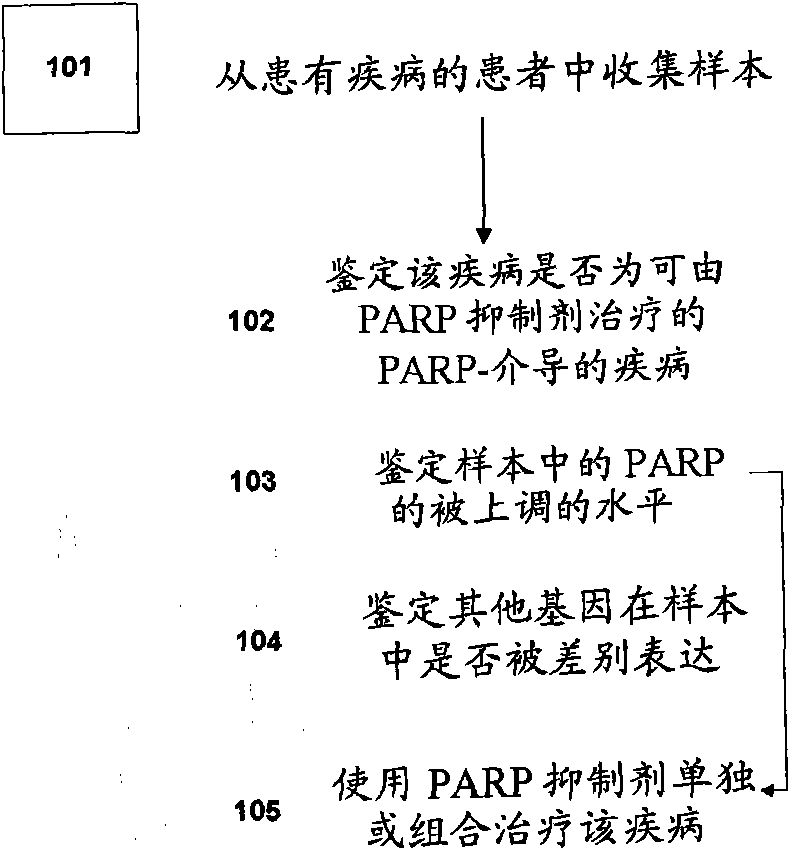
Methods of diagnosing and treating PARP-mediated diseases
A disease-mediated technology, applied in the direction of biochemical devices and methods, diseases, metabolic diseases, etc., that can address the incidence of therapy resistance and its prevention Insufficiently studied, unsuccessfully detailed delineation of cancer formation Different known Molecular pathway interactions and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0535] Gene array testing has been widely used to monitor mRNA expression in many biomedical research fields. The high-density oligonucleotide array technology allows researchers to monitor tens of thousands of genes in a single hybridization experiment, because they are expressed differently in tissues and cells. The expression profile of mRNA molecules of genes is obtained by combining the intensity information from the probes in the probe set, which consists of 11-20 probe pairs of oligonucleotides of 25 bp in length, to inquire about the difference in genes Part of the sequence.
[0536] Affymetrix Human Genome genechips (45,000 gene transcripts covering 28,473 UniGene clusters) were used to evaluate gene expression. Approximately 5 μg of total RNA from the sample was labeled using a high-yield transcription labeling kit, and the labeled RNA was hybridized, washed, and scanned (according to the manufacturer's instructions) (Affymetrix, Inc., Santa Clara, CA). Affymetrix Mic...
