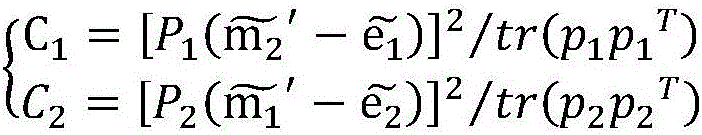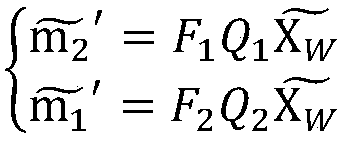Patents
Literature
38results about How to "Reduce outliers" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
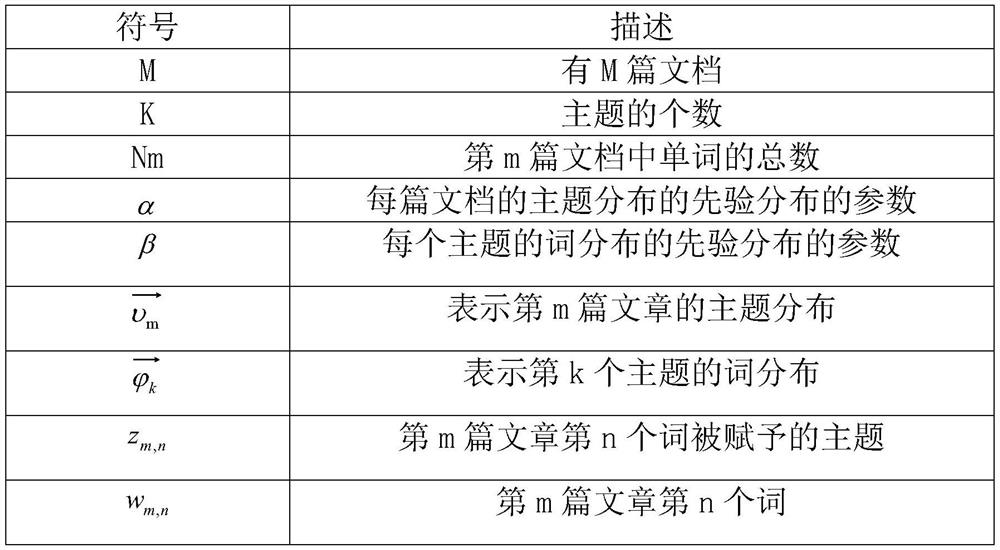
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
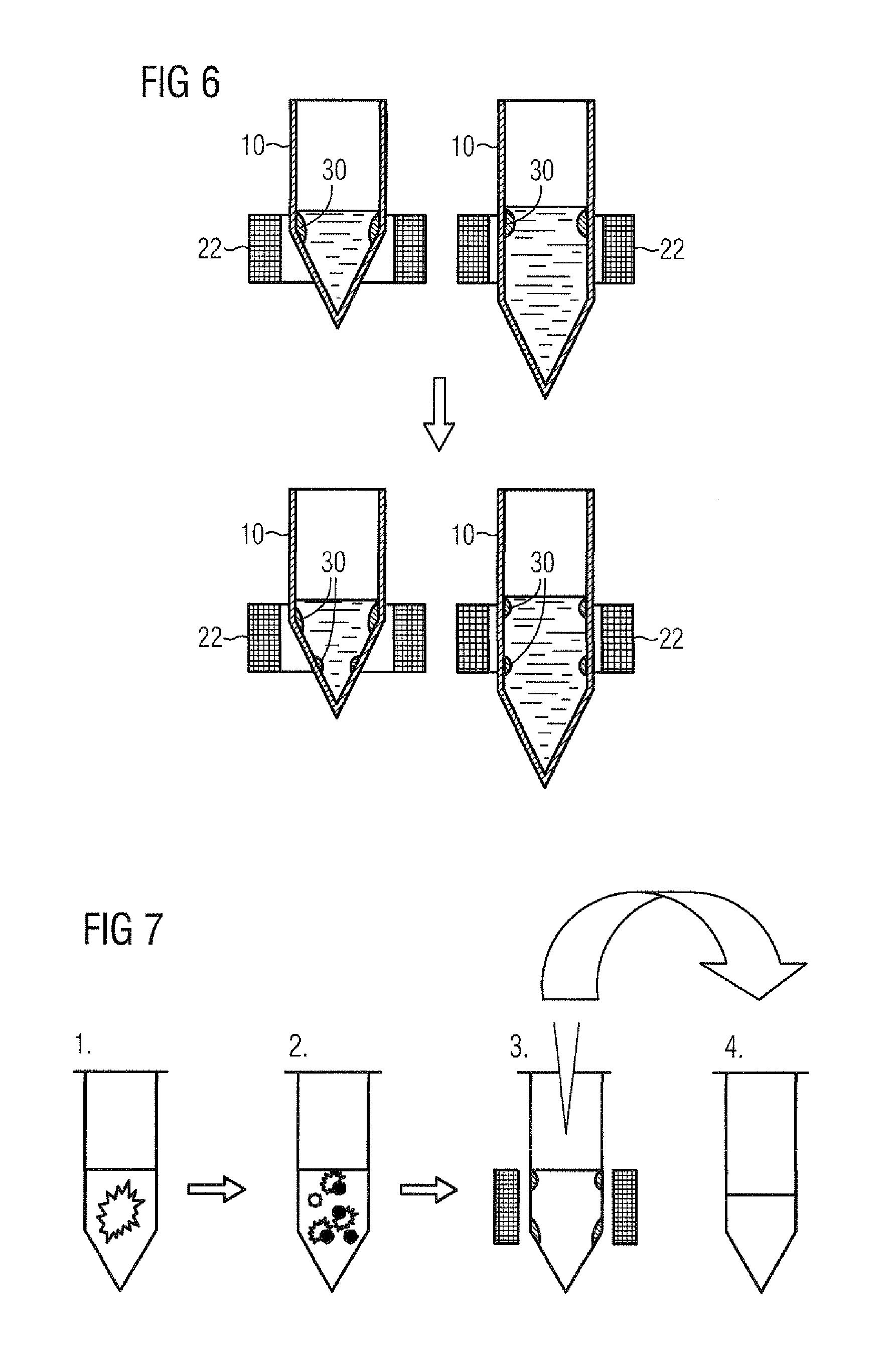
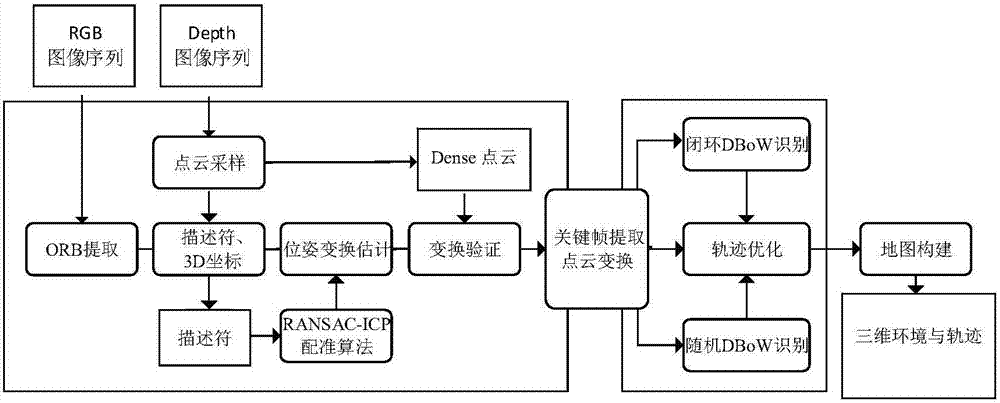
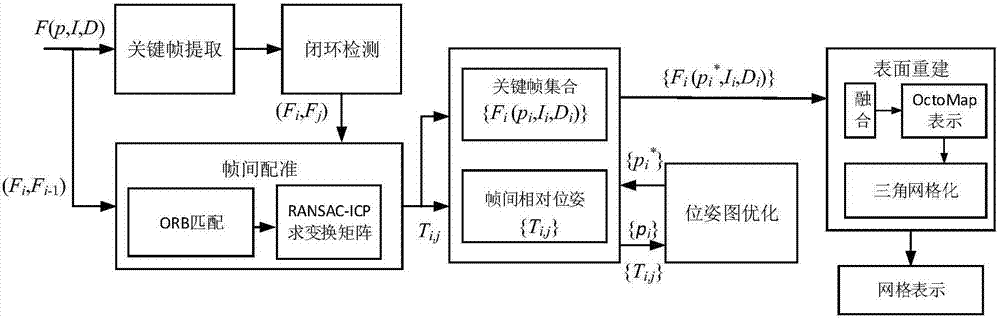
ORB key frame closed-loop detection SLAM method capable of improving consistency of position and pose of robot
InactiveCN105856230AGuaranteed positioning accuracyGuaranteed accuracyProgramme controlProgramme-controlled manipulatorClosed loopImaging Feature
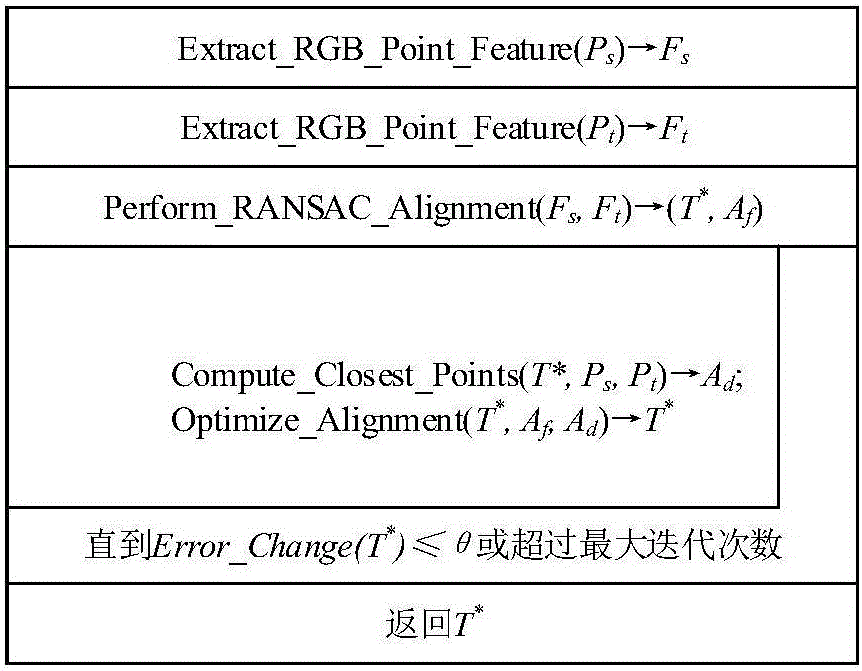
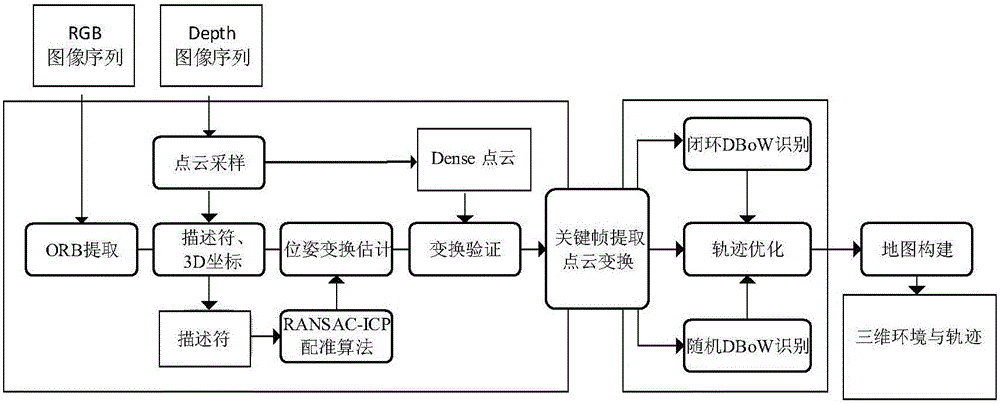
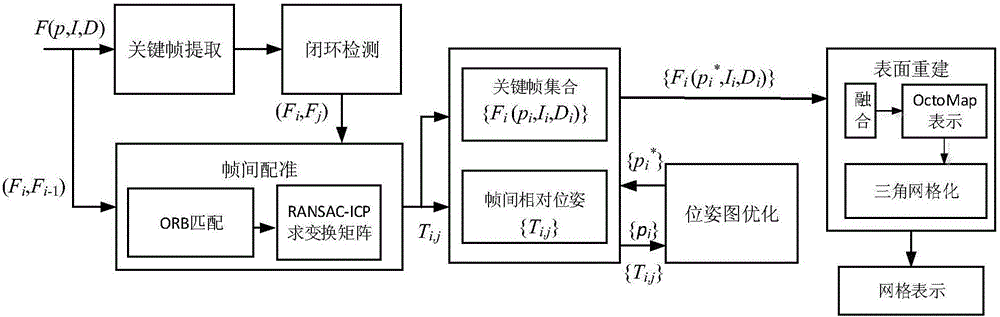
The invention discloses an ORB key frame closed-loop detection SLAM method capable of improving the consistency of the position and the pose of a robot. The ORB key frame closed-loop detection SLAM method comprises the following steps of, firstly, acquiring color information and depth information of the environment by adopting an RGB-D sensor, and extracting the image features by using the ORB features; then, estimating the position and the pose of the robot by an algorithm based on RANSAC-ICP interframe registration, and constructing an initial position and pose graph; and finally, constructing BoVW (bag of visual words) by extracting the ORB features in a Key Frame, carrying out similarity comparison on the current key frame and words in the BoVW to realize closed-loop key frame detection, adding constraint of the position and pose graph through key frame interframe registration detection, and obtaining the global optimal position and pose of the robot. The invention provides the ORB key frame closed-loop detection SLAM method with capability of improving the consistency of the position and the pose of the robot, higher constructing quality of an environmental map and high optimization efficiency.
Owner:简燕梅
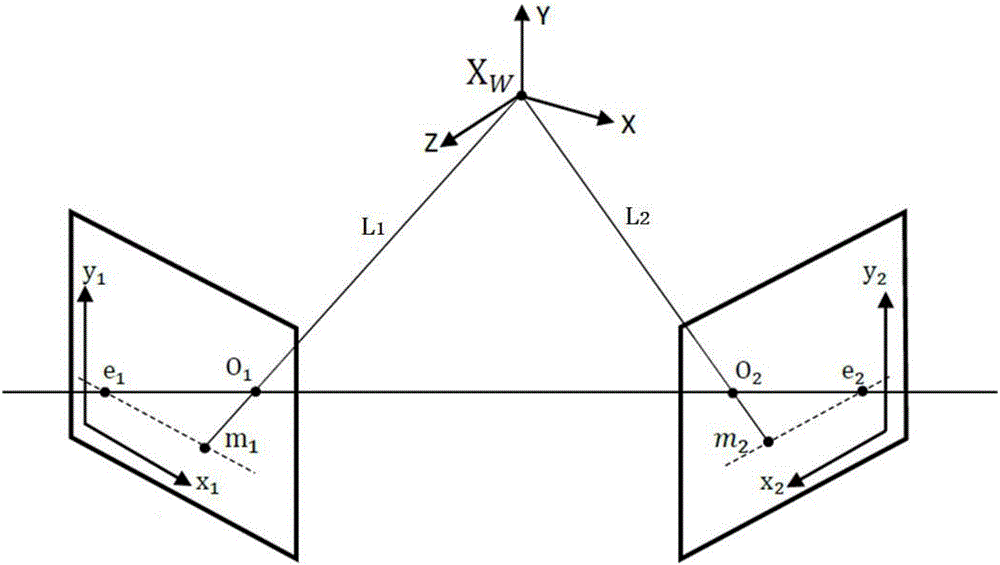
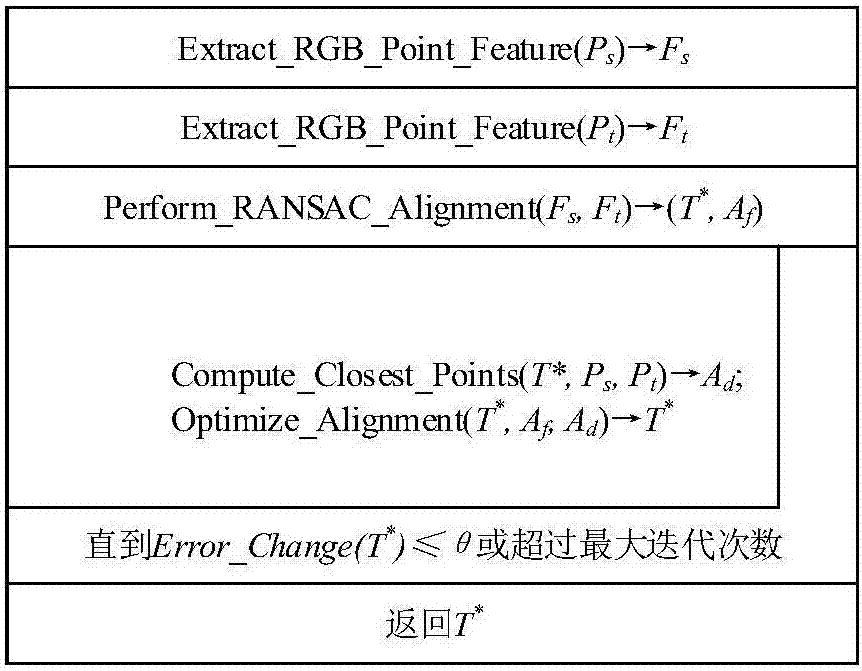
Binocular three-dimensional reconstruction method
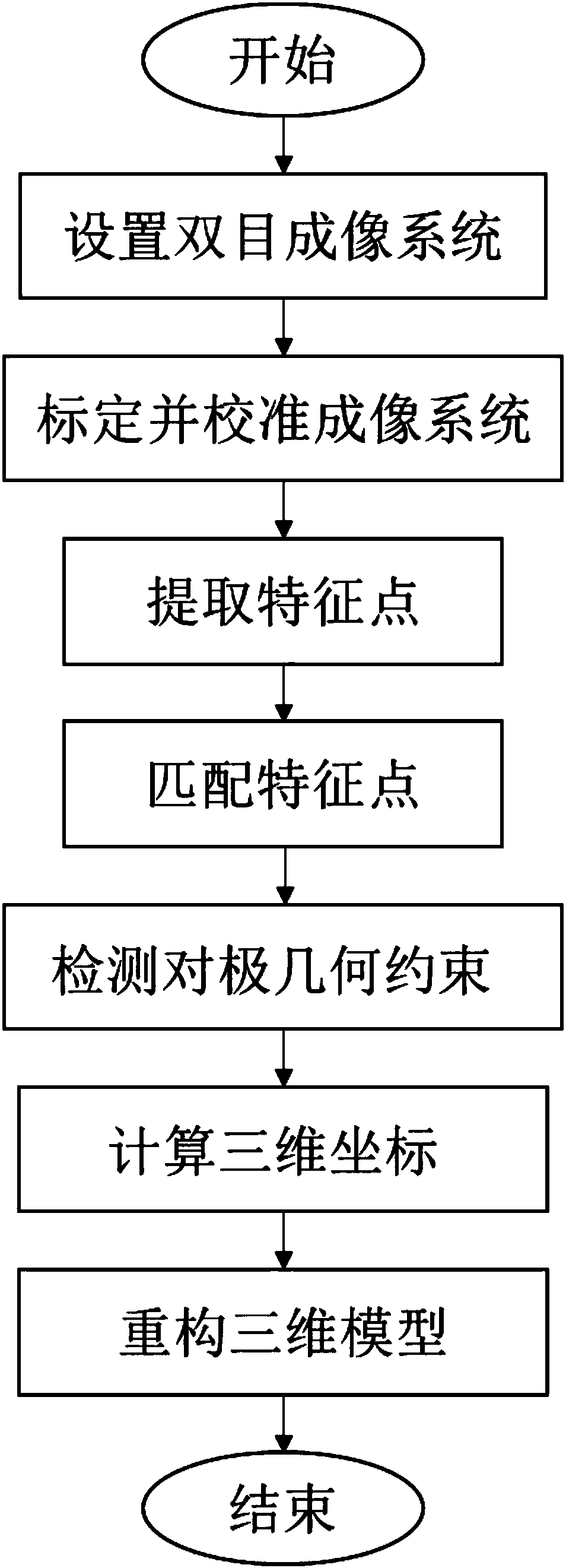
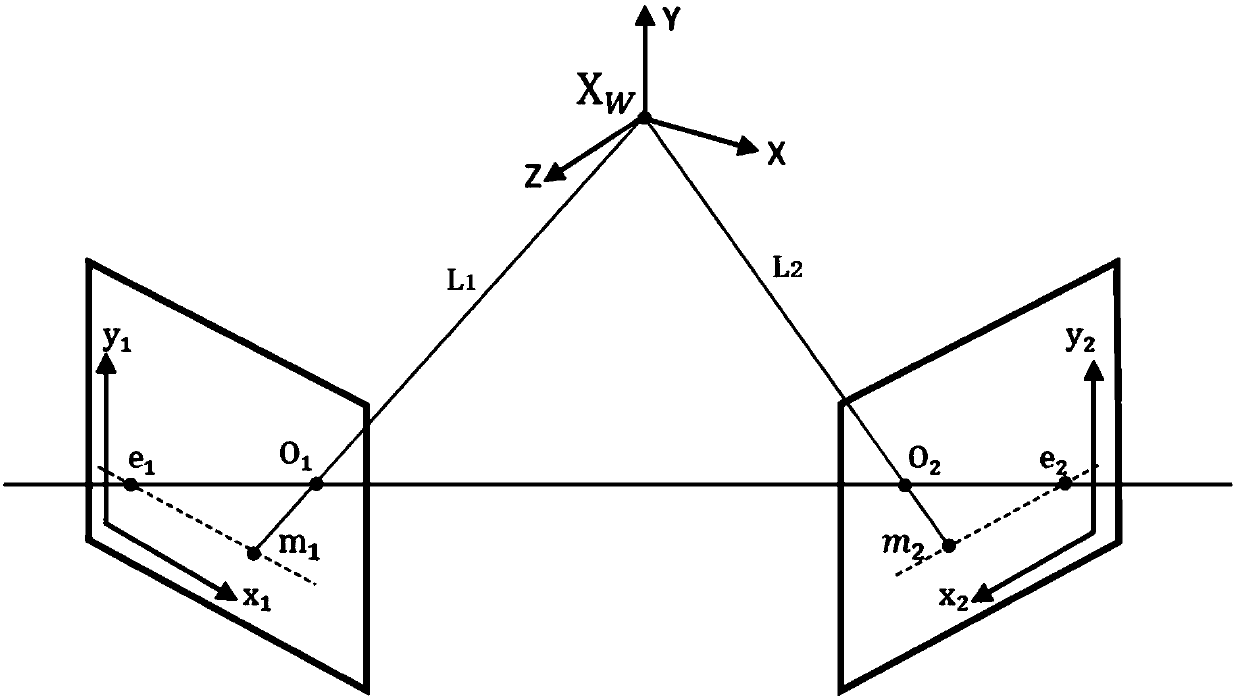
ActiveCN105894574AReduce false match rateReduce outliersDetails involving processing steps3D modellingFeature extractionReconstruction method
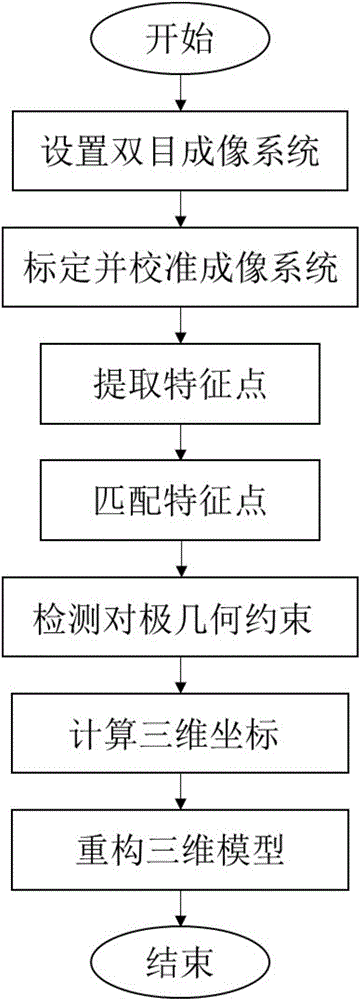
The invention discloses a binocular three-dimensional reconstruction method comprising the following steps of: 1) acquiring an image of an object to be reconstructed by two image acquisition apparatuses with the same model in order to obtain a left image and a right image; 2) calibrating the image acquisition apparatuses by a chessboard method, computing the internal and external parameter and a lens distortion coefficient, processing the left image and the right image to remove distortion in the images; 3) extracting features from the two processed images in the step 2) to obtain the feature points of the two images; 4) matching the feature points by using the feature points of the two images in the step 3) to obtain feature point pairs; 5) performing epipolar geometric constraint detection on the obtained feature point pairs to remove mismatched feature point pairs; and 6) computing three-dimensional coordinates of corresponding points, in a world coordinate system, of the obtained feature points by using the reserved feature point pairs. The binocular three-dimensional reconstruction method may reduce abnormal points and acquires an accurate three-dimensional reconstruction model.
