Universal metabarcoding amplification primers for freshwater fish mitochondria 12S and application method thereof
A technology of macro-barcode and amplification primers, which is applied in the field of 12S universal macro-barcode amplification primers for freshwater fish mitochondria, which can solve the problems of low DNA concentration in free fish, difficult sequencing of primer amplification products, and poor amplification ability of fish communities problems such as low degeneracy, strong amplification preference, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] (1) List of common primer pairs for fish mitochondrial 12S gene metabarcode
[0052] Table 1 The size of the fragments amplified by the combination of upstream and downstream primers
[0053]
Embodiment 2
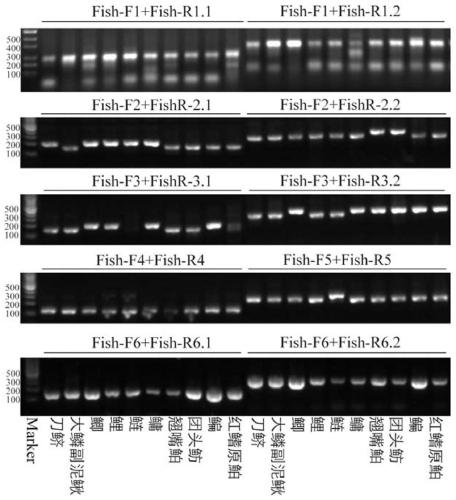
[0055] The fish samples were collected in Zhushan Bay (120.036° east longitude, 31.373° north latitude) and Miaogang (120.461° east longitude, 31.002° north latitude) in Taihu Lake, Jiangsu Province. Ground cages and gillnets were used to catch fish on site, and the collected fish samples were taken to the laboratory for species identification and separation. A total of 10 species of fish were collected (red-finned bream, bream, bream, bream, bighead carp, silver carp, common carp, crucian carp, large scale loach, sword anchovy), and the DNA of each fish sample was extracted separately. Fish-F1 and Fish-R1.1 (Fish-R1.2) primer combination, Fish-F2 