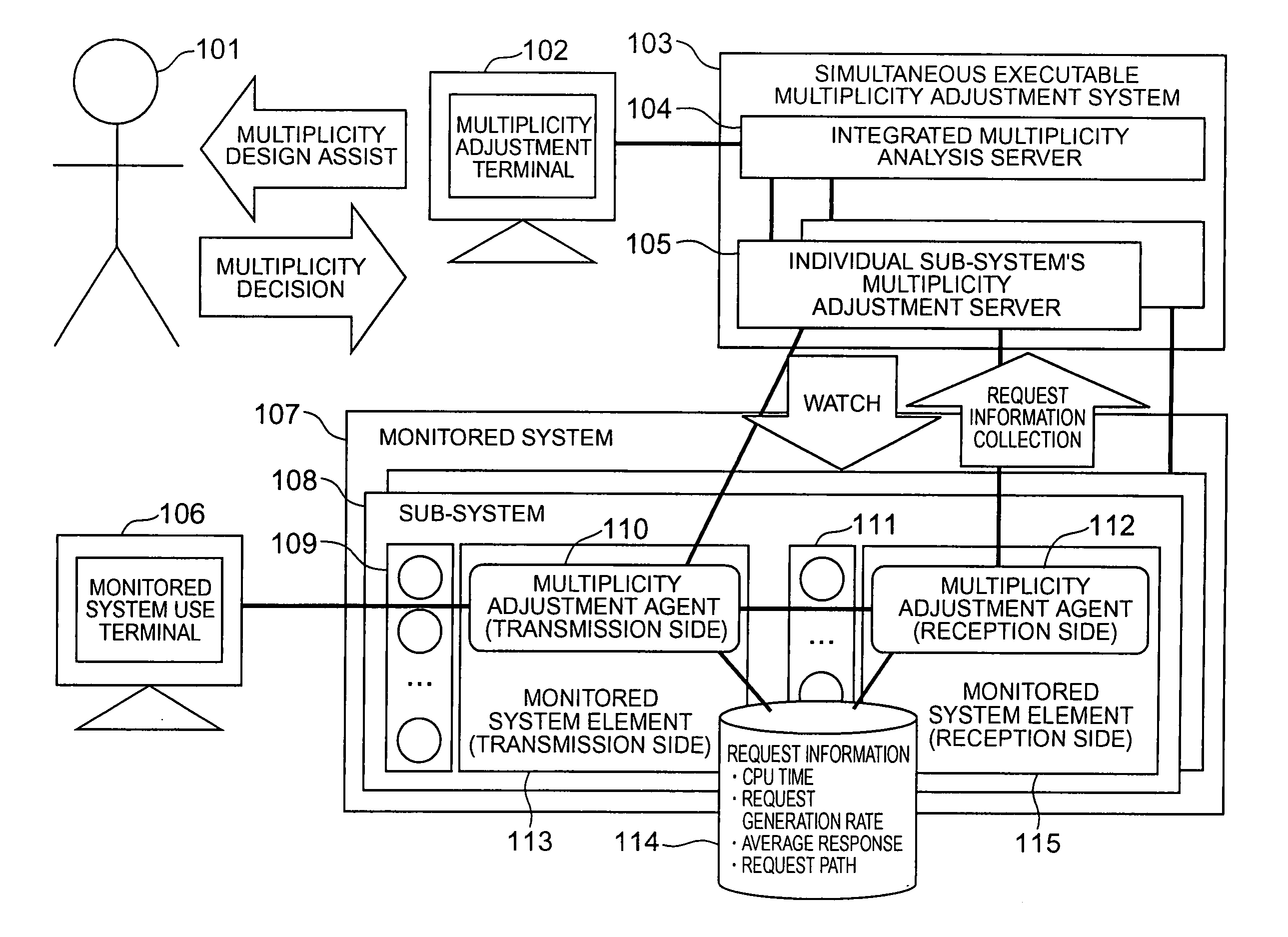
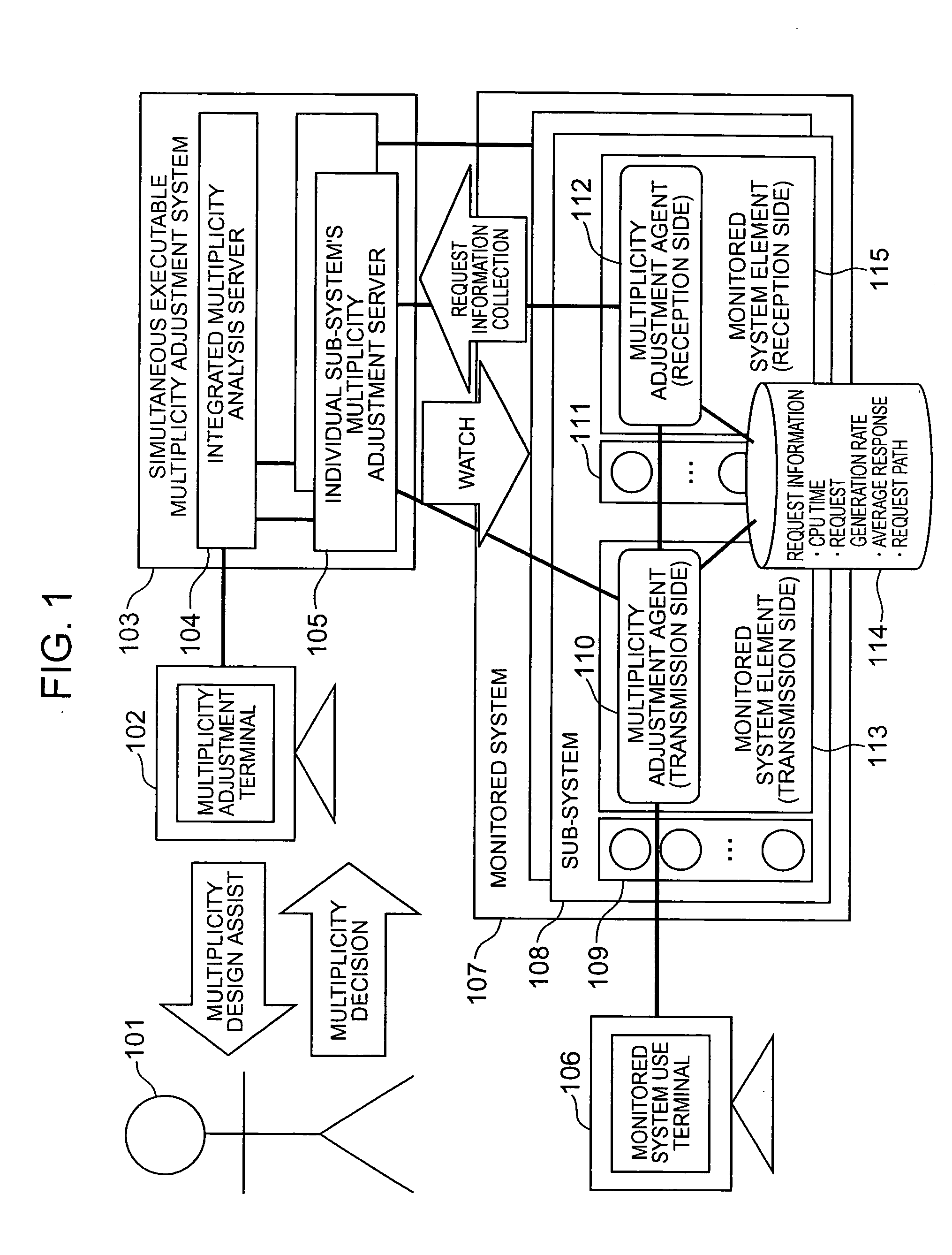
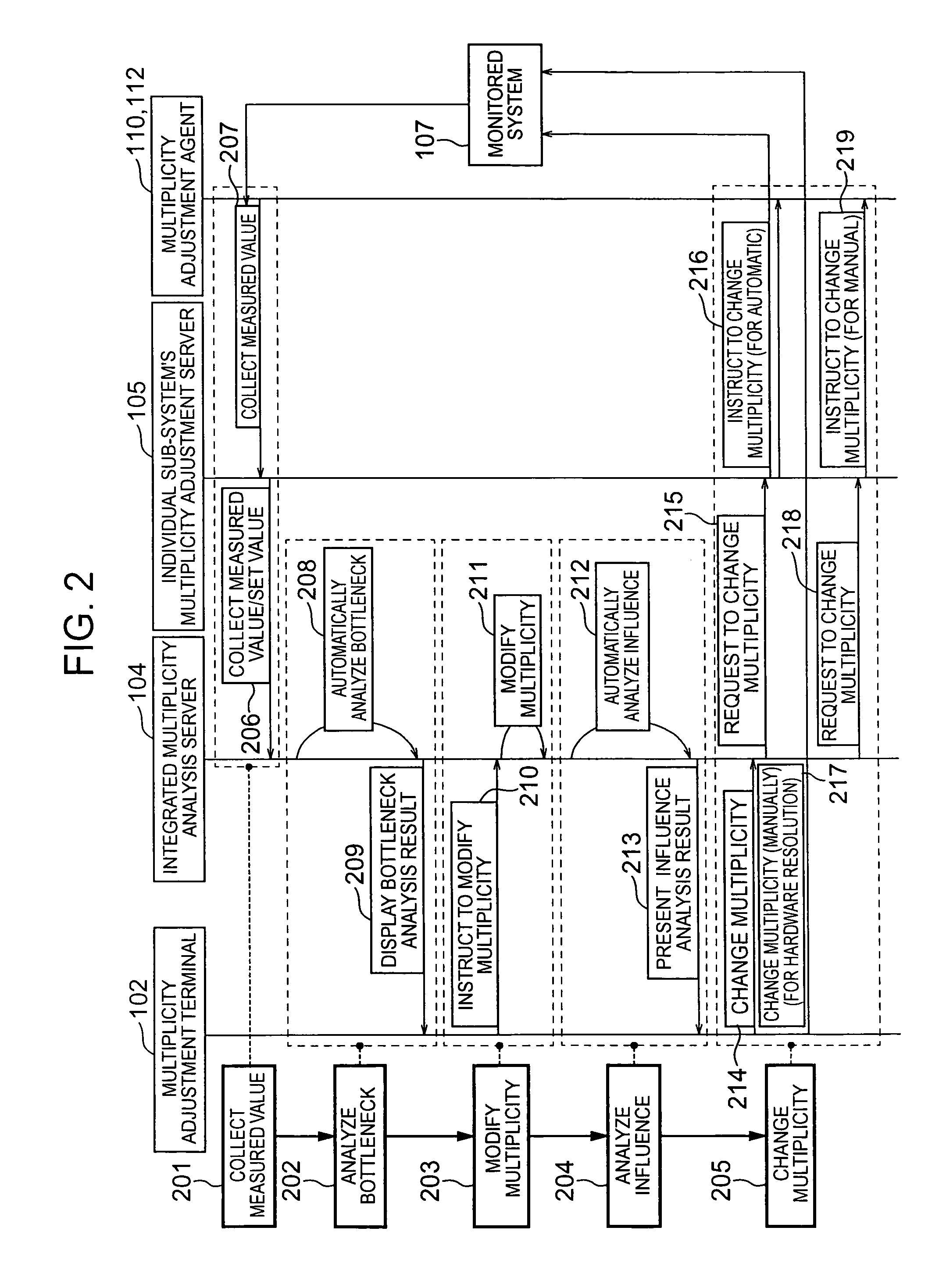
Multiplicity adjustment system and method
InactiveUS20060165000A1Safe and stable operationError preventionTransmission systemsComputer scienceDiversity analysis
A multiplicity adjustment agent collects request information database from a sub-system including an element of a system to be monitored. An individual sub-system's multiplicity adjustment server sorts and merges request information collected from each sub-system for each element / request path. An integrated multiplicity analysis server calculates a necessary multiplicity on the basis of the request information.