Owner:HUIZHOU FRONT OPTOELECTRONIC TECH CO LTD
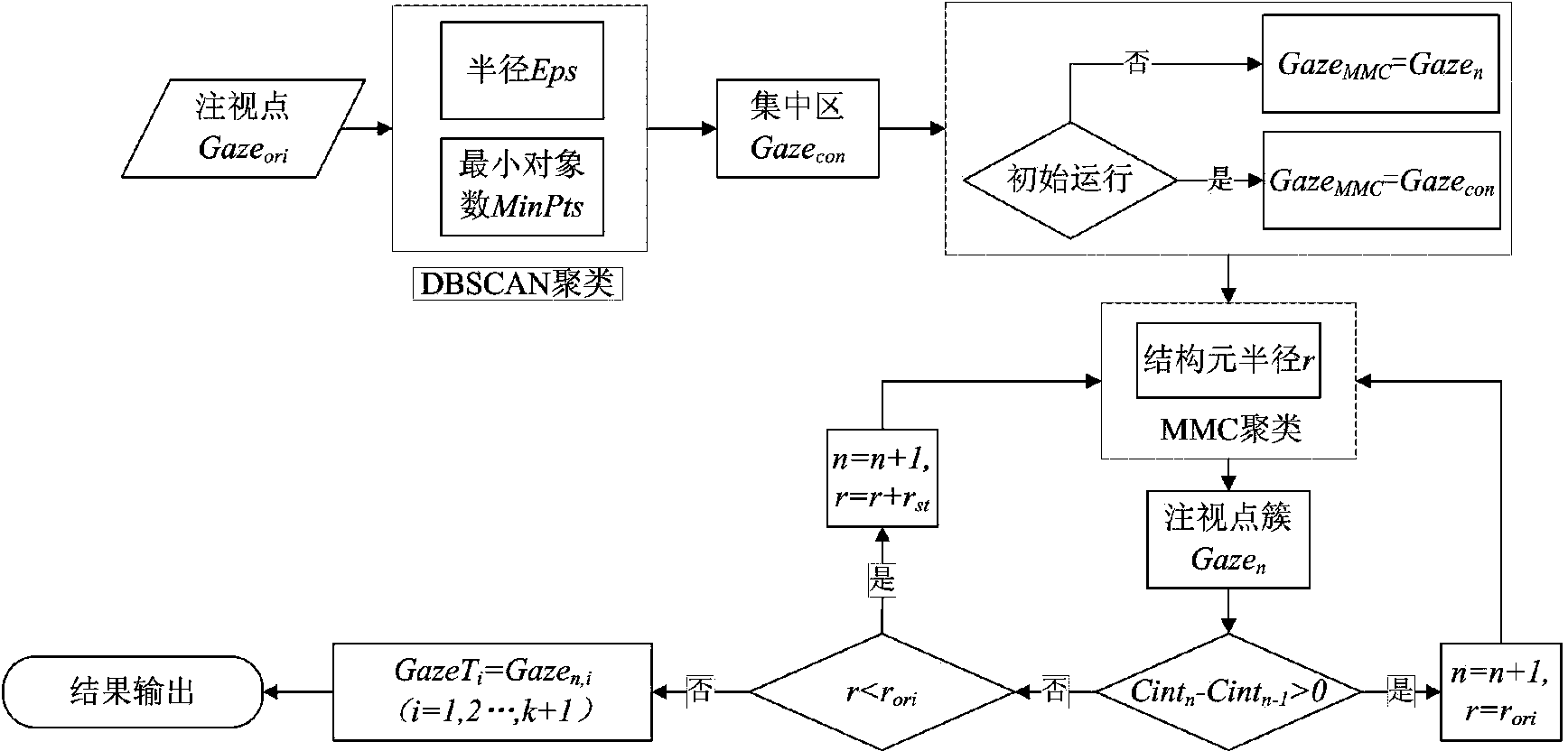
Driver fixation point clustering method based on density clustering method and morphology clustering method
InactiveCN103903276AReduce outliersOvercome clustering deficienciesImage analysisCharacter and pattern recognitionDriver/operatorDensity based
The invention provides a driver fixation point clustering method based on a density clustering method and a morphology clustering method, and belongs to the field of typical density clustering methods and mathematic morphology clustering methods. The driver fixation point clustering method includes the steps of putting forward a density method and mathematic morphology method combined self-adaption DBSCAN-MMC method, applying the method to driver fixation point clustering, setting the value of the Eps through fixation point structure parameters, obtaining an initial point set of MMC clusters through the DBSCAN, determining the number of the clusters, reducing outliers produced through DBSCAN clusters through the self-adaption MMC clusters, and completing clustering oriented to driver fixation areas. According to the method, the advantages of irregular shape clustering of the DBSCAN and the MMC are fully used, the defects of the two clustering methods are overcome, the clustering effect is superior to the clustering effect of the conventional DBSCAN clustering method and the conventional MMC clustering method when the driver fixation areas are divided, and the driver fixation clustering quality is improved.
Owner:JILIN UNIV
Differentially expressed gene identification method based on combined constraint non-negative matrix factorization
ActiveCN107016261AEffective decomposition resultsEfficient Sparse Decomposition ResultsSpecial data processing applicationsData setAlgorithm
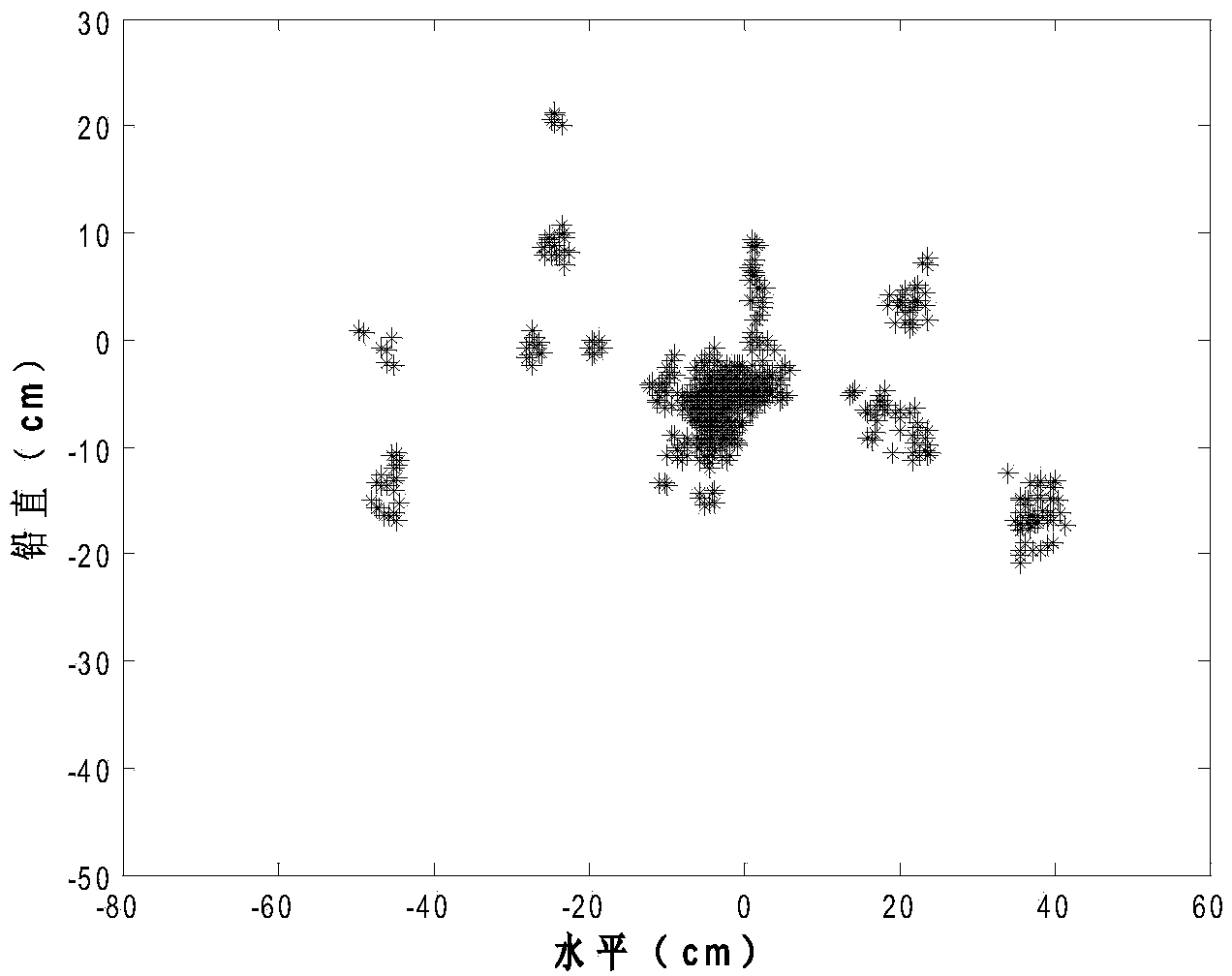
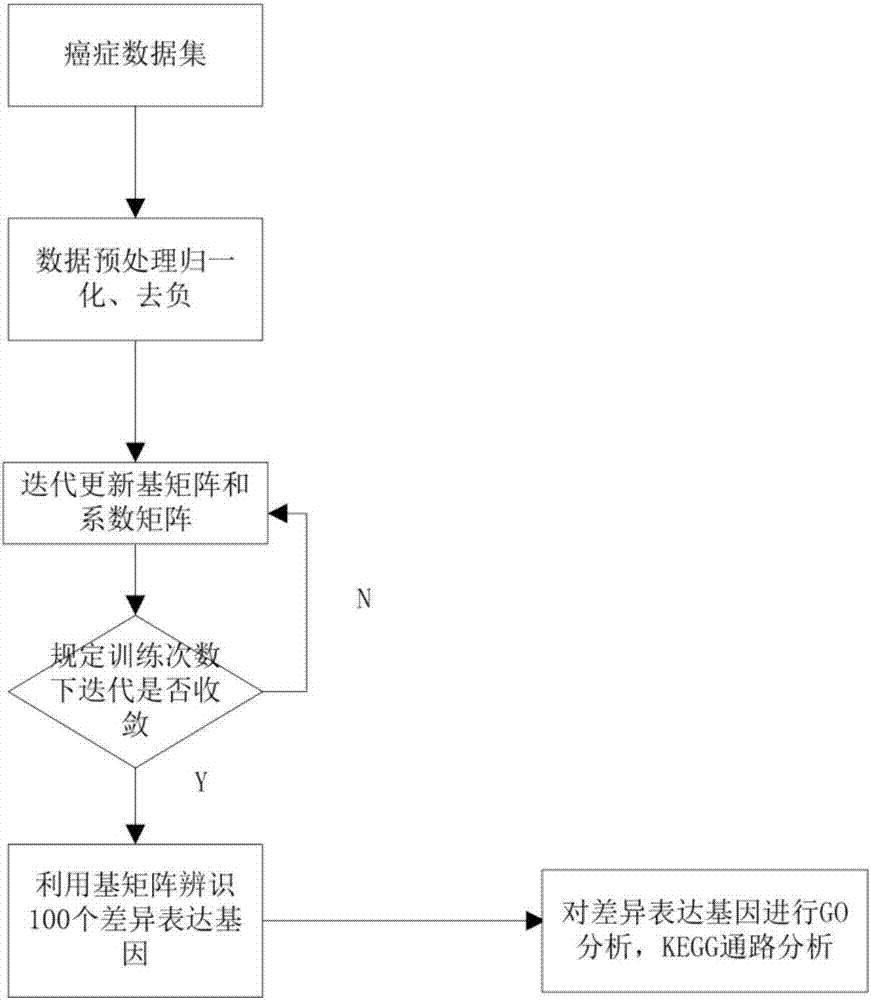
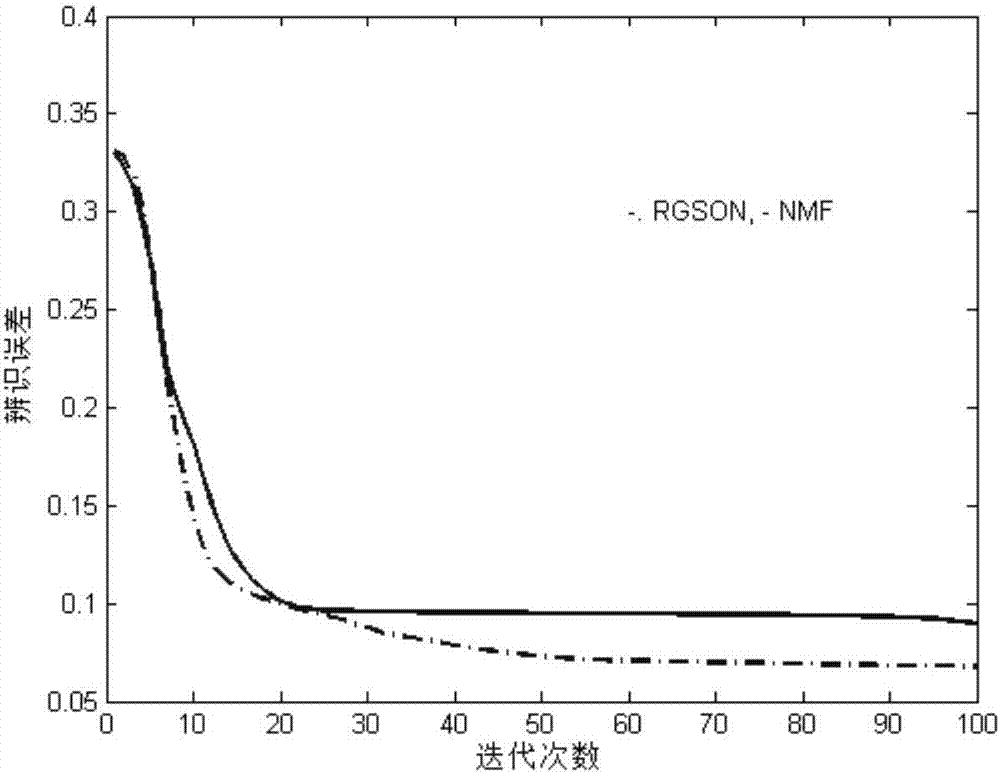
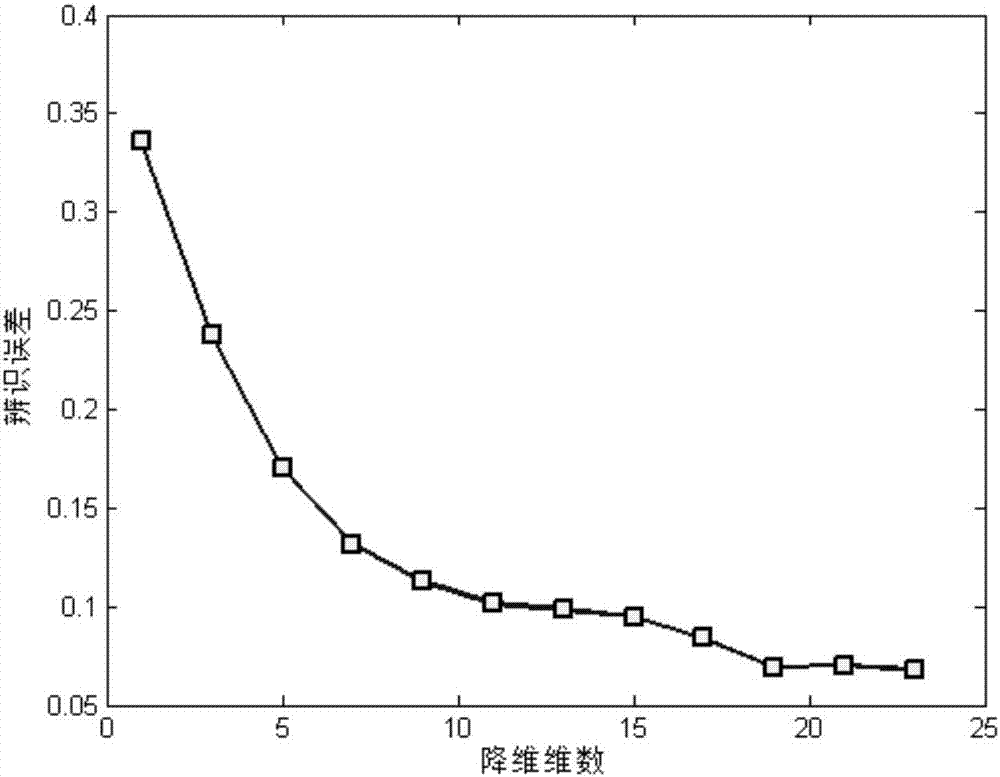
The invention discloses a differentially expressed gene identification method based on combined constraint non-negative matrix factorization. The method comprises the following steps of 1, representing a cancer-gene expression data set with a non-negative matrix X, 2, constructing a diagonal matrix Q and an element-full matrix E, 3, introducing manifold learning in the classical non-negative matrix factorization method, conducting orthogonal-constraint sparseness and constraint on a coefficient matrix G, and obtaining a combined constraint non-negative matrix factorization target function, 4, calculating the target function, and obtaining iterative formulas of a basis matrix F and the coefficient matrix G, 5, conducting semi-supervision non-negative matrix factorization on the non-negative data set X, and obtaining the basis matrix F and the coefficient matrix G after iteration convergence, 6, obtaining an evaluation vector (the formula is shown in the description), sorting elements in the evaluation vector (the formula is shown in the description) from large to small according to the basis matrix F, and obtaining differentially expressed genes, 7, testing and analyzing the identified differentially expressed genes through a GO tool. The identification method can effectively extract the differentially expressed genes where cancer data is concentrated, and be applied in discovering differential features in a human disease gene database. The identification method has important clinical significance for early diagnosis and target treatment of diseases.
