Method for detecting diversity influence of pinkflower clover on constructed wetland substrate microorganisms and application of method
A technology of artificial wetlands and detection methods, which is applied in the measurement/inspection of microorganisms, biochemical equipment and methods, biostatistics, etc., and can solve problems such as high construction costs, small hydraulic loads, and weak processing capabilities
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] 1 Materials and methods
[0034] 1.1 Sample Preparation and Collection
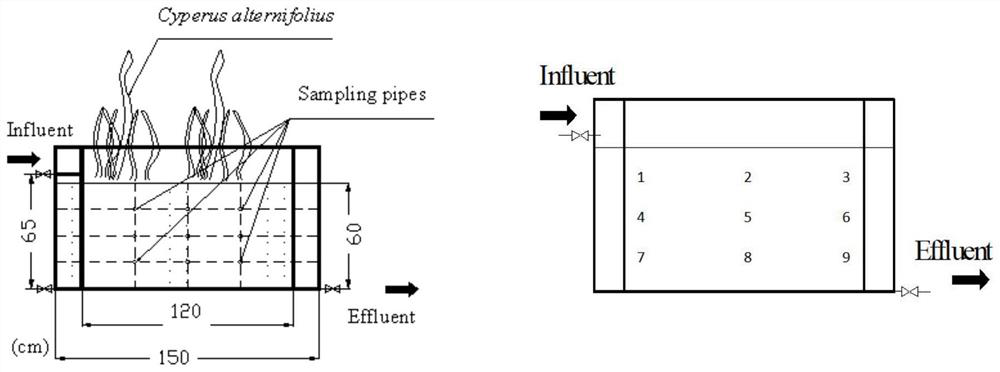
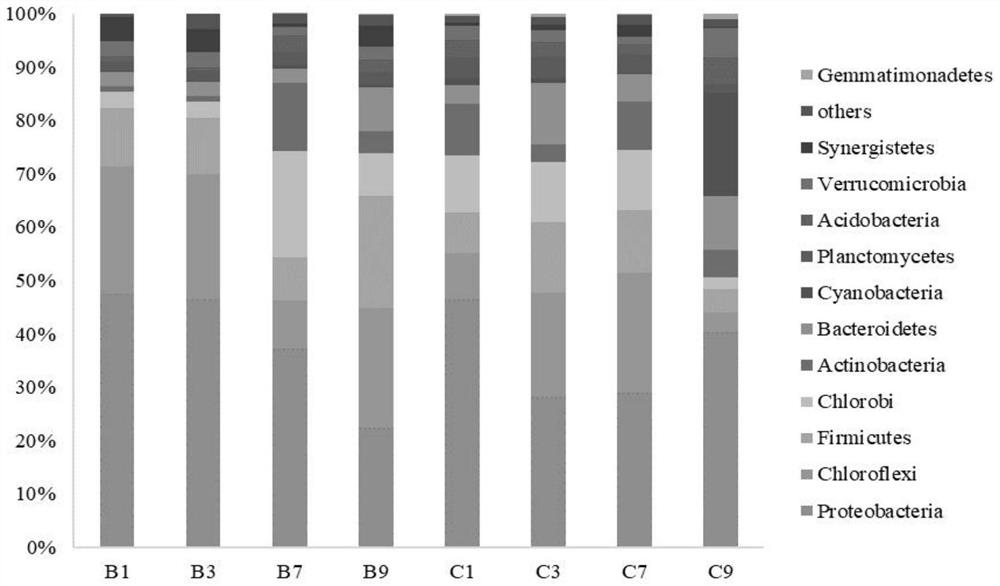
[0035] Artificial horizontal subsurface wetlands are constructed of PVC boards and filled with gravel (diameter 4-6mm) to form a filler matrix, the size of which is as follows figure 1 shown. A perforated tube is installed on the side of the device to facilitate sampling, and the collected sample is a gravel surface matrix. The planting density of windmill grass is about 25 plants per square meter, and the water flow direction is from left to right. Four areas 1, 3, 7, and 9 were selected as the sampling sites for this experiment. At the same time, groups were formed according to whether the windmill grass was planted or not. The unplanted windmill grass group was named group B: B1, B3, B7, and B9. The group of planting windmill grass was named as C group: C1, C3, C7, C9.
[0036] 1.2 Detection process
[0037] Take 1 g of sample and use a DNA extraction kit (OMEGA Soil DNA Kit) according t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com