Owner:HITACHI LTD
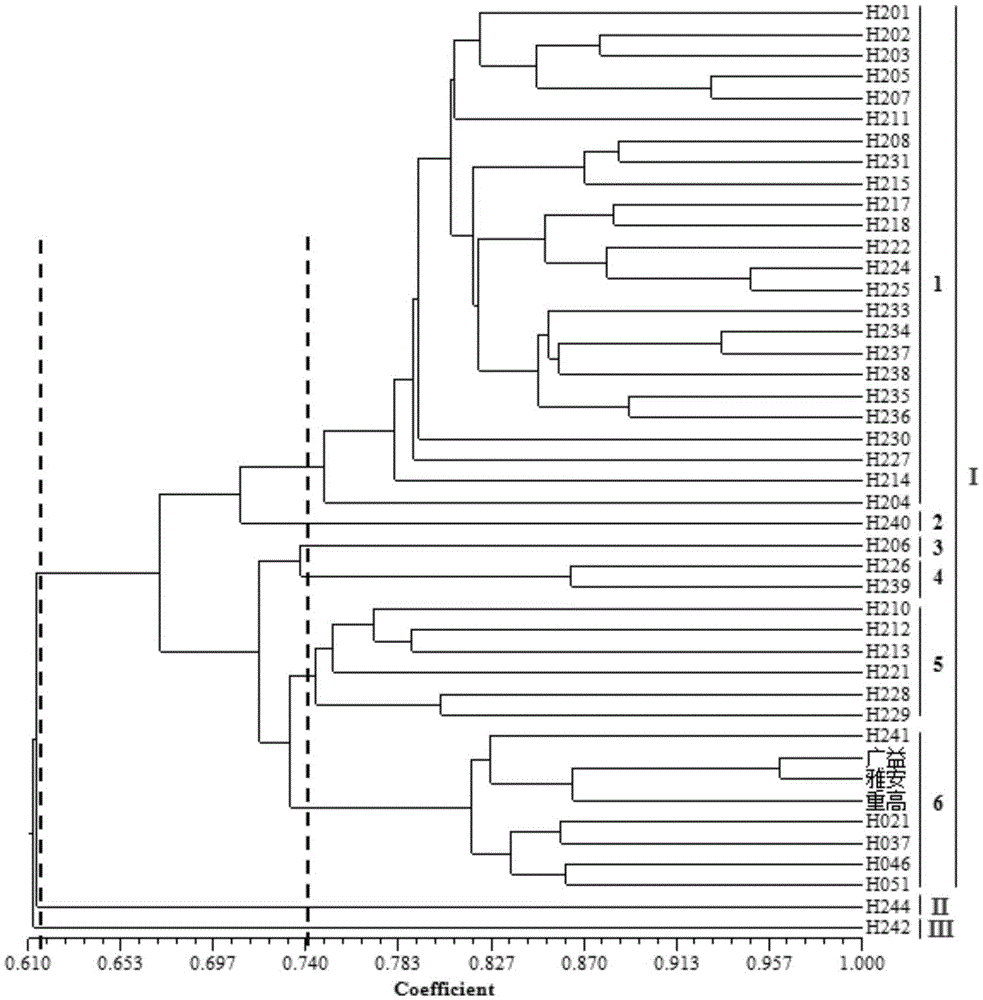
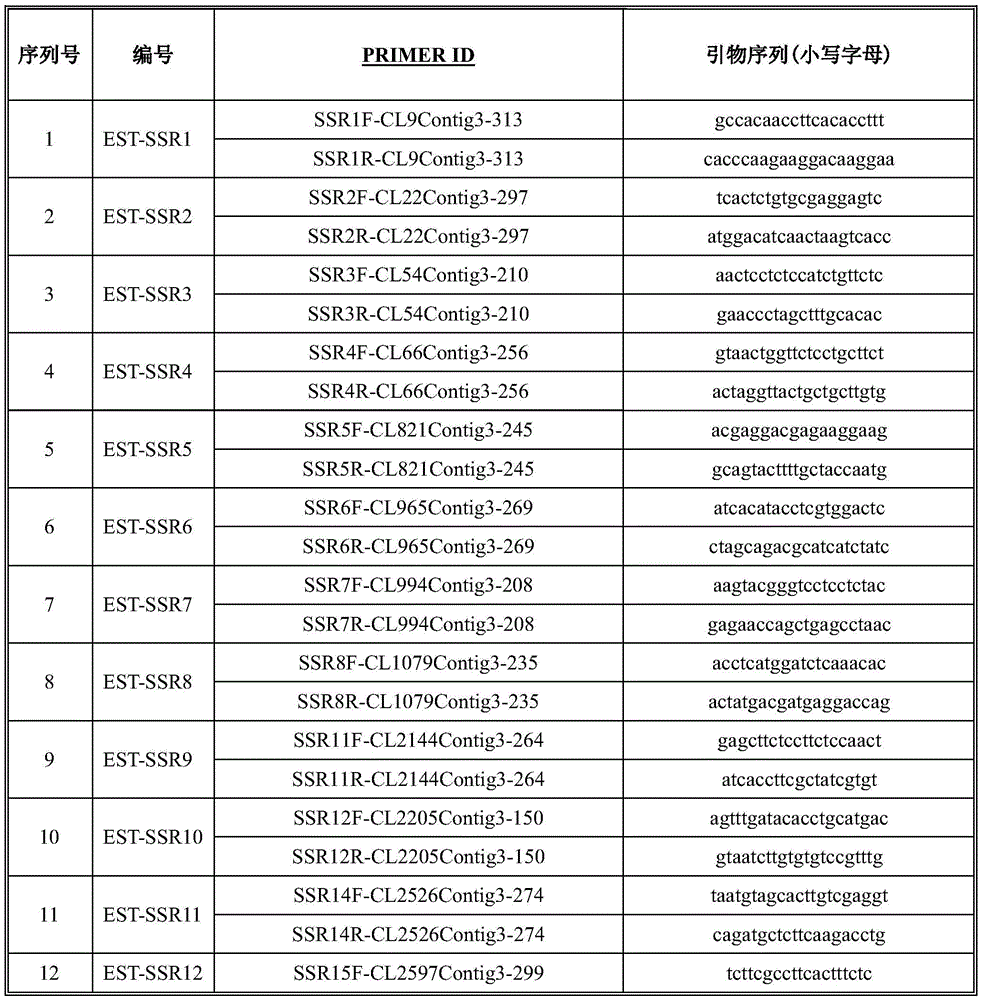
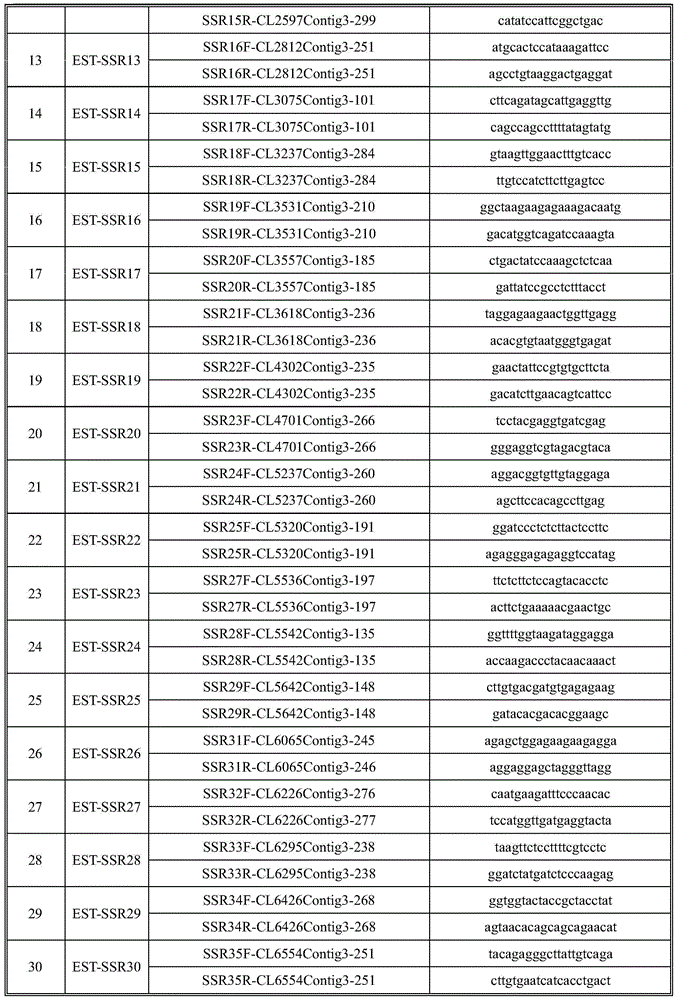
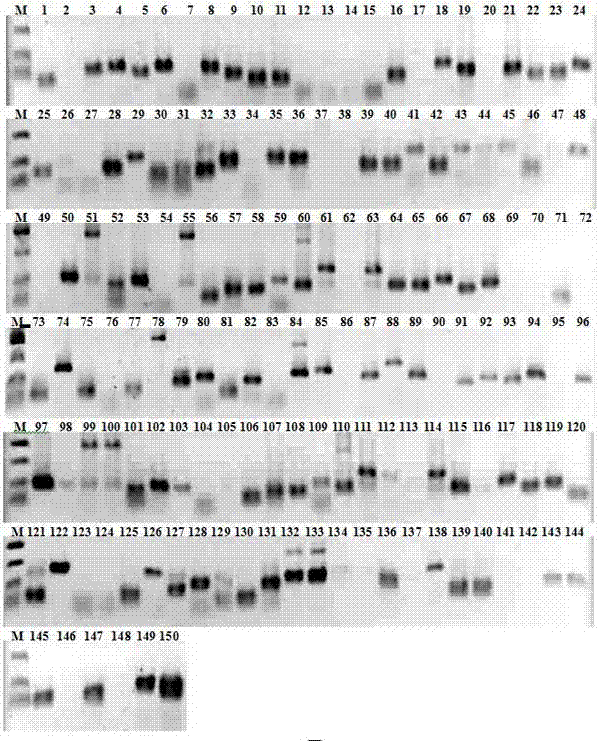
Application of EST-SSR marker primer combination and screening method in germplasm genetic diversity analysis of dwarf type and sprawl type of peas
InactiveCN105039506AQuick analysisAccurate analysisMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyElectrophoreses
The invention discloses an EST-SSR marker primer combination, which includes 12 pairs of EST-SSR available primers. A screening method of the EST-SSR marker primer combination comprises the steps of SSR locus screening in an EST sequence obtained through sequencing of a transcriptome of pea, primer design, PCR amplification, capillary electrophoresis detection to a product and marker screening. The EST-SSR marker primer combination is applied in germplasm genetic diversity analysis of dwarf type and sprawl type of peas. The EST-SSR marker primer combination can be used for quickly and accurately distinguishing different germplasm materials of peas and analyzing the genetic relationship, thereby providing a basis of oriented seed breeding of the varieties.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
Method for rapidly detecting microsatellite markers of Charybdis feriatus
InactiveCN103305611BFast detection methodThe detection method is accurateMicrobiological testing/measurementGenomic DNAGenotype
The invention relates to a method for rapidly detecting microsatellite markers of Charybdis feriatus. The method comprises the following steps of: (1) extracting a genomic DNA (Deoxyribose Nucleic Acid) of the Charybdis feriatus; (2) obtaining a gene sequence containing microsatellite repeats according to the functional gene sequence of the Charybdis feriatus in a GeneBank database; (3) designing a microsatellite marker primer; (4) implementing PCR (Polymerase Chain Reaction) amplification on the genomic DNA of different individuals of the Charybdis feriatus; and (5) detecting denaturing polyacrylamide gel electrophoresis of PCR products. The method has the advantages of rapidness, accuracy, sensitivity and the like, and can intuitively