Process for aligning targeted nucleic acid sequencing data
A target sequence and sequence technology, applied in the field of nucleic acid sequencing data for comparison and targeting, can solve problems such as hindering output, complicating workflow, and computing power requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0103] Example 1 - Example system implementing off-target match detection
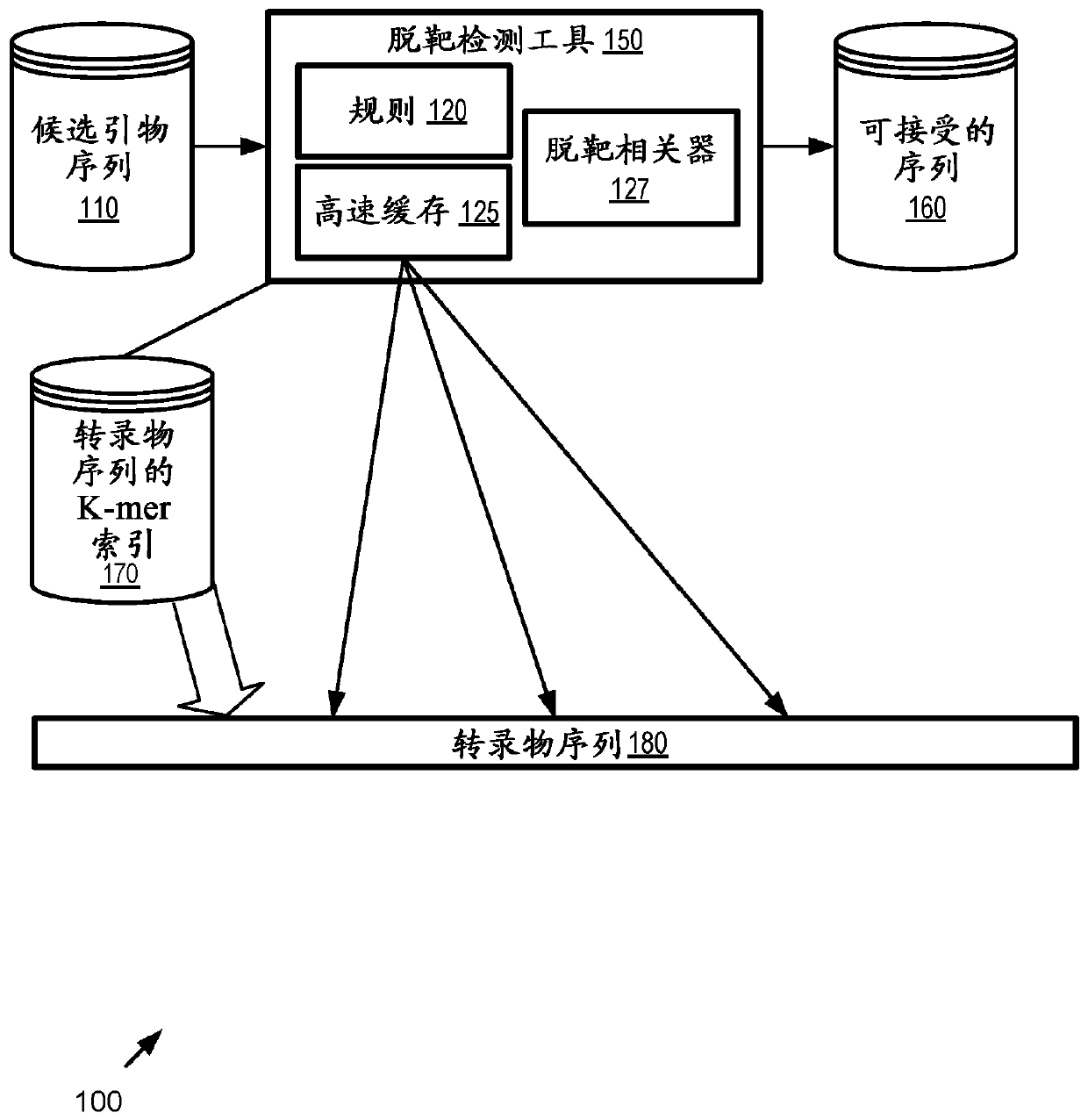
[0104] figure 1 is a block diagram of an example system 100 that implements off-target match detection for generating revised reference genome sequences from transcript sequences 180. In any instance herein, a string may take the form of a sequence of characters representing a string of values. Although referred to herein as a "string," the internal representation may take the form of a string, array, or other data structure. A character can take the form of a character or a code representing the character.
[0105] In this embodiment, a plurality of candidate primer sequences 110 are received as input by the off-target detection tool 150 . As described herein, these candidate primer sequences 110 may take the form of primer pairs that target specific positions on the transcript sequence 180 representing the plus strand of a transcript sequence transcribable from a reference genome as described he...
