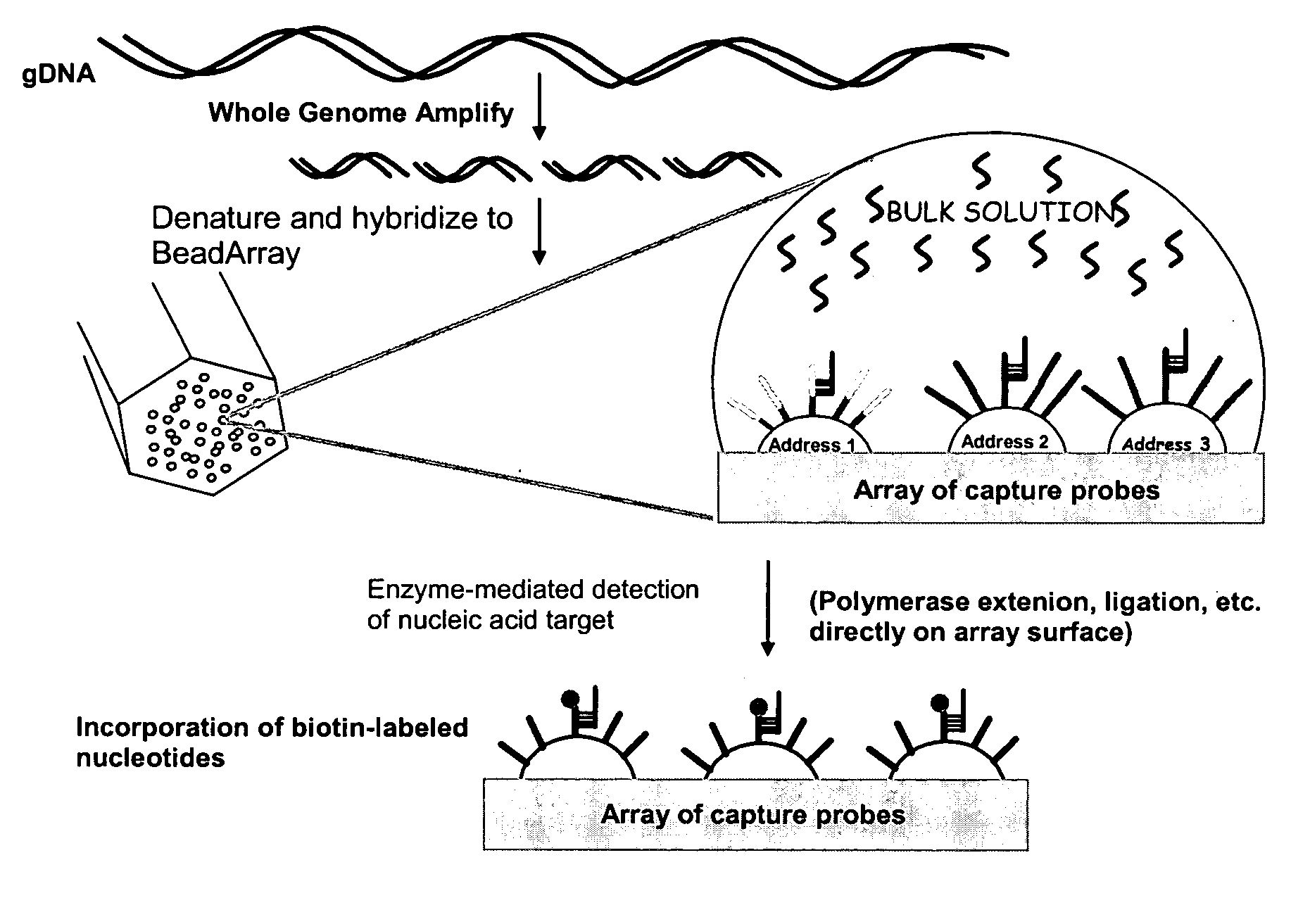
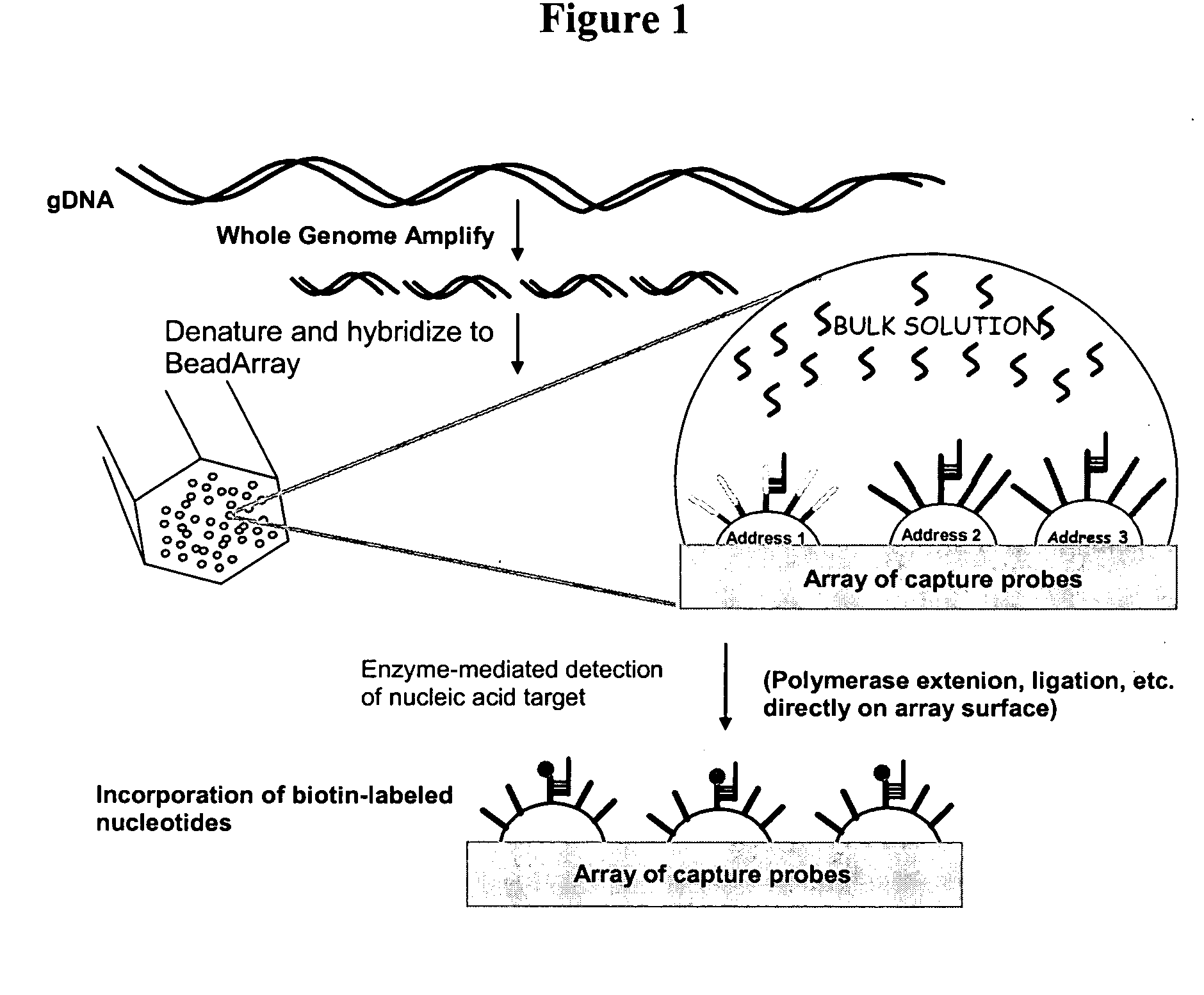
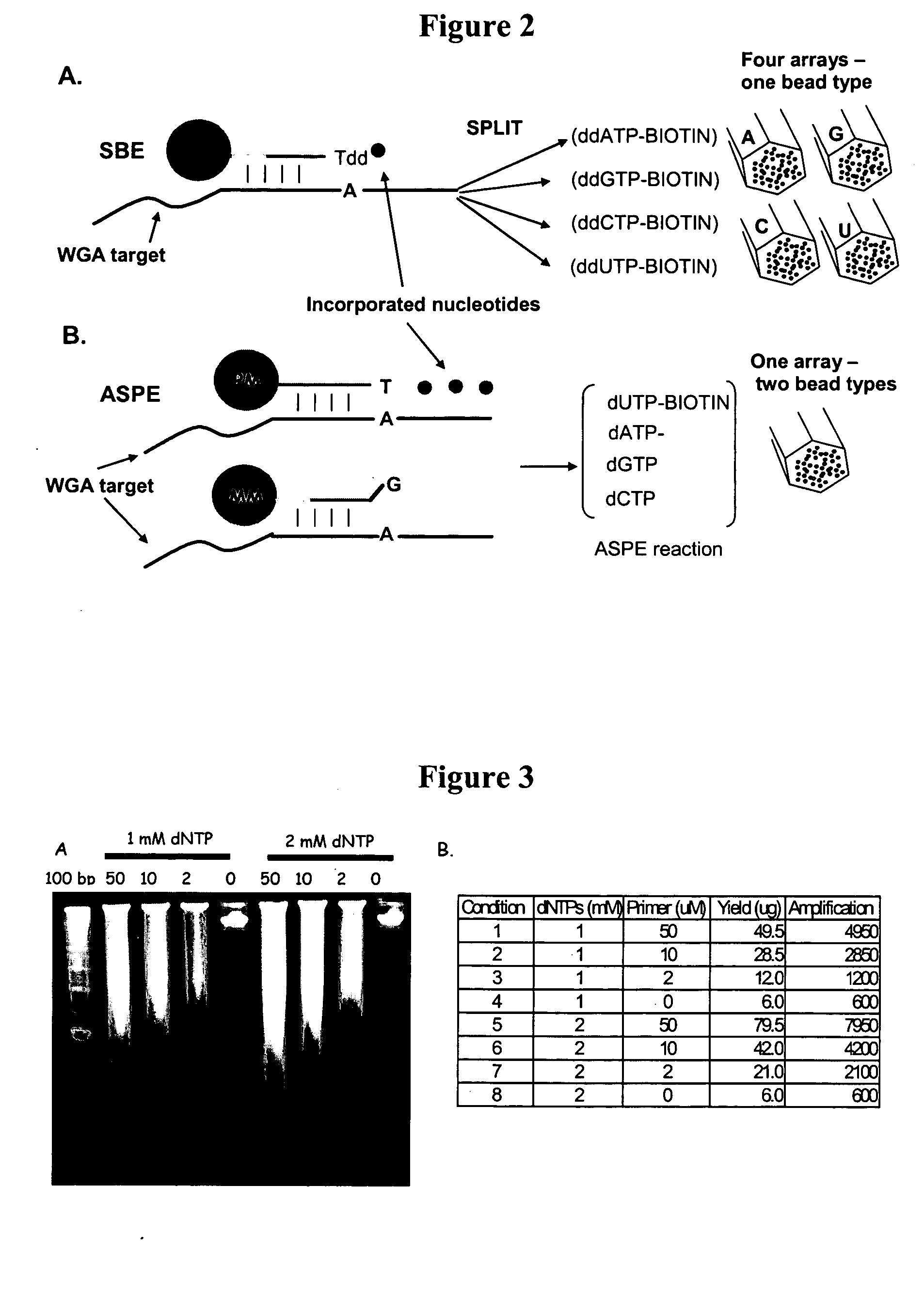
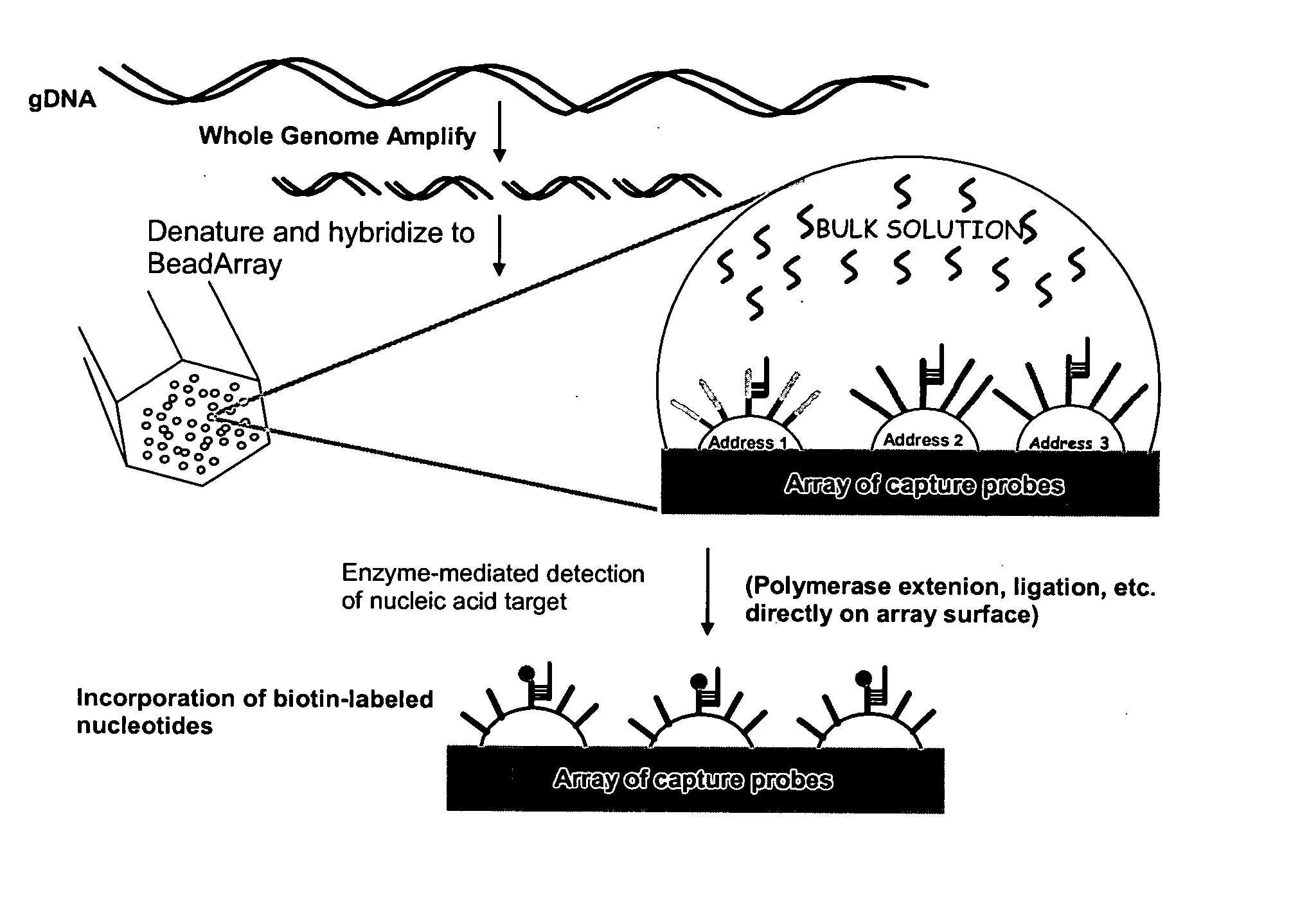
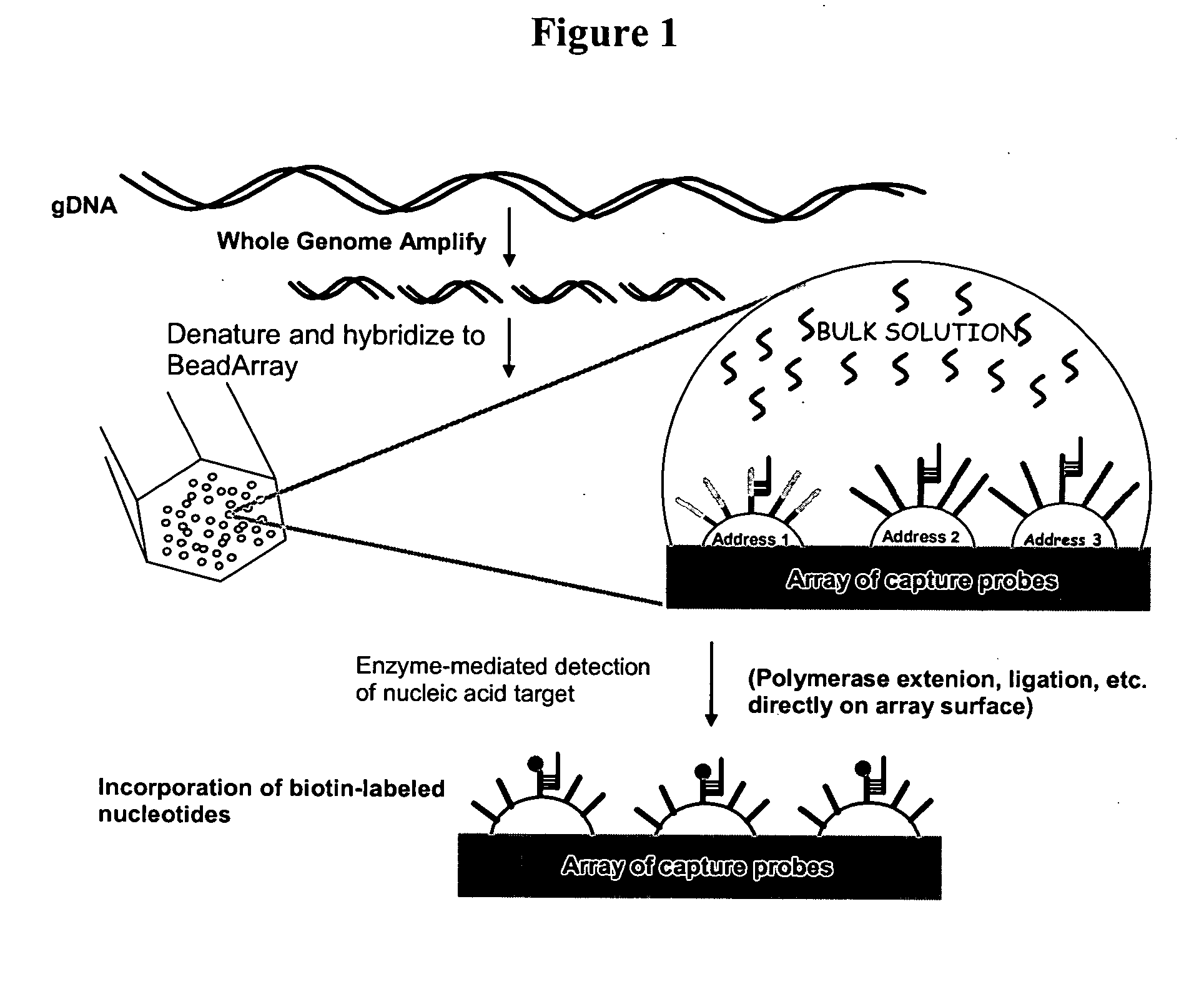
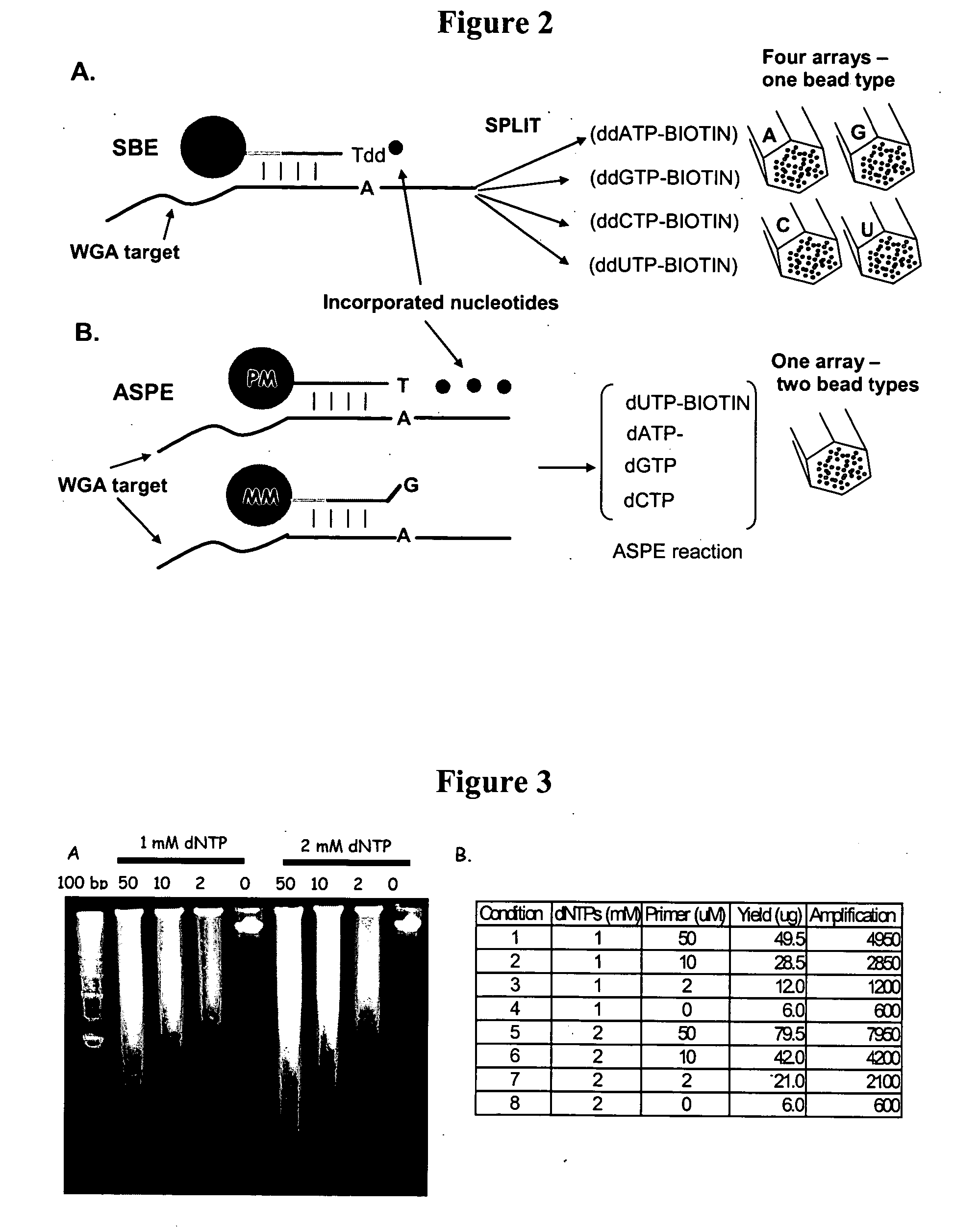
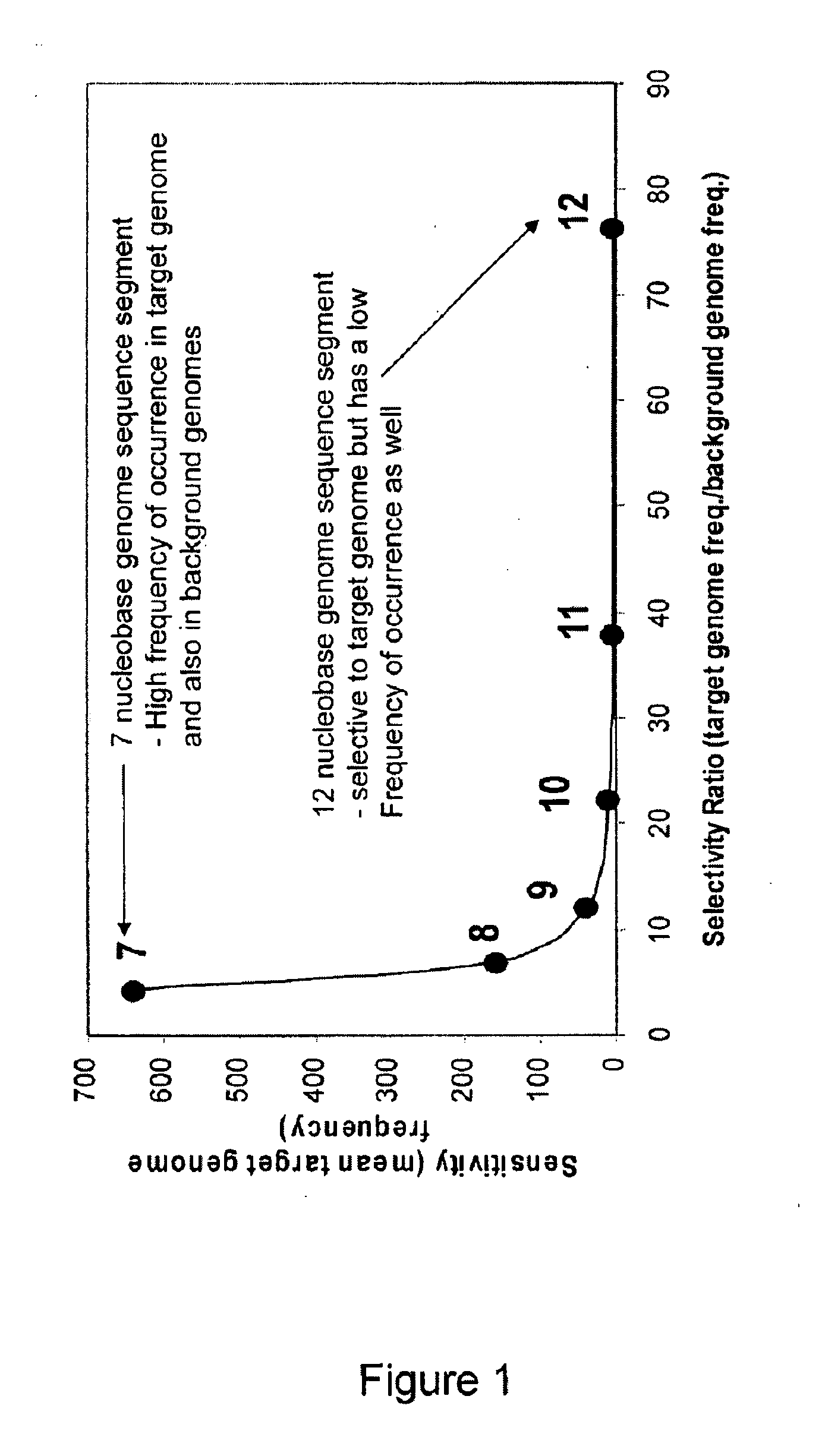
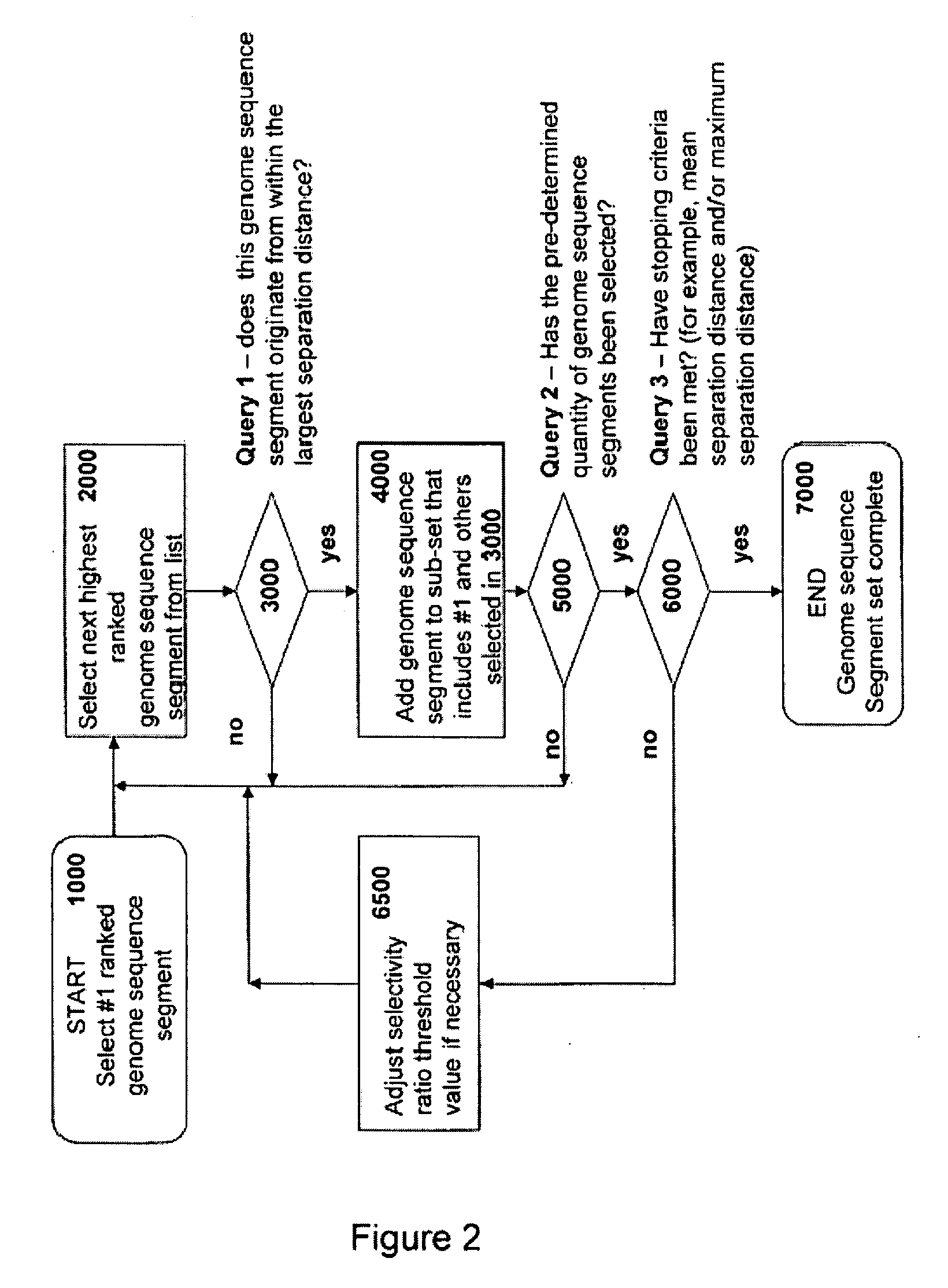
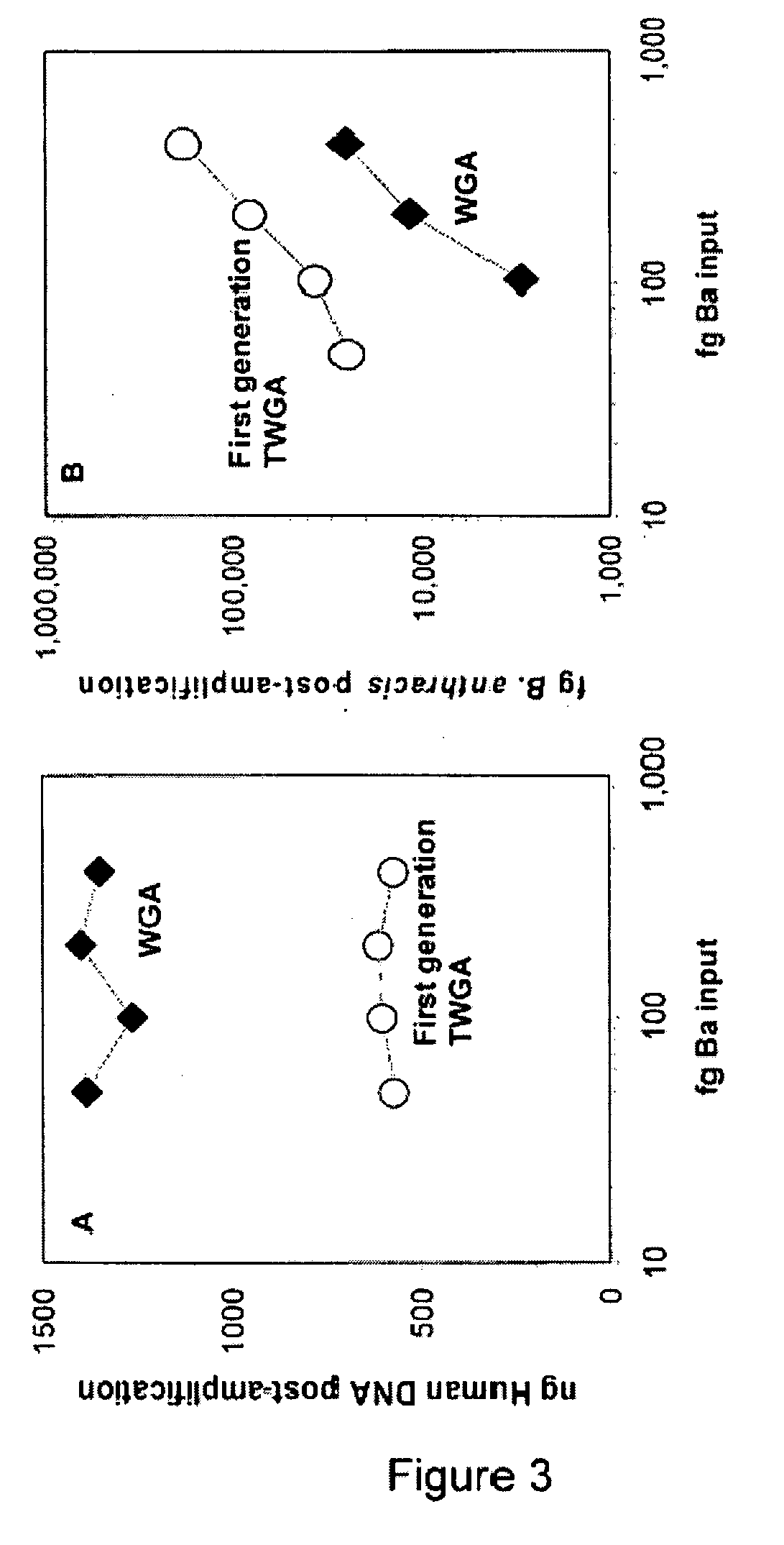
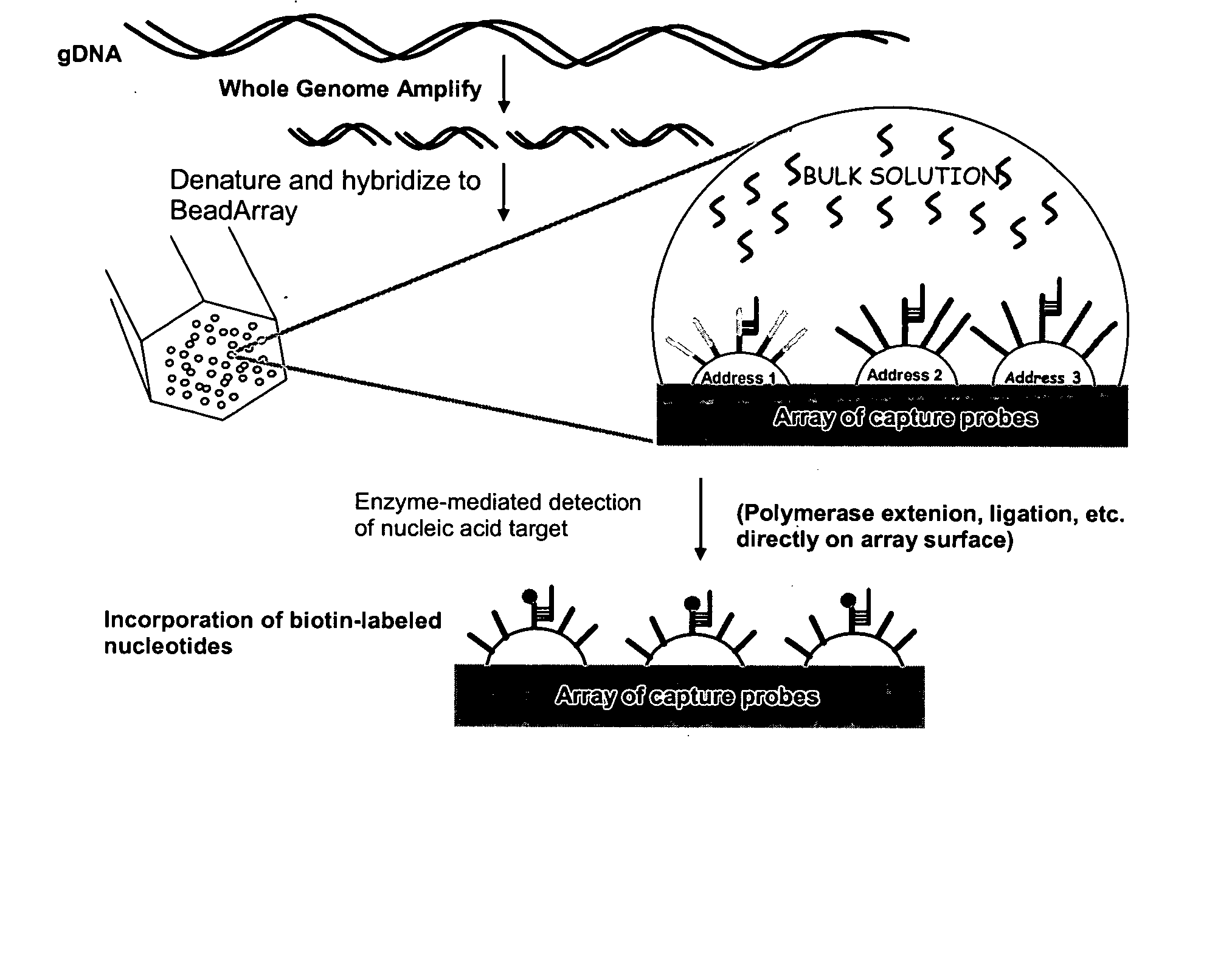
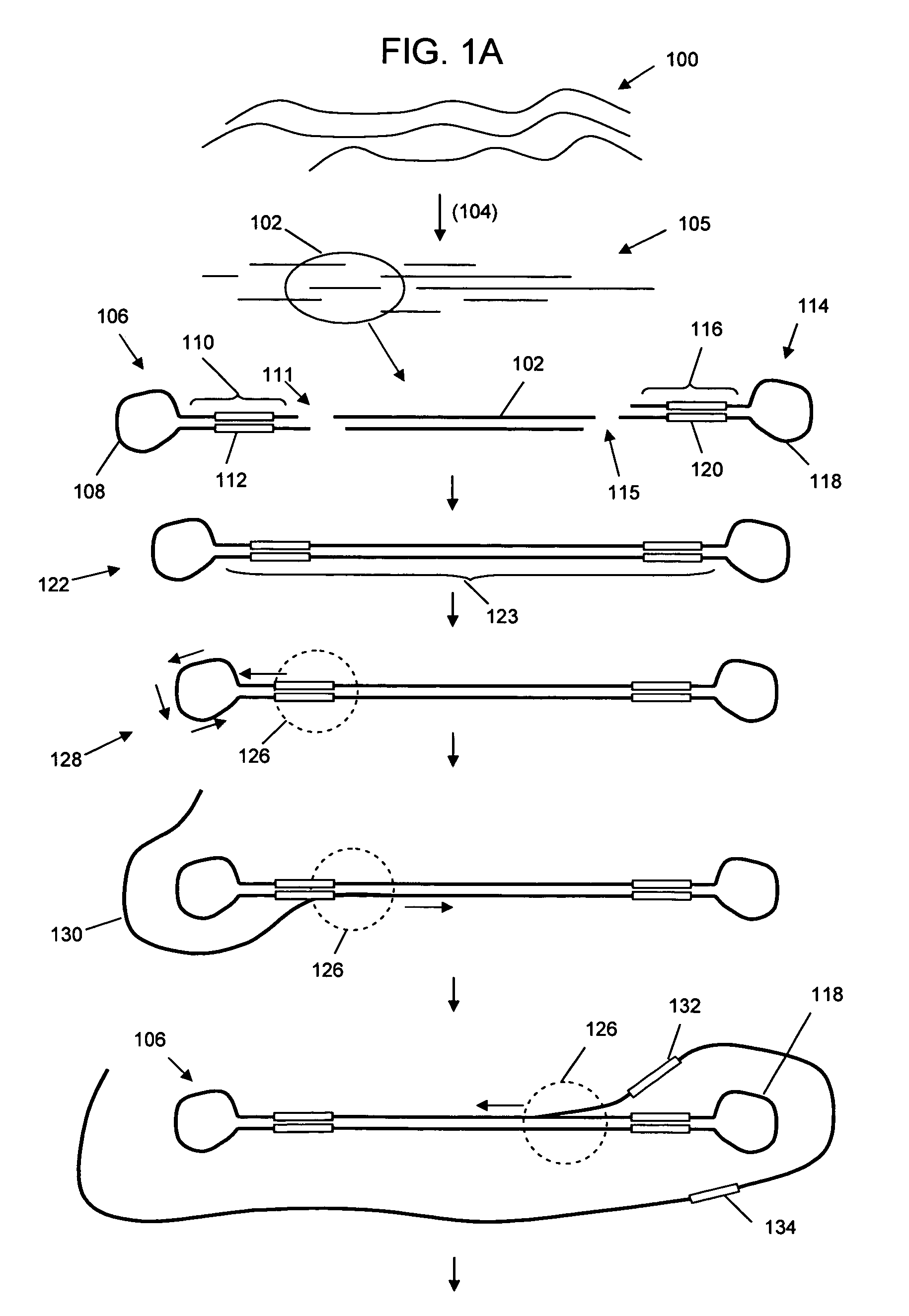
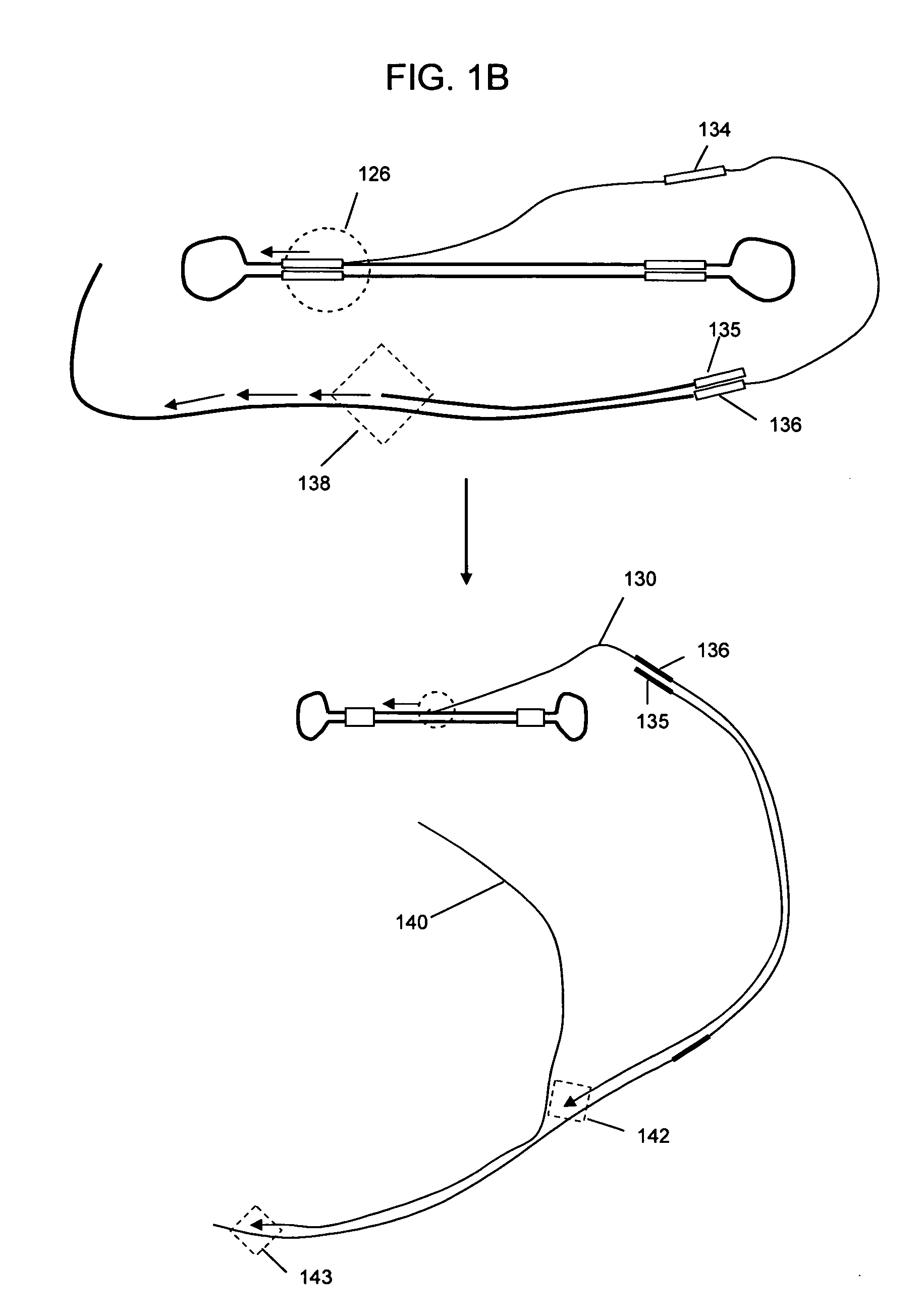
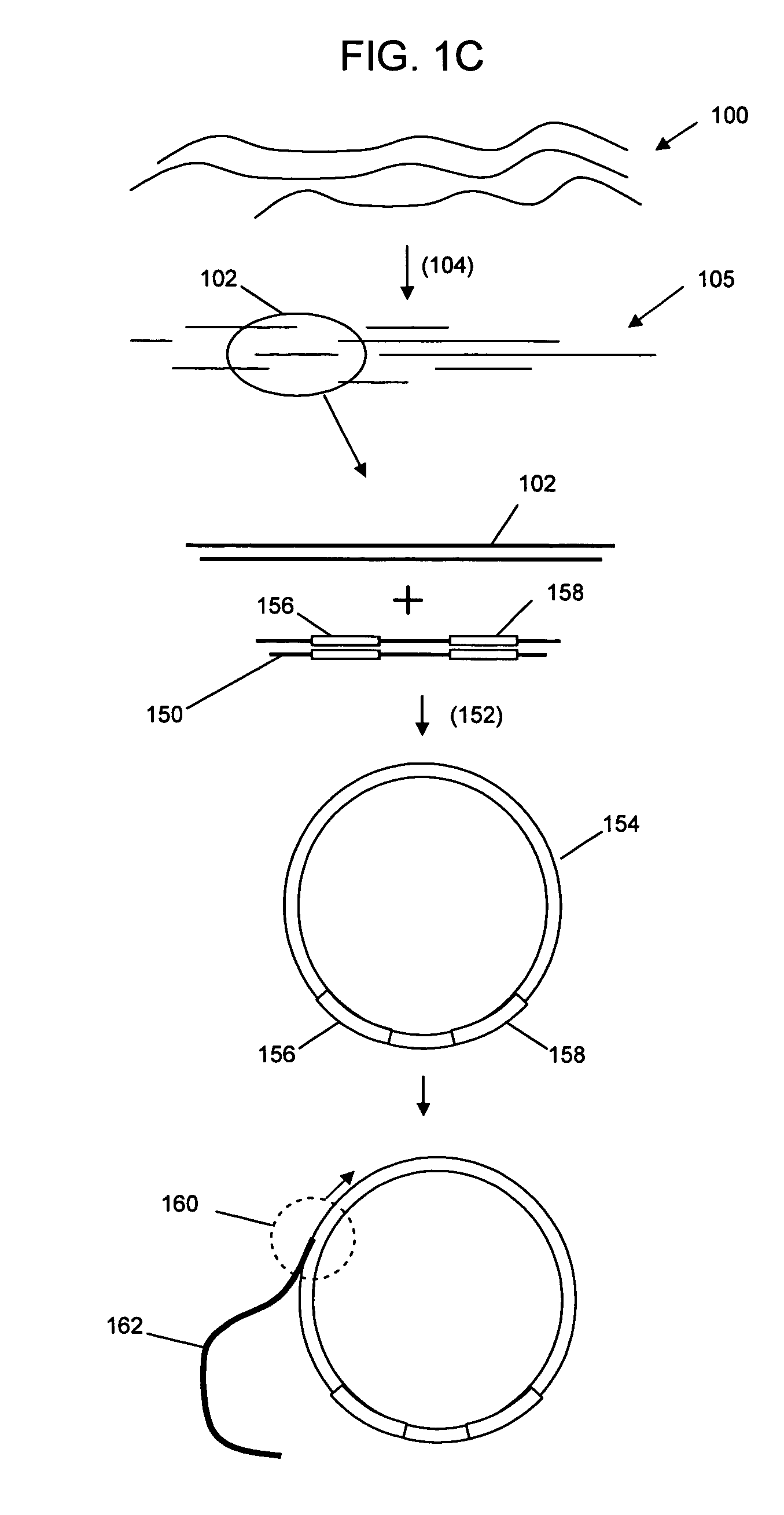
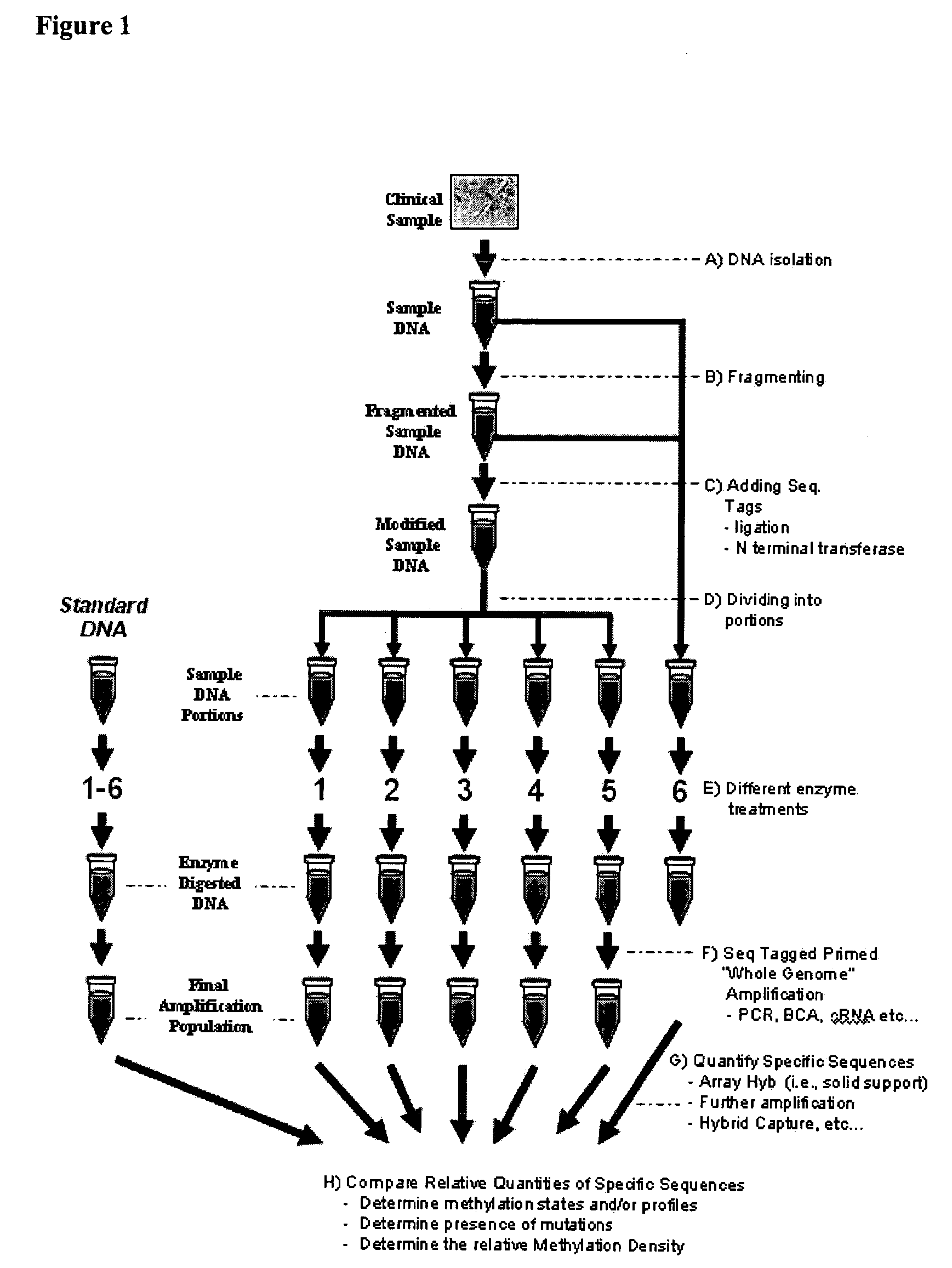
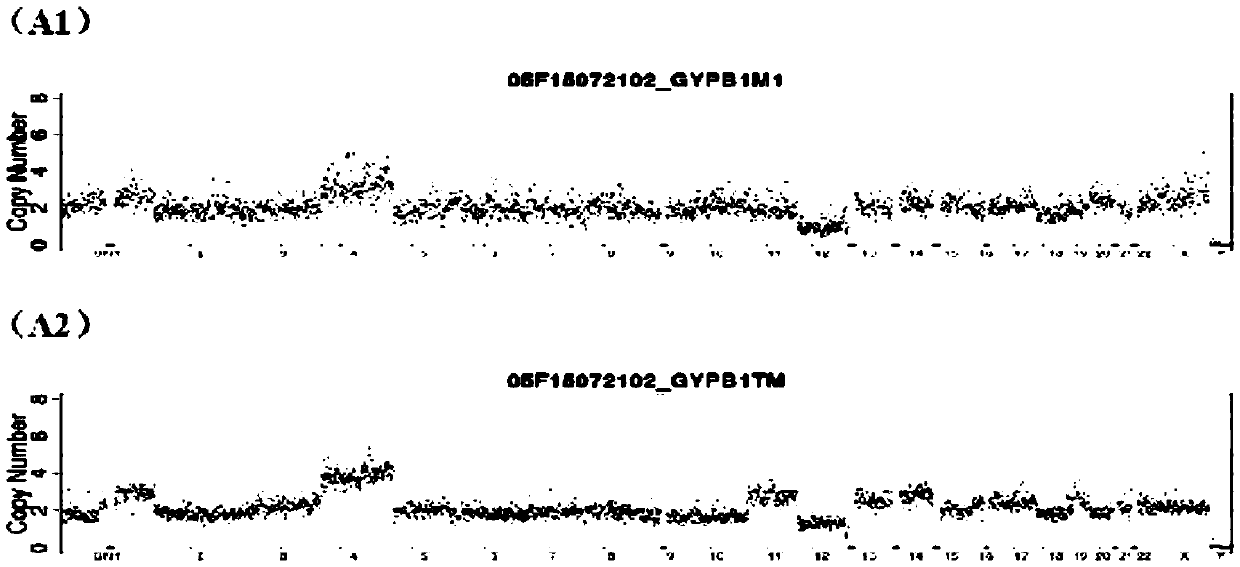
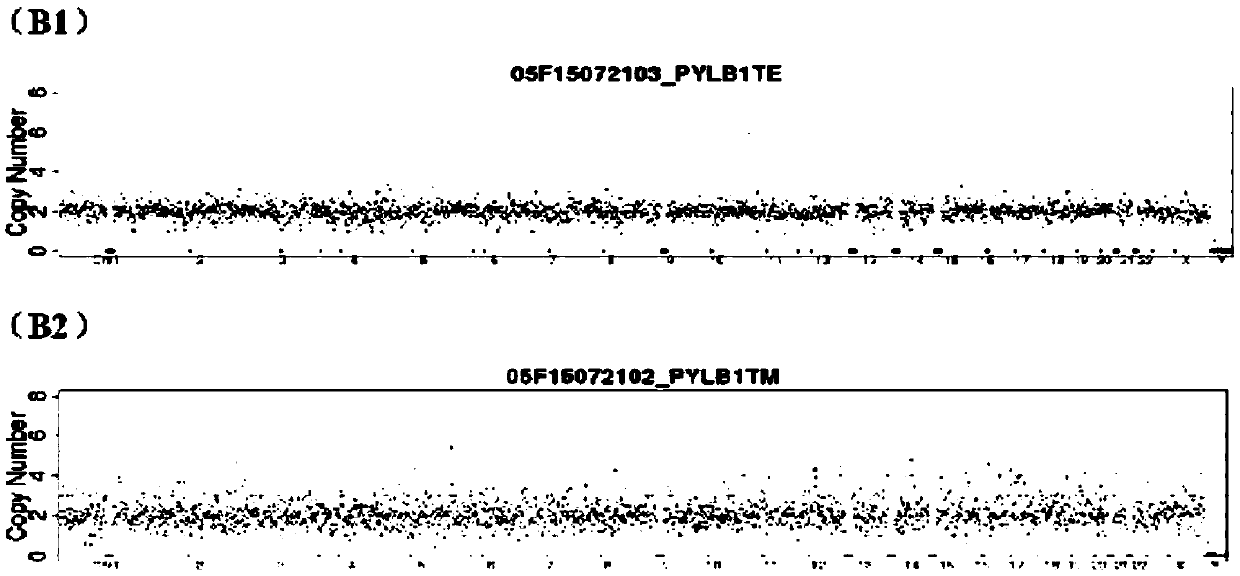
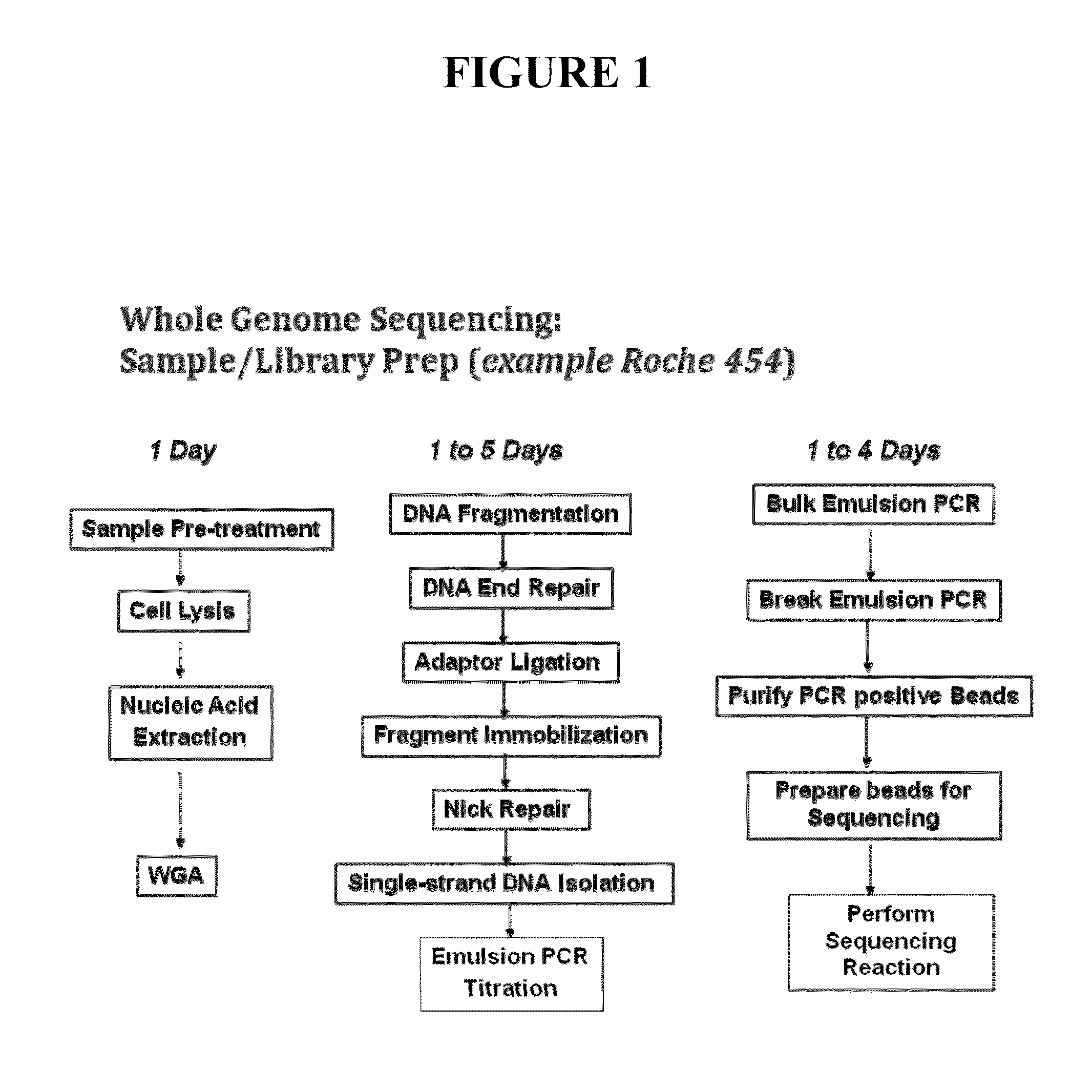
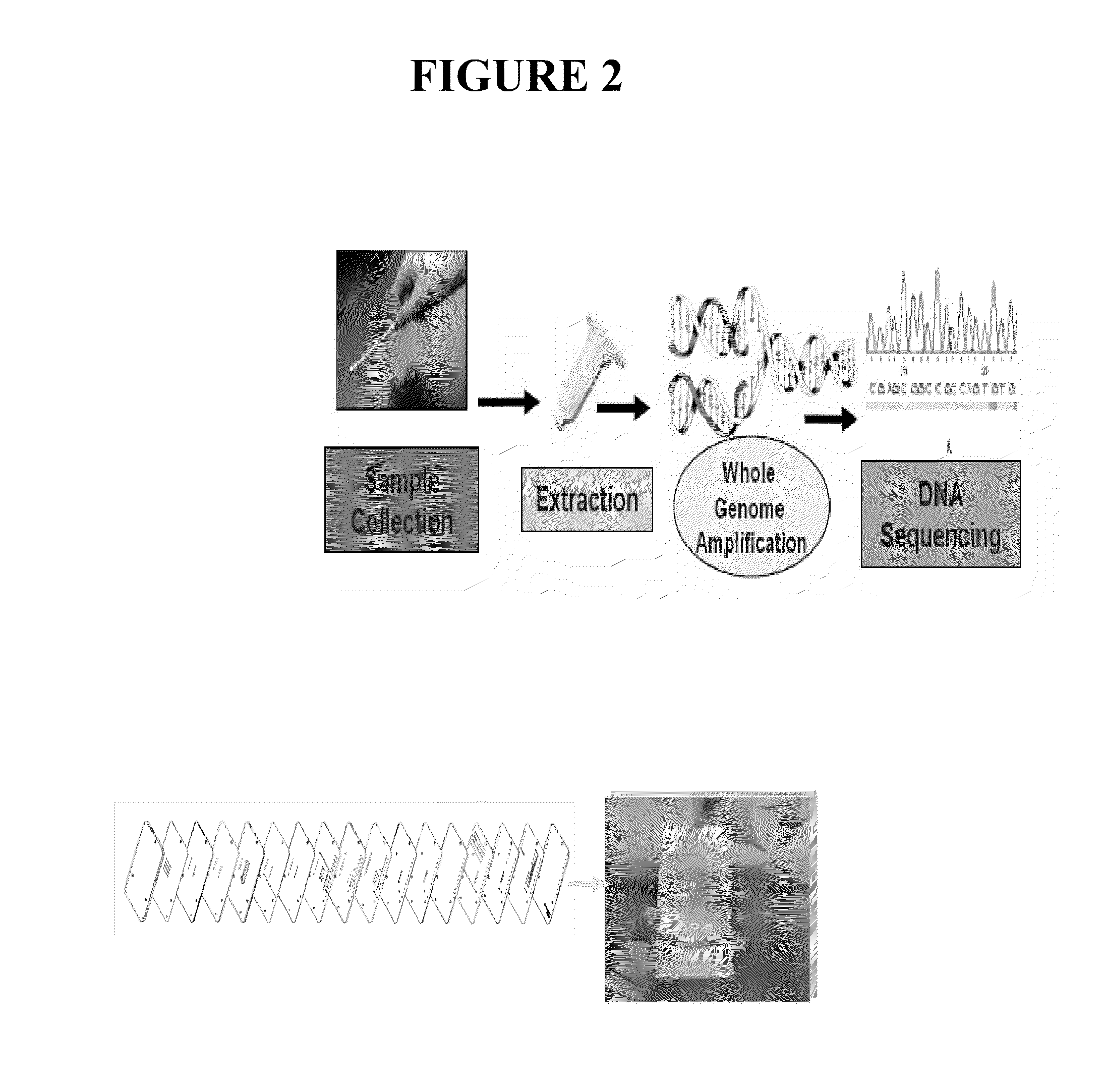
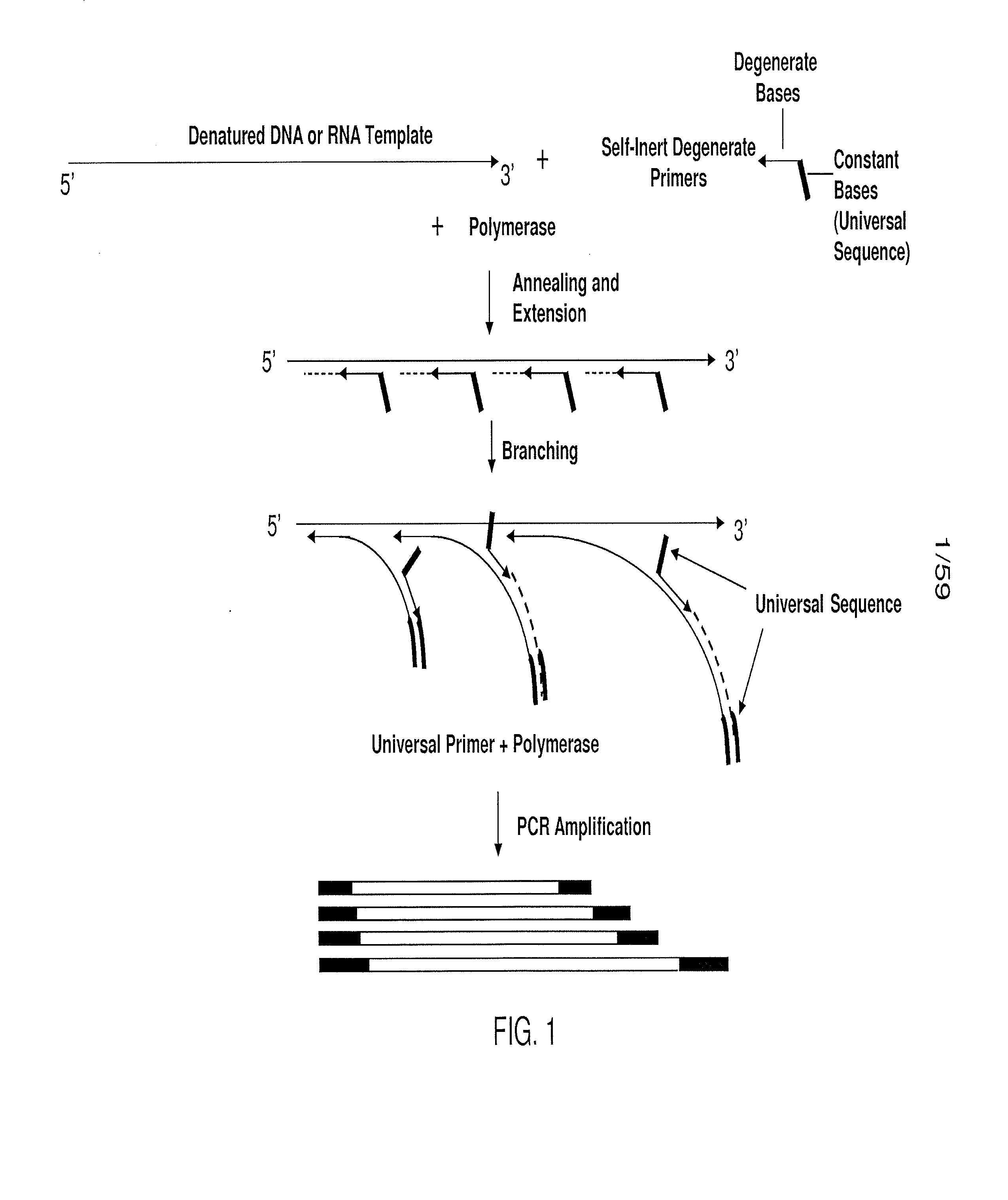
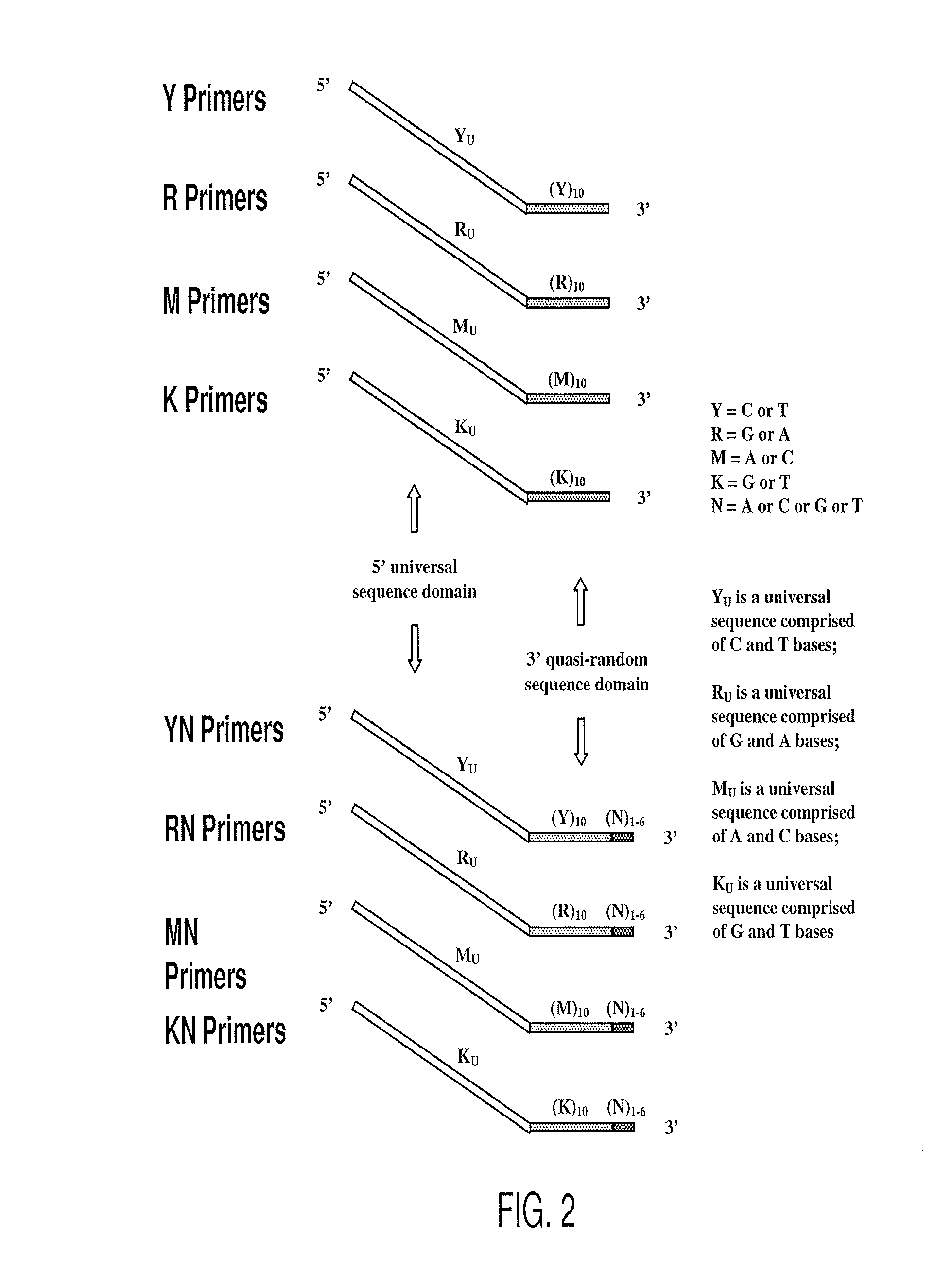
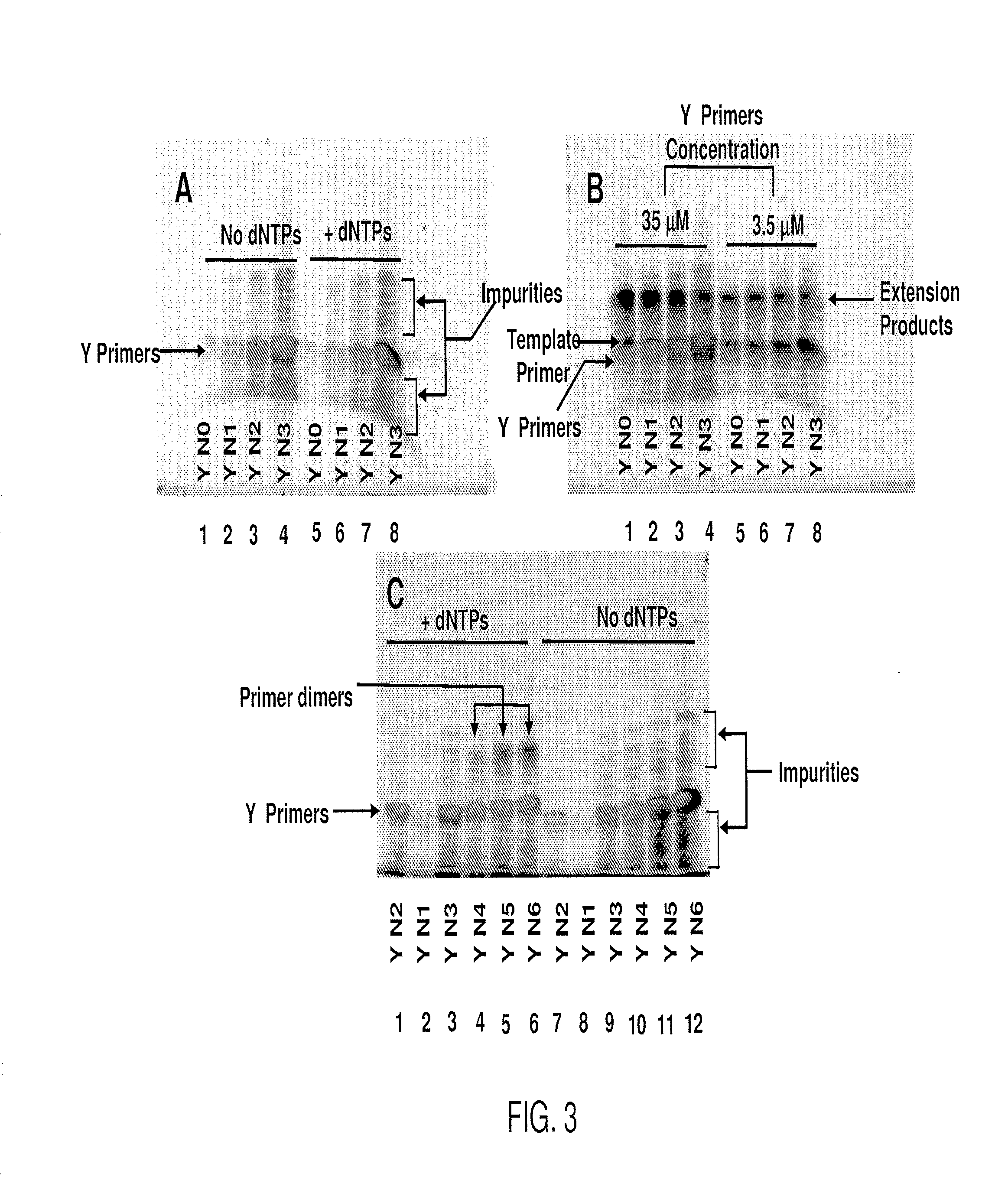
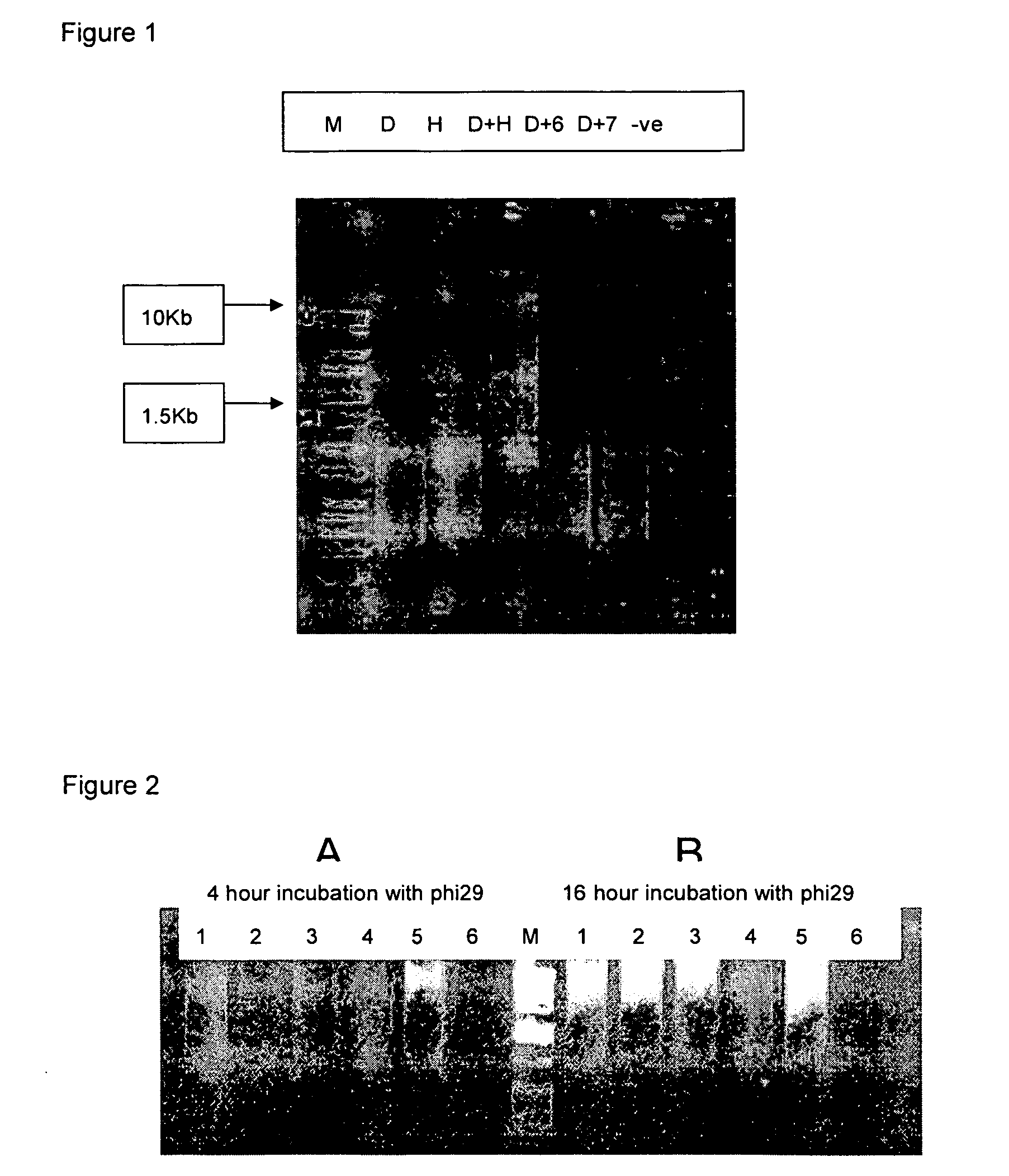
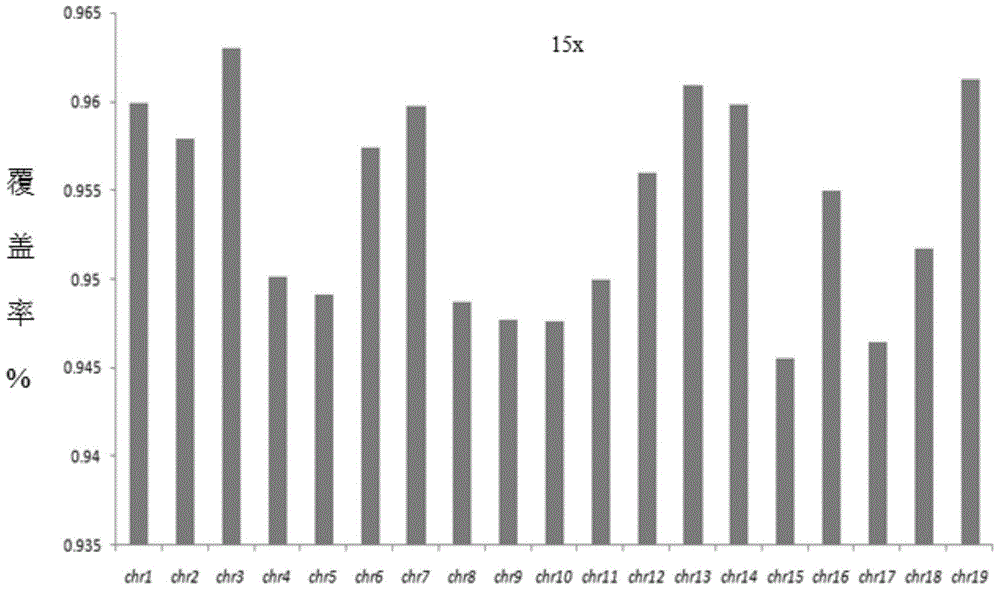
