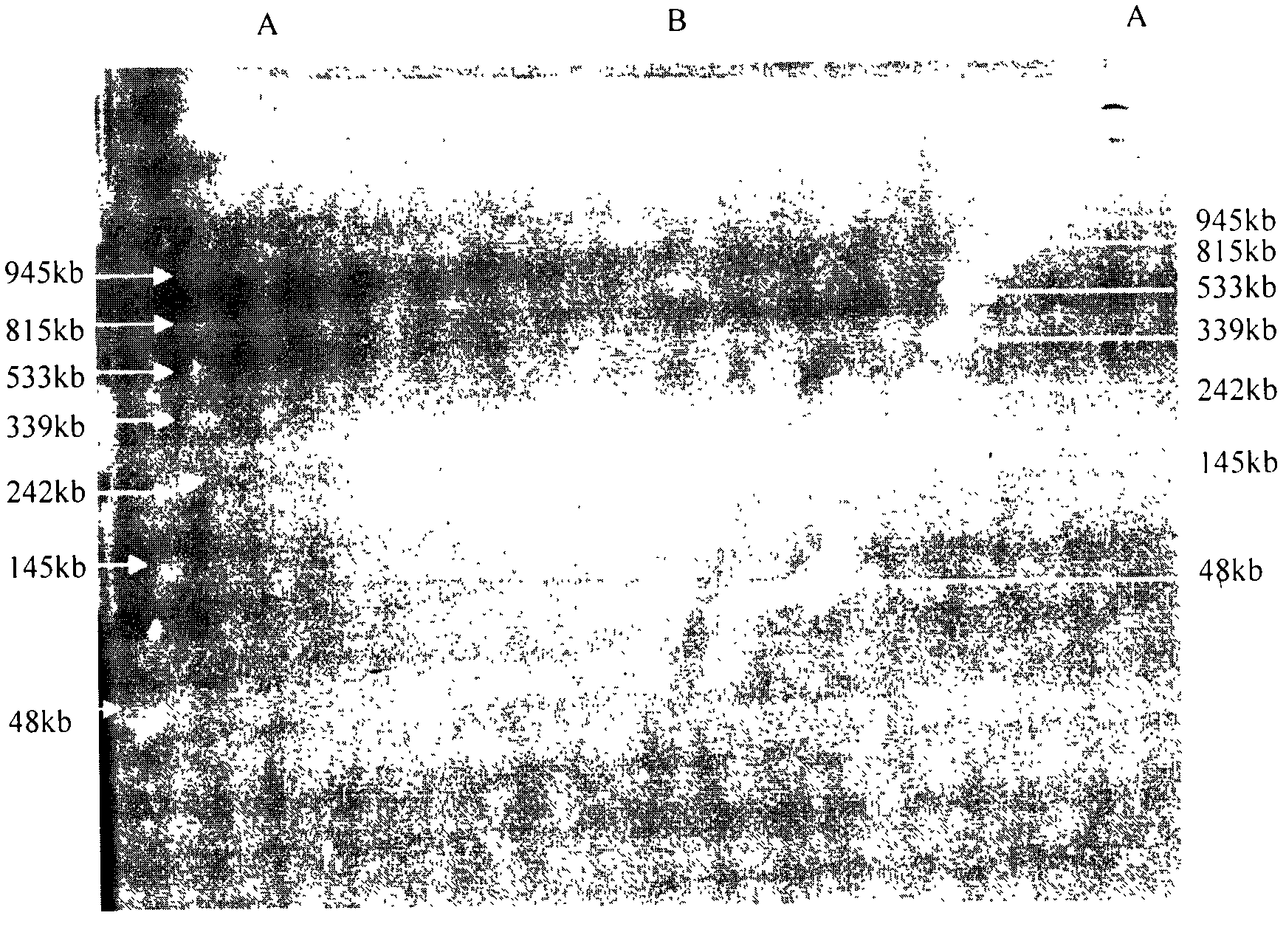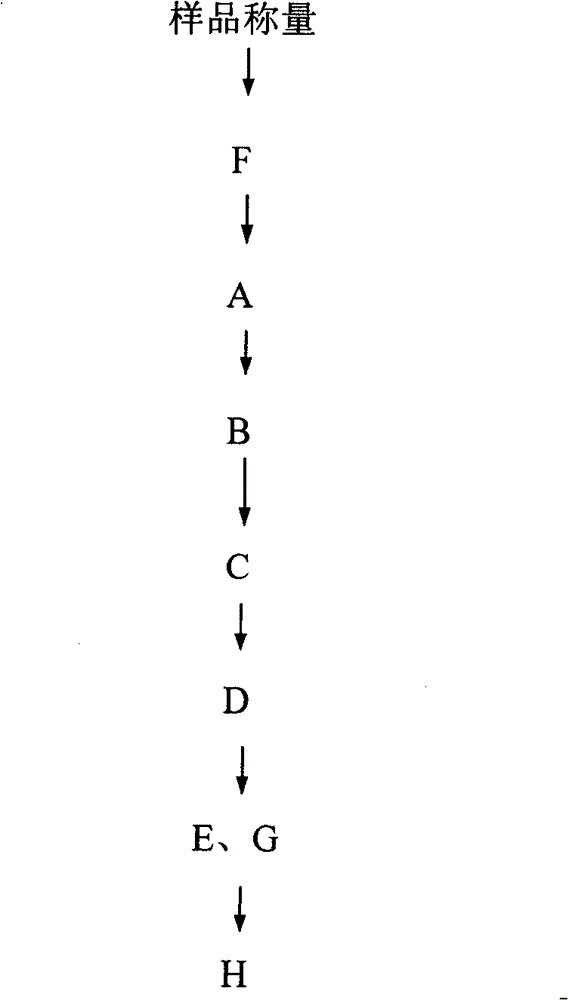Patents
Literature
30 results about "Soil dna" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
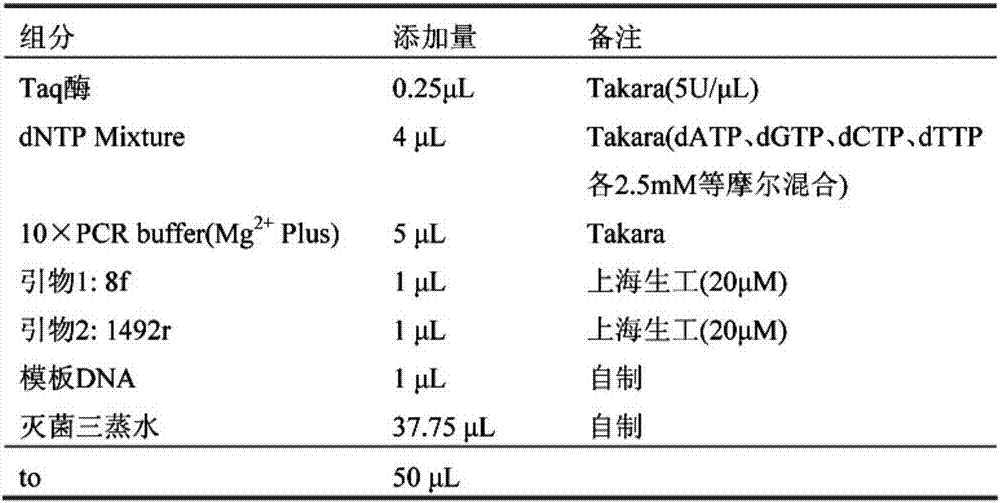
NucleoSpin Soil can process up to 500 mg (wet weight) of starting material. Depending on the individual sample, typical yields are in the range of 2 to 10 µg of DNA. The eluted DNA is ready to use (undiluted) for subsequent reactions such as PCR, restriction analysis, etc. Cat.
Small quality fast extraction method for soil total DNA
InactiveCN1990863AEasy to operateMicrobiological testing/measurementDNA preparationHigh concentrationSoil microbiology
The invention discloses a method for fast extracting the total soil DNA. It takes soil as raw material, shaking with quartz sand and silicon dioxide powder, cracking with cetyltrimethylammonium bromide for DNA releasment, the released DNA can be absorbed by diatomite under weak acid or netural pH condition and high concentration thiocyanate ion, and the DNA will be eluted by low salty buffer solution or deionized water under weak alkaline condition. The extracted DNA with enough purity and amount can be used for PCR and endonuclease reaction. The main steps comprise cell cracking, DNA extraction and DNA purification. The invention is characterized by the simple, fast and easy method, and high purity and low cost extracted DNA. The extracted DNA can be used for kit production. The soil total DNA can be extracted in half hour for microbiological and molecular biological research.
Owner:HUAZHONG AGRI UNIV
Method for analyzing generating trend of aflatoxin B1 in peanut meal by multiple PCR (Polymerase Chain Reaction) technology
InactiveCN103103258APrimer sequence specificity is highImprove reliabilityMicrobiological testing/measurementMicroorganism based processesMicroorganismEnzyme Gene
The invention discloses a method for analyzing generating trend of aflatoxin B1 in peanut meal by a multiple PCR (Polymerase Chain Reaction) technology. The method belongs to the field of microbial and molecular biology and comprises the following steps of: extracting the genome DNA (Deoxyribonucleic Acid) of peanut meal through a soil DNA extraction kit; synthesizing a metabolic pathway through aflatoxin B1 to design a key enzyme gene design primer sequence; performing PCR identification for potential producing strain of aflatoxin; and guiding storage and conveyance condition control through the PCR result. The method further provides a scientific ground for dynamic change and control of aflatoxin B1 in peanut meal and has the advantages of fast speed and high reliability.