Embodiment 2
[0116] Example 2 - Example method for detecting off-target matches
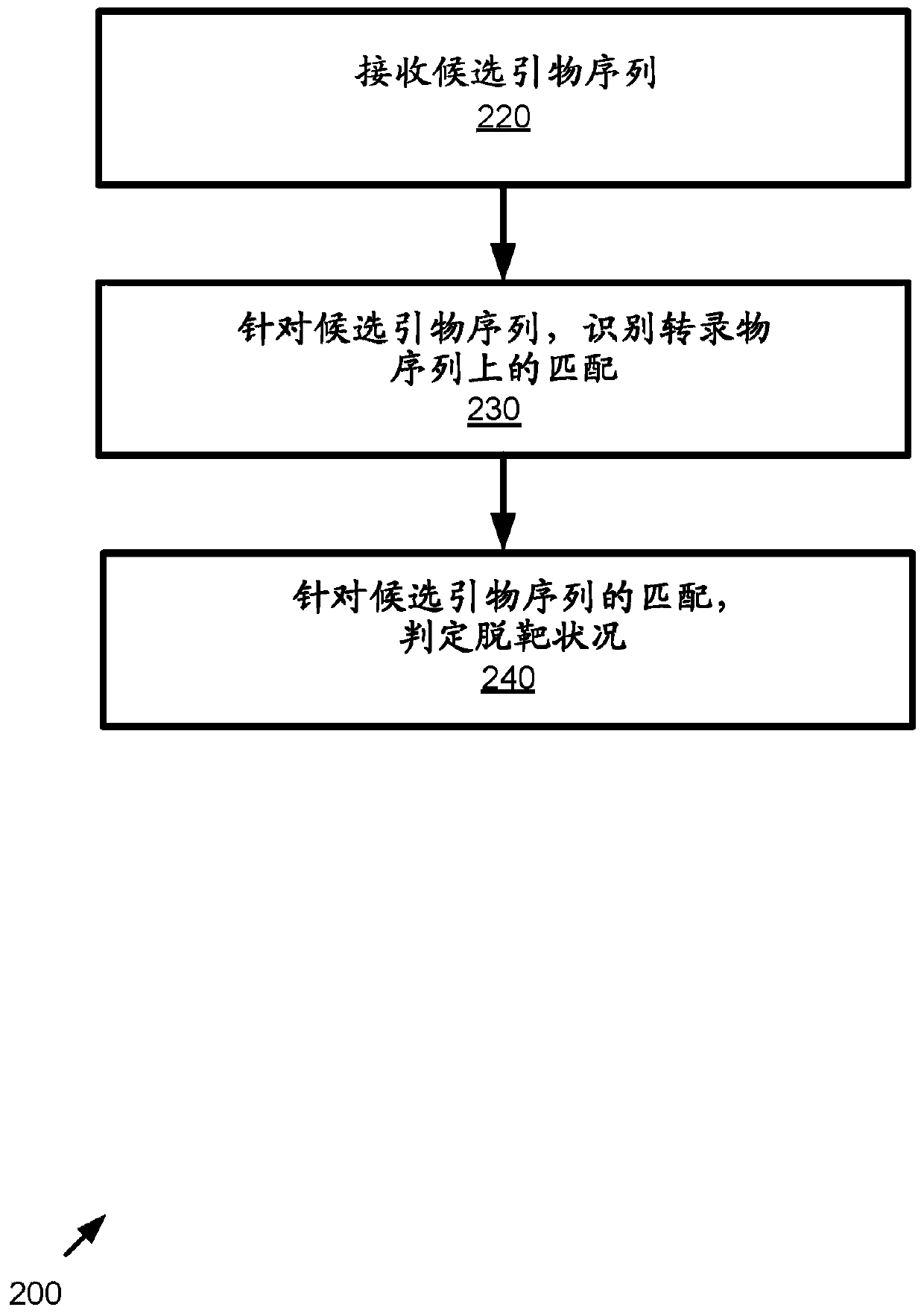
[0117] figure 2 is a flowchart of an example method 200 of implementing off-target match detection and may be performed, for example, in a figure 1 implemented in the system shown in . Multiple candidate primer sequences targeting multiple targets on the transcript sequence can be supported.
[0118] In practice, steps can be taken, such as the use of primer generation tools, etc., to generate candidate primer sequence pairs before the method begins.
[0119] At 220, candidate primer sequences are received. Candidate primer sequences can take any form described herein.
