Construction method and application of genome-wide methylation high-throughput sequencing library and
A whole-genome and sequencing library technology, applied in the field of constructing whole-genome methylation high-throughput sequencing libraries and kits for constructing whole-genome methylation high-throughput sequencing libraries, can solve problems that need to be improved
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
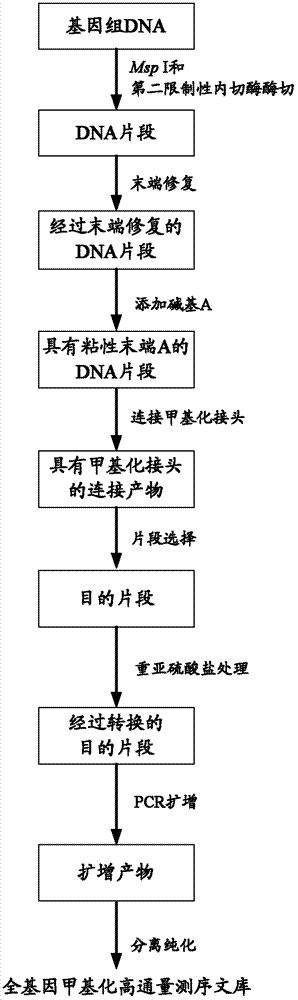
[0055] 1. Experimental process
[0056] 1. Use Msp I and ApeK I to digest genomic DNA:
[0057] 1.1 Msp I enzyme digestion: 100ng human mDC cell line whole genome DNA sample (mDC, mature dendritic cells) was digested with Msp I:
[0058] 1) Prepare the Msp I digestion reaction system in a 1.5ml centrifuge tube:
[0059]
[0060]The reaction system was reacted in a 37°C water bath for 7h. After the reaction was completed, the enzyme digestion reaction system was placed at 80° C. for 20 minutes to inactivate the restriction endonuclease Msp I.
[0061] 1.2 ApeK I digestion:
[0062] 1) Add 5 μL of ApeK I (NEB) (4,000 units / ml) directly to the restriction endonuclease-inactivated Msp I digestion reaction system
[0063] 2) The reaction system was subjected to enzyme digestion overnight (16-19h) in a water bath at 75°C.
[0064] 3) Add 1 μL of EDTA (1 mM) to the reaction system to inactivate the restriction endonuclease ApeK I.
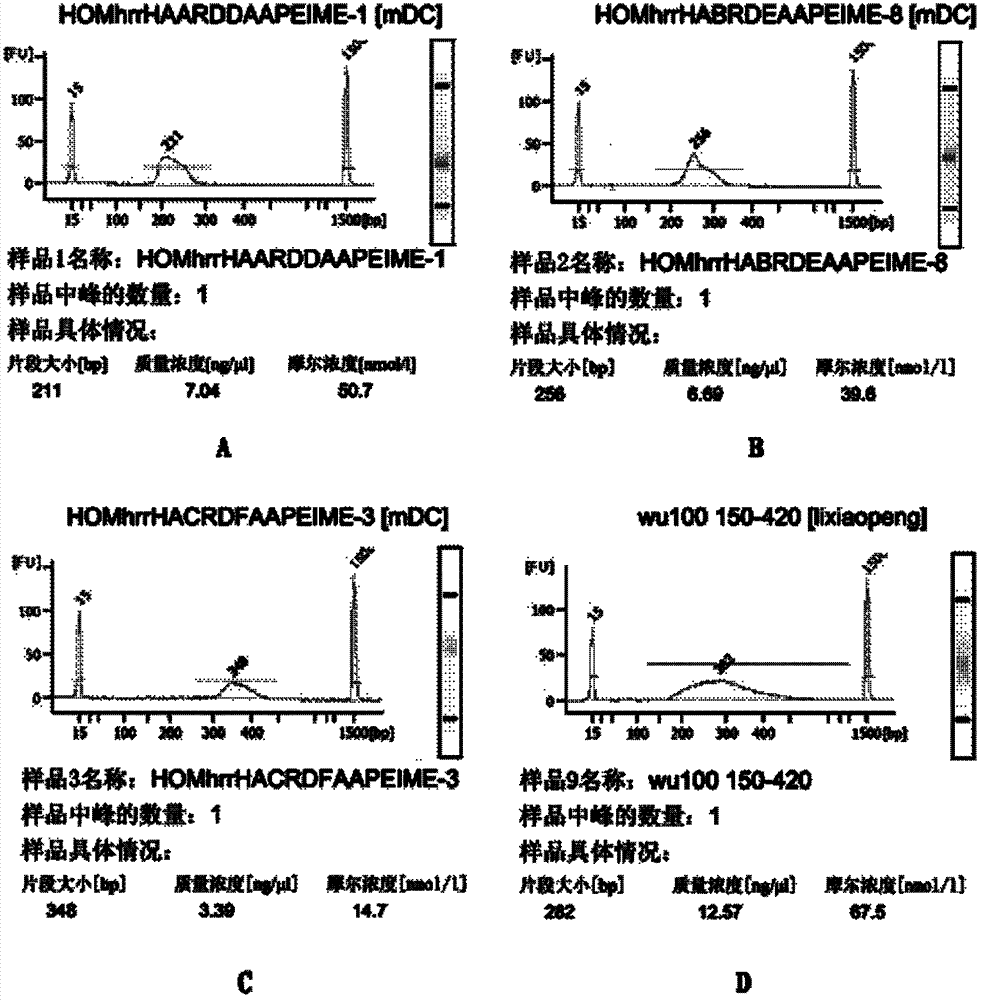
[0065] 4) The DNA in the reaction system w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com