Owner:SHANDONG LUHUA GROUP +1
Quantitative detection method for FOC race 4 from soil
InactiveCN103757093AHigh sensitivityStrong specificityMicrobiological testing/measurementFusarium oxysporumNucleic acid detection
Owner:ENVIRONMENT & PLANT PROTECTION INST CHINESE ACADEMY OF TROPICAL AGRI SCI
Method for separation and purification of large-fragment DNA from soil
InactiveCN103103180AEasy to operateOperation steps are adjustableDNA preparationMicrobe DNABiological studies
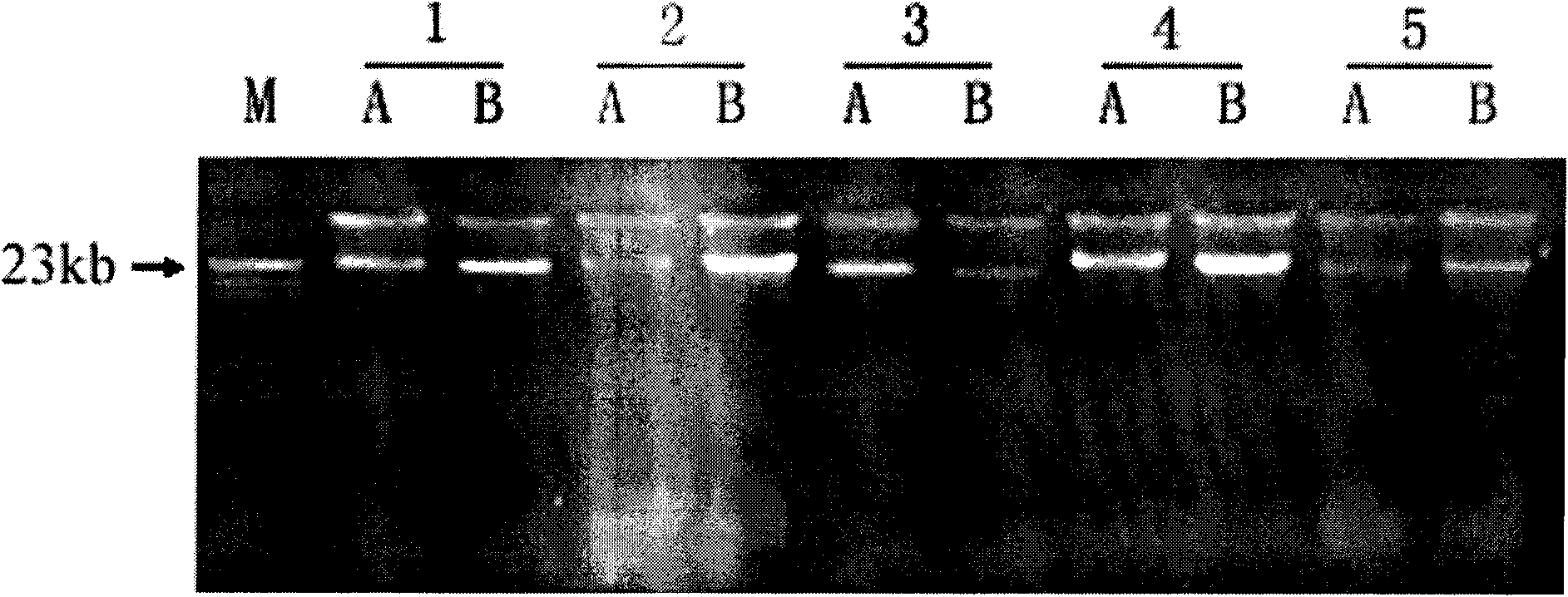
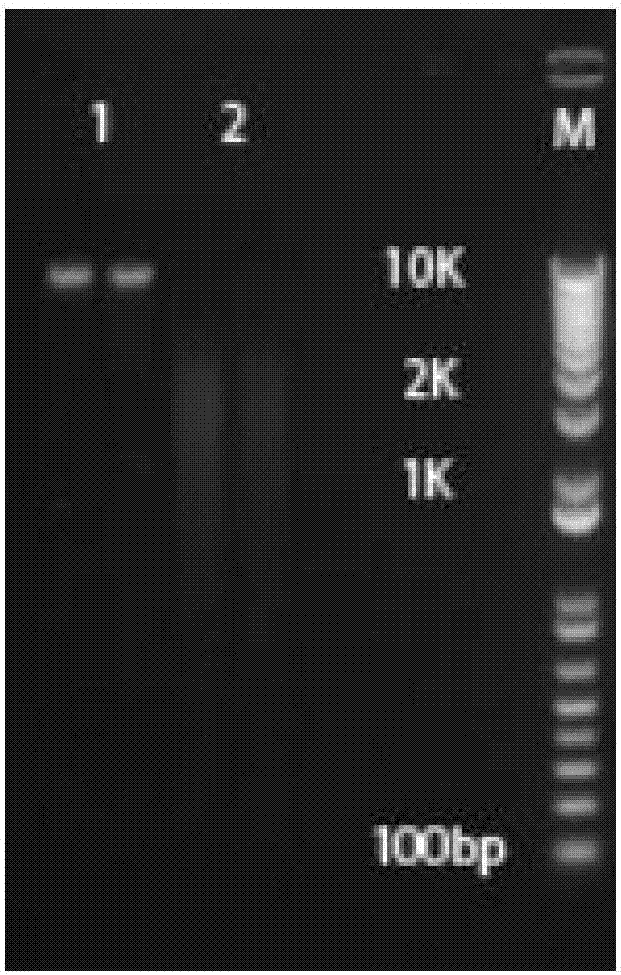
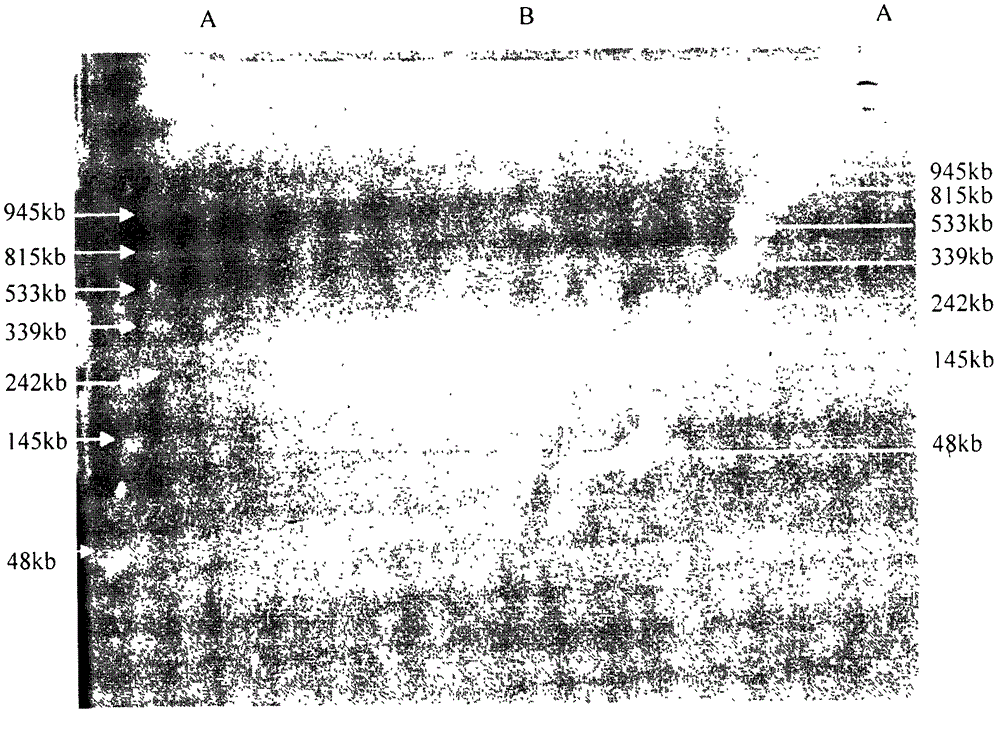
The invention belongs to the fields of soil microorganisms, biochemistry and molecular biology and relates to a method for separation and purification of a large-fragment DNA from soil. The method is used for indirect separation and purification of a large-fragment DNA having molecular weight more than 30kb from various types of soil, wherein the large-fragment DNA is used for construction of a metagenomic library, or is used for separation of soil microbial gene clusters. The method solves the problem that separation of microbial cells from soil and preparation of large-fragment soil DNA having high purity and satisfying various biological study demands are realized difficultly by the existing indirect method, and is an efficient method for extraction of soil microorganism DNA having a large fragment and high purity.
Owner:XINJIANG NORMAL UNIVERSITY
Soil DNA extracting method for evaluating diversity of microbial community of plant root system
InactiveCN101709298AEasy to operateLow costMicrobiological testing/measurementDNA preparationPlant rootsDna recovery
The invention discloses a soil DNA extracting method for evaluating diversity of a microbial community of a plant root system. In the method, pretreatment is carried out on a sample before cell splitting, extracellular DNA and humic substances are removed, and the problems of difficult removal of the humic substances and low DNA recovery rate existing in a purification step are solved. In the extraction process of the method, phenol or chloroform is not used so as to reduce harm on the health of experimenters, obtain complete DNA and molecular fragments greater than 10kb and achieve high yield. OD260 / OD230 and OD260 / OD280 of the extracted soil microorganism DNA are close to standard values and can be directly applied to molecular operation so as to evaluate the diversity of the microbial community of the plant root system.