Owner:HANGZHOU HANGENE BIOTECH CO LTD
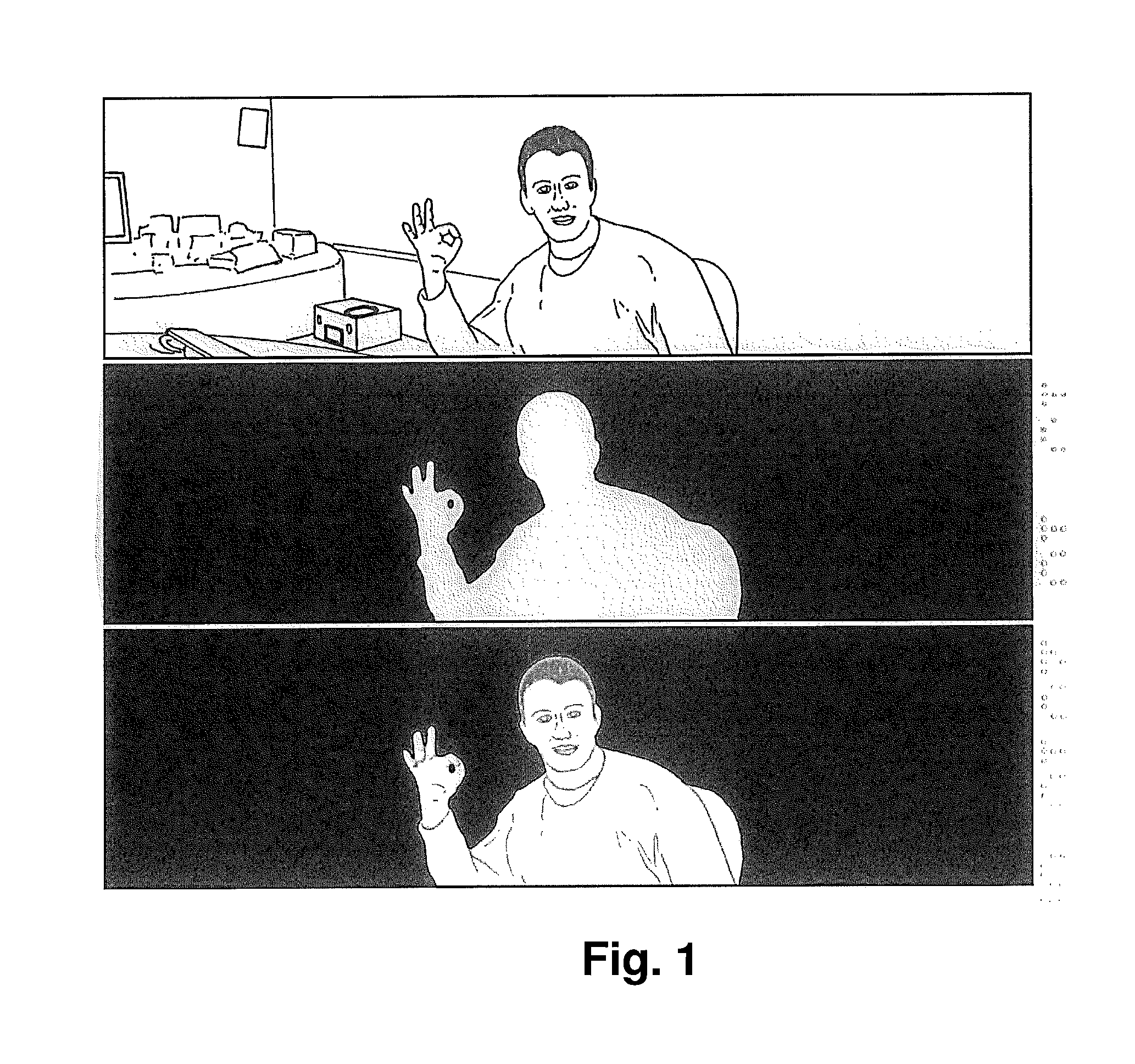
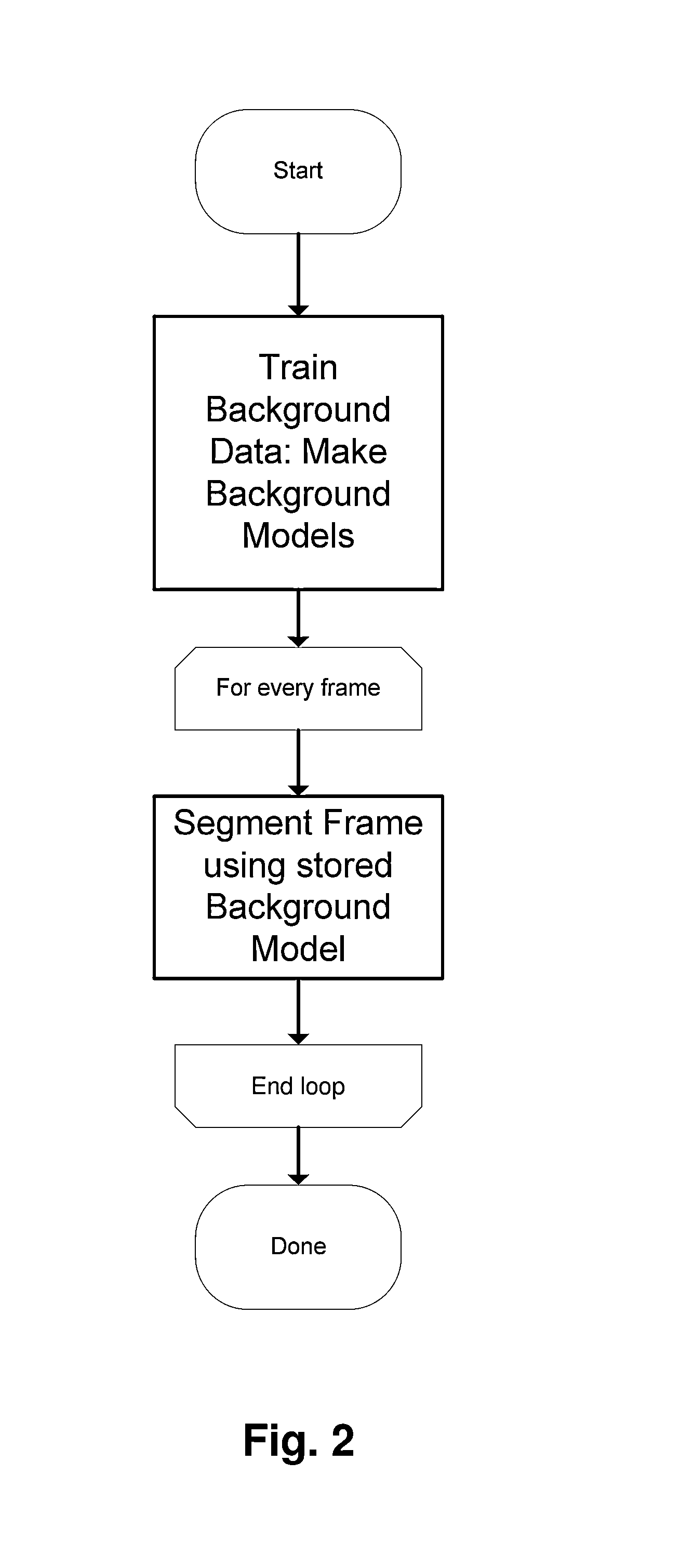
Method and system for real-time images foreground segmentation
InactiveUS20130243314A1Suppress detectionAdapt quicklyImage enhancementImage analysisPattern recognitionSpatial structure
The method comprises:generating a set of cost functions for foreground, background and shadow segmentation classes or models, where the background and shadow segmentation costs are based on chromatic distortion and brightness and colour distortion; andapplying to the pixels of an image said set of generated cost functions;The method further comprises, in addition to a local modelling of foreground, background and shadow classes carried out by said cost functions, exploiting the spatial structure of content of at least said image in a local as well as more global manner; this is done such that local spatial structure is exploited by estimating pixels' costs as an average over homogeneous colour regions, and global spatial structure is exploited by the use of a regularization optimization algorithm.The system is adapted to implement at least part of the method.
Owner:TELEFONICA SA
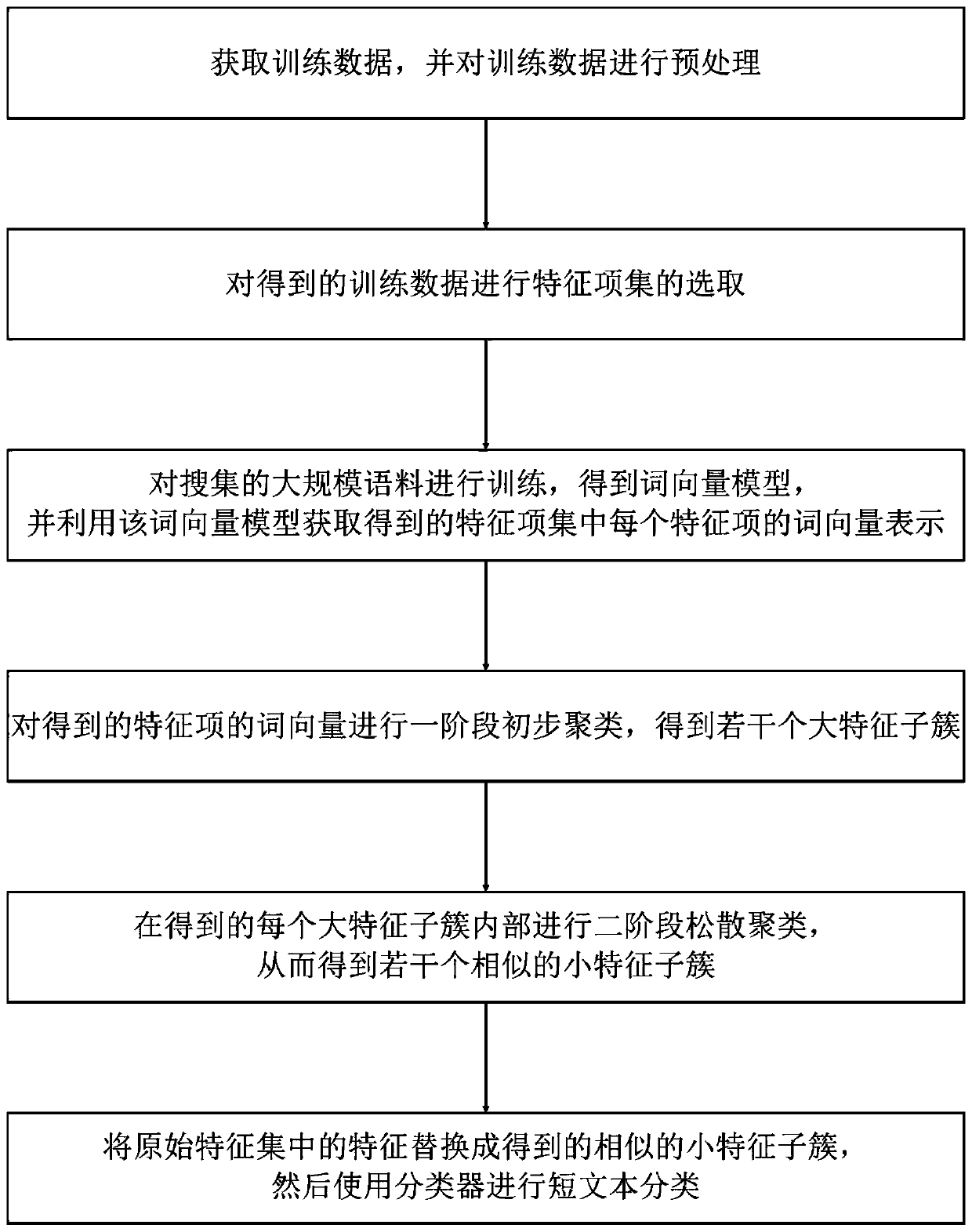
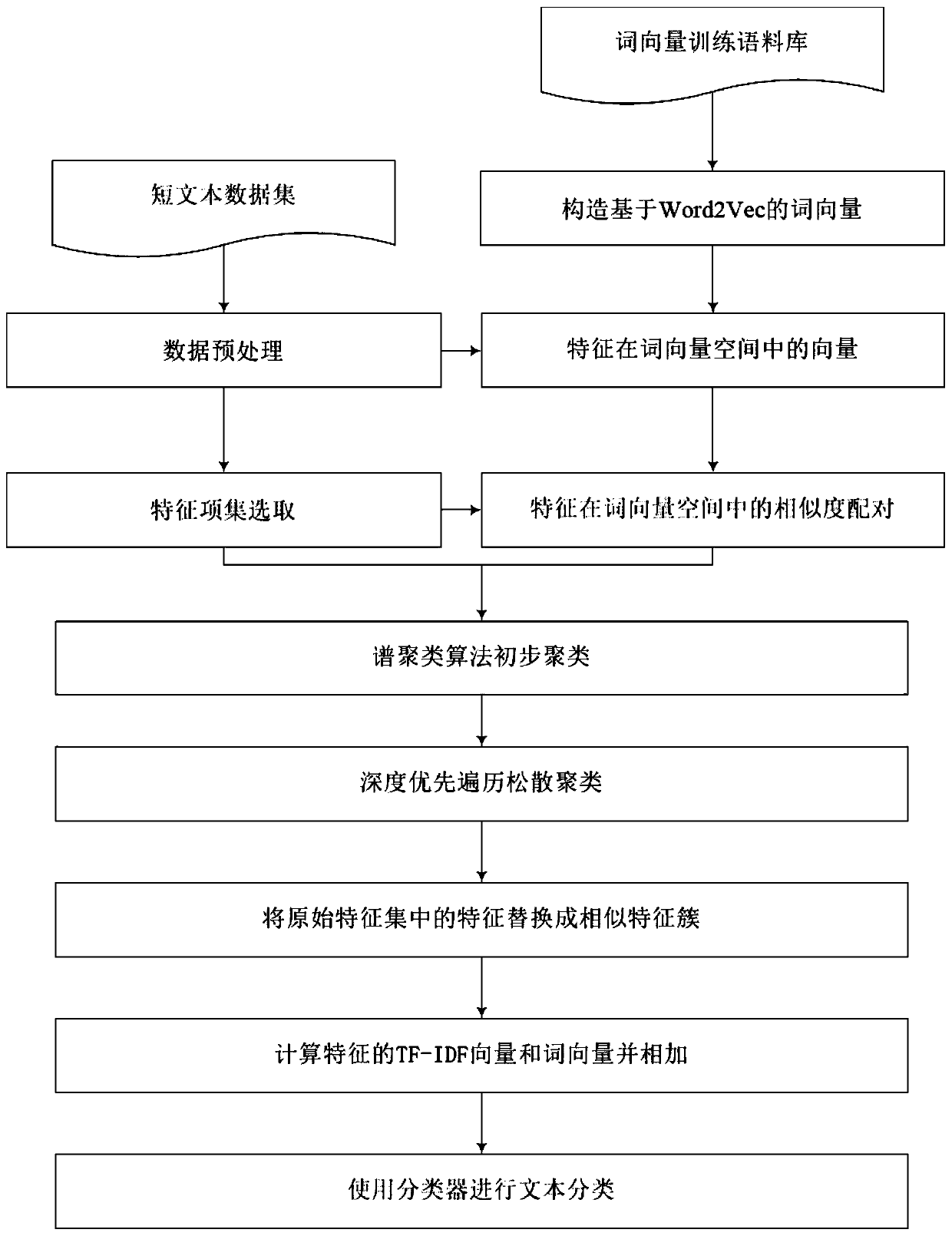
Short text-oriented optimization classification method
ActiveCN109960799AQuality improvementEnhanced Semantic Representation CapabilitiesSemantic analysisCharacter and pattern recognitionData setMicroblogging
The invention discloses a short text-oriented optimization classification method. The method comprises the following steps of: 1, obtaining an original data set and preprocessing the original data set; 2, selecting a feature item set from the preprocessed data set; 3, training the collected large-scale corpora by using a word vector tool to obtain a word vector model; 4, performing word vector representation on each feature item in the feature item set by using a word vector model, and performing primary clustering on the word vectors of the feature items to obtain a plurality of primary feature clusters; 5, performing two-stage loose clustering in each preliminary feature cluster to obtain a plurality of similar feature clusters; and 6, replacing the feature words obtained in the step 4 with the similar feature clusters obtained in the step 5, and then carrying out short text classification by using a classifier. Traditional short text classification mostly lacks semantic expression capability and is quite high in demnsion of the feature space; according to the invention, the semantic information of the short text can be expressed better, the dimension of the feature space is reduced, the precision and efficiency of short text classification are improved, and the short text classification method can be applied to short text classification tasks in various fields, such as spamshort message classification and microblog topic classification.
Owner:长沙市智为信息技术有限公司
Method For Purification Of Nucleic Acids, Particularly From Fixed Tissue
ActiveUS20110245483A1Good suspensionImprove lysisOrganic compounds purification/separation/stabilisationSugar derivativesInterior spaceAqueous solution
The invention relates to a method for purification of nucleic acids, to a kit for performing the method according to the invention and to a new application of magnetic particles for purification of a biological sample. The method according to the invention comprises the following steps: a) accommodating of the sample in a first sample vessel in an aqueous solution and lysing of the sample under non-chaotropic conditions; suspending of first magnetic particles in the solution and inserting of the first sample vessel in a sample vessel holder, wherein the sample vessel is inserted in the annular interior space of a ring magnet associated with the sample vessel holder; separating of the solution from the magnetic particles; and isolating of the nucleic acids from the solution.
Owner:SIEMENS HEALTHCARE DIAGNOSTICS INC
Method for purification of nucleic acids, particularly from fixed tissue
ActiveUS8703931B2Low costKeep timeOrganic compounds purification/separation/stabilisationSugar derivativesAqueous solutionParticle separation
