Flax SSR molecular marker and application thereof
A molecular marker and molecular marker-assisted technology, which is applied in the direction of DNA/RNA fragments, microbial measurement/inspection, recombinant DNA technology, etc., can solve the problems of SSR molecular marker development that have not been reported, and achieve stable amplification and band clear effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0113] Example 1 The acquisition of flax genome-wide SSR loci
[0114] First, NCBI downloaded the whole genome sequence information of flax
[0115] ( https: / / www.ncbi.nlm.nih.gov / assembly / GCA_000224295.2 ), further use MISA software (MIcroSAtellite identification tool)
[0116] (http: / / pgrc.ipk-gatersleben.de / misa / ) Detection of microsatellite and composite microsatellite loci in the flax genome sequence. The detection parameters adopted are 2-6, 3-5, 4-5, 5-5, 6-5 (the number of repetitions of the 2-base repeating unit motif>=6 times; the number of repetitions of the 3-base repeating unit>=5 ; and so on.); when the distance between two microsatellites was less than 100bp, a composite microsatellite was formed, and 28571 SSR loci were detected in the whole flax genome.
Embodiment 2
[0117] Embodiment 2 flax SSR primer design and synthesis
[0118] According to the SSR detection results of MISA, using Primer3
[0119] (http: / / pgrc.ipk-gatersleben.de / misa / primer3.html) software default parameters (length 100bp-300bp; Tm value 55°C-60°C, primer length 20-25bp) for primer design, try to avoid primer two Polymer (dimer), hairpin structure (hairp-in), mismatch (false primer), etc. In the design results, the 3 pairs of primers with the highest scores were selected for subsequent experiments, and a total of 71184 pairs of SSR primers were obtained. Then 96 pairs of primers were randomly selected and synthesized by Hunan Qingke Biological Company.
Embodiment 3
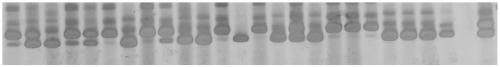
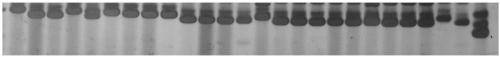
[0120] The amplification of embodiment 3 SSR primers and the genetic diversity analysis of 26 flax germplasm resources
[0121] The TPS method was used to extract 26 flax germplasms (see Table 1), and the specific method can be referred to (Zhang Youchang et al., 2016). Then use DNA to measure the DNA concentration of each germplasm, and finally dilute to the working solution 30ng / μL. The SSR reaction system was 10 μL, which included 50 μM dNTPs, 0.2 μM primers, 0.5U Taq polymerase (TaKaRa, Dalian) and 30ng of DNA template. The PCR reaction was carried out in a LongGeneA200 gene thermal cycler. The overall reaction system includes: PCR reaction is carried out with rTaq DNA polymerase from Takara Company, and the reaction system is as follows (10 μL): DNA template (10ng / μL) 2.0 μL, primer (2 pmol / μL) 1 μL, 10×Buffer 1.0 μL, dNTP ( 1 mM) 0.3 μL, rTaq (5U / μL) 0.1 μL, ddH2O 5.4 μL. The PCR amplification program includes: specific procedures are as follows: pre-denaturation at 9...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com