Lathyrus quinquenervius EST-SSR primer group developed based on transcriptome sequencing, method and application
A technology of transcriptome sequencing and lathyrus, applied in the field of molecular markers and bioinformatics, can solve the problem of lack of EST-SSR primers and achieve low-cost results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
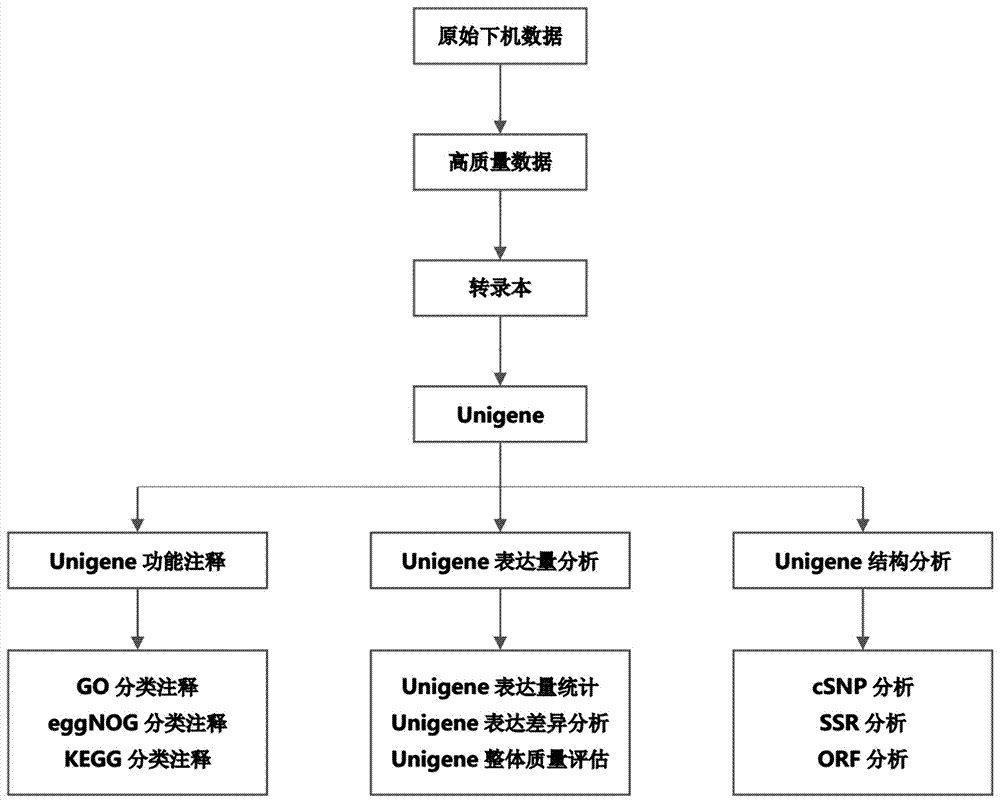
[0026] Example 1: RNA-Seq analysis and SSR primer design
[0027] 1. Determination of transcriptome data
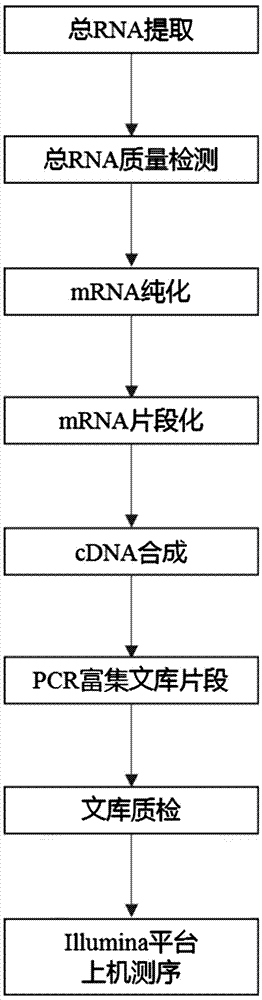
[0028] Using the Tianenze column plant RNAout kit, the total RNA of the roots, stems, and leaves of the two varieties of Lathyrus RQ23 and RQ36 at the seedling stage was used to purify the mRNA by capturing the mRNA with oligo dT magnetic beads; using fragmentation buffer containing divalent cations The captured mRNA was broken into 200-300 nt fragments, the first cDNA strand was synthesized using random primers containing 6 bases and fragmented mRNA, and then the second cDNA strand was synthesized using DNA polymerase I and RNase H. The product was purified by QIAquick PCR Extraction Kit, eluted with EB buffer, and the overhanging end was repaired by exonuclease and polymerase. Add "A" to the 3' end of the cDNA fragment and connect it with the sequencing adapter, use AMPure XPbeads to purify the adapter-added system, then perform PCR amplification, and finally use 2% ag...
Embodiment 2
[0047] Example 2 Verification of SSR primers
[0048]43 Lathyrus germplasm resources from different sources recorded in Table 2 were used for planting, and 2 g of leaves at the seedling stage were collected respectively, and the leaves were pulverized by a high-throughput pulverizer, and Shanghai Sangong DNA extraction kit (Ezup column type) was used for planting. Plant Genomic DNA Extraction Kit, B518261-0050) for leaf DNA extraction, DNA quality detection and concentration determination on a microplate reader, and DNA was diluted to a final concentration of 30ng / μL, and stored at -20°C. Using 43 materials DNA and 284 randomly selected pairs of SSR primers to carry out PCR amplification to verify the polymorphism of the primers. The PCR reaction system was 10 μL in total, including 1.5 μL (30ng / μL) of genomic DNA, 5 μL of 2× Taq PCR MasterMix (Tiangen, KT:121221), and 2 μL (0.002nmol / μL) of forward and reverse primers in total. The PCR amplification reaction program was: pre...
Embodiment 3
[0052] Example 3: Construction of 43 Fingerprints of Lathyrus Germplasm Resources
[0053] Using 284 pairs of primers to PCR amplification results of DNA from 43 Lathyrus germplasm resources, 87 pairs of polymorphic SSR markers were selected (see Table 3 for 87 pairs of polymorphic SSR markers), and 87 pairs of primers for 43 The polymorphism scanning data of these materials was used to screen the best markers to distinguish 43 Lathyrus germplasms by IDAnalysis 4.0 software, and finally 8 effective markers were obtained to distinguish 43 Lathyrus germplasms.
[0054] The PCR amplification and polyacrylamide gel electrophoresis results of 43 materials were read by 8 pairs of primers. The 8 pairs of primers can amplify different band patterns respectively, among which SSR No.013 is 6 band patterns, which are labeled separately 1-6; SSR No.253 is 5-band type, respectively marked as 1-5; SSR No.58 is 5-band type, respectively marked as 1-5; SSR No.203 is 4-band type, respectively ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com