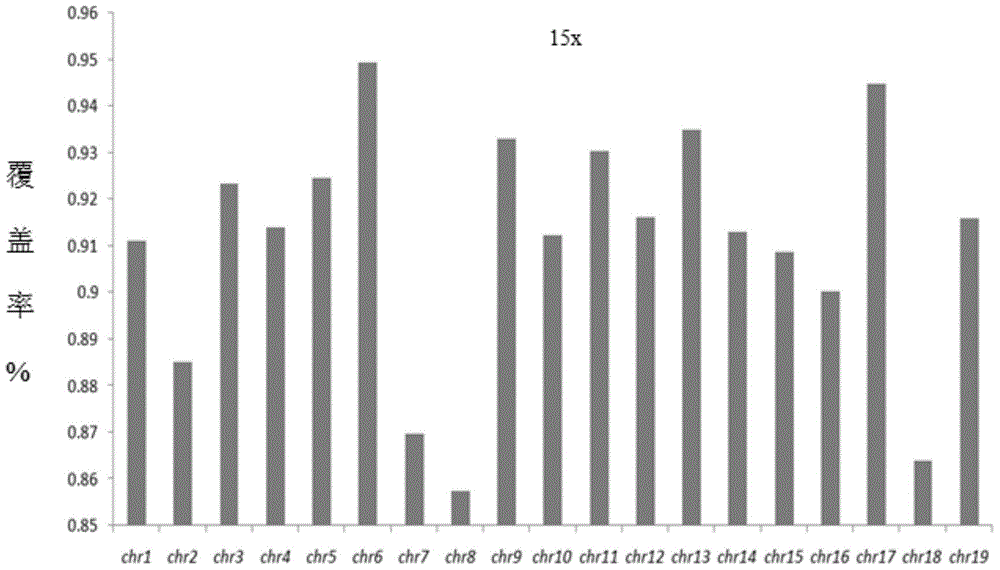
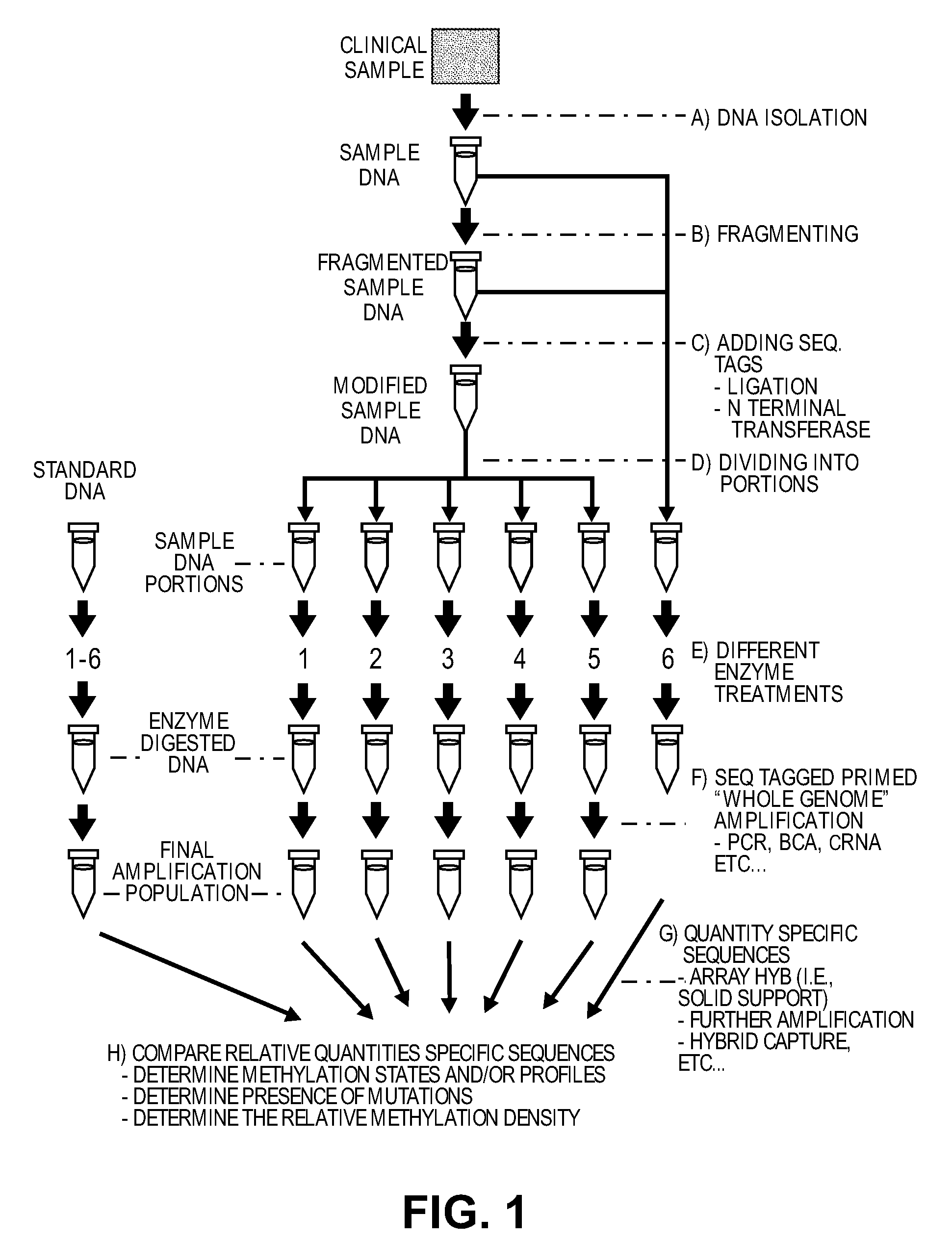
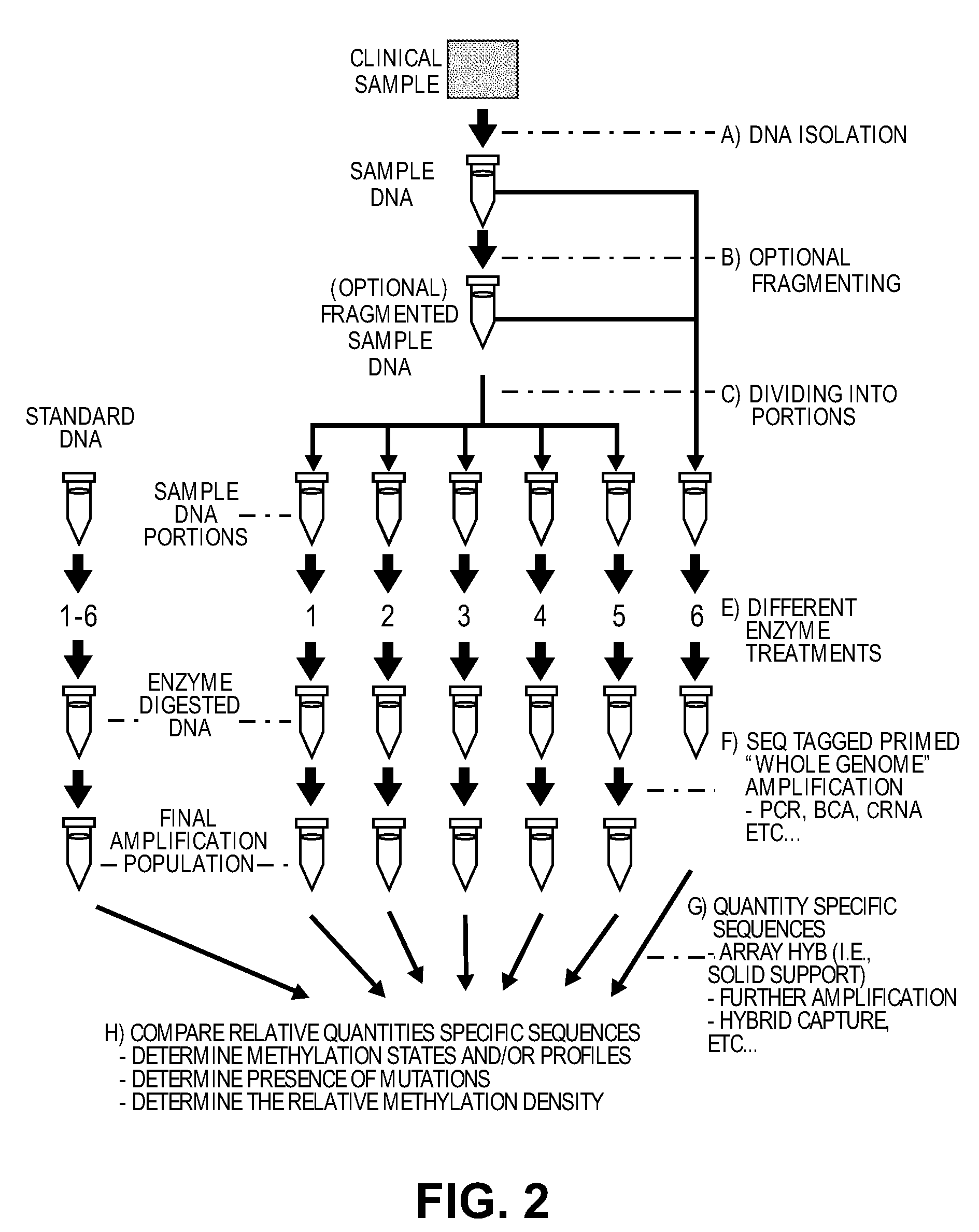
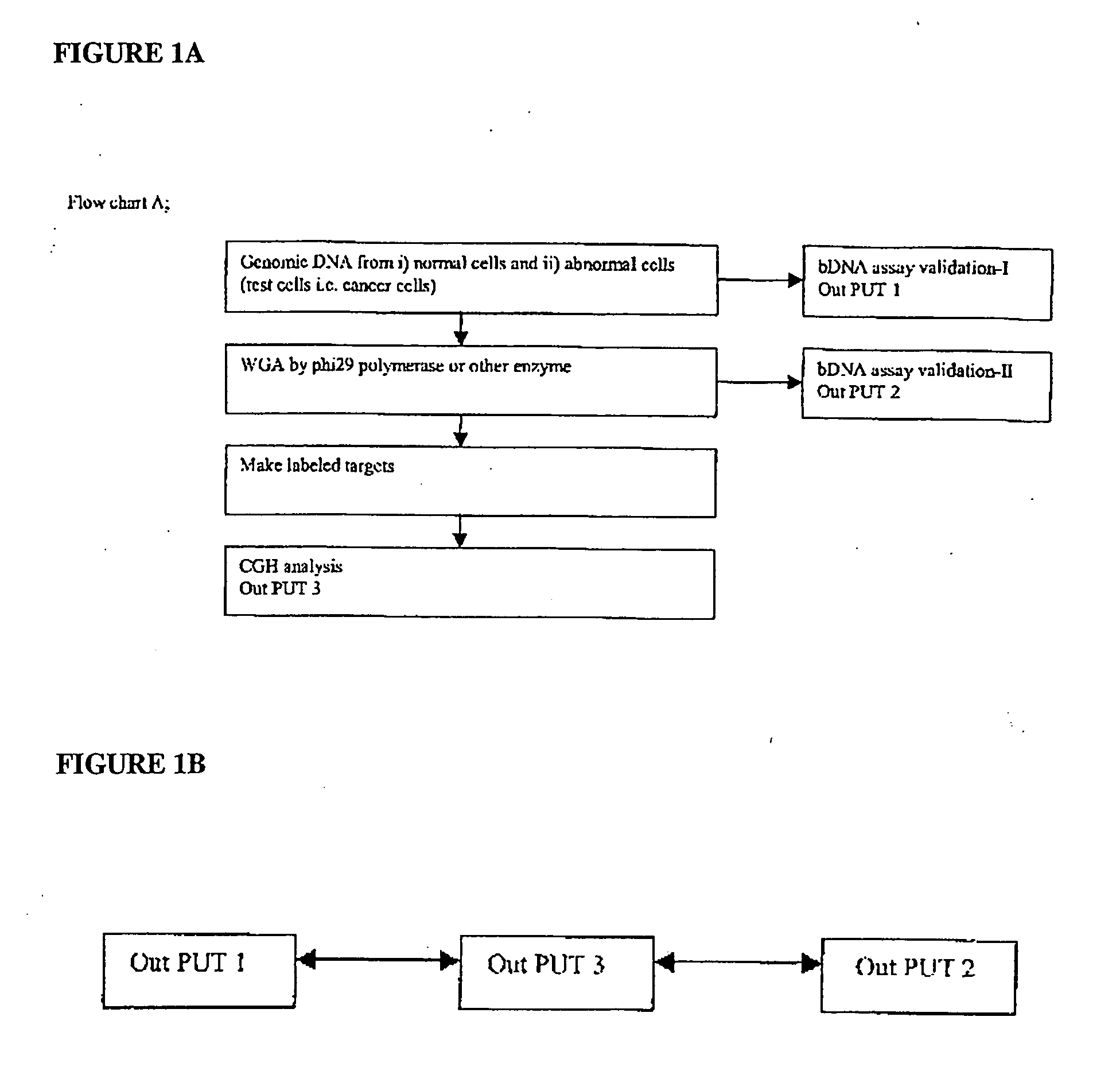
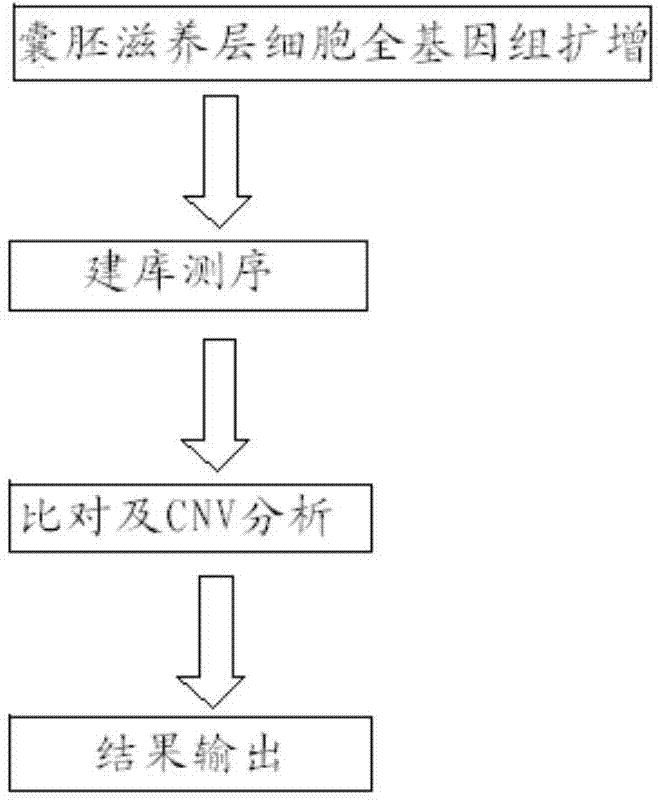
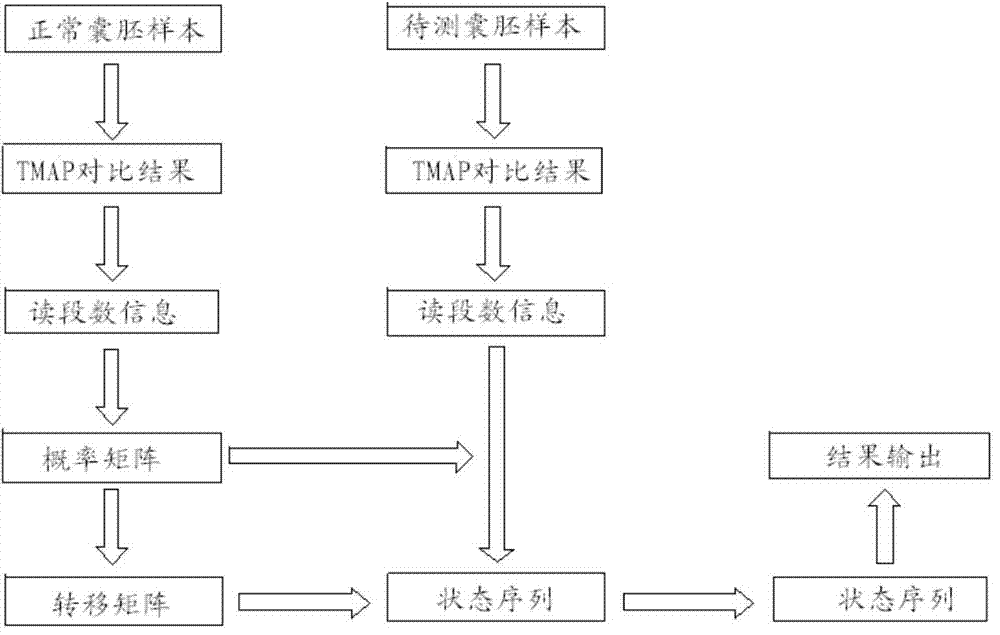
Patents
Literature
154 results about "Whole Genome Amplification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Any technique designed to amplify a limited genomic DNA sample so as to generate a new sample that is indistinguishable from the original but with a higher DNA concentration.
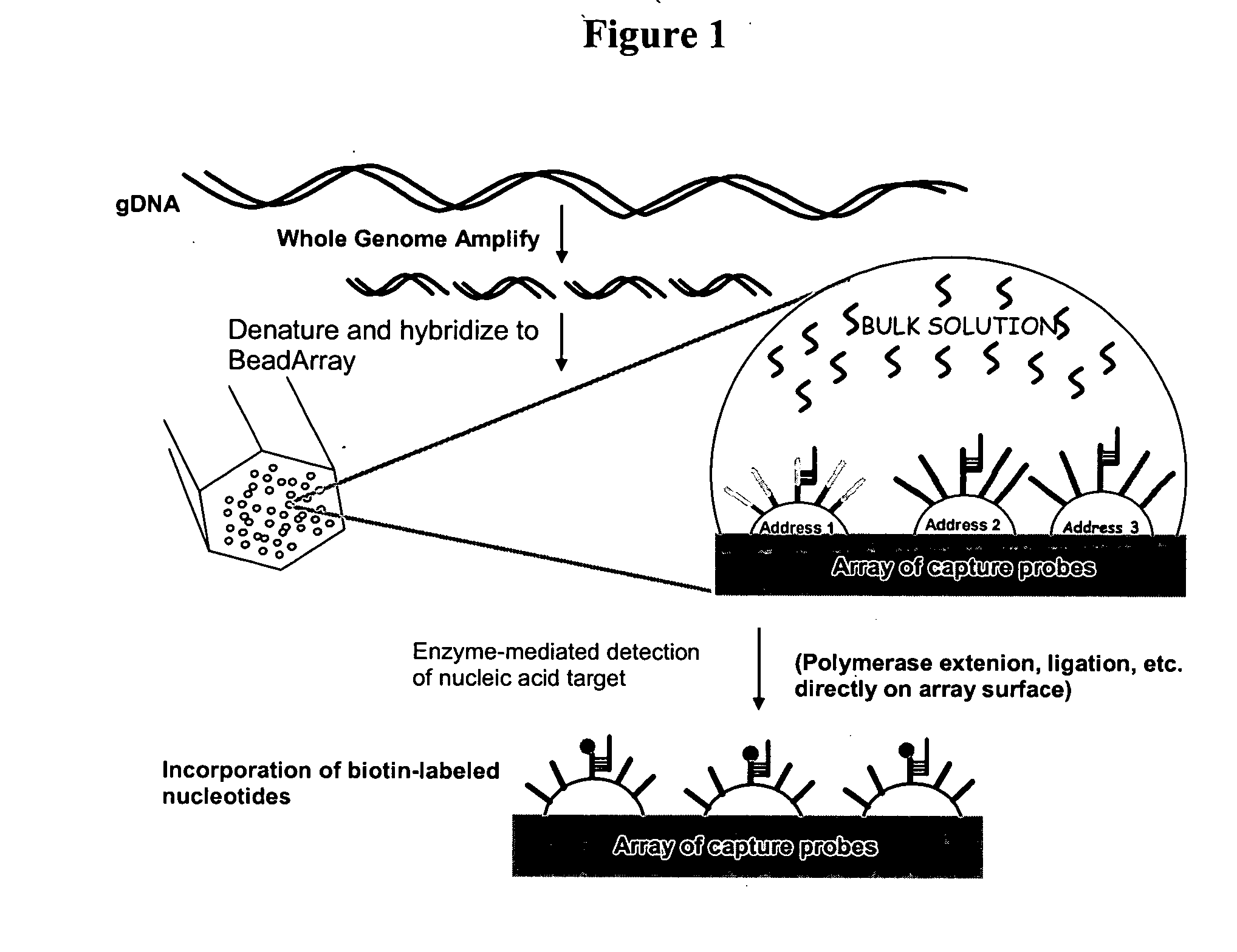
Methods and compositions for whole genome amplification and genotyping
InactiveUS20050181394A1Material nanotechnologyMicrobiological testing/measurementGenomic SegmentGenotyping
This invention provides methods of amplifying genomic DNA to obtain an amplified representative population of genome fragments. Methods are further provided for obtaining