Biomarker combination for molecular typing and/or prognosis prediction of muscle-invasive bladder cancer and application of biomarker combination
A technology of biomarkers and molecular typing, applied in the fields of bioinformatics, microbial determination/examination, biochemical equipment and methods, etc. Low requirements and the effect of meeting the needs of large sample analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
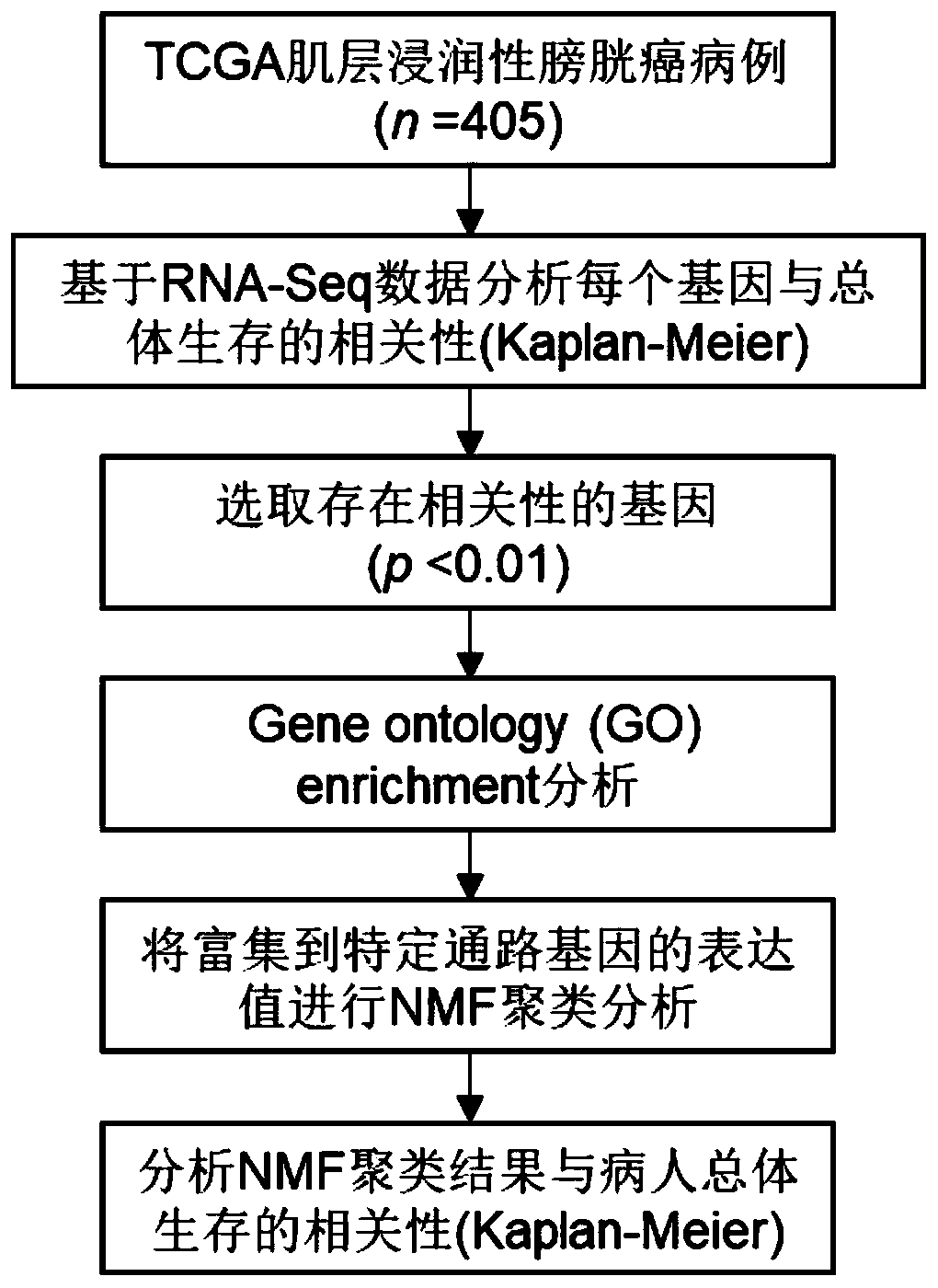
[0036] This embodiment is a screening method for a combination of biomarkers for molecular typing and / or prognosis prediction of muscle invasive bladder cancer, such as figure 1 As shown, the screening process of the biomarker combination gene set is as follows:
[0037] Download the clinical data of muscle-invasive bladder cancer cases (405 cases) and the corresponding tumor tissue RNA-Seq transcriptome data from the TCGA (The Cancer Genome Atlas) database (https: / / portal.gdc.cancer.gov), select The downloaded data type is the FPKM (fragments per kilobase of transcript per million fragments mapped) value standardized by the Upper Quartile method.
[0038] In the RNA-Seq data, genes with a median expression value of 0 were removed, and then the correlation of each gene with overall survival (OS) was analyzed. For each gene, according to whether it is greater than or less than the median expression value, patients are divided into high-expression group and low-expression group...