Owner:SHANGHAI ACAD OF AGRI SCI
High-throughput absolute quantification method for soil bacteria
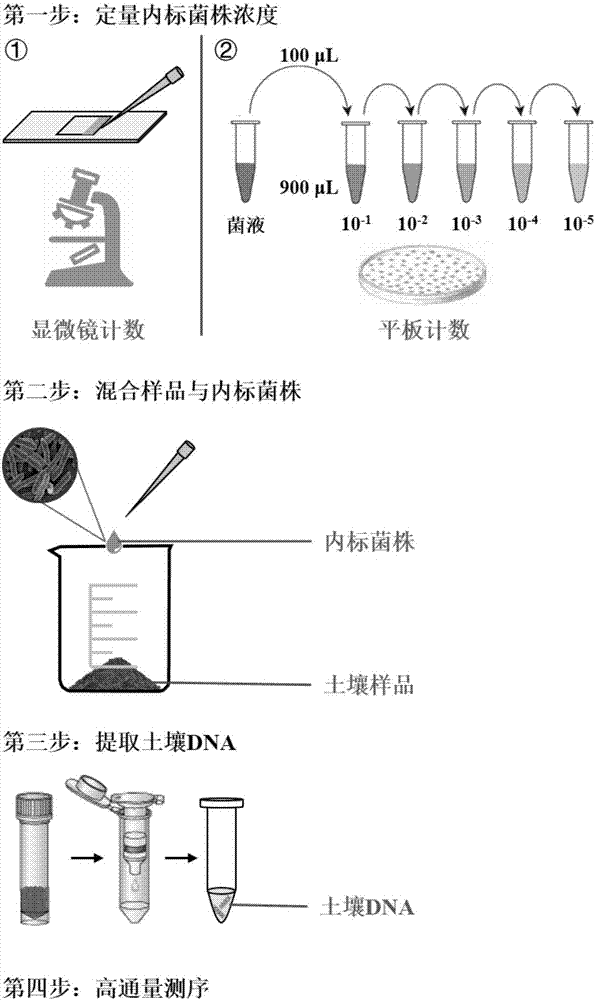
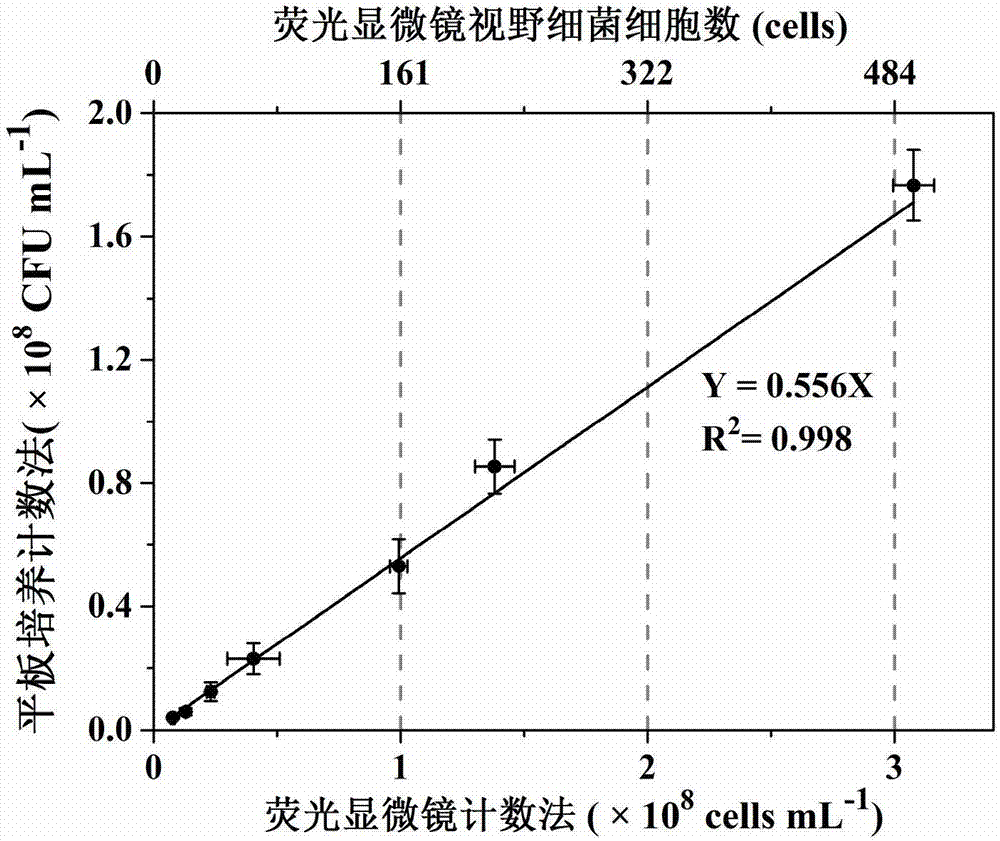
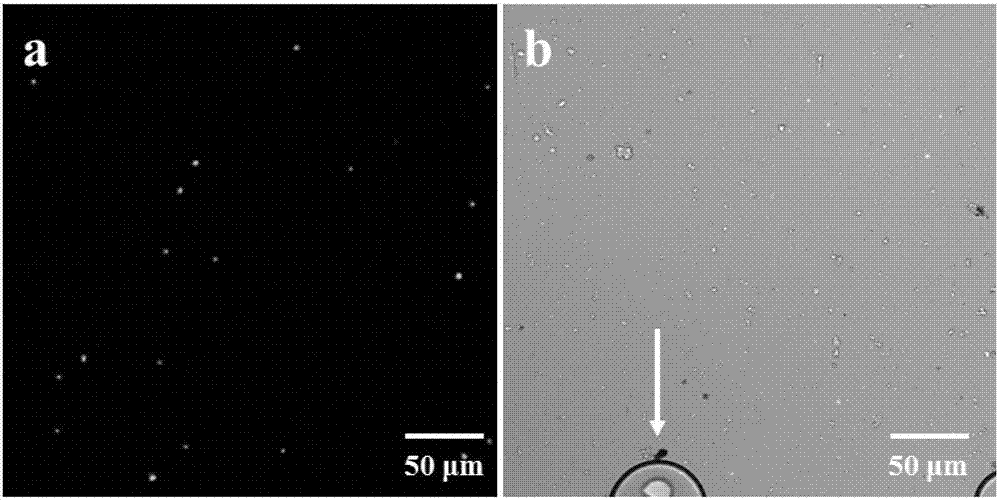
ActiveCN107190055AAchieve high-throughput absolute quantificationSimple methodMicrobiological testing/measurementInternal standardCommunity structure
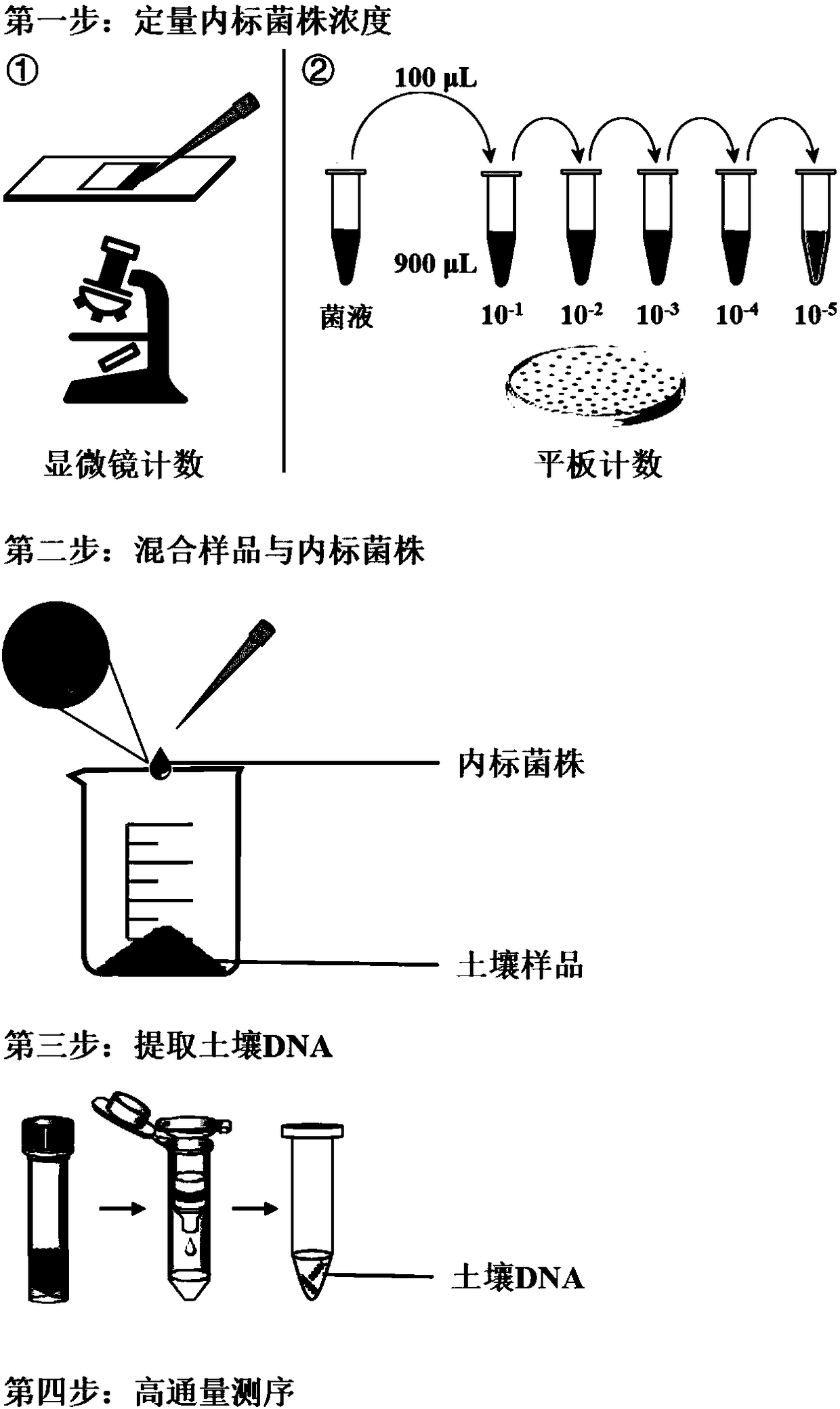
The invention discloses a high-throughput absolute quantification method for soil bacteria. The high-throughput absolute quantification method includes steps of whirling and re-suspending E. coli HTAQ-GFP thalli in sterile water and regulating bacteria suspension in ultraviolet spectrophotometers OD <600nm=1.0>; 2), absolutely quantifying internal standard strains HTAQ-GFP and acquiring the absolute content of internal standard strains E. coli HTAQ-GFP in bacteria solution; 3), adding the bacteria solution with the internal standard strains HTAQ-GFP to soil samples and uniformly stirring and mixing the bacteria solution and the soil samples with one another; 4), extracting soil DNA (deoxyribonucleic acid) and carrying out high-throughput sequencing on amplicons of 16S rRNA [16S ribosomal RNA (ribonucleic acid)] genes to obtain classification composition information of bacterial community structures and corresponding relative abundance; 5), acquiring the total quantities of native bacteria in soil and the absolute contents of various classification units according to the absolute contents of the internal standard strains HTAQ-GFP and the relative abundance of Escherichia in high-throughput sequencing results.