The invention relates to a method for purification of nucleic acids, to a kit for performing the method according to the invention and to a new application of magnetic particles for purification of a biological sample. The method according to the invention comprises the following steps: a) accommodating of the sample in a first sample vessel in an aqueous solution and lysing of the sample under non-chaotropic conditions; suspending of first magnetic particles in the solution and inserting of the first sample vessel in a sample vessel holder, wherein the sample vessel is inserted in the annular interior space of a ring magnet associated with the sample vessel holder; separating of the solution from the magnetic particles; and isolating of the nucleic acids from the solution.
Owner:SIEMENS HEALTHCARE DIAGNOSTICS INC
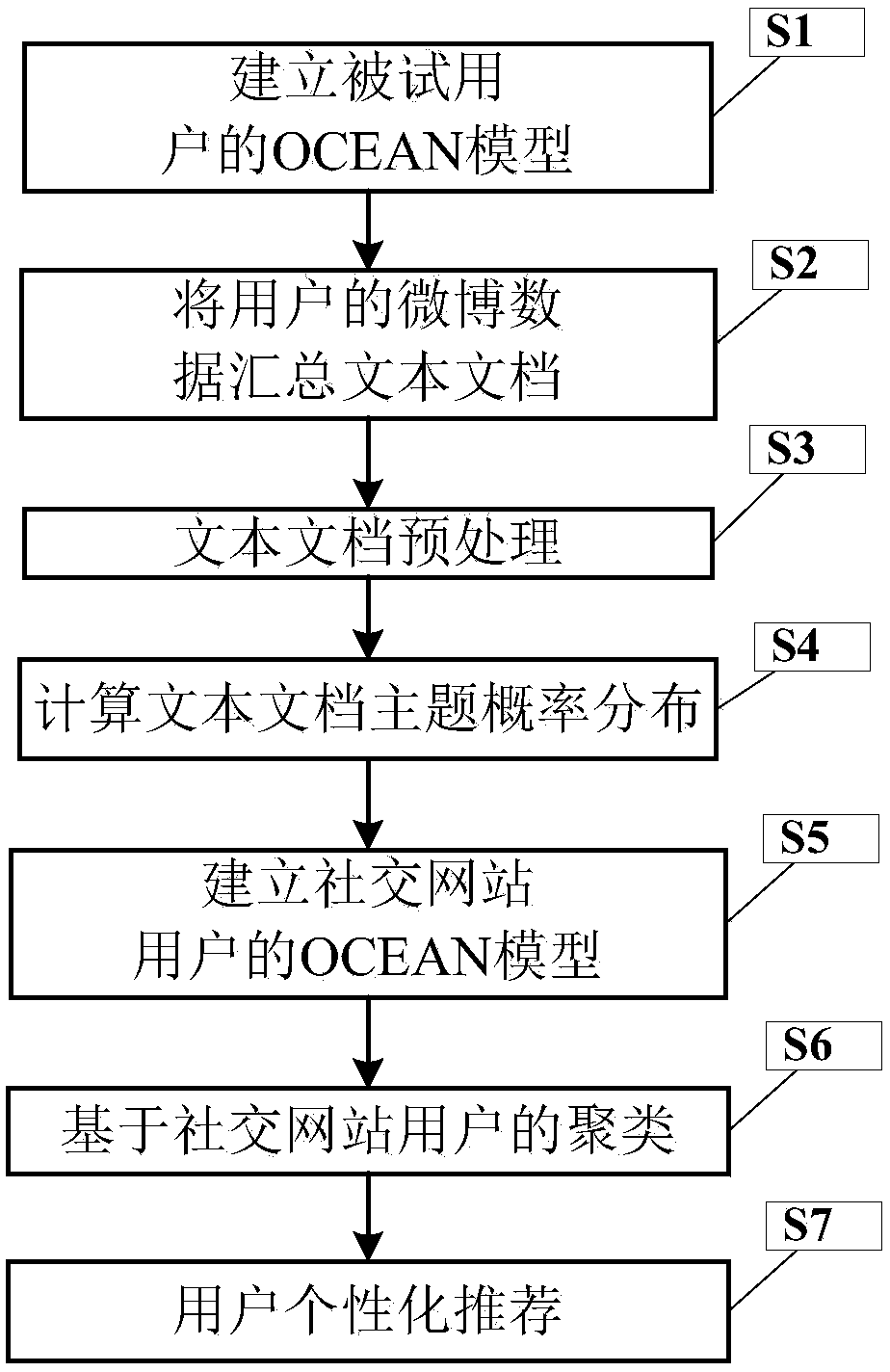
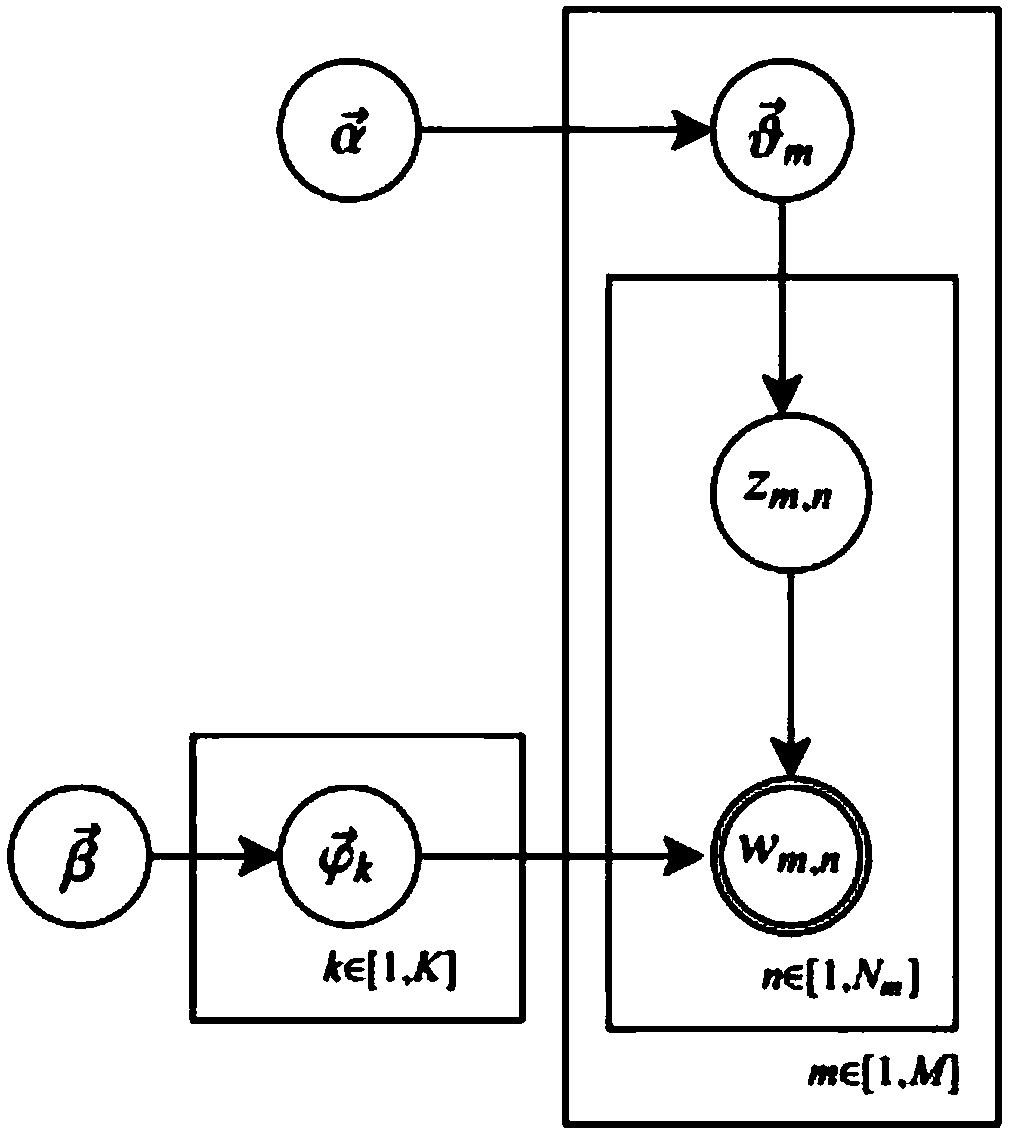
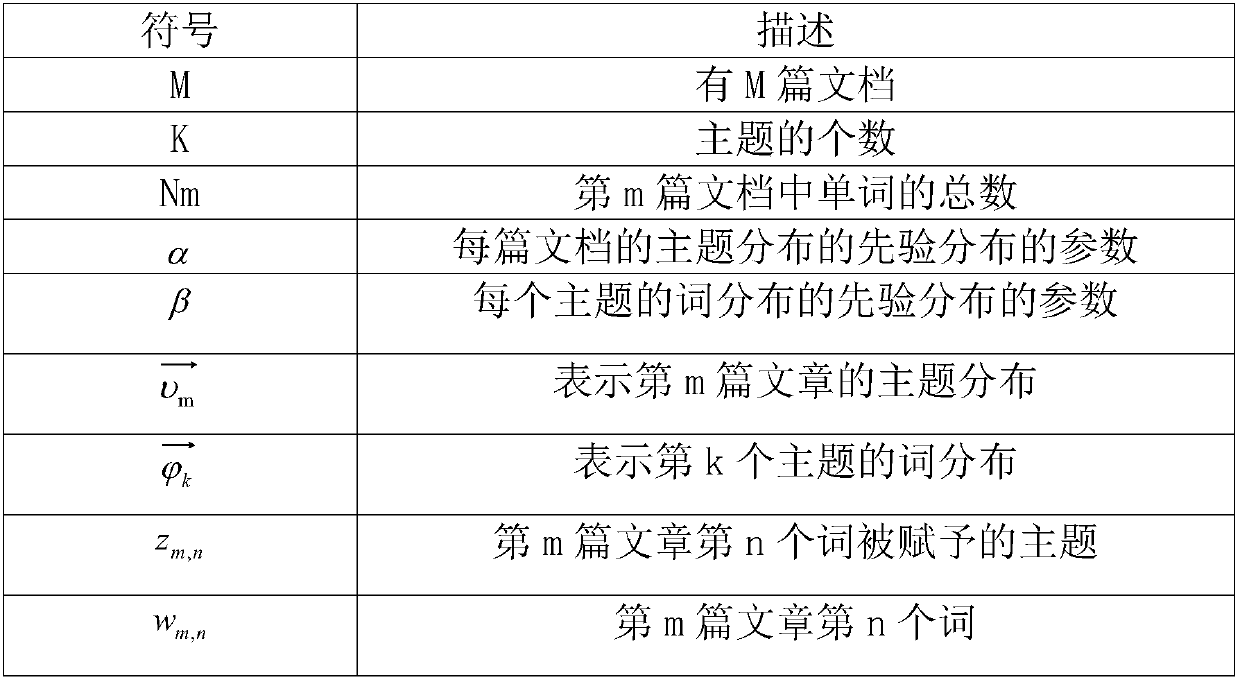
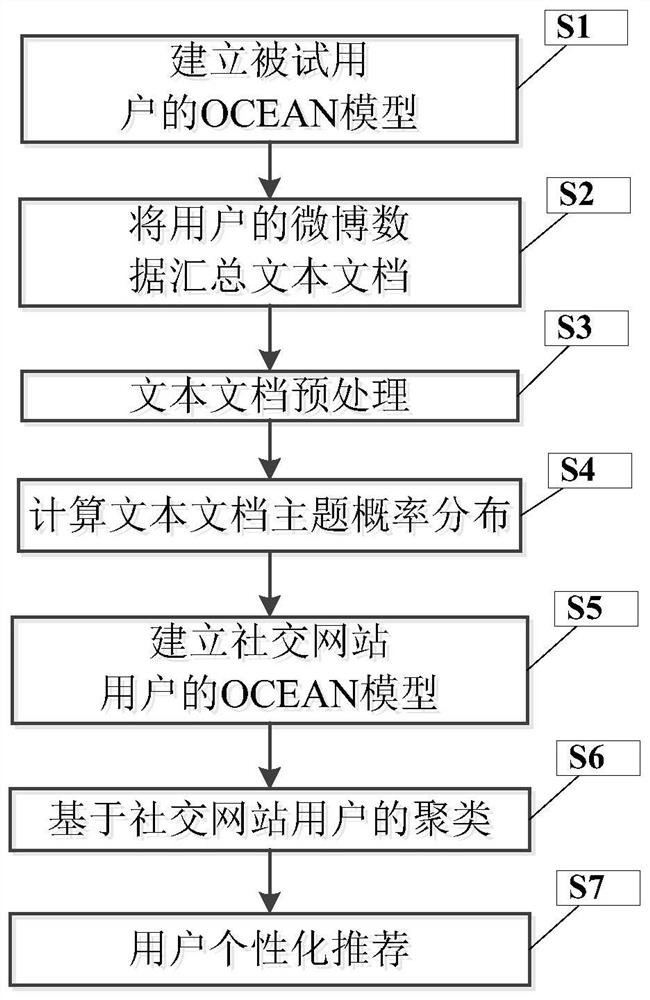
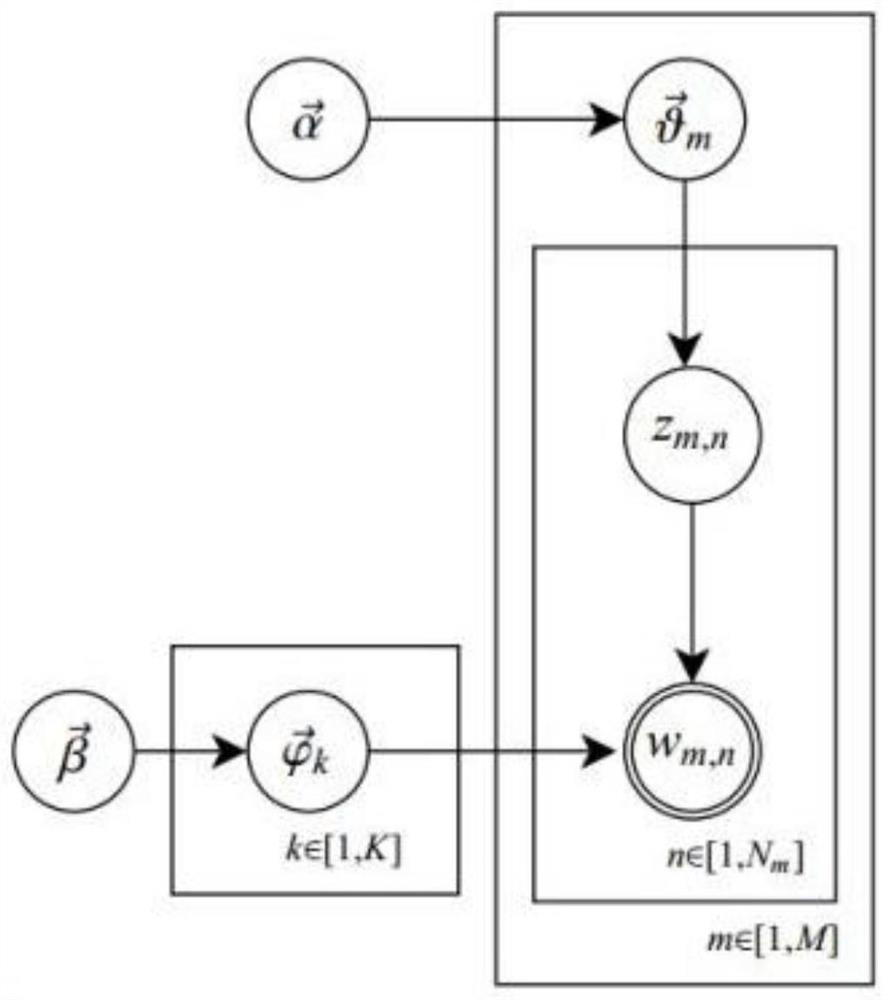
Personalized recommendation method based on OCEAN model
ActiveCN107895303AFit psychologyImprove accuracyBuying/selling/leasing transactionsSpecial data processing applicationsPersonalizationMicroblogging
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA
TDOA (Time difference of Arrival)-IMU (Inertial Measurement Unit) data adaptive fusion positioning device and method
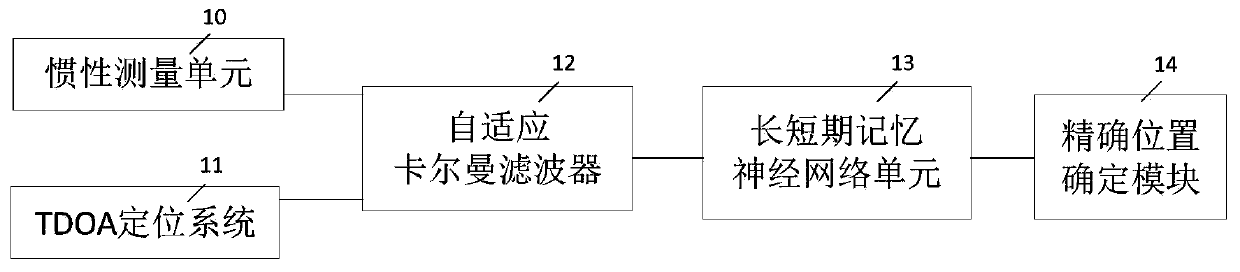
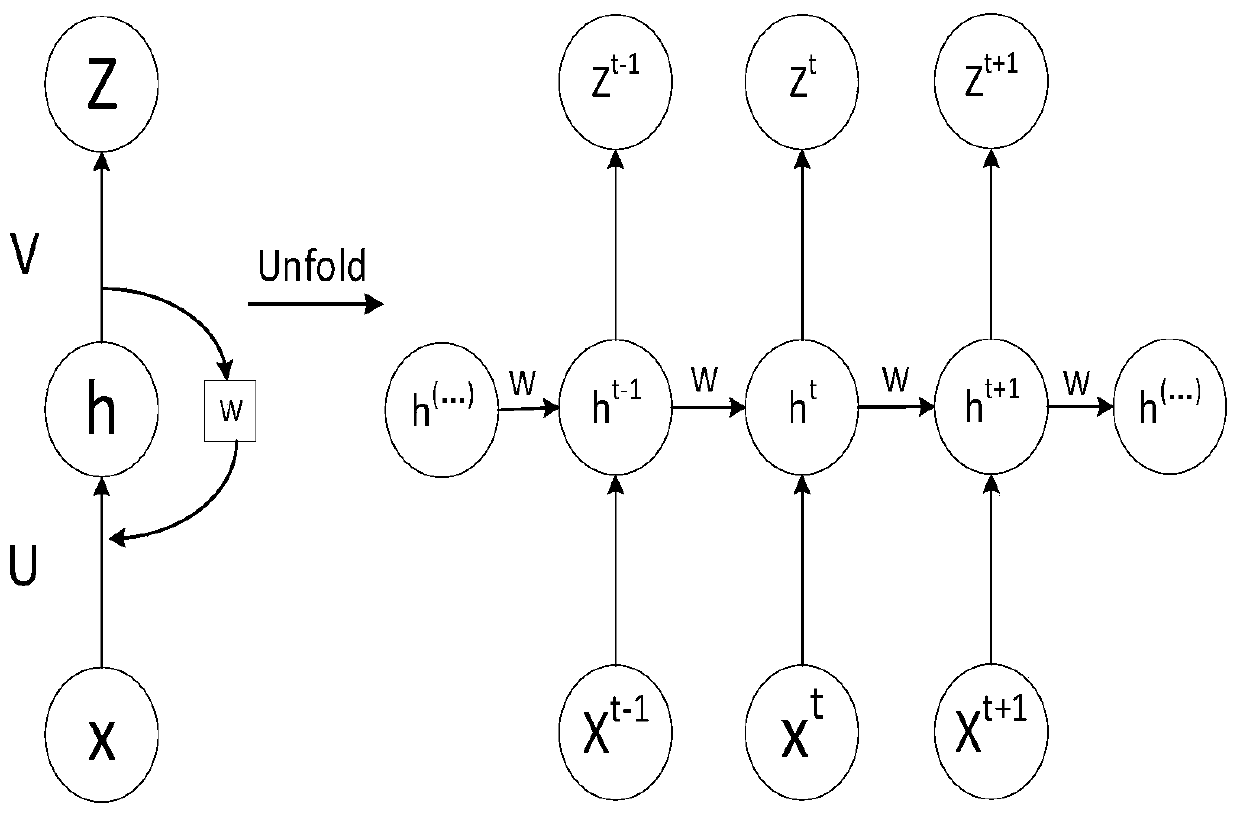
InactiveCN110208740AHigh positioning accuracyReduce glitchesNavigation by terrestrial meansNavigation by speed/acceleration measurementsNetwork modelPositioning system
The invention discloses a TDOA (Time difference of Arrival)-IMU (Inertial Measurement Unit) data adaptive fusion positioning device and method. The device comprises an inertial measurement unit used for acquiring components of the speed and the acceleration of a measured object relative to a geographic coordinate system; a TDOA positioning system used for preliminarily positioning the measured object so as to obtain position information of the measured object; an adaptive Kalman filter used for filtering a TDOA positioning system according to an adaptive Kalman algorithm, and correcting the position information acquired by the TDOA positioning system; a long short-term memory neural network unit used for predicting the position of the measured object according to a long short-term memory neural network model; and an accurate position determination module used for carrying out weighted averaging on the position information corrected by the adaptive Kalman filter and the position information predicted by the long short-term memory neural network unit, and an average value is a positioning result. The TDOA-IMU data adaptive fusion positioning device and the method provided by the invention can improve the positioning accuracy.
Owner:BEIJING SMARTCHIP MICROELECTRONICS TECH COMPANY +3
Method for fast testing contents of chemical ingredients in Eucalyptus urophylla*E. tereticornis wood
InactiveCN106383094ARapid determinationAvoid damageMaterial analysis by optical meansAdditive ingredientMathematical model
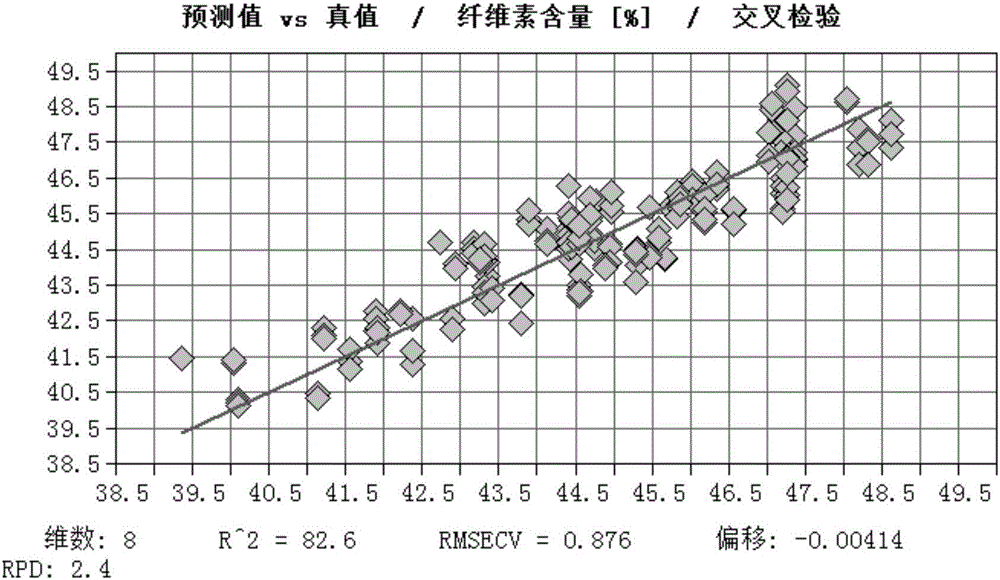
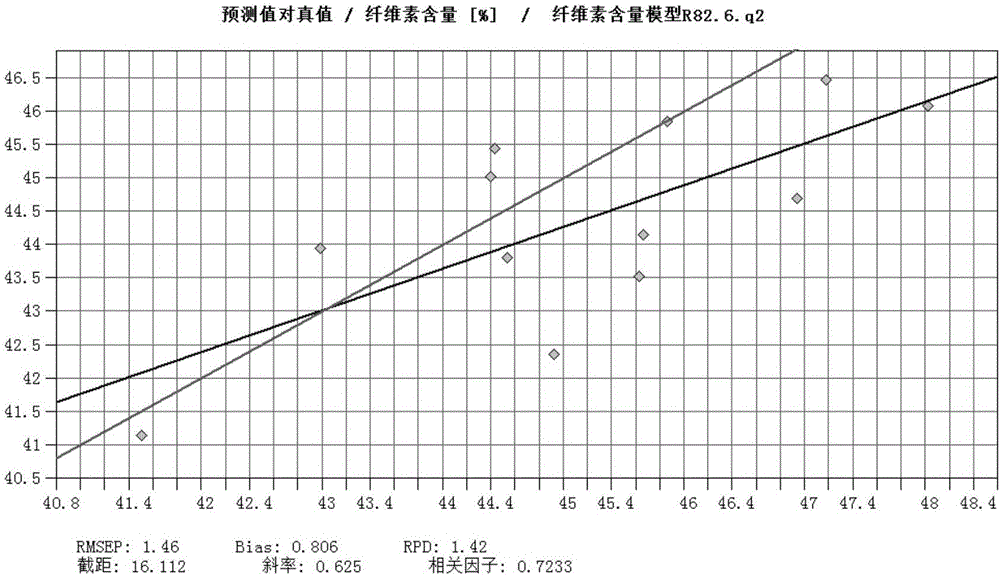
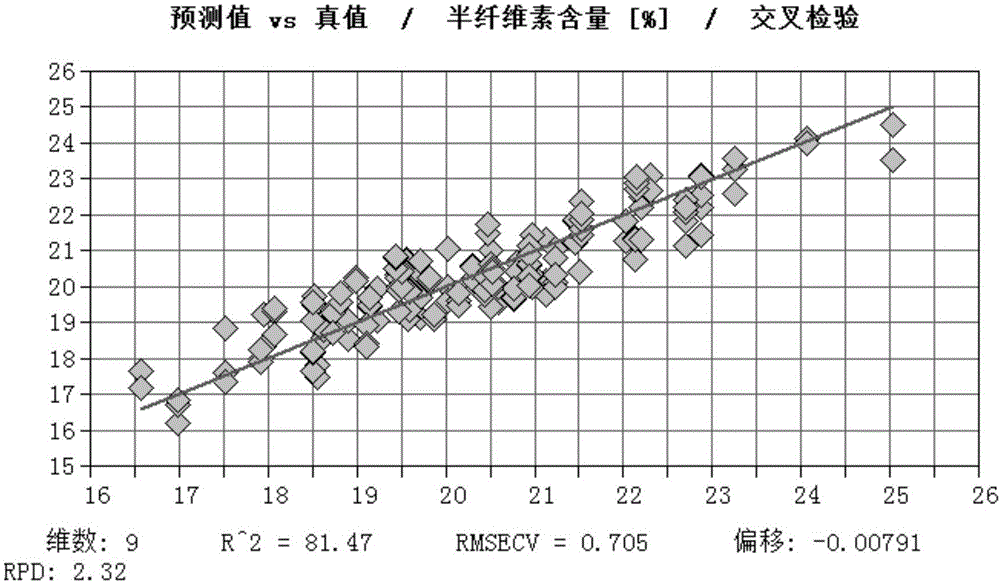
The invention discloses a method for fast testing the contents of chemical ingredients in Eucalyptus urophylla*E. tereticornis wood. According to the method, a mathematical model is built by using the existing near infrared spectral feature absorption peak of an Eucalyptus urophylla*E. tereticornis wood sample and the relationship between contents of each chemical ingredient, and the goal of fast testing the content of other wood chemical ingredients to be tested in Eucalyptus urophylla*E. tereticornis wood is achieved. The method comprises the following steps of (1) sample sampling; (2) sample crushing and near infrared spectrum collection; (3) sample wood chemical ingredient standard measurement; (4) spectrum treatment and modeling; (5) model inspection; (6) model application. The method for fast testing the contents of ingredients of cellulose, hemicelluloses and lignin of the Eucalyptus urophylla*E. tereticornis wood has the advantages that the implementation is simple, convenient and fast; the accuracy and the reliability are high; the test cost can be obviously reduced; the tree damage and the chemical medicine consumption are reduced, so that the goal of testing the contents of the chemical ingredients in a great amount of Eucalyptus urophylla*E. tereticornis wood can be achieved.