amplified genomic DNA representations of a desired complexity. The invention further provides methods for simultaneously detecting large numbers of typable loci for an amplified representative population of genome fragments. Accordingly the methods can be used to genotype individuals on a genome-wide scale.
Owner:ILLUMINA INC
Methods and compositions for whole genome amplification and genotyping
This invention provides methods of amplifying genomic DNA to obtain an amplified representative population of genome fragments. Methods are further provided for obtaining amplified genomic DNA representations of a desired complexity. The invention further provides methods for simultaneously detecting large numbers of typable loci for an amplified representative population of genome fragments. Accordingly the methods can be used to genotype individuals on a genome-wide scale.
Owner:ILLUMINA INC
Targeted whole genome amplification method for identification of pathogens
InactiveUS20100035232A1Organic active ingredientsTime-of-flight spectrometersNucleic acid sequencingNucleic acid sequence
The methods disclosed herein relate to methods and compositions for amplifying nucleic acid sequences, more specifically, from nucleic acid sequences of pathogens by targeted whole genome amplification.
Owner:IBIS BIOSCI
Methods and compositions for whole genome amplification and genotyping
This invention provides methods of amplifying genomic DNA to obtain an amplified representative population of genome fragments. Methods are further provided for obtaining amplified genomic DNA representations of a desired complexity. The invention further provides methods for simultaneously detecting large numbers of typable loci for an amplified representative population of genome fragments. Accordingly the methods can be used to genotype individuals on a genome-wide scale.
