Targeted trapping and sequencing kit for lung cancer-related 285 genes and application of targeted trapping and sequencing kit
A targeted capture and kit technology, applied in the field of kits, can solve the problems of wrong sequencing results, high price, unsatisfactory detection results in unknown regions, etc., achieve high throughput, meet maximum use, and reduce clinical testing costs Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1: Preparation of Lung Cancer Related 285 Gene Profile Detection Probe Kit
[0052] 1. Identify lung cancer-related driver genes or targeted drug resistance genes: Search the PUBMED database through the Internet, check the latest literature and unpublished data of the research team, and make statistics on the pathogenesis, chemoradiotherapy, targeted drug resistance and metastasis of lung cancer in the Chinese population. Related target genes or their parallel, adjacent pathway genes, identified 285 genes included in the kit (Table 2).
[0053] Table 2
[0054] JAK2
ABL2
ERCC3
TSHR
USP9X
SUFU
SOX2
RHBDF2
BRCA2
PTPRD
CDC73
LRP1B
KDM6A
SMC3
EIF4A2
SRSF2
RB1
CDKN2A
ELF3
ACVR2A
AKT1
GATA1
FGFR2
AXIN1
BUB1B
ERCC5
FANCG
MDM4
NFE2L2
STK11
KDM5C
PDYN
TSC2
LTK
SMARCB1
PAX5
H3F3A
PMS1
GNA11 ...
Embodiment 2
[0058] Detection using the probe designed in Example 1 includes the following steps: fragmenting the genomic DNA to obtain DNA fragments; connecting the DNA fragments with sticky ends to Adapters to obtain ligated products; using the specific The ligation product is hybridized and captured by the sex probe to obtain the target fragment; the target fragment hybridized with the probe is