Owner:ZHEJIANG UNIV
Simple, efficient and cheap method for purifying forest soil sample DNA
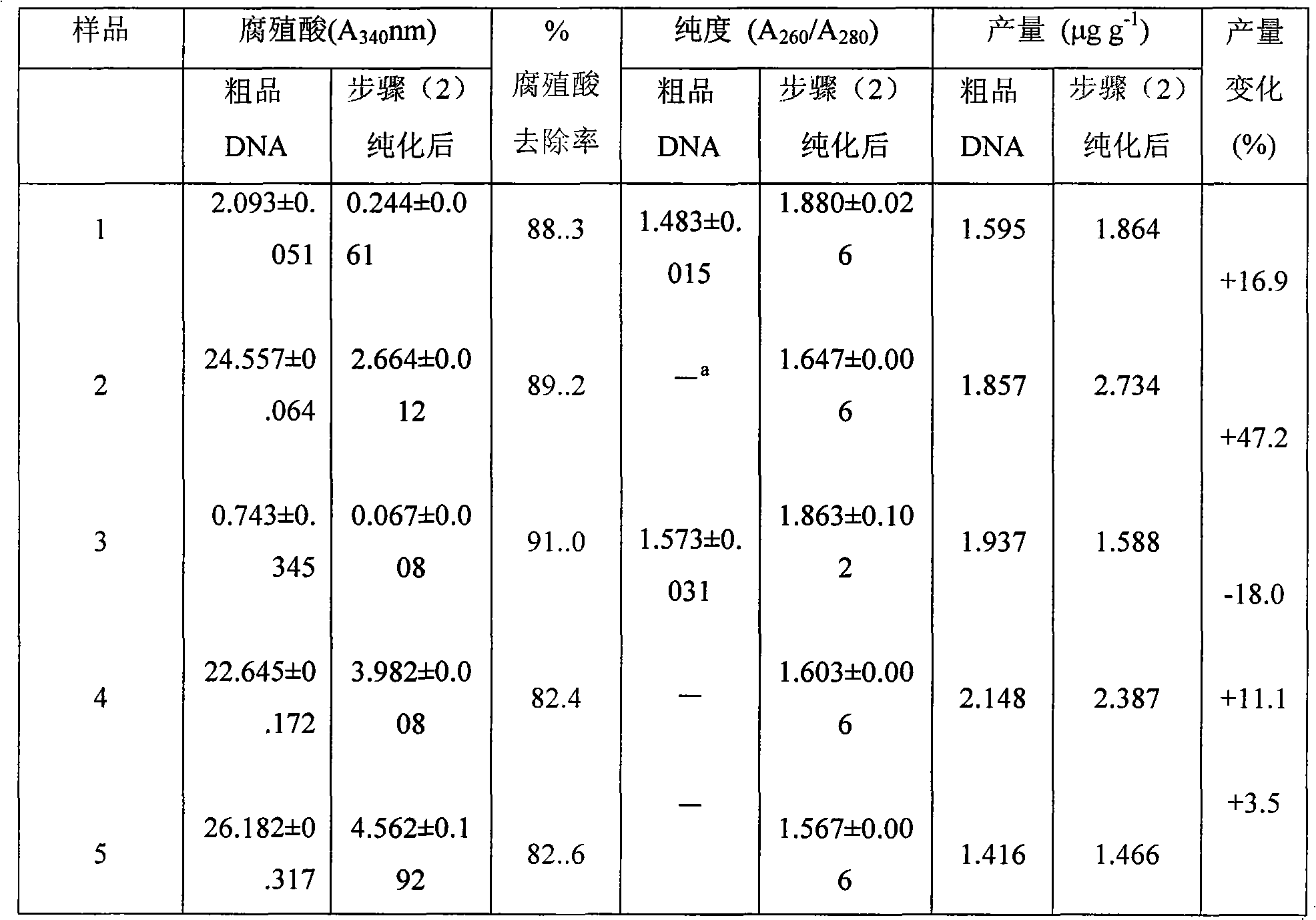
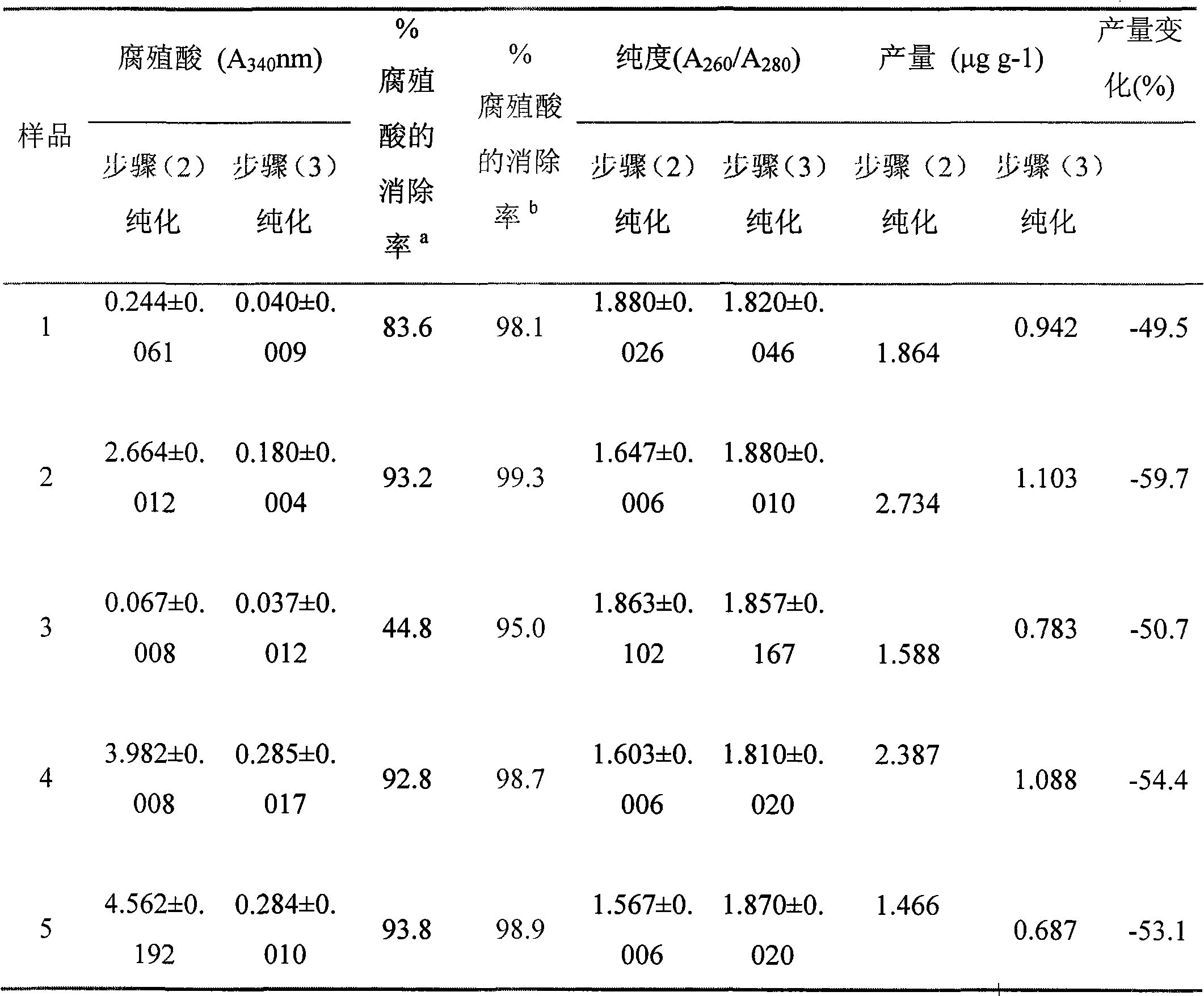
The invention provides a method for purifying microorganism DNA in forest soil, which comprises the following steps: (1) obtaining the rough DNA of a microorganism by a conventional method; (2) dissolving the rough DNA by extracted buffer solution; after the rough DNA is subjected to water bath and isovolumetric chloroform-isoamyl alcohol solution extraction, adding isopropanol which is 0.6 time of the volume of supernatant into the supernatant to precipitate DNA sediments; washing by 70 percent of pre-cooled alcohol and dissolving in the TE buffer solution; and (3) further purifying the initially purified DNA by a silica gel column. The method can remove most of impurities in a rough product of the extracted soil microorganism DNA and obtain the DNA with high purity and can be directly used for the conventional polymerase chain reaction (PCR) which is quite sensitive for an inhabiting matter. The invention has simple operation, low cost, less time consumption and wide application range and can be used for purifying the soil DNA in various types after other prior soil direct extraction methods.
Owner:JISHOU UNIVERSITY
Method for extracting microbial total DNA from soil being low in biomass and enriched in humus
InactiveCN107475249AGuaranteed fidelityIncrease productionMicrobiological testing/measurementDNA preparationWater bathsA-DNA
The invention discloses a method for extracting microbial total DNA from soil being low in biomass and enriched in humus. The method disclosed by the invention comprises the following steps: (1) grinding a soil sample which is low in biomass and enriched in humus by liquid nitrogen so as to split microbial cells in the sample; (2) further splitting the microbial cells in water baths with different temperatures by using a DNA extraction buffer solution, protease K and 20% of SDS, carrying out degeneration on proteins, and centrifuging; (3) precipitating DNA through isopropanol in a low-temperature state so as to obtain a crude DNA product; and (4) purifying the crude DNA product obtained in the step (3) by using a QIAGEN soil DNA extraction kit, thereby obtaining the microbial total DNA, wherein the pH value of the DNA extraction buffer solution is 8.0, and the DNA extraction buffer solution comprises 0.1M of NaH2PO4, 0.1M of Na2HPO4, 0.1M of EDTA (Ethylene Diamine Tetraacetic Acid), 0.1M of Tris-HCl, 1.5M of NaCl and 1% of CTAB (Cetyltrimethyl Ammonium Bromide); and the concentration of the protease K is 10mg / mL. According to the method disclosed by the invention, high-efficiency high-mass soil DNA extraction can be performed, and the extracted DNA is high in purity and excellent in integrity and has excellent application prospects.