Owner:ILLUMINA INC
Single cell nucleic acid analysis
The present invention provides methods for analysis of genomic DNA and / or RNA from small samples or even single cells. Methods for analyzing genomic DNA can entail whole genome amplification (WGA), followed by preamplification and amplification of selected target nucleic acids. Methods for analyzing RNA can entail reverse transcription of the desired RNA, followed by preamplification and amplification of selected target nucleic acids.
Owner:FLUIDIGM CORP
Amplification and analysis of whole genome and whole transcriptome libraries generated by a DNA polymerization process
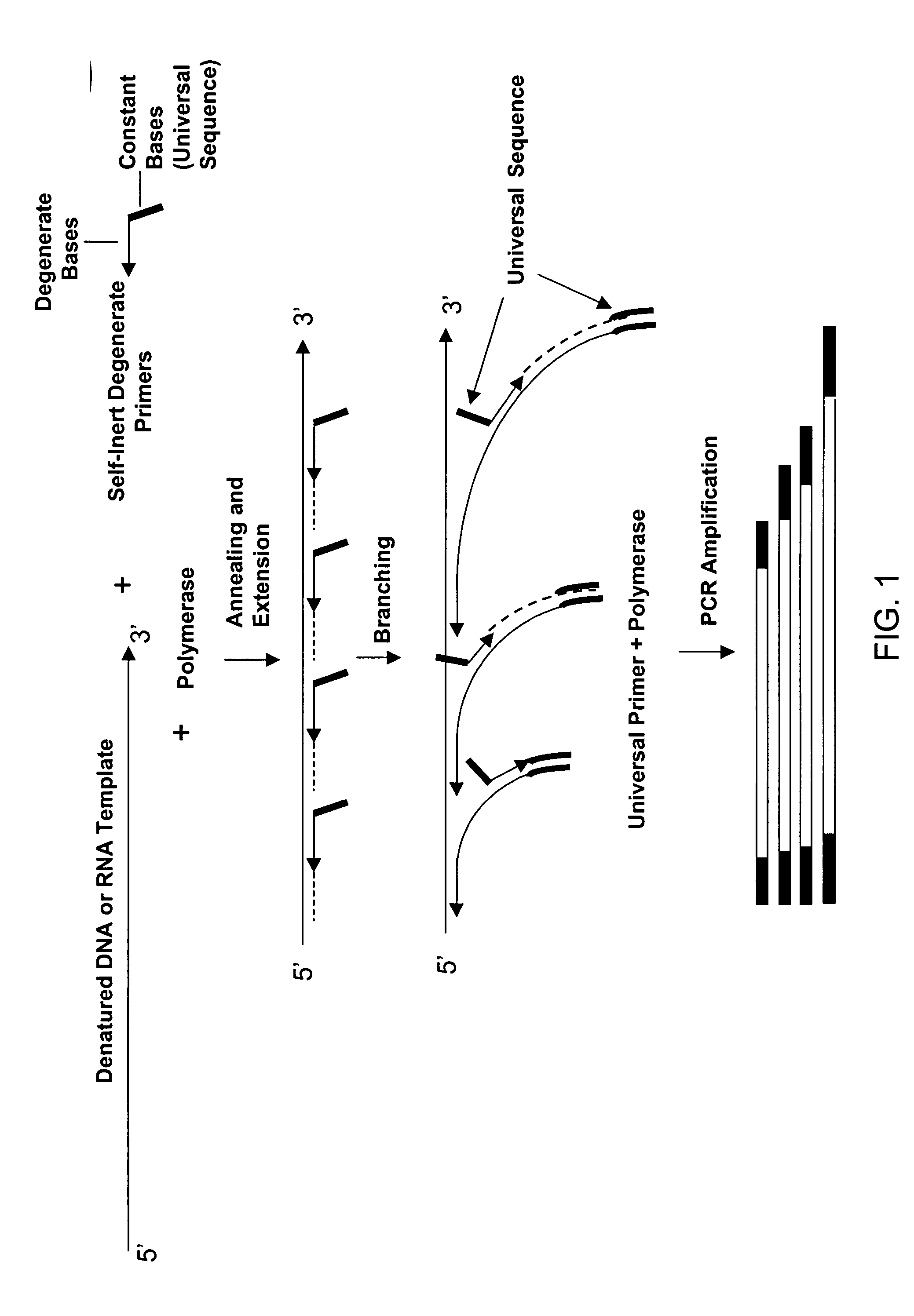
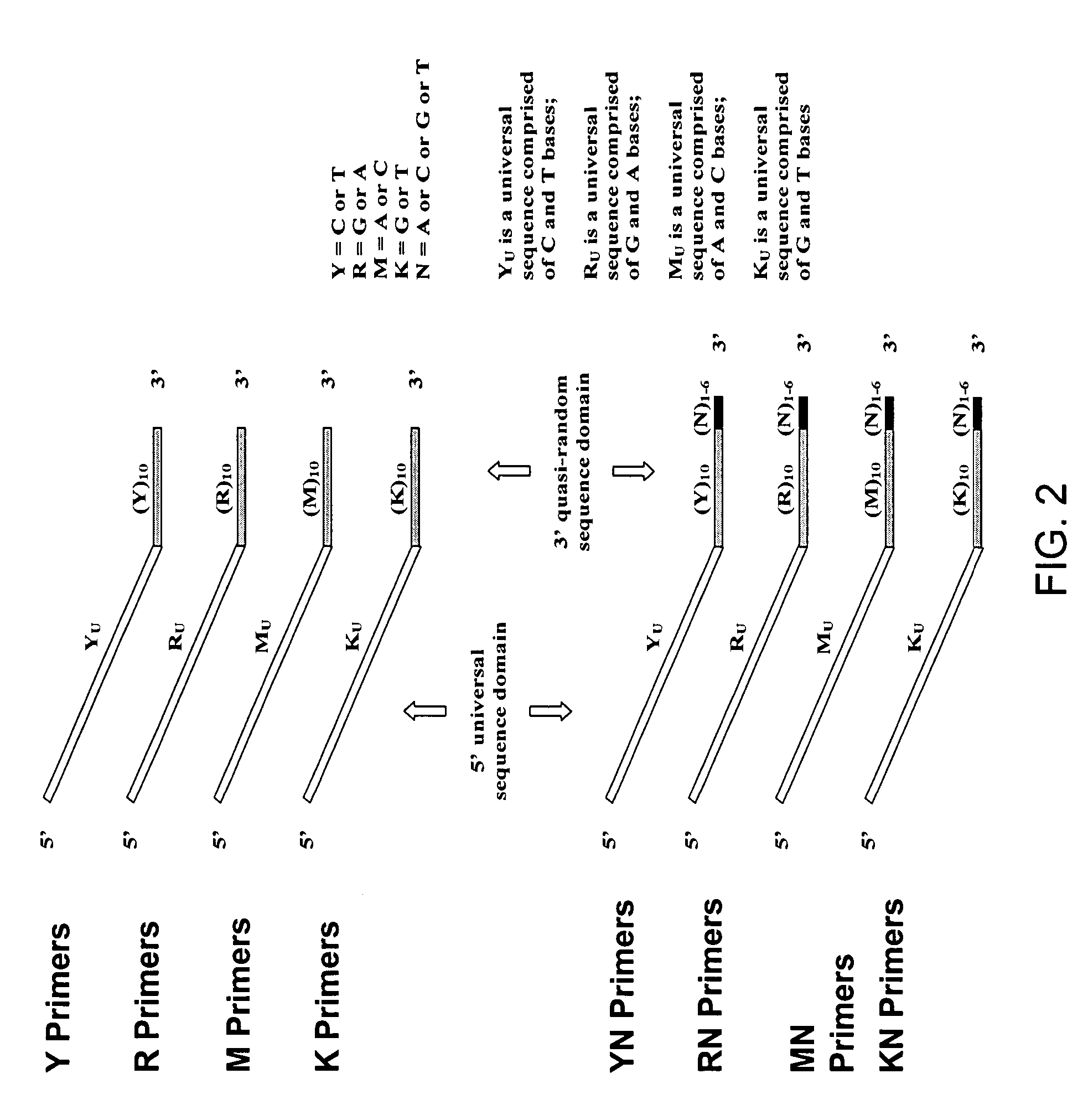
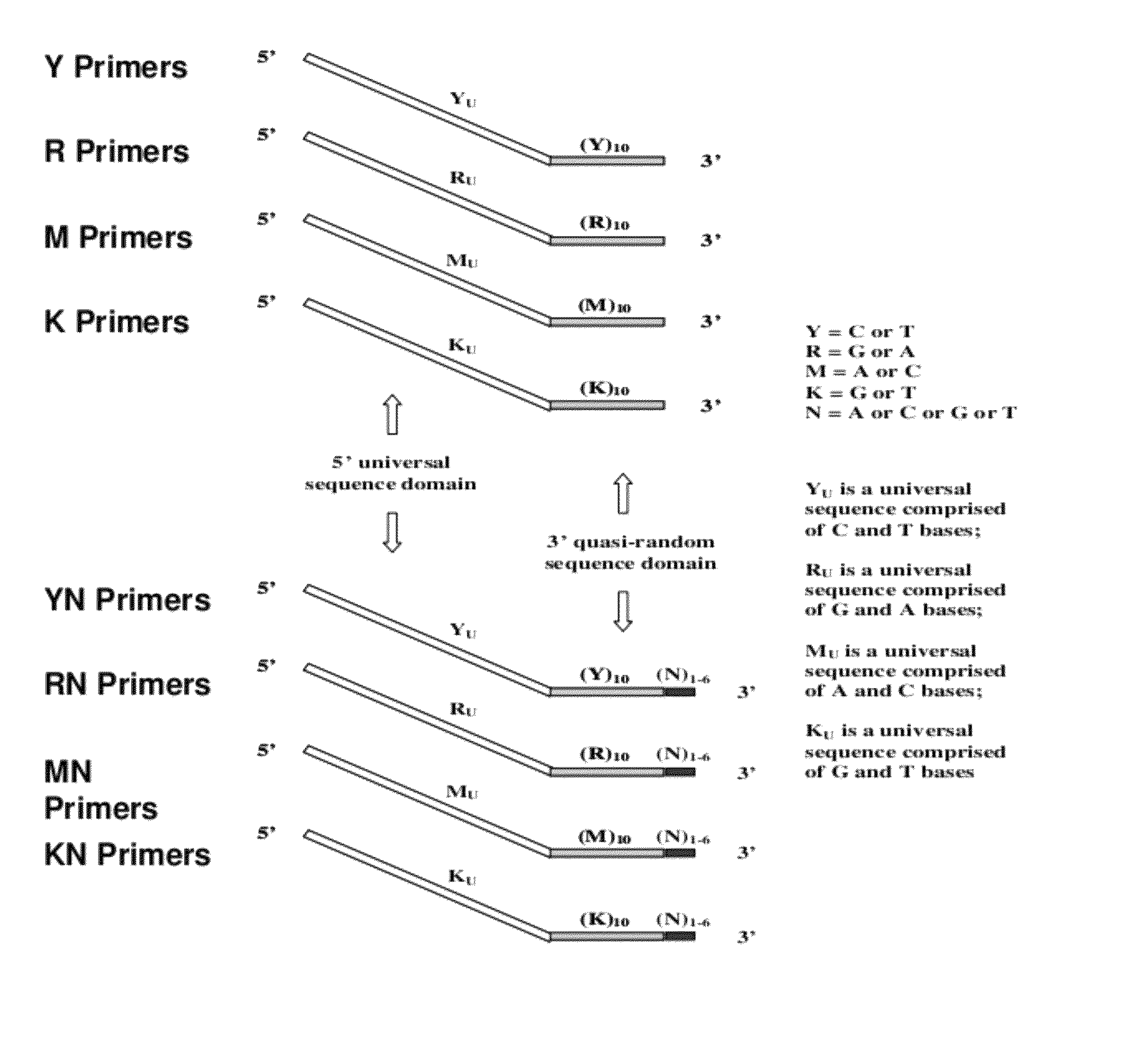
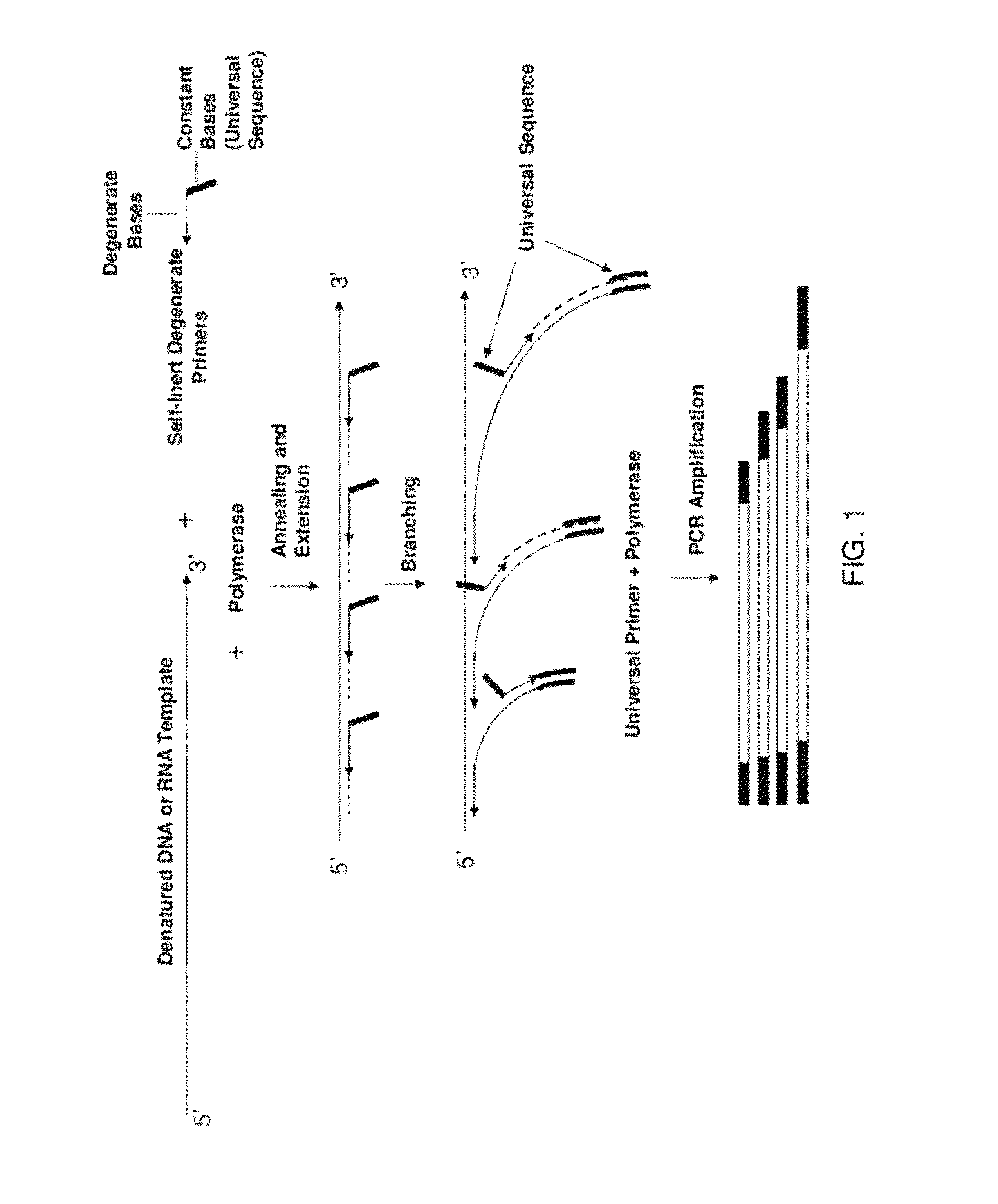
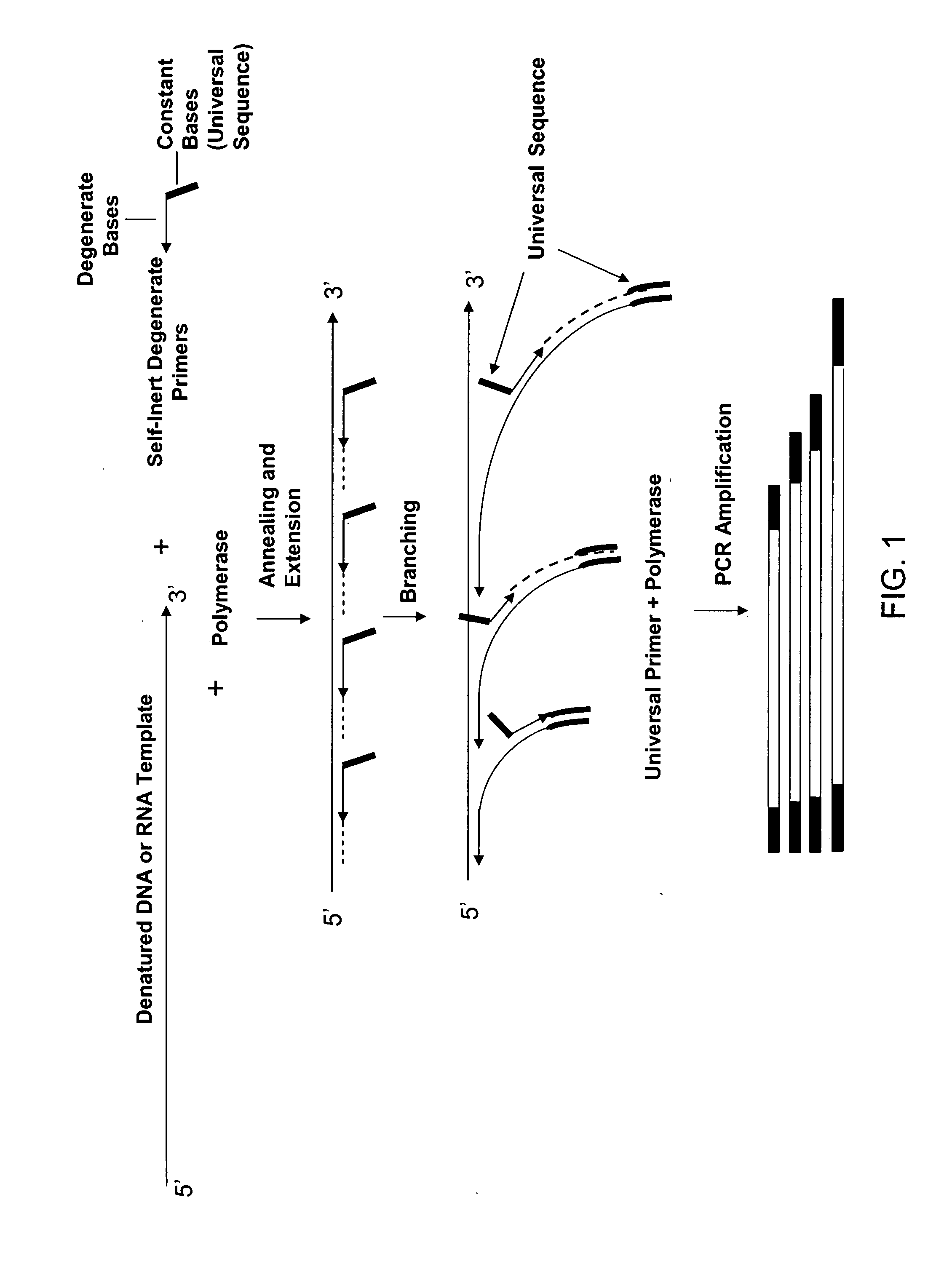
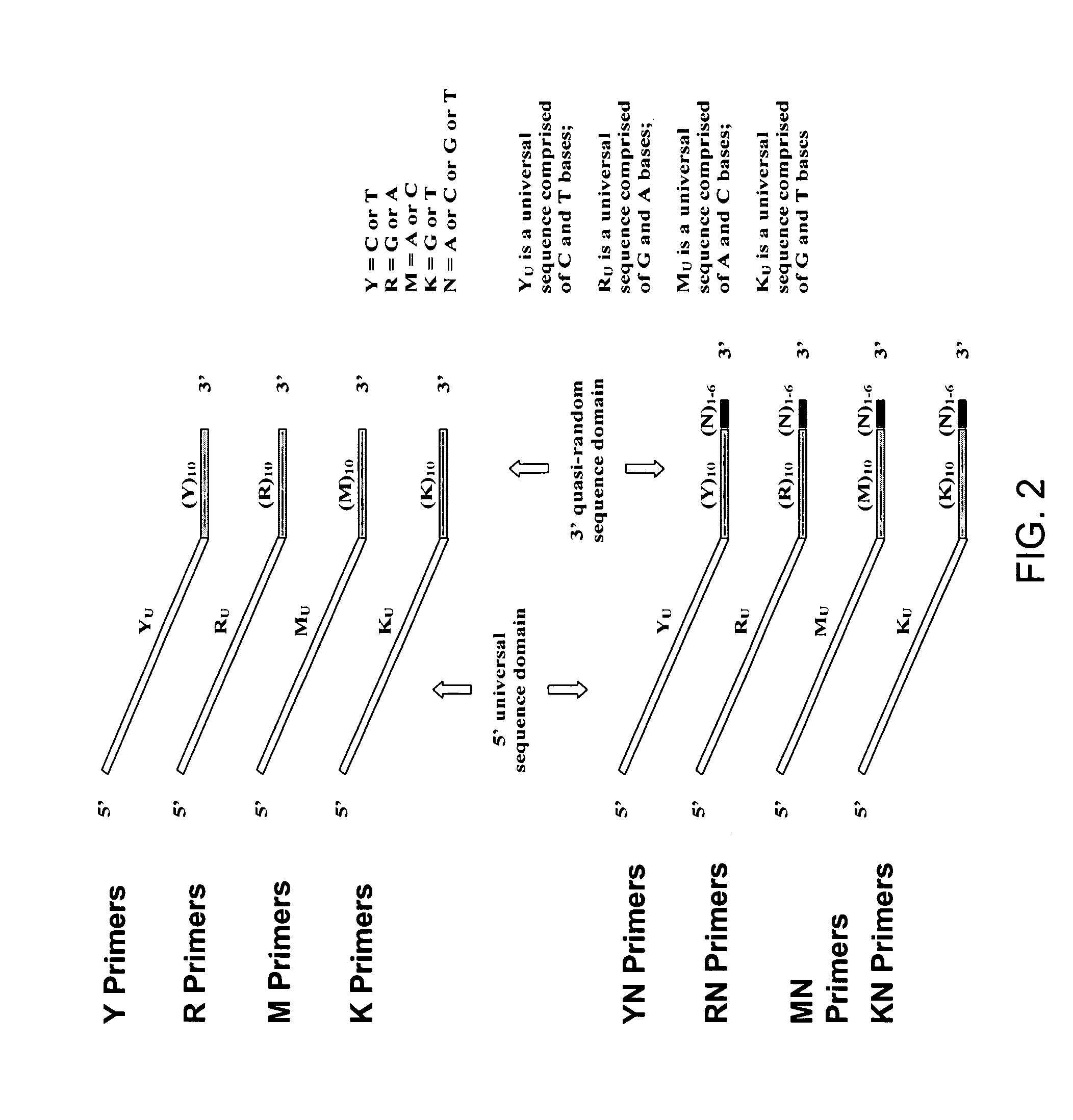
The present invention regards a variety of methods and compositions for whole genome amplification and whole transcriptome amplification. In a particular aspect of the present invention, there is a method of amplifying a genome comprising a library generation step followed by a library amplification step. In specific embodiments, the library generating step utilizes specific primer mixtures and a DNA polymerase, wherein the specific primer mixtures are designed to eliminate ability to self-hybridize and / or hybridize to other primers within a mixture but efficiently and frequently prime nucleic acid templates.
Owner:TAKARA BIO USA INC
Amplification and analysis of whole genome and whole transcriptome libraries generated by a DNA polymerization process
ActiveUS8206913B1Increase success rateEnhance single-copy sensitivityMicrobiological testing/measurementFermentationPolymerase LA-DNA
The present invention regards a variety of methods and compositions for whole genome amplification and whole transcriptome amplification. In a particular aspect of the present invention, there is a method of amplifying a genome comprising a library generation step followed by a library amplification step. In specific embodiments, the library generating step utilizes specific primer mixtures and a DNA polymerase, wherein the specific primer mixtures are designed to eliminate ability to self-hybridize and / or hybridize to other primers within a mixture but efficiently and frequently prime nucleic acid templates.
Owner:TAKARA BIO USA INC
Selective genome amplification
The invention provides methods and compositions for amplifying selected polynucleotides, especially selected subsets of restriction fragments. Generally, methods of the invention are implemented by ligating adaptors containing at least one promoter sequence to such fragments under conditions that promote the formation of closed single stranded or double stranded structures, which are capable of serving as cyclical templates for transcription.
