Mulberry EST-SSR (simple sequence repeat) molecular markers, and core primer group and application thereof
A technology of molecular markers and core primers, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve problems such as limiting mulberry molecular genetics research, and achieve clear amplification results and good repeatability , Amplify the effect of stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1: Development of EST-SSR core primer set based on mulberry transcriptome sequence
[0053] 1. Design and synthesis of EST-SSR primers in the mulberry transcriptome sequence
[0054] The MISA software was used to search all the SSR sites in the mulberry unigenes database of the mulberry transcriptome. The length of the SSR motif repeating unit retrieved was 2 to 6 nucleotides, which were set to 2, 3, 4, 5, The minimum number of search repetitions for 6 nucleotides is 6, 5, 5, 4, and 4 times, and the length of the flanking sequence of the SSR site is ≥ 150 bp. According to the searched SSR site and flanking sequence, use Primer3v2.3.4 (http: / / primer3.sourcego-rge.net) software to design specific primers, the design criteria are: the primer length is 18-25bp, the annealing temperature is 57-63°C, the GC content is 40-70%, and the length of the PCR product is 100- 300bp, select SSR sites that can be compared with the NCBI non-redundant protein database to synthes...
Embodiment 2
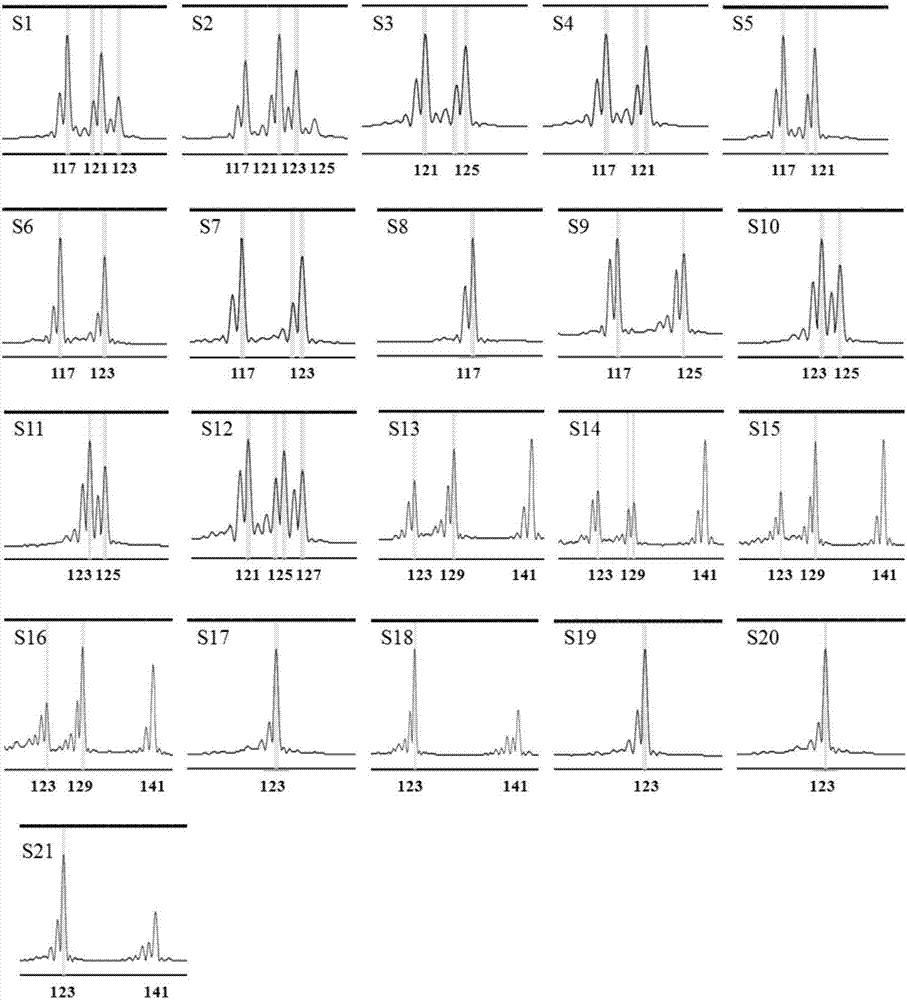
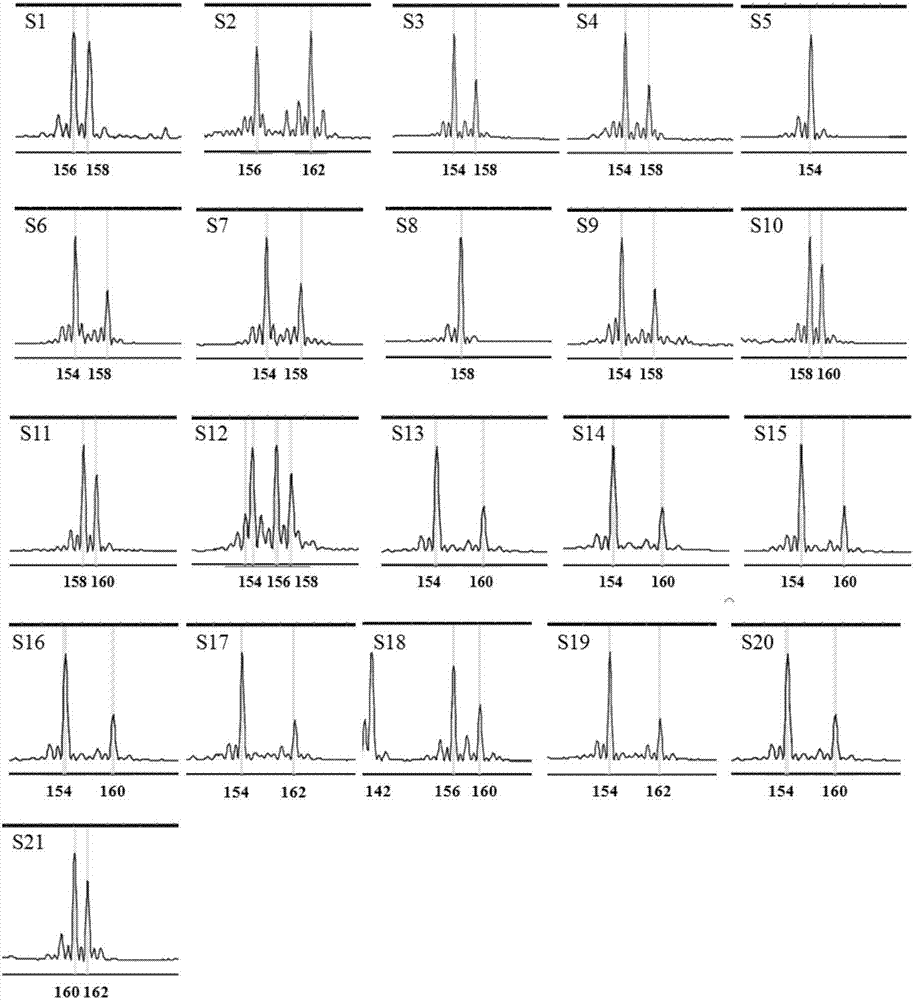
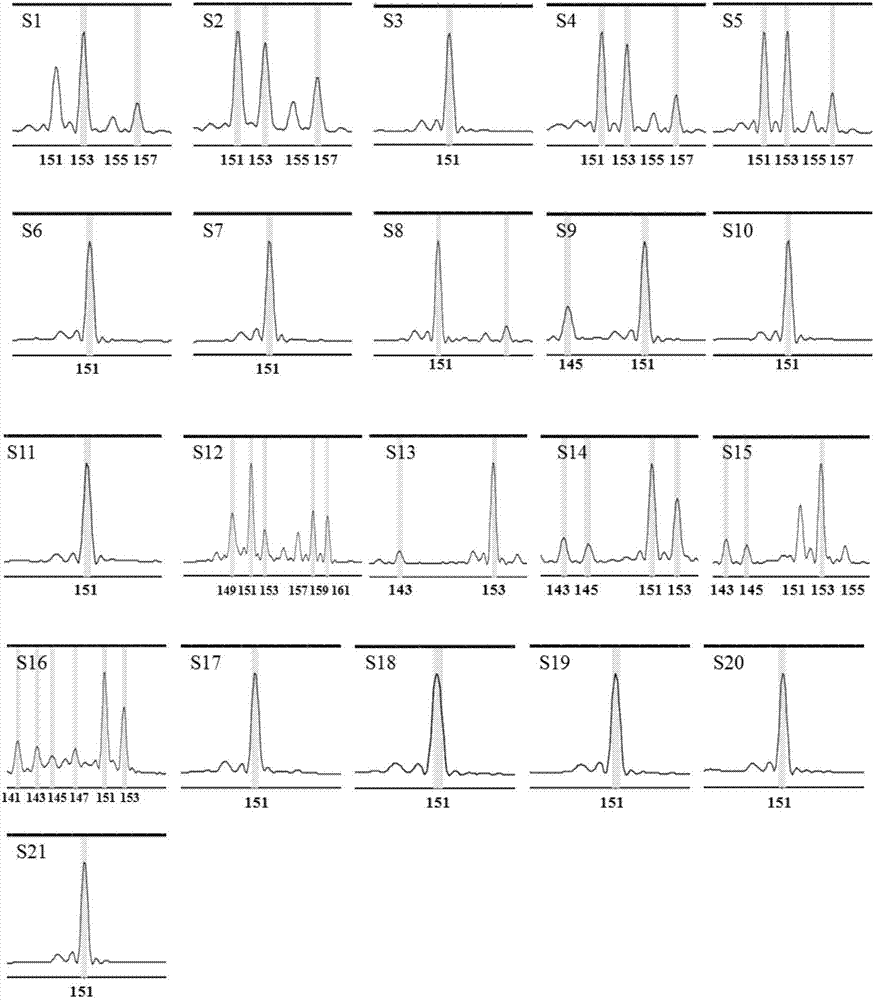
[0078] Embodiment 2: the application of mulberry EST-SSR core primer set
[0079] 1. DNA extraction
[0080] Take the improved CTAB method to extract the DNA of the genomes of the above-mentioned 21 mulberry varieties (including 11 Guangdong mulberry varieties, 5 white mulberry germplasms, 4 Mengmulberry germplasms and 1 long-fruit mulberry germplasms); the specific steps are the same as in the examples 1 step 2.
[0081] 2. PCR amplification
[0082] Using the DNA extracted in step 1 as a template, use the 6 pairs of EST-SSR core primers developed in Example 1 (see Table 1), divide them into 3 groups according to the combination in Table 1, and perform multiplex PCR.
[0083] The three sets of EST-SSR molecular marker core primer set PCR reaction system are as follows:
[0084] Reaction system I: 50ng / μL DNA template 0.5μL, 10×PCR buffer (without Mg 2+ ) 2.5μL, 25mM MgCl 2 2.0 μL, 10 mM dNTPs 0.5 μL, 5U / μL Tap DNA polymerase 0.2 μL, 10 μM MulSSR5053 upstream primer 0.1 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com