Owner:广东美格基因科技有限公司
Soil DNA sample preparation kit and preparation method thereof
InactiveCN101235350AEasy to useThe result is stableBioreactor/fermenter combinationsBiological substance pretreatmentsChain typeReagent bottle
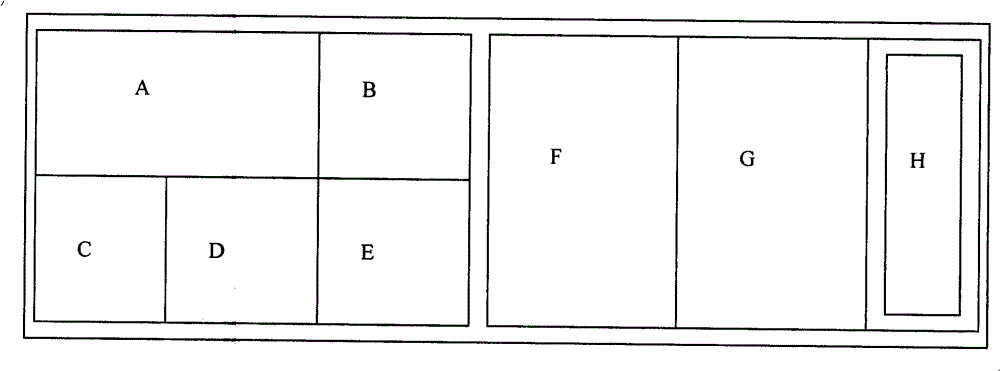
The invention discloses a soil DNA sample preparation reagent box and a manufacturing method, the reagent box comprises a box body and a baffle board, a reagent bottle A which is filled with solution I, a reagent bottle B which is filled with solution II, a reagent bottle C which is filled with solution III, a reagent bottle D which is filled with solution IV, a reagent bottle E which is filled with suspension liquid, a centrifuge tube F which is filled with acid cleaning glass bead, a mini filter G and a PCR(chain type polyase reaction) pipe H. A reagent box which is designed through the technical proposal of the invention has the advantages of simple employment and stable result, which can effectively save time and cost of users.
Owner:SHANGHAI ACADEMY OF ENVIRONMENTAL SCIENCES
Method for extracting DNA in soil
InactiveCN107228787AReduce lossesReduce physical injuryPreparing sample for investigationPhosphateElectrophoresis
The invention discloses a method for extracting DNA in soil. The method comprises the following steps: lysing a soil sample so as to obtain lysate by adopting an SDS (Sodium Sulfate)-lysozyme method; uniformly mixing the obtained lysate and 5*SDS Loading Buffera according to a ratio, adding the mixture into the obtained albumin glue comb holes; performing electrophoresis according to the conventional method, slightly taking out the albumin glue comb for staining after electrophoresis is ended; cutting off a silver target strip after the silver target strip appears, putting the target strip into a dialysis bag filled with 2 mL of PBS (Phosphate Buffer Solution), placing in a horizontal electrophoresis tank filled with a proper amount of 1*Tris-Gly electrophoretic buffer solution, and placing the horizontal electrophoresis tank on ice for performing electrophoresis; and placing the obtained dialysis bag in a small beaker filled with sterilized PBS, dialyzing at the temperature of 4 DEG C overnight, and collecting the supernatant in the dialysis bag on the next day, thereby obtaining the soil DNA extract. According to the method disclosed by the invention, the operating steps are simple, the DNA lost in the operation is less, and the obtained DNA amount is stable.
Owner:GANSU UNIV OF CHINESE MEDICINE
Method for preparing DNA Marker in nested PCR-DGGE (Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis) detection of AMF (Arbuscular Mycorrhizal Fungi) community diversity
InactiveCN102146370AShorten experiment timeDNA preparationArbuscular mycorrhizal fungiComputational biology
The invention discloses a method for preparing a DNA (Deoxyribonucleic Acid) Marker in a nested PCR-DGGE (Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis) detection of AMF (Arbuscular Mycorrhizal Fungi) community diversity. A technical scheme disclosed by the invention comprises the steps of extracting a soil DNA, carrying out DGGE analysis, cloning and transforming, verifying the position of a strip on a DGGE photographic film, making a nested PCR-DGGE Marker of AMF community diversity, carrying out PCR amplification by using NS31-GC and GLoI (Glyoxalase I) as primers, analyzing a result in combination with sequences and naming for use. The invention has an important application value in a Nested PCR-DGGE study of an AMF community structure, is beneficial to reduction of the experiment time, and is especially suitable for experiment process with large experiment sample quantity. According to the invention, a series of DNA Markers for studying the AMF community can be developed so that the nested PCR-DGGE technology is more suitable, faster and more accurate in the study on an AMF molecular community.
Owner:NORTHWEST A & F UNIV
A kind of soil dna extraction method
InactiveCN107228787BReduce lossesReduce physical injuryPreparing sample for investigationPhosphateElectrophoresis
Owner:GANSU UNIV OF CHINESE MEDICINE
Method for extracting DNA of cyst parasites of heterodera glycines
InactiveCN105154433AExtraction is fast, accurate and efficientDiversity understandingDNA preparationCystCommunity structure
The invention relates to a method for extracting DNA of cyst parasites of heterodera glycines and belongs to the nucleic acid extraction technique. According to the method, the problems that the community structure and the diversity information related to the parasites cannot be accurately provided because of loss in the current extraction process of DNA of the cyst parasites of heterodera glycines, the consumption of cysts is high, and the extraction time is long are mainly solved. The method comprises the following steps: (1) preparing fresh cysts of heterodera glycines, carrying out surface sterilization, and soaking by virtue of deionized water, so as to obtain a cyst sample; and (2) extracting DNA of the cyst sample by virtue of a soil DNA extraction kit. The extraction method has the beneficial effects that the consumption of the cysts is low, the steps are simple, the operation is easy, the use is convenient, the DNA is rapidly, accurately and efficiently extracted, and the extraction method is convenient to popularize and apply; the extracted DNA can be applied to microbial diversity analysis such as denaturing gel gradient electrophoresis and can be further applied to the extraction of DNA of similar parasitic microbes, so that the diversity and the types of the cyst parasites can be comprehensively understood, and the problem that microorganisms which are difficult to culture cannot be authenticated is solved.
Owner:NORTHEAST INST OF GEOGRAPHY & AGRIECOLOGY C A S
A kind of soil dna extraction kit and the method for extracting soil dna
InactiveCN106011130BEffective crackingEfficient removalMicrobiological testing/measurementDNA preparationBuffer solutionDNA
The invention discloses a soil DNA extraction kit and a soil DNA extraction method, wherein the soil DNA extraction kit is composed of the following six solutions: solution A: a mixed solution of Tris, EDTA, SDS and NaCl and with pH value of 6.4; solution B: 4M of KCl solution; solution C: mixed solution of Tris and GuHCl and with pH value of 6.7; solution D1: a mixed solution of Tris, EDTA and GuHCl and with pH value of 7.7; solution D2: mixed solution of Tris and ethanol and with pH value of 6.7; and solution E: 10mM of Tris-HCl buffer solution and with pH value of 8.0-8.5. The kit can extract high-quality DNA suitable for technical operation in modern molecular biology from soil easily and quickly, and the method has the advantages of simplicity, quick speed, economy, high quality and the like.
Owner:HUAIBEI NORMAL UNIVERSITY
Soil DNA extraction method for analyzing microbial community structure of polycyclic aromatic hydrocarbon polluted land
InactiveCN107254464ASimple extraction methodEasy to operateDNA preparationCommunity structureDna integrity
The invention discloses a soil DNA extraction method for analyzing the microbial community structure of a polycyclic aromatic hydrocarbon polluted land. The method comprises the following specific steps: 1, preparation of a reagent; 2, extraction of a general DNA. The extraction method is simple, convenient to operate, high in applicability and high in safety. For the characteristics of severe polycyclic aromatic hydrocarbon pollution, long-term pollution history, high humus content and the like in soil, firstly, beta-mercaptoethanol with an anti-oxidization effect at a volume ratio of 1.5 percent is added into a soil pretreatment buffer solution to reduce influence of impurities in the soil on DNA extraction; then a relatively mild method for crushing microbial cells through mechanical force of glass beads is adopted, so that the DNA is fully released; furthermore, no high shear force is caused on the DNA, so that the extracted genome DNA is relatively high in intactness.
Owner:BEIJING MUNICIPAL RES INST OF ENVIRONMENT PROTECTION
Soil DNA indirect extraction method for evaluating diversity of plant root system microflora
The invention discloses a soil DNA indirect extraction method for evaluating the diversity of a plant root system microflora. Because the mean density of bacteria is far smaller than the mean density of soil minerals, a sample is pretreated before cell lysis, and microbial cells are separated from the soil sample, thereby avoiding the problems of difficult humus removal and low DNA recovery rate occurring in the purification process. In the extraction process, the invention does not use phenols or chloroform, the damage to the health of an experimenter is reduced, the obtained DNA is complete, a molecular segment is larger than 10kb, and the yield is high. The OD260 / OD230 and the OD260 / OD280 of the extracted soil microbe DNA approach to a standard value, and the soil DNA indirect extraction method can be directly applied to molecular operation so as to evaluate the diversity of the plant root system microflora.
Owner:SHANGHAI ACAD OF AGRI SCI
Method for separation and purification of large-fragment DNA from soil
InactiveCN103103180BEasy to operateOperation steps are adjustableDNA preparationBiological studiesGenomic library
The invention belongs to the fields of soil microorganisms, biochemistry and molecular biology and relates to a method for separation and purification of a large-fragment DNA from soil. The method is used for indirect separation and purification of a large-fragment DNA having molecular weight more than 30kb from various types of soil, wherein the large-fragment DNA is used for construction of a metagenomic library, or is used for separation of soil microbial gene clusters. The method solves the problem that separation of microbial cells from soil and preparation of large-fragment soil DNA having high purity and satisfying various biological study demands are realized difficultly by the existing indirect method, and is an efficient method for extraction of soil microorganism DNA having a large fragment and high purity.
