Cancer gene expression profile data identification method based on integration of extreme learning machines
An extreme learning machine, tumor gene technology, applied in the field of computer analysis of tumor gene expression profiling data, which can solve the problems of high noise and high variation data analysis of gene expression profiling data, and few samples of gene expression profiling data.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
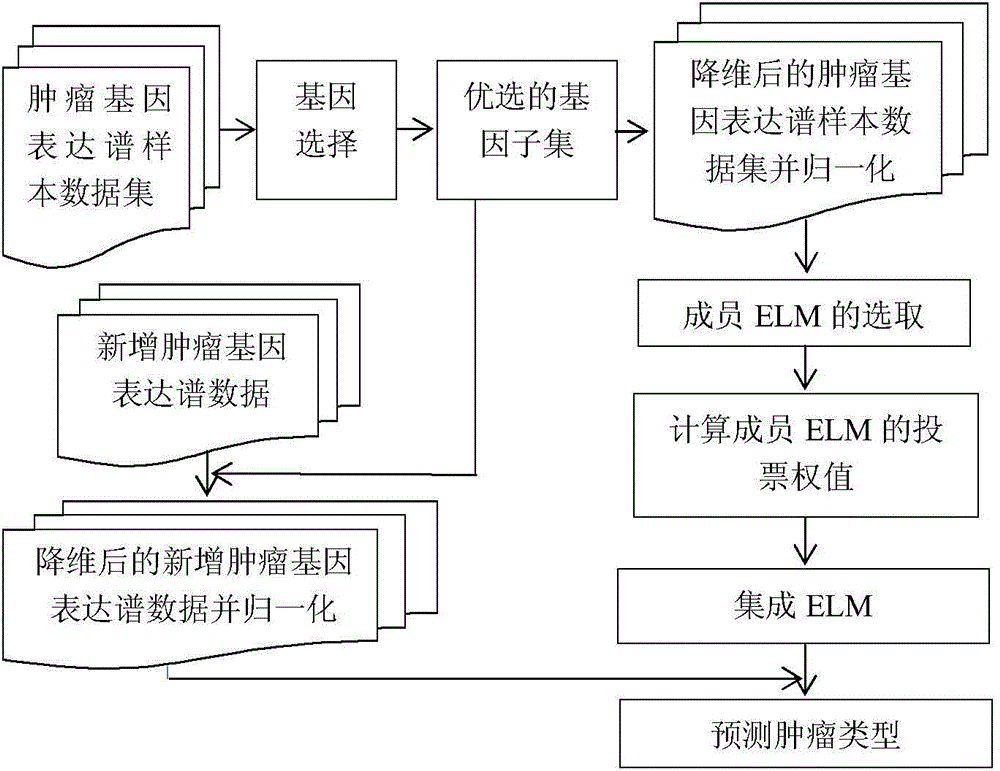
[0049] A method for identifying tumor gene expression profile data based on an integrated extreme learning machine, including the selection of member ELMs and the integration steps between member ELMs, the ELM (Extreme learning machine) in the present invention is an extreme learning machine, and the present invention specifically includes the following steps :