Owner:AGENCY FOR SCI TECH & RES
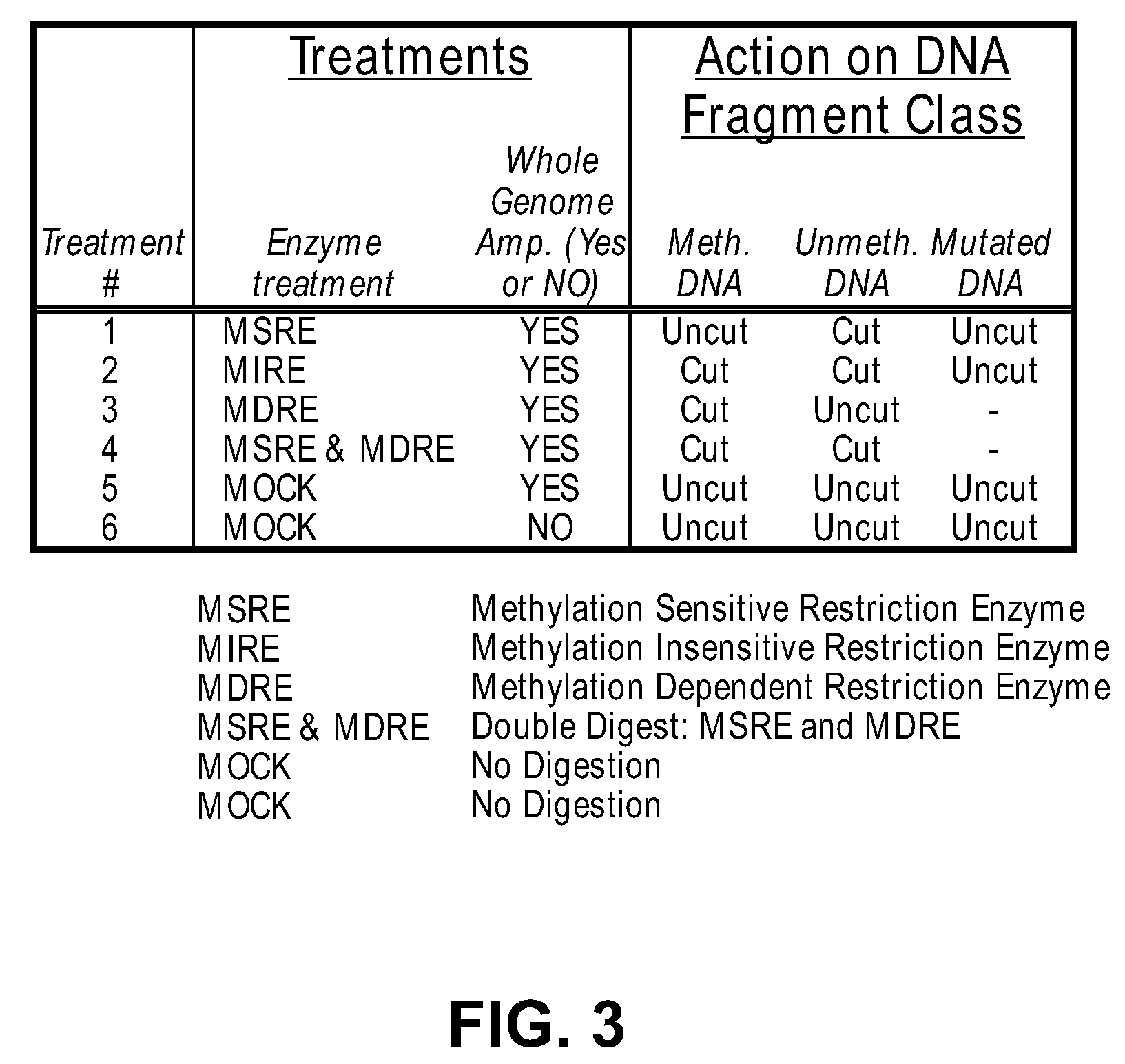
Differential enzymatic fragmentation by whole genome amplification
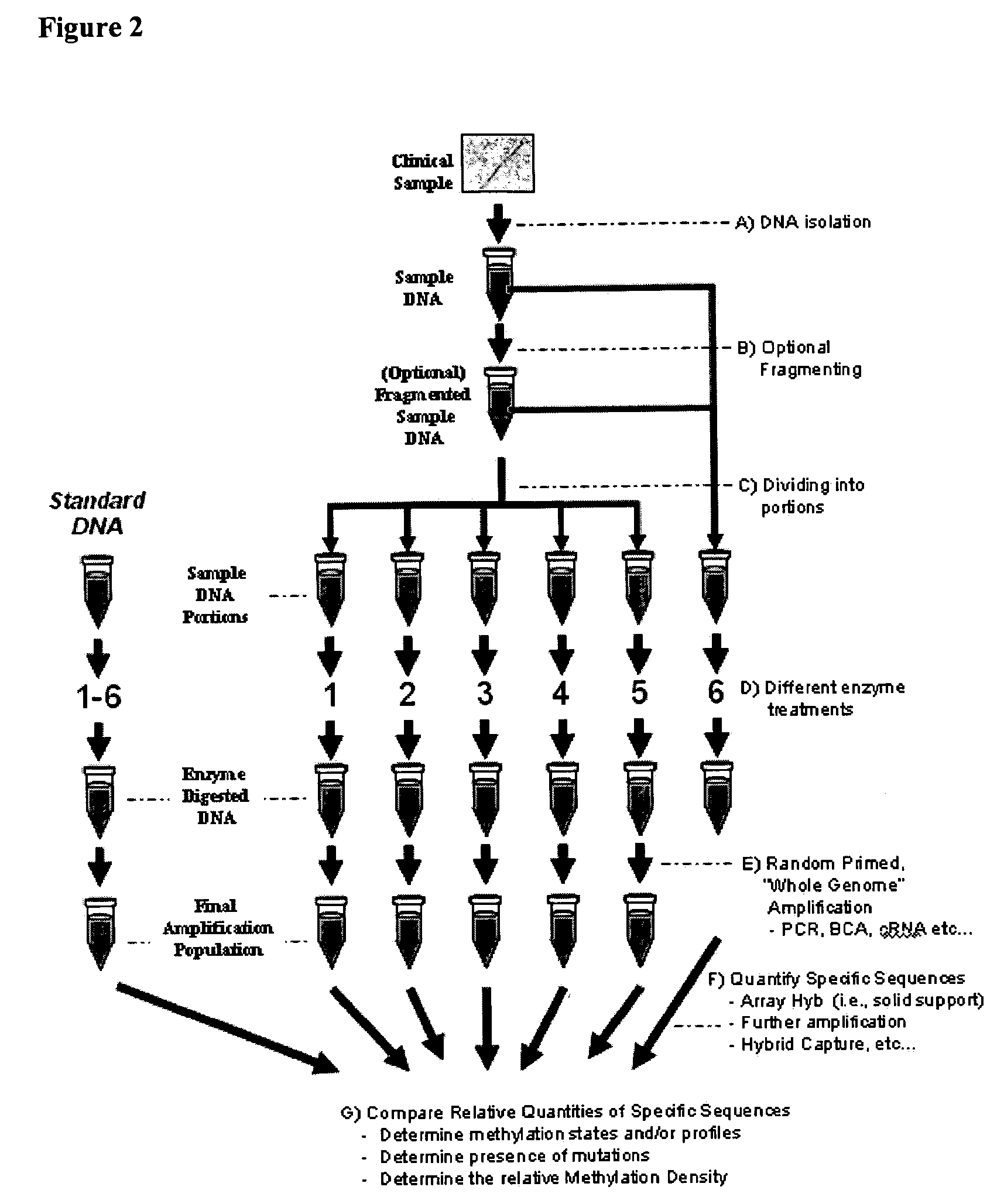
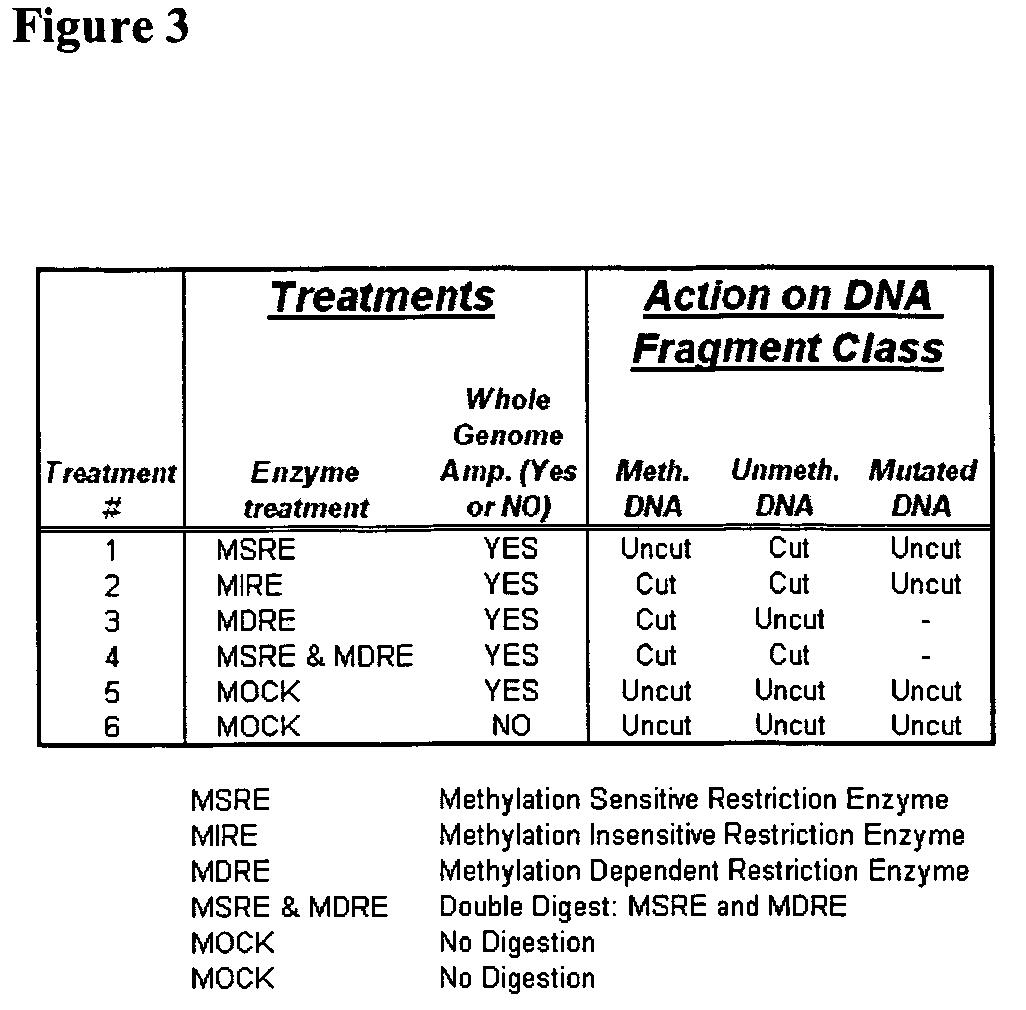
The present invention provides methods for detecting the presence of methylation at a locus within a population of nucleic acids.
Owner:ORION GENOMICS
Amplification and analysis of whole genome and whole transcriptome libraries generated by a DNA polymerization process
InactiveUS20070054311A1High throughput formatNucleotide librariesMicrobiological testing/measurementDna polymerasenPolymerase L
The present invention regards a variety of methods and compositions for whole genome amplification and whole transcriptome amplification. In a particular aspect of the present invention, there is a method of amplifying a genome comprising a library generation step followed by a library amplification step. In specific embodiments, the library generating step utilizes specific primer mixtures and a DNA polymerase, wherein the specific primer mixtures are designed to eliminate ability to self-hybridize and / or hybridize to other primers within a mixture but efficiently and frequently prime nucleic acid templates.
Owner:KAMBEROV EMMANUEL +6
Single-cell nucleic acid analysis
The present invention provides methods for analysis of genomic DNA and / or RNA from small samples or even single cells. Methods for analyzing genomic DNA can entail whole genome amplification (WGA), followed by preamplification and amplification of selected target nucleic acids. Methods for analyzing RNA can entail reverse transcription of the desired RNA, followed by preamplification and amplification of selected target nucleic acids.
Owner:FLUIDIGM CORP
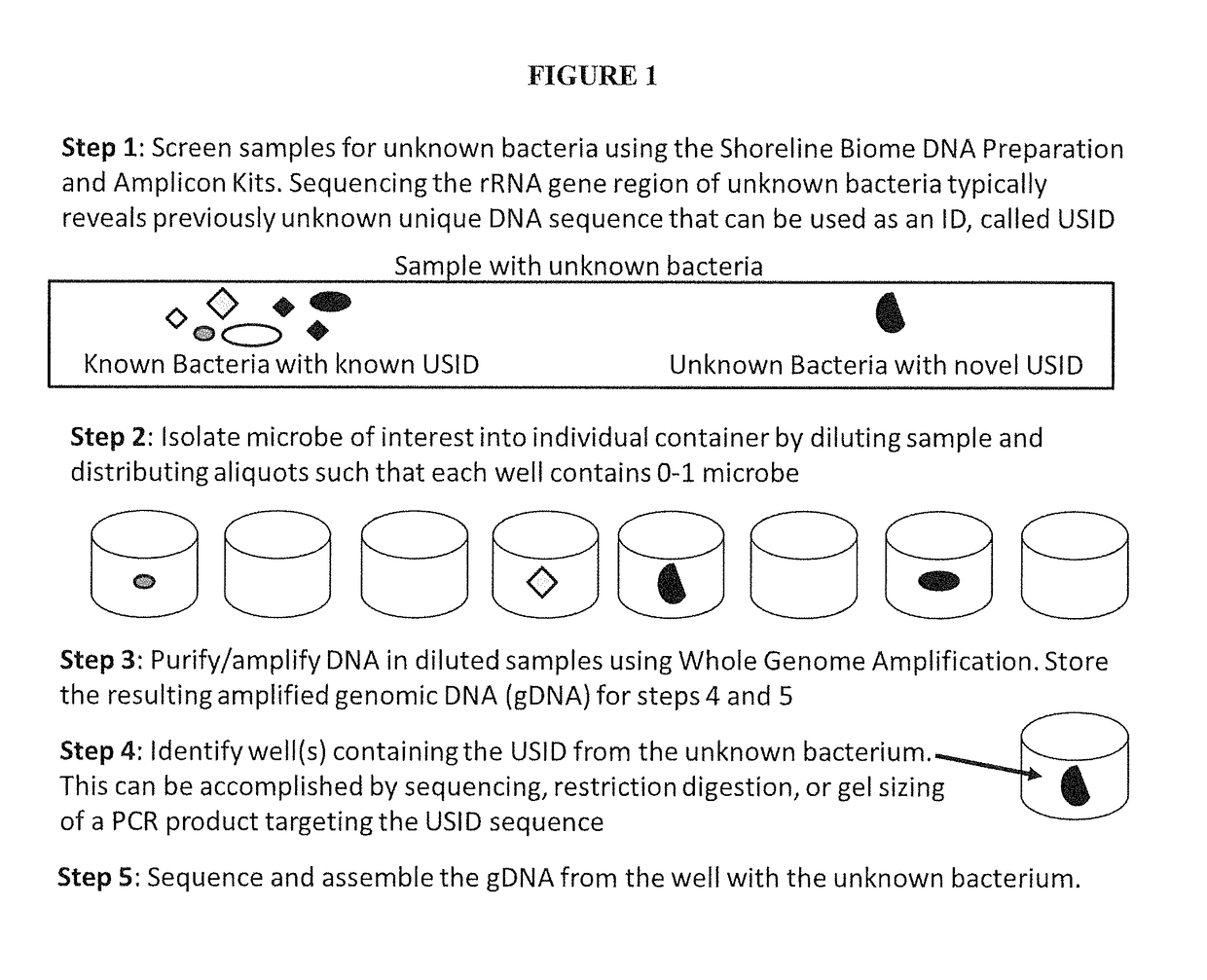
High Throughput Method for Identification and Sequencing of Unknown Microbial and Eukaryotic Genomes from Complex Mixtures
Disclosed are methods for screening biological samples for the presence unknown microbes, such as bacteria and archaea or unknown eukaryotes using rRNA gene sequences or other highly conserved genetic regions, across multiple biological samples using a unique sequence tag (barcode) corresponding to the sample. The screening process tracks the unknown microbe or eukaryote in a diluted sample where the DNA has been prepared using whole genome amplification. The whole genome of the unknown microbe or eukaryote is then sequenced and assembled.
Owner:SHORELINE BIOME LLC
Micro fluidic chip for sorting and whole genome amplification of single cell
PendingCN106065391AEnables in situ fluorescence identificationAchieve crackingBioreactor/fermenter combinationsBiological substance pretreatmentsControl layerEngineering
The invention provides a micro fluidic chip for sorting and whole genome amplification of a single cell. The micro fluidic chip comprises a four-layer structure, wherein the four layers are stacked together in order and mutually sealed, and the structure comprises the following layers from top to bottom: a top cover, a micro valve control layer, a valve plate layer and a channel layer; and the bottom of the micro fluidic chip is externally provided with a magnet. The invention also provides a system for screening and identifying cells as well as amplification and analysis of a single cell, and the system is matched with the chip. The micro fluidic chip and the corresponding detection system can be used for sorting target cells from a large amount of cells quickly, simply and cheaply, and amplifying and analyzing whole genome DNA of a single cell therein.
Owner:THE NAT CENT FOR NANOSCI & TECH NCNST OF CHINA
Method for detecting embryo chromosome abnormality by utilizing blastula culture solution
ActiveCN105368936AAvoid lostAvoid damageMicrobiological testing/measurementMedical automated diagnosisEmbryoNon invasive
Owner:XUKANG MEDICAL SCI & TECH (SUZHOU) CO LTD
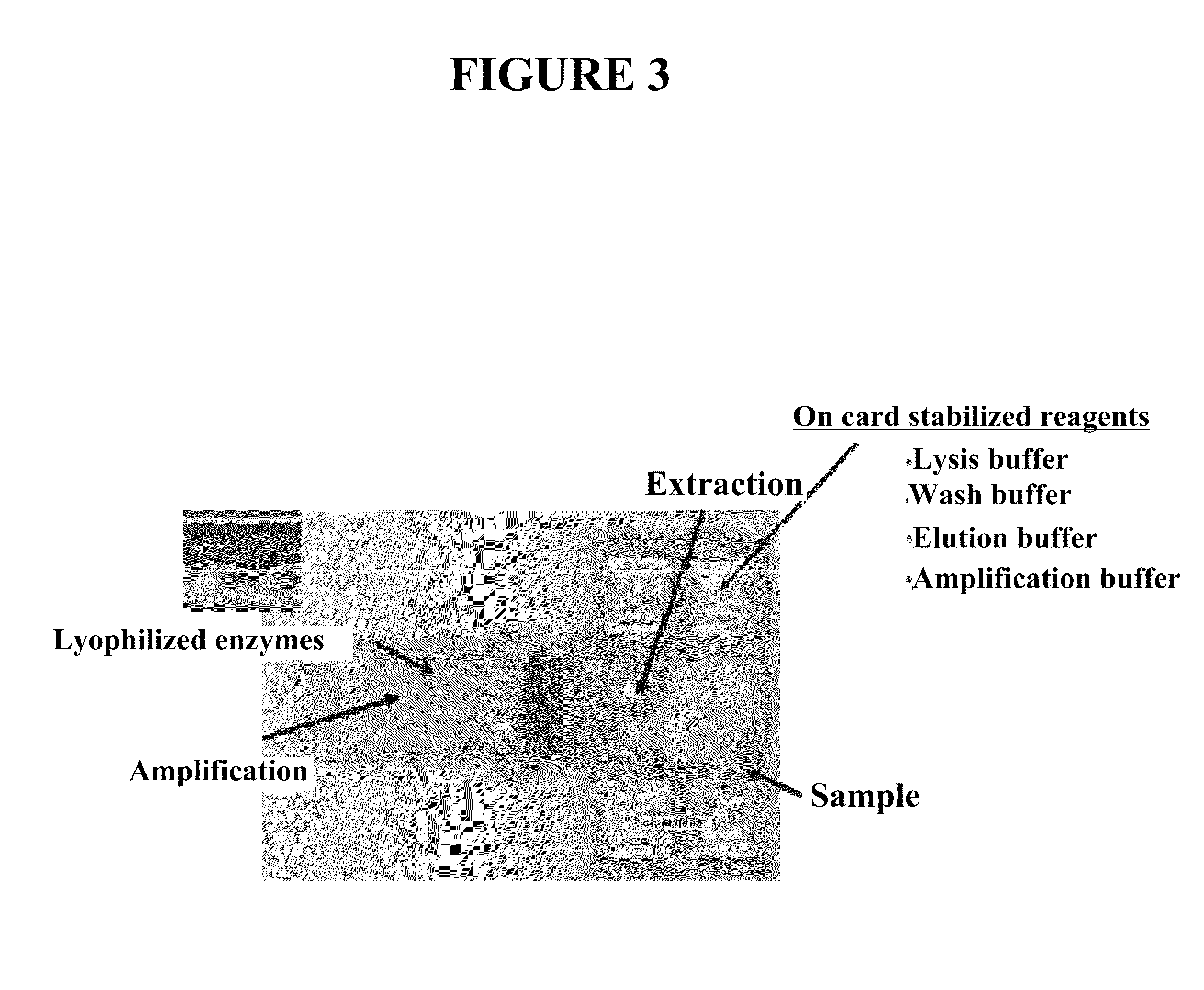
Integrated sample preparation systems and stabilized enzyme mixtures
InactiveUS20110281776A1Heating or cooling apparatusMicrobiological testing/measurementRoom temperatureSugar
The present invention provides integrated sample preparation systems and stabilized enzyme mixtures. In particular, the present invention provides microfluidic cards configured for processing a sample and generating DNA libraries that are suitable for use in sequencing methods (e.g., next generation sequencing methods) or other suitable nucleic acid analysis methods. The present invention also provides stabilized enzyme mixtures containing an enzyme (e.g., an enzyme used in whole genome amplification), BSA, and a sugar. Such enzyme mixtures may be lyophilized and stored at room temperature without significant loss of enzyme activity for months.
Owner:IBIS BIOSCI
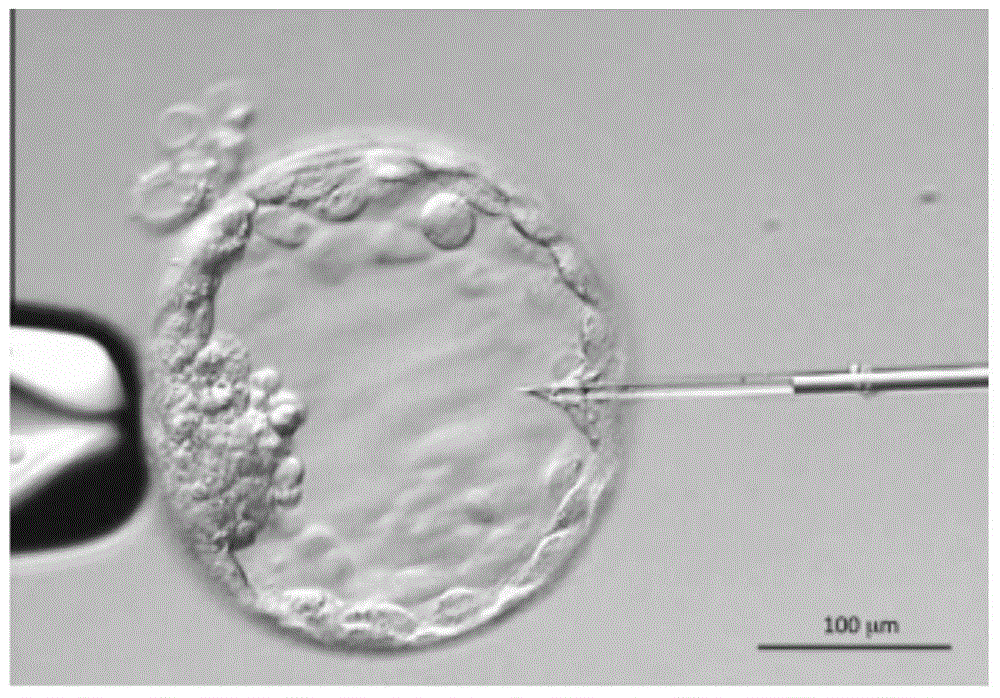
Method for detecting embryo chromosome abnormalities by using blastula-stage embryo cells
InactiveCN104711362AAvoid harmA large amountMicrobiological testing/measurementHigh cellCell separation
The invention discloses a method for performing genome amplification by using embryo blastula-stage cells, performing chromosome detection on the preimplantational embryo by combining a high-flux sequencing technique and screening out the chromosome normal embryo. The method can comprehensively and completely analyze the genetic variation information of the embryo genome, thereby instructing the preimplantational embryo selection, reducing the hereditary diseases and enhancing the success rate of test tube babies. The method comprises the following steps: blastula-stage trophocyte separation; genome amplification; DNA (deoxyribonucleic acid) segmentation; and Proton library establishment, mounting sequencing and sequencing data analysis. By using the blastula-stage embryo to perform trophocyte separation detection, the method avoids the injuries of cleavage-stage cell separation to the embryo, obtains higher cell quantity than the cleavage stage, and enhances the success rate and amplification effect of genome amplification. After the blastula-stage embryos are subjected to the natural elimination process, the high-quality blastula-stage embryo is selected for detection, thereby saving the cost.
Owner:SUZHOU BASECARE MEDICAL DEVICE CO LTD
Substantially non-self complementary primers
InactiveUS20130085083A1Reduced background amplificationConvenient clinical diagnosisNucleotide librariesMicrobiological testing/measurementPolymerase LA-DNA
Owner:RUBICON GENOMICS INC
Methods for genome amplification
A method for whole genome amplification comprising (a) treating genomic DNA with a modifying agent which modifies cytosine bases but does not modify 5′-methyl-cytosine bases under conditions to form single stranded modified DNA; (b) providing a population of random X-mers of exonuclease-resistant primers capable of binding to at least one strand of the modified DNA, wherein X is an integer 3 or greater; (c) providing polymerase capable of amplifying double stranded DNA, together with nucleotides and optionally any suitable buffers or diluents to the modified DNA; and (d) allowing the polymerase to amplify the modified DNA.