Owner:RES INST OF TROPICAL FORESTRY CHINESE ACAD OF FORESTRY
Sample clustering and feature recognition method based on integrated non-negative matrix factorization
ActiveCN110826635AImprove performanceImprove robustnessCharacter and pattern recognitionICT adaptationAlgorithmOmics
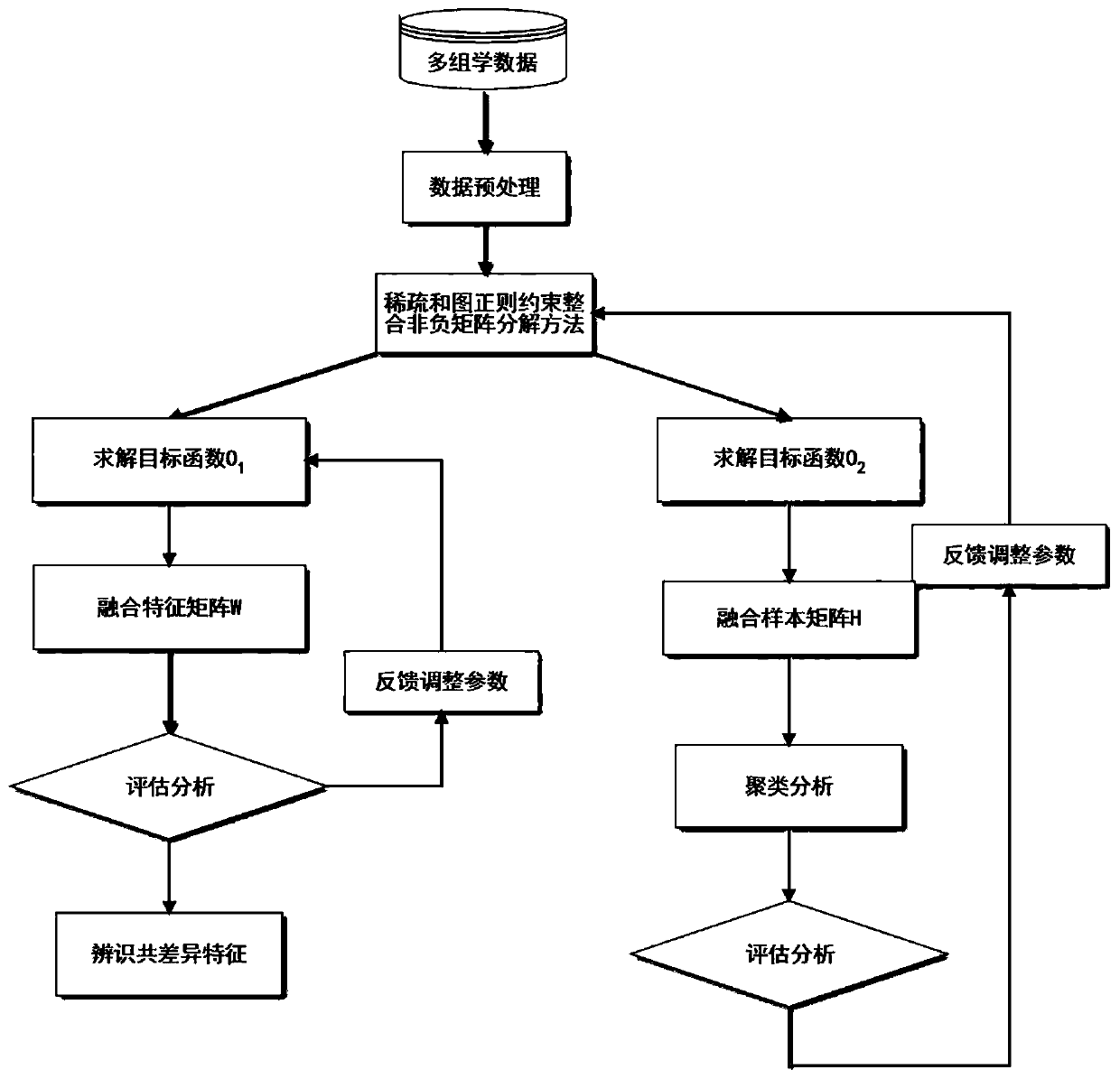
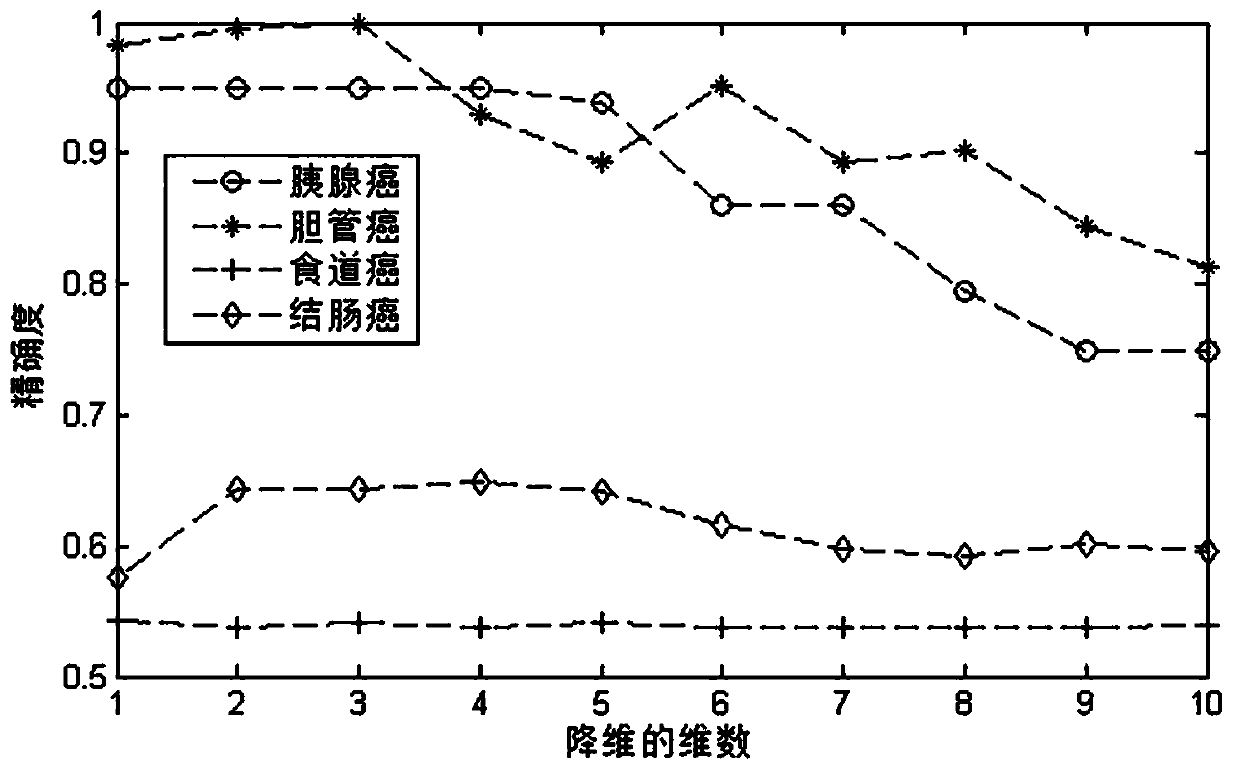
The invention discloses a sample clustering and feature recognition method based on integrated non-negative matrix factorization. The method comprises: 1, X = {X1, X2... XP} representing multi-view data composed of P different omics data matrixes of the same cancer; 2, constructing a diagonal matrix Q; 3, introducing graph regularization and sparse constraints into the integrated non-negative matrix factorization framework to obtain target functions O1 and O2; 4, solving the target function O1 to obtain a fusion feature matrix W and a coefficient matrix HI; solving the target function O2 to obtain a feature matrix WI and a fusion sample matrix H; 5, constructing an evaluation vector according to the fusion feature matrix W, and identifying common difference features according to the vector; 6, performing functional explanation on the identified common difference characteristics by using GeneCards; and 7, performing sample clustering analysis according to the fusion sample matrix. According to the method, the complementary and difference information of the multiple omics data can be fully utilized to identify the common difference characteristics, clustering analysis can be carriedout on the sample data provided by the multiple omics data, and a calculation method basis is provided for integrated research of different types of omics data.
Owner:QUFU NORMAL UNIV
Processing depth data of a three-dimensional scene
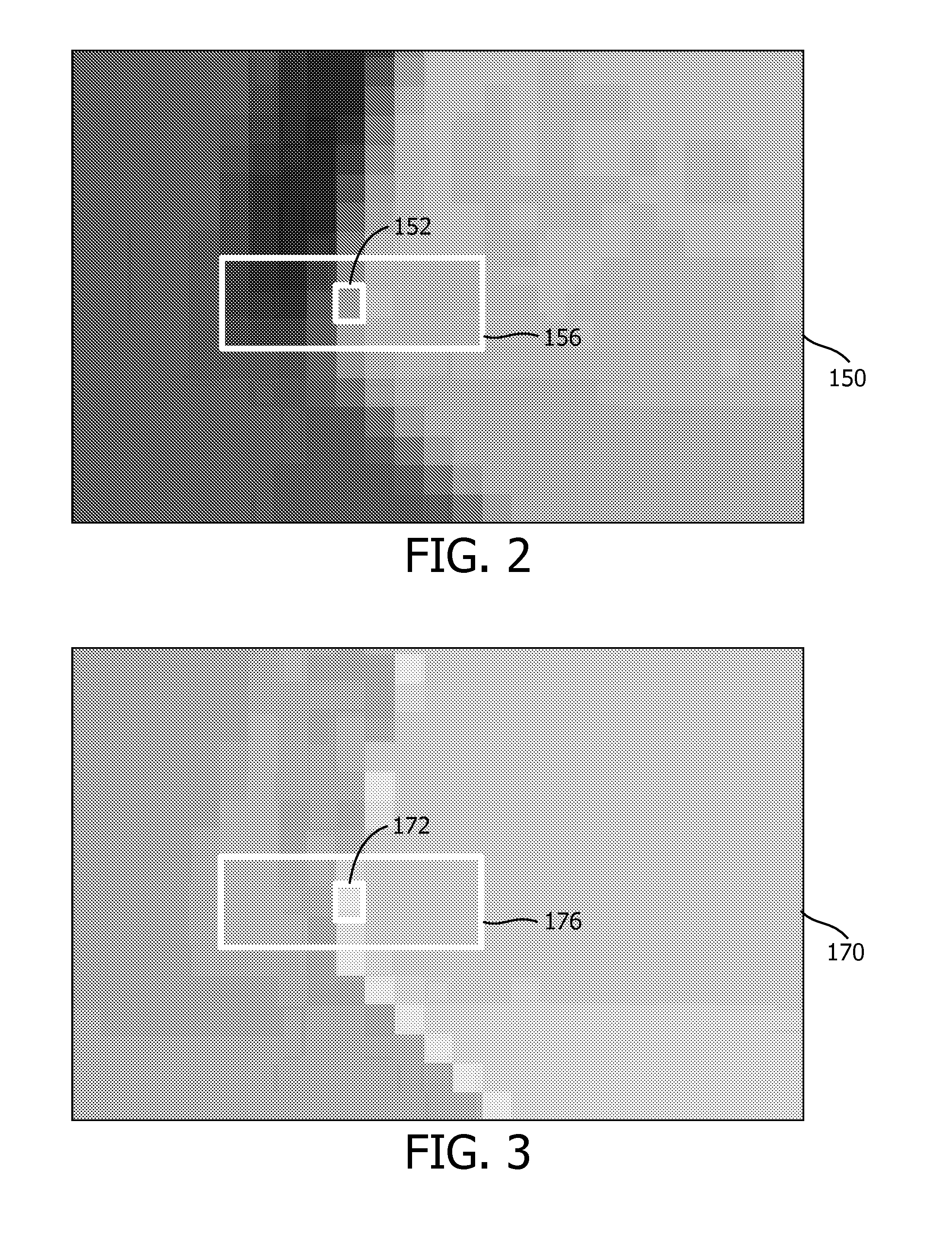
InactiveUS20130321571A1Improved reduction of artifactReduce outliersSteroscopic systemsPattern recognition3d image
The invention relates to a signal processing device (100) for processing three-dimensional [3D] image data, the 3D image data comprising two-dimensional [2D] image data (150) and thereto corresponding 2D depth data (170), and the signal processing device comprising an input (120) for receiving the 2D image data and the 2D depth data, a first outlier detector (160) for establishing a statistical depth deviation (178) of a depth value from other depth values in a first spatial neighborhood of the 2D depth data, a second outlier detector (140) for establishing a statistical image deviation (158) of an image value from other image values in a second spatial neighborhood of the 2D image data, the depth value representing a depth of the image value, and a depth value generator (180) for, in dependence on the statistical depth deviation and the statistical image deviation, providing a replacement depth value (190) when the statistical depth deviation exceeds the statistical image deviation, the replacement depth value having a lower statistical depth deviation than the depth value.
Owner:KONINKLIJKE PHILIPS ELECTRONICS NV
Multi-beam sounding signal processing method and device
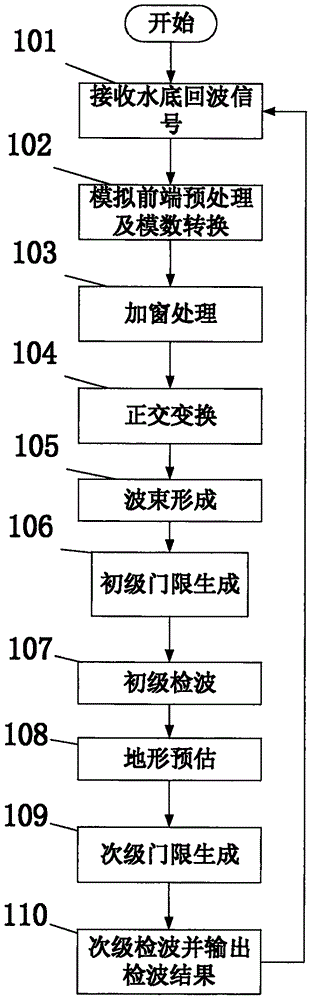
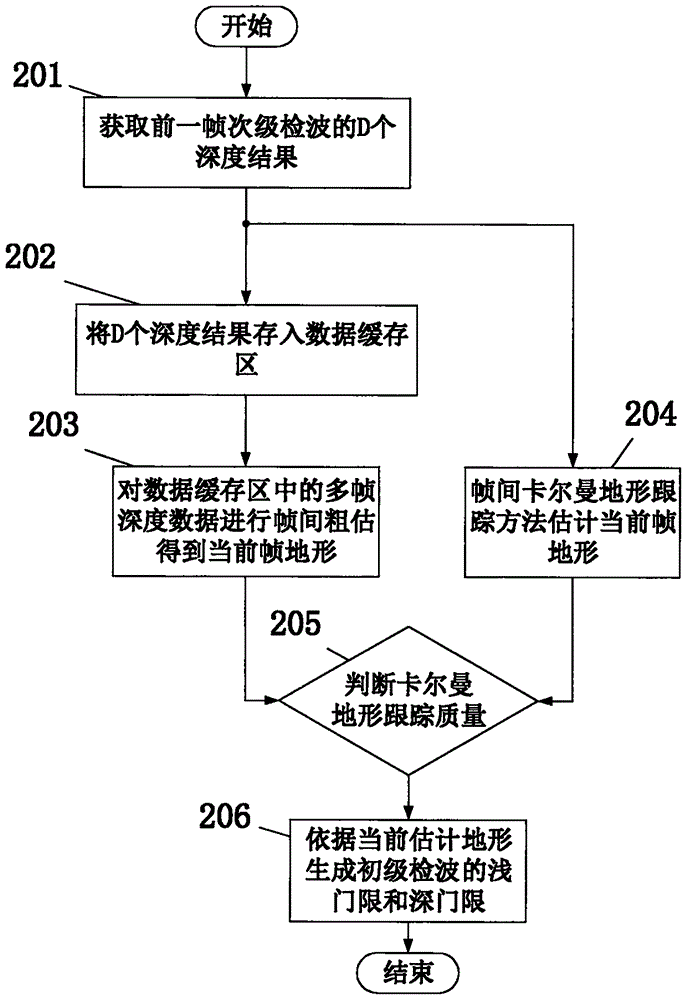
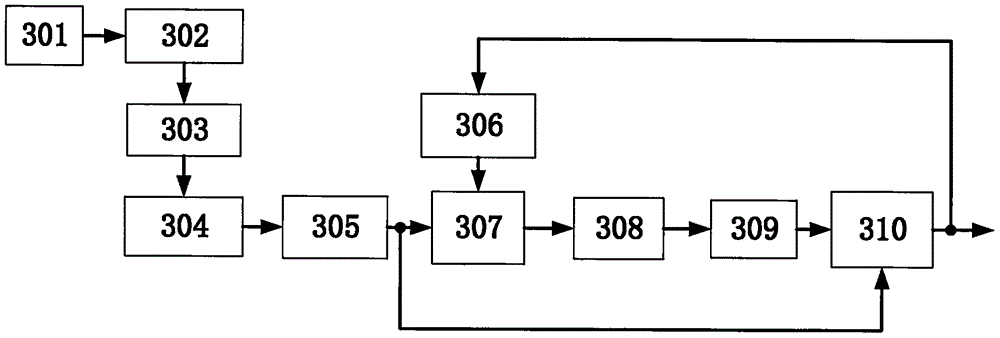
InactiveCN106526601AEffective automatic trackingEliminate distractionsWater resource assessmentAcoustic wave reradiationTerrainEngineering
The invention discloses a multi-beam sounding signal processing method and device. The device comprises a receiving transducer array, an analog front-end preprocessing unit, an analog-to-digital conversion unit, a multi-channel data preprocessing unit, a beam forming unit, a primary threshold generation unit, a primary detection unit, a terrain estimation unit, a secondary threshold generation unit, and a secondary detection unit, wherein the primary threshold generation unit carries out Kalman terrain tracking according to a previous frame terrain result so as to generate the detection threshold of a current frame terrain, at the same time the terrain estimation unit, the secondary threshold generation unit and the secondary detection unit are orderly connected to realize secondary detection, and the accuracy of terrain detection is effectively improved. According to the multi-beam sounding signal processing method and device, the terrain detection can be automatically carried out in an actual underwater terrain survey, the frequent manual threshold adjustment operation in a conventional method is reduced, the field operation and office data processing workload of the multi-beam sounding device are reduced greatly, and the method and the device can be widely applied to the field of underwater topographic survey.
Owner:BEIJING HYDRO TECH MARINE TECH CO LTD
Method of determining physical properties of Eucalyptus cloeziana
InactiveCN106248613AAvoid damageRapid determinationMaterial analysis by optical meansCompressive strengthPhysical property
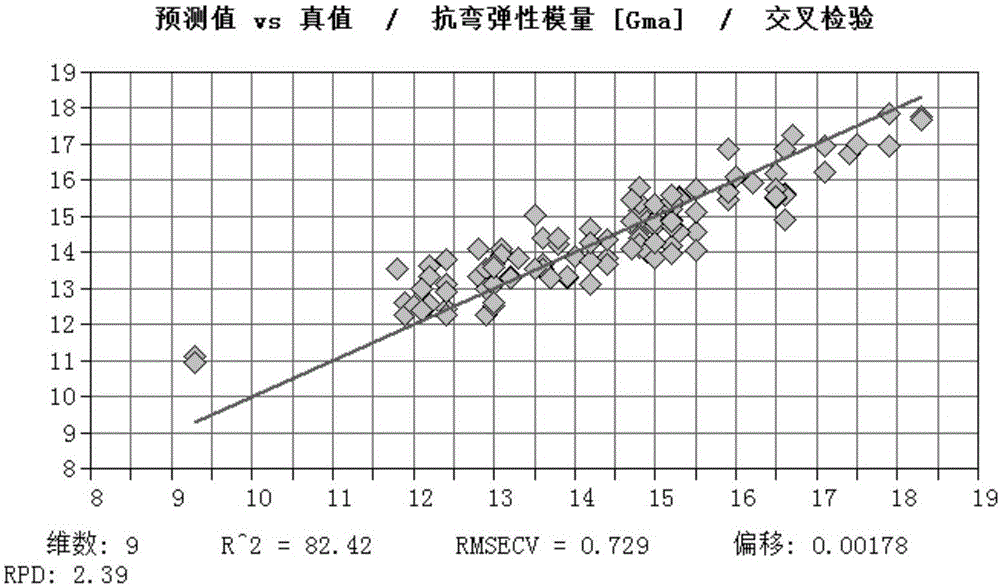
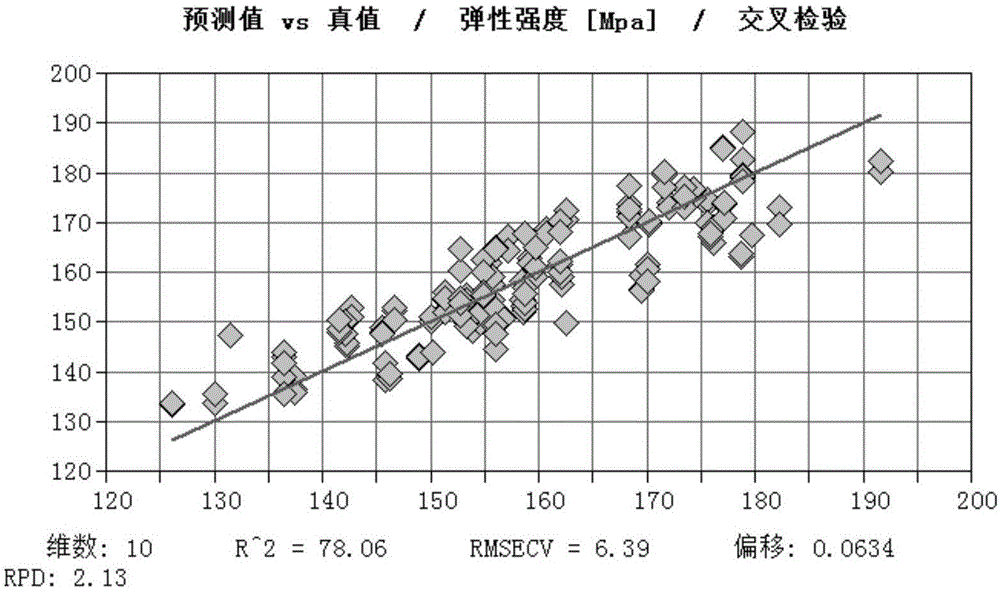
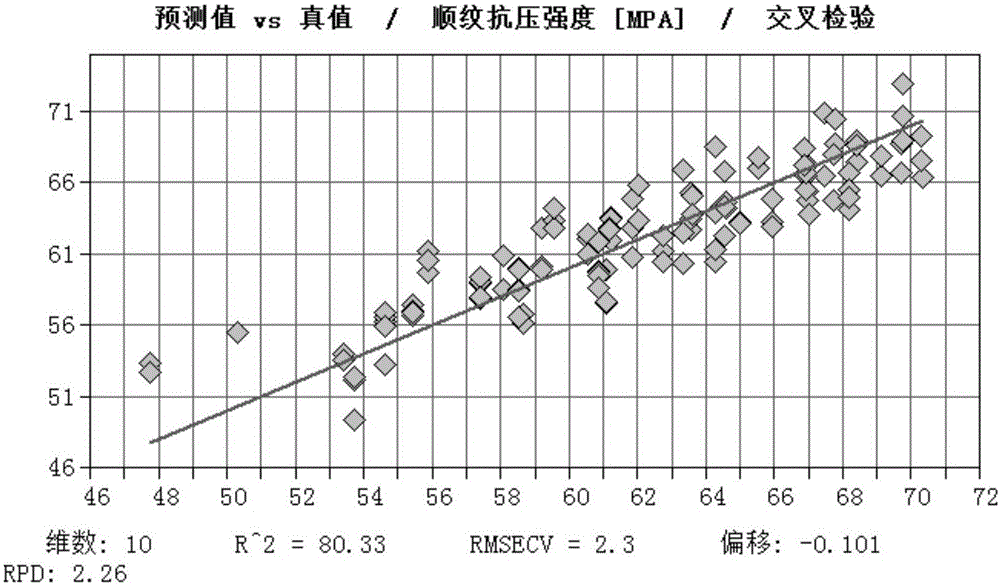
The invention discloses a method of determining physical properties of Eucalyptus cloeziana, comprising the steps of (1), collecting a sawdust sample; (2), subjecting the sawdust sample to near-infrared spectral scanning; (3), standardly determining various physical properties of wood; (4), carrying out spectral pretreatment and establishing and optimizing a model; (5), carrying out model outside inspection and selection; (6), applying the model. The method of determining modulus of elasticity, bending strength and parallel-to-grain compressive strength of Eucalyptus cloeziana according to the invention not only can provide significantly improved operating efficiency and greatly saved manpower, material and financial resources, can also ensure reduced damage to woods and give full play to the advantages of near-infrared rapid analysis, and therefore, quick determination is achieved for property indexes of large batch of wood samples of Eucalyptus cloeziana.
Owner:RES INST OF TROPICAL FORESTRY CHINESE ACAD OF FORESTRY +1
Data detection method, device and equipment and storage medium
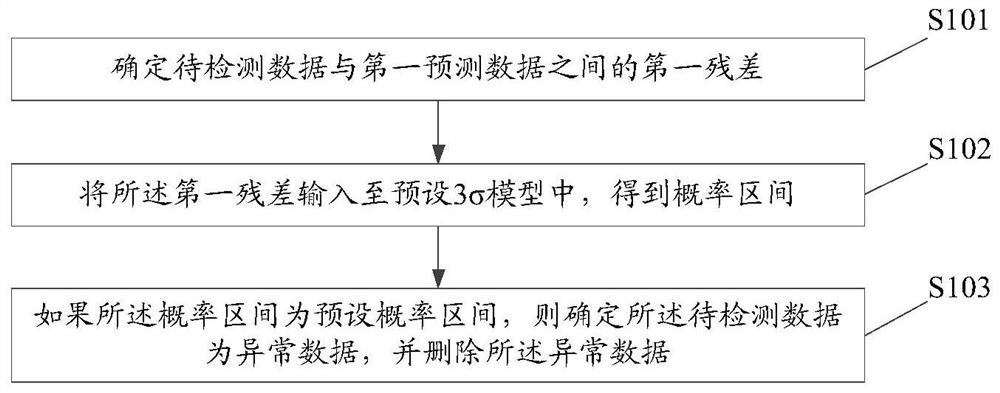
PendingCN112182056AReduce limitationsAccurate detectionDigital data information retrievalNeural architecturesTime segmentSigma model
An embodiment of the invention discloses a data detection method and device, equipment and a storage medium. The method comprises the steps: determining a first residual error between to-be-detected data and first prediction data, wherein the first prediction data is data obtained by predicting first historical data in a first preset historical time period by using a data prediction model; inputting the first residual error into a preset 3 sigma model to obtain a probability interval; if the probability interval is a preset probability interval, determining that the to-be-detected data is abnormal data, and deleting the abnormal data, wherein the influence of an abnormal value and noise can be reduced to adapt to more different data models, and the use limitation of the data detection method is reduced.
