Whole-genome 50KSNP chip for apostichopus japonicus breeding and application
A whole-genome, japonicus imitation technology, applied in the fields of bioinformatics, molecular breeding, functional genomics, and molecular biology, can solve the problems of lack of liquid-phase chips, etc., and achieve good typing effect, good targeting, and accuracy sex high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Example 1: Development of a genome-wide 50kSNP chip of imitation sea cucumber
[0086] 1. Construction of imitation japonicus sample population
[0087] In different sea areas of Liaoning and Shandong, 500 imitation sea cucumber samples were randomly selected, and the tissues were taken and placed in 95% ethanol and brought back to the laboratory for preservation.
[0088] 2. Genome-wide SNP typing of sea cucumber
[0089] 2.1 DNA extraction
[0090] ①Add 500ul STE lysis buffer (100mM NaCl; 10mM Tris-Cl, pH8.0; 1mM EDTA, pH8.0), 50ul 10% SDS, 3.5ul proteinase K (20mg / ml), 16ul to a 1.5ml tube RNase A (100mg / ml), add about 0.1g of scallop adductor muscle, chop it up, grind it with a grinding rod, grind it until flocculent, treat it at 56°C for about 2 hours, during this period, mix it upside down every 30mins, and finally clarify the lysate state.
[0091] ②Add 500ul Tris saturated phenol, 100ul chloroform / isoamyl alcohol (24:1), shake gently for 20min, and centrifug...
Embodiment 2
[0149] Example 2: Application of 50K SNP chip of japonicus japonicus in molecular breeding
[0150] In order to verify the application effect of japonicus japonicus chips in imitation japonicus japonicus molecular breeding, we used 50K chips to detect samples from Russia (30), Dalian (100), and Shandong (100), and carried out molecular breeding:
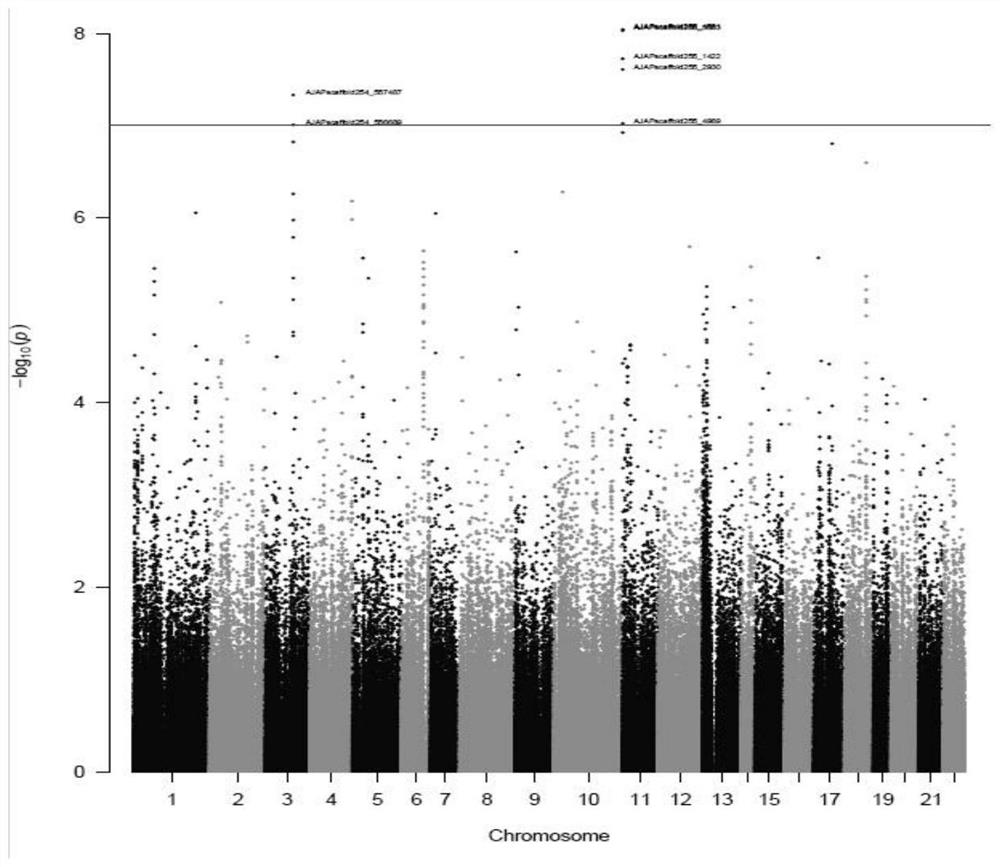
[0151] (1) Genetic background analysis of different geographical populations; (2) Genome-wide association analysis (GWAS) of the growth-related number of japonicus japonicus spines
[0152] 1. The role of whole-genome breeding chip of sea cucumber in the analysis of genetic background of different populations of sea cucumber:
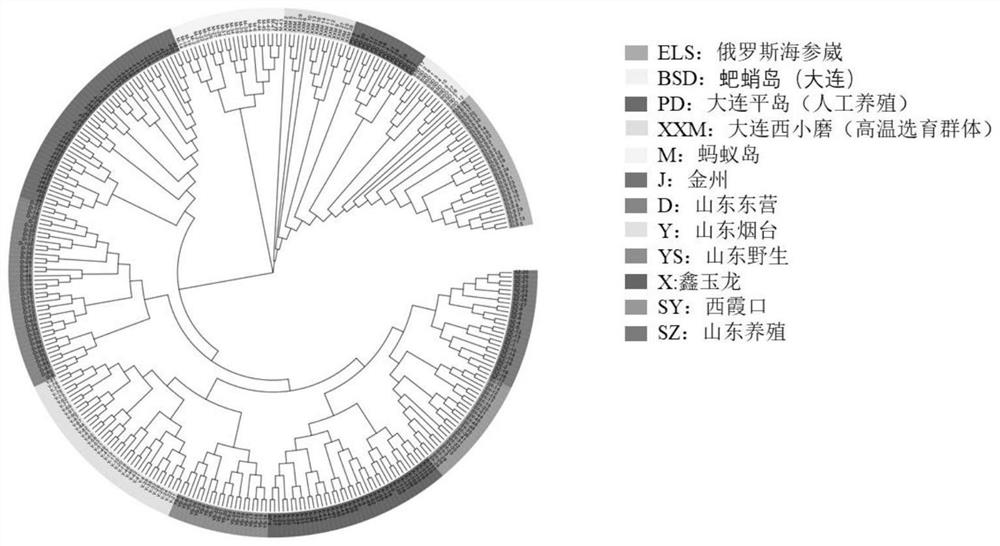
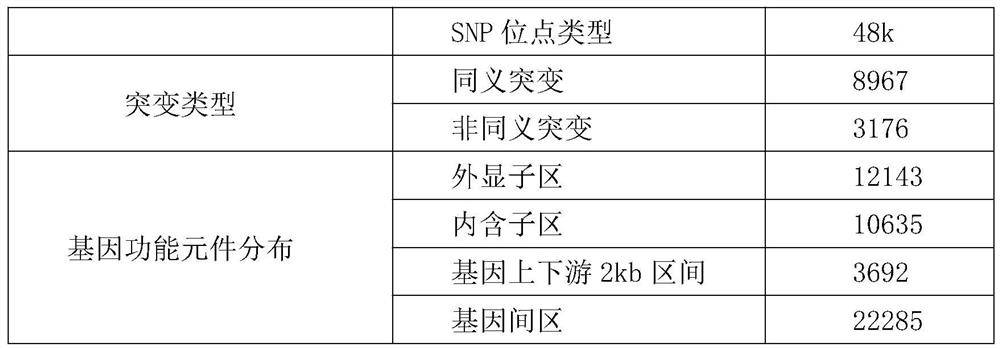
[0153] Screen the loci with a population typing rate greater than 90% and a minimum allele frequency greater than 0.05, and obtain 46,232 genotype information of 46,232 high-quality loci, and use the typing data to perform individual cluster analysis of A. japonicus, such as figure 1 It shows that the positio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com