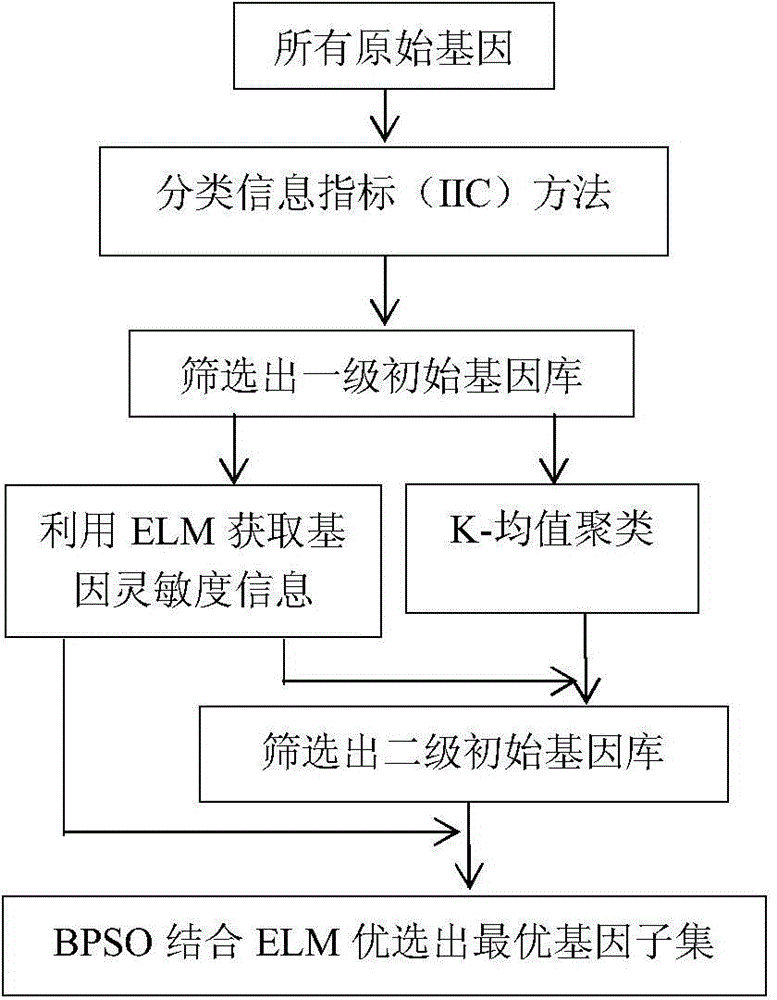
[0050] Step 1: Preprocessing of tumor gene expression profile data sets, including gene selection and normalization of tumor expression profile data;
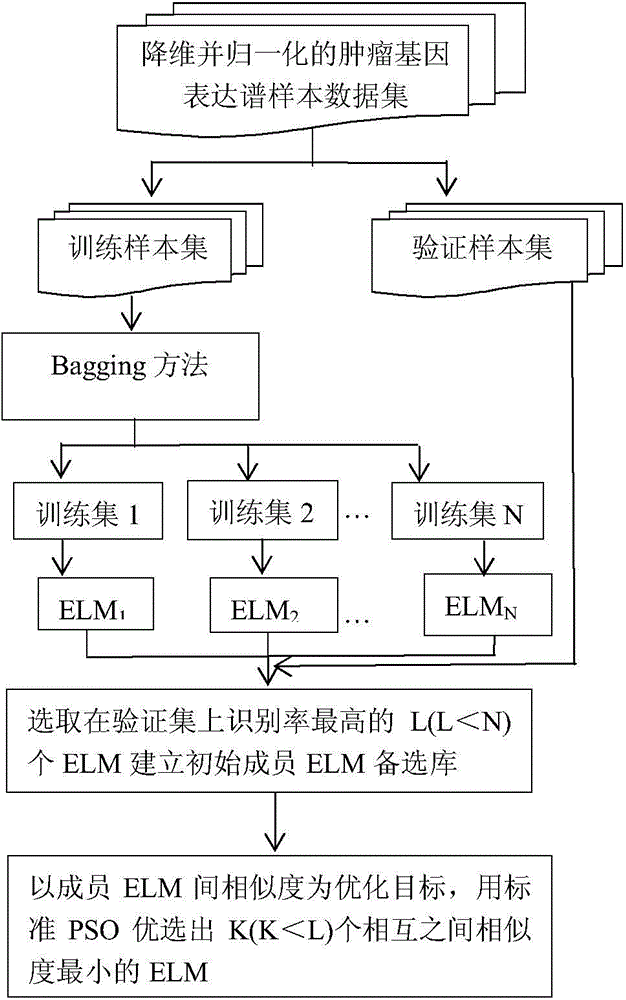
[0051] Step 2: Generate N sample sets according to a certain proportion through the Bagging method for the data set obtained in step 1, and then generate N training sets and verification sets according to a certain proportion of these N sample sets;
[0052] Step 3: Learn to generate N extreme learning machines on the N training sets in step 2, and select the highest L ELMs (L
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com