Embodiment 2
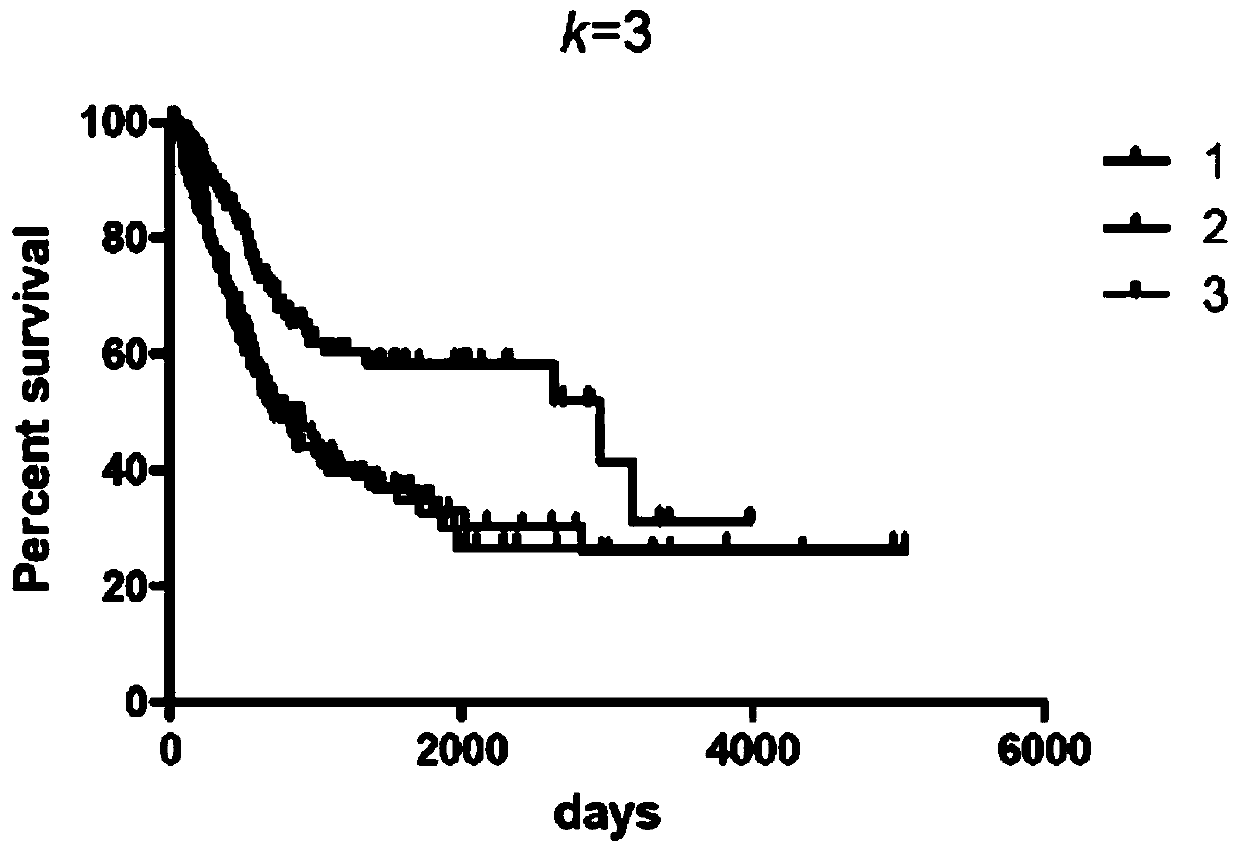
[0051] In this embodiment, the human expression profile chip data of 71 cases of muscle-invasive bladder cancer are taken as an example, and the data set and non-negative matrix factorization analysis method described in the present invention are used to predict the survival and prognosis of bladder cancer. The details are as follows:
[0052] The transcriptome data and patient overall survival data of tumor specimens were downloaded from the GEO database (accession number GSE48277), the data type was human expression profiling chip data, and the specimen type was fresh frozen tissue.
[0053] From the transcriptome data, the expression values of FGF10, TP53INP1, DDR2, MYC, CDC73, IGF1, PLA2G1B, SKI, FN1, EGFR, PPARG, PDGFRA, PDGFD, GAS6, PDGFC, FNTB and CCNB1 genes were extracted to establish an expression matrix.
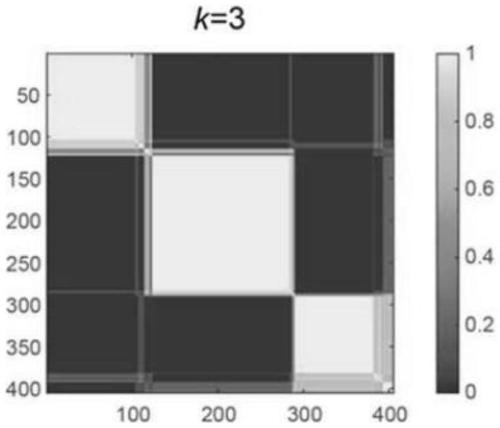
[0054] Import the expression matrix into the NMF software in Matlab, preset the grouping parameter range from 1 to 7 (k=1 to k=7), and run the analysis program; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com