detect the genotype of different individuals of the Charybdis feriatus, so that a polymorphism map of genetic variation of the Charybdis feriatus on a functional gene microsatellite site is obtained quickly; the microsatellite markers can be applied to the fields of analysis on genetic variation and population genetic diversity of the Charybdis feriatus.
Owner:EAST CHINA SEA FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
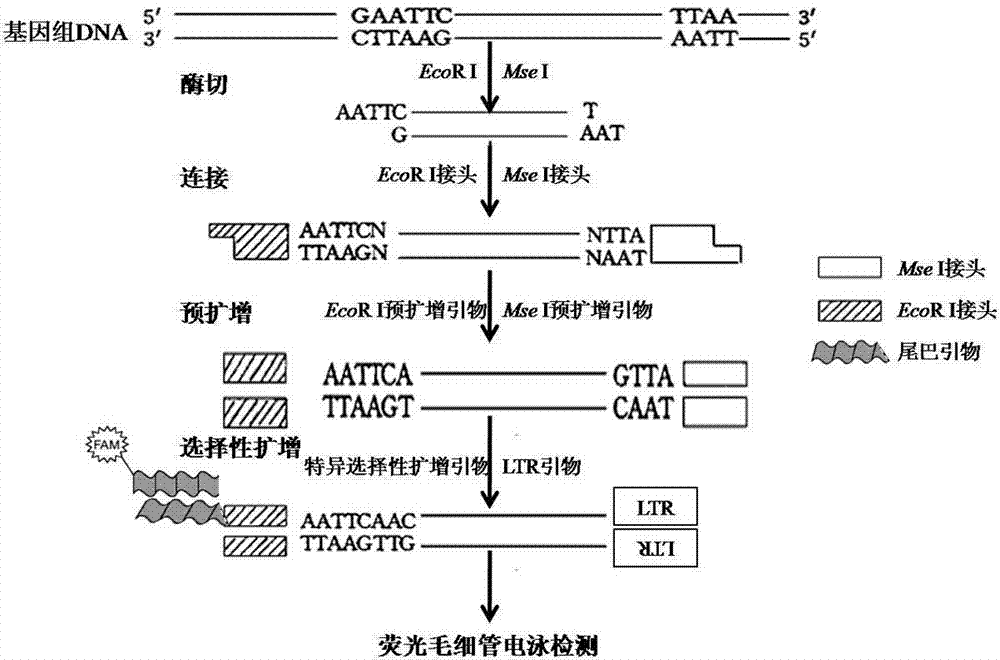
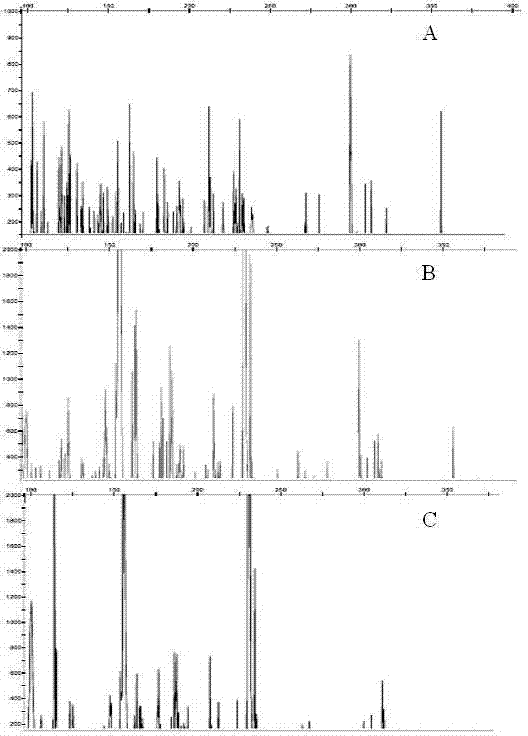
Peach SSAP (Source Service Access Point) molecular marker primer combination, molecular marker combination and application of molecular marker combination in analysis on genetic diversity of peach varieties
ActiveCN103966210AGood repeatabilityHigh polymorphismMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyElectrophoreses
The invention discloses a peach SSAP (Source Service Access Point) molecular marker primer combination, a peach SSAP molecular marker combination and an application of the peach SSAP molecular marker combination in analysis on the genetic diversity of peach varieties, wherein the peach SSAP molecular marker primer combination comprises an LTR (Long Terminal Repeat) primer, a selective amplification primer and a tail primer; the peach SSAP molecular marker combination comprises ten molecular markers JY01, JY02, JY03, JY04, JY05, JY06, JY07, JY08, JY09 and JY10. Due to the design of a peach retrotransposon LTR sequence primer, a selective amplification product is proved to be clear and abundant in amplification strip and have high efficiency, reliability and practicability through fluorescent capillary electrophoresis detection; in addition, a selective amplification PCR (Polymerase Chain Reaction) system is optimized, and a tail sequence is added, so that the traditional selective amplification PCR system is improved, and the cost for carrying out relevant researches by using the molecular markers is reduced. The SSAP molecular marker combination disclosed by the invention has relatively high polymorphism in the plurality of peach varieties, comprises stably existing markers and can be used for peach variety identification and genetic diversity analysis.