Owner:HUMAN GENETIC SIGNATURES PTY LTD
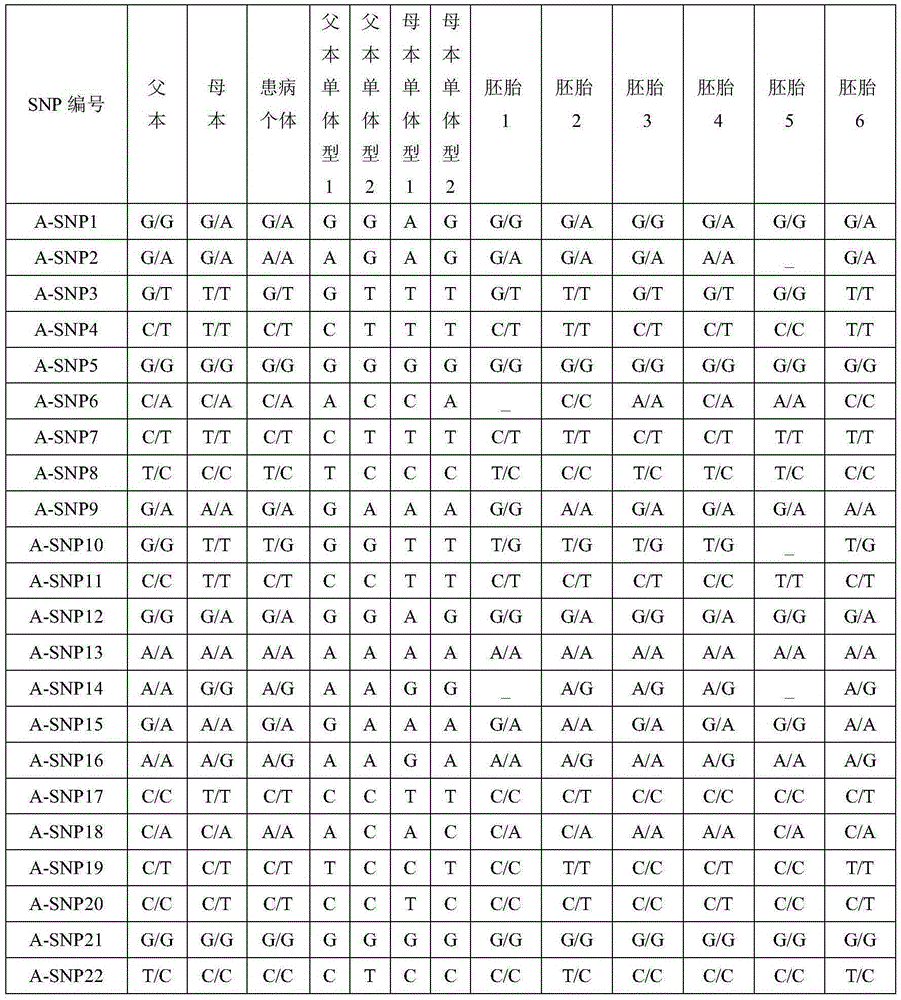
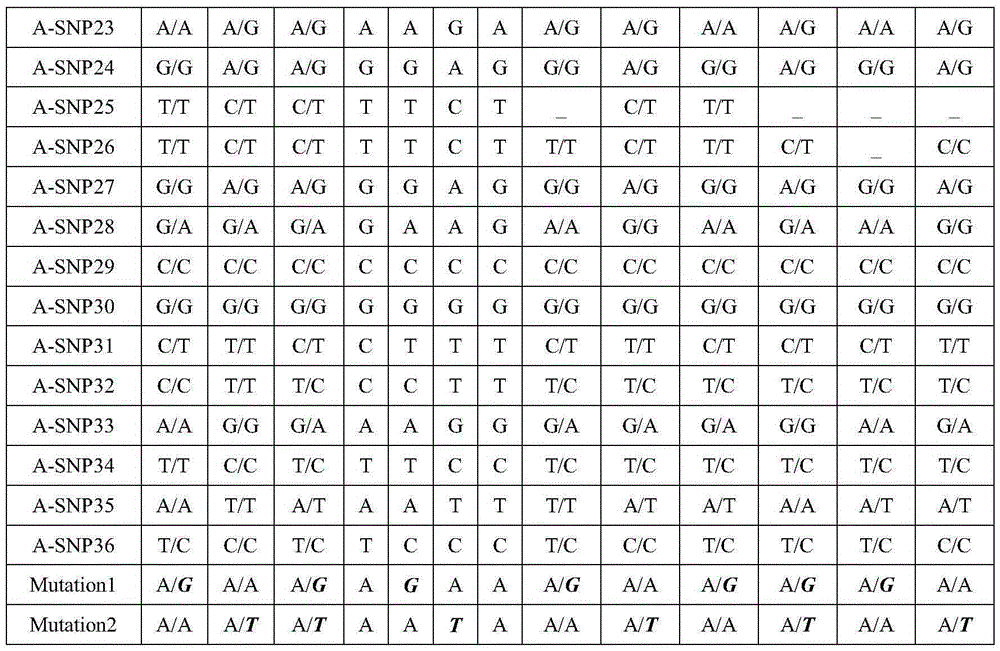
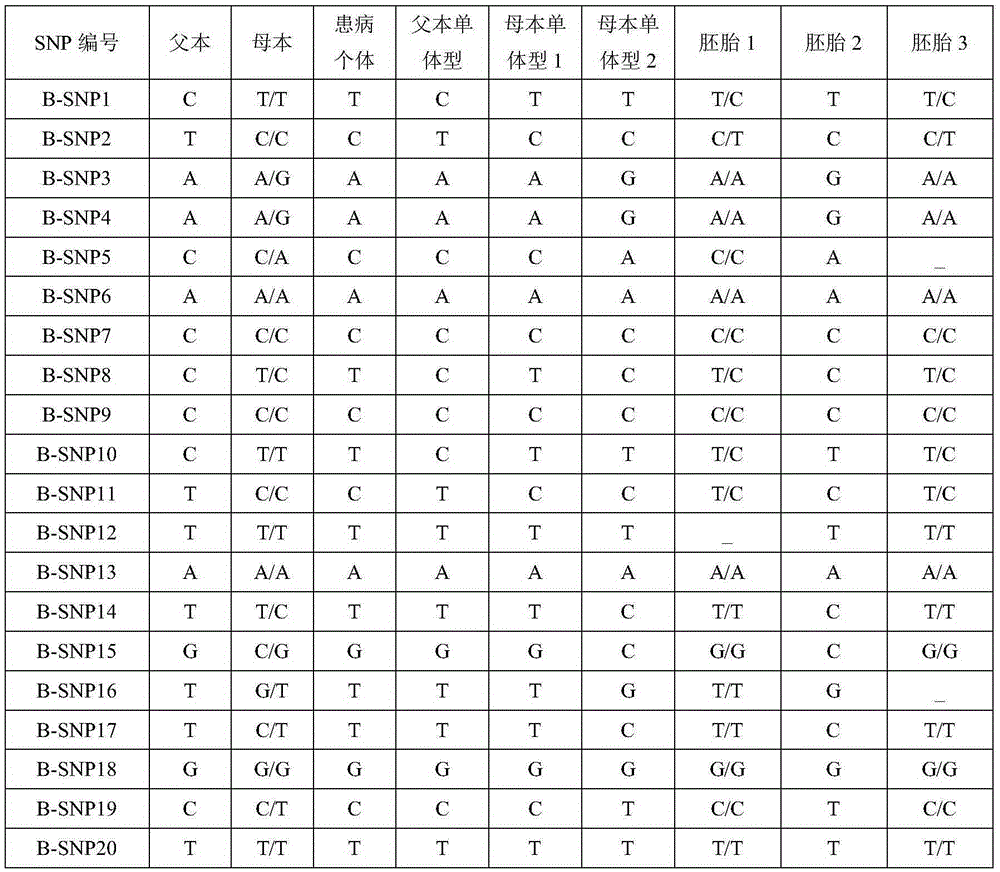
Method for conducting SNP-haplotype analysis by means of multiplex PCR technology
PendingCN105385755AEfficient determinationAccurately determineMicrobiological testing/measurementWork cycleEmbryo
The invention discloses a method for conducting SNP-haplotype analysis by means of a multiplex PCR technology. Designed primers are combined into one tube for single primer amplification when synthesis is conducted, and meanwhile whole genome amplification is completed through an MALBAC technology; when multiplex PCR amplification is conducted, a touch down PCR amplification program is adopted, high-throughput sequencing and reasonable data analysis are combined, only one set of SNP detection is needed, different schemes do not need to be designed according to different objects, the working cycle is shortened, and the cost is reduced. In addition, according to the method for conducting the SNP-haplotype analysis by means of the multiplex PCR technology, family and embryonic genome SNP information is captured, embryo SNP information can be effectively and accurately determined, SNP-haplotype analysis is conducted to determine whether an embryo carries mutation sites of a genetic gene or not, and the defects that in the prior art, the technological cost is high, the cycle is long, and the application range is small are effectively overcome.
Owner:YIKON GENOMICS SHANGHAI CO LTD +1
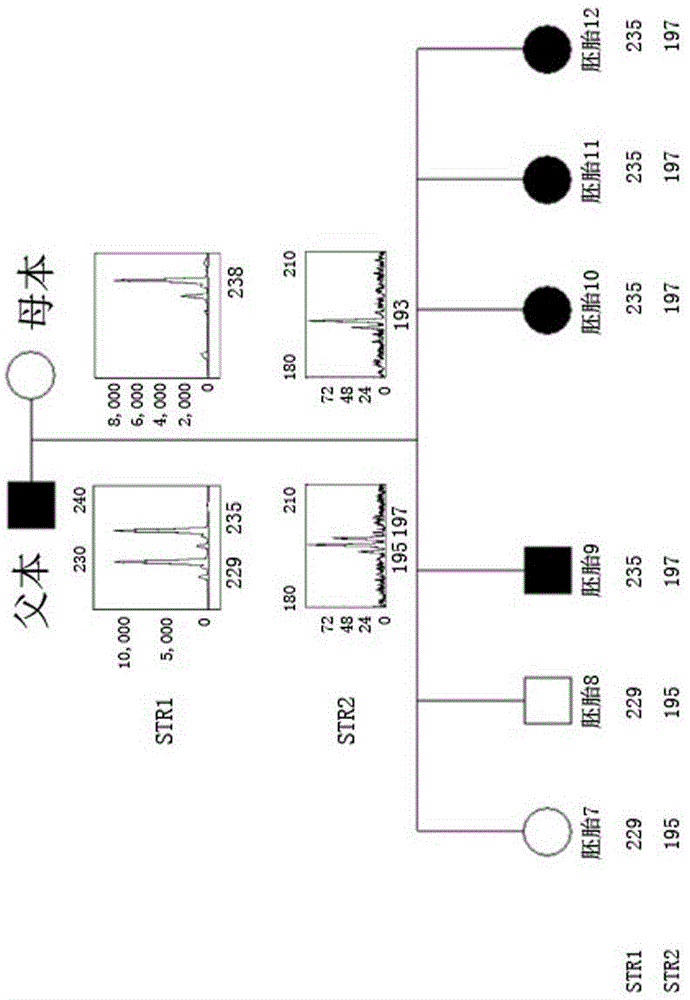
Method for simultaneously completing gene locus, chromosome and linkage analysis
ActiveCN105543339AEasy to operateStrong practical feasibilityMicrobiological testing/measurementEmbryoRecurrent abortion
The invention relates to a method for simultaneously completing gene locus, chromosome and linkage analysis. The method concretely and mainly comprises the following steps: collecting an embryo cell sample, amplifying a whole genome, amplifying a target gene mutation locus, establishing a whole genome and target gene mutation locus library, carrying out high flux sequencing, and carrying out data analysis. Multiple-item comprehensive detection is completed through one step by combining a whole genome amplification technology with the high flux sequencing, so respective detection of single-gene genetic disease mutation site, chromosome diseases and linkage analysis through using multiple methods and multiple steps is avoided. The method provided by the invention provides favorable conditions for a tiny amount of a sample, can be used for PGD detection to determine whether an embryo carries a pathogenic gene and chromosome copy number abnormity or not, is also suitable for genetic screening of embryos of recurrent abortion older women, and realizes multi-item detection of a plurality of single samples through one step. The method has the advantages of simple operation, short period and strong feasibility, so promotion and application of the method are facilitated.
Owner:SHANGHAI XUKANG MEDICAL TECH CO LTD +2
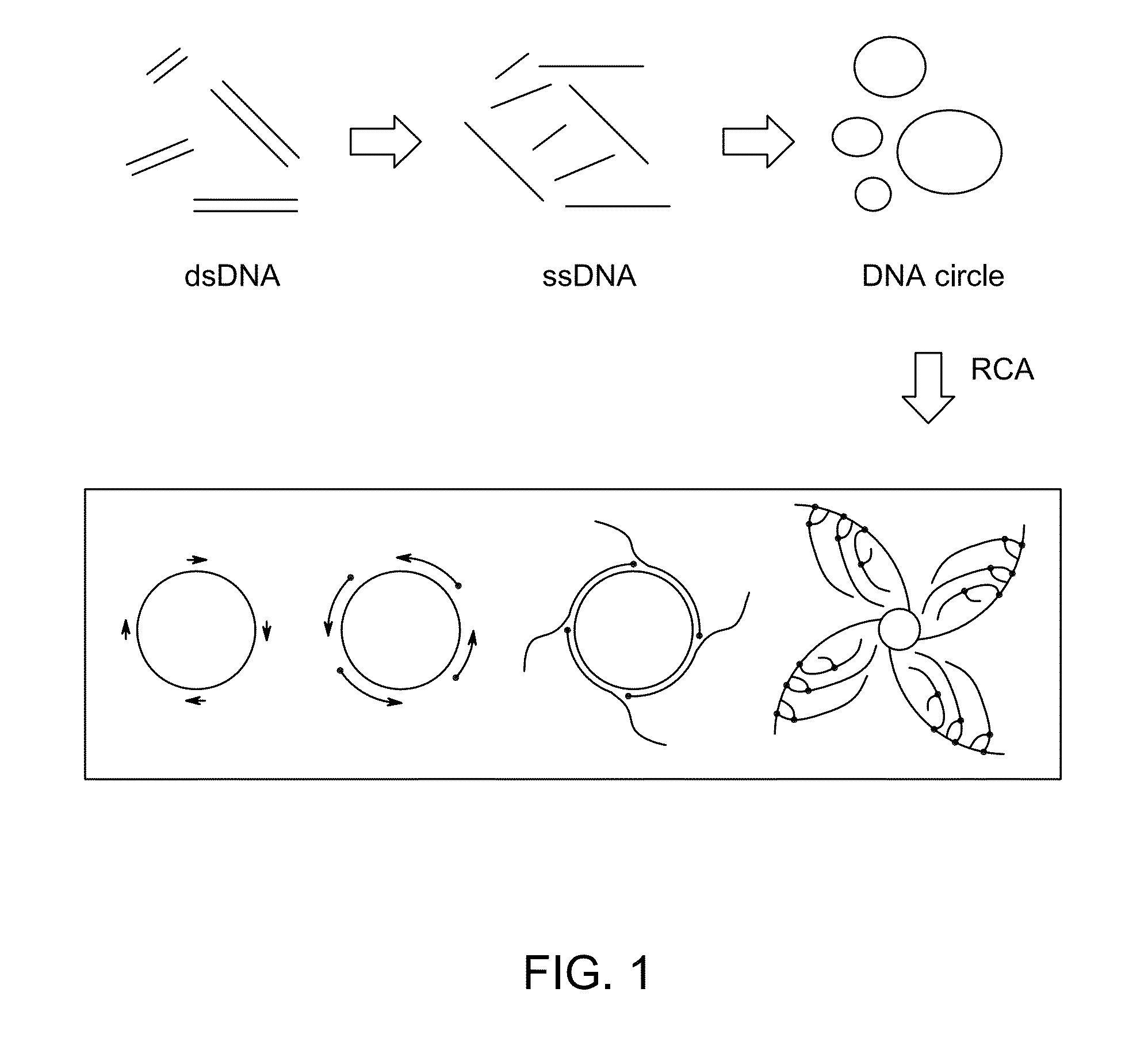
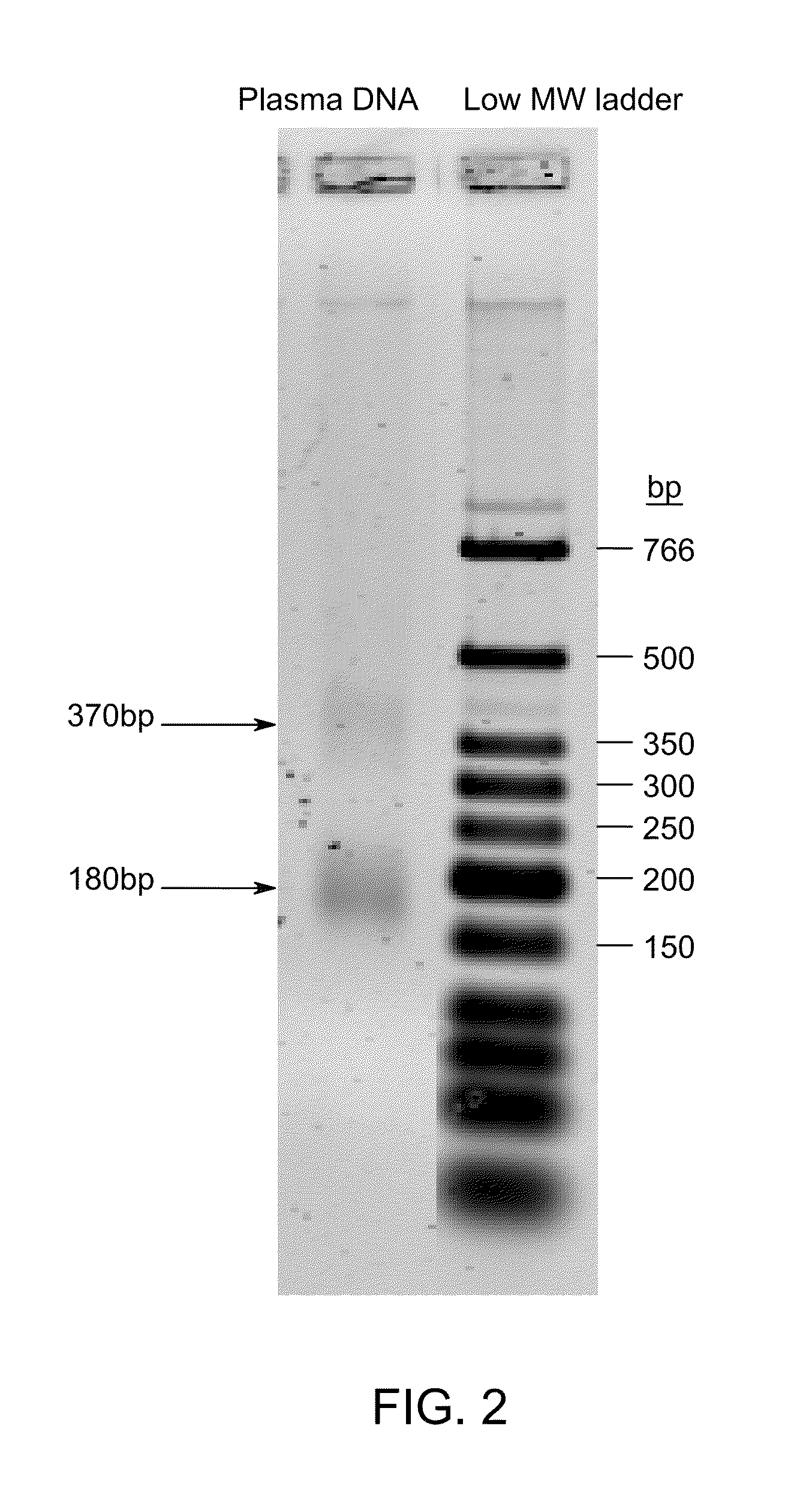
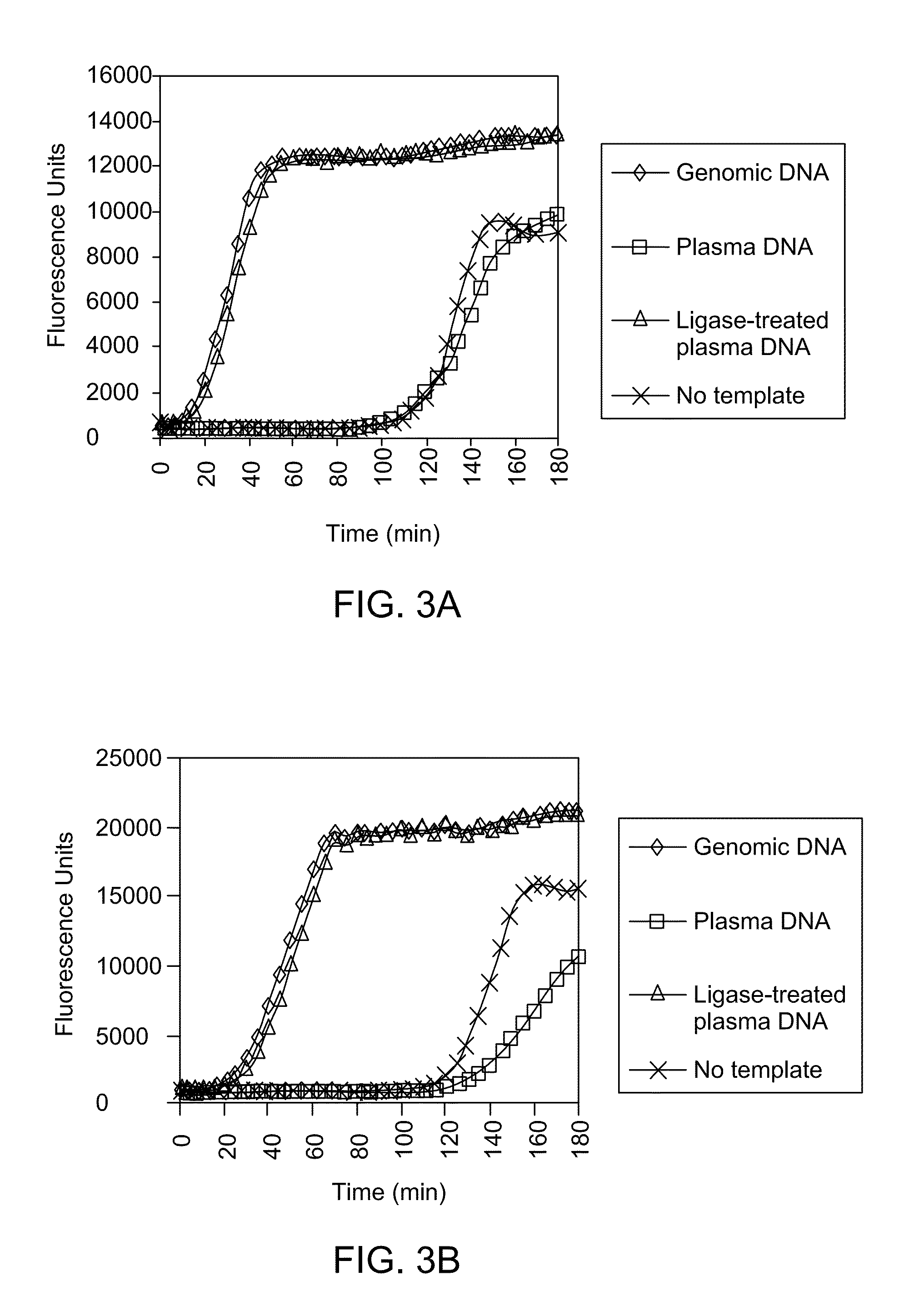
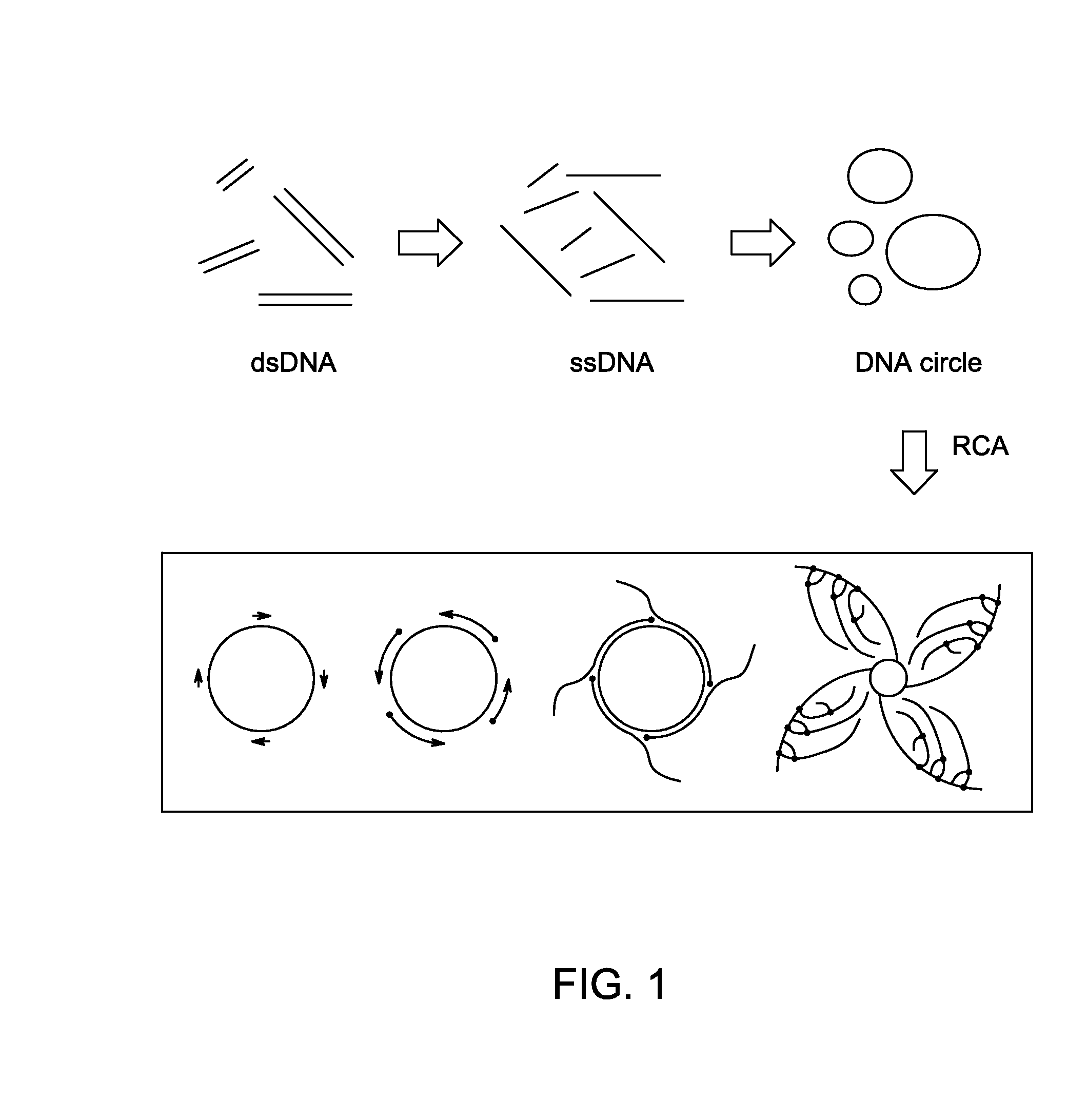
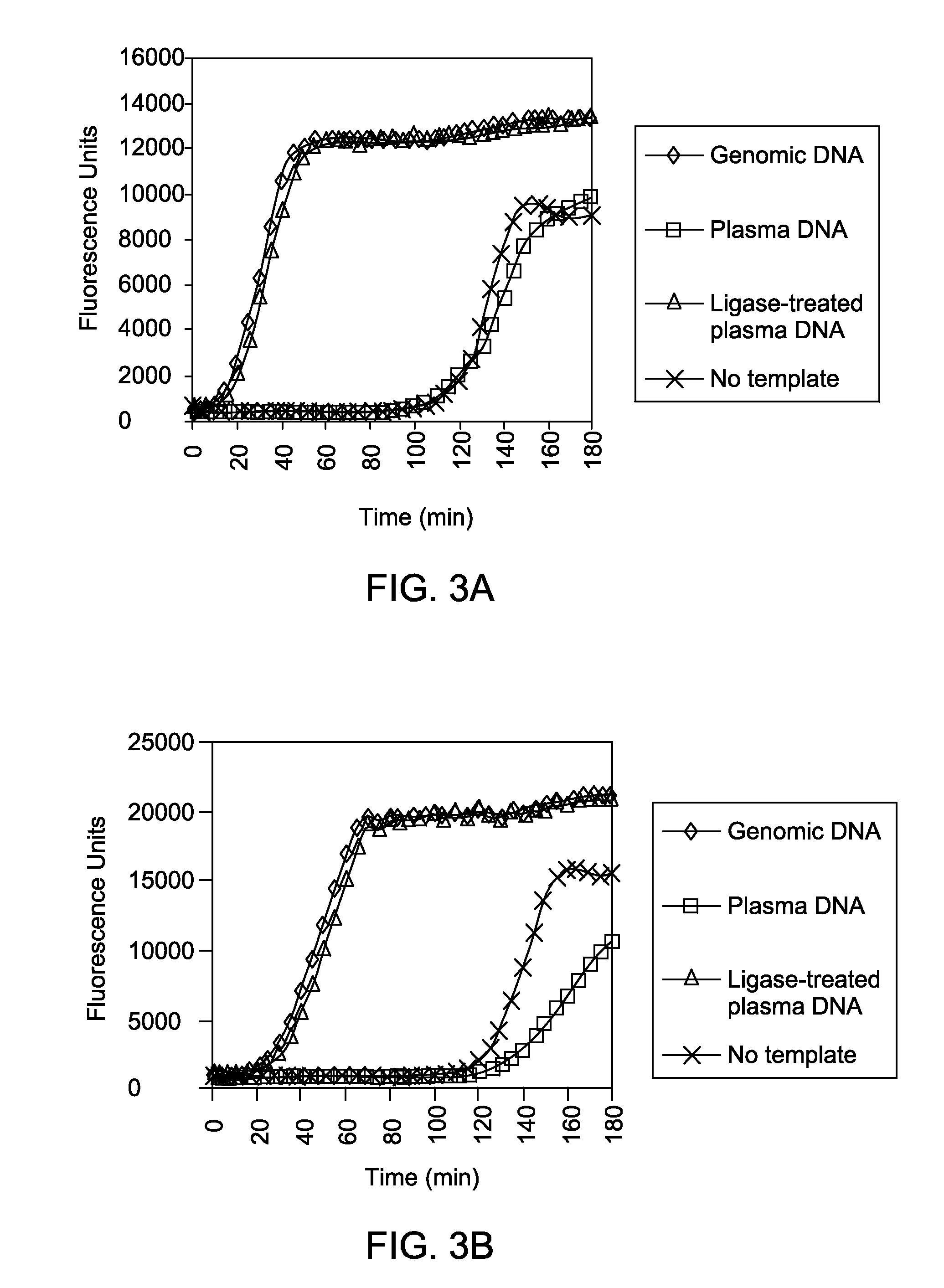
Ligase-assisted nucleic acid circularization and amplification
ActiveUS9217167B2Microbiological testing/measurementFermentationSingle strand dnaWhole Genome Amplification
Provided herein are methods for generation and amplification of a single-stranded DNA circle in a single reaction vessel from a linear DNA without any intervening purification steps. The single-stranded DNA circle is generated via a template-independent single-stranded DNA ligation. Whole-genome amplification of circulating nucleic acids extracted from blood is provided. Kits for performing the disclosed methods are also provided.