Owner:XINJIANG NORMAL UNIVERSITY
Soil DNA extraction kit and soil DNA extraction method
InactiveCN106011130AEffective crackingEfficient removalMicrobiological testing/measurementDNA preparationBuffer solutionDNA
The invention discloses a soil DNA extraction kit and a soil DNA extraction method, wherein the soil DNA extraction kit is composed of the following six solutions: solution A: a mixed solution of Tris, EDTA, SDS and NaCl and with pH value of 6.4; solution B: 4M of KCl solution; solution C: mixed solution of Tris and GuHCl and with pH value of 6.7; solution D1: a mixed solution of Tris, EDTA and GuHCl and with pH value of 7.7; solution D2: mixed solution of Tris and ethanol and with pH value of 6.7; and solution E: 10mM of Tris-HCl buffer solution and with pH value of 8.0-8.5. The kit can extract high-quality DNA suitable for technical operation in modern molecular biology from soil easily and quickly, and the method has the advantages of simplicity, quick speed, economy, high quality and the like.
Owner:HUAIBEI NORMAL UNIVERSITY
Method for Quantitative Detection of Banana Fusarium wilt Race 4 from Soil
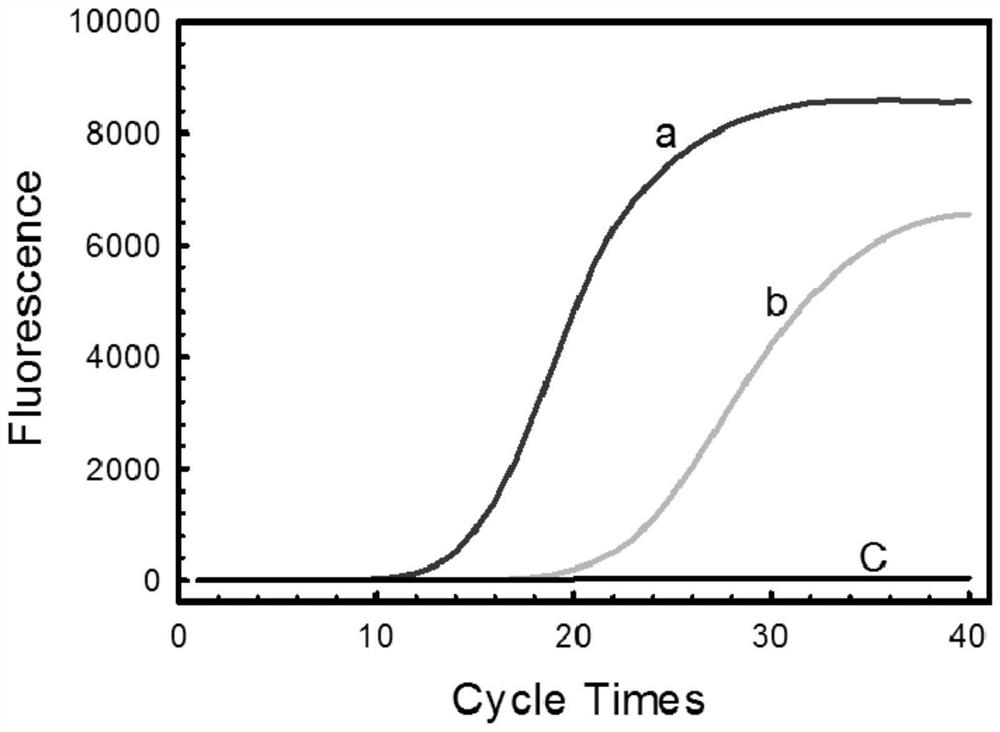
InactiveCN103757093BImprove toleranceCapable of chain displacementMicrobiological testing/measurementLesser floricanFluorescence
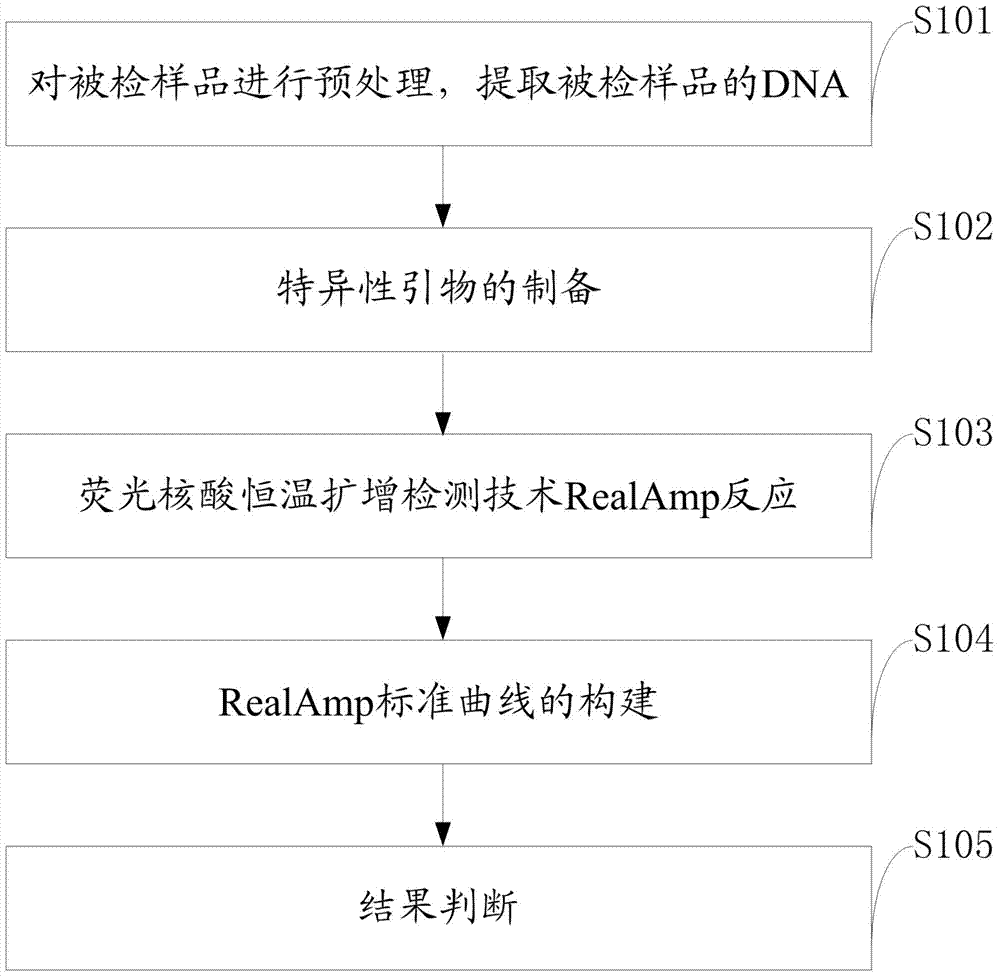
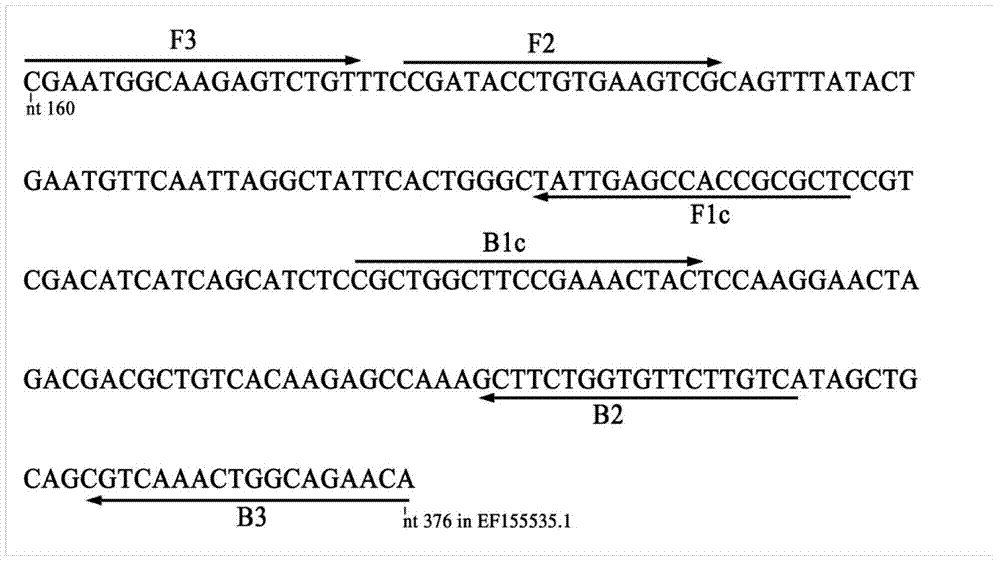
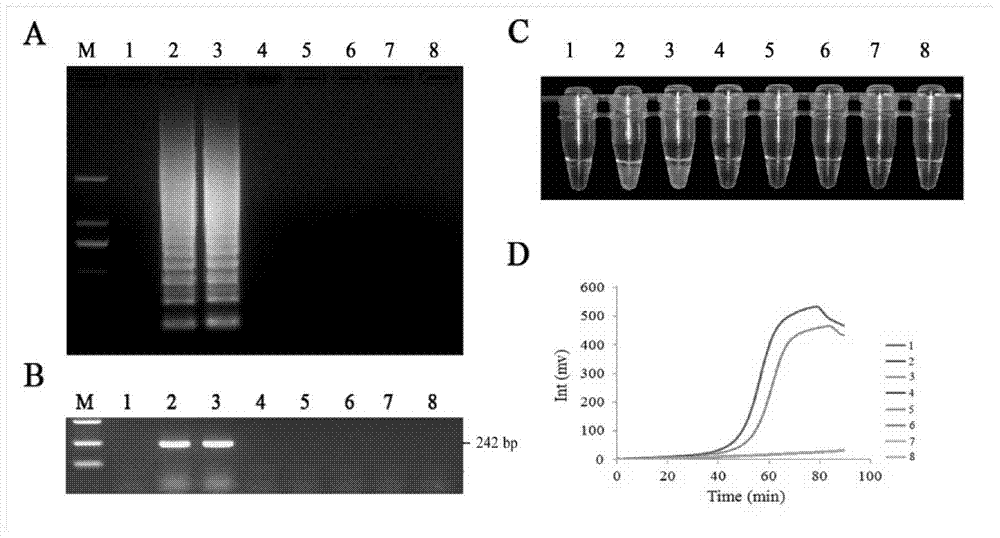
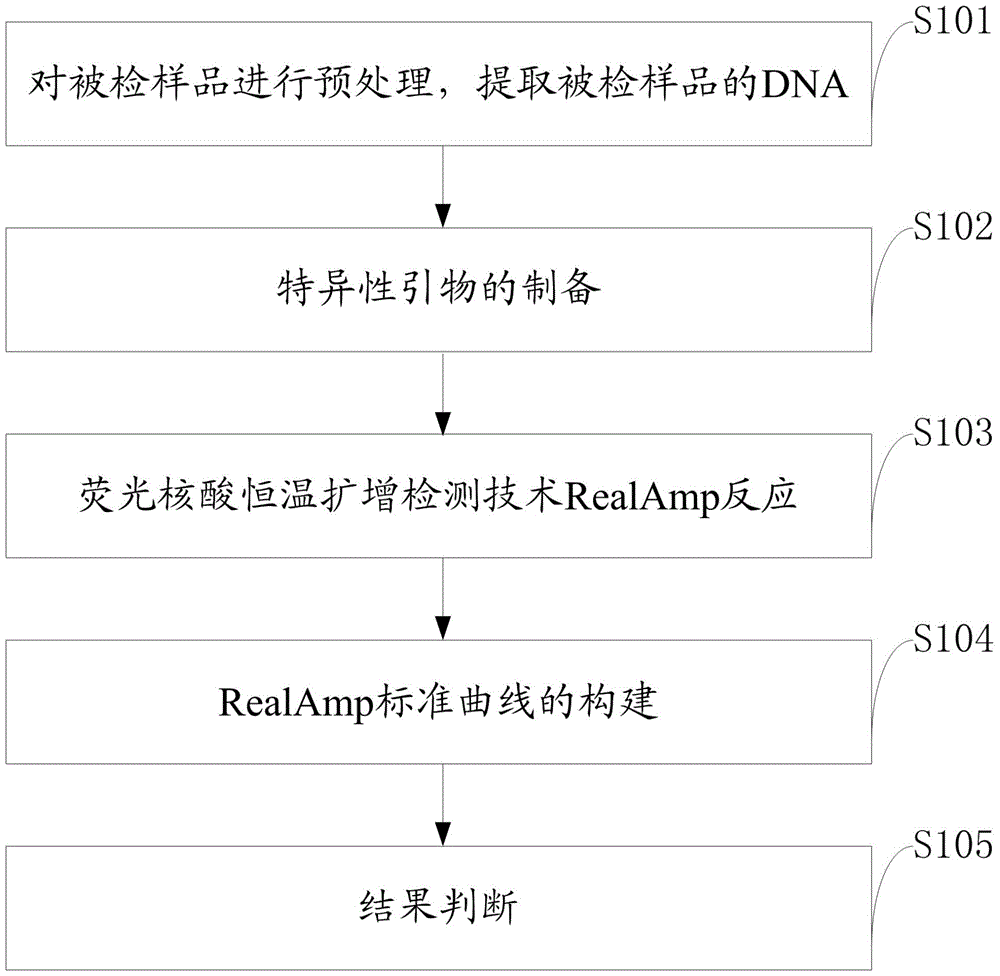
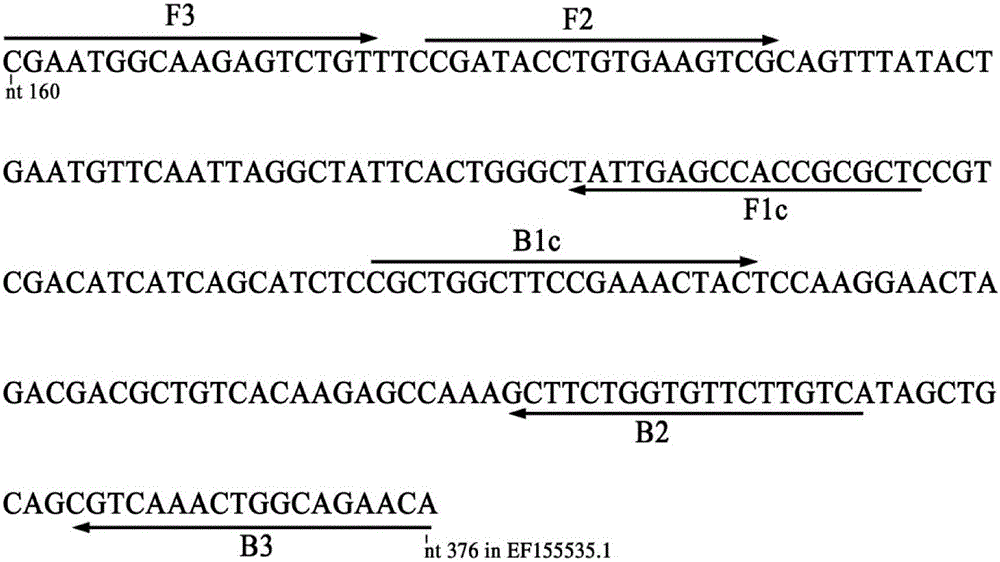
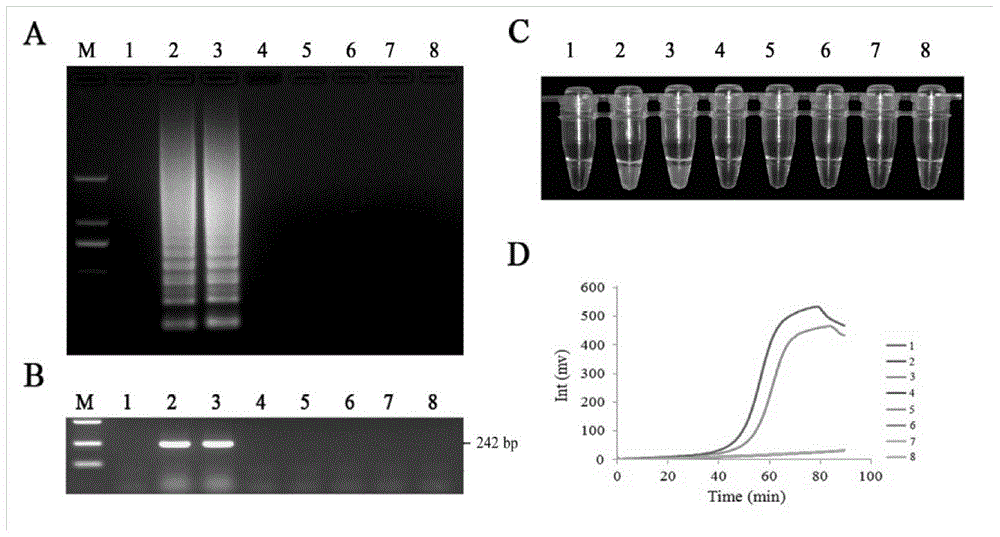
The invention discloses a quantitative detection method for FOC (Fusarium oxysporum f. sp. cubense (E.F. Smith) Snyd. & Hans.) race 4 from the soil. The method comprises the following steps: pretreating a sample to be detected and extracting DNA of the sample to be detected; preparing specific primers; conducting fluorescent nucleic acid isothermal amplification detection technique RealAmp reaction; constructing a RealAmp standard curve; and determining the result. The real-time fluorescent nucleic acid isothermal amplification detection technology provided by the invention is a novel nucleic acid detection technology combining a new generation of isothermal nucleic acid amplification technology with the real-time fluorescent detection technology, and has the advantages of high sensitivity, high specificity, low pollution and stable reaction. RealAmp can quantitatively detect pathogens; because of the characteristics of LAMP, Bst DNA polymerase has chain exchange capacity and good tolerance of inhibiting substances contained in the soil DNA extracts; the method carries out detection work on the soil, avoids cover opening and influence by contaminants and conducts on-site soil sample detection; the method for analysis and determination of reaction products is very simple; therefore, the method is suitable for wide application.