[0120] At 230, for candidate primer sequences, matches on transcript sequences are identified. The match determination may include applying a plurality of rules as described herein. For example, multiple candidate matching conditions can be identified on the transcript sequence (eg, by means of matching rules as described herein). F...
Embodiment 3
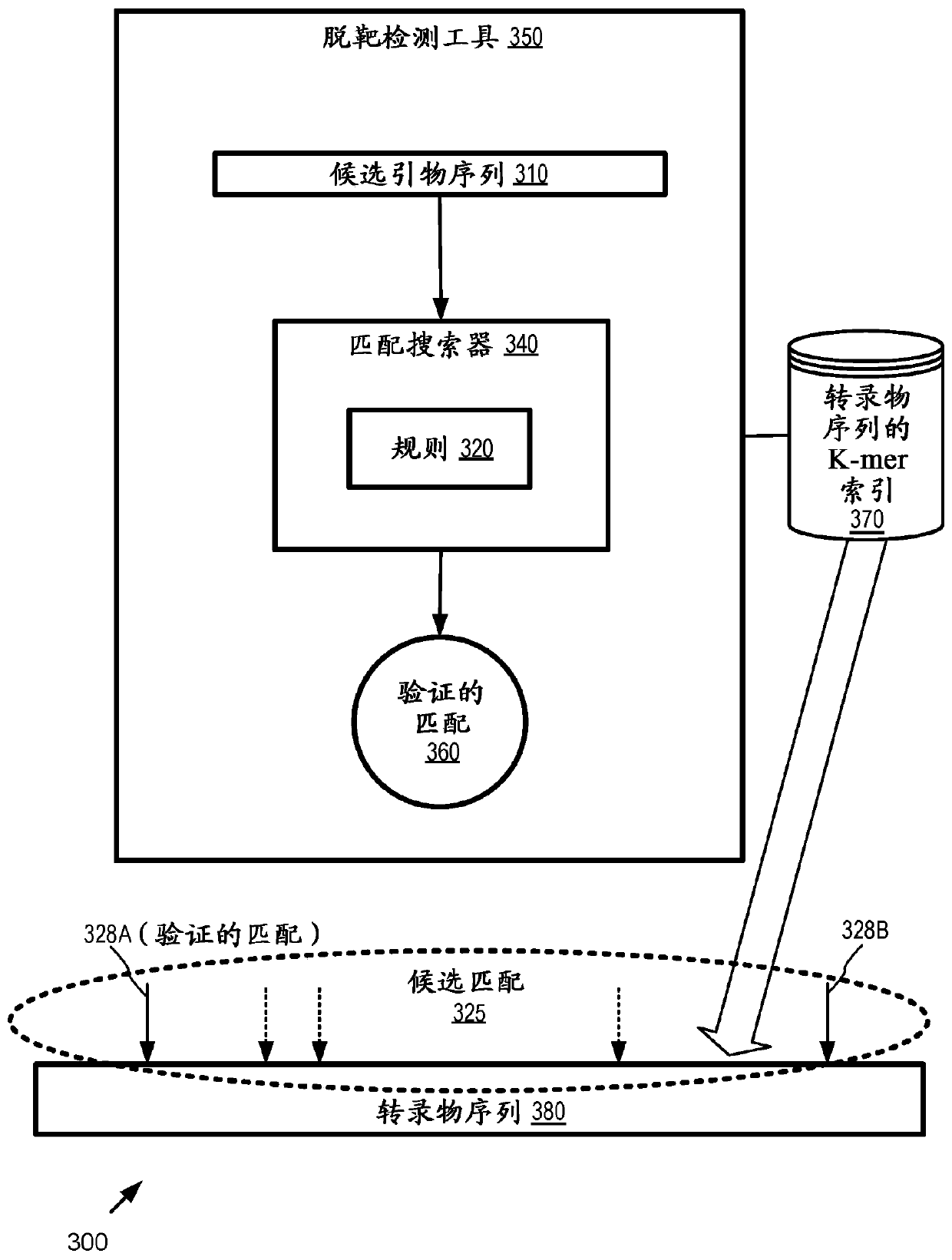
[0127] Example 3 - Example off-target match detection
[0128] In any of the examples herein, an off-target match can take the form of a pair of candidate primer sequences (eg, from the original primer pair or two different primer pairs) that match at adjacent positions as described herein. In practice, adjacent positions may be on two different (eg, one original, one reverse complementary to the original) transcript sequences as described herein; by taking the reverse complement of the candidate primer sequence and incorporating it into the candidate Of the primer sequences, a single transcript sequence can be used to perform the calculations. Detection of such off-target matches can be used to determine whether candidate primer sequences are acceptable or unacceptable, as described herein. Candidate primer sequences (and their primer pairs) that exceed the off-target match status threshold may be considered unacceptable.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com