Owner:GLOBAL LIFE SCI SOLUTIONS OPERATIONS UK LTD
In vitro fertilization
InactiveUS20100160717A1Microbiological testing/measurementVeterinary instrumentsBiotechnologyMedicine
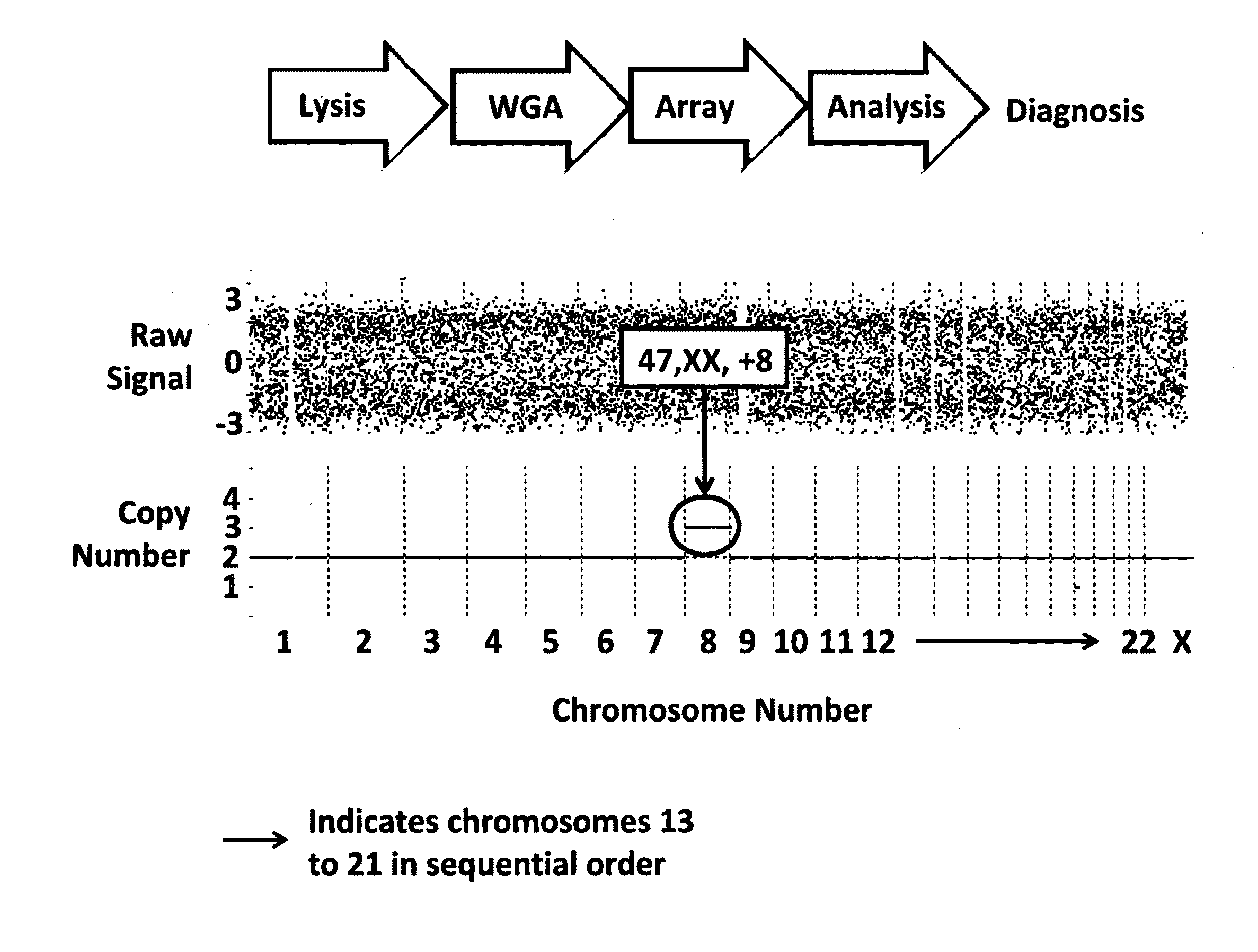
Methods of in vitro fertilization wherein said method includes preimplantation genetic diagnosis of all 24 chromosomes of an IVF embryo comprising whole genome amplification and SNP-based microarray analyses are disclosed.
Owner:SCOTT JR RICHARD T +1
Method of whole genome amplification with reduced artifact production
ActiveUS7955795B2Reduced and undetectable levelQuality improvementMicrobiological testing/measurementFermentationPolymerase LNucleic acid sequencing
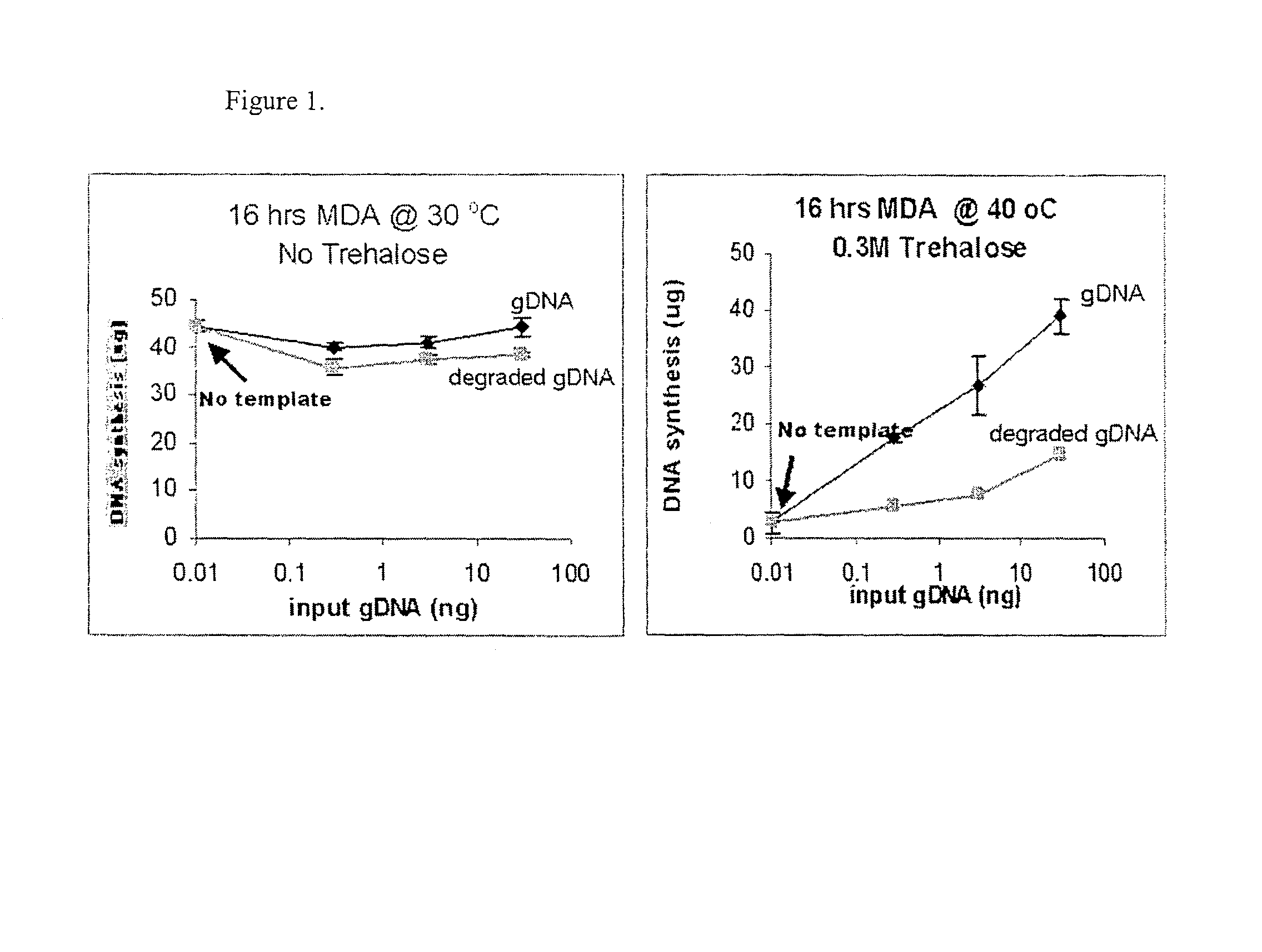
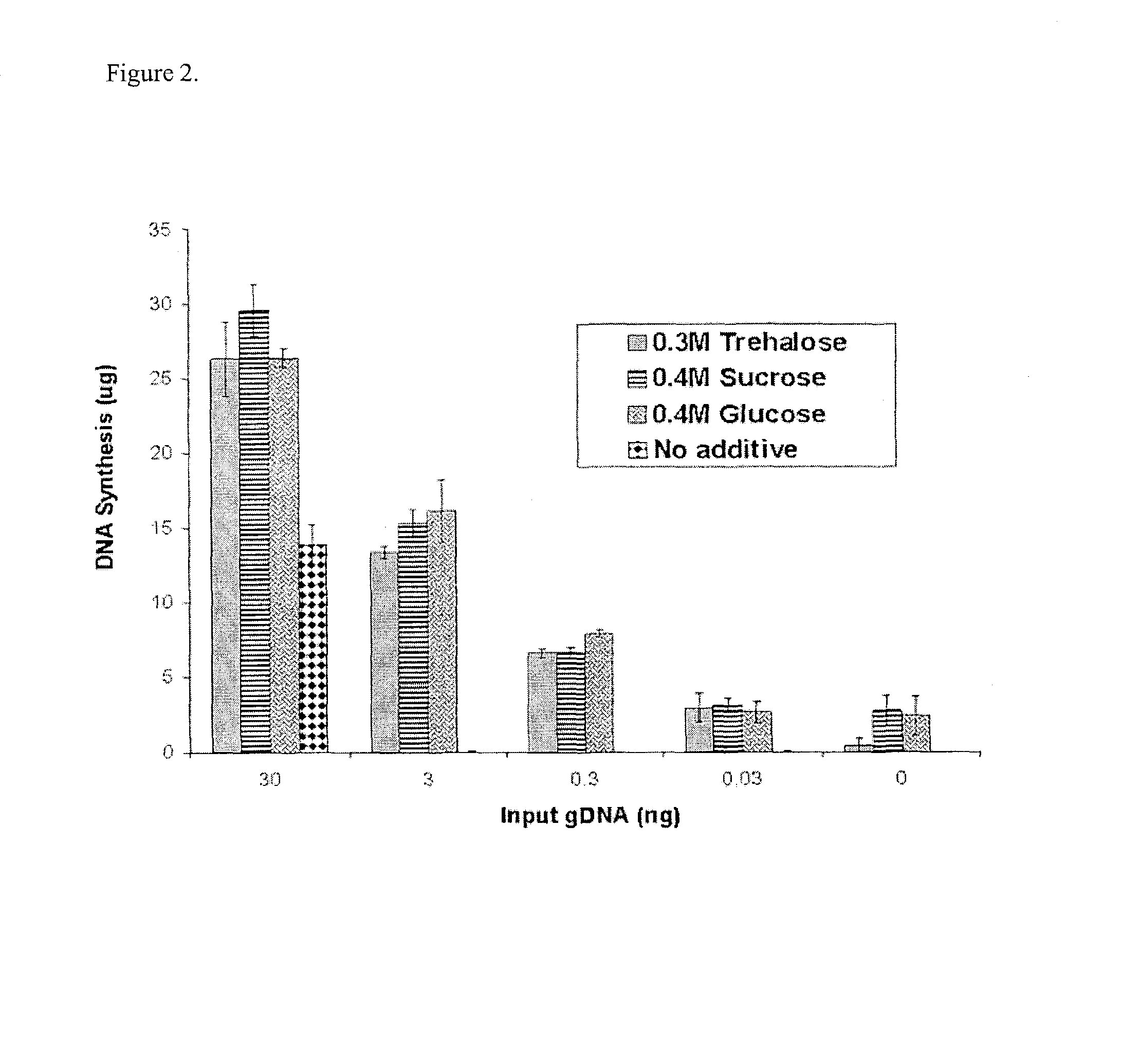
Disclosed are compositions and methods for amplification of nucleic acid sequences of interest with greater efficiency and fidelity. The disclosed method relates to isothermal amplification techniques, such as Multiple Displacement Amplification (MDA), where the generation of DNA artifacts is decreased or eliminated. Generally, this can be accomplished by carrying out the reaction at elevated temperature. In particularly useful embodiments of the method, sugars and / or other additives can be used to stabilized the polymerase at high temperature. It has been discovered that generation of high molecular weight artifacts, in an isothermal amplification procedure, is substantially reduced or eliminated while still allowing the desired amplification of input DNA by carrying out the reaction at a higher temperature and, optionally, in the presence of one or more additives. It also has been discovered that isothermal amplification reactions can produce amplification products of high quality, such as low amplification bias, if performed at a higher temperature and, optionally, in the presence of one or more additives.
Owner:QIAGEN GMBH
Method for detecting embryonic chromosome abnormality by virtue of blastochyle free DNA
InactiveCN104450923AProbability of small developmental abnormalitiesSimple and fast operationMicrobiological testing/measurementFragment sizeEmbryo
The invention relates to a method for detecting embryonic chromosome abnormality by virtue of blastochyle free DNA. The method comprises the following steps: acquiring blastochyle free DNA, detecting the blastochyle DNA, carrying out whole genome amplification of the free DNA, analyzing a product of the whole genome amplification, implementing fragmenting treatment on genome DNA, carrying out quantitative analysis and fragment size analysis on fragmented target DNA, constructing a library, sequencing by virtue of a computer and analyzing biological information. By virtue of high-throughput sequencing, the method disclosed by the invention can be used for overcoming shortcomings of a conventional DNA analysis method which is merely used for researching partial region of a single cell genome, and is capable of completely analyzing the genetic information of the single cell genome; the method is simple and convenient to operate, time-saving and efficient; meanwhile, by using the blastochyle free DNA as a detection sample, the method is convenient and safe to sample, so that the probability of later embryonic development abnormality is reduced and embryo is protected from being influenced in later development.
Owner:SUZHOU BASECARE MEDICAL DEVICE CO LTD
Differential enzymatic fragmentation by whole genome amplification
The present invention provides methods for detecting the presence of methylation at a locus within a population of nucleic acids.
Owner:ORION GENOMICS
Ligase-assisted nucleic acid circularization and amplification
ActiveUS20150031086A1Microbiological testing/measurementFermentationSingle strand dnaWhole Genome Amplification
Provided herein are methods for generation and amplification of a single-stranded DNA circle in a single reaction vessel from a linear DNA without any intervening purification steps. The single-stranded DNA circle is generated via a template-independent single-stranded DNA ligation. Whole-genome amplification of circulating nucleic acids extracted from blood is provided. Kits for performing the disclosed methods are also provided.
Owner:GLOBAL LIFE SCI SOLUTIONS OPERATIONS UK LTD
Quantification of amplified nucleic acids
InactiveUS20060172314A1Validate lack of biasReduce complexityMicrobiological testing/measurementFermentationSignal amplificationEnzyme
The invention relates to methods and kits for quantification of amplified nucleic acids, such as genomic DNA. The methods and kits can be used to assess bias in an amplification procedure such as a whole genome amplification procedure. In one aspect, the method comprises performing an enzyme-based amplification procedure and validating the results of the procedure using a signal amplification method.
Owner:AGILENT TECH INC
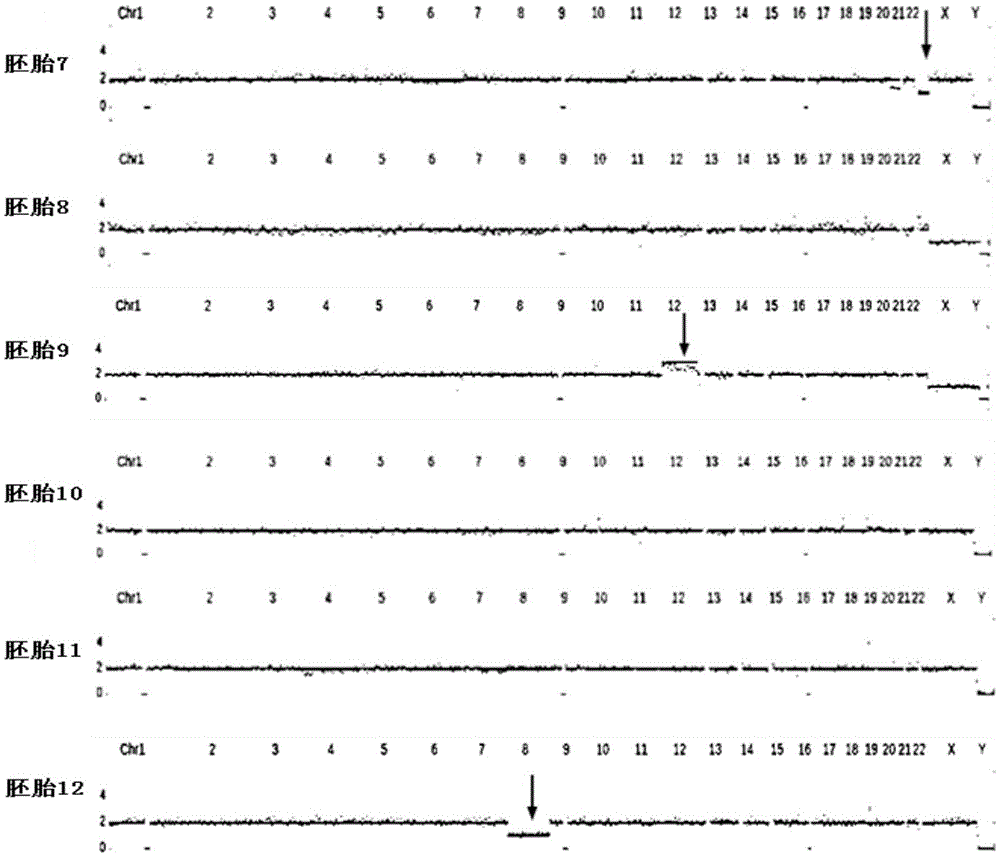
Method for detecting chromosome microdeletion and micro-duplication of human embryo
ActiveCN104745718AHigh sensitivityLow costMicrobiological testing/measurementEmbryoChromosome microdeletion
The invention relates to a method for detecting chromosome microdeletion and micro-duplication of a human embryo. The method comprises the following steps of performing whole genome amplification on cells cultured in vitro, interrupting DNA (deoxyribonucleic acid) molecules, and sequencing DNA fragments to obtain sequencing reads; comparing the sequencing reads with a reference sequence, and positioning the sequencing reads on the reference sequence; screening non-repeated areas of the reference sequence, and reserving the non-repeated areas; establishing a matrix of read number in windows through normal samples, analyzing the data of the normal samples, performing statistics on the read number of all the windows in the non-repeated areas, and establishing a probability matrix of the read number and chromosome enpeoids; calculating the copy number, i.e., the A / B / C state, of loci; selecting m continuous loci, i.e., the A state, as micro-duplication loci, and selecting m continuous loci, i.e., the C state, as microdeletion loci; contrasting the micro-duplication loci and the microdeletion loci with the existing CNV (copy number variation) and disease database, performing basic gene annotation and gene function analysis which relates to deletion parts, and annotating with a microdeletion syndrome disease type.
Owner:BEIJING ZHONGYI KANGWEI MEDICAL INSTR
Single-cell genome analysis method and kit
ActiveCN102533960AEfficient analysisQuality improvementMicrobiological testing/measurementHousekeeping geneResearch strategies
The invention discloses a single-cell genome analysis method and a kit. The method comprises the following steps of: a, acquiring a complete cell genome DNA (Deoxyribonucleic Acid); b, carrying out single-cell whole genome amplification; c, carrying out quantitative detection and qualitative detection, wherein the qualitative determination comprises that single-cell whole genome amplification product is determinated by adopting a Housekeeping Gene detection method. The kit contains a specific primer of the single cell Housekeeping Gene. By adopting the method disclosed by the invention, genetic variation information of a single cell genome can be comprehensively and completely analyzed, and an effective research strategy can be provided for the single cell genome of a brand-new specie; meanwhile, qualitative and quantitative detection screening steps are introduced, and many unqualified amplification samples can be removed, thus quality of a downstream upper computer sequencing library is controlled and unnecessary waste can be greatly avoided.
Owner:BGI SHENZHEN CO LTD +1
Methods, systems and devices for multiple single-cell capturing and processing using microfluidics
InactiveUS20130302884A1Bioreactor/fermenter combinationsHeating or cooling apparatusReal-Time PCRsMRNA Sequencing
Methods, systems, and devices are described for multiple single-cell capturing and processing utilizing microfluidics. Tools and techniques are provided for capturing, partitioning, and / or manipulating individual cells from a larger population of cells along with generating genetic information and / or reactions related to each individual cell. Different capture configurations may be utilized to capture individual cells and then processing each individual cell in a multi-chamber reaction configuration. Some embodiments may provide for specific target amplification, whole genome amplification, whole transcriptome amplification, real-time PCR preparation, copy number variation, preamplification, mRNA sequencing, and / or haplotyping of the multiple individual cells that have been partitioned from the larger population of cells. Some embodiments may provide for other applications. Some embodiments may be configured for imaging the individual cells or associated reaction products as part of the processing. Reaction products may be harvested and / or further analyzed in some cases.
Owner:FLUIDIGM CORP
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com