Embodiment 2
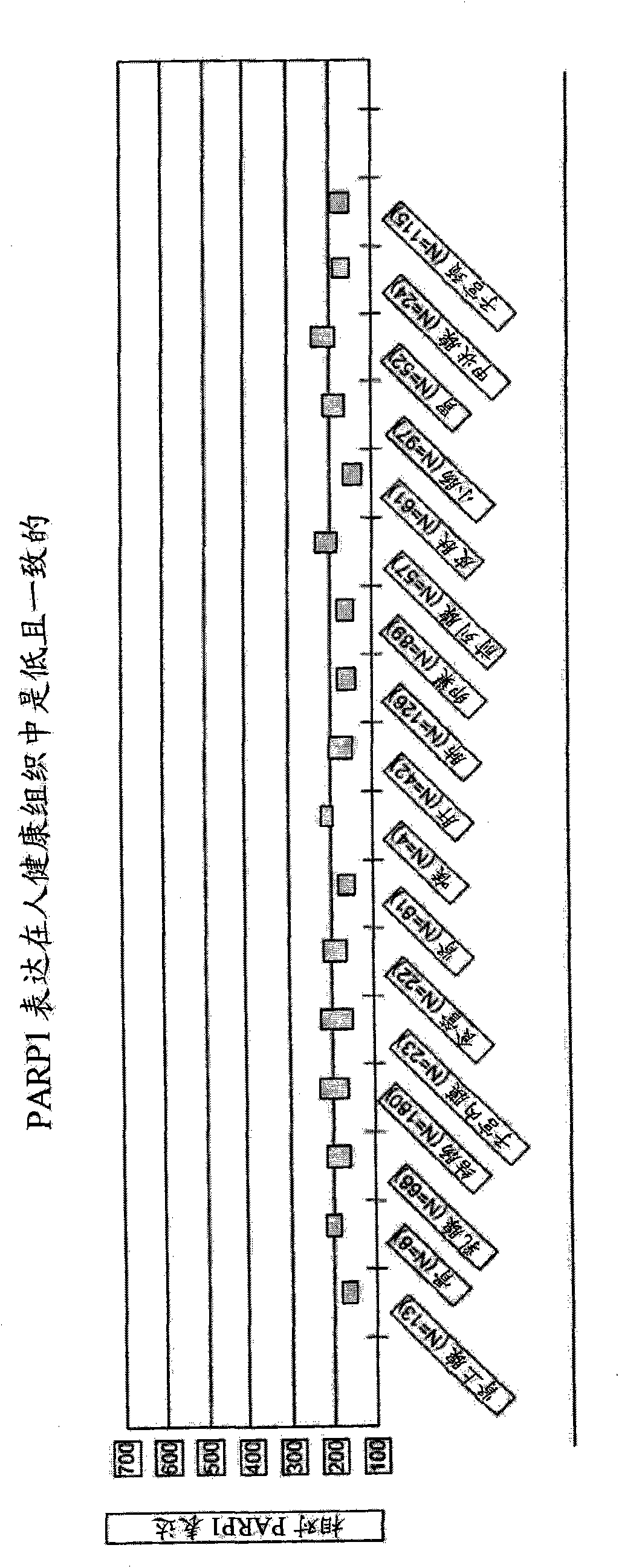
[0538] Up-regulation of PARP1mRNA in normal and tumor tissues
[0539] Research design and materials and methods
[0540] Tissue samples: Normal and cancer tissue samples were collected in the United States or the United Kingdom. The samples are collected as part of normal surgical procedures and are quickly frozen within 30 minutes of resection. The samples are transported at -80°C and stored at -170 to -196°C in the vapor phase of liquid nitrogen until processing. Perform medical pathological examination and confirmation of samples to be analyzed. Combined with the initial diagnosis report, observe the H&E-stained slides obtained from the adjacent part of the tissue, and classify the samples according to the diagnosis category. During the examination of the slide by the pathologist, the visually estimated percentage of tissue involved in the tumor was recorded, and the fraction of malignant nucleated cells was noted. Auxiliary studies such as ER / PR and Her-2 / neu expression st...
Embodiment 3
[0555] Co-expression of PARP1 Mrna and other targets in normal and cancer tissues
[0556] The PARP1 gene is represented by a single probe set with the identifier "208644_at" on the HG-U133A array. Other genes, such as BRCA1, BRCA2, RAD51, MRE11, p53, PARP2 and Mucin 16 are represented by their respective information probe sets in the HG-U133A / B array set. The probe sets corresponding to the 7 genes in the analysis of ovarian cancer samples are listed in Table XXXIII.
[0557] Table XXXIII: Comparison genes and their corresponding HG-U133A / B probe set ID
[0558] Gene symbol
title
Fragment name
BRCA1
204531_s_at
BRCA2
214727_at
MRE11A
MRE11 meiotic recombination 11 homolog A (Saccharomyces cerevisiae)
205395_s_at, 242456
MUC16
Mucin 16, cell surface association
220196_at
PARP2
Poly(ADP ribose) polymerase family, member 7
204752x_at, 214086_s_at, 215773_x_at
RAD51
RAD51 homolog (RecA homolog, Esche...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com