purified, then PCR amplified, separated and purified, and the obtained amplified product constitutes the high-throughput sequencing library. The library was sequenced by the IonProton next-generation sequencing platform, and all the variations of the 285 genes detected in the samples were determined by analysis. The more specific steps are as follows (the operation of the kit is carried out according to the instructions):
[0059] 1. Sequencing library construction
[0060] (1) Prepare sample DNA (concentration greater than 20ng / μl, total amount greater than 200ng).
[0061] (2) Fragmentation o...
Embodiment 3
[0117] Detection of lung cancer patient tissue samples using the kit of Example 1
[0118] 1. Validation Select 13 human lung adenocarcinoma tissue samples (from Guangdong Provincial People's Hospital) whose mutations have been detected by direct sequencing. Each sample requires 200ng of DNA for detection. The operation steps and conditions of the kit of the present invention are the same as the specific steps in Example 2.
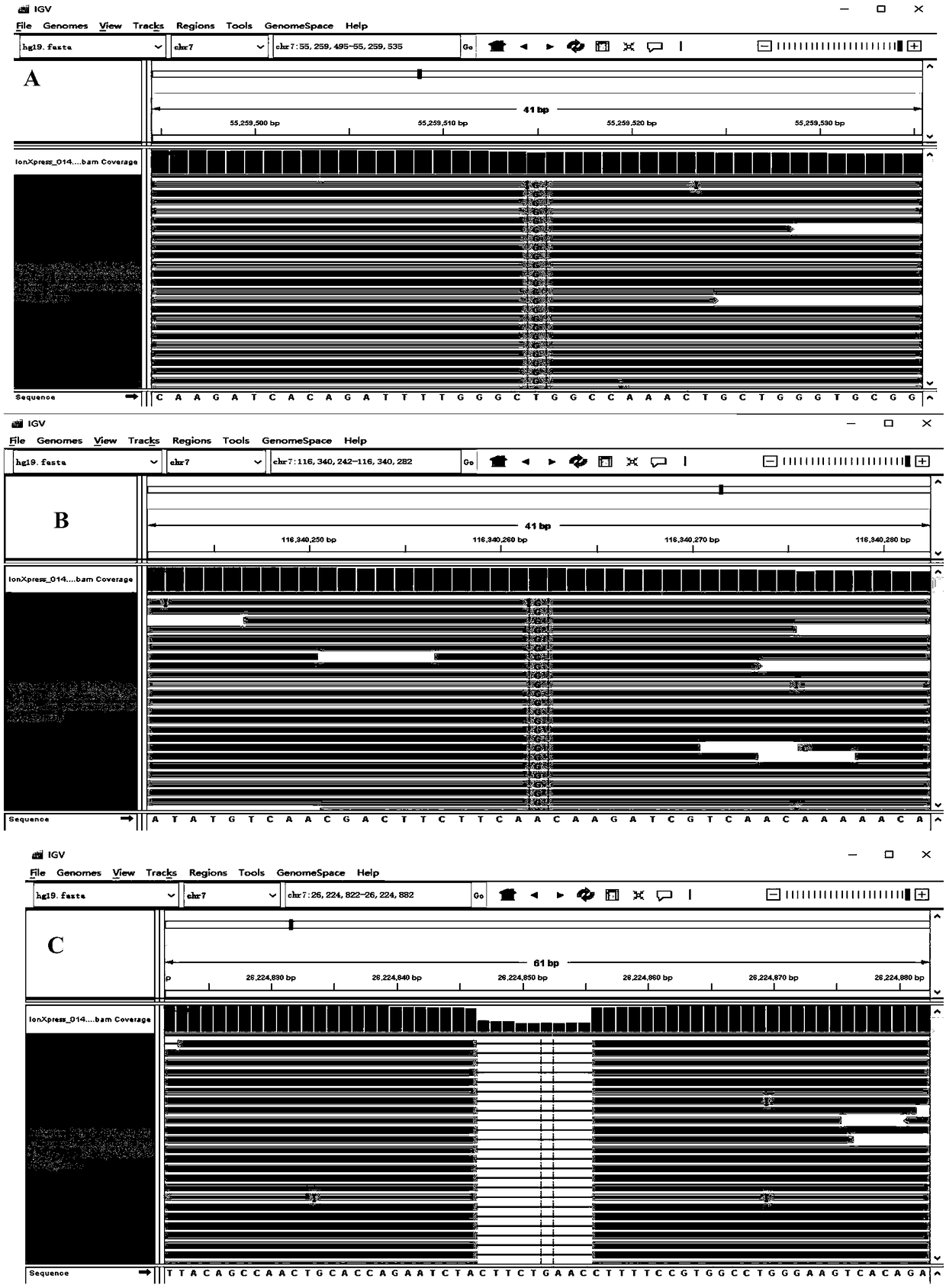
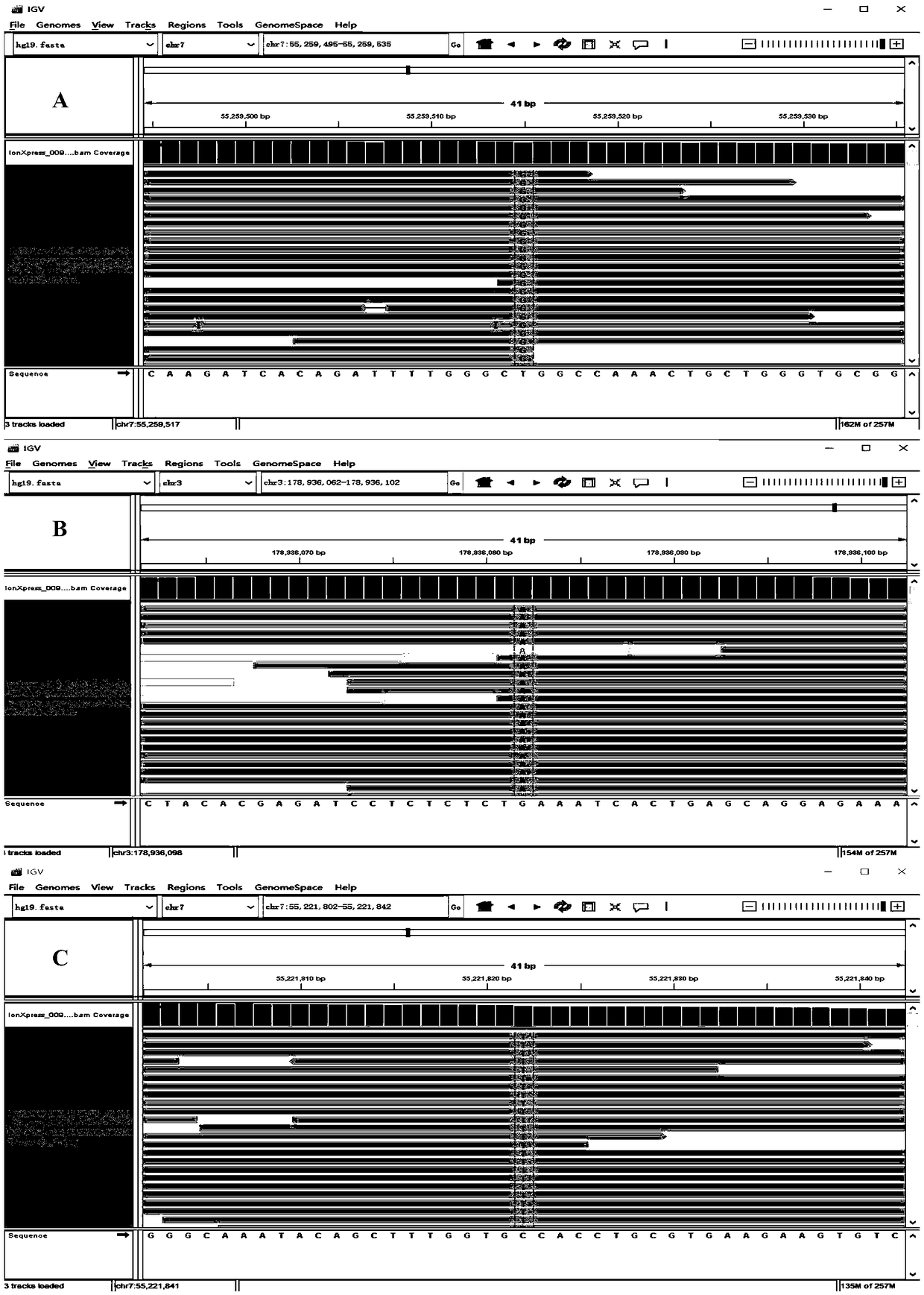
[0119] 2. 13 cases of lung cancer tissue samples were detected by the kit of the present invention, and compared with the detection results of the direct sequencing method (see Table 6). Using the kit of the present invention in these 13 tissue samples was completely consistent with the detection results of the direct sequencing method.
[0120] The verification result of the test kit of the present invention compared with the direct sequencing method of table 6
[0121]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com