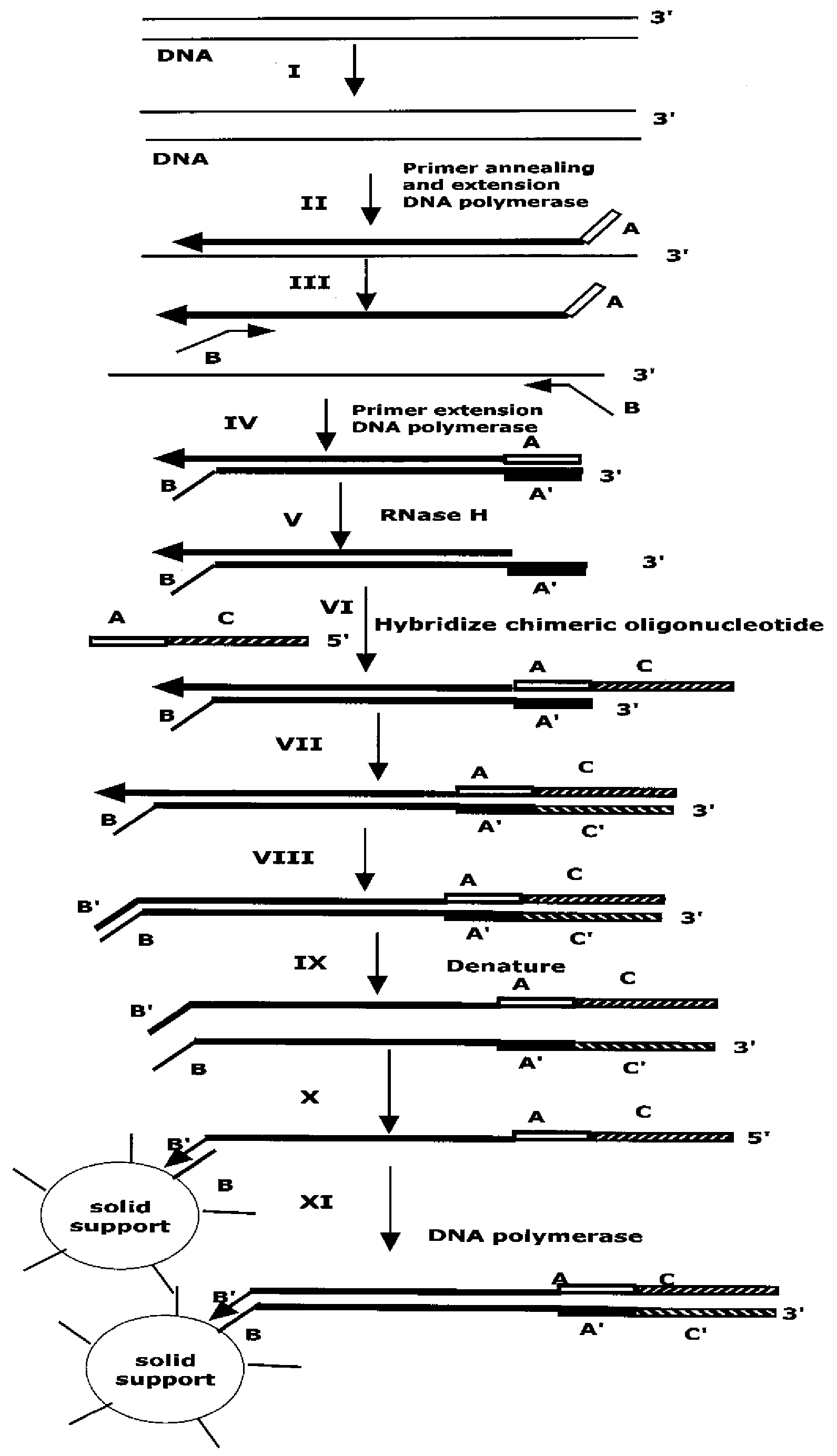
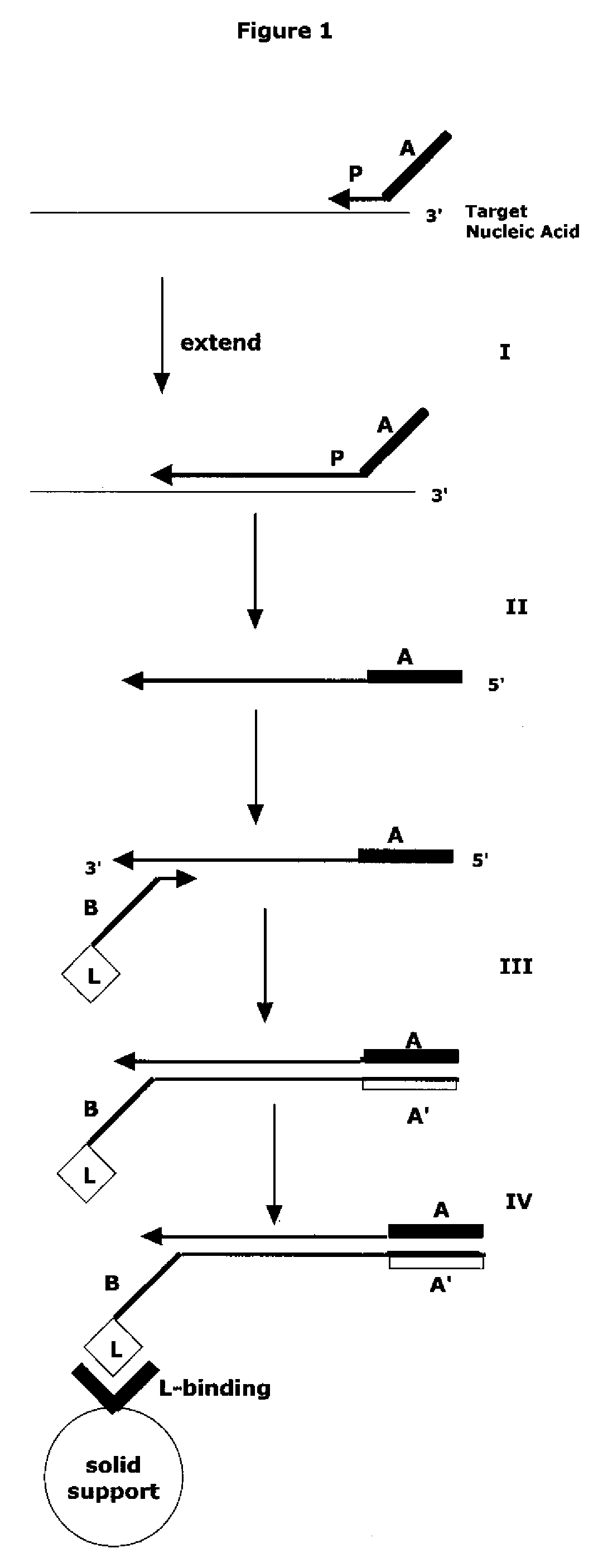
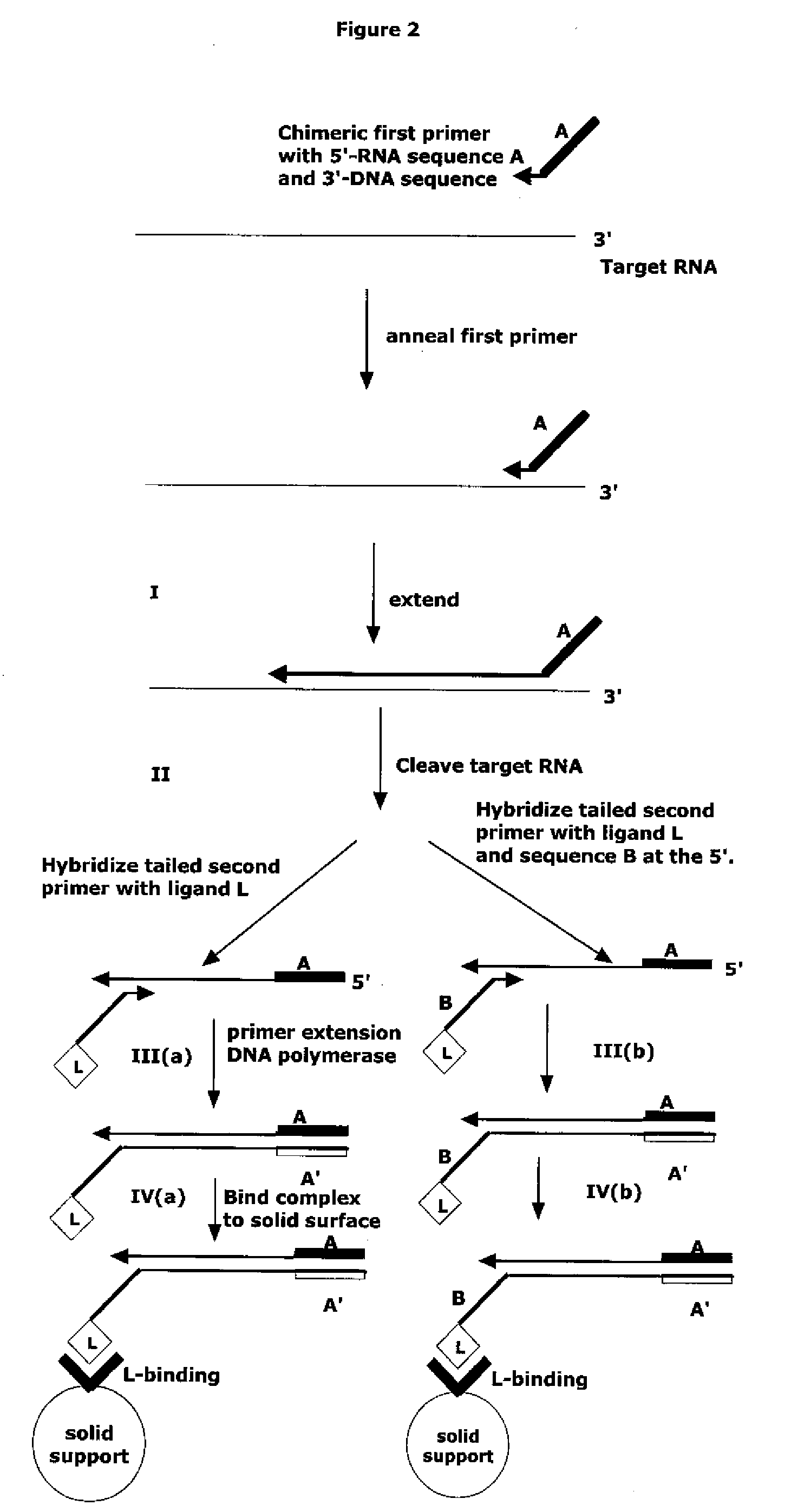
Method for Archiving and Clonal Expansion
a clonal expansion and archiving technology, applied in the field of archiving and clonal expansion, can solve the problems of forensic and paleoarcheology work being severely limited by the size of nucleic acid sample samples, affecting the ability to carry out large-scale analysis of multiple parameters, and affecting the ability to start materials
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Clonal Expansion of RNA
Step 1: Synthesis of First Primer Extension Product
[0595]100 ng of an RNA template is provided. The provided RNA template is produced from a biological specimen using a commercially available kit (i.e. Qiagen RNeasy) according to the manufacturer's instructions. A first primer extension reaction mixture is assembled comprising a first primer consisting of a 3′ annealing sequence, a portion of which is DNA, and a 5′ tail sequence (A), a portion of which is RNA and the following reagents in a total volume of 10 μl:
100 ng of RNA template
20 pmol of primer
0.5 μl dNTPs (25 mM)
0.1 μl RNasin
0.1 μl DTT
[0596]2 μl 5×AMV reverse transcriptase reaction buffer