Owner:ENVIRONMENT & PLANT PROTECTION INST CHINESE ACADEMY OF TROPICAL AGRI SCI
A detection kit and detection method for Fusarium wilt of cabbage
InactiveCN103361423BStrong specificityImprove practicalityMicrobiological testing/measurementActive enzymePositive control
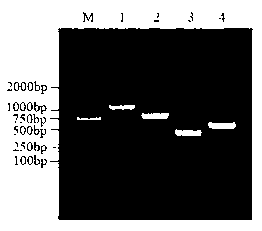
The invention discloses a detection kit for cabbage oxysporum. A reagent of the kit comprises a detection primer; the detection primer comprises 1 mul of upstream primer Foc-R and 1 mul of downstream primer Foc-S both of which the concentration is 10 pmol / mul, 2.0 mul of 10*PCR (Polymerase Chain Reaction) Easy Taq buffer solution, 0.5mul of 10mM dNTPs (diethyl-nitrophenyl thiophosphates), 0.4 mul of Taq polymerase of which the active enzyme concentration is 5U / ml, 1mul of cabbage oxysporum positive control DNA (deoxyribonucleic acid), and 20 mul of ultrapure water of which the weight concentration is not smaller than 99%; the sequence of Foc-R is 5'-TCAATGATAGTGACAAGGGTTT-3', and the sequence of the Foc-S is 5'-AATTTGCTGTGATAGGTGGAT-3'. The invention further discloses a detection method which comprises the following steps of: (1) extracting the DNA of a plant tissue or soil; (2) carrying out PCR amplification on the DNA; and (3) carrying out electrophoretic separation on the amplified product, and after separation, judging the results according to the size of the amplified product under a UV (ultraviolet) lamp after the product is colored by ethidium bromide. The kit and the method for quick molecular detection of cabbage oxysporum are high in accuracy, simple and convenient to operate, strong in specificity and high sensitivity.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
Soil DNA sample preparation kit and preparation method thereof
InactiveCN101235350BEasy to useThe result is stableBioreactor/fermenter combinationsBiological substance pretreatmentsChain typeReagent bottle
The invention discloses a soil DNA sample preparation reagent box and a manufacturing method, the reagent box comprises a box body and a baffle board, a reagent bottle A which is filled with solution I, a reagent bottle B which is filled with solution II, a reagent bottle C which is filled with solution III, a reagent bottle D which is filled with solution IV, a reagent bottle E which is filled with suspension liquid, a centrifuge tube F which is filled with acid cleaning glass bead, a mini filter G and a PCR(chain type polyase reaction) pipe H. A reagent box which is designed through the technical proposal of the invention has the advantages of simple employment and stable result, which can effectively save time and cost of users.