Owner:JIANGSU ACAD OF AGRI SCI
Method for preparing microbe bacteria liquid for treating black-odor riverway
InactiveCN106676022AConducive to maintaining stable growthSmall growth rateBacteriaBiological water/sewage treatmentLiquid mediumNutrient solution
The invention relates to a method for preparing microbe bacteria liquid for treating black-odor riverway. The method comprises the following steps: collecting mud at bottom of the riverway, performing microflora diversity analysis on the mud through a high-flux sequencing experiment, performing primary screening, taking mud at the bottom of the riverway, inoculating the mud into a aseptic nitrogen-removal bacterium enrichment nutrient solution, inoculating the mud into fresh enrichment nutrient solution, detecting the ammonia nitrogen and nitrate nitrogen content in the medium, taking mixing bacteria liquid with less accumulation of nitrogen as the primarily screened flora having nitrogen removal effect; optimizing enrichment screening, inoculating the fresh optimized enrichment medium into the primarily screened nitrogen-removal bacteria liquid, adding a trace element solution in the mixing bacteria liquid, inoculating the mixing bacteria liquid added with the trace element into the liquid medium which takes calcium carbonate and sodium citrate as carbon sources, uniformly preparing an uniform breeding diluents by the mixing bacteria liquid, inoculating the material to beef extract peptone nutrient agar, uniformly distributing the materials in a flat, and culturing the material.
Owner:DANYANG SHANGDE BIOTECH CO LTD
Sargassum fusiform line differentiation and classification method
A method provided by the present invention is applicable to the aspects such as sargassum fusiform germplasm resource analysis, artificially cultured sargassum fusiform group line classification, elite seed and pedigree seed near-source genetic relationship analysis and manual seedling screening by using sexual reproduction of sargassum fusiform. The method mainly comprises the operating steps of sargassum fusiform history feature and sargassum fusiform mature sporophyte morphological feature main component morphological measurement, sargassum fusiform mature sporophyte morphological feature correlation mathematical model analysis, sargassum fusiform mature sporophyte morphological feature main component mathematical model analysis, sargassum fusiform mature sporophyte morphological feature main component cluster mathematical model analysis, and sargassum fusiform mature sporophyte line differentiation and classification determination and the like. The present invention may provide a scientific differentiation and classification method for the aspects such as sargassum fusiform germplasm resource investigation, sargassum fusiform mature sporophyte group diversity analysis, sargassum basic scientific research and experimental sample screening, sargassum fusiform introduced seed morphological feature stability analysis, manual elite seed differentiation and screening by using sexual reproduction of sargassum fusiform and asexual reproduction excellent plant rhizoid storage.
Owner:温州市洞头区水产科学技术研究所 +1
Flax SSR molecular marker and application thereof
InactiveCN109517925AEfficient constructionStable amplificationMicrobiological testing/measurementDNA/RNA fragmentationHigh densityBiology
The invention relates to the technical field of plant variety identification and breeding, in particular to a building method of flax SSR fingerprint chromatography. According to the method, through MISA software, a flax genome is searched for to obtain 28751 SSR sites, Primer 5.0 is used for designing 71184 pairs of primers, and 26 flax genetic resources are used for screening out the 11 pairs ofSSR markers rich in polymorphism: RM4-5, RM4-7, RM6-2, RM6-6, RM6-7, RM7-1, RM7-3, RM8-1, RM9-4, RM13-5 and RM15-1. The SSR primers which are developed based on the flax genome have the advantages ofbeing stable in amplification, clear in electrophoretic band, rich in polymorphism and the like, and the building method of the flax SSR fingerprint chromatography can be effectively used for the study fields of flax genetic resource genetic diversity analysis, high-density chromatography building, variety purity and authenticity identification, molecular marker-assisted breeding and the like.
Owner:INST OF BAST FIBER CROPS CHINESE ACADEMY OF AGRI SCI
Molecular marker for japonica rice genetic diversity analysis and authentication method for molecular marker
InactiveCN103789308AMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyGenetic divergence
The invention relates to a molecular marker for japonica rice genetic diversity analysis and an authentication method for the molecular marker. The molecular marker comprises 32 pairs of SSR markers covering 12 chromosomes of paddy rice; each of the fourth, sixth and ninth chromosomes has one marker, each of the fifth, seventh, eighth and tenth chromosomes has two markers; each of the second, third and twelfth chromosomes has three markers; and each of the first and eleventh chromosomes has six markers. The authentication method comprises the steps of a, DNA extraction; b, PCR (polymerase chain reaction) amplification: a PCR reaction system is 15 microlitres and contains 50mM of KCl, 10mM of Tris-HCl(pH8.8), 0.1 percent of Triton-X, 1.5mM of MgCl2, 200mu M of each of NTPs, 0.2mu M of each primer, 5 percent (v / v) dimethyl sulfoxide and 0.5UT of aq DNA polymerase; c, PCR product detection; and d, statistical analysis. According to the molecular marker for japonica rice genetic diversity analysis and the authentication method for the molecular marker, the genetic diversity of a japonica rice material to be detected and genetic difference can be detected, and the molecular markers for the japonica rice genetic diversity analysis are screened.
Owner:FARMING & CULTIVATION RES INST OF HEILONGJIANG ACADEMY OF AGRI SCI
Mulberry EST-SSR (simple sequence repeat) molecular markers, and core primer group and application thereof
ActiveCN107142324AGood polymorphismStable amplificationMicrobiological testing/measurementDNA/RNA fragmentationFluorescenceTechnological system
The invention discloses mulberry EST-SSR (simple sequence repeat) molecular markers, and a core primer group and application thereof. A large amount of SSR molecular markers are developed by performing transcriptome sequencing and sequence information process on mulberries, 6 groups of polymorphic markers are screened from the SSR molecular markers, and a technological system according with SSR marker fluorescence detection is established. Through amplification and detection on the DNA of 21 mulberry varieties, the 6 groups of polymorphic markers have rich primer amplification polymorphism and high repeatability, the genetic diversity analysis result is identical to morphological morus plant classification, and the 6 groups of polymorphic markers are new markers existing stably and can be applied to mulberry germplasm resource genetic diversity analysis, variety identification, genetic map construction and molecular marker assistant breeding.
Owner:SERICULTURE & AGRI FOOD RES INST GUANGDONG ACAD OF AGRI SCI
Specific primer system of EST (expressed sequence tag)-SSR (simple sequence repeat) molecular markers for Pleurotus ostreatus and application of specific primer system
InactiveCN102181559AEasy to manageImprove the level of comprehensive utilizationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceResearch efficiency
The invention relates to a specific primer system of EST (expressed sequence tag)-SSR (simple sequence repeat) molecular markers for Pleurotus ostreatus and application of the specific primer system, belonging to the technical field of molecular biology. The specific primer system of EST-SSR molecular markers for Pleurotus ostreatus comprises 4 pairs of specific primers for EST-SSR molecular markers. The invention also provides the application of the specific primer system of EST-SSR molecular markers in the genetic diversity analysis and the germplasm resource identification of the Pleurotusostreatus. The specific primer system disclosed by the invention is used for establishing a core germplasm bank of the Pleurotus ostreatus, has convenience for use, can simply and rapidly carry out the genetic diversity analysis of germplasm resources, germplasm resource identification, genetic map construction and research on functional genes; and compared with a traditional method, the invention can greatly shorten an analytical cycle and increase research efficiency.
Owner:INST OF AGRI RESOURCES & ENVIRONMENT SHANDONG ACADEMY OF AGRI SCI
Universal metabarcoding amplification primers for freshwater fish mitochondria 12S and application method thereof
ActiveCN109943645AImprove scalabilityIncrease heightMicrobiological testing/measurementInvasive species monitoringBiotechnologyBarcode
The invention discloses universal metabarcoding amplification primers for freshwater fish mitochondria 12S and an application method thereof, and belongs to the field of biotechnology. The 16 metabarcoding amplification primers are designed according to the gene sequences of common freshwater fish mitochondria 12S, including 6 upstream primers and 10 downstream primers; the primer pairs have widecoverage on fish communities and high amplification efficiency, the primer combinations can amplify products with the length of 70bp to 350bp and are compatible with different high-throughput sequencing platforms, the multi-primer combination method can reduce non-specific amplification and increase the PCR success rate, and the primers can be widely applied to fish species identification, fish diversity analysis and other research as characteristic barcode sequences.