Owner:CHINA MOBILE SUZHOU SOFTWARE TECH CO LTD +1
Network reconstruction method and device
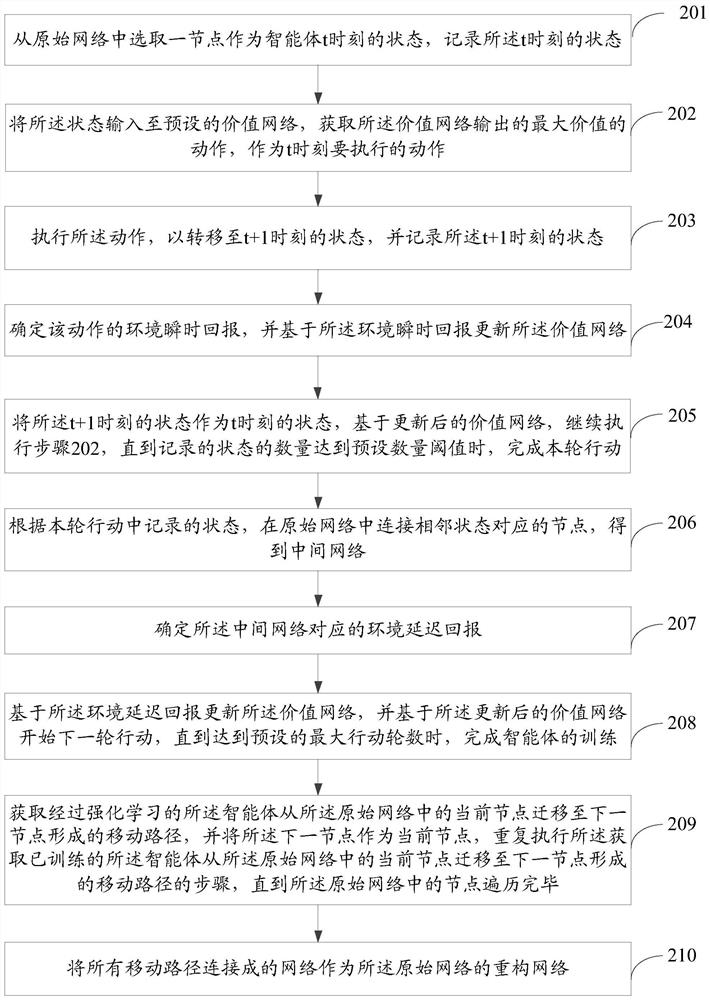
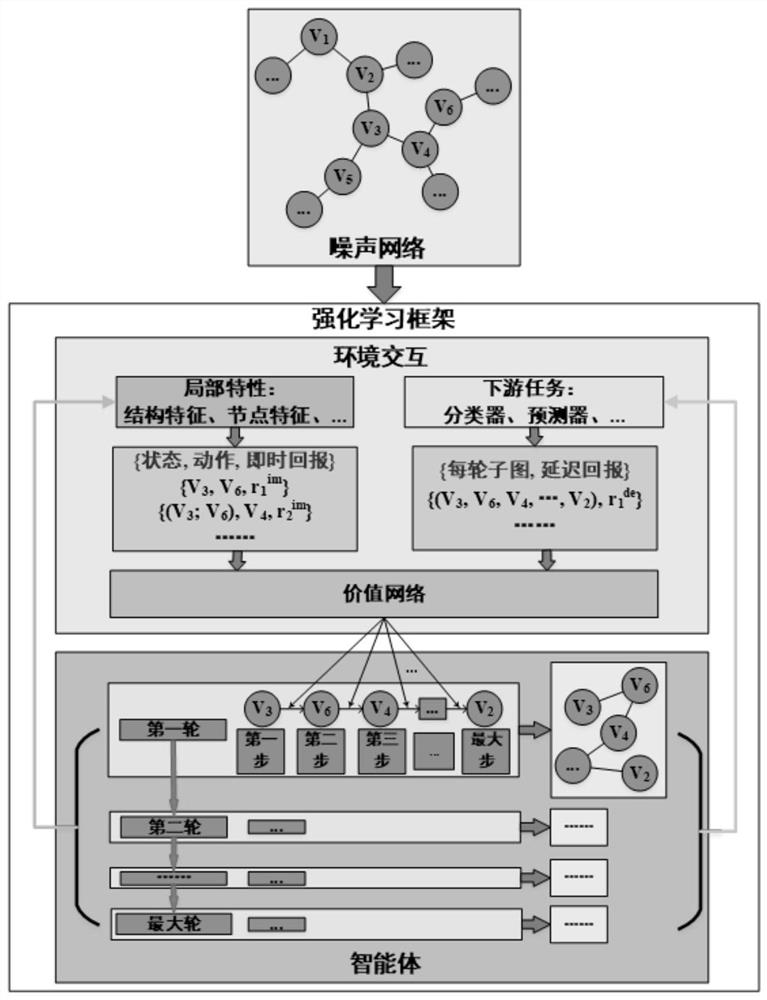
PendingCN112149835AReduce noiseReduce outliersCharacter and pattern recognitionMachine learningPathPingTopology information
The invention provides a network reconstruction method and device, and the method comprises the steps: representing the state and action of an intelligent agent through a node in an original network,and carrying out the reinforcement learning of the intelligent agent in the original network based on environment return information, enabling the intelligent agent to learn structural topology information in the original network and rules for removing noise information in the original network; and obtaining a moving path formed by migrating the agent subjected to reinforcement learning from a current node to a next node in the original network, taking the next node as the current node, and repeatedly executing the step of obtaining the moving path until the nodes in the original network are traversed; taking the network formed by connecting all the moving paths as the reconstruction network of the original network, wherein the reconstruction network can effectively reduce noise and abnormal edges of the original network while reserving an important topological structure of the original network, and meanwhile some potential relation edges are added.
Owner:HANGZHOU HIKVISION DIGITAL TECH
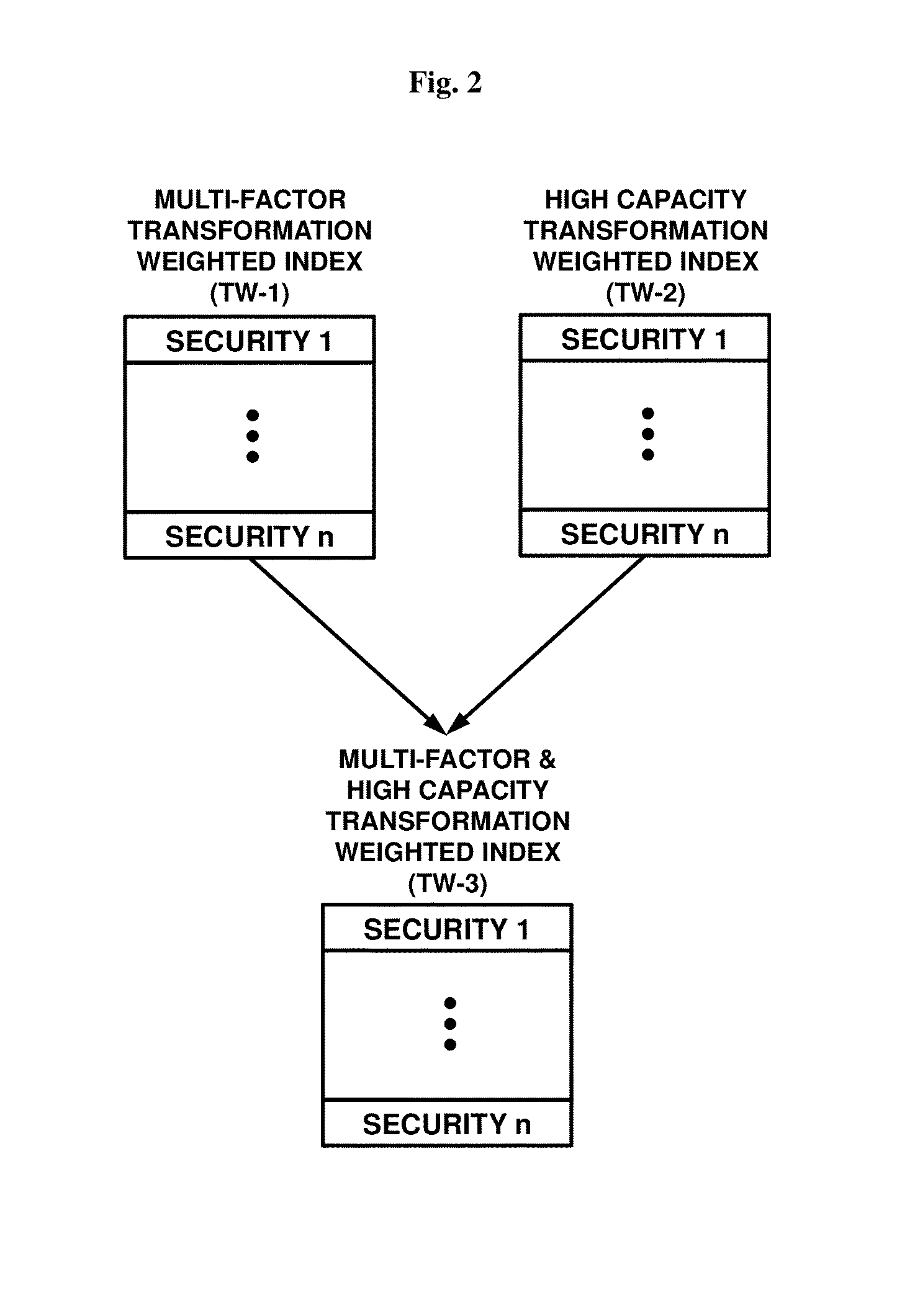
Transformation weighted indexes offering concentrated multi-risk factor exposure
InactiveUS20140108299A1Reduce decreaseSimplify the management processFinanceData transformationData mining
Computer-based systems, software, and computer-implemented methods for creating an index of securities based upon various data transformations of risk factor metrics regarding entities or securities associated with the entities and weighting each index member in proportion to its combined transformed weighting value.
Owner:GRAHAM INVESTMENT MANAGEMENT INC
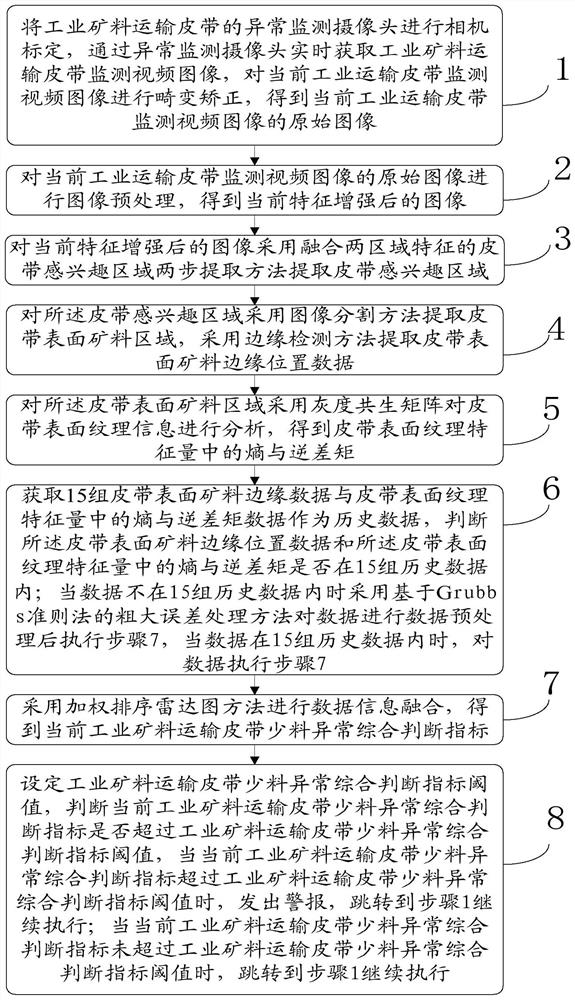
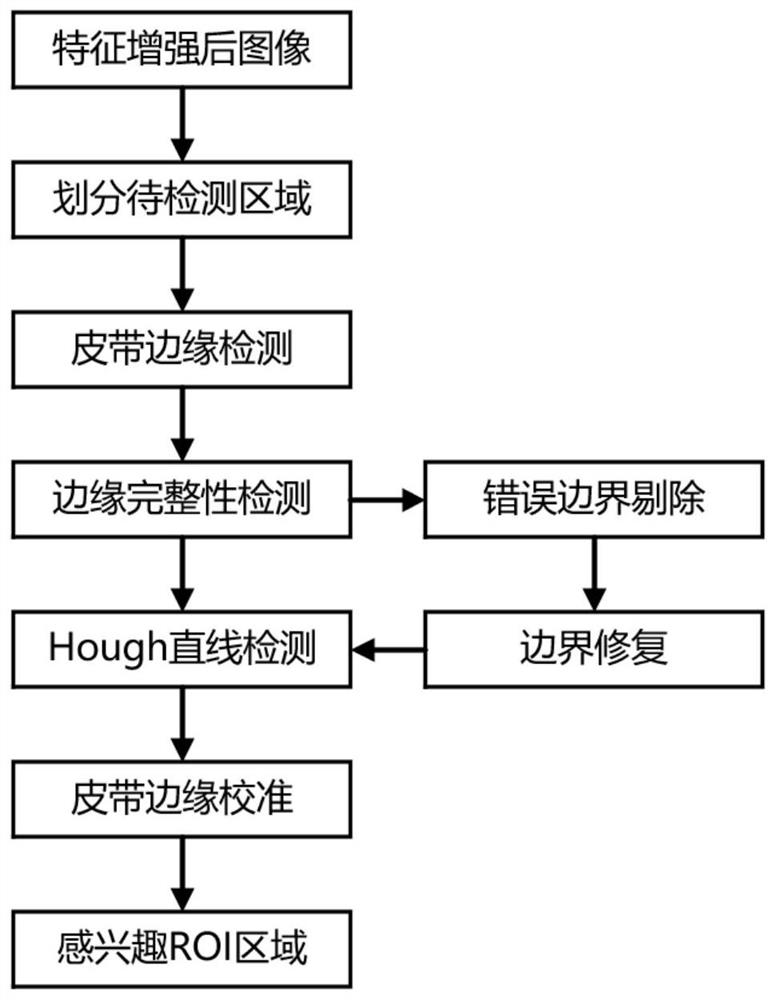
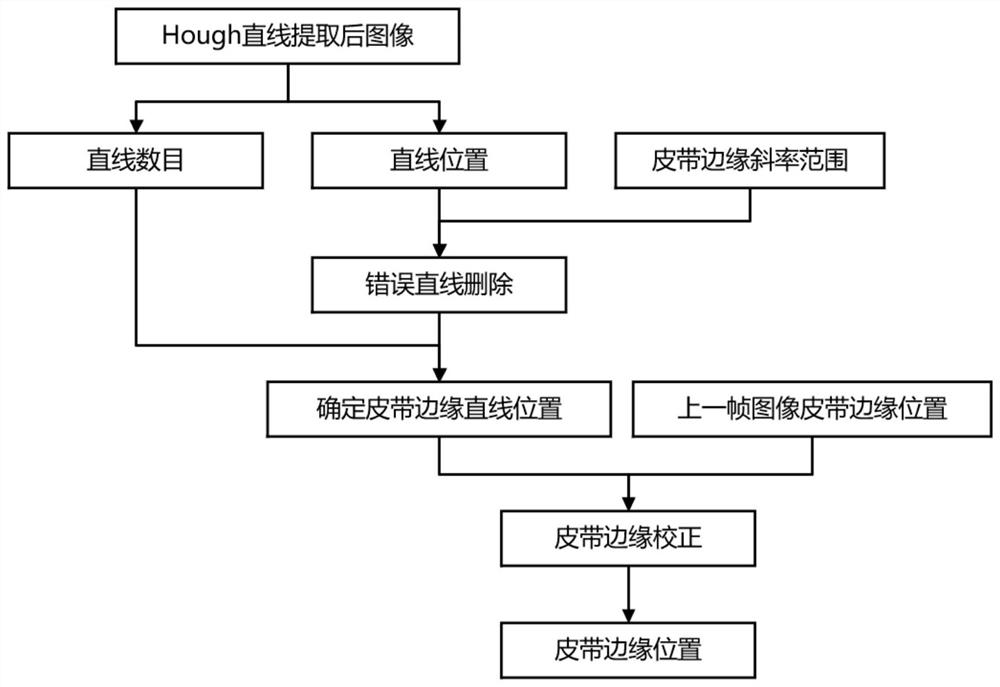
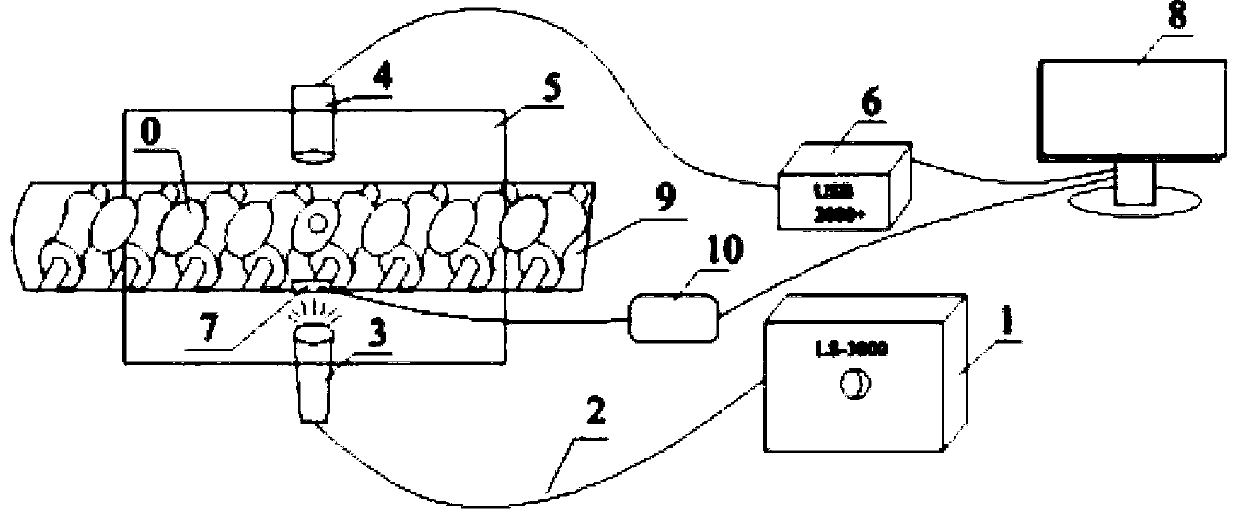
Industrial mineral aggregate conveying belt material shortage abnormity monitoring method based on machine vision
ActiveCN113283339ALow material abnormality monitoringReduce outliersImage analysisCharacter and pattern recognitionMachine visionMining engineering
The invention provides an industrial mineral aggregate conveying belt material shortage abnormity monitoring method based on machine vision, and the method comprises the steps: obtaining an industrial conveying belt monitoring video image, carrying out the feature enhancement of an abnormity monitoring image in a complex environment, employing a belt region-of-interest two-step extraction method fusing the features of two regions, extracting a belt region-of-interest, adopting an edge detection method to extract the edge of the mineral aggregate on the surface of the belt, adopting a gray-level co-occurrence matrix to analyze the texture information of the surface of the belt, and adopting a gross error processing method based on a Grubbs criterion method to carry out data preprocessing on the extracted image feature quantity; and fusing the belt surface texture information and the mineral aggregate edge information by adopting a weighted sorting radar map method, and judging the material shortage abnormity of the industrial mineral aggregate conveying belt. According to the method, the material shortage abnormity of the industrial mineral aggregate conveying belt can be judged faster, more accurately and more comprehensively, and the method has great significance in improving the production efficiency and reducing the abnormal influence range.