Owner:SHANGHAI ACADEMY OF ENVIRONMENTAL SCIENCES
A product for classification of cadmium-resistant vegetable varieties
The invention discloses a product which can be used for classification of cadmium-resistant vegetable varieties. In a specific embodiment, the present invention finds the difference in the abundance of g__MND1 in different cluster groups by extracting and sequencing the soil DNA of cadmium-resistant peppers in different cluster groups, thereby providing a new idea for the classification of cadmium-resistant vegetable varieties. The invention also provides a method for screening substances that improve the cadmium-resistant ability of vegetables.
Owner:GUIZHOU SERICULTURE RES INST GUIZHOU PEPPER RES INST +1
Simple, efficient and cheap method for purifying forest soil sample DNA
The invention provides a method for purifying microorganism DNA in forest soil, which comprises the following steps: (1) obtaining the rough DNA of a microorganism by a conventional method; (2) dissolving the rough DNA by extracted buffer solution; after the rough DNA is subjected to water bath and isovolumetric chloroform-isoamyl alcohol solution extraction, adding isopropanol which is 0.6 time of the volume of supernatant into the supernatant to precipitate DNA sediments; washing by 70 percent of pre-cooled alcohol and dissolving in the TE buffer solution; and (3) further purifying the initially purified DNA by a silica gel column. The method can remove most of impurities in a rough product of the extracted soil microorganism DNA and obtain the DNA with high purity and can be directly used for the conventional polymerase chain reaction (PCR) which is quite sensitive for an inhabiting matter. The invention has simple operation, low cost, less time consumption and wide application range and can be used for purifying the soil DNA in various types after other prior soil direct extraction methods.
Owner:JISHOU UNIVERSITY
A high-throughput method for absolute quantification of soil bacteria
ActiveCN107190055BConvenient researchReliable across samplesMicrobiological testing/measurementInternal standardCommunity structure
The invention discloses a high-throughput absolute quantification method for soil bacteria. The high-throughput absolute quantification method includes steps of whirling and re-suspending E. coli HTAQ-GFP thalli in sterile water and regulating bacteria suspension in ultraviolet spectrophotometers OD <600nm=1.0>; 2), absolutely quantifying internal standard strains HTAQ-GFP and acquiring the absolute content of internal standard strains E. coli HTAQ-GFP in bacteria solution; 3), adding the bacteria solution with the internal standard strains HTAQ-GFP to soil samples and uniformly stirring and mixing the bacteria solution and the soil samples with one another; 4), extracting soil DNA (deoxyribonucleic acid) and carrying out high-throughput sequencing on amplicons of 16S rRNA [16S ribosomal RNA (ribonucleic acid)] genes to obtain classification composition information of bacterial community structures and corresponding relative abundance; 5), acquiring the total quantities of native bacteria in soil and the absolute contents of various classification units according to the absolute contents of the internal standard strains HTAQ-GFP and the relative abundance of Escherichia in high-throughput sequencing results.
Owner:ZHEJIANG UNIV
Product for cadmium-tolerant vegetable variety classification
The invention discloses a product for cadmium-tolerant vegetable variety classification. In the specific embodiment of the invention, the abundance difference of the g_MND1 in different clustering groups is found by extracting the soil DNA of the cadmium-tolerant peppers in different clustering groups and sequencing, so a new thought is provided for cadmium-tolerant vegetable variety classification. The invention further provides a method for screening the substance for improving the cadmium tolerance of the vegetables.
Owner:GUIZHOU SERICULTURE RES INST GUIZHOU PEPPER RES INST +1
Small quality fast extraction method for soil total DNA
InactiveCN100441686CEasy to operateMicrobiological testing/measurementDNA preparationHigh concentrationEndonuclease
The invention discloses a method for fast extracting the total soil DNA. It takes soil as raw material, shaking with quartz sand and silicon dioxide powder, cracking with cetyltrimethylammonium bromide for DNA releasment, the released DNA can be absorbed by diatomite under weak acid or netural pH condition and high concentration thiocyanate ion, and the DNA will be eluted by low salty buffer solution or deionized water under weak alkaline condition. The extracted DNA with enough purity and amount can be used for PCR and endonuclease reaction. The main steps comprise cell cracking, DNA extraction and DNA purification. The invention is characterized by the simple, fast and easy method, and high purity and low cost extracted DNA. The extracted DNA can be used for kit production. The soil total DNA can be extracted in half hour for microbiological and molecular biological research.
Owner:HUAZHONG AGRI UNIV
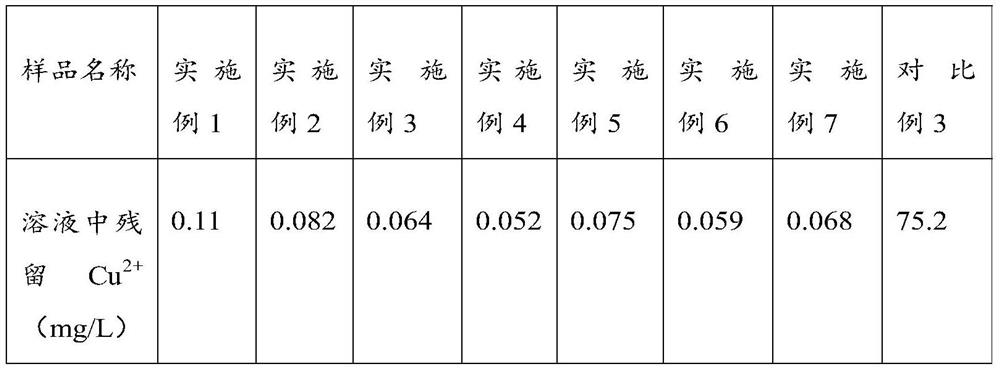
Solid-phase metal ion chelating agent as well as preparation method and application thereof
ActiveCN113045684AHigh activityEfficient removalProcess efficiency improvementDNA preparationGraft reactionGlucan
The invention discloses a solid-phase metal ion chelating agent as well as a preparation method and application thereof, and relates to the technical field of chelating agents. The solid-phase metal ion chelating agent is obtained by carrying out esterification grafting reaction on ethylenediamine tetraacetic anhydride and glucan; and preferably, the grafting rate is 20 to 30 percent based on the percentage of the carboxyl group. The prepared product can effectively remove metal ions in a biological sample, can effectively improve the activity of nucleic acid in a soil sample, especially an extreme soil sample, and can be applied to soil DNA extraction.
Owner:山西奥睿基赛生物科技有限公司
Detection kit and detection method for cabbage oxysporum
InactiveCN103361423AStrong specificityImprove practicalityMicrobiological testing/measurementPositive controlActive enzyme
The invention discloses a detection kit for cabbage oxysporum. A reagent of the kit comprises a detection primer; the detection primer comprises 1 mul of upstream primer Foc-R and 1 mul of downstream primer Foc-S both of which the concentration is 10 pmol / mul, 2.0 mul of 10*PCR (Polymerase Chain Reaction) Easy Taq buffer solution, 0.5mul of 10mM dNTPs (diethyl-nitrophenyl thiophosphates), 0.4 mul of Taq polymerase of which the active enzyme concentration is 5U / ml, 1mul of cabbage oxysporum positive control DNA (deoxyribonucleic acid), and 20 mul of ultrapure water of which the weight concentration is not smaller than 99%; the sequence of Foc-R is 5'-TCAATGATAGTGACAAGGGTTT-3', and the sequence of the Foc-S is 5'-AATTTGCTGTGATAGGTGGAT-3'. The invention further discloses a detection method which comprises the following steps of: (1) extracting the DNA of a plant tissue or soil; (2) carrying out PCR amplification on the DNA; and (3) carrying out electrophoretic separation on the amplified product, and after separation, judging the results according to the size of the amplified product under a UV (ultraviolet) lamp after the product is colored by ethidium bromide. The kit and the method for quick molecular detection of cabbage oxysporum are high in accuracy, simple and convenient to operate, strong in specificity and high sensitivity.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
Soil DNA extracting method for evaluating diversity of microbial community of plant root system
InactiveCN101709298BEasy to operateLow costMicrobiological testing/measurementDNA preparationPlant rootsDna recovery
The invention discloses a soil DNA extracting method for evaluating diversity of a microbial community of a plant root system. In the method, pretreatment is carried out on a sample before cell splitting, extracellular DNA and humic substances are removed, and the problems of difficult removal of the humic substances and low DNA recovery rate existing in a purification step are solved. In the extraction process of the method, phenol or chloroform is not used so as to reduce harm on the health of experimenters, obtain complete DNA and molecular fragments greater than 10kb and achieve high yield. OD260 / OD230 and OD260 / OD280 of the extracted soil microorganism DNA are close to standard values and can be directly applied to molecular operation so as to evaluate the diversity of the microbial community of the plant root system.
Owner:SHANGHAI ACAD OF AGRI SCI
A Soil DNA Extraction Method Directly Used to Analyze Microbial Community Structure
InactiveCN101514339BEasy to operateImprove applicabilityMicrobiological testing/measurementDNA preparationBiologySulphate sodium
The invention discloses a soil DNA extraction method directly used for analyzing the microbial community structure, which belongs to the technical field of microbiology and molecular biology. Weigh a certain amount of soil sample into a centrifuge tube, add phosphate buffer, vortex, and centrifuge to separate the washed soil sample. Add lysate to washed soil samples and mix well. Then add proteinase K and small glass beads, add sodium dodecyl sulfate solution after shaking, vortex for 5 minutes, shake in a water bath for 1 hour, and centrifuge to separate the supernatant. The KSCN solution was added to the collected supernatant, and the supernatant was separated by centrifugation after shaking in a water bath for 15 min. Add the collected supernatant to the spin column, then centrifuge to discard the liquid. Add washing solution to the centrifugal purification column, and centrifuge to discard the liquid. Then add the eluent to the centrifugal purification column, place it in a water bath for 2 minutes, collect the liquid in the centrifuge tube after centrifugation, which is the purified soil microbial DNA solution, and store it at -20°C for later use. The invention has the characteristics of simple operation of the extraction method and strong applicability, and the extracted soil microbial DNA can be directly applied to molecular operations to analyze the structure of the soil microbial community.
Owner:BEIJING NORMAL UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com