Owner:南京易基诺环保科技有限公司
Penaeus japonicus molecule marking method and application
InactiveCN105002267AShorten the breeding periodImprove efficiencyMicrobiological testing/measurementGermplasmA-DNA
The invention relates to a penaeus japonicus molecule marking method and application. The penaeus japonicus molecule marking method comprises the steps of extracting a DNA genome, performing PCR amplification and product identification and is applied to diversity analysis of screened penaeus japonicus germplasm resources, genetic diversity analysis, parentage assignment, genetic research of molecular population, genetic map establishment, important economic character positioning, functional gene research and assisting of penaeus japonicus molecule genetic breeding or cultivation. By means of the penaeus japonicus molecule marking method, a genetic variation polymorphic map of a genetic marker gene locus of the penaeus japonicus can be rapidly obtained, the method is simple, convenient and quick, and a result can be visually observed. The penaeus japonicus molecule marking method is mainly applied to genetic marking, genealogical identification and genetic map establishment of a penaeus japonicus colony and is rapid in detection, low in cost and wider in application range.
Owner:ZHEJIANG MARINE DEV RES INST
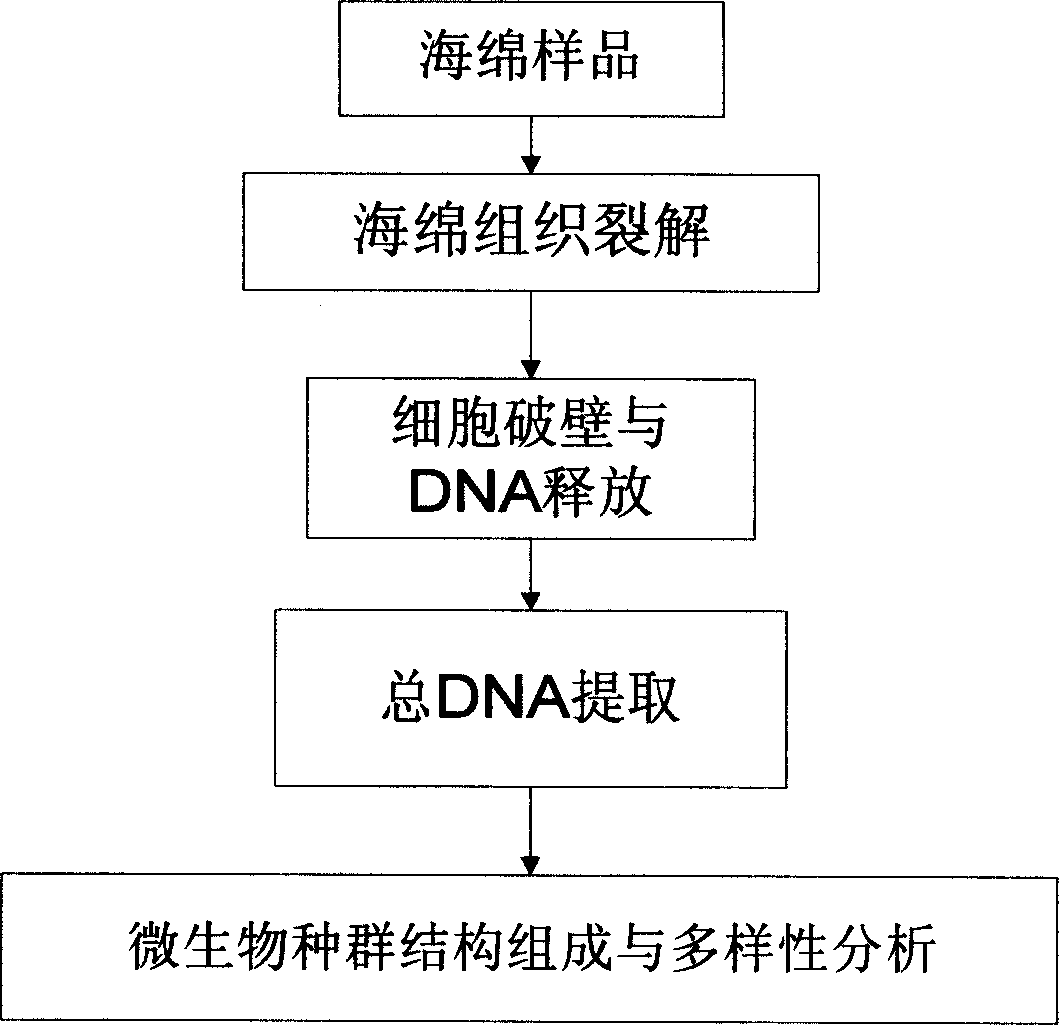
Fast extraction method of adnascent microbe community total DNA of sponge
Owner:SHANGHAI JIAO TONG UNIV
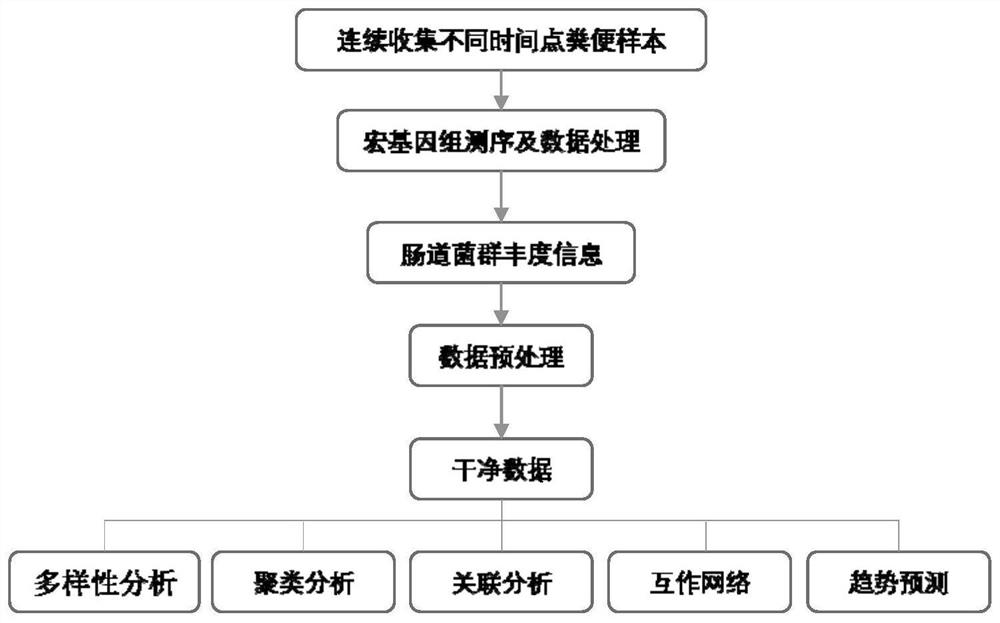
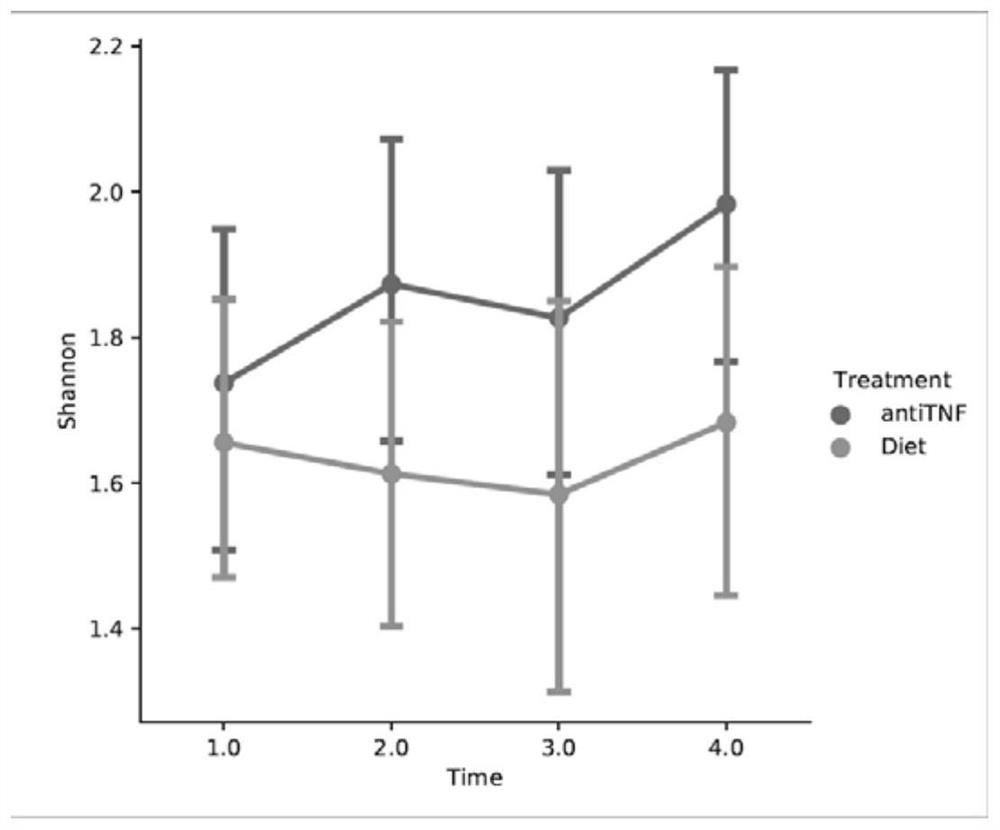
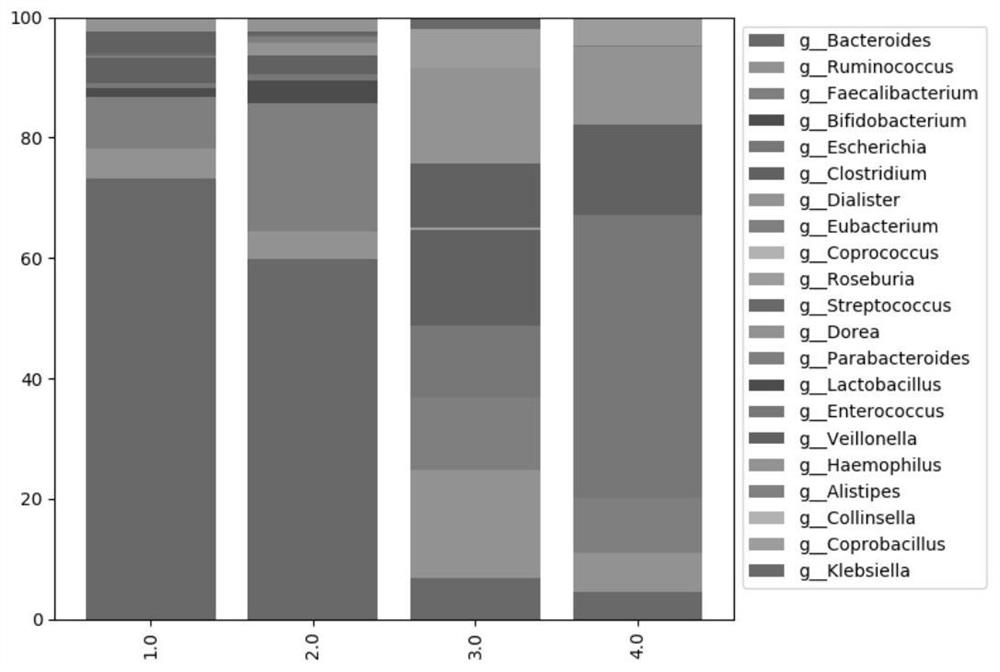
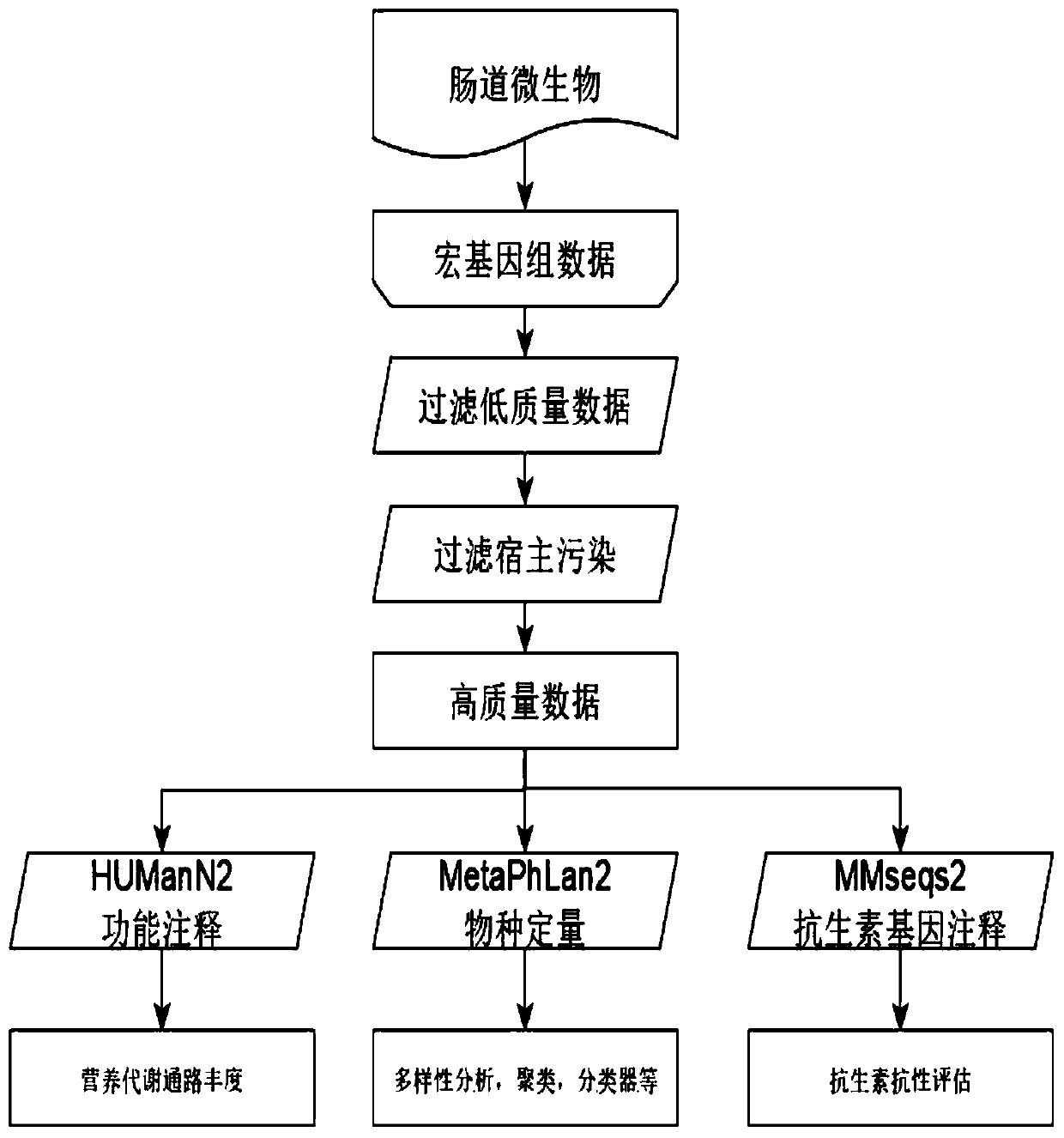
Multi-time-sequence intestinal flora data analysis process control method
ActiveCN112151118AImprove scientific research work efficiencyReduce research costsHealth-index calculationData visualisationEngineeringSequence clustering
The invention discloses a multi-time-sequence intestinal flora data analysis process control method. The analysis process mainly comprises the following stages: sample acquisition, intestinal flora data preprocessing, diversity analysis, (sequential sequence) clustering analysis, correlation analysis, bacterial colony interaction network construction and single strain change trend (sequence) prediction. A user inputs a file and corresponding parameters according to the requirements of a process file, and the system automatically analyzes data and outputs the corresponding file and a visualization result. Scientific researchers, including researchers who do not understand data analysis, can efficiently complete a set of standardized intestinal flora data analysis process based on a time sequence, and a final result is obtained. Therefore, the purposes of improving scientific research work efficiency and reducing scientific research cost are achieved.
Owner:康美华大基因技术有限公司
ISSR molecular marker method for genetic diversity analysis of hedyotis diffusa
ActiveCN107354217ABelt stabilityClear bandMicrobiological testing/measurementDNA/RNA fragmentationGenomic DNAA-DNA