and Fish-R2.1 (Fish-R2.2) primer combination, Fish-F3 and Fish-R3.1 ( Fish-R3.2) primer combination, Fish-F4 and Fish-R4 primer combination, Fish-F5 and Fish-R5 primer combination, Fish-F6 and Fish-R6.1 (Fish-R6.2) primer pair extraction For PCR amplification of fish DNA, the molar ratio of the combination of upstream primers and do...
Embodiment 3
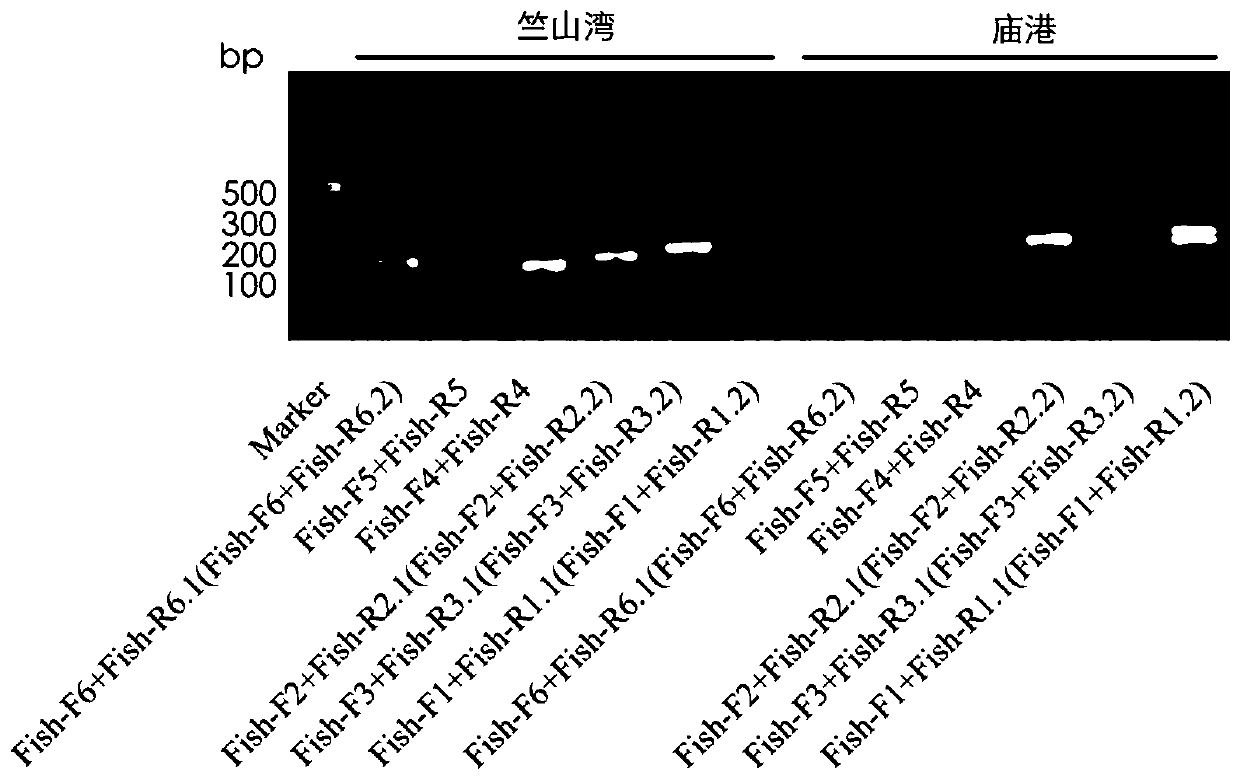
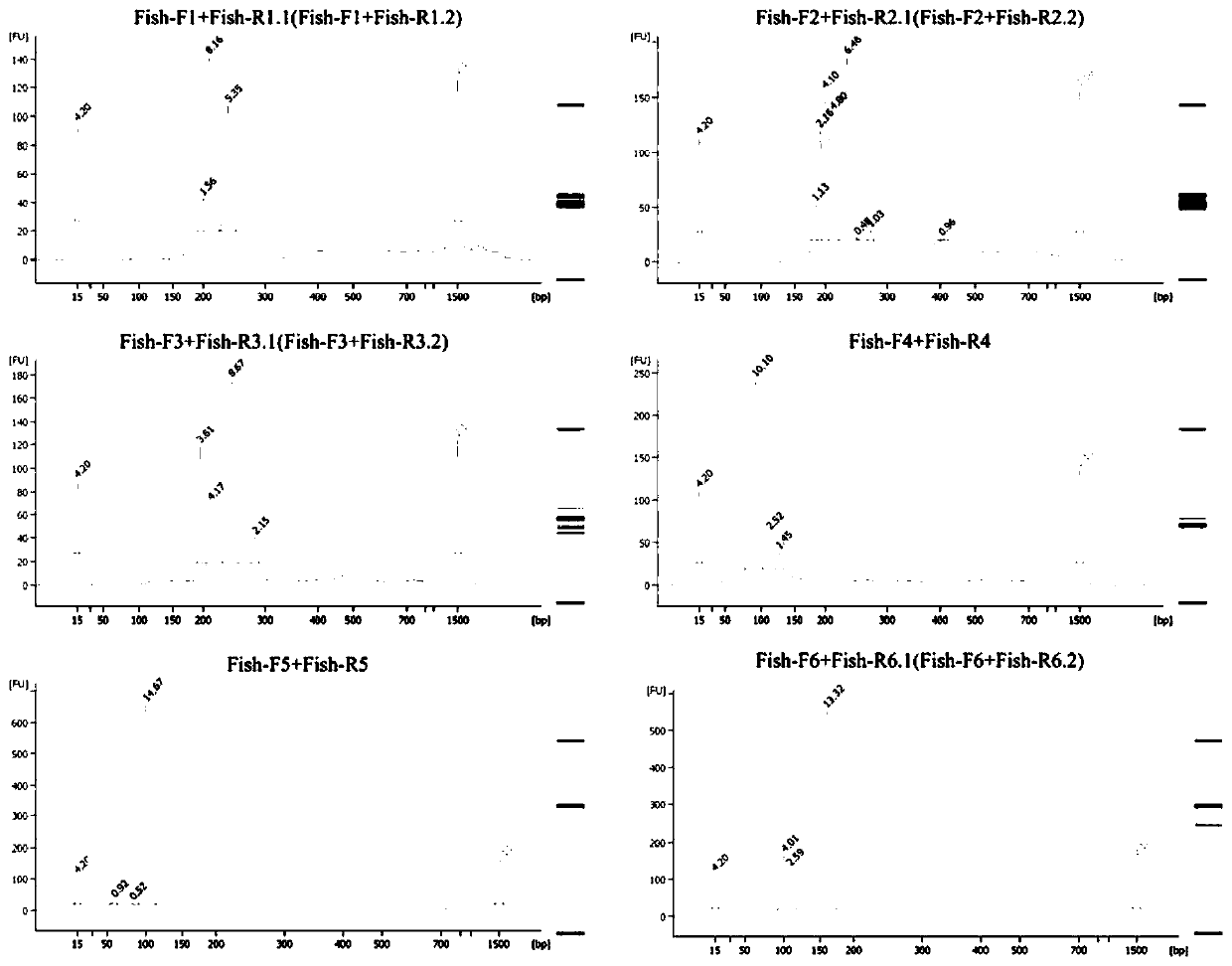
[0059] In Zhushan Bay (120.036° east longitude, 31.373° north latitude) and Miaogang (120.461° east longitude, 31.002° north latitude), 0.5-1L of surface water was collected with a 1L sampling bottle, and the glass fiber filter with a pore size of 0.22μm was used in the laboratory. Membrane filtration to remove water, according to The Water DNA Kit (OMEGA, CHINA) standard operation was used to extract the environmental DNA retained in the filter membrane, and PCR amplification was performed on the filter membrane DNA using the mitochondrial 12S primers and reagents in Example 2. PCR amplification products were detected by 2% agarose electrophoresis ( figure 2 shown). The PCR product was recovered by tapping the gel through the gel recovery kit (MinElute Gel Extraction Kit), using Qubit TM dsDNA HS Assay Kits were used for accurate quantification, and the purified PCR products were analyzed for fragment size using Bioanalyser 2100 (Agilent Technologies, USA). The analysis ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com