DEPC treated water to 10 μl total volume
[0597]The reaction mixture is incubated for 2 min at 75° C., and then cooled to 37° C. 1 μl AMV reverse transcriptase (USB 70041Y, 15U / μl) is added to each reaction and the reaction mixture is further incubated at this temperature for 60 min. The resulting product is a first prim...
example 2
Diagnosis and Prognosis of Cancer
[0606]A suggested course of treatment can be determined by RNA expression analysis of a tumor biopsy. A needle biopsy is performed on a subject to obtain tissue from the suspicious mass for further analysis. The biopsied tissue recovered from the subject is processed to extract and purify total RNA using a commercially available Qiagen RNeasy kit according to the manufacturer's instructions.
[0607]500 pg of total RNA representing at least a portion of the transcriptome of the biopsied material is amplified by the methods of the present invention as described briefly herein. To the RNA in a reaction mixture is added: 100 pmol of a first primer comprising random first primer and a poly dT first primer, a 5′ segment and a 3′ segment, a portion of the 5′ segment comprising RNA, and a portion of the 3′ segment comprising DNA. The 3′ DNA segment of the random first primer further comprises an annealing sequence that comprises random hexamers. The 5′ RNA seg...
example 3
Personal Genomics
[0616]An individual is tested by a personal genomics business using the methods of the present invention for single nucleotide polymorphisms (SNPs) within the BRCA1, BRCA2, p53, MPO, NAT1, NAT2, and ras coding regions that are related to increased risks for specific types of cancer.
[0617]The individual supplies a small sample of tissue (i.e. a cheek swab) to the personal genomics business. Genomic DNA from the sample of tissue is isolated using a commercially available kit (i.e. Promega's Wizard® Genomic DNA Purification Kit), according to the manufacturer's protocol.
[0618]1 to 10 ng of purified genomic DNA is used to clonally amplify the sequences corresponding to the genomic regions with known, cancer related, SNPs of the BRCA1, BRCA2, p53, MPO, NAT1, NAT2, and ras genes on a solid support (i.e. a bead). The target sequences are clonally amplified by isothermal linear amplification using the steps shown in FIG. 13 steps I(b), II(b), and III(b); FIG. 17 steps IV to...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Volume | aaaaa | aaaaa |
| Area | aaaaa | aaaaa |
| Magnetism | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com