The invention provides an ISSR molecular marker method for genetic diversity analysis of hedyotis diffusa, and belongs to the technical field of molecular biology. The method comprises the following steps of selecting four primers with clear background, good amplification effect and high polymorphism, amplifying genomic DNAs of the hedyotis diffusa by using ISSR-PCR reaction, analyzing stripes according to an ISSR-PCR result, establishing a DNA fingerprint of the hedyotis diffusa, and constructing a genetic resource database of the hedyotis diffusa. According to the method of the invention, a new method for identification of variety of the hedyotis diffusa and analysis of genetic diversity is created, so as to effectively solve the problem of poor representativeness of a database in the construction of the genetic marker database in the past, and also solve the problem that the existing marking technology has a defect in the hedyotis diffusa genetic resources; the method has the advantages of high polymorphism, good repeatability, low cost, and simple testing procedure, and is not affected by an environment.
Owner:GUANGXI UNIV OF CHINESE MEDICINE
Method for rapidly detecting microsatellite markers of Charybdis feriatus
InactiveCN103305611AFast detection methodThe detection method is accurateMicrobiological testing/measurementGenomic DNAGenotype
The invention relates to a method for rapidly detecting microsatellite markers of Charybdis feriatus. The method comprises the following steps of: (1) extracting a genomic DNA (Deoxyribose Nucleic Acid) of the Charybdis feriatus; (2) obtaining a gene sequence containing microsatellite repeats according to the functional gene sequence of the Charybdis feriatus in a GeneBank database; (3) designing a microsatellite marker primer; (4) implementing PCR (Polymerase Chain Reaction) amplification on the genomic DNA of different individuals of the Charybdis feriatus; and (5) detecting denaturing polyacrylamide gel electrophoresis of PCR products. The method has the advantages of rapidness, accuracy, sensitivity and the like, and can intuitively detect the genotype of different individuals of the Charybdis feriatus, so that a polymorphism map of genetic variation of the Charybdis feriatus on a functional gene microsatellite site is obtained quickly; the microsatellite markers can be applied to the fields of analysis on genetic variation and population genetic diversity of the Charybdis feriatus.
Owner:EAST CHINA SEA FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
Group of cabbage mustard InDel molecular markers as well as development method and application thereof
ActiveCN110791550AIncrease success rateHigh polymorphismMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyNucleotide
The invention provides a group of cabbage mustard InDel molecular markers as well as a development method and an application thereof, which relate to the technical field of molecular markers. The cabbage mustard InDel molecular marker provided by the invention comprises primer pairs corresponding to 60 sites, and the nucleotide sequences of the primer pairs are shown as SEQ ID NO. 1 to SEQ ID NO.120. The marker developed by the invention has the characteristics of simple operation, good stability and high polymorphism; the primer pairs are good in stability, are uniformly distributed in 9 linkage groups, can be used for positioning of important agronomic trait genes of cabbage mustard, genetic diversity analysis, fingerprint construction, genome-wide association analysis and genetic linkage map construction or molecular marker-assisted selective breeding, and can improve the working efficiency.