Owner:CENT SOUTH UNIV
An online nondestructive detection method for blood-spotted eggs based on spectral amplitude space conversion
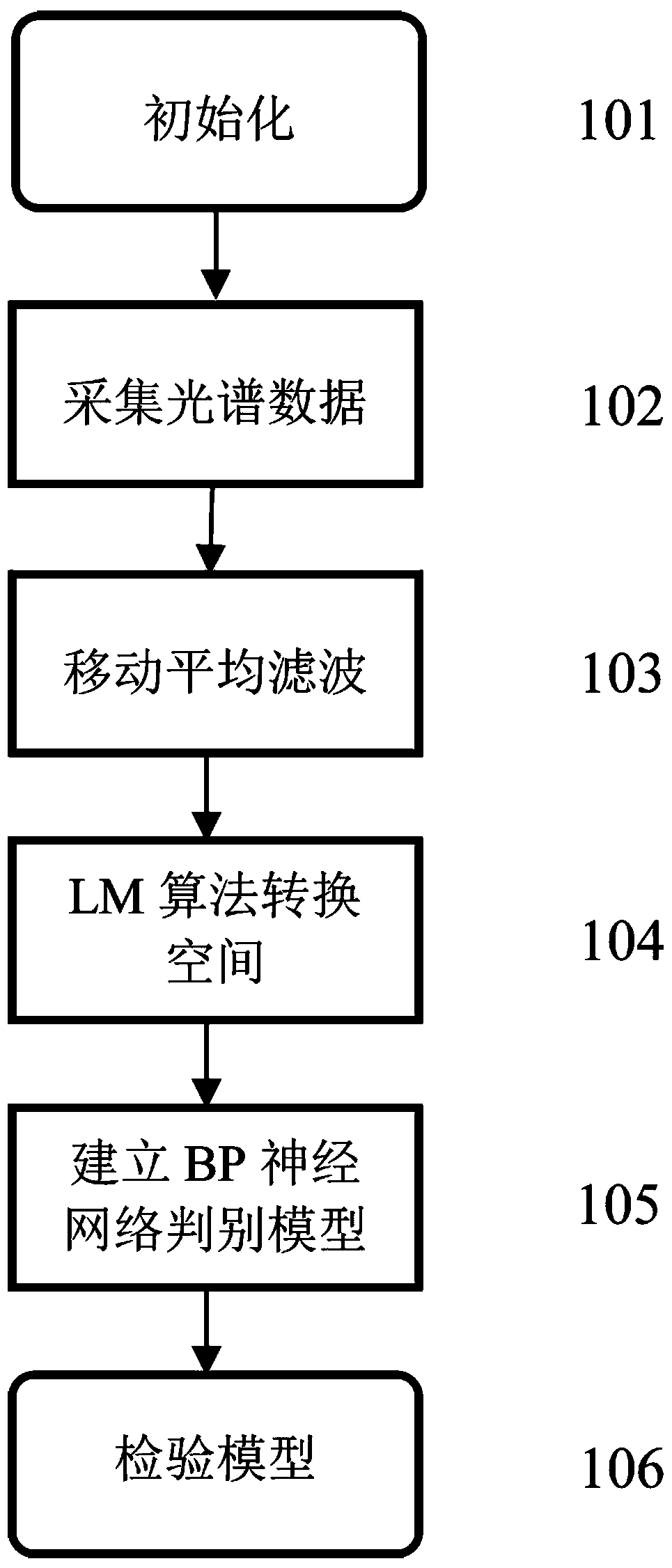
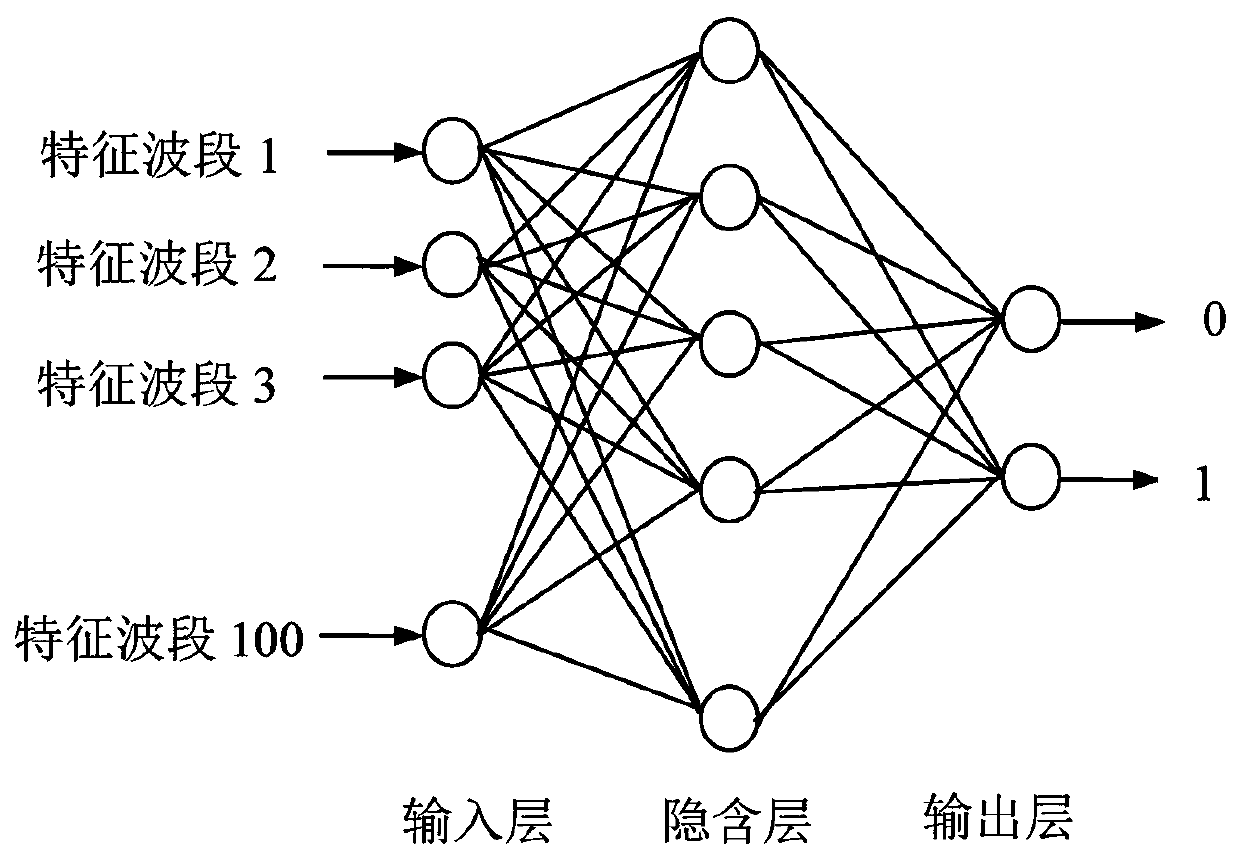
ActiveCN107064021BRealize online detectionReduce volatilityImage analysisOptically investigating flaws/contaminationSpectral transmissionNon destructive
The invention relates to the technical field of egg quality dynamic detection, in particular to a blood spot egg online nondestructive detection method based on spectrum amplitude value space conversion. The method mainly comprises the following steps of collecting the spectral transmission rate of each egg sample through a blood spot egg online detection system to obtain spectral data; performing preprocessing on the spectral data; in order to reduce the interference caused by the speed, the batch, the instrument and the environment in the experiment process, converting spectral signals of the preprocessed data into an artificially specified amplitude value space by using an LM algorithm; obtaining the converted processing data; building a blood spot egg BP neural network online discriminating model; inspecting the model. The invention provides the intelligent nondestructive detection method; the precision and the applicability of the model are improved; high automation and intelligence are realized in the whole detection process; the wide market prospects are realized.
Owner:HUAZHONG AGRI UNIV
CPS delay jitter buffering method
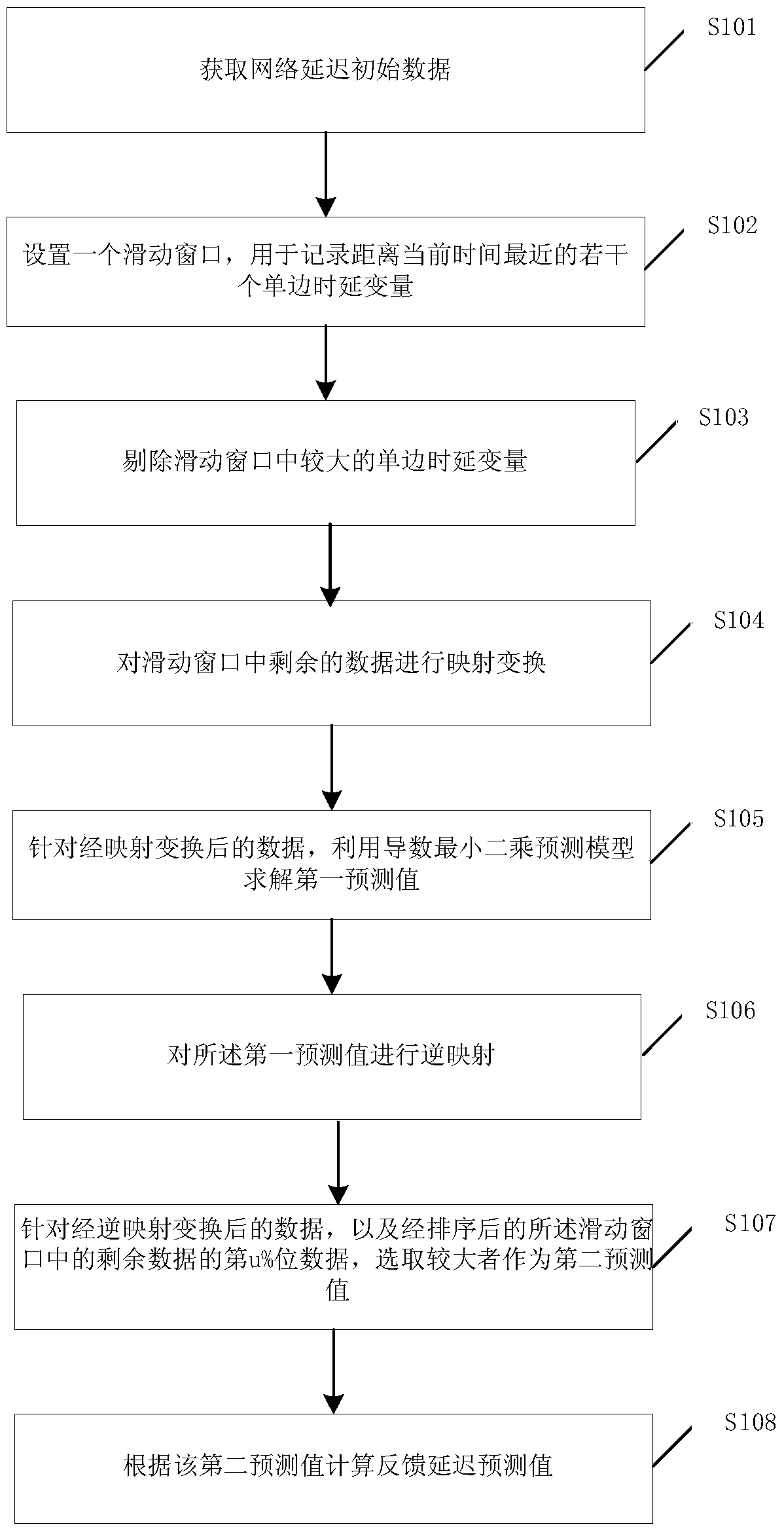
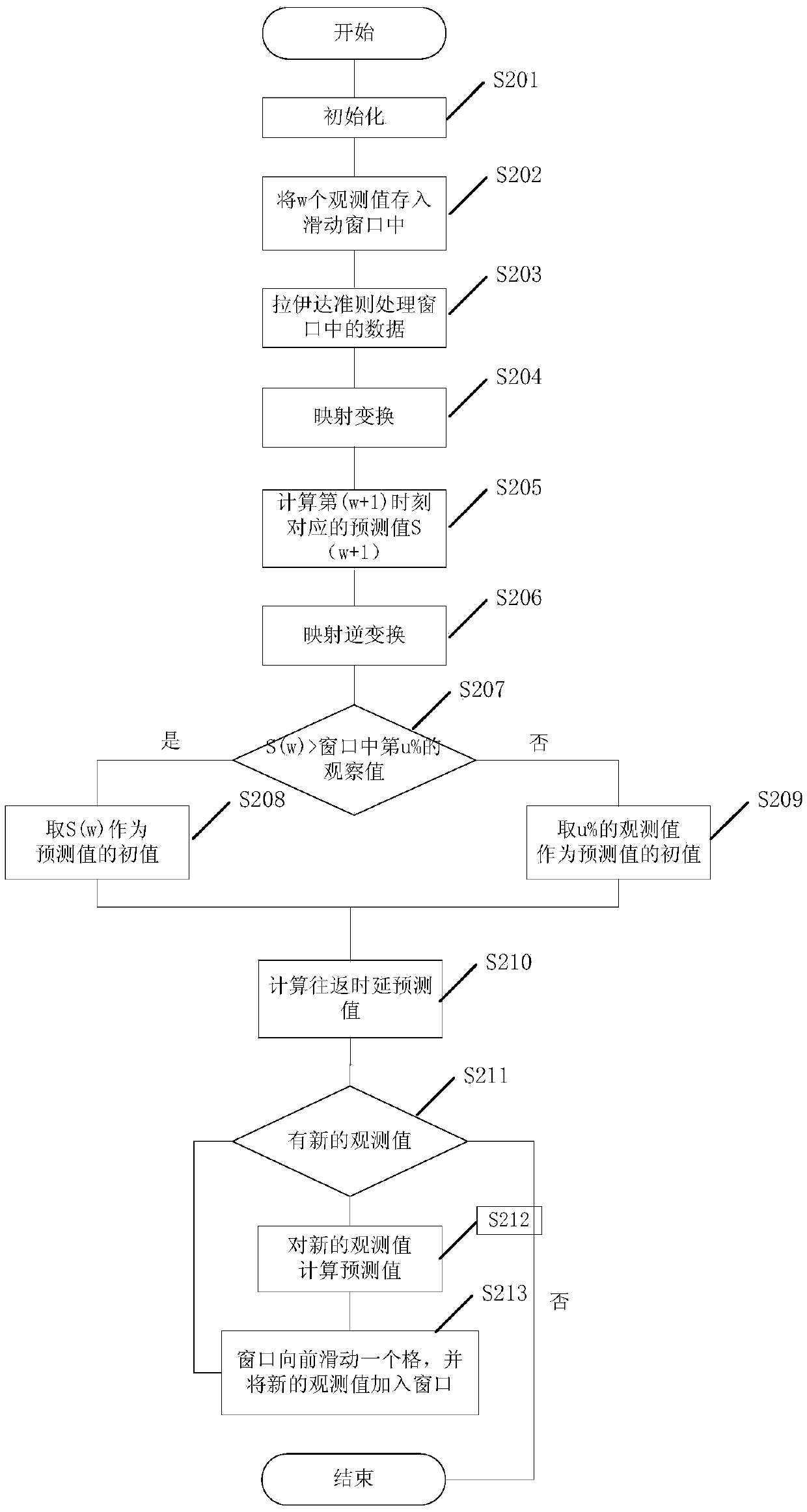
InactiveCN105517141AReduce outliersPrecisely predict changes in the magnitude of delay jitterSynchronisation arrangementNetwork topologiesSlide windowLeast squares
The invention provides a CPS delay jitter buffering method. The CPS delay jitter buffering method comprises the steps of obtaining network delay initial data; setting a sliding window used for recording multiple unilateral delay variables which are closest to the current time; removing relatively big unilateral delay variables in the sliding window and performing mapping transformation for the residual data in the sliding window; aiming at the data after the mapping transformation, solving a first prediction value by utilizing a derivative least square prediction model; performing inverse mapping for the first prediction value; aiming at the data after the inverse mapping and the u%-bit data of the residual data in the sliding window after sequencing, selecting the bigger one as a second prediction value, wherein the u% is arranged based on an experience; and calculating a feedback delay prediction value based on the second prediction value. According to the CPS delay jitter buffering method, the size of a feedback delay zone can be set reasonably, and network jitter can be smoothed.
Owner:SPACE STAR TECH CO LTD
A slurrying process of diatomite filter for lithium battery copper foil system
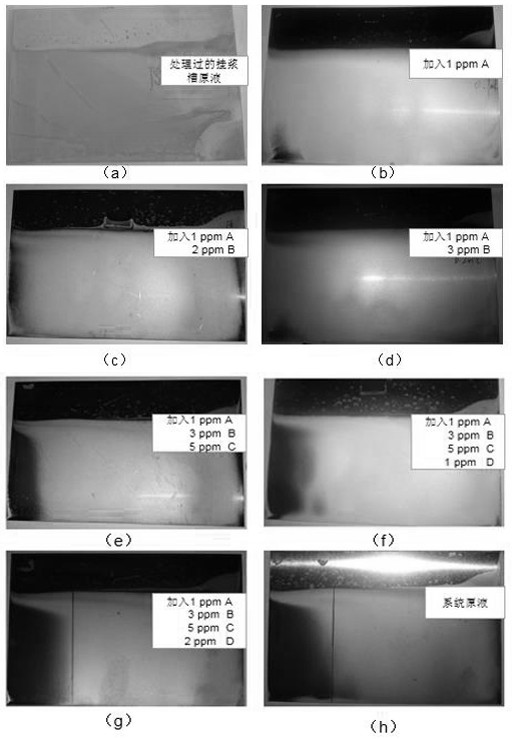
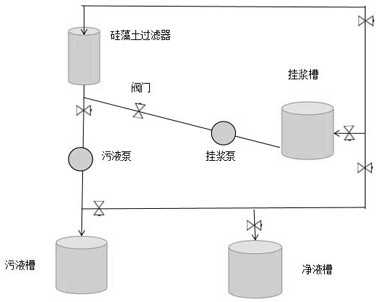
ActiveCN112516673BImprove yield rateSmall fluctuationProcess efficiency improvementFiltration circuitsElectrolytic agentCellulose
The application belongs to the technical field of electrolytic copper foil production technology, and in particular relates to a slurry-hanging process of a diatomite filter for a lithium battery copper foil system. This application uses the electroplating experiment to determine the concentrations of different additives A, B, C, and D in the copper foil electrolyte to be coated, wherein A is HEC hydroxyethyl cellulose, B is HP sodium alcohol thiopropane sulfonate, and C is PN polyethyleneimine alkyl salt, D is BSP sodium phenyl dithiopropane sulfonate. By adding an appropriate concentration of additives used in the initial stage of production into the electrolyte to be treated, the fluctuation of the solute system in the slurrying system is effectively reduced, and the abnormal amount during the slurrying period is reduced. The verification results show that the glossiness of the copper foil after using the process of this application is compared with that before the coating. The impact of the process on the production of electrolytic copper foil can improve the yield of copper foil.
Owner:LINGBAO WASON COPPER FOIL
Seismic interval velocity determination method and device
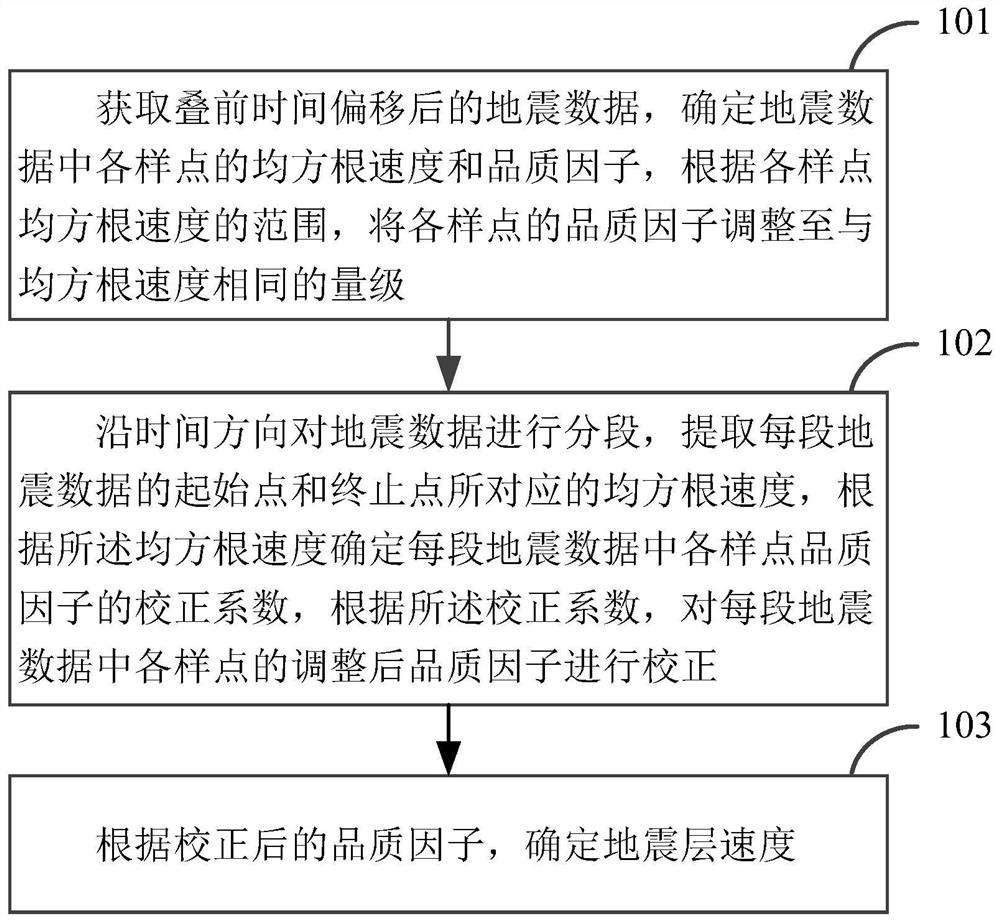
ActiveCN111781643AReduce outliersExact parameter valueSeismic signal processingTime domainGeophysics
The invention discloses a seismic interval velocity determination method and device, and the method comprises the steps: obtaining seismic data after pre-stack time migration, determining the root-mean-square velocity and quality factor of each sample point in the seismic data, and adjusting the quality factor of each sample point to the magnitude same with that of the root-mean-square velocity according to the range of the root-mean-square velocity of each sample point; segmenting the seismic data along the time direction; extracting root-mean-square velocities corresponding to a starting point and an ending point of each section of seismic data, determining a correction coefficient of the quality factor of each sample point in each section of seismic data according to the root-mean-square velocities, and correcting the adjusted quality factor of each sample point in each section of seismic data according to the correction coefficient; and determining the seismic interval velocity according to the corrected quality factor. According to the method, high-precision seismic interval velocity can be obtained according to the corrected quality factor, and a foundation is laid for obtaining accurate results of time domain-depth domain conversion and pre-stack depth migration.
Owner:PETROCHINA CO LTD
A method of personalized recommendation based on ocean model
ActiveCN107895303BFit psychologyImprove accuracyDigital data information retrievalBuying/selling/leasing transactionsPersonalizationData mining
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA
A Binocular 3D Reconstruction Method
ActiveCN105894574BReduce false match rateReduce outliersDetails involving processing steps3D modellingCamera lensFeature extraction
The invention discloses a binocular three-dimensional reconstruction method comprising the following steps of: 1) acquiring an image of an object to be reconstructed by two image acquisition apparatuses with the same model in order to obtain a left image and a right image; 2) calibrating the image acquisition apparatuses by a chessboard method, computing the internal and external parameter and a lens distortion coefficient, processing the left image and the right image to remove distortion in the images; 3) extracting features from the two processed images in the step 2) to obtain the feature points of the two images; 4) matching the feature points by using the feature points of the two images in the step 3) to obtain feature point pairs; 5) performing epipolar geometric constraint detection on the obtained feature point pairs to remove mismatched feature point pairs; and 6) computing three-dimensional coordinates of corresponding points, in a world coordinate system, of the obtained feature points by using the reserved feature point pairs. The binocular three-dimensional reconstruction method may reduce abnormal points and acquires an accurate three-dimensional reconstruction model.
