Hybridization buffer solution for hybrid capture of DNA sequencing library
A DNA sequencing and hybridization capture technology, applied in the field of molecular biology, can solve the problems of expensive hardware equipment, limited number of chips, high chip cost, and achieve the effects of simple formula, improved efficiency and simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0012] Below in conjunction with specific embodiment the present invention is described in further detail:
[0013] The hybridization buffer used for DNA sequencing library hybridization capture of the present invention includes the following components in parts by volume:
[0014]
[0015] The hybridization solution of the present invention only needs a thermocycler routinely used in a laboratory, and the hybridization temperature of 65 degrees is suitable for commonly used single-stranded nucleotide capture probes with a length of 80-120 bases.
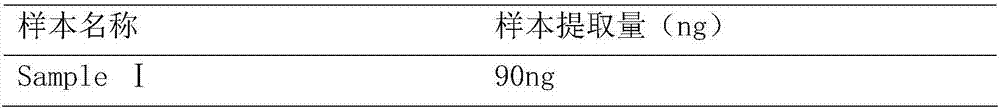
[0016] The hybridization solution formula and hybridization method of the present invention are applicable to the capture and enrichment of genomic DNA sequencing libraries prepared by various methods (such as Illumina kit, NEB kit, KAPA kit, etc.). And can simultaneously enrich multiple library samples in one hybridization reaction. No less than 250ng of DNA is required for the total input library sample.
[0017] By using the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com