Owner:INST OF VEGETABLES GUANGDONG PROV ACAD OF AGRI SCI
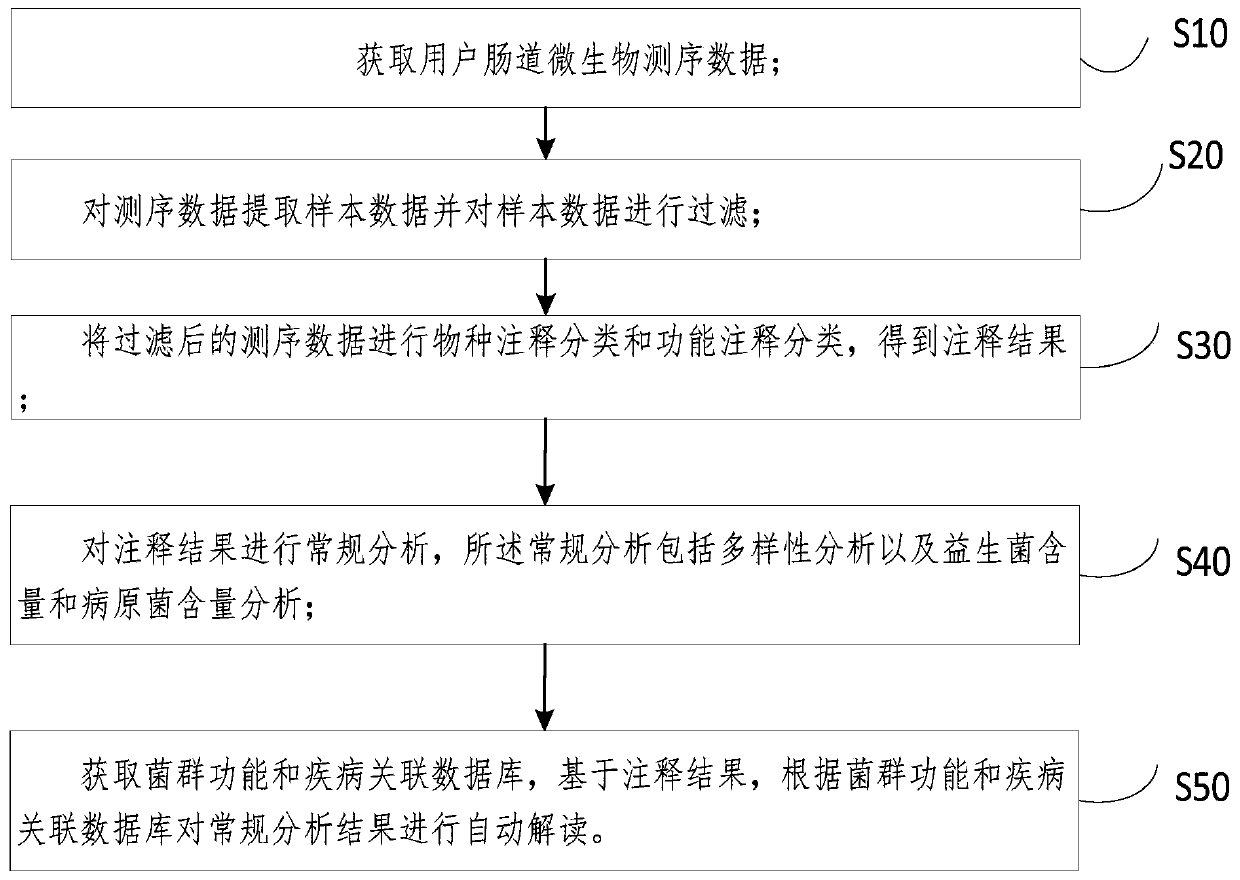
Enteric microorganism detection data analysis method, automatic interpretation system and medium
PendingCN111462819ARelieve pressureVarious formsBiostatisticsSequence analysisIntestinal microorganismsFlora
The invention discloses an enteric microorganism detection data analysis method, an automatic interpretation system and a medium. The method comprises the following steps: acquiring enteric microorganism sequencing data of a user; extracting sample data from the sequencing data, and filtering the sample data; performing species annotation classification and function annotation classification on the filtered sequencing data to obtain an annotation result; performing conventional analysis on the annotation result, wherein the conventional analysis comprises diversity analysis and probiotics content and pathogenic bacteria content analysis; and obtaining a flora function and disease association database, and based on the annotation result, automatically interpreting the conventional analysisresult according to the flora function and disease association database. According to the method, analysis and interpretation can be automatically executed, so that the working pressure is reduced, the generated analysis results are diversified in form and good in readability, the requirements of users are met, process and batch operations are conveniently carried out, and the workload of manual interpretation is small, visual and convenient.
Owner:康美华大基因技术有限公司
EST-SSR primer group developed on basis of transcriptome sequences of hemarthria compressa and hemarthria altissima and application of EST-SSR primer group
InactiveCN105586338AA large amountPolymorphism richMicrobiological testing/measurementDNA/RNA fragmentationGermplasmGenetics
The invention discloses an EST-SSR primer group developed on the basis of transcriptome sequences of hemarthria compressa and hemarthria altissima. The EST-SSR primer group comprises 52 pairs of primers, and nucleotide sequences of the primers are represented as sequence tables SEQUENCE ID NO.1-104. The invention further relates to an application of the EST-SSR labeled primers in the aspects of genetic diversity analysis of hemarthria sibirica and transferability of the hemarthria sibirica in gramineae species. The 52 pairs of primers in the EST-SSR primer group developed on the basis of the transcriptome sequences of hemarthria compressa and hemarthria altissima have the advantages of abundant polymorphism, stable amplification, good repeatability, convenience in counting and the like, can be effectively applied to research fields such as hemarthria sibirica germ resource genetic diversity analysis, variety identification, fingerprinting construction, molecular marker assisted breeding and the like and are applied to correlational research of other gramineae forage species.
Owner:SICHUAN AGRI UNIV +1
SSR primer group developed on basis of zucchini transcriptome sequence and application of SSR primer group
ActiveCN107312868AImproved geneticsPolymorphism richMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGermplasm
The invention discloses an SSR primer group developed on the basis of a zucchini transcriptome sequence and application of the SSR primer group in identification of a genetic relationship and a hereditary character of zucchini, and belongs to the technical field of molecular biology. The SSR primer group comprises 35 pairs of primers, and nucleotide sequences of the first pair of primers to the 35th pair of primers are as shown in SEQ ID No. 1 to SEQ ID No. 70. The obtained SSR primer group has the advantages of abundant polymorphism, good repeatability, stable amplification, clear electrophoretic bands and the like. Meanwhile, because the primers are from zucchini expression genes and can reflect variation of a functional domain in a genome, diversity of genetic germplasm of loofah can be distinguished well. The primer group can be used for zucchini material resource genetic diversity analysis, genetic relationship identification, DNA finger-print construction and the like.
Owner:CROP RES INST OF FUJIAN ACAD OF AGRI SCI
SSR core primer group developed based on whole genome data of tartary buckwheat and application thereof
ActiveCN110195126AMicrobiological testing/measurementDNA/RNA fragmentationPolygonum fagopyrumAgricultural science
The invention discloses an SSR core primer group developed based on a whole genome sequence of tartary buckwheat and an application thereof. A development process includes the steps: (1) acquiring thetartary buckwheat genome sequence; (2) searching the genome sequence containing SSR; (3) designing an SSR primer; and (4) screening the SSR primer. The SSR core primer group comprises 12 pairs of primers having nucleotide sequences shown in SEQ No.1-24. The SSR core primer group provided by the invention has the advantages of uniform distribution, abundant polymorphism, good amplification reproducibility and clear band shape in the tartary buckwheat genome, and is suitable for detection of a common denatured polyacrylamide gel electrophoresis detection platform and a capillary fluorescence detection platform, and can be applied to tartary buckwheat genetic diversity analysis, variety identification, DNA fingerprint spectrum construction, molecular marker assisted breeding and other research fields.
Owner:山西省农业科学院农作物品种资源研究所
Method for obtaining nucleotide sequence in V3 region of 16S rRNA genes of bacteria and special primer thereof
ActiveCN101792760AReduce experiment costShort experiment cycleMicrobiological testing/measurementDNA/RNA fragmentationNucleotide sequencingGel electrophoresis
The invention discloses a method for obtaining a nucleotide sequence in the V3 region of 16S rRNA genes of bacteria and a special primer thereof. The special primer is a specific primer pair consisting of nucleotides having a sequence 17 and a sequence 19 in a sequence table. The method comprises the following steps of: (1) carrying out PCR amplification by using a universal primer and using the genome DNA of the bacterium as a template, wherein the universal primer is a primer pair consisting of nucleotides having a sequence 17 and a sequence 18 in the sequence table; (2) carrying out PCR amplification by using the specific primer and using the PCR product of the step (1) as a temple; (3) sequencing the PCR product in the step (2) by using a sequencing primer, wherein the nucleotide sequence of the sequencing primer is shown as the sequence 20 in the sequence table. The primer pair aiming at the V3 region of 16S rRNA genes for bacterial diversity analysis provided by the invention can be directly used for sequencing without carriers, which is beneficial to reducing the experimental cost, shortening the experimental period and enhancing the accuracy of the experiment. The specific primer pair and the method are particularly suitable for bacterial diversity test of environmental sample aiming at the V3 region of 16S rRNA genes by using PCR-DGGE (PCR-Denaturing Gradient Gel Electrophoresis) technology.
