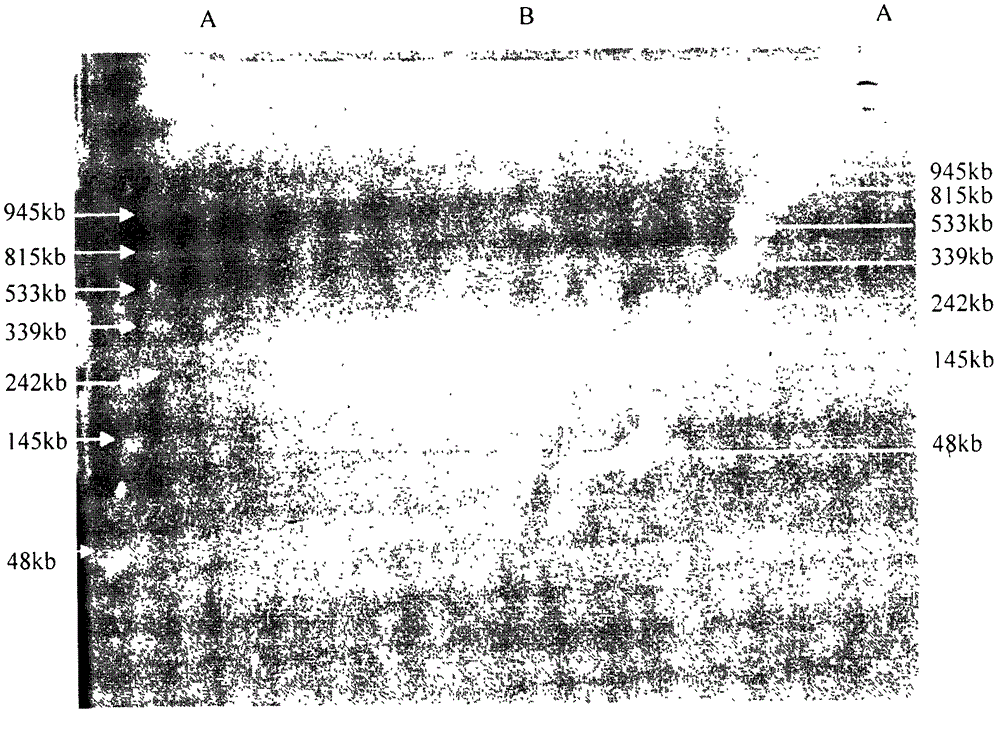
Method for separation and purification of large-fragment DNA from soil
A large fragment and soil technology, applied in the fields of biochemistry, molecular biology, and soil microorganisms, can solve the problems of insufficient competence, inability to generate DNA fragments, and increase the complexity of the living environment of microorganisms, and achieve the effect of strong operability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Embodiment one: the acquisition of soil microorganisms
[0030]1. Collect fresh soil samples and pass through an 80-mesh copper sieve.
[0031] 2. Weigh 100 g of the sieved soil sample and add 250 ml of ice-cooled distilled water, stir 3 times with a vortex stirrer under ice bath, each time for 30 seconds, with an interval of 30 seconds.
[0032] 3. Centrifuge at 700-1000 rpm for 5 minutes at 4°C and collect the supernatant.
[0033] 4. The pellet was resuspended twice and centrifuged, and all supernatants were combined.
[0034] 5. Centrifuge at 10,000 rpm at 4°C for 5 minutes, discard the supernatant, and resuspend the pellet with 10 ml of phosphate buffer.
[0035] 6. Carefully add 10 ml of Nycodenz (1.3 g / ml) solution to the bottom of the soil suspension, and centrifuge at 10,000 rpm for 30 minutes at 4°C.
[0036] 7. Carefully collect the white cell layer between the Nycodenz-soil suspension, add more than 4 times the volume of phosphate buffer at 10,000 rpm, an...
Embodiment 2
[0042] Example 2: Embedding of microbial cells in low melting point agarose
[0043] 1. Prepare 1% low-melting point agarose solution. The type of solution used is easily consistent with the type of microorganism obtained by resuspension and separation. After heating and dissolving, store it in a water bath at 45 degrees Celsius for later use.
[0044] 2. Adjust the concentration of the obtained soil microbial bacterial liquid with a serum counting plate to reach 2*10 8 cells / ml, mix the above-mentioned 1% low-melting point agarose solution with an equal volume of the adjusted bacterial solution, and quickly fill it into a 2mm*1.5mm*5mm strip module, and place the module in After cooling at 4°C, take it out and store it in an EDTA solution containing 1 micromolar concentration for later use.
[0045] Precautions:
[0046] 1. Adjust the bacterial solution to a suitable concentration. If the concentration is too high, it is not conducive to the in situ lysis of microorganism...
Embodiment 3
[0048] Embodiment three: in situ cracking of microorganisms
[0049] 1. Take out the spare strips in the EDTA solution with a concentration of 1 micromolar, place them in 4 ℃ pre-cooled distilled water for soaking, and shake slowly at the same time, change the distilled water 3 times during the process, and soak in the lysate for the last time.
[0050] 2. After taking out the gel strip, react with the lysate containing 1 mg / ml lysozyme at 37 degrees Celsius for 1 hour while shaking slowly. The volume of the lysate is at least 10 times that of the gel. During this period, lysozyme can be added or the lysate containing lysozyme can be replaced.
[0051] 3. After the cracked strips are taken out, rinse gently with 4°C pre-cooled distilled water several times, and then soak in protease solution for a period of time. Then use at least 4 times the volume of the gel block to degrade the protein with a protease solution containing 1 mg / ml protease at 55 degrees Celsius for more than...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com