Owner:HUIZHOU FRONT OPTOELECTRONIC TECH CO LTD
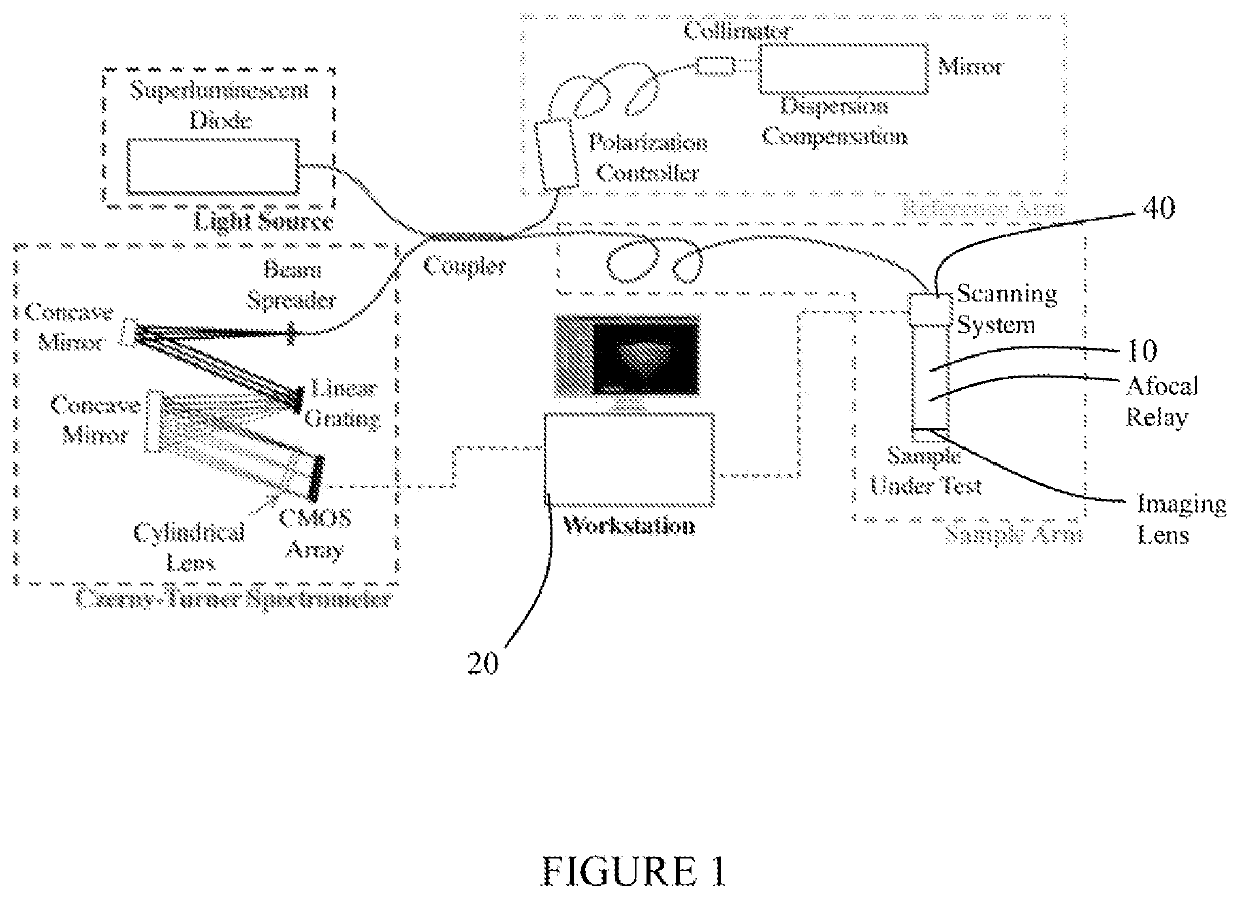
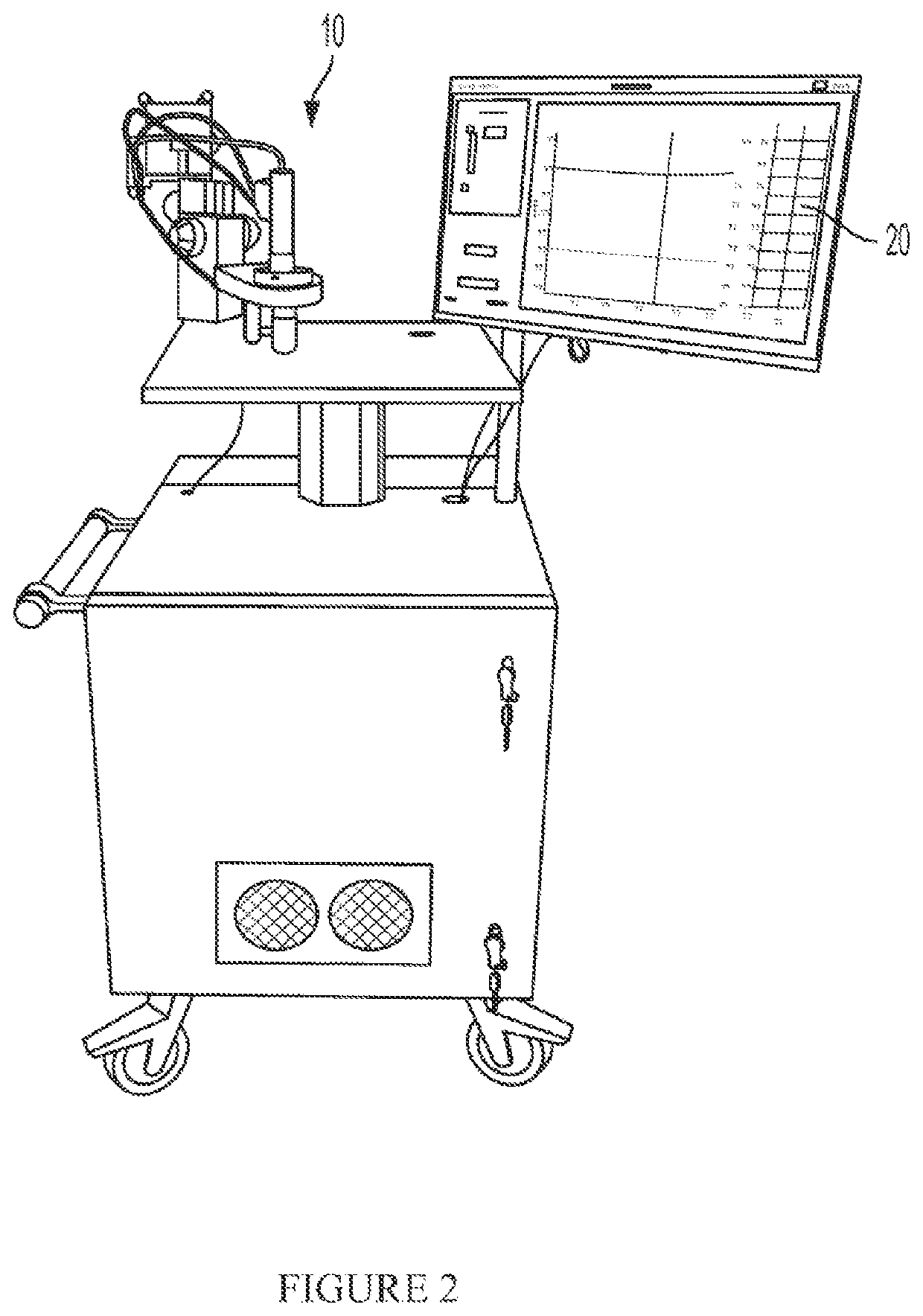
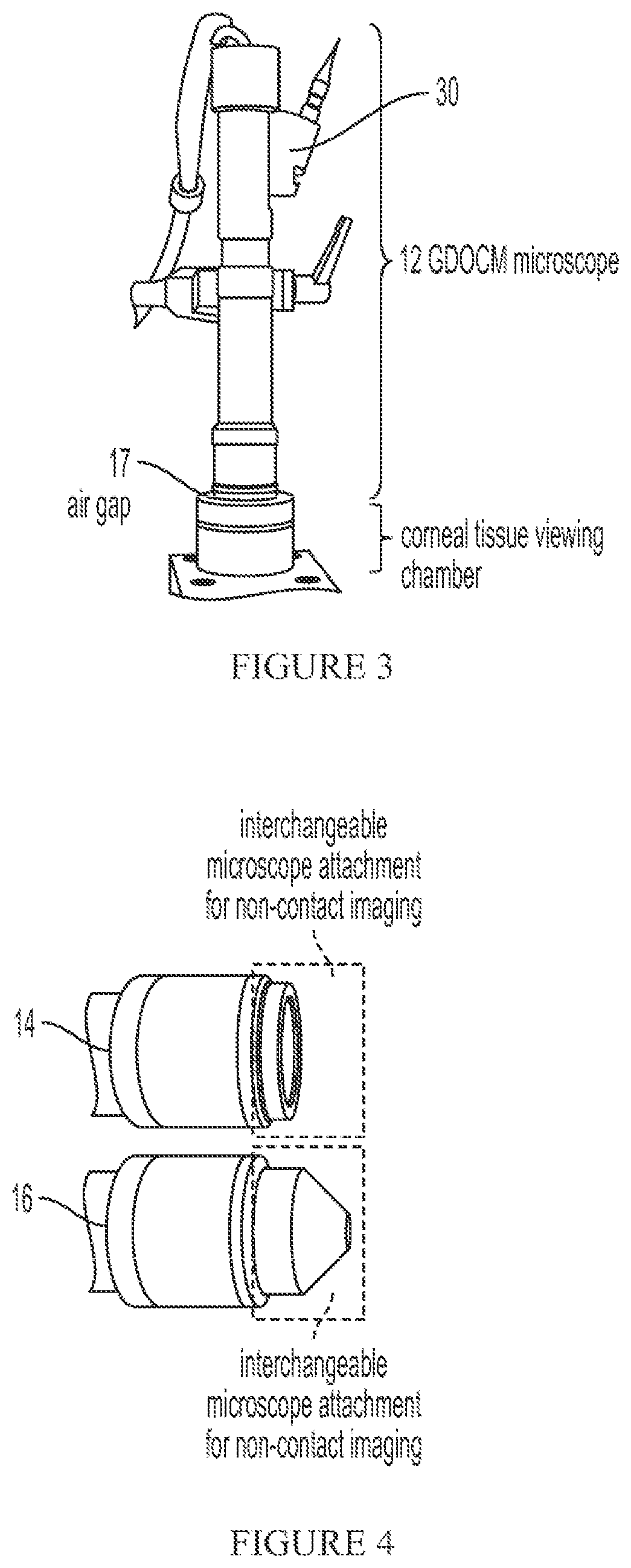
Three dimensional corneal imaging with gabor-domain optical coherence microscopy
ActiveUS11094064B2Improve accuracyReduce biasImage enhancementReconstruction from projectionRadiologyMicroscope
A system for non-contact imaging of corneal tissue stored in a viewing chamber using Gabor-domain optical coherence microscopy (GDOCM), wherein a 3D numerical flattening procedure is applied to the image data to produce an at least substantially artifact-free en face view of the endothelium.
Owner:LIGHTOPTECH CORP
A slam method for closed-loop detection of orb keyframes that can improve robot pose consistency
InactiveCN105856230BGuaranteed positioning accuracyGuaranteed accuracyProgramme controlProgramme-controlled manipulatorClosed loopImaging Feature
The invention discloses an ORB key frame closed-loop detection SLAM method capable of improving the consistency of the position and the pose of a robot. The ORB key frame closed-loop detection SLAM method comprises the following steps of, firstly, acquiring color information and depth information of the environment by adopting an RGB-D sensor, and extracting the image features by using the ORB features; then, estimating the position and the pose of the robot by an algorithm based on RANSAC-ICP interframe registration, and constructing an initial position and pose graph; and finally, constructing BoVW (bag of visual words) by extracting the ORB features in a Key Frame, carrying out similarity comparison on the current key frame and words in the BoVW to realize closed-loop key frame detection, adding constraint of the position and pose graph through key frame interframe registration detection, and obtaining the global optimal position and pose of the robot. The invention provides the ORB key frame closed-loop detection SLAM method with capability of improving the consistency of the position and the pose of the robot, higher constructing quality of an environmental map and high optimization efficiency.
Owner:简燕梅
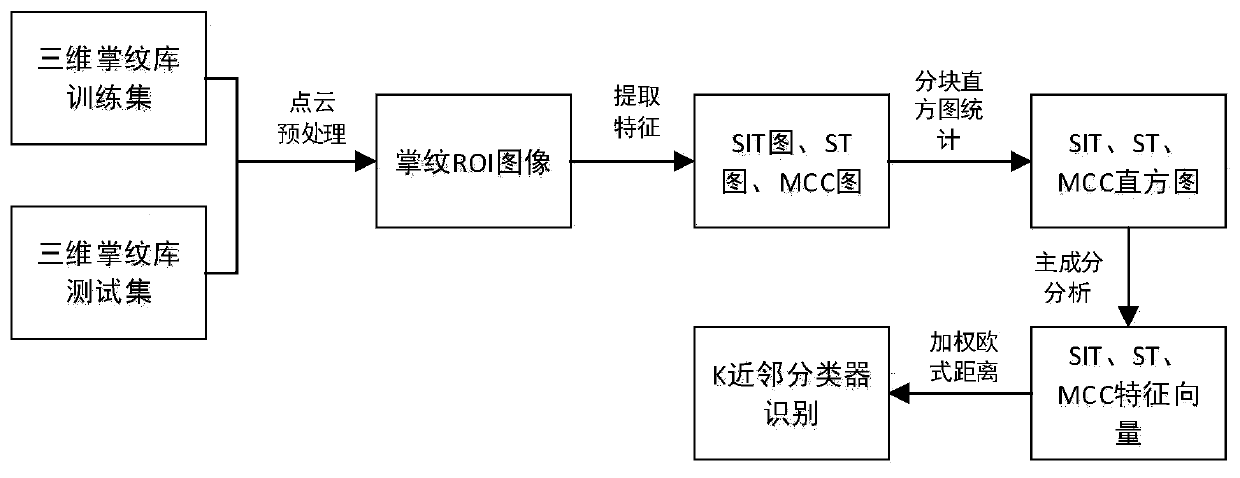
Three-dimensional palmprint recognition method integrating multiple features and principal component analysis
PendingCN111160306AReduce outliersReduce noiseMatching and classificationThree-dimensional object recognitionPalm printFeature Dimension
The invention provides a three-dimensional palmprint recognition method integrating multiple features and principal component analysis, which extracts geometrical features based on classification coding of shape indexes, self-adaptive correction competition coding and curved surface type feature coding, and effectively reduces the influence on the recognition rate caused by wrong distribution dueto threshold classification. Competitive coding is improved, so that the direction characteristics of the three-dimensional palmprint are expressed more accurately, block histogram statistics and principal component analysis are utilized to reduce the feature dimension and remove redundant information, so that the features are more robust to micro displacement and the discrimination is improved, Euclidean distances of the three features are used as a matching distance by weighted fusion, so that the precision and robustness of the algorithm are effectively improved, and meanwhile, a K nearestneighbor algorithm is used for realizing classification, so that the influence of abnormal values and noise in the data is effectively reduced, and the comparison time is shortened.
Owner:SOUTHEAST UNIV
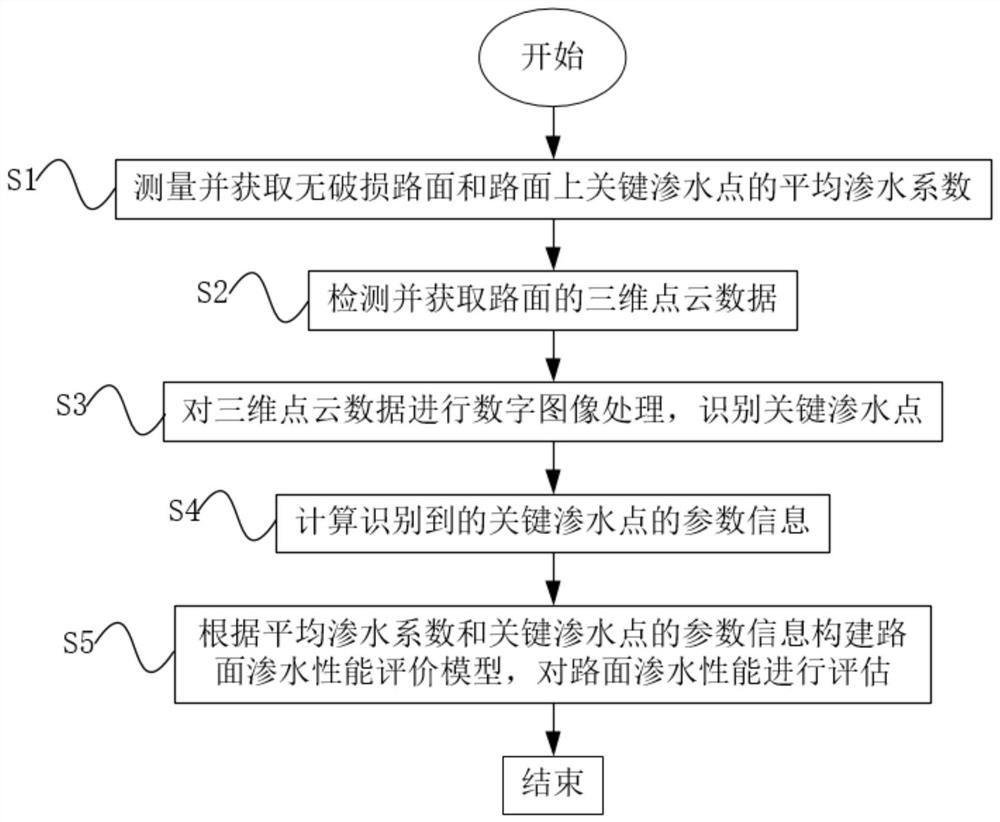
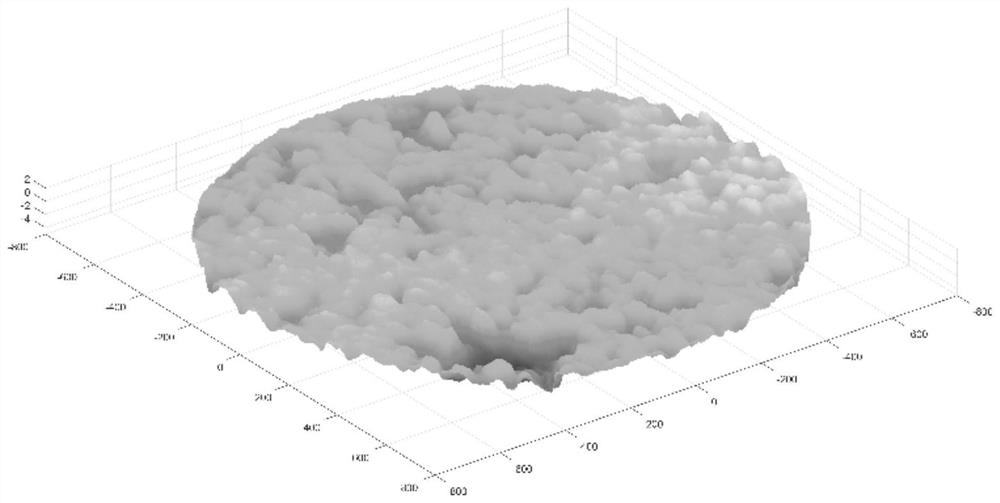
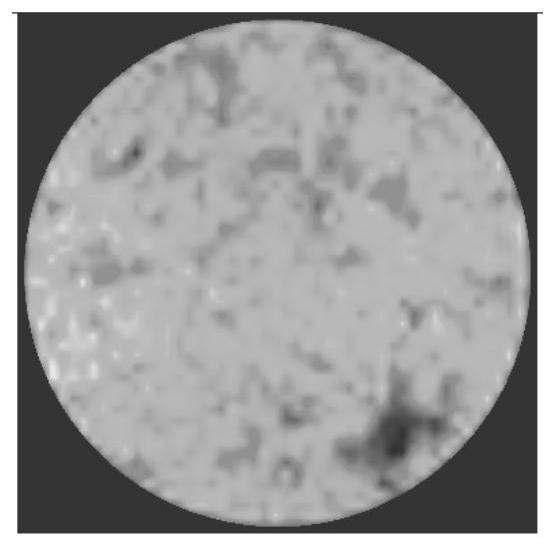
Road surface water seepage performance evaluation method based on key water seepage point identification
ActiveCN112927204AReduce risk of damageReduce constraintsImage enhancementImage analysisSoil sciencePoint cloud
The invention relates to a pavement water seepage performance evaluation method based on key water seepage point identification. The method comprises the following steps: S1, measuring and acquiring an average water seepage coefficient of an undamaged pavement and key water seepage points on the pavement; S2, detecting and acquiring three-dimensional point cloud data of a road surface; S3, performing digital image processing on the three-dimensional point cloud data, and identifying key water seepage points; S4, calculating parameter information of the identified key water seepage points; and S5, constructing a pavement water seepage performance evaluation model according to the average water seepage coefficient and the parameter information of the key water seepage points, and evaluating the pavement water seepage performance. Compared with the prior art, the method has the advantages that a new information source is introduced for traditional water seepage detection, the risk that an asphalt pavement is damaged by water is relieved, a brand new view angle is provided for preventive maintenance, the existing pavement does not need to be damaged, multiple data points are obtained and collected, the evaluation accuracy is high, environmental constraint is small, daily use can be met, and the overall water seepage performance of the pavement can be better evaluated.
Owner:TONGJI UNIV
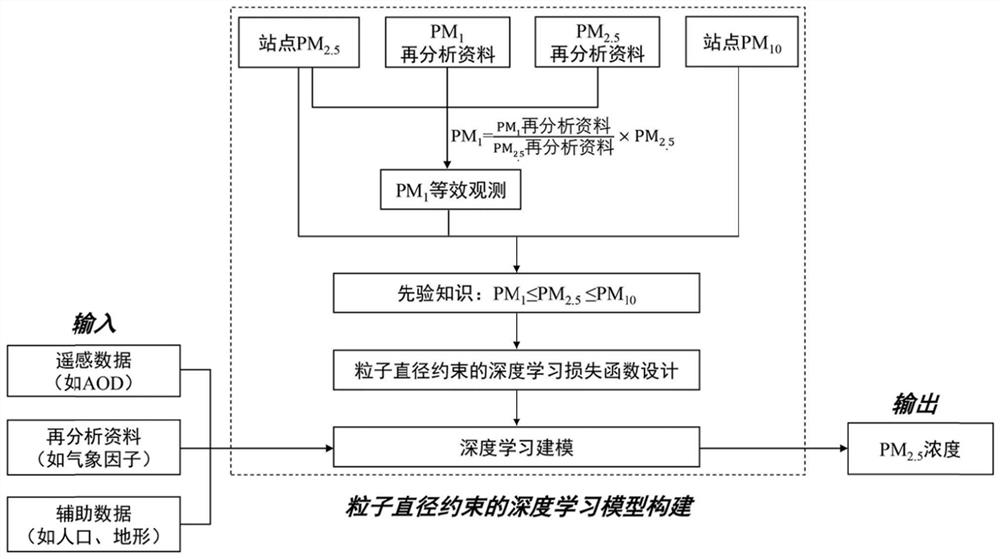
Particle diameter constrained PM2.5 deep learning remote sensing estimation method
PendingCN114334027AReduce outliersImprove robustnessMolecular entity identificationNeural architecturesObservation dataAtmospheric sciences
The invention discloses a particle diameter constrained PM2.5 deep learning remote sensing estimation method, which comprises the following steps of: constructing a deep neural network model based on satellite remote sensing aerosol optical thickness (AOD), reanalysis data, ground station observation data and the like, and establishing constraint conditions by utilizing a priori relationship (namely PM2.5 concentration is more than or equal to PM1 concentration and less than or equal to PM10 concentration) of particle diameters of PM1, PM2.5 and PM10; and on the basis of the constructed constraint term of the deep neural network loss function, the PM2.5 deep learning remote sensing estimation method with the particle diameter constraint is provided. According to the method, the phenomena of high-value underestimation and low-value overestimation in deep learning PM2.5 estimation can be relieved, the precision of PM2.5 remote sensing estimation is improved, and the method can be widely applied to the technical field of PM2.5 concentration remote sensing estimation.
Owner:SUN YAT SEN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com