Owner:FEED RESEARCH INSTITUTE CHINESE ACADEMY OF AGRICULTURAL SCIENCES
Diversity analysis method based on chemical structure with CPU (Central Processing Unit) acceleration
InactiveCN102436545ADiversity analysis worksSolve the time-consuming problem of diversity analysisSpecial data processing applicationsChemical structureTheoretical computer science
The invention relates to a diversity analysis method based on a chemical structure with CPU (Central Processing Unit) acceleration. The method comprises the following steps of: (a) reading data in chemical structure linked lists in a query library and an inquired library in memory equipment into a main memory; (b) respectively resolving the data as a tree topology sub map set of chemical environment coding of the query library and the inquired library, and storing the tree topology sub map set in the main memory; (c) respectively comparing the tree topology sub map set of chemical environment coding of the query library and the inquired library with a tree topology sub map template, respectively generating binary data for the query library and the inquired library, and storing the binary data in the main memory; (d) transmitting the binary data in the query library and the inquired library to frame caching from the main memory; (e) reading the binary data in the query library and the inquired library from the frame caching by the CPU, and calculating both similarity value by the CPU; (f) transmitting the similarity value to the main memory from the frame caching; (g) reading the similarity value from the main memory and outputting the similarity value to the memory equipment by the CPU.
Owner:KMS MEDITECH
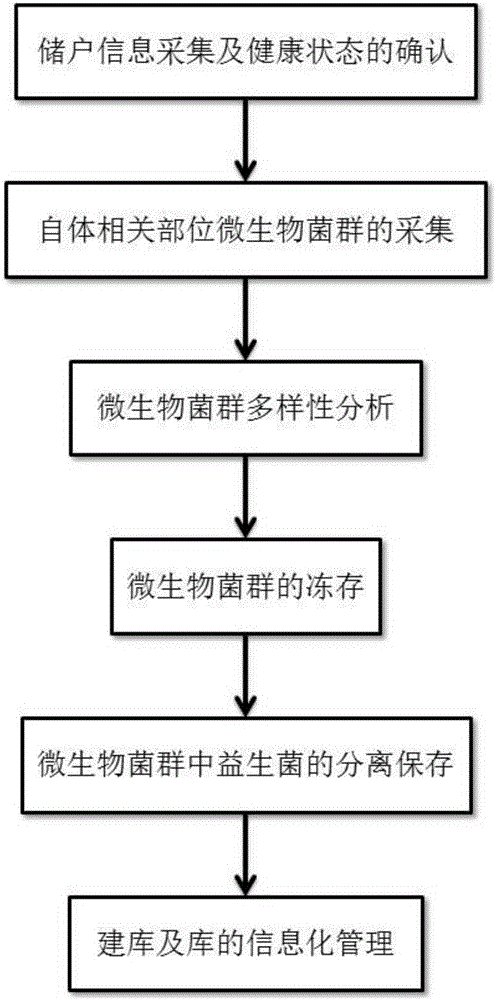
Method for establishing flora transplanting flora libraries from microorganisms in autologous health statuses
PendingCN106192020AMicrobiological testing/measurementMicroorganism based processesDiseaseMicroorganism
The invention discloses a method for establishing flora transplanting flora libraries from microorganisms in autologous health statues. The method includes operation steps of 1, acquiring saver information and confirming the health statues; 2, acquiring microbial floras at autologous related positions; 3, analyzing the diversity of the microbial floras; 4, storing the microbial floras; 5, separately storing probiotics in the microbial floras; 6, establishing the libraries and carrying out information management on the libraries. The method has the advantages that transplanting for disease treatment, health protection and the like can be carried out by the aid of the microorganisms stored in the autologous health statues, and the microorganisms are high in psychological receptivity and free of immunological rejection or ethical controversy as compared with microorganisms transplanted from other people; the autologous microbial flora libraries of healthy people are established by the aid of the method, and accordingly foundations can be provided for future personalized precision medical treatment, health protection and the like.
Owner:BEIHANG UNIV
SSR (Simple Sequence Repeat) primesr of platygaster robiniae buhl and duso and application thereof to population genetic diversity analysis
ActiveCN108588234AMicrobiological testing/measurementDNA/RNA fragmentationPolymorphism analysisPopulation evolution
The invention discloses SSR (Simple Sequence Repeat) primers of platygaster robiniae buhl and duso and application thereof to population genetic diversity analysis. The invention designs according tomitochondrial genome data of the platygaster robiniae buhl and duso to obtain more than 130 thousand of SSR sequences and screens out 37 pairs of SSR specific primers which are relatively stable and have excellent polymorphism from the SSR sequences. The invention further discloses application of the 37 pairs of SSR specific primers in the aspects of population genetic diversity analysis, population evolution analysis and polymorphic analysis of the platygaster robiniae buhl and duso, construction of fingerprint spectrum of the platygaster robiniae buhl and duso and the like. The SSR primers and the application thereof have the important value for developing natural enemy class group population genetic diversity research of the platygaster robiniae buhl and duso, mastering the source and the population differentiation condition of the platygaster robiniae buhl and duso and the like.
Owner:INST OF FOREST ECOLOGY ENVIRONMENT & PROTECTION CHINESE ACAD OF FORESTRY
Bare sand land vegetation quick recovery method
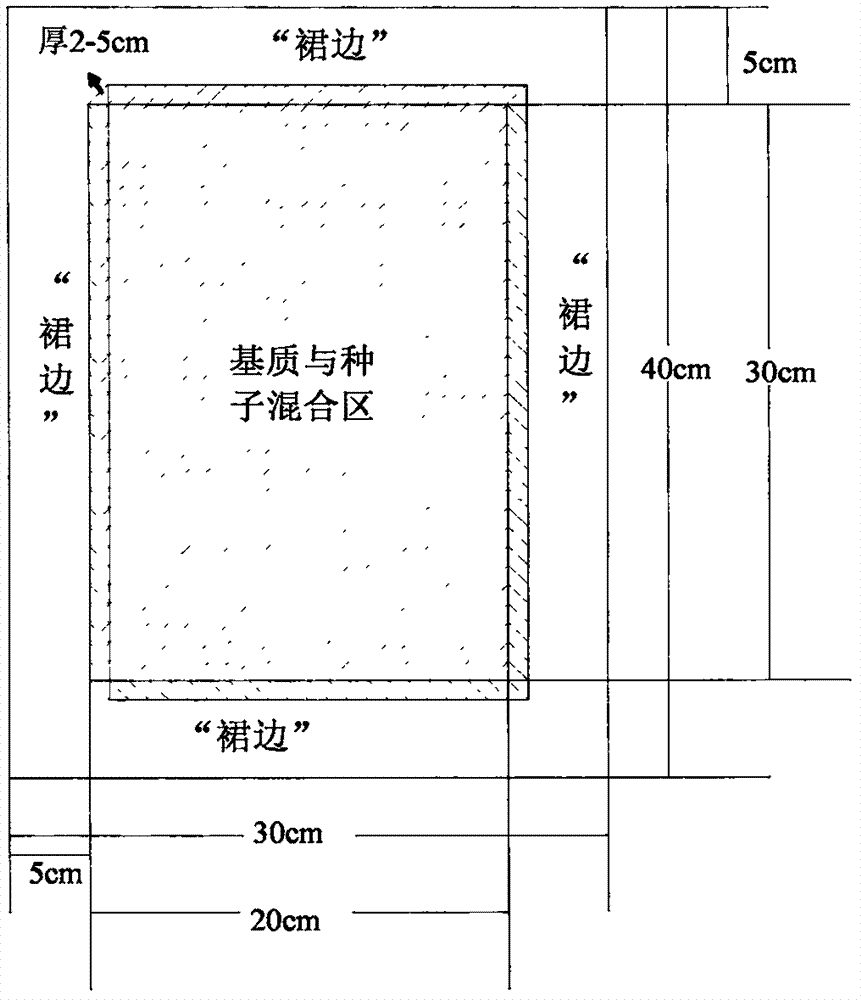
InactiveCN107172906AGuaranteed adaptabilityNot easy to dieCalcareous fertilisersBio-organic fraction processingRecovery methodInvestigation methods
The invention discloses a bare sand land vegetation quick recovery method. The method comprises the first step of selecting seeds, the second step of preparing a tile-type vegetation bag, wherein the second step comprises the procedures of a, conducting mixed matching on multiple plant seeds according to the proportion of the degree of dominance; b, uniformly mixing organic compost and sand according to the volume ratio of 1:3, and adopting the mixture as a seed growing matrix; c; sowing seeds on the growing matrix to prepare the tile-type vegetation bag. Compared with the prior art, through a phytocoenology investigation method, diversity analysis is conducted, seed selection is conducted according to the calculation of the degree of dominance, a natural or secondary growth plant community structure in a controlled region is simulated, according to a plant community natural matching rule, seed configuration is conducted on multiple selected species according to the degree of dominance, the adaptability of the seed in a local environment is guaranteed, the seeds are not prone to death, and the vegetation recovery effect is thus not influenced. By precisely calculating the size of the vegetation bag, the vegetation bag is convenient to deliver and lay, the vegetation bag can be entirely buried by wind and sand within a short time, the water and fertilizer are retained, and the vegetation bag is quickly degraded.
Owner:周金星
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com